Background: Ara h 8 is hypothesized to be responsible for cross-reactivity between birch pollen and peanut.
Results: The Ara h 8 crystal structure is very similar to Bet v 1 from birch, and it has a similar ligand-binding function.
Conclusion: Ara h 8 is most likely responsible for cross-reactivity between birch pollen and peanut.
Significance: A structural and functional understanding of cross-reactivity between Bet v 1 and Ara h 8 was obtained.
Keywords: Allergen, Antigen, Crystal Structure, Flavonoids, Fluorescence, Peanut
Abstract
The incidence of peanut allergy continues to rise in the United States and Europe. Whereas exposure to the major allergens Ara h 1, 2, 3, and 6 can cause fatal anaphylaxis, exposure to the minor allergens usually does not. Ara h 8 is a minor allergen. Importantly, it is the minor food allergens that are thought to be responsible for oral allergy syndrome (OAS), in which sensitization to airborne allergens causes a Type 2 allergic reaction to ingested foods. Furthermore, it is believed that similar protein structure rather than a similar linear sequence is the cause of OAS. Bet v 1 from birch pollen is a common sensitizing agent, and OAS results when patients consume certain fruits, vegetables, tree nuts, and peanuts. Here, we report the three-dimensional structure of Ara h 8, a Bet v 1 homolog. The overall fold is very similar to that of Bet v 1, Api g 1 (celery), Gly m 4 (soy), and Pru av 1 (cherry). Ara h 8 binds the isoflavones quercetin and apigenin as well as resveratrol avidly.
Introduction
Peanuts are high in protein and oil content. Accordingly, they are consumed widely around the world. Unfortunately, the protein components can be potent allergens with fatal anaphylaxis as a possible result of consumption. Unlike other food allergies, allergy to peanuts usually persists into adulthood. In the United States and Europe, peanut allergy affects 1–2% of adults (1).
Thirteen proteins have been identified as allergens in peanuts. Ara h 1, 2, 3, and 6 are considered major allergens because they are recognized by the IgE of a majority of allergic patients. Ara h 5, 7, 8, 9, 10, 11, and 12/13 are considered minor allergens and are not thought to be the causative agents in most of the life-threatening allergic reactions (anaphylaxis). Ara h 8 has been shown to have cross-reactivity with IgE antibodies to Bet v 1 from birch pollen and may be the cause of oral allergy syndrome (OAS)4 in birch allergic patients (2, 3).
OAS is an allergic phenomenon in which a person allergic to tree pollen consumes food and has a reaction primarily in the oral cavity (4). This has been described at length, with various fruits and vegetables inducing the symptoms. For some of these, the protein responsible has been identified. The structure of Bet v 1 has been determined and is unusual because it does not have a globular, hydrophobic core (5). Rather, a curved, seven-stranded β-sheet with a long α-helix lying across it comprises the protein fold. Several of the proteins from food that cause OAS have also had their structures determined, and in each case the structures are highly similar to Bet v 1; however, the primary amino acid sequences are very different (6–10). The Bet v 1-like superfamily of proteins includes a class of proteins called PR-10, for pathogenesis-related class 10. Among these proteins, Ara h 8 has the lowest sequence identity to Bet v 1 (48%).
In efforts to identify a function for Bet v 1, two theories have emerged. There were early reports that it has RNase activity (11, 12). Since then, several other related proteins have been shown to possess RNase activity, including birch PR-10c (13) and lupin LaPR-10 (14). The Bet v 1 family of proteins is homologous to the START domain of the human MLN64 protein that binds steroids (15). Bet v 1 and its homologs from various species have been shown to bind small hydrophobic compounds. Mogensen et al. (16) showed that flavonoids, cytokinins, and fatty acids bind to Bet v 1. PR-10c from birch is related to Bet v 1 and binds a variety of biologically important ligands (17). Recently, the structures of different isoforms of Bet v 1 in complex with deoxycholate, naringenin, kinetin, NDSB-256, or the molten globule probe 8-anilino-1-napthalenesulfonic acid (ANS) were determined (PDB codes 4A83, 4A87, 4A85, 4A8G, and 4A80, respectively) (18, 19). Additionally, Fernandes et al. (20) reported a co-crystal structure of LIPR-102B from Lupinus luteus (yellow lupine) with three zeatin molecules bound. A structurally similar protein, VrCSBP from Vigna radiata (mung bean) was also found to bind zeatin (PDB code 2FLH) (21).
Based on these reports, we decided to initiate the structural and functional characterization of Ara h 8. The structure was determined using x-ray crystallography and refined to 1.6 Å. Purified Ara h 8 was found to bind ANS, some isoflavones, some fatty acids, and resveratrol. These findings are reported here.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
The sequence of Ara h 8 was taken from Mittag et al. (2) (accession number AY328088.1), back-translated into DNA sequence, and optimized for expression in Escherichia coli using programs from the Lasergene software suite (DNASTAR, Madison, WI). Short extensions were added to the coding region to facilitate cloning into pET9a that included a NdeI site overlapping the start codon and a BamHI site downstream of the stop codon. This sequence was synthesized and cloned into pUC57 by EZBiolab (Carmel, IN). The synthetic insert was cloned into pET9a as a NdeI/BamHI fragment generating pET-A8.
pET-A8 was transformed into BL21(DE3) E. coli cells. Cells were grown in Luria-Bertani broth (LB) at 37 °C to an optical density (600 nm) of 0.5 and then induced with isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mm. Following induction, the cells were incubated overnight with shaking at 20 °C. Cells were harvested by centrifugation for 10 min at 11,000 × g and 4 °C and stored at −20 °C. Protein was then purified in one of two ways. In protocol 1, cells were resuspended in 5 ml of Q-column buffer (QCB; 50 mm Tris-Cl, pH 8.3, 1 mm EDTA, 50 mm NaCl) supplemented with 1 mm DTT and 1 mm PMSF per g of wet weight cells. Once the cells were resuspended, 200 μl of lysozyme (50 mg/ml) was added. Ultrasound was used to reduce the viscosity, and the crude lysate was cleared by centrifugation for 15 min at 32,000 × g and 4 °C. The supernatant was passed through a 50-ml High-Q column (Macro Prep, Bio-Rad). The unbound material plus a 25-ml wash were pooled and brought to 60% saturation with ammonium sulfate. Following a 30-min incubation, the solution was cleared by centrifugation at 32,000 × g and 4 °C for 15 min. The supernatant was loaded onto a 50-ml tertiary butyl column (Bio-Rad) and washed with QCB plus 60% ammonium sulfate until the absorbance of the eluate at 280 nm was 0. Protein was eluted with a linear gradient from 60 to 0% ammonium sulfate in QCB. Following the tertiary butyl column, protein was passed through a Superdex 200 column attached to an ÅKTA FPLC gel filtration system (GE Healthcare) in a buffer containing 10 mm Tris-HCl, pH 7.5, and 150 mm NaCl. Subsequently, fractions containing the protein were pooled and concentrated to 15 mg/ml.
In protocol 2, cells were resuspended in 5 ml of QCB supplemented with 1 mm DTT and 1 mm PMSF per g of wet weight cells. Once the cells were resuspended, 200 μl of lysozyme (50 mg/ml) was added. Ultrasound was used to reduce the viscosity, and the crude lysate was cleared by centrifugation for 15 min at 32,000 × g and 4 °C. The supernatant was heated to 70 °C in a water bath, held at that temperature for 10 min, and then chilled in an ice-water bath. Insoluble material was cleared by centrifugation at 32,000 × g and 4 °C for 15 min. The supernatant was passed through a 50-ml High-Q column (Macro Prep, Bio-Rad). The unbound material plus a 25-ml wash were pooled and brought to 60% saturation with ammonium sulfate. Following a 30-min incubation, the solution was cleared by centrifugation at 32,000 × g and 4 °C for 15 min. The supernatant was loaded onto a 50-ml tertiary butyl column (Bio-Rad) and washed with QCB plus 60% ammonium sulfate until the absorbance of the eluate at 280 nm was 0. Protein was eluted with a linear gradient from 60 to 0% ammonium sulfate in QCB. Following elution, fractions containing protein were pooled and concentrated to 11 mg/ml.
In both protocols, pooled fractions were used for crystallization and ligand binding studies. The concentration of Ara h 8 was determined spectroscopically at 280 nm using an extinction coefficient of 10,430 m−1.
Crystallization, Data Collection, and Processing
Crystallization experiments were performed at 293 K using the hanging drop vapor diffusion method and NeXtal plates (Qiagen, Chatsworth, CA). A solution of recombinant protein (11–15 mg/ml) was mixed with well solution (20% (w/v) PEG 3350, 200 mm magnesium formate for the apo-structure; 0.1 m MES, pH 6.5, 25% (w/v) PEG 2000 methyl ether for the MES-bound structure; 0.1 m Tris-HCl, pH 8.5, 0.2 m lithium sulfate, 30% (w/v) PEG 4000 for the epicatechin-bound Ara h 8; or 0.1 m Tris, pH 8.0, 0.2 m lithium sulfate, 30% (w/v) PEG 4000 for the protein purified with the additional heating step) in a 1:1 ratio. Crystals were cryoprotected with well solution and cooled in liquid N2. Soaking experiments were performed with epicatechin, where ligand was added in form of a powder to the drop containing the crystal. Data were collected from single crystals at 100 K at the SBC 19-ID and SBC 19-BM beamline at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL). Data were processed with the HKL-2000 software package (22). Data collection statistics are reported in Table 1.
TABLE 1.
Crystallographic data and refinement statistics for Ara h 8
Values in parenthesis refer to the highest resolution shell. AU, asymmetric unit.
| PDB 4M9B (apo) | PDB 4M9W (MES) | PDB 4MA6 (epicatechin) | PDB 4MAP (heated) | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 0.98 | 0.98 | 0.98 | 0.98 |
| Unit cell (Å) | a = 40.9, b = 88.0, c = 42.9 | a = 40.2, b = 87.5, c = 42.3 | a = 40.6, b = 89.0, c = 42.7 | a = 40.9, b = 87.1, c = 42.9 |
| Unit cell (degrees) | α = γ = 90, β = 96.8 | α = γ = 90, β = 95.8 | α = γ = 90, β = 96.6 | α = γ = 90, β = 96.7 |
| Space group | P21 | P21 | P21 | P21 |
| Solvent content (%) | 45 | 45 | 43 | 45 |
| Protein chains in AU | 2 | 2 | 2 | 2 |
| Resolution range (Å) | 50.0–1.60 | 50.0–1.95 | 50.0–2.0 | 50.0–1.90 |
| Highest resolution shell (Å) | 1.63-1.60 | 1.98-1.95 | 2.03-2.00 | 1.93-1.90 |
| Unique reflections | 39,702 (1979) | 21,429 (1081) | 20,539 (1049) | 23,735 (1192) |
| Redundancy | 3.7 (3.7) | 4.1 (3.9) | 4.1 (3.7) | 3.1 (3.0) |
| Completeness (%) | 99.7 (99.9) | 100 (99.6) | 99.9 (99.0) | 99.6 (99.8) |
| Rmerge (%) | 0.045 (0.691) | 0.052 (0.364) | 0.050 (0.566) | 0.048 (0.360) |
| Average I/σ(I) | 38.1 (2.3) | 35.9 (2.9) | 25.0 (2.3) | 29.4 (2.8) |
| Refinement | ||||
| R (%) | 18.0 | 20.5 | 19.2 | 19.7 |
| Rfree (%) | 22.2 | 24.8 | 24.8 | 25.2 |
| Mean B value (Å2) | 30.7 | 42.9 | 37.7 | 36.0 |
| B from Wilson plot (Å2) | 25.5 | 33.4 | 30.7 | 27.6 |
| Root mean square deviation bond lengths (Å) | 0.02 | 0.02 | 0.02 | 0.02 |
| Root mean square deviation bond angles (degrees) | 2.1 | 1.8 | 1.9 | 1.9 |
| No. of amino acid residues | 312 | 312 | 312 | 312 |
| No. of water molecules | 351 | 137 | 155 | 209 |
| Ramachandran plot | ||||
| Most favored regions (%) | 94.2 | 93.4 | 93.4 | 93.8 |
| Additional allowed regions (%) | 5.0 | 6.6 | 6.2 | 5.4 |
Structure Determination and Refinement
The apo Ara h 8 structure was solved by molecular replacement using pathogenesis-related protein LLPR10.1B from yellow lupine (PDB code 1IFV) as a start model. The MES-bound structure, epicatechin-bound structure, and heated protein structures were determined using molecular replacement with the apo Ara h 8 structure, reported here, as a start model. Molecular replacement was performed with HKL-3000 (23) integrated with MOLREP (24) and selected programs from the CCP4 package (25). Models were rebuilt using COOT (26) and refined with REFMAC (27). TLS was used in the final stages of refinement, and TLS groups were determined using the TLSMD server (28). MOLPROBITY (29) and ADIT (30) were used for structure validation. Metal binding sites were validated using the CheckMyMetal server.5 Refinement statistics are summarized in Table 1.
Ligand Binding-Fluorescence Assay
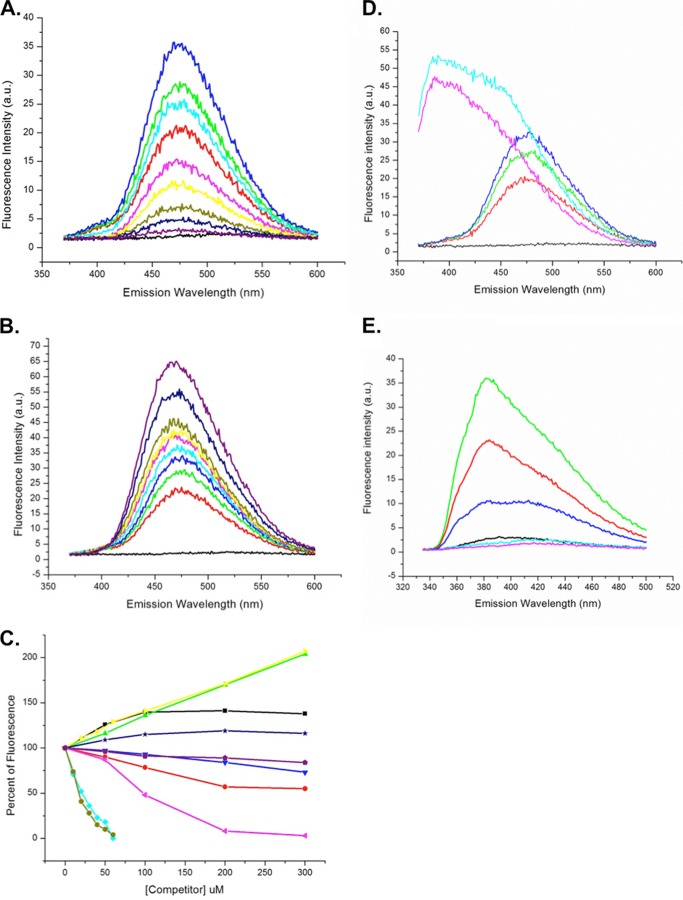
Ara h 8 was tested for ligand binding by displacement of fluorescent ANS as described by Mogensen et al. (16). Briefly, ANS was dissolved in 1 ml of DMSO and diluted to 100 ml with MilliQ water, and its concentration was determined spectrophotometrically, using an extinction coefficient of 4990 m−1 at 350 nm. Fluorescence experiments were performed on a Shimadzu RF-5301 PC spectrophotometer. Base-line fluorescence of the initial sample (2.5 ml) of ANS diluted to 40 μm in 50 mm phosphate buffer, pH 7.4, was measured by excitation at 350 nm and then recording emission from 370 to 600 nm. Recombinant purified Ara h 8 was added stepwise to a final concentration of 40 μm, and fluorescence was measured as before. For ligand binding experiments, potential Ara h 8 ligands were dissolved in an appropriate solvent (water, ethanol, or DMSO) to a concentration of 10 mm and added to the Ara h 8-ANS solution in increments (generally from 10 to 60 μm or from 50 to 300 μm, depending on the change in signal observed), and fluorescence was recorded after each addition. An appropriate volume of ANS and Ara h 8 were added as ligand concentration increased to maintain a concentration of 40 μm for each compound. To ensure that the ligand solvent did not have a denaturing effect on Ara h 8, titrations were performed with ethanol and DMSO alone (data not shown). The potential ligands chosen for testing were a combination of physiologically relevant (e.g. resveratrol, epicatechin, and apigenin) and irrelevant compounds (e.g. Good's biological buffers: MOPS, MES, PIPES, HEPES, and CAPS). The latter were included because a MES molecule was observed in the hydrophobic pocket in one of the early structures. For the data presented in Fig. 6E, resveratrol was substituted for ANS, and the assay was performed as described above. The fluorescence intensity was plotted against wavelength in Origin (Microcal, Northampton, MA). The height of the peak was designated as maximal fluorescence signal and was used to calculate the percentage increase or decrease of the signal compared with the peak height from 40 μm ANS, 40 μm Ara h 8 alone. The percentage change from at least two independent experiments was averaged.
FIGURE 6.
Ligand binding by Ara h 8. A, titration of 40 μm ANS (black) with Ara h 8: 20 μm (red), 30 μm (green), and 40 μm (blue). Also shown is titration of 40 μm ANS, 40 μm Ara h 8 with quercetin: 10 μm (cyan), 20 μm (pink), 30 μm (yellow), 40 μm (olive), 50 μm (navy), and 100 μm (purple). B, titration of 40 μm ANS (black) with Ara h 8: 20 μm (red), 30 μm (green), and 40 μm (blue). Titration of 40 μm ANS, 40 μm Ara h 8 with daidzein: 20 μm (cyan), 40 μm (pink), 60 μm (yellow), 100 μm (olive), 200 μm (navy), and 300 μm (purple). C, compilation of fluorescence data. 100% was the value of fluorescence for 40 μm ANS, 40 μm Ara h 8. The values for increases or decreases in fluorescence for each concentration of ligand are shown relative to the 100% value. Shown are progesterone (black), myristic acid (red), arachidic acid (green), caffeic acid (blue), quercetin (cyan), genistein (pink), daidzein (yellow), apigenin (olive), stigmasterol (navy), and zeatin (purple). D, titration of 40 μm ANS (black) with Ara h 8: 20 μm (orange), 30 μm (green), and 40 μm (blue). Shown is titration of 40 μm ANS, 40 μm Ara h 8 with resveratrol 50 μm (pink) and 100 μm (cyan). E, fluorescence of resveratrol. Shown is titration of 50 μm resveratrol (cyan) with Ara h 8: 10 μm (red) and 20 μm (green). Also shown is titration of 50 μm resveratrol, 20 μm Ara h 8 with 10 μm quercetin (dark blue) and 20 μm quercetin (light blue).
Sequence Analysis
The Ara h 8 sequence was used as a query in PSI-BLAST (31) searches of the non-redundant database with the expectation value threshold for the retrieval of related sequences set to 10−30. The sequences were clustered using CD-HIT (32) with a 95% identity threshold; thus, the sequences with 95% or above identity to the representative sequences were removed. To identify and visualize subgroups of closely related sequences, we used CLANS (cluster analysis of sequences (33)). CLANS uses the p values of highly scoring segment pairs obtained from an N × N BLAST search to compute attractive and repulsive forces between each sequence pair.
Other Computational Calculations
Binding cavity volume was calculated with CASTp (34). The SSM algorithm (35) in COOT was used to superpose protein models. All figures containing protein structures were prepared with PyMOL (36). The ConSurf server was used to map sequence conservation on the Ara h 8 structure (37).
RESULTS
Clustering Analysis of the Closest Ara h 8 Homologs
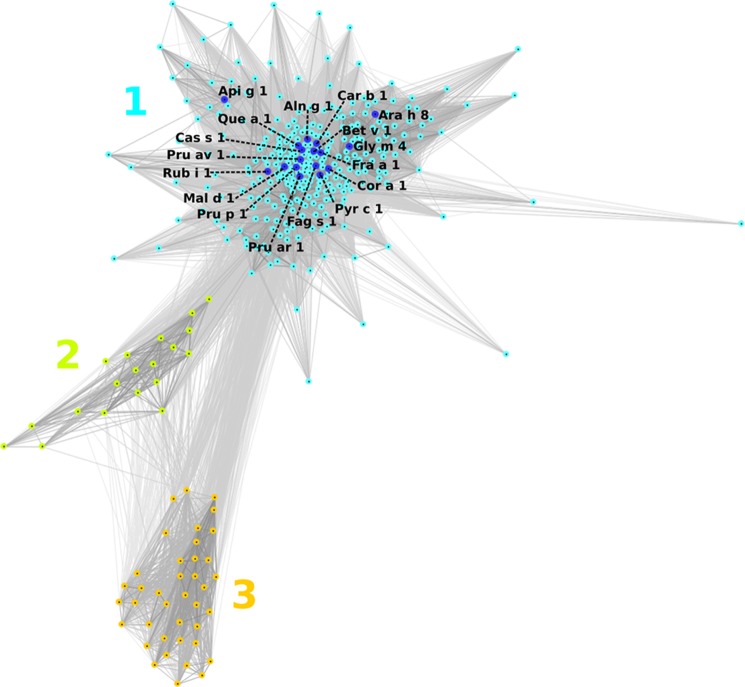
Sequences of the closest Ara h 8 homologs were clustered based on their pair-wise BLAST similarity scores using CLANS (33). Because the sequences were selected on a very restrictive threshold of 10−30, they all show high similarity (Fig. 1). Cluster 1 is composed of sequences of proteins from eudicots, most of which include known allergens. Cluster 2 is composed of sequences of proteins from gymnosperms: mainly from Pinus and Picea species and pathogenesis-related protein PsemI from Pseudotsuga menziesii but also one uncharacterized sequence from Populus trichocarpa (other sequences form P. trichocarpa grouped in cluster 1). Cluster 3 comprises proteins of different species of monocots, including Lolium, Zea, Triticum, Oryza, and others.
FIGURE 1.
Two-dimensional projection of the CLANS clustering results. Proteins are indicated by dots. Lines indicate sequence similarity detectable with BLAST and are colored by a spectrum of shades of gray according to the BLAST p value (black, p < 10–200; light gray, p < 10–35). Sequences of known allergens are indicated by dark blue dots. 1–3, clusters 1–3.
Structural Analysis of Ara h 8
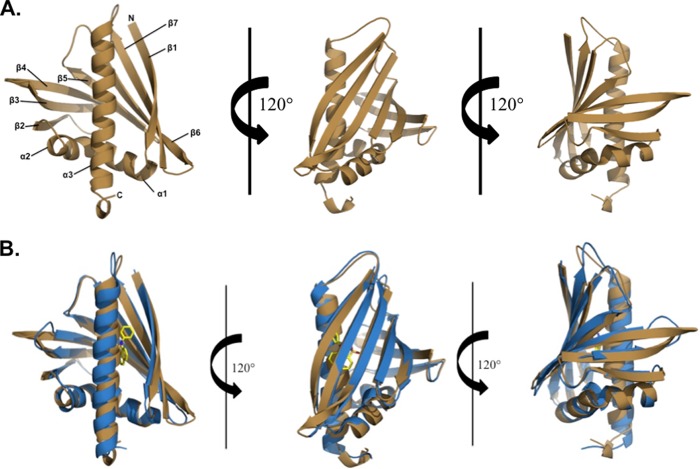
The Ara h 8 molecule is composed of 157 residues (residues 2–157 are visible in the electron density map) that are arranged by three α-helices that flank the seven-stranded anti-parallel β-sheet (Fig. 2A). Located between the β1 and β2 strands are the two shorter helices (α1 and α2), whereas the longest helix (α3) is located at the C terminus of the protein. Ara h 8 has an overall fold very similar to Bet v 1 despite a sequence identity of 48%. When superposed, the root mean square deviation (RMSD) value between Ara h 8 and Bet v 1 is 1.7 Å over 146 aligned Cα atoms (Fig. 2B). Protein purified by both protocols crystallized in a few different crystallization conditions, all of which resulted in crystals of the same form with similar conformations. The Cα atoms of the MES-bound structure, epicatechin, or heated protein structure superposed on the apo-structure have RMSD values of 0.2, 0.4, or 0.1 Å, respectively, over 150 aligned Cα atoms. The largest differences between the apo-structure and the MES-bound structure are found in α3 (residues 133–149) at the C terminus of the protein, whereas the largest change between the apo-structure and the epicatechin-bound structure is at the turn connecting β5 and β6. It is apparent that the changes are due to the binding of the ligands. There are no significant differences between the structures of the apoprotein and the protein that was purified with heating. The central part of the Ara h 8 molecule contains a large cavity with an approximate volume of 1240 Å3. This binding cavity is slightly smaller than that of Bet v 1 but significantly larger than Act d 11, a member of the Bet v 1 superfamily (38). Similar to that of Bet v 1 and Bet v 1 homologs, the walls of the cavity are composed of both α-helical and β-sheet parts of the protein (Fig. 3). The pocket is composed of 30 residues, six of which are charged.
FIGURE 2.
A, overall three-dimensional structure of Ara h 8 at three positions showing the backbone folding pattern with secondary structural elements. Ara h 8, like Bet v 1, has three α-helices flanking the seven-stranded β-sheet. B, superposition of Ara h 8 (shown in brown) and Bet v 1 (shown in blue). The ligand, ANS, is shown in atom type colors and is bound to Bet v 1 (PDB code 4A80). The overall fold of the two allergens is very similar, with three α-helices and seven β-strands, a root mean square deviation value of 1.7Å, and a sequence identity of 48%.
FIGURE 3.
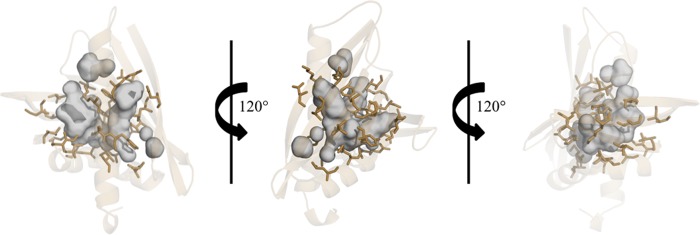
Cavities in Ara h 8. Residues that form the large central cavity are shown as sticks in brown. The cavities are shown as a surface in gray.
In addition, Ara h 8 structures revealed the presence of a metal-binding site, which is formed by residues connecting helix α2 and strand β2. The metal was identified as sodium, which is consistent with previous structural studies of PR-10 proteins (39). For example, an observed Na+ binding site overlaps very well with one of the sodium binding sites found in L1PR-10.2B (PDB code 3E85) originating from yellow lupine (40).
DALI (41) identified a pathogenesis-related protein, LLPR10.1B, from L. luteus (PDB code 1IFV, RMSD value 1.2 Å over 148 aligned Cα atoms); a pathogenesis-related protein, SPE16, from Pachyrhizus erosus (PDB codes 1TW0 and 1TXC; RMSD values of 1.3 Å over 151 or 149 aligned Cα atoms, respectively); and a pathogenesis-related protein, LLPR10.1A, from L. luteus (PDB code 1ICX, RMSD value 1.7 Å over 149 aligned Cα atoms) as having the most similar structures to Ara h 8. These three proteins are followed by Bet v 1 (PDB codes 4A88, 4A87, 1QMR, 4A85, 1BV1 and 4A80; RMSD values 1.6–1.8 Å over 146 aligned Cα atoms). Other PR-10-related allergens with determined structures (Api g 1, Dau c 1, Fra a 1, Gly m 4, and Pru av 1) also show high structural and significant sequence similarities to Ara h 8. Pru av 1 (PDB code 1H2O), Api g 1 (PDB code 2BK0), and Dau c 1 (PDB code 2W0L) superpose with Ara h 8 with lower RMSD values (1.8–1.9 Å over 150, 143, or 142 aligned Cα atoms, respectively) than Gly m 4 (PDB code 2K7H; RMSD 2.1 Å over 152 aligned Cα atoms) and Fra a 1 (PDB code: 2LPX; RMSD 2.2 Å over 141 aligned Cα atoms). The most significant structural differences between Bet v 1, Bet v 1 homologs (Api g 1, Dau c 1, Fra a 1, and Pru av 1), and Ara h 8 are related to the conformation of β3, β4, and the loop that connects them. In the case of Ara h 8, the main chain of the loop that connects β3 and β4 is rotated out toward the surface of the protein, whereas the loop in Bet v 1 and Bet v 1-related allergens is rotated toward α3. This loop region, however, is consistent between Ara h 8 and Gly m 4 despite the higher RMSD value (2.1 Å) (Fig. 4). Furthermore, a BLAST search with the Ara h 8 sequence against the Protein Data Bank identifies Gly m 4, the soybean allergen (PDB code 2K7H), as the most similar protein to Ara h 8 in terms of sequence with 68% sequence identity and 84% sequence similarity. The largest structural differences between the two correspond to helix α2 and strand β2 of Ara h 8. In Ara h 8, the main chain of this region is rotated toward helix α3, whereas Gly m 4 has this region rotated out toward the protein surface.
FIGURE 4.
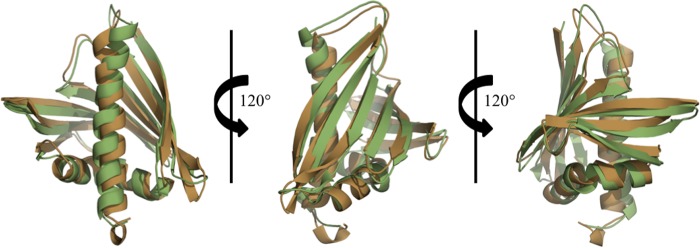
Superposition of Ara h 8 (shown in brown) and Gly m 4 (shown in green). Despite the largest RMSD value when superposed, BLAST identified Gly m 4, the soybean allergen, (PDB code 2K7H) as the most similar protein to Ara h 8 in terms of sequence with 68% sequence identity and 84% sequence similarity.
Act d 11 is an allergen that is immunologically associated with Bet v 1-like allergens but is not a PR-10 protein (38). It is a kiwifruit allergen that belongs to the major latex protein/ripening-related protein family (MLP/RRP). The PR-10 family and MLP/RRP family both belong to the Bet v 1 superfamily despite low sequence identity (<25%) between the two protein families (42). D'Avino et al. (43) were able to show that Act d 11 is able to inhibit the binding of IgE to Bet v 1, Cor a 1, Dau c 1, and Mal d 1. These results indicate that these allergens, despite being from separate protein families, share some epitopes (43). Currently, there are seven protein structures of Act d 11 deposited in the PDB (PDB entries 4IGV, 4IGW, 4IGX, 4IGY, 4IH0, 4IH2, and 4IHR). These seven structures superposed on apo-Ara h 8 have RMSD values between 2.2 and 2.5 Å over 123 aligned Cα atoms and have a sequence identity of only about 16%. The largest structural differences between apo Ara h 8 and the Act d 11 structures correspond to the regions that connect the β-sheets and α-helices. The loop that connects β3 to β4, β5 to β6, β6 to β7, and β7 to α3 have the largest degree of variation from Ara h 8. All forms of Act d 11 appear to be more compact than Ara h 8. The previously mentioned varying loop regions of Ara h 8 are all found “flipped out” toward the surface of the protein, whereas these regions of Act d 11 are all “flipped in” toward the core of the protein.
Structure of Ara h 8 Complexes
The structure of Ara h 8 in complex with the MES molecule was determined to 1.95 Å resolution. Despite Ara h 8 crystallizing with two molecules per asymmetric unit cell, electron density reveals that MES (which is present at 100 mm in the crystallization medium) is bound only in subunit B with full occupancy. It is stabilized within the binding pocket through various hydrogen bonding interactions. The oxygen atom of the MES ring moiety is coordinated to a water molecule, whereas the oxygen atoms of the phosphate tail are coordinated to Lys53, Tyr80, and His68. The Ara h 8-epicatechin structure was determined to 2.0 Å resolution. Similar to the MES-bound structure, epicatechin is only bound in one subunit; however, epicatechin is only bound in subunit A with full occupancy. It is stabilized within the pocket through several hydrogen bonding interactions as well as a few noncovalent interactions. The dihydroxyphenyl moiety of epicatechin hydrogen bonds to the side chain of Thr30 as well as to two waters that act as bridges to the side chains of His68 and Asp27. The fused rings of epicatechin coordinate to another water that acts as a bridge to His68, whereas the hydroxyl groups strengthen the stabilization of epicatechin by hydrogen bonding to the side chain hydroxyl group of Tyr82, to the backbone oxygen of Lys138, and to a bridging water molecule. Noncovalent interactions may be taking place from the side chains of Lys138, Ile57, and Leu142 to epicatechin, helping to further stabilize the ligand within the binding pocket.
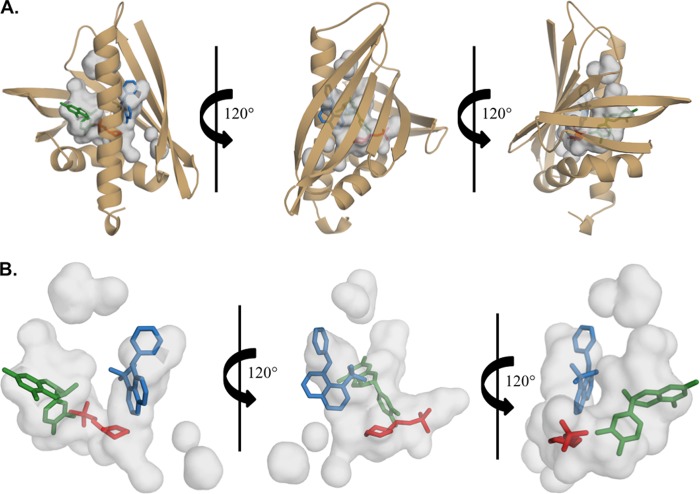
Although MES and epicatechin are bound in different subunits, when superposed, it is apparent that they bind in different positions within the same pocket. Furthermore, when superposed with the Bet v 1 structure (PDB entry 4A80), an additional position within the pocket is revealed. Fig. 5A shows the superposition of Ara h 8-MES, Ara h 8-epicatechin, and Bet v 1-ANS. This superposition reveals the possibility of ligands being able to bind in three different positions within the same pocket. Moreover, it is also possible that more than one ligand may be able to bind simultaneously (Fig. 5B).
FIGURE 5.
Cavities in Ara h 8. Shown is the main cavity, which is assumed to be the binding pocket of Ara h 8 with MES (shown in red), epicatechin (shown in dark green), and ANS (shown in blue) from Bet v 1 (PDB code 4A80). The pocket is large enough to allow for ligands to bind in three different positions.
Ligand Binding
Based on various reports that Bet v 1 and its homologs bind small molecules (17, 18, 20, 21), we tested Ara h 8 for a similar function. ANS has long been known to fluoresce when bound to hydrophobic surfaces on proteins (44). We surveyed an array of potential physiologically relevant ligands, as well as some others. Fig. 6A shows representative data for a ligand, quercetin, which binds and displaces ANS. ANS (40 μm) in buffer without protein very weakly fluoresces with an emission maximum around 515 nm. Titration of this mixture with Ara h 8 (20, 30, and 40 μm) shows stepwise, dramatic increases in fluorescence and a shift in emission maximum to 475 nm. Subsequent titration of the equimolar ANS/Ara h 8 mixture (40 μm) with quercetin (10–100 μm) shows a strong inhibition of the fluorescence, indicating competitive displacement of ANS from the hydrophobic pocket of Ara h 8. Representative data of a compound (daidzein) that had the effect of increasing the ANS fluorescence are shown in Fig. 6B. The characteristic, stepwise increase in ANS fluorescence was observed upon the addition of increasing concentrations of Ara h 8 to 40 μm ANS. Surprisingly, when daidzein was titrated in (20–300 μm), fluorescence increased. We interpret this phenomenon to indicate that daidzein does bind in the hydrophobic pocket but does not displace ANS. This phenomenon was observed with Bet v 1 and was thought to be due to increasing the quantum yield of the bound ANS (16). Fig. 6C shows a compilation of the data for the compounds that affected the fluorescence of ANS. Apigenin and quercetin had the strongest displacement activity, whereas genistein, myristic acid, caffeic acid, and zeatin had weaker displacement activity. Daidzein and arachidic acid had the strongest enhancement activity, whereas progesterone and stigmasterol were weaker. Resveratrol was also tested in this assay, but it fluoresced on its own. Fig. 6D shows data from an attempted competition assay of ANS with resveratrol. It is clear that resveratrol displaced ANS because the ANS-specific fluorescence decreased, and the resveratrol fluorescence showed strongly with an absorbance maximum shift to 380 nm. Fig. 6E shows data for a titration of 50 μm resveratrol with Ara h 8. The compound by itself fluoresced weakly with a maximum around 400 nm. The addition of Ara h 8 caused a blue shift similar to ANS and an increase in fluorescence indicating direct binding of resveratrol to the protein. The resveratrol binding site was tested with quercetin, which competed with ANS binding in the hydrophobic pocket. As seen in Fig. 6E, quercetin also competed with resveratrol binding, indicating that resveratrol binds to the same place as ANS. The structures of the compounds that bound are shown in Fig. 7A. Fig. 7B is a list of the other compounds that were tested but had no effect. Good's buffers were used because a MES molecule was observed in one of the early structures.
FIGURE 7.
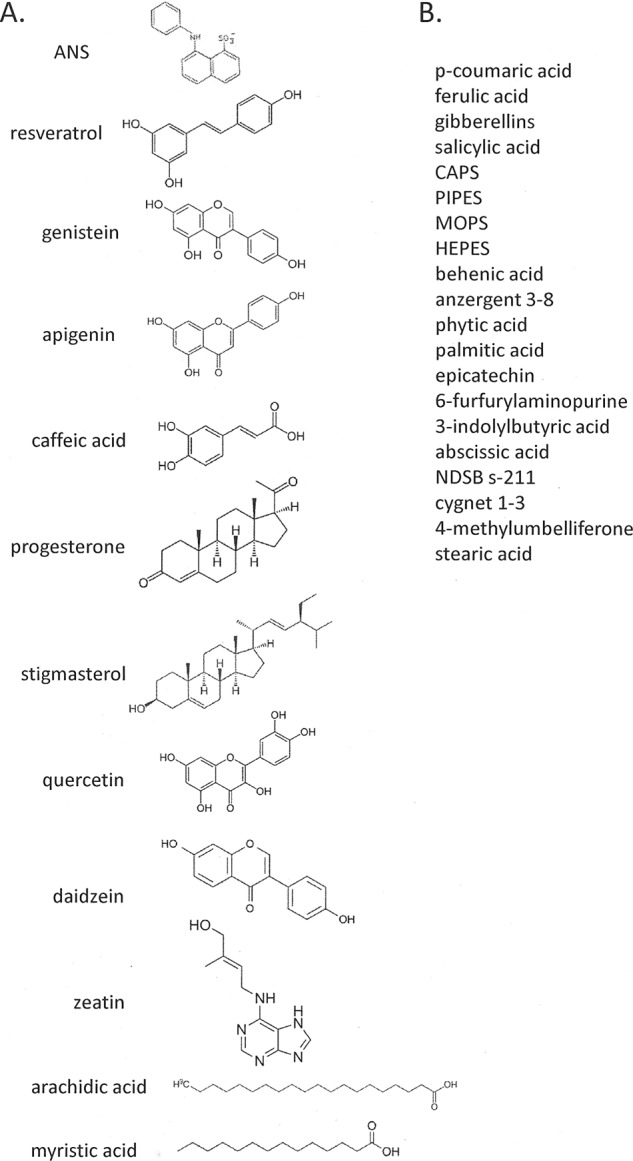
Ligands tested in fluorescence assays. A, ligand names and structures of compounds that bound Ara h 8 and affected ANS fluorescence. B, compound names that were tested but did not affect ANS fluorescence.
DISCUSSION
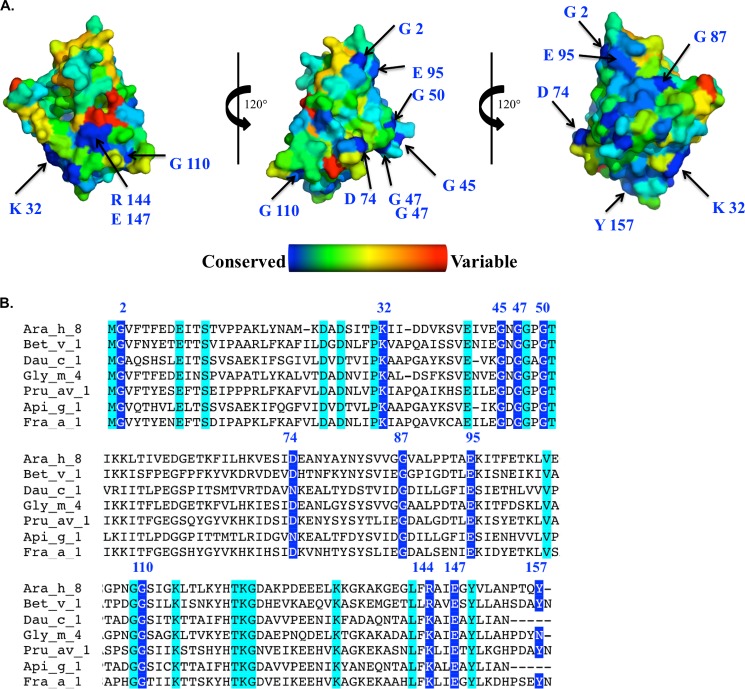
Oral allergy syndrome is defined as an IgE-mediated, immediate allergic reaction that is confined to the oral mucosa. Symptoms may include vascular edema, stinging, and itching of the tongue, palate, lips, and pharynx. These symptoms manifest when a patient who is allergic to pollen consumes a specific food, usually a member of the Rosaceae family. Although more distantly related, legumes (soy, mungbean, and peanut) have been reported to have cross-reactivity in pollen-allergic patients (2, 45, 46). The proteins that are responsible for OAS have been labeled panallergens (47). Panallergens are thought to be involved in vital cellular processes and accordingly have similar structures although the species may be quite distant evolutionarily (47). Fig. 8 shows a surface representation of the Ara h 8 molecule, indicating conservation among Ara h 8-related protein sequences as well as a sequence alignment indicating identical residues and those residues conserved among PR-10 allergens that have available structures in the PDB. It was demonstrated that the Bet v 1 dominant IgE-binding epitope includes Glu45 (48). This residue not only is conserved in Ara h 8 (Glu44), but it also starts the longest stretch of residues (Glu44–Lys54 according to Ara h 8 numbering) that is identical in both these allergens. This region corresponds to the so-called “P-loop,” the highly conserved region that is characteristic of the allergenic PR-10 proteins (37). This region was already shown to be involved in cross-reactivity between Bet v 1 and its homologs (49, 50). There are two other longer regions of the sequence that are common to Bet v 1 and Ara h 8. These include Tyr80–Val84, which is only partially exposed, and Lys118–Asp124. The second of these regions together with conserved fragment Gly2-Phe3 and residues Thr93, Glu95, and Lys96 form a large solvent-exposed patch on the allergen's surface that also may play a role in cross-reactivity between Bet v 1 and Ara h 8. The presence of these large conserved regions may explain why Ara h 8 is the main cross-reactive allergen for patients that have combined birch pollen and peanut allergy (2, 51, 52). In addition, residues Thr11, Ser12, Glu140, Arg144, Glu147, Leu151, and Ala152, which are conserved in both Ara h 8 and Bet v 1, form a relatively large, solvent-exposed patch on the surface of these allergens. Some of these residues were identified as part of the IgE epitope, with Glu147 (Ara h 8 numbering) being the most critical (53).
FIGURE 8.
A, surface representation of Ara h 8 showing conserved residues derived from an alignment of 500 Ara h 8-related protein sequences. The most conserved residues are shown in blue, whereas the most variable residues are shown in red. B, multiple-sequence alignment of Ara h 8. The most conserved residues are highlighted in blue, and their position is indicated on the structures above. Residues highlighted in cyan are conserved among PR-10 allergens for which there are available structures in the Protein Data Bank.
The Bet v 1-like proteins represent a class of proteins called PR-10 (for pathogenesis-related class 10, whose expression increases in response to pathogen attack (39). The function of the Bet v 1 family of proteins has been investigated. Initially, there were reports of RNase activity (11, 12). Since then, several other related proteins have been shown to possess RNase activity, including birch PR-10c (13) and lupin LaPR-10 (14). We tested our recombinant Ara h 8 for RNase activity and found none (data not shown). Some PR-10 proteins have been shown to be up-regulated in response to microbial invasion and have antimicrobial activity (reviewed in Ref. 39). Ara h 8 showed no growth inhibition activity against various Gram-negative and positive bacteria, S. cerevisiae and A. flavus (data not shown).
The Bet v 1 family of proteins is structurally homologous to the START domain of the human MLN64 protein (15) that has been shown to bind one molecule of cholesterol (54). Bet v 1 and its homologs from various species have been shown to bind small hydrophobic compounds (reviewed in Ref. 39). Mogensen et al. (16) showed that flavonoids, cytokinins, and fatty acids bind to Bet v 1. PR-10c from birch is related to Bet v 1 and binds a variety of biologically important ligands (17). Recently, the structures of different isoforms of Bet v 1 in complex with deoxycholate, naringenin, kinetin, NDSB-256, or the molten globule probe ANS were determined (PDB codes 4A83, 4A87, 4A85, 4A8G, and 4A80, respectively) (18, 19). Additionally, Fernandes et al. reported a co-crystal structure of LIPR-102B from L. luteus (yellow lupine) with three zeatin molecules bound (20). A structurally similar protein, VrCSBP from V. radiata (mung bean), was also found to bind zeatin (PDB code 2FLH) (21). Our structural results show details of Ara h 8-epicatechin interactions, and it is the first structure of a PR-10-related protein in complex with this flavonoid. Prior to our studies, the Bet v 1-naringenin complex was the only one to have its structure determined in complex with a flavonoid (PDB code 4A87), and it was the only structure demonstrating binding of flavonoid to a PR-10 protein. Superposition of Ara h 8-epicatechin and Bet v 1-naringenin structures reveals that the binding sites for these flavonoids are different and are located in non-overlapping parts of the large ligand-binding cavities of Ara h 8 and Bet v 1. Recently, it was also shown that Fra a 1 crystallized in the presence of (+)-catechin; however, a structure is not yet available (55).
All structures reported here have electron density for the entire molecule with the exception of the start methionine in both subunits. The overall fold of Ara h 8 is similar to Bet v 1, which includes three α-helices that flank the seven-stranded anti-parallel β-sheet. Reported here are four structures of recombinant Ara h 8, two of which are native protein purified by different methods (see “Experimental Procedures”) and two with bound ligands (MES or epicatechin). When comparing the all structures to the apo form of the protein, the RMSD values are between 0.1 and 0.4 Å. There are no significant differences between the structures of the apoprotein and the protein that was purified with heating; however, the heated protein structure only contains one sodium atom, whereas all other structures contain two (one in each subunit). It demonstrates that Ara h 8 has significant thermostability that may allow the allergen to stay intact during food processing. The largest differences between the apo-structure and the MES-bound structure are found in α3 at the C terminus of the protein, whereas the largest change between the epicatechin-bound structure and the apo-structure is at the turn connecting β5 and β6. It is apparent that these changes are due to the ligands being bound. Our observations provide additional evidence for significant plasticity and unusual ligand binding properties of PR-10-related proteins that can be compared with the properties of mammalian albumins.
We have shown here that ANS and several biologically relevant compounds can bind to Ara h 8. The most avidly bound are apigenin, quercetin, daidzein, resveratrol, and arachidic acid. The first three of these are phytoestrogen flavonoids, which in plants serve a number of functions, including but not limited to UV protection, defense against microbial attack, and signaling during development (reviewed in Refs. 56 and 57). The fact that the PR-10 class of proteins and some flavonoids are up-regulated in response to damage or microbial invasion suggests that Ara h 8 may serve as a delivery vector for flavonoids. Mogensen et al. (58) showed that Bet v 1 can bind and permeabilize membranes, suggesting a mechanism for delivery of bound molecules from the cytoplasm to the extracellular space.
It is interesting to note that apigenin and daidzein had opposite effects in the ANS assay because their structures are so similar. The different substitution of the phenolic group is probably behind this observation. Resveratrol, a competitor for ANS, was not only shown to displace ANS but was also shown to compete with ANS competitors (see Fig. 6E). It is also interesting that epicatechin showed no activity in the assay but can clearly bind, as observed in the co-crystal. A closer inspection of the ligand-binding cavity reveals a large pocket capable of binding ligands in different positions or capable of binding more than one ligand simultaneously (see Fig. 5). The superposition of Bet v 1 containing ANS and Ara h 8 indicates where ANS may bind within the Ara h 8 binding site. From this we can extrapolate that ANS and resveratrol most likely bind in the same position within the pocket, whereas ANS and epicatechin may be able to bind simultaneously. These observations would explain why a loss of fluorescence in the presence of resveratrol is seen and why epicatechin is bound in the crystal structure, but a loss of fluorescence is not seen in the ANS displacement assay. Kofler et al. (19) determined that Bet v 1 was able to bind multiple ligands simultaneously, which lends support for our hypothesis that Ara h 8 can do so as well. In summary, we have shown that PR-10 ligand-binding has to be investigated by several different experimental methods because only such an approach provides a full picture of the unusual promiscuity and complexity of the interactions of these proteins with small molecular ligands.
Acknowledgments
We thank Drs. Ken Ehrlich and Steve Boue for critical review of the manuscript. The structural results shown here are derived from work performed at Argonne National Laboratory at the Structural Biology Center of the Advanced Photon Source. Argonne is operated by the University of Chicago Argonne, LLC, for the United States Department of Energy Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
This work was supported by the United States Department of Agriculture, Agricultural Research Service, and internal funds from the University of South Carolina.
The atomic coordinates and structure factors (codes 4M9B, 4M9W, 4MA6, and 4MAP) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Zheng, H., Chordia, M. D., Cooper, D. R., Chruszcz, M., Müller, P., Sheldrick, G. M., and Minor, W. (2014) Validation of metal-binding sites in macromolecule structures with the CheckMyMetal web server. Nat. Protoc., in press.
- OAS
- oral allergy syndrome
- ANS
- 8-anilino-1-napthalenesulfonic acid
- PDB
- Protein Data Bank
- QCB
- Q-column buffer
- RMSD
- root mean square deviation.
REFERENCES
- 1. Liu A. H., Jaramillo R., Sicherer S. H., Wood R. A., Bock S. A., Burks A. W., Massing M., Cohn R. D., Zeldin D. C. (2010) National prevalence and risk factors for food allergy and relationship to asthma. Results from the National Health and Nutrition Examination Survey 2005–2006. J. Allergy Clin. Immun. 126, 798–806.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mittag D., Akkerdaas J., Ballmer-Weber B. K., Vogel L., Wensing M., Becker W.-M., Koppelman S. J., Knulst A. C., Helbling A., Hefle S. L., Van Ree R., Vieths S. (2004) Ara h 8, a Bet v 1-homologous allergen from peanut, is a major allergen in patients with combined birch pollen and peanut allergy. J. Allergy Clin. Immunol. 114, 1410–1417 [DOI] [PubMed] [Google Scholar]
- 3. Riecken S., Lindner B., Petersen A., Jappe U., Becker W.-M. (2008) Purification and characterization of natural Ara h 8, the Bet v 1 homologous allergen from peanut, provides a novel isoform. Biol. Chem. 389, 415–423 [DOI] [PubMed] [Google Scholar]
- 4. Kondo Y., Urisu A. (2009) Oral allergy syndrome. Allergol. Int. 58, 485–491 [DOI] [PubMed] [Google Scholar]
- 5. Gajhede M., Osmark P., Poulsen F. M., Ipsen H., Larsen J. N., Joost van Neerven R. J., Schou C., Løwenstein H., Spangfort M. D. (1996) X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nat. Struct. Biol. 3, 1040–1045 [DOI] [PubMed] [Google Scholar]
- 6. Schirmer T., Hoffimann-Sommergrube K., Susani M., Breiteneder H., Marković-Housley (2005) Crystal structure of the major celery allergen Api g 1. Molecular analysis of cross-reactivity. J. Mol. Biol. 351, 1101–1109 [DOI] [PubMed] [Google Scholar]
- 7. Pasternak O., Biesiadka J., Dolot R., Handschuh L., Bujacz G., Sikorski M. M., Jaskolski M. (2005) Structure of a yellow lupin pathogenesis-related PR-10 protein belonging to a novel subclass. Acta Crystallogr. D Biol. Crystallogr. 61, 99–107 [DOI] [PubMed] [Google Scholar]
- 8. Biesiadka J., Bujacz G., Sikorski M. M., Jaskolski M. (2002) Crystal structures of two homologous pathogenesis-related proteins from yellow lupine. J. Mol. Biol. 319, 1223–1234 [DOI] [PubMed] [Google Scholar]
- 9. Schweimer K., Sticht H., Nerkamp J., Boehm M., Breitenback M., Vieths S., Rösch. P. (1999) NMR spectroscopy reveals common structural features of the birch pollen allergen Bet v 1 and the cherry allergen Pru a 1. Appl. Magn. Reson. 17, 449–464 [Google Scholar]
- 10. Berkner H., Neudecker P., Mittag D., Ballmer-Weber B. K., Schweimer K., Vieths S., Rösch P. (2009) Cross-reactivity of pollen and food allergens. Soybean Gly m 4 is a member of the Bet v 1 superfamily and closely resembles yellow lupine proteins. Biosci. Rep. 29, 183–192 [DOI] [PubMed] [Google Scholar]
- 11. Bufe A., Spangfort M. D., Kahlert H., Schlaak M., Becker W.-M. (1996) The major birch pollen allergen, Bet v 1, show ribonuclease activity. Planta 199, 413–415 [DOI] [PubMed] [Google Scholar]
- 12. Swoboda I., Hoffmann-Sommergruber K., O'Ríordáin G., Scheiner O., Heberle-Bors E., Vicente O. (1996) Bet v 1 proteins, the major birch pollen allergens and members of a family of conserved pathogenesis-related proteins, show ribonuclease activity in vitro. Physiol. Plant. 96, 433–438 [Google Scholar]
- 13. Koistinen K. M., Kokko H. I., Hassinen V. H., Tervahauta A. I., Auriola S., Käranlampi S. O. (2002) Stress-related RNase PR-10c is post-translationally modified by glutathione in birch. Plant Cell Environ. 25, 707–715 [Google Scholar]
- 14. Bantignies B., Séguin J., Muzac I., Dédaldéchamp F., Gulick P., Ibrahim R. (2000) Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant. Mol. Biol. 42, 871–881 [DOI] [PubMed] [Google Scholar]
- 15. Tsujishita Y., Hurley J. H. (2000) Structure and lipid transport mechanism of a StAR-related domain. Nat. Struct. Biol. 7, 408–414 [DOI] [PubMed] [Google Scholar]
- 16. Mogensen J. E., Wimmer R., Larsen J. N., Spangfort M. D., Otzen D. E. (2002) The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J. Biol. Chem. 277, 23684–23692 [DOI] [PubMed] [Google Scholar]
- 17. Koistinen K. M., Soininen P., Venäläinen T. A., Häyrinen J., Laatikainen R., Peräkylä M., Tervahauta A. I., Kärenlampi S. O. (2005) Birch PR-10c interacts with several biologically important ligands. Phytochemistry 66, 2524–2533 [DOI] [PubMed] [Google Scholar]
- 18. Marković-Housley Z., Degano M., Lamba D., von Roepenack-Lahaye E., Clemens S., Susani M., Ferreira F., Scheiner O., Breiteneder H. (2003) Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function at a plant steroid carrier. J. Mol. Biol. 325, 123–133 [DOI] [PubMed] [Google Scholar]
- 19. Kofler S., Asam C., Eckhard U., Wallner M., Ferreira F., Brandstetter H. (2012) Crystallographically mapped ligand binding differs in high and low IgE binding isoforms of birch pollen allergen Bet v 1. J. Mol. Biol. 422, 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernandes H., Pasternak O., Bujacz G., Bujacz A., Sikorski M. M., Jaskolski M. (2008) Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin. J. Mol. Biol. 378, 1040–1051 [DOI] [PubMed] [Google Scholar]
- 21. Pasternak O., Bujacz G. D., Fujimoto Y., Hashimoto Y., Jelen F., Otlewski J., Sikorski M. M., Jaskolski M. (2006) Crystal structure of Vigna radiata cytokinin-specific binding protein in complex with zeatin. Plant Cell 18, 2622–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) HKL-3000. The integration of data reduction and structure solution, from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 24. Vagin A., Teplyakov A. (1997) MOLREP. An automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 25. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Painter J., Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 29. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang H., Guranovic V., Dutta S., Feng Z., Berman H. M., Westbrook J. D. (2004) Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 60, 1833–1839 [DOI] [PubMed] [Google Scholar]
- 31. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST. A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li W., Jaroszewski L., Godzik A. (2001) Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17, 282–283 [DOI] [PubMed] [Google Scholar]
- 33. Frickey T., Lupas A. (2004) CLANS. A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 [DOI] [PubMed] [Google Scholar]
- 34. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) CASTp. Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 36. DeLano W., S. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 37. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010. Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chruszcz M., Ciardiello M. A., Osinski T., Majorek K. A., Giangrieco I., Font J., Breiteneder H., Thalassinos K., Minor W. (2013) Structural and bioinformatics analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins. Mol. Immunol. 56, 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fernandes H., Michalska K., Sikorski M., Jaskolski M. (2013) Structural and functional aspects of PR-10 proteins. FEBS J. 280, 1169–1199 [DOI] [PubMed] [Google Scholar]
- 40. Fernandes H., Bujacz A., Bujacz G., Jelen F., Jasinski M., Kachlicki P., Otlewski J., Sikorski M. M., Jaskolski M. (2009) Cytokin-induced structural adaptability of Lupinus luteus PR-10 protein. FEBS J. 276, 1596–1609 [DOI] [PubMed] [Google Scholar]
- 41. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Osmark P., Boyle B., Brisson N. (1998) Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins. Plant Mol. Biol. 38, 1243–1246 [DOI] [PubMed] [Google Scholar]
- 43. D'Avino R., Bernardi M. L., Wallner M., Palazzo P., Camardella L., Tuppo L., Alessandri C., Breiteneder H., Ferreira F., Ciardiello M. A., Mari A. (2011) Kiwifruit Act d 11 is the first member of the ripening-related protein family identified as an allergen. Allergy 66, 870–877 [DOI] [PubMed] [Google Scholar]
- 44. Goldberg M. E., Semisotnov G. V., Friguet B., Kuwajima K., Ptitsyn O. B., Sugai S. (1990) An early immunoreactive folding intermediate of the tryptophan synthase β2 subunit is a “molten globule”. FEBS Lett. 263, 51–56 [DOI] [PubMed] [Google Scholar]
- 45. Mittag D., Vieths S., Vogel L., Becker W.-M., Rihs H.-P., Helbling A., Wüthrich B., Ballmer-Weber B. K. (2004) Soybean allergy in patients allergic to birch pollen. Clinical investigation and molecular characterization of allergens. J. Allergy Clin. Immun. 113, 148–154 [DOI] [PubMed] [Google Scholar]
- 46. Mittag D., Vieths S., Vogel L., Wagner-Loew D., Starke A., Hunziker P., Becker W.-M., Ballmer-Weber B. K. (2005) Birch pollen-related food allergy to legumes. Identification and characterization of the Bet v 1 homologue in mungbean (Vigna radiate), Vig r 1. Clin. Exp. Allergy 35, 1049–1055 [DOI] [PubMed] [Google Scholar]
- 47. Hauser M., Roulias A., Ferreira F., Egger M. (2010) Panallergens and their impact on the allergic patient. Allergy Asthma Clin. Immunol. 6, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spangfort M. D., Mirza O., Ipsen H., Van Neerven R. J., Gajhede M., Larsen J. N. (2003) Dominating IgE-binding epitope of Bet v 1, the major allergen of birch pollen, characterized by x-ray crystallography and site-directed mutagenesis. J. Immunol. 171, 3084–3090 [DOI] [PubMed] [Google Scholar]
- 49. Mittag D., Batori V., Neudecker P., Wiche R., Friis E. P., Ballmer-Weber B. K., Vieths S., Roggen E. L. (2006) A novel approach for investigation of specific and cross-reactive IgE epitopes on Bet v 1 and homologous food allergens in individual patients. Mol. Immunol. 43, 268–278 [DOI] [PubMed] [Google Scholar]
- 50. Neudecker P., Schweimer K., Nerkamp J., Scheurer S., Vieths S., Sticht H., Rösch P. (2001) Allergic cross-reactivity made visible. J. Biol. Chem. 276, 22756–22763 [DOI] [PubMed] [Google Scholar]
- 51. Asarnoj A., Ostblom E., Ahlstedt S., Hedlin G., Lilja G., van Hage M., Wickman M. (2010) Reported symptoms to peanut between 4 and 8 years among children sensitized to peanut and birch pollen. Results from the BAMSE birth cohort. Allergy 65, 213–219 [DOI] [PubMed] [Google Scholar]
- 52. Vereda A., van Hage M., Ahlstedt S., Ibañez M. D., Cuesta-Herranz J., van Odijk J., Wickman M., Sampson H. A. (2011) Peanut allergy. Clinical and immunologic differences among patients from 3 different geographic regions. J. Allergy Clin. Immunol. 127, 603–607 [DOI] [PubMed] [Google Scholar]
- 53. Hecker J., Diethers A., Schulz D., Sabri A., Plum M., Michel Y., Mempel M., Ollert M., Jakob T., Blank S., Braren I., Spillner E. (2012) An IgE epitope of Bet v 1 and fagales PR10 proteins as defined by a human monoclonal IgE. Allergy 67, 1530–1537 [DOI] [PubMed] [Google Scholar]
- 54. Watari H., Arakane F., Moog-Lutz C., Kallen C. B., Tomasetto C., Gerton G. L., Rio M.-C., Baker M. E., Strauss J. F., 3rd (1997) MLN64 contains a domain with homology to the steriodogenic acute regulatory protein (StAR) that stimulates steriodogenesis. Proc. Natl. Acad. Sci. U.S.A. 94, 8462–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Casañal A., Zander U., Dupeux F., Valpuesta V., Marquez J. A. (2013) Purification, crystallization, and preliminary x-ray analysis of the strawberry allergens Fra a 1E and Fra a 3 in the presence of catechin. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69, 510–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Samanta A., Das G., Das S. K. (2011) Role of flavonoids in plants. Int. J. Pharm. Sci. Tech. 6, 12–35 [Google Scholar]
- 57. Falcone Ferreyra M.L., Rius S.P., Casati P. (2012) Flavonoids. Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mogensen J. E., Ferreras M., Wimmer R., Petersen S. V., Enghild J. J., Otzen D. E. (2007) The major allergen from birch tree pollen, Bet v 1, binds and permeablizes membranes. Biochemistry 46, 3356–3365 [DOI] [PubMed] [Google Scholar]