Background: JMJD1A specifically demethylates mono- and dimethyl-histone H3K9.
Results: Two active sites of JMJD1A homodimer cooperatively demethylate dimethylated H3K9.
Conclusion: Homodimerization of JMJD1A is essential for appropriate control of histone methylation state.
Significance: The novel demethylation mechanism of JMJD1A will help in understanding the histone code.
Keywords: Chromatin Histone Modification, Enzyme Mechanisms, Histone Methylation, Mass Spectrometry (MS), Surface Plasmon Resonance (SPR), JMJD1A, Chromatin Biology, Dimerization, Histone Lysine Demethylase
Abstract
Post-translational histone methylation is a dynamic and reversible process that is involved in the spatio-temporal regulation of gene transcription and contributes to various cellular phenotypes. Methylation of histone H3 at lysine 9 (H3K9), which is generally a transcriptional repression mark, is demethylated by H3K9-specific demethylases, leading to transcriptional activation. However, how multiple demethylases with the same substrate specificity differ in their chromatin targeting mechanisms has not been well understood. Unlike other H3K9-specific demethylases, it has been reported that JMJD1A likely forms a homodimer, but a detailed mode of dimerization and the possible link between structure and enzymatic activity have remained unresolved. Here, we report the structure-function relationship of JMJD1A in detail. First, JMJD1A forms a homodimer through its catalytic domains, bringing the two active sites close together. Second, increasing the concentration of JMJD1A facilitates efficient production of unmethylated product from dimethyl-H3K9 and decreases the release of the monomethylated intermediate. Finally, substituting one of the two active sites with an inactive mutant results in a significant reduction of the demethylation rate without changing the affinity to the intermediate. Given this evidence, we propose a substrate channeling model for the efficient conversion of dimethylated H3K9 into the unmethylated state. Our study provides valuable information that will help in understanding the redundancy of H3K9-specific demethylases and the complementary activity of their unique structures and enzymatic properties for appropriate control of chromatin modification patterns.
Introduction
Post-translational histone modification is an important component of the epigenome, and is involved in the regulation of gene transcription in the chromatin context. Histone lysine methylation is known to contribute to human diseases, such as cancer, neuropsychiatric disorders, inflammation, and metabolic disorders (1). The methylation state of lysine residues can be mono, di, or tri. These different states provide information as part of the “histone code,” in conjunction with other modifications such as phosphorylation and acetylation, for fine-tuning gene expression in response to specific signals. Methylation is reversibly regulated by S-adenosylmethionine-dependent methyltransferases, the flavin-dependent demethylases, and the Jumonji family of iron and 2-oxoglutarate-dependent demethylases (1). Jumonji C (JmjC)2 domain-containing demethylases show differential specificities toward various histone lysine residues and also demonstrate methyl group number specificity, suggesting a fine tuning role for demethylases.
Methylation of Lys-9 in histone H3 (H3K9) is generally involved in transcriptional repression and heterochromatin formation (2). High-resolution mapping of histone modification patterns has shown that di- and trimethylations of Lys-9 (H3K9me2 and H3K9me3) are enriched in the transcriptional start sites of silenced genes, whereas H3K9me1 is present in the promoter regions of active genes (3). These findings suggest that the methylation state of H3K9 would be an important mark denoting the transcriptional status of a given gene. Within the last decade, various H3K9-specific modifying enzymes and recognition proteins, including JMJD1A/KDM3A/JHDM2A, have been isolated (2, 4–6). In particular, despite the similar substrate specificity of these demethylating enzymes, the differences in their kinetic properties and chromatin targeting mechanisms have remained unresolved.
JMJD1A is capable of demethylating H3K9me1 and H3K9me2 specifically, but not H3K9me3 (7). Unlike other H3K9me2/me1-specific demethylases, such as PHF8 and KIAA1718 (8), JMJD1A likely forms a homodimer (7), but a detailed model of dimerization and the possible link between its structure and enzymatic activity are unknown. There are accumulating in vivo evidence on the importance of JMJD1A, such as its involvement in spermatogenesis (9, 10), and metabolic disorders (11, 12). JMJD1A is up-regulated under hypoxic conditions (13–18), and may be a good therapeutic target against various types of cancer (18–22). However, there is little information thus far about the molecular basis of its enzymatic activity. Enzymological studies can provide valuable information in elucidating the biological role of JMJD1A, and understanding the catalytic mechanism of JMJD1A compared with other H3K9-specific demethylases.
In this study, we investigated the structure-activity relationship of JMJD1A in detail and demonstrated how it functions as a homodimer in the sequential removal of H3K9me2 methylation. Using quantitative data during demethylating processes from formaldehyde dehydrogenase-coupled assays, surface plasmon resonance (SPR) binding assays, and MALDI-TOF mass spectrometry analyses, we investigated H3K9 demethylation by JMJD1A homodimer. Based on its kinetic properties, we propose a substrate channeling mechanism by which the two active sites cooperatively demethylate two methyl groups.
EXPERIMENTAL PROCEDURES
Plasmids
A plasmid encoding human JMJD1A(1–1321) was kindly provided by Dr. Mimura (17). Strep-FLAG (SF) tag (23) and Myc-HA tag were generated by annealing complementary DNA oligos and subcloned into pcDNA3.1(+) vector. Then these tags were inserted into pcDNA3-JMJD1A to generate pcDNA3-SF-JMJD1A and pcDNA3-Myc-HA-JMJD1A. JMJD1A was also inserted into the pcDNA4-Myc-His vector to generate pcDNA4-JMJD1A-Myc-His. pcDNA3-SF-JMJD1A (H1120Y), pcDNA3-Myc-HA-JMJD1A (H1120Y), and deletion mutants (623–717, 1058–1281, 718–889, 890–1057, Δ623–717, and Δ623–1321) were generated by PCR. The Δ890–1321 deletion mutant was generated by blunting two EcoRI sites and self-ligation. All the constructs generated through PCR were verified by sequencing.
Cell Culture and Transient Transfection
293T cells were maintained in DMEM supplemented with 10% FBS and antibiotics at 37 °C. The plasmids were transfected using Lipofectamine 2000 (Invitrogen) following standard procedures.
Blue Native-PAGE, SDS-PAGE, and Immunoblotting
For Blue Native-PAGE, Native-PAGETM Novex BisTris gels were purchased from Invitrogen, and the sample preparation and electrophoresis were performed according to the manufacturer's instructions. For SDS-PAGE, samples were boiled for 5 min at 95 °C in SDS-PAGE sample buffer containing 125 mm Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, and 0.01% bromphenol blue. Samples were loaded onto 7% gels. For immunoblotting analysis, the gel was electroblotted to a PVDF membrane, which was blocked in 2% skim milk. Primary antibodies used were anti-FLAG M2 (Sigma), anti-HA (Roche), and anti-Myc (Santa Cruz). Secondary detection was carried out using anti-mouse (GE Healthcare) and anti-rat (Santa Cruz) antibodies. Band intensity and relative molecular weights were determined using LAS3000 ImageGauge software (Fuji-film, Japan).
Immunoprecipitation
Forty-eight hours after being transfected with plasmids, 293T cells were lysed in lysis buffer containing 20 mm Tris-HCl, pH 8.0, 1% Nonidet P-40, 150 mm NaCl, and a protease inhibitor mixture (Roche Applied Science). The insoluble material was removed by centrifugation at 15,000 × g for 25 min, and the soluble fraction was incubated with an anti-FLAG M2 antibody (Sigma) or an anti-Myc antibody (Santa Cruz) for 1 h. The immune complexes were pulled down with protein A-Sepharose (GE Healthcare) and washed three times with lysis buffer. Samples were eluted with SDS-PAGE sample buffer and analyzed by immunoblotting.
Formaldehyde Dehydrogenase-coupled Demethylase Assay
Coupled fluorescent demethylase assays were performed in a reaction buffer containing 50 mm HEPES-KOH, pH 8.0, 50 μm Fe(NH4)2(SO4)2, 1 mm 2-oxoglutarate, 2 mm ascorbate, 500 μm NAD+, 0.025 units of formaldehyde dehydrogenase, 0.01% Tween 20, 100 nm JMJD1A, and variable concentrations of K9-methylated H3 peptide (residues 1–15). Production of NADH was monitored (excitation 355 nm/emission 460 nm) and readings were taken over 20 cycles with a cycle time of 3 min. An NADH standard curve was used to convert fluorescence to a concentration of formed product. The initial 9 min were used to calculate initial velocities, which were graphed against substrate concentration. Michaelis-Menten parameters were determined by non-linear least squares fitting using GraphPad Prism 6.
Surface Plasmon Resonance (SPR) Measurements
All SPR experiments were carried out using a Biacore T200 system using a Series S Sensorchip SA (GE Healthcare). All sensorgrams were recorded at 10 °C using a flow rate of 30 μl/min. FLAG tag affinity purified SF-JMJD1A and SF-JMJD1A (H1120Y) were captured (∼3000 response units) on parallel channels of the chip. The dissociation constant (KD) value of the peptide-JMJD1A interaction was estimated by 5 injections for 1 min of K9-modified H3 peptides at 0.64, 1.6, 4, 10, and 25 μm in buffer containing 50 mm HEPES-KOH, 50 mm NaCl, 100 μm NiCl2, and 1 mm 2-oxoglutarate, pH 7.8. All data were corrected for nonspecific binding and buffer shifts by double subtracting binding responses collected from a blank reference cell and a buffer injection over an SF-JMJD1A or SF-JMJD1A (H1120Y) captured flow cell.
Purification of JMJD1A Dimer and in Vitro Demethylase Assay
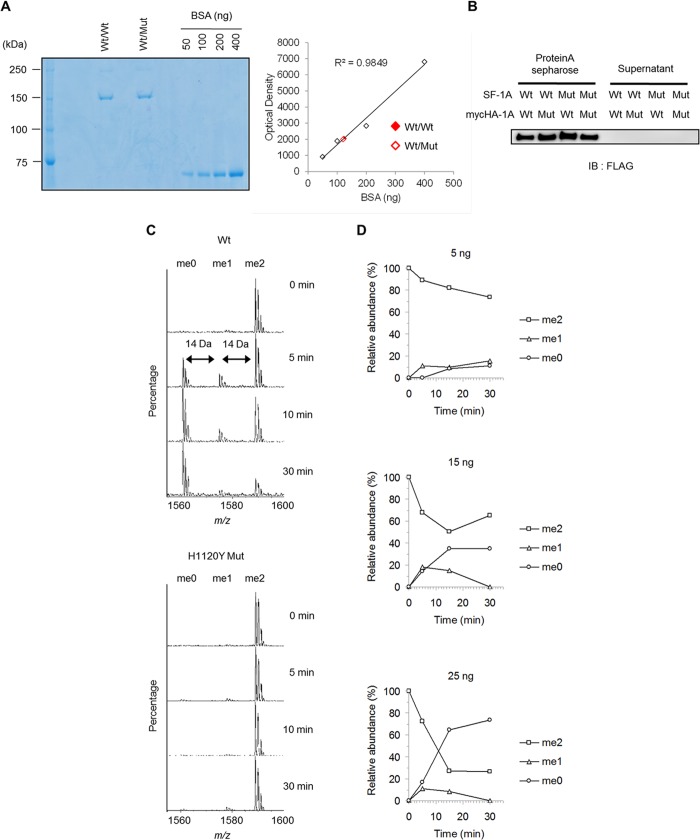
For purification of JMJD1A homodimer and heterodimer by combination of wild-type and H1120Y mutant, SF-tagged and Myc-HA-tagged plasmids were cotransfected into 293T cells. Three days after transfection, cells were washed with phosphate-buffered saline (PBS) and lysed with lysis buffer (50 mm HEPES-KOH, pH 7.9, 3 mm MgCl2, 100 mm NaCl, 1 mm EGTA, and 0.5% Nonidet P-40) containing phosphatase inhibitor mixture (Sigma) and protease inhibitor mixture (Roche Applied Science). The cell lysates were incubated with M2 α-FLAG-agarose (Sigma) for 2 h at 4 °C. The beads were washed with lysis buffer three times and eluted with 200 μg/ml of FLAG peptide. The eluted fraction was then subjected to immunoprecipitation using anti-Myc antibody. The immune complexes were pulled down with protein A-Sepharose and washed three times with 50 mm HEPES-KOH, pH 7.9, 100 mm NaCl. To measure the amount of immobilized dimer, aliquots were eluted with SDS sample buffer and analyzed by SDS-PAGE with bovine serum albumin (BSA) as a standard (Fig. 4A). We quantified the amount of JMJD1A proteins with a standard curve by plotting the optical density obtained by ImageJ analysis for each BSA standard versus its amount in nanograms (Fig. 4A). In vitro demethylase assays were performed using varying amounts of the immunoprecipitated dimer in a reaction buffer containing 50 mm HEPES-KOH, pH 8.0, 50 μm Fe(NH4)2(SO4)2, 1 mm 2-oxoglutarate, 2 mm ascorbate, and 1 μm K9-methylated H3 peptide (residues 1–15) in a final volume of 20 μl. After gentle rotation at 37 °C, we collected 5-μl supernatants at the desired time points and mixed them with 5 μl of 2.5% (v/v) trifluoroacetic acid (TFA) to stop the reaction. The solution was desalted through a C-tip300 (AMR Inc.), mixed with α-cyano-4-hydroxycinnamic acid, and then analyzed with a MALDI-TOF AXIMA performance (Shimadzu).
FIGURE 4.
Dose-dependent di-demethylation by JMJD1A homodimer. A, two-step purified SF-1A/Myc-HA-1A dimer was analyzed by SDS-PAGE with bovine serum albumin (BSA) as a standard (left). Standard curve by plotting the optical density obtained by ImageJ analysis for each BSA standard versus its amount in nanograms (right). B, stability of the immobilized JMJD1A homodimer. Immunoblotting analysis to rule out the generation of monomer units during the demethylase assay. C, mass spectrometry analysis of immobilized wild-type (upper) and H1120Y mutant (lower) JMJD1A demethylase activity using 1 μm dimethyl-K9 peptide (ARTKQTARK[me2]STGGKA) as a substrate. D, 1 μm H3K9me2 peptide was incubated with varying amounts of wild-type JMJD1A homodimer. Reaction time course of demethylase assay using H3K9me2 as a substrate.
RESULTS
JMJD1A Forms a Homodimer
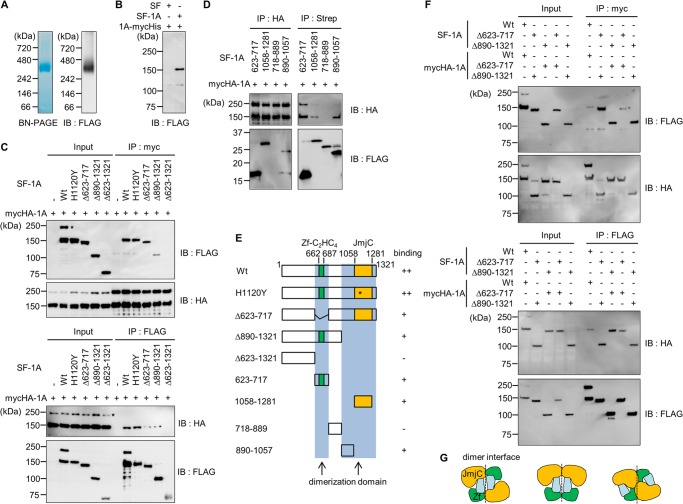
Based on fractionation of HeLa cell nuclear extracts by gel-filtration, JMJD1A likely forms a homodimer (7). To test whether JMJD1A forms a homodimer, we affinity purified Strep-FLAG-tagged (SF) JMJD1A using streptactin-Sepharose, and performed Blue Native-PAGE. This technique can be used to determine the native protein masses and oligomeric states and to identify physiological protein-protein interactions (24). Blue Native-PAGE analysis revealed a 300-kDa band (Fig. 1A, left), which we also observed by immunoblotting with anti-FLAG antibody (Fig. 1A, right). Given that the monomer size of JMJD1A is about 150 kDa, this result indicated homodimerization of JMJD1A. Furthermore, we co-expressed SF-tagged and Myc-His-tagged JMJD1A in 293T cells to validate the interaction of these two proteins. SF-JMJD1A was co-immunoprecipitated with anti-Myc antibody after affinity purification by streptactin-Sepharose (Fig. 1B). These data strongly suggested that JMJD1A forms a homodimer.
FIGURE 1.
JMJD1A forms a homodimer through its catalytic domains. A, the purified recombinant SF-tagged JMJD1A from 293T cells was applied to Blue Native-PAGE (left) followed by immunoblotting with an anti-FLAG antibody (right). B, 293T cells were co-transfected with SF- and Myc-His-tagged JMJD1A followed by streptactin tag affinity purification and anti-Myc immunoprecipitation. Immunoblotting (IB) was performed with anti-FLAG antibody. C, Myc-HA-JMJD1A was coexpressed with indicated SF-tagged proteins followed by immunoprecipitation and immunoblotting with the indicated antibodies. Asterisks indicate possible JMJD1A dimers. D, Myc-HA-JMJD1A was coexpressed with the indicated SF-tagged proteins followed by purification with anti-HA-agarose or streptactin-Sepharose, and immunoblotting with the indicated antibodies. E, schematic representation of the wild-type and mutant JMJD1A proteins and the position of the observed dimerization domain. F, SF-tagged proteins were coexpressed with Myc-HA-tagged proteins followed by immunoprecipitation (IP) and immunoblotting with the indicated antibodies. G, mode of dimerization in trans (left), cis (center), and complexed (right).
JMJD1A Forms a Homodimer through Its Catalytic Domain
To gain more insight into the dimer interface of JMJD1A, we prepared various deletion mutants as shown in Fig. 1E, and investigated dimer formation with immunoprecipitation followed by immunoblot analysis. To date, mutational studies indicate that a JmjC domain and a zinc finger (ZF) in JMJD1A are required for its enzymatic activity (7, 25). H1120 is also involved in demethylation, which is necessary for binding of Fe(II) (7). Deletion of the ZF or JmjC domain caused the reduction of dimerization efficiency (Fig. 1C). Conversely, when co-expressing each domain with full-length JMJD1A, both the ZF and JmjC domains interacted with full-length JMJD1A (Fig. 1D). Using mutant proteins, we finally narrowed down the interaction domain to amino acids 623–717 and 890–1321 (Fig. 1E). Furthermore, we examined whether the mode of interaction between ZF and JmjC occurs in trans, cis, or a complexed mode using deletion mutants (Fig. 1F). The results indicated that each domain in each monomer interacted with both the ZF and JmjC domains in another monomer (Fig. 1F). Thus, the binding mode of homodimerization is complexed (Fig. 1G, right), bringing the two active sites close together.
Two Active Sites of a JMJD1A Dimer Are Required for Effective Demethylation
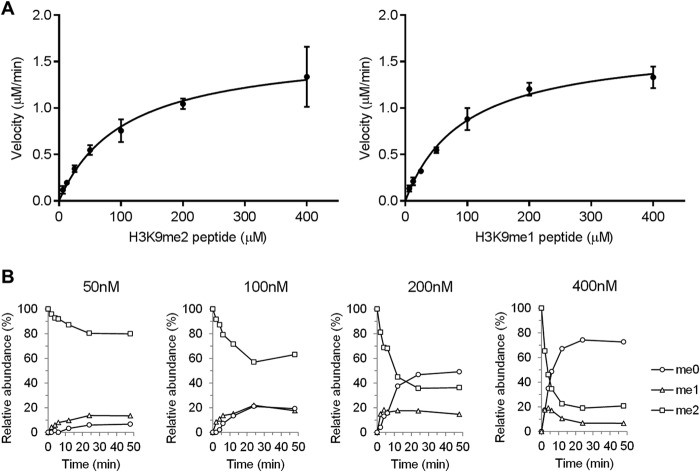
To examine the relationship between homodimerization and demethylase activity, we performed a demethylation assay using K9-methylated H3 peptide substrates and analyzed the demethylation products by formaldehyde dehydrogenase counterscreen and mass spectrometry. Using a fluorescent assay that follows the formation of the demethylation byproduct formaldehyde, demethylation of di- and monomethylated H3K9 peptides was analyzed (Fig. 2A). The catalytic efficiency kcat/Km of intermediate me1 is slightly higher than substrate me2, but almost the same (Table 1). Next we used mass spectrometry to measure the extent of demethylation of the dimethylated substrate as a function of time (Fig. 2B). Surprisingly, throughout the reaction the level of the intermediate substrate H3K9me1 remained low, at most 22%. These data are most consistent with a processive mechanism. However, monomethylated intermediate is also released above the enzyme concentration. Therefore, the processivity is not obligate. SPR analysis suggested that all peptides of H3K9me2, H3K9me1, and H3K9me0 showed similar dissociation constant (KD) against JMJD1A (Fig. 3, Table 2). The KD for each peptide from JMJD1A is less than the Km, indicating that the Km reflects more than just the true affinity of the enzyme for each peptide. Taken together, these results suggested partially processive whereas having a distributive mechanism for multiple demethylations.
FIGURE 2.
Kinetic analysis of JMJD1A-mediated demethylation of peptides. A, Michaelis-Menten plots of initial velocity as a function of the concentration of H3K9me2 (left) and H3K9me1 (right). Experiments were done in triplicate and are represented as mean ± S.D. B, mass spectrometry analysis of JMJD1A demethylase activity using 12.5 μm dimethyl-K9 peptide as a substrate and indicated amounts of JMJD1A.
TABLE 1.
Kinetic analysis of JMJD1A-mediated demethylation of peptides
Kinetic characterization of JMJD1A using H3K9me1 or H3K9me2 as a substrate. Experiments were done in triplicate and are represented as mean ± S.D.
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | μm min−1 | min−1μm−1 | |
| H3K9me1 | 95.2 ± 10.5 | 16.9 ± 0.71 | 0.178 ± 0.021 |
| H3K9me2 | 106.1 ± 11.5 | 16.5 ± 0.71 | 0.155 ± 0.018 |
FIGURE 3.
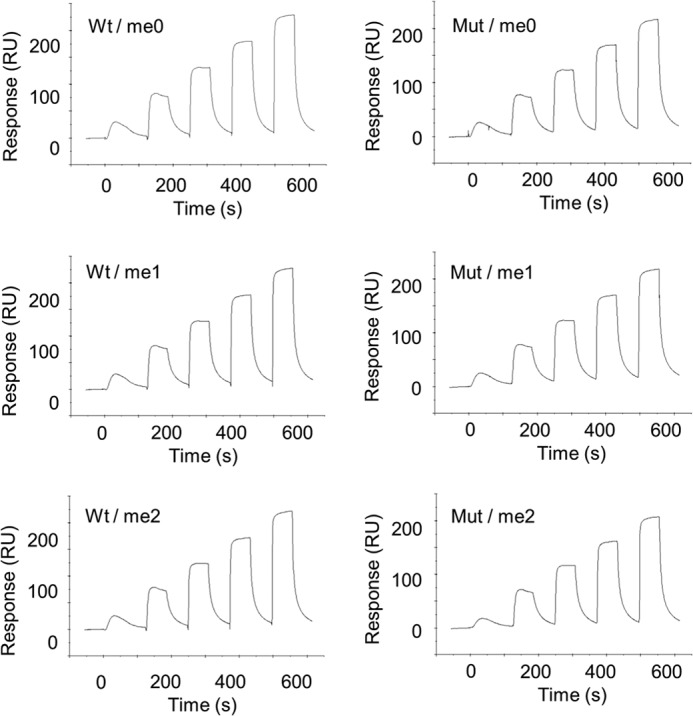
Single-cycle kinetics data reflecting the interaction of SA-captured JMJD1A with histone peptides. Six panels of wild-type (left) and H1120Y mutant (right) show experimental sensorgrams. Each run reflects five injections of analyte (H3K9me2, me1, me0) at 0.64, 1.6, 4, 10, and 25 μm. Analyte injections lasted for 60 s each and were separated by 60-s dissociation phases.
TABLE 2.
Effect of methylation on H3K9 peptides in binding to wild-type and H1120Y mutant JMJD1A
The KD values were derived using Biacore T200 by equilibrium state analyses. Values are mean ± S.D. for triplicates.
| Protein |
KD |
||
|---|---|---|---|
| H3K9me0 | H3K9me1 | H3K9me2 | |
| μm | |||
| WT | 3.32 ± 0.29 | 3.59 ± 0.02 | 3.56 ± 0.13 |
| Mut | 3.49 ± 0.05 | 3.55 ± 0.04 | 3.48 ± 0.13 |
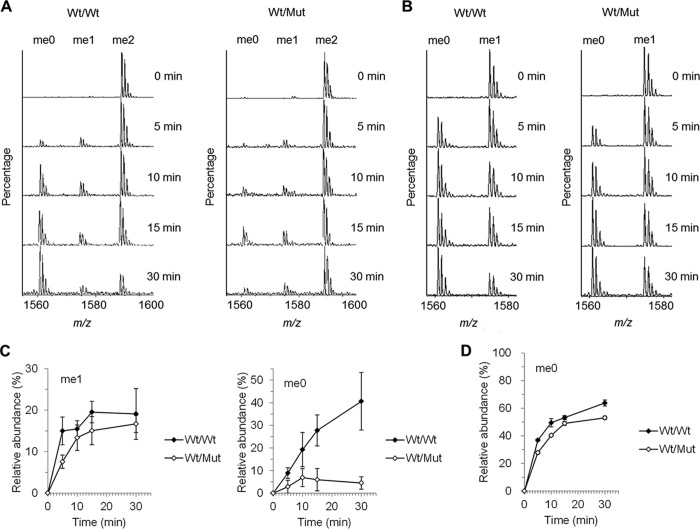
To address whether the proximal two active sites of the JMJD1A homodimer are important for its enzymatic activity, we performed a demethylase assay using a heterodimer between the wild-type and an enzymatic activity deficient mutant (H1120Y). We performed a two-step purification procedure with FLAG M2-agarose and immunoprecipitation by anti-Myc antibody to obtain immobilized FLAG-tagged/Myc-tagged WT/WT and WT/Mut dimers (Fig. 4A). To rule out the possibility that the low activity of the WT/Mut heterodimer was caused by monomerization, we performed immunoblot analysis after the demethylase assay. As shown in Fig. 4B, monomer units were not observed in the supernatant of the reaction mixture. This indicated that the homodimer of JMJD1A is stable. Time-dependent demethylation was successfully observed (Fig. 4C). Fig. 4D shows that the demethylation efficiency of the H3K9me2 peptide depends on the amount of immobilized JMJD1A homodimer, as in Fig. 2B. Furthermore, rapid accumulation of the non-methylated product relative to the formation of the monomethylated intermediate was observed, even in the presence of an excess of the dimethylated substrate, as in Fig. 2B. Time-dependent demethylation was observed using WT/WT and WT/Mut dimers (Fig. 5A). Two pseudo first-order rate constants for two demethylation steps were obtained through this analysis, k1, k2, and k1′, respectively (Table 3). Notably, the relative abundance of me0 was significantly decreased in WT/Mut compared with WT/WT (Fig. 5, A and C, Table 3). So we speculated that homodimerization of JMJD1A is important for efficient two-step demethylation. Based on our hypothesis, we performed a demethylase assay using the H3K9me1 peptide as a substrate to observe the one-step reaction. As expected, the WT/Mut heterodimer showed almost the same activity as the WT/WT homodimer (Fig. 5, B and D).
FIGURE 5.
Two active sites of the JMJD1A dimer are required for efficient demethylation. A, MALDI-TOF-MS analysis of the reaction products using 1 μm of a H3 peptide, which was dimethylated at K9, and 20 ng of JMJD1A homodimer (left) or heterodimer (right). The demethylation reaction was stopped at different time points after starting the reaction, and reaction products were analyzed by MALDI-TOF-MS. B, the same experiment as in A using monomethylated K9 peptide as a substrate. C, reaction time course of demethylated products using H3K9me2 as a substrate. D, reaction time course of demethylated products using H3K9me1 as a substrate. Values are mean ± S.E. for four independent experiments.
TABLE 3.
Demethylation activity of JMJD1A homodimer and heterodimer
Pseudo first-order rate constants for two demethylation steps of a JMJD1A homodimer and a heterodimer. k1 indicates the conversion of me2 into me1, k2 and k1′ indicate the conversion of me1 into me0, respectively. Relative abundance of demethylated products were after a 30-min reaction.
| Pseudo first-order rate constants |
Relative abundance after 30 min |
|||||
|---|---|---|---|---|---|---|
| me2a | me2a | me1a | me2a | me2a | me1a | |
| k1, min−1 | k2, min−1 | k1′, min−1 | me1, % | me0, % | me0, % | |
| WT/WT | 0.150 | 0.089 | 0.369 | 19.1 | 40.6 | 63.8 |
| WT/Mut | 0.076 | 0.029 | 0.278 | 16.7 | 4.5 | 53.1 |
a Substrate.
DISCUSSION
It has been shown that there exist several H3K9-specific demethylases (2, 7, 8), but the differences in their chromatin targeting mechanisms have remained unresolved. In this study, we have investigated the relationship between enzymatic activity and homodimerization of JMJD1A, which specifically demethylates H3K9me2/me1 (7). Using recombinant JMJD1A proteins, we have demonstrated that JMJD1A forms a homodimer through its catalytic domains. We speculated that homodimerization of JMJD1A is important for its enzymatic activity. Therefore, we measured the demethylase activity of the JMJD1A homodimer. Combined with SPR data, the results indicated that two-step demethylation occurs in a continuous fashion, i.e. partially processive while having a distributive.
Furthermore, if one active site in the JMJD1A dimer is mutated, the enzymatic activity of two-step demethylation is significantly decreased (Fig. 5C). Taken together, our findings suggest the following model (Fig. 6). First, the initial conversion of H3K9me2 into H3K9me1 occurs at one active site of the dimer. After the first demethylation step is finished, allosteric regulation of substrate channeling occurs, and monomethylated substrate binding occurs at the second site. Then, the conversion of H3K9me1 into H3K9me0 takes place at the second site.
FIGURE 6.
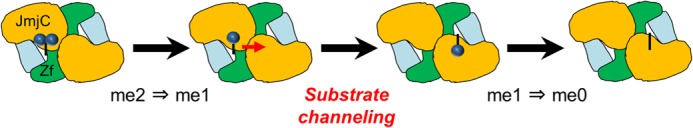
Model of two-step demethylation. Each site in the dimer has an equal chance for the initial binding. Once that happens, it triggers a conformational change. After the conversion of me2 into me1 at the first site, allosteric regulation of intermediate binding occurs at the second site. Then, me1 is converted to me0.
Such a rapid two-step demethylation is not observed in H3K9me3 demethylase, JMJD2A (26). The dimethylated peptide exhibited a 6-fold increase in Km and a 2-fold decrease in turnover number compared with the trimethylated peptide (26). These data clearly showed that JMJD2A has a distributive mechanism. The comparison of kinetic properties between JMJD1A and JMJD2A strongly suggest that JMJD1A has a distinct mechanism, i.e. partially processive. As the other examples, protein arginine methyltransferases, which catalyze methyl transfer from S-adenosyl-l-methionine to the guanidium side chain of arginine residues, are partially processive enzymes without release of intermediate between the methylation steps, but not in an obligate fashion as the intermediate is also released (27). Because protein arginine methyltransferases form a ring-like dimer, it is conceivable that binding of the substrate within this ring-like structure could facilitate the processive dimethylation of arginine residues by allowing the product of the first methylation reaction to enter the active site of the second molecule of the dimer without releasing the substrate from the ring (28). Partially processive demethylation by two active sites of the JMJD1A homodimer would provide a rationale for the homodimeric structure.
Substrate channeling not only enhances the reaction rates, but also prevents substrate competition and protects unstable intermediates (29, 30). This mechanism has the following biological significance. Unlike other JmjC domain-containing histone demethylases, JMJD1A has no reader domains such as PHD or Tudor that contribute to specific combinatorial recognition of multiple histone marks (1, 31). Therefore, if a JMJD1A dimer releases an intermediate-monomethylated substrate at a specific nucleosomal site, it would be difficult to rebind efficiently to the same enzyme in view of the vast array of possible alternative substrates in nearby chromatin. We think that homodimerization of JMJD1A enables the appropriate alteration of the chromatin structure by cooperatively demethylating two methyl groups of H3K9 to the null state. Furthermore, non-dissociation of the JMJD1A dimer from intermediate H3K9me1 could have in vivo functional significance. If JMJD1A releases a monomethylated intermediate, and methylated lysine-binding proteins such as G9a (4) or GLP ankyrin repeats (6) were to bind the residue prior to formation of the final product, inappropriate alteration of the chromatin structure might occur by recruiting different proteins that discriminate among varying degrees of lysine methylation, forming inappropriate complexes. This would impair the coding inherent in the H3K9 methylation status.
Although JMJD1A, PHF8, and KIAA1718 catalyze demethylation of the same substrate, their reaction mechanisms are quite different. Unlike JMJD1A, the other known dimethylated-H3K9 demethylases, PHF8 and KIAA1718, have not been reported to form a homodimer, nor do they catalyze processive two-step demethylation (8). Efficient demethylation of H3K9me2/1 by PHF8 requires the association of H3K4me3 with its PHD domain, facilitating H3K9me2/1 demethylation via a cis binding mode in which both methyl marks are recognized within the same N-terminal tail of histone H3 (8). Thus we consider that each enzyme might have a distinct function that regulates the methylation status of H3K9 to flexibly respond to complex signals, adding complexity to the strict control of gene expression patterns.
Despite different substrate specificities, there exist multiple demethylases that catalyze the removal of two methyl groups (31), and some of these likely form homodimers (32–34). Additional comparative structural and biochemical studies on histone demethylases are needed to fully understand their relationships in the context of nucleosomal substrates.
Methylation of histone lysine is reversibly regulated by S-adenosylmethionine-dependent methyltransferases. It has been proposed that some S-adenosylmethionine-dependent histone methyltransferases are dimers and that dimerization is essential for its enzymatic activity (35–39). Based on a structural, biophysical, and enzymatic study, Wei et al. demonstrated that viral SET protein functions as a dimer and performs H3K27 methylation catalysis one active site at a time. They suggest a “walking” mechanism by which viral SET acts all by itself to globally methylate host H3K27 (38). If such a mechanism would exist in JMJD1A, neither the continuous two-step demethylation as shown in Fig. 2B nor a dramatic decrease of the relative abundance of me0 in WT/Mut as shown in Fig. 5C would be observed. Thus, a different mechanism appears to exist in dimeric viral SET for efficient dimethylation activity.
JMJD1A is a member of a diverse family of iron and 2-oxoglutarate-dependent oxygenases, including JmjC-domain containing histone demethylases (40). There exist not only monomeric but also dimeric forms of 2-oxoglutarate oxygenases (40). Factor inhibiting HIF, which hydroxylates an asparagine residue in HIF-α, is dimeric (41–43). Disruption of dimerization by a single point mutation causes loss of catalytic activity (44). γ-Butyrobetaine hydroxylase also forms a homodimer, where elements from each monomer are involved in substrate binding (45). However, with respect to the homodimers described above, substrate channeling mediated by two proximal active sites has not been identified. Thus, the mechanism proposed seems to be unique for histone demethylases among oxygenase family proteins.
Future structural analysis should be directed toward further understanding of how the two active sites of the JMJD1A homodimer allosterically convert a dimethylated substrate into an unmethylated state. Because JMJD1A preferentially forms a stable homodimer, it is difficult to compare kinetic profiles between the dimer and monomer. To further support the channeling model, the detection of a monomethylated intermediate that binds to the inactive mutant in the WT/Mut heterodimer is important, for example, using cross-linking and tandem mass spectrometry (46). X-ray crystallography and high-resolution NMR spectroscopy (47) will provide data crucial in understanding the channeling mechanism in detail. At the same time, a systematic proteomics approach also may be useful to gain further insights into the possible mechanisms of substrate recognition and region-specific demethylation (48). It may be able to identify the structural basis for the discrepancy observed between the molar ratio of H3K9me2/JMJD1A and the low levels of H3K9me1 intermediate observed (Figs. 2B and 4D).
To our knowledge, this is the first report for mammalian histone demethylases in which dimerization has been enzymatically characterized. Our study provides useful information for designing anti-cancer agents that specifically inhibit JMJD1A among several post-translational histone modifiers. In addition, the WT/Mut heterodimer will be a useful tool for studying a snapshot of the monomethylated state in live cells. Both knockdown of JMJD1A and Ni(II) treatment (49) resulted in JMJD1A activity inhibition, but both of these experiments enabled the inhibition of two monomer units simultaneously. Therefore, we cannot observe the impact of one-step demethylation from H3K9me2 into H3K9me1 by JMJD1A on cellular events.
In summary, our study demonstrates that JMJD1A forms a homodimer through its catalytic domains, and the two active sites of the JMJD1A dimer are essential for effective conversion of dimethylated H3K9 into the null methylation state. Our enzymatic study illuminates how JMJD1A functions as a dimer and uniquely catalyzes two-step demethylation among several H3K9 specific demethylases. Thus, our work identifies the novel functional significance of the JMJD1A homodimer.
Acknowledgments
We thank Drs. Tatsuhiko Kodama, Juro Sakai, Takeshi Inagaki, Motoaki Seki, and Shigeru Morikawa for helpful comments, and Saori Kawanabe and Aya Nakayama for technical assistance.
This work was supported by a grant for the New Energy and Industrial Technology Development Organization (NEDO) (to H .A. and T. N.), Japan.
- JmjC
- Jumonji C
- JMJD1A
- Jumonji domain containing 1A
- Mut
- mutant
- ZF
- zinc finger
- SF
- Strep-FLAG
- SPR
- surface plasmon resonance
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Arrowsmith C. H., Bountra C., Fish P. V., Lee K., Schapira M. (2012) Epigenetic protein families. A new frontier for drug discovery. Nat. Rev. Drug. Discov. 11, 384–400 [DOI] [PubMed] [Google Scholar]
- 2. Krishnan S., Horowitz S., Trievel R. C. (2011) Structure and function of histone H3 lysine 9 methyltransferases and demethylases. ChemBioChem 12, 254–263 [DOI] [PubMed] [Google Scholar]
- 3. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 4. Collins R. E., Northrop J. P., Horton J. R., Lee D. Y., Zhang X., Stallcup M. R., Cheng X. (2008) The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat. Struct. Mol. Biol. 15, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nair S. S., Nair B. C., Cortez V., Chakravarty D., Metzger E., Schüle R., Brann D. W., Tekmal R. R., Vadlamudi R. K. (2010) PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-α target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 11, 438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rothbart S. B., Krajewski K., Nady N., Tempel W., Xue S., Badeaux A. I., Barsyte-Lovejoy D., Martinez J. Y., Bedford M. T., Fuchs S. M., Arrowsmith C. H., Strahl B. D. (2012) Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 19, 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 [DOI] [PubMed] [Google Scholar]
- 8. Horton J. R., Upadhyay A. K., Qi H. H., Zhang X., Shi Y., Cheng X. (2010) Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 17, 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Okada Y., Scott G., Ray M. K., Mishina Y., Zhang Y. (2007) Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450, 119–123 [DOI] [PubMed] [Google Scholar]
- 10. Liu Z., Zhou S., Liao L., Chen X., Meistrich M., Xu J. (2010) Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J. Biol. Chem. 285, 2758–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tateishi K., Okada Y., Kallin E. M., Zhang Y. (2009) Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Inagaki T., Tachibana M., Magoori K., Kudo H., Tanaka T., Okamura M., Naito M., Kodama T., Shinkai Y., Sakai J. (2009) Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells 14, 991–1001 [DOI] [PubMed] [Google Scholar]
- 13. Beyer S., Kristensen M. M., Jensen K. S., Johansen J. V., Staller P. (2008) The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283, 36542–36552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wellmann S., Bettkober M., Zelmer A., Seeger K., Faigle M., Eltzschig H. K., Bührer C. (2008) Hypoxia up-regulates the histone demethylase JMJD1A via HIF-1. Biochem. Biophys. Res. Commun. 372, 892–897 [DOI] [PubMed] [Google Scholar]
- 15. Pollard P. J., Loenarz C., Mole D. R., McDonough M. A., Gleadle J. M., Schofield C. J., Ratcliffe P. J. (2008) Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1α. Biochem. J. 416, 387–394 [DOI] [PubMed] [Google Scholar]
- 16. Sar A., Ponjevic D., Nguyen M., Box A. H., Demetrick D. J. (2009) Identification and characterization of demethylase JMJD1A as a gene up-regulated in the human cellular response to hypoxia. Cell Tissue Res. 337, 223–234 [DOI] [PubMed] [Google Scholar]
- 17. Mimura I., Nangaku M., Kanki Y., Tsutsumi S., Inoue T., Kohro T., Yamamoto S., Fujita T., Shimamura T., Suehiro J., Taguchi A., Kobayashi M., Tanimura K., Inagaki T., Tanaka T., Hamakubo T., Sakai J., Aburatani H., Kodama T., Wada Y. (2012) Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol. Cell Biol. 32, 3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krieg A. J., Rankin E. B., Chan D., Razorenova O., Fernandez S., Giaccia A. J. (2010) Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1α enhances hypoxic gene expression and tumor growth. Mol. Cell Biol. 30, 344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uemura M., Yamamoto H., Takemasa I., Mimori K., Hemmi H., Mizushima T., Ikeda M., Sekimoto M., Matsuura N., Doki Y., Mori M. (2010) Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer. In vivo identification from hypoxic tumor cells. Clin. Cancer Res. 16, 4636–4646 [DOI] [PubMed] [Google Scholar]
- 20. Cho H. S., Toyokawa G., Daigo Y., Hayami S., Masuda K., Ikawa N., Yamane Y., Maejima K., Tsunoda T., Field H. I., Kelly J. D., Neal D. E., Ponder B. A., Maehara Y., Nakamura Y., Hamamoto R. (2012) The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int. J. Cancer 131, E179–189 [DOI] [PubMed] [Google Scholar]
- 21. Yamada D., Kobayashi S., Yamamoto H., Tomimaru Y., Noda T., Uemura M., Wada H., Marubashi S., Eguchi H., Tanemura M., Doki Y., Mori M., Nagano H. (2012) Role of the hypoxia-related gene, JMJD1A, in hepatocellular carcinoma. Clinical impact on recurrence after hepatic resection. Ann. Surg. Oncol. 19, S355–364 [DOI] [PubMed] [Google Scholar]
- 22. Osawa T., Tsuchida R., Muramatsu M., Shimamura T., Wang F., Suehiro J., Kanki Y., Wada Y., Yuasa Y., Aburatani H., Miyano S., Minami T., Kodama T., Shibuya M. (2013) Inhibition of histone demethylase JMJD1A improves anti-angiogenic therapy and reduces tumor associated macrophages. Cancer Res. 73, 3019–3028 [DOI] [PubMed] [Google Scholar]
- 23. Gloeckner C. J., Boldt K., Schumacher A., Roepman R., Ueffing M. (2007) A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 7, 4228–4234 [DOI] [PubMed] [Google Scholar]
- 24. Wittig I., Braun H. P., Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 25. Brauchle M., Yao Z., Arora R., Thigale S., Clay I., Inverardi B., Fletcher J., Taslimi P., Acker M. G., Gerrits B., Voshol J., Bauer A., Schübeler D., Bouwmeester T., Ruffner H. (2013) Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS One 8, e60549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shiau C., Trnka M. J., Bozicevic A., Ortiz Torres I., Al-Sady B., Burlingame A. L., Narlikar G. J., Fujimori D. G. (2013) Reconstitution of nucleosome demethylation and catalytic properties of a Jumonji histone demethylase. Chem. Biol. 20, 494–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith B. C., Denu J. M. (2009) Chemical mechanisms of histone lysine and arginine modifications. Biochim. Biophys. Acta 1789, 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osborne T. C., Obianyo O., Zhang X., Cheng X., Thompson P. R. (2007) Protein arginine methyltransferase 1. Positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry 46, 13370–13381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miles E. W., Rhee S., Davies D. R. (1999) The molecular basis of substrate channeling. J. Biol. Chem. 274, 12193–12196 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y. H. (2011) Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol. Adv. 29, 715–725 [DOI] [PubMed] [Google Scholar]
- 31. Pedersen M. T., Helin K. (2010) Histone demethylases in development and disease. Trends Cell Biol. 20, 662–671 [DOI] [PubMed] [Google Scholar]
- 32. Chen Z., Zang J., Whetstine J., Hong X., Davrazou F., Kutateladze T. G., Simpson M., Mao Q., Pan C. H., Dai S., Hagman J., Hansen K., Shi Y., Zhang G. (2006) Structural insights into histone demethylation by JMJD2 family members. Cell 125, 691–702 [DOI] [PubMed] [Google Scholar]
- 33. Shin S., Janknecht R. (2007) Diversity within the JMJD2 histone demethylase family. Biochem. Biophys. Res. Commun. 353, 973–977 [DOI] [PubMed] [Google Scholar]
- 34. Lee J., Thompson J. R., Botuyan M. V., Mer G. (2008) Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 15, 109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Manzur K. L., Farooq A., Zeng L., Plotnikova O., Koch A. W., Sachchidanand, Zhou M. M. (2003) A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3. Nat. Struct. Biol. 10, 187–196 [DOI] [PubMed] [Google Scholar]
- 36. Zhang X., Cheng X. (2003) Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eskeland R., Czermin B., Boeke J., Bonaldi T., Regula J. T., Imhof A. (2004) The N-terminus of Drosophila SU(VAR)3–9 mediates dimerization and regulates its methyltransferase activity. Biochemistry 43, 3740–3749 [DOI] [PubMed] [Google Scholar]
- 38. Wei H., Zhou M. M. (2010) Dimerization of a viral SET protein endows its function. Proc. Natl. Acad. Sci. U.S.A. 107, 18433–18438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng Y., Frazier M., Lu F., Cao X., Redinbo M. R. (2011) Crystal structure of the plant epigenetic protein arginine methyltransferase 10. J. Mol. Biol. 414, 106–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McDonough M. A., Loenarz C., Chowdhury R., Clifton I. J., Schofield C. J. (2010) Structural studies on human 2-oxoglutarate dependent oxygenases. Curr. Opin. Struct. Biol. 20, 659–672 [DOI] [PubMed] [Google Scholar]
- 41. Dann C. E., 3rd, Bruick R. K., Deisenhofer J. (2002) Structure of factor-inhibiting hypoxia-inducible factor 1. An asparaginyl hydroxylase involved in the hypoxic response pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 15351–15356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elkins J. M., Hewitson K. S., McNeill L. A., Seibel J. F., Schlemminger I., Pugh C. W., Ratcliffe P. J., Schofield C. J. (2003) Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α. J. Biol. Chem. 278, 1802–1806 [DOI] [PubMed] [Google Scholar]
- 43. Lee C., Kim S. J., Jeong D. G., Lee S. M., Ryu S. E. (2003) Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau. J. Biol. Chem. 278, 7558–7563 [DOI] [PubMed] [Google Scholar]
- 44. Lancaster D. E., McNeill L. A., McDonough M. A., Aplin R. T., Hewitson K. S., Pugh C. W., Ratcliffe P. J., Schofield C. J. (2004) Disruption of dimerization and substrate phosphorylation inhibit factor inhibiting hypoxia-inducible factor (FIH) activity. Biochem. J. 383, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leung I. K., Krojer T. J., Kochan G. T., Henry L., von Delft F., Claridge T. D., Oppermann U., McDonough M. A., Schofield C. J. (2010) Structural and mechanistic studies on γ-butyrobetaine hydroxylase. Chem. Biol. 17, 1316–1324 [DOI] [PubMed] [Google Scholar]
- 46. Watson A. A., Mahajan P., Mertens H. D., Deery M. J., Zhang W., Pham P., Du X., Bartke T., Zhang W., Edlich C., Berridge G., Chen Y., Burgess-Brown N. A., Kouzarides T., Wiechens N., Owen-Hughes T., Svergun D. I., Gileadi O., Laue E. D. (2012) The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4. J. Mol. Biol. 422, 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Selenko P., Frueh D. P., Elsaesser S. J., Haas W., Gygi S. P., Wagner G. (2008) In situ observation of protein phosphorylation by high-resolution NMR spectroscopy. Nat. Struct. Mol. Biol. 15, 321–329 [DOI] [PubMed] [Google Scholar]
- 48. Bartke T., Borgel J., Dimaggio P. A. (2013) Proteomics in epigenetics. New perspectives for cancer research. Brief. Funct. Genomics 12, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen H., Giri N. C., Zhang R., Yamane K., Zhang Y., Maroney M., Costa M. (2010) Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers. J. Biol. Chem. 285, 7374–7383 [DOI] [PMC free article] [PubMed] [Google Scholar]