Background: The conserved EF-hand (EF) domain is necessary for active phospholipase.
Results: EF binds to anionic phospholipid-containing vesicles; EF mutations introduced into PLC δ1 reduce activity not recoverable with added PIP2.
Conclusion: EF-hand domain aids substrate binding in the active site when the protein is membrane-anchored.
Significance: This may be the function of the EF-hand domain in other PLC enzymes as well.
Keywords: Enzymes, Glycerophospholipid, Membrane Enzymes, NMR, Phospholipase C, Phospholipid Vesicle, Site-directed Mutagenesis
Abstract
Recombinant EF-hand domain of phospholipase C δ1 has a moderate affinity for anionic phospholipids in the absence of Ca2+ that is driven by interactions of cationic and hydrophobic residues in the first EF-hand sequence. This region of PLC δ1 is missing in the crystal structure. The relative orientation of recombinant EF with respect to the bilayer, established with NMR methods, shows that the N-terminal helix of EF-1 is close to the membrane interface. Specific mutations of EF-1 residues in full-length PLC δ1 reduce enzyme activity but not because of disturbing partitioning of the protein onto vesicles. The reduction in enzymatic activity coupled with vesicle binding studies are consistent with a role for this domain in aiding substrate binding in the active site once the protein is transiently anchored at its target membrane.
Introduction
Eukaryotic phosphoinositide-specific phospholipase C (PLC)2 enzymes play an important role in cellular signaling by catalyzing the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2). The hydrolysis generates two second messengers, inositol 1,4,5-trisphosphate (IP3) and diacylglycerol. IP3 mobilizes intracellular calcium from the endoplasmic reticulum (1), and diacylglycerol activates protein kinase C (2, 3). These signal transduction cascades are essential for many cellular events, such as cell growth, proliferation, fertilization, and secretion. Approximately 13 distinct isoforms of PLC have been described in mammals, and they are grouped into six subfamilies based on sequence homology as follows: PLCβ(1–4), PLCγ(1,2), PLCδ(1,3,4), PLCϵ(1), PLCζ(1), and PLCη(1,2). These subfamilies differ in structure and mode of regulation. All members of the PLC family exhibit a multidomain structure that comprises the catalytic X and Y domains, the protein kinase C conserved region 2 (C2) domain, EF-hand domain, and pleckstrin homology (PH) domain (except for the sperm-specific PLCζ, which does not have a PH domain). In addition to the core conserved regions, some isoforms also contain subfamily-specific domains that contribute to their specific regulatory mechanisms. For example, the Src homology domains in PLCγ are important for its regulation by tyrosine kinase-coupled receptors (4, 5).
Phospholipase C δ1 (PLC δ1), widely expressed in various cell types (6), is one of the smallest PLC isoforms that contain all the core conserved domains (Fig. 1). The crystal structures of the isolated PH domain and the PH domain deletion variant of PLC δ1 have been solved separately (7, 8). The PH domain of PLC δ1 specifically binds to PIP2 and its headgroup IP3 (9), and the affinity for the former allows the PH domain to tether the PLC enzyme to the plasma membrane, where the catalysis is carried out in a processive manner (10). The C2 domain of PLC δ1 has been shown to form a ternary complex with calcium and phosphatidylserine (PS), which activates the enzyme (10). The EF-hand domain of PLC δ1 consists of four consecutive EF-hand motifs, pairwise distributed in two lobes, exhibiting a characteristic helix-loop-helix topology (7). The EF-hand motifs, found in all PLC isoforms, are necessary to express full PLC activity, as evidenced by the abolition of PLC activity resulting from the deletion of EF-hand residues (11, 12). Based on the structural similarities between the PLC δ1 EF-hand and other calcium-binding EF-hand proteins such as calmodulin, it has been suggested that the EF-hand domain of PLC δ1 might serve a regulatory role through calcium binding. Elsewhere, interactions of the EF-hand domain with free fatty acids have also been suggested (13, 14). Additionally, numerous EF-hand motif-containing proteins have been reported to be involved in lipid binding. For example, diacylglycerol kinase-α, a multidomain enzyme, contains the EF-hand motifs and is regulated by lipids and calcium (15, 16).
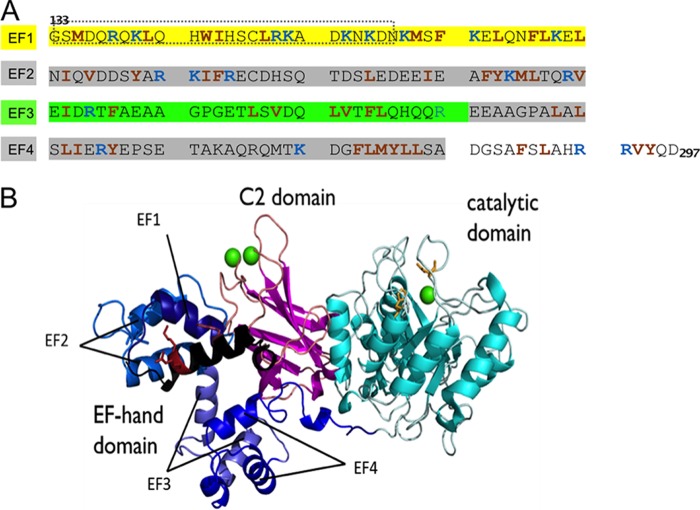
FIGURE 1.
A, amino acid sequence (133–297) of EF-hand domain of human PLC δ1; red and blue residues are cationic and hydrophobic, respectively. The region of EF-1 in the dotted lines is not visible in the crystal structure. B, crystal structure of rat PLCδ1 (Δ1–132) complexed with calcium (Protein Data Bank code 1DJI). The initial part of the EF-hand domain, missing in the crystal structure, is modeled as black helices; the rest of the EF-hand domain is shown in different shades of blue; the catalytic domain is shown in cyan, with active site residues His-311 and His-356 side chains shown as orange sticks; the C2 domain is magenta; calcium ions are shown as green spheres. The red sticks in EF-1 represent the side chains of Trp-144, Arg-150, and Lys-151.
In this report, we characterize the interactions of the isolated EF-hand domain of PLC δ1 with model membranes. Mutagenesis of specific cationic and hydrophobic residues in the separate EF-hand domain has modest effects on vesicle binding but significant effects on the activity of full-length PLC δ1. The results suggest that the EF-hand domain, particularly the first EF-hand unit, aids the interfacial binding step where substrate occupies the active site. Conserved residues in other PLC EF-hand domains suggest this may be the primary function of this structural unit in all mammalian PLC enzymes.
EXPERIMENTAL PROCEDURES
Chemicals
1,2-Dimyristoyl-sn-glycerol-3-phosphocholine,1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine (d54-DMPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphate (DOPA), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), l-α-phosphatidylinositol (bovine liver) (PI), l-α-phosphatidylinositol-4,5-bisphosphate (porcine brain) (PIP2), porcine brain sphingomyelin (SM), and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissaminerhodamine B sulfonyl) (rhodamine-PE, or Rho-PE) were purchased from Avanti Polar Lipids, Inc. Cholesterol (CH), thrombin, Triton X-100, and d-glucose 6-phosphate were purchased from Sigma. Chloramphenicol, ampicillin, and isopropyl 1-thio-β-d-galactopyranoside were purchased from American Bioanalytical Inc. Succinimidyl 6-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-hexanoate was obtained from Invitrogen. The spin-labeling reagent (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl), methanethiosulfonate (MTSL), was purchased from Toronto Research Chemicals Inc. All other chemicals were reagent grade.
Construction of EF-hand Domain Mutations
The plasmid containing the full-length rat PLC δ1 gene, a gift from Dr. Matilda Katan (Cancer Research UK Centre for Cell and Molecular Biology, Chester Beatty Laboratories, Institute of Cancer Research, London, UK) (17), and the plasmid containing the human PLC δ1 EF-hand domain gene (13) were inserted into the pGEX-2T vector with an N-terminal glutathione S-transferase (GST) tag for ease in purification. All mutations of the PLC genes were carried out by QuikChange methodology, using the site-directed mutagenesis kit from Stratagene and complementary primers from Operon. The mutated gene sequences were confirmed by Genewiz.
Overexpression and Purification of Proteins
The plasmids (either containing the gene for PLCδ1 or the isolated EF-hand domain) were transformed into Escherichia coli BL21-Codonplus (DE3)-RIL cells. Overexpression of the proteins followed protocols used for a bacterial PI-PLC (18). After addition of isopropyl 1-thio-β-d-galactopyranoside (0.8 mm), the cell suspension was incubated for 20 h at 16 °C. Cells harvested by centrifugation were either stored at −20 °C for later use or lysed immediately after resuspension in PBS buffer, pH 7.4, by sonication on ice. The PLC proteins were purified by affinity chromatography using glutathione-Sepharose 4B resin (GE Healthcare). After application of the crude E. coli lysate to the column and washing with PBS buffer to remove nonspecifically bound impurities, thrombin was added to the resin, and the mixture was gently shaken at 4 °C for ∼18 h to cleave the GST tag. The PLC protein was eluted from the resin with PBS buffer. The concentration of the proteins was determined by the absorption at 280 nm using an extinction coefficient calculated by the ProtParam software or by DC protein assay (Bio-Rad).
rEF (19.1 kDa) and mutated rEF proteins were well folded and exhibited similar CD profiles (data not shown). CD spectra were acquired on an AVIV 420 spectrometer (Biomedical Inc.) as described previously (13).
Spin Labeling the EF-hand Domain
Purified rEF or mutants with a single cysteine (C148S and C188S) were stored at 8–10 mg/ml in PBS buffer with 2 mm DTT, pH 7.4. Prior to reaction with MTSL, the solutions were passed through a Micro Bio-Spin 6 column (Bio-Rad) equilibrated with PBS to remove the DTT. The protein was mixed with ∼5-fold excess of MTSL compared with the total number of cysteines. Excess MTSL was removed by three consecutive Micro Bio-Spin 6 columns equilibrated with 10 mm borate, pH 7.8 (in D2O for 1H NMR experiments). CD analysis of the spin-labeled proteins showed that they were the same as unlabeled rRF in terms of secondary structure and thermal stability.
Preparation of Vesicles
Dry films prepared from organic stocks of the desired component lipids were lyophilized for at least 3 h, rehydrated with buffer, and then dispersed by vortexing. LUVs were prepared by at least 20 passages of the lipid solution through polycarbonate membranes (0.1 μm pore diameter) using a Mini-extruder (Avanti Polar Lipids, Inc.). SUVs were prepared by intermittent probe sonication (20–30 s per interval) of the lipid solution in an ice bath using a Branson Sonifier Cell Disrupter until the vesicle solution became translucent.
Centrifugation-Filtration Vesicle Binding Assay for rEF
SUV samples ranging from 0.025 to 6 mm total lipid were mixed with a fixed amount of rEF (14–16 μm) in 20 mm Tris, 0.1 mm EGTA, pH 7.5, in the absence or presence of 0.15 m NaCl. The same amount of rEF in the absence of vesicles served as a control. After 15 min of incubation at room temperature, the protein/SUV mixtures were centrifuged through an Amicon Ultra 0.5-ml centrifugal filter with a molecular mass cutoff of 100 kDa (Millipore). Free protein eluted through the membrane was quantified by SDS-PAGE or by DC protein assay (Bio-Rad). The gels were imaged by EAGLE EYE from Stratagene, and the rEF intensities were quantified by ImageJ. The fraction of free protein (Ef/ET, where ET is the total amount of protein, obtained from the control incubated without vesicles) at each SUV concentration was calculated. The fraction of EF protein bound to vesicles, Eb/ET, was then calculated as (1 − Ef/ET), and the apparent dissociation constant (KD) was estimated as follows: Eb/ET = fmax[PL]/([PL] + KD), where fmax is the maximum fraction bound.
FRET Vesicle Binding Assay for PLC δ1
FRET was used to monitor binding of 6-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)hexanoate)-labeled PLC δ1 (labeled on the N-terminal amino group with succinimidyl 6-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)hexanoate) according to the manufacturer's suggested protocol. The protein had a labeling ratio of 90 ± 20%, determined by comparing the absorption of the protein at 280 nm with that of the probe at 465 nm. The PLC δ1 was kept at a fixed concentration (0.5 μm), and vesicles containing the phospholipids POPC/DOPMe or POPC/DOPG (1:1 and 7:3), with ∼0.5 mol % Rho-PE, were titrated into the protein solution at 25 °C. The excitation was set at 465 nm with a 2-mm slit width, and the fluorescence emission was measured at 535 nm with a slit width of 5 nm. The decrease in donor fluorescence (Fd − Fda)/Fd, where Fd and Fda are the fluorescence intensities of the donor PLCδ1 in the absence and presence of Rho-PE-labeled vesicles, was used to estimate an apparent KD.
PLC δ1 Enzymatic Activity
The specific activity of PLC δ1 toward PI was measured by monitoring myo-inositol 1,2-(cyclic)-phosphate (cIP) and inositol 1-phosphate (I-1-P) generation at fixed time points using 31P NMR spectroscopy (at 242.8 MHz on a Varian VNMRS 600 spectrometer without 1H decoupling). The substrate PI was embedded in physiological membrane mimicking LUVs (19), with a total lipid concentration of 10 or 0.5 mm. The amount of PLC δ1 added (1–2 μg) was adjusted so that less than 20% of the PI was hydrolyzed within 30 min. The reaction was carried out in 50 mm HEPES buffer with 0.5 mm CaCl2, 100 mm NaCl, and 0.1 mg/ml BSA, pH 7.5, unless otherwise specified. When the total lipid concentration was 5 or 10 mm, 300 or 400 μl of enzyme/vesicle mixture was incubated at 37 °C. The reaction was quenched by adding 10 vol % acetic acid, 1 mm EDTA, and a few drops of Triton X-100. Samples were then stored at −20 °C before NMR analysis. Prior to NMR measurement, samples were subjected to the following treatment: (i) additional Triton X-100 was added when necessary to clarify the solution, ensuring solubilization of all phospholipid components; (ii) the sample pH was adjusted to 6.1–6.5 by adding NaOH, so that the 31P peak for bulk phosphodiester (PE, PC, SM, PS, and PI that was not hydrolyzed) was well resolved from the I-1-P peak (whose chemical shift varies depending on pH); (iii) 200 μl of D2O was added to each sample. The cIP and I-1-P peaks were integrated and compared with the integral for the phosphodiester peak.
When the total lipid concentration was 0.5 mm, 5 ml of the enzyme/LUV mixture with 0.1 μmol of glucose 6-phosphate added as an internal standard was extracted with chloroform/methanol (2:1) to remove lipids (PE, PC, SM, CH, and PI that was not hydrolyzed). The aqueous solution, containing products cIP and I-1-P, as well as internal standard, was frozen and lyophilized overnight. The dried solids were dissolved in D2O. The cIP and I-1-P peaks were integrated and compared with the standard. In all the PLC δ1 assays, both the intermediate product cIP and the final product I-1-P were observed. The specific activity was calculated based on the sum of both products.
EF-hand Vesicle Binding Monitored by 1H NMR Spectroscopy
EF proteins (rEF or spin-labeled rEF, C148S, or C188S) were titrated into POPC or POPC/DOPA (1:1) SUVs in 10 mm borate, pH 7.8, in D2O, and 1H NMR spectra were acquired at 37 °C on a Varian VNMRS 600 spectrometer. Line widths of the acyl chain bulk methylenes (1.4 ppm) and terminal methyl groups (1.0 ppm) were measured at half-height. Errors in measuring the line widths for those two resonances were typically ±2 Hz in a given titration experiment. The line widths for the same resonances when titrated with unlabeled rEF were consistent with a measurement error of that magnitude. The exact concentrations of the proteins were determined after the NMR experiments using DC protein assay.
High Resolution Field Cycling 31P NMR
The apparatus for carrying out high resolution field cycling spin-lattice relaxation (R1 is the spin-lattice relaxation rate and is equal to 1/T1) studies has been described in detail (20). Briefly, the current system is capable of moving the sample, attached to a push-rod that is driven by a linear motor, to a position up to the top of the main magnet and back down to the probe in ∼350 ms. The magnetic fields at positions between the probe and above the superconducting magnet vary from 10 T to nearly zero. This device allows us to prime the spins in the probe for 2–4 s, then shuttle the sample to a defined lower field where the nuclear spins can relax for a variable time, and then shuttle it back into the probe for application of a pulse and read-out of the magnetization lost as a function of the delay time at the lower field. High field read-out is critical for maintaining chemical shift differences when there are multiple nuclei of interest, in this case 31P for DOPA and POPC in the same SUV.
For phospholipids, the dipolar contribution of nearby protons to relaxation of 31P is small at high fields but is easily measured below 2 T. Theory describing the field dependence of R1 provides a way to extract correlation times and limiting relaxation rates for different types of motions (21, 22). In the field cycling experiments reported here, electron spin labels, each containing an unpaired electron spin radical that can be a potent contributor to the R1 rate, are introduced onto the protein. Their effects on the 31P nuclei are much larger than nearby protons and can be measured over distances to 25 Å or more (depending on protein and phospholipid concentrations). The closer the spin label is to the 31P, the stronger the paramagnetic relaxation enhancement (PRE). The magnetic interaction between 31P and unpaired electron is modulated by the tumbling of the vesicles in solution, and this is sensed at low magnetic fields, less than 0.03 T for small unilamellar vesicles. R1 for the phospholipids in the vesicles at different low fields is measured in the absence and again in the presence of spin-labeled rEF. If the phospholipid and the protein stay associated for about 1–2 μs, then there will be a distance-dependent PRE. This can vary for different phospholipids in a mixture if the protein has discrete binding sites for the different lipids (23). The field dependence of the extra paramagnetic relaxation enhancement or PRE is fit to Equation 1,
where RP-e(0) is the low field limit of ΔR1; ωP is the angular frequency of 31P; τP-e is the correlation time of the dipolar relaxation of 31P by the nitroxide unpaired electron. The c term represents either a limiting chemical shift anisotropy contribution or a faster discrete dipolar interaction when at the lower fields (ωP2τP-e2 <1). For a single spin label attached to a protein, an estimate of the distance between the 31P and the spin label can be obtained (23, 24) as shown in Equation 2.
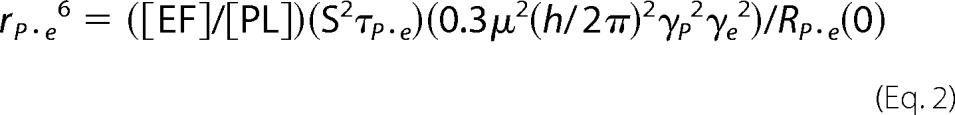 |
Because rEF has two cysteine residues and both are labeled, we used the approach to compare how effectively spin-labeled rEF relaxes PA and PC in the same vesicle. Field cycling profiles and evaluating the ΔR1 attributed to the presence of the spin label can be used to evaluate a distance for the phospholipids with the single cysteine mutants, either C148S or C188S, of the EF-hand domain.
RESULTS
Isolated EF-hand Domain of PLC δ1 Binds to Anionic Phospholipid-containing SUVs
The isolated EF-hand domain of PLC δ1 has previously been expressed and shown to interact with free fatty acids (13, 14). To further explore its binding partners, interaction of rEF with various phospholipid species was examined by a centrifugation-filtration vesicle binding assay. Initial binding experiments using intrinsic fluorescence changes or a FRET assay were compromised by light scattering at the higher phospholipid concentrations so that the bulk partitioning assay using centrifugation-filtration to separate free and bound rEF was deemed the best choice.
rEF has virtually no affinity for POPC SUVs (Fig. 2A). However, with POPA in the vesicles, the amount of the free rEF decreased proportional to the amount of SUVs present (Fig. 2B). The same trend was observed when SUVs containing other anionic phospholipids (PS or PG) were mixed with rEF. The variation in the amount of rEF partitioned onto SUVs was fit with a hyperbolic curve to generate an apparent dissociation constant, KD (two examples are given in Fig. 2C). This is not a true dissociation constant but an apparent KD value that provides a way of comparing affinities of rEF for various model membrane surfaces. As listed in Table 1, rEF bound to SUVs containing a 0.5-mol fraction anionic phospholipid (PA, PS, and PG) with moderate KD values (on the order of 0.1–0.2 mm) (Table 1). In the presence of 0.15 m NaCl, the KD values were decreased slightly compared with those in the absence of NaCl, indicating that the binding of rEF to SUVs containing anionic phospholipids is not solely electrostatic but that there is a hydrophobic component as well. The affinity of rEF for LUVs of POPA/POPC (1:1), although less accurate, was comparable with that for SUVs.
FIGURE 2.
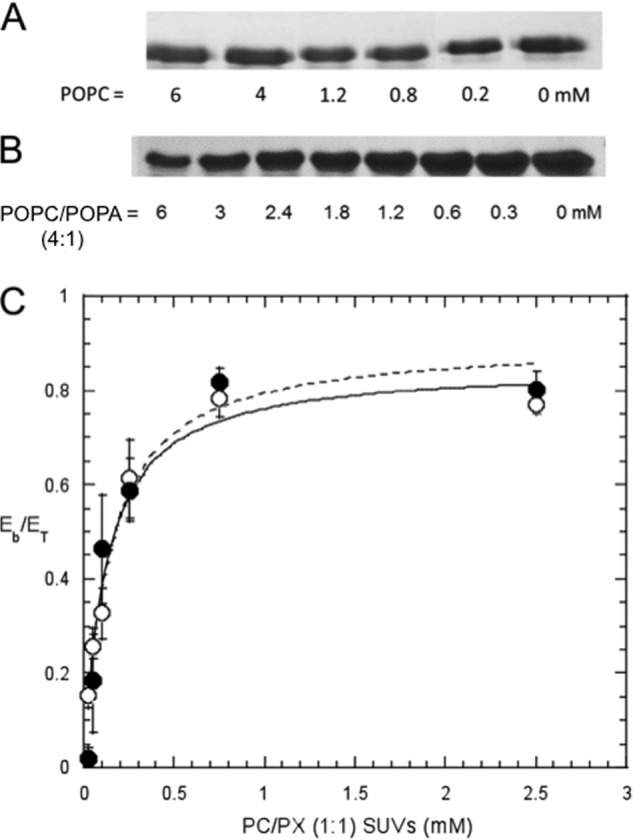
SDS-PAGE showing free EF as a function of added vesicles. A, POPC SUVs; B, POPC/POPA (4:1) SUVs. Bulk phospholipid concentrations are indicated below each lane. C, fraction of EF bound (Eb/ET) as a function of total lipid concentration is shown for PC/PS (●) or PC/PA (○) (1:1) SUVs. The lines are the least square fit with an apparent KD = 0.14 ± 0.04 mm for PC/PS and 0.12 ± 0.02 mm for PC/PA. The error bars indicate the standard deviation of at least four repeats.
TABLE 1.
Apparent dissociation constants for EF-hand domain binding to anionic phospholipid-containing SUVs in the absence or presence of 0.15 m NaCl and 0.5 mm CaCl2
The buffer was 20 mm Tris, 0.1 mm EGTA, pH 7.5, in the absence or presence of NaCl and CaCl2.
| SUVs | NaCl | CaCl2 | Apparent KDa |
|---|---|---|---|
| mm | |||
| POPC/DOPA (1:1) | − | − | 0.12 ± 0.02 |
| + | − | 0.087 ± 0.028 | |
| − | + | 0.63 ± 0.17 | |
| POPC/1,2-dioleoyl-sn-glycero-3-phospho-l-serine (1:1) | − | − | 0.14 ± 0.04 |
| + | − | 0.085 ± 0.046 | |
| − | + | 0.14 ± 0.05 | |
| POPC/DOPG (1:1) | − | − | 0.25 ± 0.04 |
| + | − | 0.11 ± 0.09 |
a This is the total concentration of phospholipids present for half of maximum binding. Errors were determined from the hyperbolic fit to the data.
It should be noted that this binding occurred in the absence of calcium and with 0.1 mm EGTA present. The presence of 0.5 mm Ca2+ did not dramatically affect binding of rEF to SUVs containing PS (Table 1). However, there was loss of binding to PC/PA SUVs with Ca2+ present. No calcium ion is found associated with the EF-hand domain that is visible in crystal structures of PLC δ1, even though the enzyme is soaked in calcium or its analogs (7, 25–27). The lack of requirement for calcium, taken together with the selective affinity for anionic phospholipids, suggests that basic residues of the EF-hand domain, rather than its acidic residues in coordination with calcium, are likely to contribute to the binding of the EF-hand domain to the membrane surfaces.
Binding of PLC δ1 to Vesicles
For comparison with the vesicle binding of the isolated EF-hand domain, binding of full-length PLC δ1 to vesicles was also examined but with a FRET assay. Two-component LUVs, POPC/DOPMe and POPC/DOPG, were examined (not PS because this could interact with the C2 domain if traces of Ca2+ were present). The vesicles purposely had no PIP2 as well as no Ca2+ to avoid complications from engaging other noncatalytic domains in the partitioning of the enzyme to the bilayer surface. At a mole fraction of anionic phospholipid equal to 0.5, the apparent KD values were 0.05 ± 0.01 and 0.15 ± 0.04 mm for vesicles with DOPG and DOPMe, respectively. When the content of the anionic phospholipid was reduced (mole fraction = 0.3), the KD values for PLC δ1 increased to 0.40 ± 0.14 and 0.25 ± 0.03 mm for the same two phospholipids. Interestingly, these KD values for full-length PLC δ1 binding to binary component LUVs are within a factor of 2 of the rEF binding to binary component SUVs.
Vesicle Bound rEF Orientation from 1H NMR Line Widths
Although the centrifugation-filtration vesicle binding assay proves to be an effective method to compare affinities of the EF-hand domain for various model membrane surfaces, it does not offer information on vesicle dynamics, orientation of the isolated domain on the vesicle surface, or differentiate the interaction of the protein with a specific type of phospholipid in mixed component membranes.
The binding studies clearly suggest that anionic phospholipid is necessary for bulk partitioning of rEF onto SUVs. To explore how rEF binds to SUVs, we used conventional 1H NMR to look at the line widths of the acyl chain resonances of the anionic phospholipid in vesicles with a chain perdeuterated PC, d54-DMPC, as a function of the amount of added rEF spin-labeled on one or the other of the cysteine residues (Cys-148 or Cys-188) in the first or second EF-hand. In the d54-DMPC/POPA system, only the PA acyl chain resonances are observed. The experimental temperature was set at 37 °C to keep the acyl chains of both components in these SUVs in a fluid state. As shown in Fig. 3, when up to 45 μm (0.86 mg/ml) of unlabeled rEF was titrated into 4 mm d54-DMPC/POPA (1:1) SUVs, the changes in 1H resonance line width were small with less than a 5 Hz increase in line width at the highest amount of protein used.
FIGURE 3.

Effect of spin-labeled EF-hand domain mutants on line widths of acyl chain 1H resonances in 4 mmd54-DMPC/POPA (1:1) SUVs. Spin-labeled EF-hand domain C148S was specifically labeled on Cys-188, (CH2)n (○), and ω-CH3 (Δ); spin-labeled EF-hand domain C188S was specifically labeled on Cys-148, (CH2)n (●), and ω-CH3 (▴). Control line widths for the same SUVs mixed with unlabeled protein are also shown: (CH2)n (×); ω-CH3 (+).
With spin-labeled rEF C188S (nitroxide on Cys-148) or C148S (nitroxide on Cys-188), more pronounced line broadening was observed (Fig. 3). Dose-dependent increases in line width were observed upon addition of the spin-labeled proteins. If the same amount of spin-labeled protein was titrated into pure POPC SUVs, no increase in line width was observed, consistent with the lack of binding of the protein to PC. However, there was a difference with the two spin-labeled proteins. The spin label located on Cys-148 appeared to have a larger paramagnetic effect on the acyl chain line widths than when the spin label was attached to Cys-188. The errors in measuring the line widths are less than the changes in titrations with unlabeled protein (<4 Hz), so the difference in the effect of the two spin-labeled proteins is real. This could suggest the following: (i) there is a discrete membrane-binding site(s) on rEF that is closer to Cys-148, in EF-1, than to Cys-188, in EF-2, or (ii) the orientation of the bound EF-hand protein is such that EF-1 is closer to the membrane than EF-2.
High Resolution Field Cycling 31P Detects Phospholipid/rEF Association and an Average rP-e
Another way to assay for specific effects of proteins on phospholipids is to use high resolution field cycling to monitor the spin-lattice 31P relaxation rates (R1) of the phospholipid headgroups to compare the two differently spin-labeled rEFs (23). The field dependence of R1 can provide information on where phospholipids bind relative to the spin label site. If measurements are made at very low fields (<0.03 T), the PRE of R1 also reflects a minimum lifetime of phospholipids interacting with the protein; very fast nanosecond interactions would not contribute much to the relaxation at these low fields, whereas microsecond (or longer) lifetimes for rEF-phospholipid interactions, which would be comparable with the overall tumbling time for an SUV (22), should show an increase at very low fields for phospholipids in small vesicles depending on where the spin label is with respect to the phospholipid (23). If the EF-hand domain binds to vesicles primarily by binding an individual anionic phospholipid, then in mixed POPC/DOPA SUVs the spin-labeled rEF should lead to a larger PRE for DOPA than for POPC. If, once attracted to the surface by electrostatics, the protein is anchored so that there are similar interactions with both phospholipids on the microsecond time scale (as would be the case with glycerol and acyl chain interactions of the lipids with the proteins), then the effects of the spin-labeled protein should be the same for both phospholipids.
Fig. 4, A and B, shows the dependence of 31P R1 on magnetic fields between 0.03 and 11.7 T for DOPA in the outer monolayer (which can be distinguished from DOPA on the inner leaflet of SUVs (21)) and POPC in SUVs with spin-labeled rEF (containing two spin-labeled cysteines, Cys-148 and Cys-188). Both DOPA in the outer monolayer and POPC show enhanced relaxation compared with the control SUVs (DOPA/POPC SUVs mixed with the same amount of unlabeled rEF). The magnitudes of the effects in this field range are about the same. The correlation time for this dispersion is ∼10 ns. With 0.5 mg/ml spin-labeled rEF (26 μm) and 6 mm of each phospholipid, the relaxation enhancement is large and about the same for both phospholipids, although it is difficult to obtain accurate values because this dispersion overlaps partially with the very low field one, and the R1 values rapidly increase above 25 s−1 below 0.02 T. With a reduced concentration of enzyme (0.1 mg/ml or 5.2 μm) and DOPA/POPC SUVs with 10 mm of each phospholipid, there is an increase in the limiting ΔR1 for the 10-ns dispersion of 0.4–0.5 s−1. The value is similar for both phospholipids.
FIGURE 4.
Dependence of the 31P spin-lattice relaxation rate, R1, for PA and PC in small vesicles with spin-labeled rEF added. In all plots the gray line represents the behavior for each phospholipid with unlabeled rEF added. The mid-to-high field region is shown in A and B, where the spin-labeled rEF was either 0.1 mg/ml (PA (●) and PC (■), 6 mm of each) or 0.5 mg/ml (PA (○) and PC (□), 5 mm each). The very low field region is shown in C and D for 0.2 mg/ml spin-labeled rEF and 10 mm each of PA and PC.
The R1 profile below 0.03 was compared with that for the phospholipids in the presence of unlabeled protein (Fig. 4, C and D, in this case with 0.2 mg/ml labeled rEF and 10 mm of each phospholipid). It is clear that both phospholipids are similarly relaxed, although with a slightly larger effect for DOPA. Because there are two spin labels attached to rEF, it is difficult to extract any phospholipid 31P-nitroxide electron distances.
The two mutated rEFs that have a single cysteine can be spin-labeled and examined in these experiments (data were only obtained down to 0.006 T for these samples). As seen in Fig. 5, the relaxation enhancement from spin-labeled C188S (0.5 mg/ml, 26 μm) is similar for both phospholipids (10 mm each) in the vesicle. However, at the concentrations of protein and phospholipid used, spin-labeled C148S has little effect at low fields. This indicates that spin-labeled Cys-148 in the first EF-hand of the domain is responsible for most of the relaxation of both DOPA and POPC. This result confirms the placement of rEF on the vesicles that was proposed above from the increased acyl chain proton line widths; the first EF-hand region is closer than the second indicating that rEF is binding in a manner that resembles its orientation in full-length PLC δ1.
FIGURE 5.
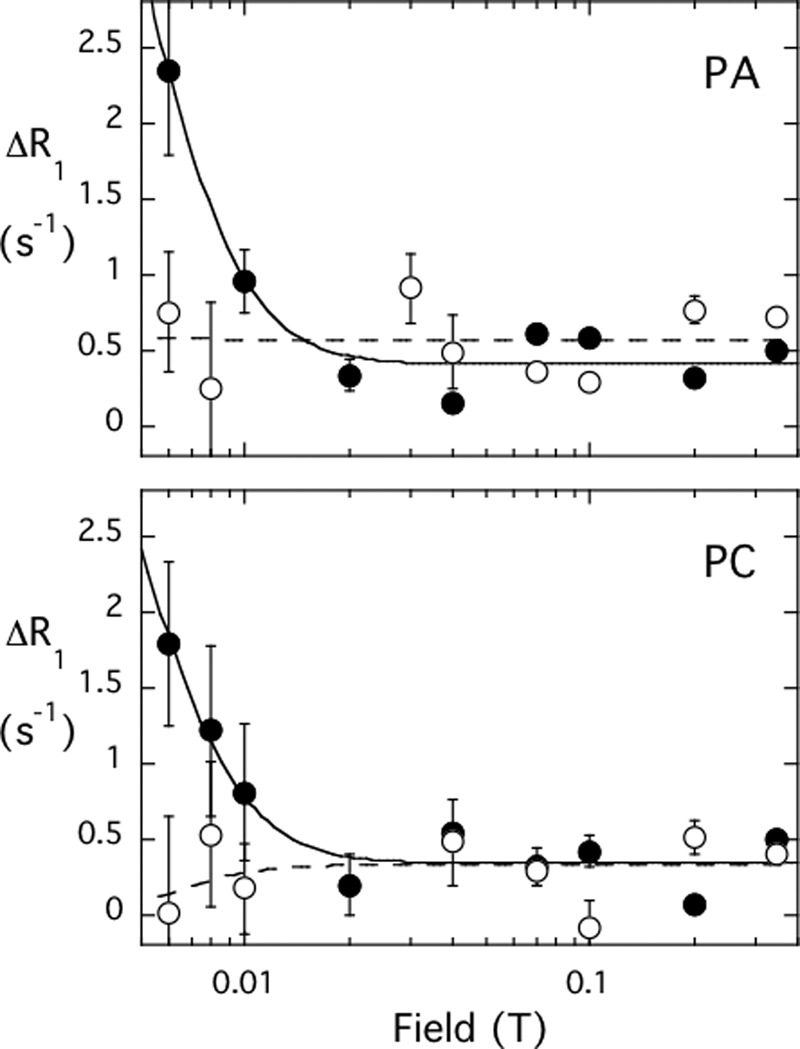
Dependence of the paramagnetic enhancement of 31P R1 for PA/PC (1:1) vesicles caused by the presence of 0.5 mg/ml spin-labeled C148S (open symbols) or spin-labeled C188S (filled symbols). The vesicles contained 10 mm each of PA (● and ○) and PC (■ and □). Note that for this amount of protein and phospholipid, the spin label on Cys-188 (i.e. labeled C148S) has virtually no effect at low field for either phospholipid, although the spin label attached to Cys-148 (labeled C188S) has a larger enhancement. The error bars represent the error in the fit to obtain R1 for each field strength.
If one uses the RP-e(0) and τP-e extracted from these data with the single cysteine mutants and assumes that ⅔ of each phospholipid is on the outside of these small vesicles, the average rP-e from the label on Cys-148 is 14.5 Å for DOPA and 15.9 Å for POPC. The much smaller RP-e for DOPA with the spin label on Cys-188 leads to a distance of 21 Å. Given the flexibility in the spin label and our lack of knowledge on whether the N-terminal peptide of EF-1 is helical or not, the difference in distances for PA produced by the two spin labels are reasonable.
Because the spin label on Cys-148 dominates the relaxation rate, we can re-evaluate the increases in R1 for the nanosecond and microsecond dispersions in Fig. 4, which used spin-labeled rEF and has more points at lower fields. The distance RP-e from the very low field region is 12–13 Å for both phospholipids, a value slightly shorter than with the single label, but because there will be some contribution from the added spin label on Cys-188, the values are fairly consistent. The rP-e extracted from the mid-field dispersion, which has a correlation time ∼20 ns, is 9.4 ± 0.1 Å. This time scale likely just reflects the average proximity of the electron to the membrane surface and not a longer lived complex.
The observation that both DOPA and POPC are significantly relaxed by the nitroxide attached to Cys-148 (mutant C188S) is also consistent with a binding mechanism whereby electrostatic attraction positions rEF at the membrane surface, but hydrophobic interactions, likely insertion of protein side chains into the bilayer, also play a role in transiently anchoring the protein on the vesicles, and interactions occur with both phospholipids that are stable for at least a microsecond.
Removing Basic Residues in the EF-hand Domain Weakens Vesicle Binding Only Slightly
The affinities of the EF-hand domain for anionic phospholipids in the absence of calcium strongly suggest that basic residues of the EF-hand domain, such as Arg and Lys, are important for the interactions with the membrane surface. The first pair of EF-hand helices, EF-1, is rich in basic residues, with 23% of its amino acids Arg or Lys (Fig. 1A). Additionally, given the positions in PLC δ1 of the active site of the catalytic domain and the calcium binding regions of the C2 domain (27), as well as the topological arrangement of the catalytic core of the protein (namely EF, catalytic, and C2 domains), the N-terminal lobe of the EF-hand domain, EF-1 and perhaps EF-2, could also be near the membrane surface when the full-length PLC δ1 binds to the membrane (Fig. 1B).
To test the importance of Arg or Lys in EF-1 and EF-2, mutants with one or multiple Arg/Lys substituted with alanine were constructed. The affinities of these mutant proteins for different SUVs are listed in Table 2. Compared with rEF, all basic residue mutants showed weaker binding to the PC/PA SUVs. However, the change was not large. The KD values were slightly increased (by ∼2-fold) when one Arg or Lys was replaced (K163A, K170A, and R186A), and the extent to which the KD value was increased was similar among these single mutants; when multiple Arg/Lys were replaced (e.g. triple mutant R182A/K183A/R186A), the KD values were further increased. Once Arg-150 and Lys-151 were replaced, further substitution of neighboring lysines with alanines (triple mutant R150A/K151A/K154A and quadruple mutant R150A/K151A/K154A/K156A) did not result in a further loss of affinity for vesicles.
TABLE 2.
Apparent dissociation constants for rEF with mutations of basic residues binding to POPC/DOPA (1:1) SUVs in 20 mm Tris, 0.1 mm EGTA, pH 7.5
| EF constructs | Apparent KDa | KD (mutant)/KD (WT) |
|---|---|---|
| mm | ||
| WT | 0.12 ± 0.02 | 1.0 |
| R150A/K151A | 0.51 ± 0.04 | 4.3 |
| R150A/K151A/K154A | 0.41 ± 0.07 | 3.4 |
| R150A/K151A/K154A/K156A | 0.44 ± 0.06 | 3.7 |
| K163A | 0.20 ± 0.03 | 1.7 |
| K170A | 0.27 ± 0.08 | 2.3 |
| R186A | 0.23 ± 0.06 | 1.9 |
| R182A/K183A/R186A | 0.35 ± 0.02 | 2.9 |
a This is the total concentration of phospholipids present for half of maximum binding. Errors were determined from the hyperbolic fit to the data.
In addition to PC and PS, there are other lipids present in the plasma membrane inner leaflet in considerable amounts (e.g. cholesterol, phosphatidylethanolamine, sphingomyelin, and phosphatidylinositol). SUVs with a composition of PE/PC/PS/PI/SM/CH (34.5:10.5:21:4.5:4.5:25), similar to the plasma membrane inner leaflet (19), were also used to assess rEF binding. The mole fraction of anionic lipids (PS and PI) was 0.26 in these vesicles, compared with a mole fraction of 0.50 in the binary component vesicles. Because the EF-hand domain preferred negatively charged membranes, it was not surprising that the apparent KD value of EF for the inner plasma membrane-mimicking vesicles, 0.31 mm (Table 3), was greater than those for the binary vesicles (Table 2). The binding of rEF to LUVs prepared from this mixture was also examined. The apparent KD was 0.32 ± 0.22 mm, a value similar to that for SUVs, although the errors in using LUVs in the binding assay were much larger than using the smaller SUV particles. The lack of sensitivity to vesicle size is quite different from small bacterial PI-PLC enzymes where the binding to small vesicles is often orders of magnitude tighter than to LUVs (28).
TABLE 3.
Apparent dissociation constants for EF-hand domain mutants binding to SUVs mimicking the phospholipid composition of the inner plasma membrane
The vesicle (SUV and LUV) composition was PE/PC/PS/PI/SM/CH equal to 34.5:10.5:21:4.5:4.5:25; the buffer was 20 mm Tris, 0.1 mm EGTA, pH 7.5.
| EF constructs | Apparent KDa | KD (mutant)/KD (WT) |
|---|---|---|
| mm | ||
| WT | 0.31 ± 0.05 | 1.0 |
| WT (LUVs) | 0.33 ± 0.22 | |
| R150A/K151A/K154A | 1.03 ± 0.19 | 3.3 |
| R182A/K183A/R186A | 0.81 ± 0.19 | 2.6 |
| W144A | 1.08 ± 0.36 | 3.5 |
| L149A | 0.45 ± 0.03 | 1.5 |
| F162A | 0.31 ± 0.01 | 1.0 |
| F168A | 1.54 ± 0.34 | 5.0 |
| V176A | 0.44 ± 0.02 | 1.4 |
| Y180A | 0.54 ± 0.06 | 1.7 |
| F185A | 0.46 ± 0.10 | 1.5 |
| I201A | 0.37 ± 0.03 | 1.2 |
a This is the total concentration of phospholipids present for half of maximum binding. Errors were determined from the hyperbolic fit to the data.
Similar to the binding study with POPC/POPA SUVs, the two triple mutants, R150A/K151A/K154A and R182A/K183A/R186A, showed weaker binding (a 2–3-fold increase in KD) than rEF to the complex SUVs (Table 3). Here, as with the binary component vesicles, the largest increase in KD among these mutants was 4-fold, which is not a large change in terms of binding. This indicates that other types of forces, such as hydrophobic interactions, contribute to overall partitioning of the isolated domain onto phospholipid surfaces.
Removing Selective Hydrophobic Residues in the EF-hand Domain Weakens Vesicle Binding
In an effort to identify residues important for hydrophobic interaction with membranes, we chose a variety of hydrophobic residues (Trp, Phe, Tyr, Val, Ile, and Leu) in the N-terminal lobe of the rEF (EF-1 and EF-2) and separately mutated them to alanine. Some of these hydrophobic residues appear to be on the protein surface with the first EF-1 hand modeled into the crystal structure (Fig. 6). The apparent KD values for these mutant proteins binding to SUVs mimicking the plasma membrane inner leaflet are shown in Table 3. Except for W144A and F168A, all the other mutants showed affinities for the vesicles comparable with rEF.
FIGURE 6.
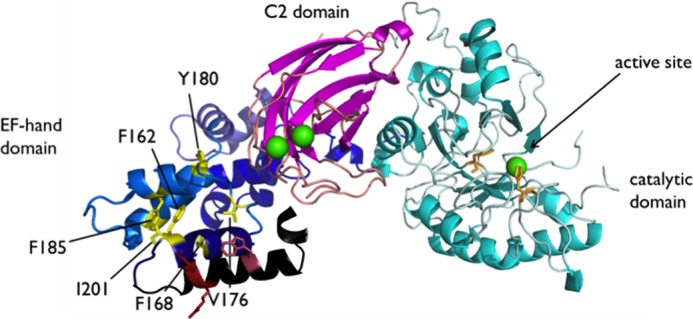
Crystal structure of rat PLC δ1 (Protein Data Bank code 1DJI) with hydrophobic residues observed in the crystal structure are marked as yellow sticks, and Trp-144 as well as Arg-150 and Lys-150, residues not observed in the crystal structure, are shown as red sticks. Leu-149, about 1 turn of the helix from Trp-144 in this model, is not shown. The view is presumably looking down from the membrane surface at an angle. The EF-hand domain is shown in black and blue; the catalytic domain is cyan with active site residues His-311 and His-356 side chains shown as orange sticks; the C2 domain is magenta, and the calcium ions are shown as green spheres.
Previous fluorescence studies of rEF (13) suggested that there was a substantial change in the environment of Trp-144 when the protein bound to fatty acids in detergent micelles. This would be consistent with a surface orientation of Trp-144 in the EF-1 helix shown in Figs. 1B and 6 (different views). Tryptophan is prone to insertion into membranes, and it could very well be the case for Trp-144. Insertion of the tryptophan side chain into membranes would assist in anchoring EF-1 onto the membranes.
In the PLC δ1 crystal structure, Phe-168 appears near the protein surface because the peptide that would constitute the first helix of EF-1 is missing. However, it is oriented away from what would be the membrane interface as suggested in Fig. 6. The weaker binding shown by F168A may be an artifact in the isolated EF-hand domain, caused by the lack of positioning restrictions imposed by other domains, or it could be indicative of a conformational change that the EF-hand domain undergoes in the presence of membranes.
Mutations in the EF-hand Domain Reduce Enzymatic Activity
Two EF mutations, single mutant W144A and double mutant R150A/K151A, both of which showed weaker binding to model membranes, were introduced into the full-length PLC δ1. The specific activities of PLC δ1 wild type and mutants were measured using PI as substrate, embedded in vesicles of various compositions and sizes (LUV or SUV), or with varied concentrations of calcium (0.5 or 0.1 mm) (Fig. 7). Although PI is not the preferred substrate for PLC δ1, kinetics are complicated with PI(4,5)P2, because that lipid also binds tightly to the PH domain and will anchor the protein to vesicles. With PI, it is easier to assess the contribution of other domains to enzyme performance.
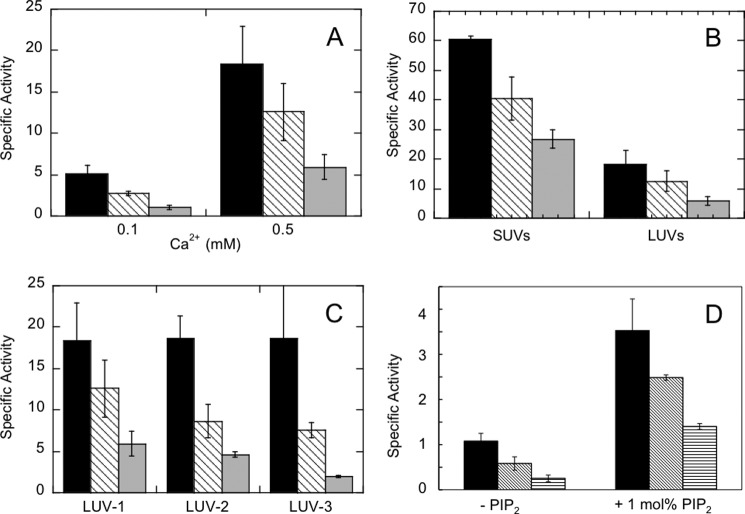
FIGURE 7.
Specific activities of PLC δ1 (black bars) and EF-hand mutant enzymes W144A (hatched bars) and R150A/K151A (gray bars) toward PI in different unilamellar vesicles. A, LUVs of 10 mm total lipids with composition PI/PE/PC/SM/CH (25.5:34.5:10.5:4.5:25) with either 0.1 or 0.5 mm Ca2+. B, LUVs or SUVs with 10 mm total phospholipid of the same vesicle composition, PI/PE/PC/SM/CH (25.5:34.5:10.5:4.5:25) and 0.5 mm Ca2+. C, LUVs with various bulk PI concentrations, molar ratio of PI, and molar ratio of anionic lipids in the presence of 0.5 mm Ca2+: LUV-1 = PI/PE/PC/SM/CH (25.5:34.5:10.5:4.5:25) with 2.55 mm PI; LUV-2 = PI/PS/PE/PC/SM/CH (12.75:12.75:34.5:10.5:4.5:25) with 1.28 mm PI; LUV-3 = PI/PC (1:1) with 2.5 mm PI. D, LUVs of the same composition in A, but with only 0.5 mm total lipid and 0.5 mm Ca2+ in the absence or presence of 1 mol % PIP2. Here the gray bars are for W144A and the vertical hatched bars are for R150A/K151A. For all specific activities, the error bars indicate the standard deviation of at least three repeats.
The vesicle compositions were designed based on what was used in the binding assays (binary vesicles and plasma membrane mimicking vesicles), with some modification. For plasma membrane composition vesicles, we increased the amount of PI (from 4.5 to 25.5 or 12.75%) while keeping the molar fraction of total anionic lipids constant (25.5%), so the PI hydrolysis cleavage products could be accurately measured by 31P NMR within a reasonable time frame. In all our assays, both the intermediate product myo-inositol 1,2-(cyclic)-phosphate (cIP) and the final hydrolysis product inositol 1-phosphate (I-1-P) were observed. This has been seen before for this enzyme acting on PI (28). PLC δ1 does not proceed by covalent catalysis, rather cIP is formed as an intermediate, and it is either released into solution or attacked by H2O with the release of I-1-P. The specific activities were calculated as the sum of both products.
The LUV system (total lipid concentration = 10 mm) composed of PI/PE/PC/SM/CH (25.5:34.5:10.5:4.5:25) in the presence of 0.5 mm Ca2+ is used as our “standard system.” As shown in Fig. 7A, when the calcium concentration was decreased from 0.5 to 0.1 mm, PLC δ1 and mutants all showed lower activities, consistent with the important role calcium plays in PLC δ1 hydrolysis. Furthermore, it suggests these mutations in the EF-hand domain did not reduce the enzyme's sensitivity toward calcium.
Next, the effect of vesicle size was examined. With the same lipid composition (PI/PE/PC/SM/CH (25.5:34.5:10.5:4.5:25)), the WT and the two mutant enzymes preferred PI presented in SUVs to LUVs (Fig. 7B). The specific activities were enhanced 3–4.5-fold. At least in the absence of PIP2, PLC δ1-catalyzed hydrolysis of PI could be enhanced by membrane curvature and/or possibly surface defects likely to occur in small vesicles.
Finally, the effects of substrate concentration and anionic lipid concentration in the vesicle surface were examined. As shown in Fig. 7C, the specific activities of PLC δ1 were equivalent for all three LUV systems with 10 mm PI/PE/PC/SM/CH (25.5:34.5:10.5:4.5:25) LUVs (denoted as “LUV-1”), 10 mm PI/PS/PE/PC/SM/CH (12.75:12.75:34.5:10.5:4.5:25) LUVs (“LUV-2”), or 5 mm PI/PC (1:1) LUVs (“LUV-3”), respectively. This indicates that at these total lipid concentrations, the activity of PLC δ1 is unaffected by the changes in the bulk concentration of PI or the interfacial mole fraction (surface concentration) of PI (comparing LUV-1 with LUV-2) or the mole fraction of all anionic lipids (comparing LUV-1 with LUV-3). Thus, at the substrate concentrations we are using, we are above the KD values for partitioning of the enzyme to the vesicle and the surface Km values for substrate PI accessing the active site.
Both mutants were unaffected by the 2-fold difference in the surface concentration of PI between different LUV systems. However, R150A/K151A, in particular, exhibited a 3-fold decrease in specific activity when comparing LUV-3 with LUV-1, even though the surface concentration of the substrate (50 mol %) was actually higher under conditions where the bulk PI concentration was the same. The lipid components missing from LUV-3 included sphingomyelin and cholesterol, which are thought to form raft-like domains in the presence of a fluid PC-matrix (29). For LUV-3, the apparent KD value for PLC δ1 is likely to be below 0.3 mm, assuming the PI in the vesicles would lead to similar bulk binding to that observed with PA in the binary vesicle binding studies and given the observation that rEF binding to SUVs and LUVs is similar (Tables 1 and 2). The apparent KD value for R150A/K151A binding to the PA/PC SUVs was 0.5 mm, well below the 10 mm total phospholipid used in the kinetic assays so that all of that enzyme should be bound to vesicles. Reduced activity in the mutated proteins likely reflects an altered affinity for the substrate in the active site. These kinetics indicate that residues Trp-144, Arg-150, and Lys-151 in the EF-hand domain play an indirect role in modulating the PI cleavage and hydrolysis activity of the full-length enzyme.
PIP2 Binding Cannot Substitute for the Loss in Relative Activity of EF-domain Mutants
Because the total lipid concentration in the vesicles used in the activity assays (10 or 5 mm) were well above the KD value for rEF or PLC δ1 partitioning onto binary component vesicles (0.14 to 0.25 mm for rEF binding to SUVs with a 1:1 anionic phospholipid to PC and 0.05 to 0.15 for PLCd1 binding to LUVs with a 1:1 anionic phospholipid), it is reasonable to assume the full-length PLC to be completely partitioned onto vesicles. This should eliminate any effects the mutations may have on the enzyme activity due to differences in bulk partitioning onto the vesicles. The fact that the mutants exhibited decreased activities (and roughly the same fraction of PLC δ1 activity) in several vesicle systems suggests that factors other than bulk binding are at play. To further examine this hypothesis, the specific activities of PLC δ1 and variants at a much lower lipid concentration, 0.5 mm, were measured (Fig. 7D). If the mutant enzymes had much larger increases in the apparent KD value for bulk binding than PLC δ1, then the differences in enzymatic activity should be larger at lower total lipid concentrations.
With the LUV-1 composition, the PLC δ1 enzymes showed much lower activities when the total lipid concentration was reduced from 10 to 0.5 mm. The specific activity for the WT enzyme was 1.1 μmol·min−1·mg−1; W144A and R150A/K151A activities were reduced to 54 and 23% of that value.
If 1 mol % PIP2 is present in the same lower concentration of LUVs, all of the proteins should be bound to the vesicles (0.5 mm total lipid) given that the isolated PH domain is reported to bind to PIP2-containing vesicles with a KD of 1.7 μm (9). PLC δ1 action on these vesicles leads to hydrolysis of PIP2 (producing a single product IP3) in addition to PI. In fact, about 40% of the PIP2 was hydrolyzed, whereas less than 20% of the much larger concentration of PI was hydrolyzed. The specific activities shown in Fig. 7D only represent PI cleavage and hydrolysis. Consistent with previous studies (27), the incorporation of PIP2 into the membranes activated PLC hydrolysis toward PI. With these vesicles and 0.5 mm total lipid concentration, the increase was 3.3-, 4.3-, and 5.5-fold for PLC δ1, W144A, and R150A/R151A, respectively. With the small amount of PIP2 present and ensuring the enzyme was bound to the vesicles, the relative activities of the mutant enzymes compared with PLC δ1 were comparable with what was seen with 10 mm total lipid, although the specific activities were still much lower than at the higher total lipid concentration. Once binding of the enzyme on the vesicle is achieved, differences in the interfacial Km value for substrate remain because the mole fraction of substrate in the interface is the same for both 10 and 0.5 mm total lipid vesicles.
DISCUSSION
Although the relationship between many of the structural features of the components of PLC δ1 and their contributions to the activity regulation has been scrutinized, the role of the EF-hand domain remains unclear. Previous reports have identified the EF-hand domain of PLC δ1 as a lipid interacting domain, highlighting its interaction with fatty acid (arachidonic acid in particular) (13, 14). However, as we show, the domain also can interact with anionic phospholipids showing little specificity for a particular phospholipid once the domain is associated with the vesicle.
Although EF-hand motifs are often associated with Ca2+ binding, there is no experimental evidence to show that this is actually the case with PLC δ1. Althoughthe loop regions of EF-3 and EF-4 do not have side chains that would act as typical calcium ligands, EF-1 and EF-2 do (7). However, the mutations of these putative calcium-binding residues (Asp-153, Asp-157, and Glu-164) do not affect enzyme activity (12), suggesting that EF-hand domain binding of calcium, even if it occurs, is not connected to the enzymatic reaction of PLC δ1. In fact, in all the known crystal structures of PLC δ1, no calcium ion is found associated with the EF-hand domain, even though the enzyme is soaked in calcium or its analogs (7, 25–27). Likewise, EF-hand domains of PLC β2 (31, 32) and β3 (33), the only other PLC EF-hand domains with known structures, are not found to be associated with calcium. In our binding assays, the isolated EF-hand domain can bind to vesicles of various compositions containing anionic phospholipid without Ca2+. Interestingly, the first EF-hand motif of the PLC δ1 is replete with cationic residues, which would exert an attractive electrostatic force on negatively charged lipids similar to what the Ca2+ bound to the domain might exert. A sequence comparison of several members from the PLCβ, -γ, and -δ subfamilies reveals that only 13 amino acids are identical or conserved outside the highly conserved X/Y catalytic domain and C2 domain, and that all of them are located in the EF-hand domain (34). Among these conserved residues are Lys-151 and Lys-170 of PLC δ1 (the rest include nine hydrophobic residues, one Gln, and one Glu/Asp). Taken together, the cationic residues in the EF-hand domain of PLC δ1, especially the well conserved Lys-151, may play an important role in the interaction of the EF-hand domain with membranes containing anionic lipids.
It is possible that the EF-hand domain is initially attracted to the anionic component of the membrane through electrostatic interactions (hence no binding occurs without anionic lipids). But once the protein is in the proximity of the membrane, the interaction of hydrophobic side chains, in particular Trp-144, also contributes to binding. Experimentally, removing key basic residues or hydrophobic residues does indeed weaken the EF binding to vesicles, although not dramatically, with a 4-fold increase in the apparent KD, the largest change for each type of mutation. This is not surprising, given that other domains of the PLC δ1 can contribute to membrane tethering of the enzyme. However, the fact that PLC δ1 loses enzymatic activity with membrane substrates when the EF-hand domain is deleted (11, 12) argues that the EF-hand domain carries significance in the regulation of the enzymatic activity of the PLC δ1.
Structural studies show that PLC δ1 has a strikingly rigid core composed of the C-terminal half of the EF-hand domain, the catalytic domain (except for residues 445–485 and 509–514), and the C2 domain (25). In contrast, the first half of the EF-hand domain is quite flexible, as evidenced by the poor ordering of this region in the crystal structures. Therefore, this region may serve as a flexible bridge between the PH domain and the rigid core of the enzyme, which, upon association with membrane lipids, primes the active site for substrate access. Anionic phospholipids PA, PG, and PS can greatly enhance PLC δ1 hydrolysis toward PI (5–15-fold increase over SUV containing PC only (10)). Although it is believed that PS is involved in enzyme binding (via the C2 domain) in a Ca2+-dependent manner, PA does not participate in Ca2+-mediated binding of the enzyme (10). In our binding studies, the isolated EF-hand domain binds to both PA and PS with comparable affinity and to PG with a slightly weaker affinity, all independent of Ca2+. Thus, the stimulation of PLC δ1 activity by PA may involve the interaction between PA and the EF-hand domain. Fatty acids are also anionic, and these can bind to the EF-hand domain (14). It was pointed out (13) that the fluorescence changes induced by arachidonic acid binding could also reflect tight binding of an anionic amphiphile and not necessarily a strict requirement for that fatty acid, in which case anionic phospholipids could produce similar effects.
The field cycling NMR experiments show that there is little difference in proximity of PC and PA to the spin label attached to the EF, consistent with a model whereby the charged surface attracts the cationic groups in the EF-1, but the Trp-44 side chain contributes to anchoring the protein to the vesicle. The increased 31P relaxation by spin-labeled rEF at very low fields does suggest that the protein interacts with both PA and PC molecules for at least 1 μs (otherwise there would be no low field PRE); hence, the insertion of hydrophobic residues immobilizes the phospholipids for at least this time scale. This is certainly suggestive of the interaction of arachidonic acid with this domain (13).
One of the effects shown for arachidonic acid binding to rEF is a reduction in the Km value for dibutyroylphosphatidylinositol (14). If the fatty acid activation is really an anionic amphiphile effect, then we can explain the reduction in specific activity for R150A/R151A (and the smaller reduction seen for W144A) as an increased interfacial Km value for substrate binding in the active site of the enzyme, not an overall loss of affinity of the protein for vesicles. This would imply that the EF-hand domain aids allosterically by either electrostatically providing a pathway for substrate to reach the catalytic domain or perhaps by helping to stabilize a critical conformation in the catalytic domain, in substrate binding in the active site (this could even involve interactions with the PH domain).
In addition to the N-terminal half of the EF-hand domain, there is another extended region of PLC δ1 that lacks interpretable electron density in crystallographic studies, namely residues 445–485 (7, 25). This region, the X/Y linker, connects the catalytic X and Y domains. Acidic residues in the X/Y linker play an autoinhibitory role in membrane binding and affect enzyme activity by regulating the access of phosphoinositide substrates to the active site (32). For PLC β2, the disordered region of the X/Y linker in its crystal structure serves as the major autoinhibitory determinant, although the ordered region appears to fine-tune access to the active site. The calcium binding regions in the C2 domain of PLC δ1 are also highly flexible. In fact, there is a lack of interpretable density for these regions in the crystal structure when metal ions are absent. It is with the binding of the metal ions that these regions become defined (25). These highly flexible regions, including the N-terminal half of the EF-hand domain, may modulate PLC δ1 activity by controlling substrate access and binding in the correct orientation in the active site.
At present, the only other crystal structures of PLC isoforms that have been solved are those of PLC β2 and β3 complexed with a regulatory protein (31–33), in which the EF-hand domains are fully defined. Although the EF-hand domains of these PLC β enzymes have not been implicated in membrane association, the topological arrangement of the PLC domains (PH, EF-hand, catalytic X/Y, and C2) as well as the regulatory protein in the complex (Rac1 for PLC β2 and Gαq for PLC β3) with respect to the interface suggest that the N-terminal lobe of the EF-hand domain in these PLCs could be in the proximity of the membrane surface when the enzymes are bound. In both PLC β structures, there is a cluster of Arg and Lys residues in the N-terminal lobe of the EF-hand domain appearing to point toward the assumed interface in a similar fashion as PLC δ1.
PLC ζ, the only isoform in the PLC family lacking a PH domain, specifically expressed in sperm, is known to induce intracellular Ca2+ oscillations and subsequently activate oocyte fertilization (35). A splicing variant of PLC ζ lacking 110 residues from the N terminus corresponding to the EF-hands 1–3 but identical to PLC ζ in EF4 and the succeeding region failed to elicit a Ca2+ spike (36). This indicates that the EF-hand domain is important for PLC ζ to induce Ca2+ oscillations (which presumably requires correct membrane binding). Of all the other isoforms, PLC ζ shares the closest homology with PLC δ1 (30). Arg-182, Lys-183, and Arg-186 of PLC δ1, which have been shown to contribute to the binding of vesicles containing anionic lipids in our study, are conserved in PLC ζ. Using regions rich in basic side chains for electrostatic orientation of a protein to acidic phospholipids in a membrane may be a common mechanism for peripheral proteins, less for absolute binding than for correct arrangement on the membrane surface.
Our work in the context of other PLC isoforms suggests that the N-terminal lobe of the EF-domain of PLCs, by interacting with the target membrane, provides a tether that facilitates proper substrate access and binding in the active site.
Acknowledgment
We thank Prof. Alfred G. Redfield for access to the high resolution field cycling device and for sealing samples.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM60418 (to M. F. R.).
- PLC
- phospholipase C
- rEF
- recombinant EF
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- IP3
- inositol 1,4,5-trisphosphate
- PH
- pleckstrin homology
- PRE
- paramagnetic relaxation enhancement
- SUV
- small unilamellar vesicle
- LUV
- large unilamellar vesicle
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- DOPA
- 1,2-dioleoyl-sn-glycero-3-phosphate
- d54-DMPC
- 1,2-dimyristoyl-d54-sn-glycero-3-phosphocholine
- MTSL
- methanethiosulfonate
- PI
- l-α-phosphatidylinositol
- DOPG
- 1,2-dioleoylphosphatidylglycerol
- DOPMe
- 1,2-dioleoylphosphatidylmethanol
- Rho-PE
- 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl)
- SM
- sphingomyelin
- CH
- cholesterol
- cIP
- myo-inositol 1,2-(cyclic)-phosphate
- PC
- phosphocholine
- PA
- phosphatidic acid
- PG
- phosphatidylglycerol
- I-1-P
- inositol 1-phosphate
- PS
- phosphatidylserine
- T
- tesla.
REFERENCES
- 1. Streb H., Irvine R. F., Berridge M. J., Schulz I. (1983) Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature 306, 67–69 [DOI] [PubMed] [Google Scholar]
- 2. Takai Y., Kishimoto A., Kikkawa U., Mori T., Nishizuka Y. (1979) Unsaturated diacylglycerol as a possible messenger for the activation of calcium-activated, phospholipid-dependent protein kinase system. Biochem. Biophys. Res. Commun. 91, 1218–1224 [DOI] [PubMed] [Google Scholar]
- 3. Kishimoto A., Takai Y., Mori T., Kikkawa U., Nishizuka Y. (1980) Activation of calcium and phospholipid-dependent protein kinase by diacylglycerol, its possible relation to phosphatidylinositol turnover. J. Biol. Chem. 255, 2273–2276 [PubMed] [Google Scholar]
- 4. Rebecchi M. J., Pentyala S. N. (2000) Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol. Rev. 80, 1291–1335 [DOI] [PubMed] [Google Scholar]
- 5. Kamat A., Carpenter G. (1997) Phospholipase C-γ1: regulation of enzyme function and role in growth factor-dependent signal transduction. Cytokine Growth Factor Rev. 8, 109–117 [DOI] [PubMed] [Google Scholar]
- 6. Suh P. G., Park J. I., Manzoli L., Cocco L., Peak J. C., Katan M., Fukami K., Kataoka T., Yun S., Ryu S. H. (2008) Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 41, 415–434 [DOI] [PubMed] [Google Scholar]
- 7. Essen L. O., Perisic O., Cheung R., Katan M., Williams R. L. (1996) Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature 380, 595–602 [DOI] [PubMed] [Google Scholar]
- 8. Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B. (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83, 1037–1046 [DOI] [PubMed] [Google Scholar]
- 9. Lemmon M. A., Ferguson K. M., O'Brien R., Sigler P. B., Schlessinger J. (1995) Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. U.S.A. 92, 10472–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lomasney J. W., Cheng H. F., Roffler S. R., King K. (1999) Activation of phospholipase C δ1 through C2 domain by a Ca2+-enzyme-phosphatidylserine ternary complex. J. Biol. Chem. 274, 21995–22001 [DOI] [PubMed] [Google Scholar]
- 11. Ellis M. V., Carne A., Katan M. (1993) Structural requirements of phosphatidylinositol-specific phospholipase C δ1 for enzyme activity. Eur. J. Biochem. 213, 339–347 [DOI] [PubMed] [Google Scholar]
- 12. Nakashima S., Banno Y., Watanabe T., Nakamura Y., Mizutani T., Sakai H., Zhao Y., Sugimoto Y., Nozawa Y. (1995) Deletion and site-directed mutagenesis of EF-hand domain of phospholipase C-δ1: effects on its activity. Biochem. Biophys. Res. Commun. 211, 365–369 [DOI] [PubMed] [Google Scholar]
- 13. Kobayashi M., Gryczynski Z., Lukomska J., Feng J., Roberts M. F., Lakowicz J. R., Lomasney J. W. (2005) Spectroscopic characterization of the EF-hand domain of phospholipase C δ1: identification of a lipid interacting domain. Arch. Biochem. Biophys. 440, 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kobayashi M., Mutharasan R. K., Feng J., Roberts M. F., Lomasney J. W. (2004) Identification of hydrophobic interactions between proteins and lipids: free fatty acids activate phospholipase C δ1 via allosterism. Biochemistry 43, 7522–7533 [DOI] [PubMed] [Google Scholar]
- 15. Jiang Y., Qian W., Hawes J. W., Walsh J. P. (2000) A domain with homology to neuronal calcium sensors is required for calcium-dependent activation of diacylglycerol kinase α. J. Biol. Chem. 275, 34092–34099 [DOI] [PubMed] [Google Scholar]
- 16. Sakane F., Yamada K., Kanoh H., Yokoyama C., Tanabe T. (1990) Porcine diacylglycerol kinase sequence has zinc finger and E–F hand motifs. Nature 344, 345–348 [DOI] [PubMed] [Google Scholar]
- 17. Ellis M. V., James S. R., Perisic O., Downes C. P., Williams R. L., Katan M. (1998) Catalytic domain of phosphoinositide-specific phospholipase C (PLC). J. Biol. Chem. 273, 11650–11659 [DOI] [PubMed] [Google Scholar]
- 18. Feng J., Wehbi H., Roberts M. F. (2002) Role of tryptophan residues in interfacial binding of phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 277, 19867–19875 [DOI] [PubMed] [Google Scholar]
- 19. Landgraf K. E., Malmberg N. J., Falke J. J. (2008) Effect of PIP2 binding on the membrane docking geometry of PKCR C2 domain: an EPR site-directed spin-labeling and relaxation study. Biochemistry 47, 8301–8316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Redfield A. G. (2012) High resolution NMR field-cycling device for full-range relaxation and structural studies of biopolymers on a shared commercial instrument. J. Biomol. NMR 52, 159–177 [DOI] [PubMed] [Google Scholar]
- 21. Roberts M. F., Redfield A. G. (2004) High resolution 31P field cycling as a probe of phospholipid dynamics. J. Am. Chem. Soc. 126, 13765–13777 [DOI] [PubMed] [Google Scholar]
- 22. Roberts M. F., Redfield A. G. (2004) Phospholipid bilayer surface configuration probed quantitatively by 31P field-cycling NMR. Proc. Natl. Acad. Sci. U.S.A. 101, 17066–17071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pu M., Orr A., Redfield A. G., Roberts M. F. (2010) Defining specific lipid binding sites for a peripheral membrane protein in situ using subtesla field-cycling NMR. J. Biol. Chem. 285, 26916–26922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng J., Goldstein R., Gershenson A., Stec B., Roberts M. F. (2013) The cation-π box is a specific phosphatidylcholine membrane targeting motif. J. Biol. Chem. 288, 14863–14873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grobler J. A., Essen L. O., Williams R. L., Hurley J. H. (1996) C2 domain conformational changes in phospholipase C-δ 1. Nat. Struct. Biol. 3, 788–795 [DOI] [PubMed] [Google Scholar]
- 26. Essen L. O., Perisic O., Katan M., Wu Y., Roberts M. F., Williams R. L. (1997) Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C. Biochemistry 36, 1704–1718 [DOI] [PubMed] [Google Scholar]
- 27. Lomasney J. W., Cheng H. F., Wang L. P., Kuan Y., Liu S., Fesik S. W., King K. (1996) Phosphatidylinositol 4,5-bisphosphate binding to the pleckstrin homology domain of phospholipase C-δ1 enhances enzyme activity. J. Biol. Chem. 271, 25316–25326 [DOI] [PubMed] [Google Scholar]
- 28. Roberts M. F., Wu Y., Perisic O., Williams R. L., Katan M. (1998) in Phosphoinositides: Chemistry, Biochemistry and Biomedical Applications (Bruzik K. S., ed) Vol. 718, pp. 137–157, American Chemical Society, Washington, D. C. [Google Scholar]
- 29. Risselada H. J., Marrink S. J. (2008) The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. U.S.A. 105, 17367–17372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Swann K., Saunders C. M., Rogers N. T., Lai F. A. (2006) PLCζ: a sperm protein that triggers Ca2+ oscillations and egg activation in mammals. Semin. Cell Dev. Biol. 17, 264–273 [DOI] [PubMed] [Google Scholar]
- 31. Jezyk M. R., Snyder J. T., Gershberg S., Worthylake D. K., Harden T. K., Sondek J. (2006) Crystal structure of Rac1 bound to its effector phospholipase C-β2. Nat. Struct. Mol. Biol. 13, 1135–1140 [DOI] [PubMed] [Google Scholar]
- 32. Hicks S. N., Jezyk M. R., Gershburg S., Seifert J. P., Harden T. K., Sondek J. (2008) General and versatile autoinhibition of PLC isozymes. Mol. Cell 31, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Waldo G. L., Ricks T. K., Hicks S. N., Cheever M. L., Kawano T., Tsuboi K., Wang X., Montell C., Kozasa T., Sondek J., Harden T. K. (2010) Kinetic scaffolding mediated by a phospholipase C-β and Gq signaling complex. Science 330, 974–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rhee S. G., Choi K. D. (1992) Multiple forms of phospholipase C isozymes and their activation mechanisms. Adv. Second Messenger Phosphoprotein Res. 26, 35–61 [PubMed] [Google Scholar]
- 35. Saunders C. M., Larman M. G., Parrington J., Cox L. J., Royse J., Blayney L. M., Swann K., Lai F. A. (2002) PLCζ: a sperm-specific trigger of Ca2+ oscillations in eggs and embryo development. Development 129, 3533–3544 [DOI] [PubMed] [Google Scholar]
- 36. Kouchi Z., Fukami K., Shikano T., Oda S., Nakamura Y., Takenawa T., Miyazaki S. (2004) Recombinant phospholipase Cζ has high Ca2+ sensitivity and induces Ca2+ oscillations in mouse eggs. J. Biol. Chem. 279, 10408–10412 [DOI] [PubMed] [Google Scholar]