Abstract
Sequence polymorphisms linked to human diseases and phenotypes in genome-wide association studies often affect non-coding regions. A single nucleotide polymorphism (SNP) within an intron of the gene encoding Interferon Regulatory Factor 4 (IRF4), a transcription factor with no known role in melanocyte biology, is strongly associated with sensitivity of skin to sun exposure, freckles, blue eyes and brown hair color. Here we demonstrate that this SNP lies within an enhancer of IRF4 transcription in melanocytes. The allele associated with this pigmentation phenotype impairs binding of the TFAP2A transcription factor which together with the melanocyte master regulator MITF, regulates activity of the enhancer. Assays in zebrafish and mice reveal that IRF4 cooperates with MITF to activate expression of Tyrosinase (TYR), an essential enzyme in melanin synthesis. Our findings provide a clear example of a non-coding polymorphism that affects a phenotype by modulating a developmental gene regulatory network.
Introduction
Human pigmentation is a complex process involving melanocytes which synthesize the pigment melanin in melanosomes, cell organelles that are transferred to neighbouring keratinocytes, where they form a cap over nuclei, thus protecting them from negative effects of ultra violet radiation (UVR). Pigmentation is not only one of the most distinguishing features of humans but also serves an important protective role. In humans, pigmentation decreases with increasing distance from the equator, presumably due to a balance between the pressure to optimize the amount of available UVR for the generation of vitamin D3 and the protection from UVR-mediated damage, which may result in increased risk of cutaneous malignancies. This has led to positive selection for less pigmented skin, hair and eyes in areas distant from the equator. The major difference between dark- and light-skinned individuals is due to differences in the number, size and density of the melanin-containing melanosomes; the number of melanocytes is roughly the same (reviewed in Sturm, (2009)).
The genetics of pigmentation is complex and involves several genes and pathways. It has been best characterized in the mouse where 171 of nearly 400 loci implicated in pigmentation have been functionally characterized (http://www.espcr.org/micemut/). These pigmentation genes affect various steps in the formation of melanocytes from the neural crest (e.g. Mgf, Kit and Mitf), the generation of components of melanosomes and pigment (e.g. pMel17/Silver and Tyr) or the transport of melanosomes along microtubules in the dendrites before delivery to adjacent keratinocytes (e.g. MyoVa, Rab27 and Mlph). Most genes involved in pigmentation are transcriptionally regulated by the bHLHZip transcription factor MITF, the master regulator of melanocytes (reviewed in Steingrimsson et al., (2004)).
Genome wide association studies (GWAS) have identified several Single Nucleotide Polymorphism (SNPs) involved in human pigmentation, including SNPs on chromosome 6 near the DUSP22, IRF4, and EXOC2 genes, none of which were previously implicated in pigmentation (Sulem et al., 2007; Han et al., 2008; Sulem et al., 2008). The SNP showing the strongest association is located in intron 4 of IRF4 (Han et al., 2008). IRF4 belongs to the interferon regulatory factors, a wing-helix-turn-helix family of transcription factors that regulate interferon (IFN)-inducible genes (Paun and Pitha, 2007). Although IRF4 does not depend on interferon stimulation, it binds to the IFN-stimulated response element (ISRE) within IFN-responsive genes (Matsuyama, 1995; Grossman et al., 1996; Escalante et al, 1998). Irf4 mutant mice completely lack germinal centers and plasma cells and show severely reduced immunoglobulin levels in the serum; no defects in pigmentation were reported (Mittrucker et al., 1997). In a subset of multiple myeloma patients, IRF4 is translocated downstream of the immunoglobulin heavy-chain regulatory regions (Iida et al., 1997; Tsuboi et al., 2000). Interestingly, in this disease, IRF4 acts as a lineage survival oncogene regardless of translocation status (Shaffer et al., 2008). Polymorphisms in the IRF4 gene are associated with chronic lymphocytic leukemia (CLL) (Di Bernardo et al., 2008) and with non-hematopoietic diseases including celiac disease (Dubois et al., 2010), and progressive supranuclear palsy (Hoglinger et al., 2011). Finally, IRF4 was recently shown to be critical for transcriptional response to nutrient availability in adipocytes (Eguchi et al., 2011).
A few reports have linked IRF4 to pigment cells. IRF4 is expressed in melanocytes in the skin and in the G361 melanoma cell line (Grossman et al., 1996) as well as in most melanomas (Sundram, 2003). Importantly, IRF4 is associated with human pigmentation (Sulem et al., 2007; Han et al., 2008) and IRF4 expression correlates with MITF expression in melanoma cells (Hoek et al. 2008). Here we show that IRF4 is involved in pigmentation, that the melanocyte master regulator MITF activates expression of the IRF4 gene and that this activation depends on the presence of the Transcription Factor Activator Protein alpha (TFAP2A). A naturally occurring sequence variant associated with human pigmentation overlaps the TFAP2A binding site, impairs binding by this transcription factor and consequently lowers induction of IRF4 expression. Together, the MITF and IRF4 proteins cooperatively activate expression of the gene encoding the pigmentation enzyme Tyrosinase (TYR). This activation depends on MITF and IRF4 binding sites in the TYR promoter. Thus, we have established a direct link between a polymorphism in an intron in the IRF4 gene and reduced expression of an enzyme essential for pigmentation.
Results
Fine mapping of the IRF4 locus implicates rs12203592 as a functional variant affecting pigmentation
Several sequence variants in the IRF4 locus have been associated with freckling, sun sensitivity, eye and hair color and with nevus counts, with rs12203592-[T/C] showing the strongest association (Sulem et al., 2007; Han et al., 2008; Duffy et al., 2010). The rs12203592-T minor allele is most common in individuals of European descent; it is not seen in sub-Saharan Africans or in East Asians (Figure S1). Analysis of sequenced vertebrate species shows that this position is occupied by C, suggesting that it is the ancestral allele.
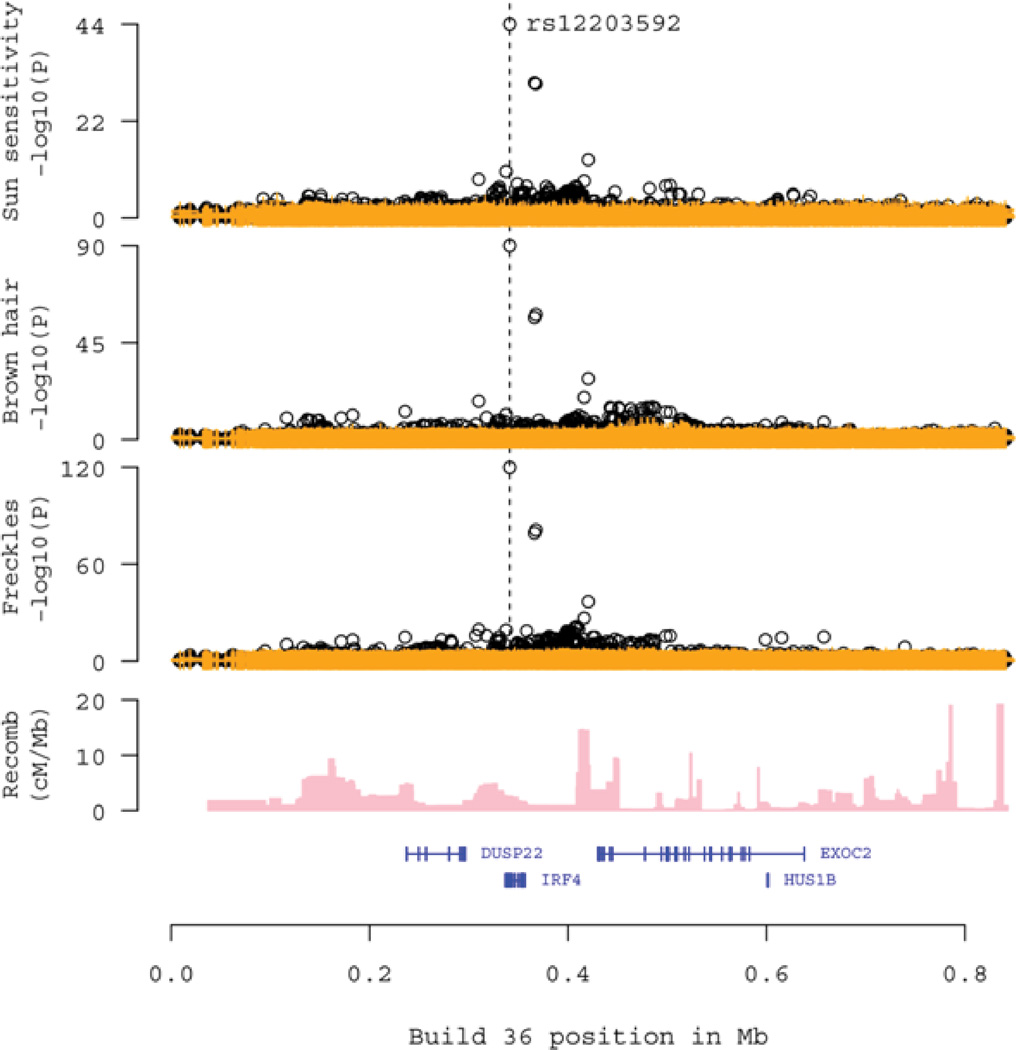
In order to fine map the pigmentation trait associations at the IRF4 locus, we used whole genome sequencing data derived from 2,230 Icelanders sequenced to an average coverage of at least 10×. This yielded approximately 38.5 million SNP and small indel (insertion or deletion) variants genome-wide, 16,280 of which were located in the region around IRF4 (0–1 Mb on chromosome 6). Using imputation assisted by long-range haplotype phasing (Kong et al., 2008; Kong et al., 2009; Holm et al., 2011), we determined the genotypes of these 16,280 variants in 95,085 individuals who had been typed using Illumina SNP chips. We then tested each variant for association with eye color, hair color, freckling and sun sensitivity, observing numerous significant signals (Figure 1). The strongest signals in the IRF4 region came from the association of rs12203592-T with presence of freckles, brown hair and high sensitivity of skin to sun exposure (Figure 1, Table S1). The second most significant variant association was for rs62389424-A with freckles (P = 1.1 × 10−81, nearly 40 orders of magnitude less significant than the corresponding signal from rs12203592-T (P = 2.0 × 10−120). The rs62389424 variant is correlated with rs12203592 with an r2 of 0.65. Two other SNPs in the region have r2 values in excess of 0.2 with rs12203592 and they also gave association signals with similar phenotypes (Table S2). When, in a multivariate analysis, the associations were conditioned on the effect of rs12203592, the signals from the three correlated SNPs became non-significant (Table S2). Indeed, once rs12203592 was taken into account in the multivariate analysis, no variant in the IRF4 region retained a significant signal after Bonferroni correction for the number of tests (Figure 1). We note in this context that the signal from the originally reported pigmentation variant at this locus, rs1540771-T (Sulem et al., 2007) was also captured by rs12203592 (Table S2). Thus, the associations at the IRF4 locus with freckles, hair color, sun sensitivity and eye color could all be accounted for by rs12203592 and no other variant detected by sequencing explains the effect with a similar level of significance.
Figure 1.
Association with pigmentation traits of variants in the IRF4 region (0–1Mb on chromosome 6) as determined by whole genome sequencing and imputation into SNP chip typed individuals. Black points show the unadjusted association values, yellow points show association values adjusted for the effect of rs12203592. The X axis is the genomic coordinate (hg18 Build 36). The Y axes are the association −log10 P values for Sun Sensitivity (yes/no, upper panel), Brown vs. Blond Hair Color (second panel), and Freckling (yes/no, third panel). The position of rs12203592 is indicated. The fourth panel shows the local recombination rates in cM/Mb. The lowest panel indicates the locations of known genes in the region, from UCSC. See also Table S1 and Figure S1.
Intron 4 of IRF4 contains a melanocyte-specific enhancer element
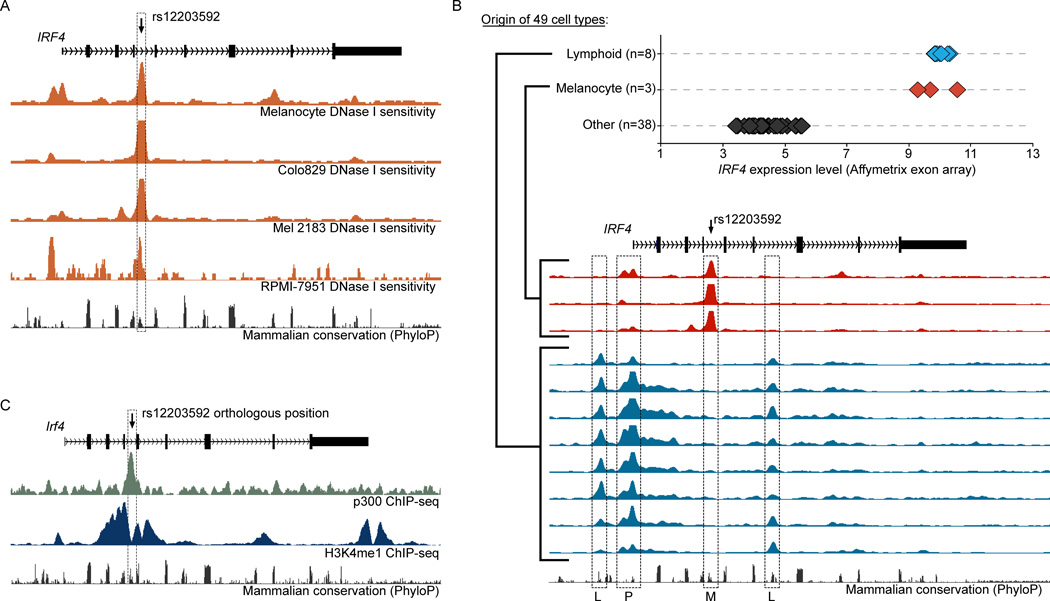
rs12203592 is located in intron 4 of the IRF4 gene, suggesting that this SNP might alter the function of a cis-regulatory element. Data from the ENCODE consortium (Thurman et al., 2012) shows that rs12203592 overlaps a peak of DNase I hypersensitivity (HS), a property of active regulatory elements, in human primary melanocytes and three human melanoma lines (Figure 2a). Moreover, rs12203592 does not overlap a DNase I HS peak in 158 of 165 (96%) non-melanocyte-derived cell types examined by ENCODE, suggesting that the regulatory activity in this region is specific to the melanocyte lineage. In a subset of 49 ENCODE cell types assayed by the Crawford group (Duke University) for both DNase I HS and global gene expression, IRF4 is expressed at high levels only in cell types of melanocyte (n=3) and lymphocyte origin (n=8). The corresponding patterns of DNase I HS in these cell types suggest that IRF4 expression in lymphocytes and melanocytes are directed by distinct sets of regulatory elements (Figure 2b). In addition, the position orthologous to rs12203592 in the mouse genome directly overlaps a melanocyte enhancer (Gorkin et al., 2012) since it is occupied by p300 and marked by H3K4me1 in the melanocyte line melan-Ink4a-Arf (Sviderskaya et al., 2002) (Figure 2c).
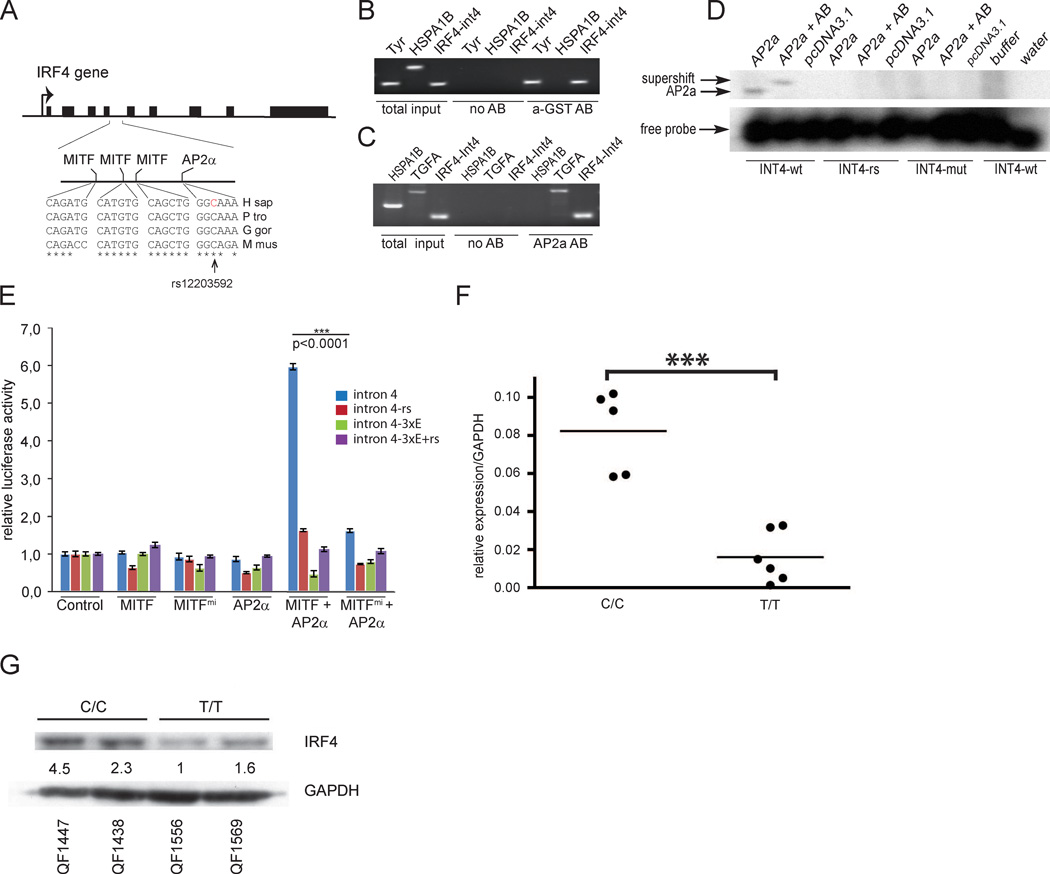
Figure 2. rs12203592 disrupts a conserved melanocyte enhancer at IRF4.
a) UCSC genome browser view of 25 KB region around IRF4 (hg19 coordinates chr6:388,750–413,750). DNase-seq signal (orange) in human epidermal melanocytes, Colo829, Mel 2183, and RPMI-7951 generated by ENCODE. b) Top: Graph shows IRF4 expression level as measured by ENCODE in 49 cell types (Duke Affymetrix exon arrays). Bottom: UCSC genome browser view of 25 KB region around IRF4 (hg19 coordinates chr6:388,750–413,750) in corresponding cell lines. Melanocyte-specific DNase HS peak overlapping rs12203592 labeled ‘M’; Lymphoid-specific DNase HS peaks labeled ‘L’; DNase HS overlapping the IRF4 promoter (labeled ‘P’). Cell types shown are (top to bottom) human epidermal melanocytes, Colo829, Mel 2183, GM12878, GM12891, GM12892, GM19238, GM19238, GM19240, GM18507, and CLL. c) UCSC genome browser view of 25 KB region around Irf4 (mm9 coordinates chr13:30,838,000–30,863,000). The ChIP-seq signals for EP300 (green) and H3K4me1 (blue) in the mouse melanocyte line melan-Ink4a-Arf from Gorkin et al., 2012.
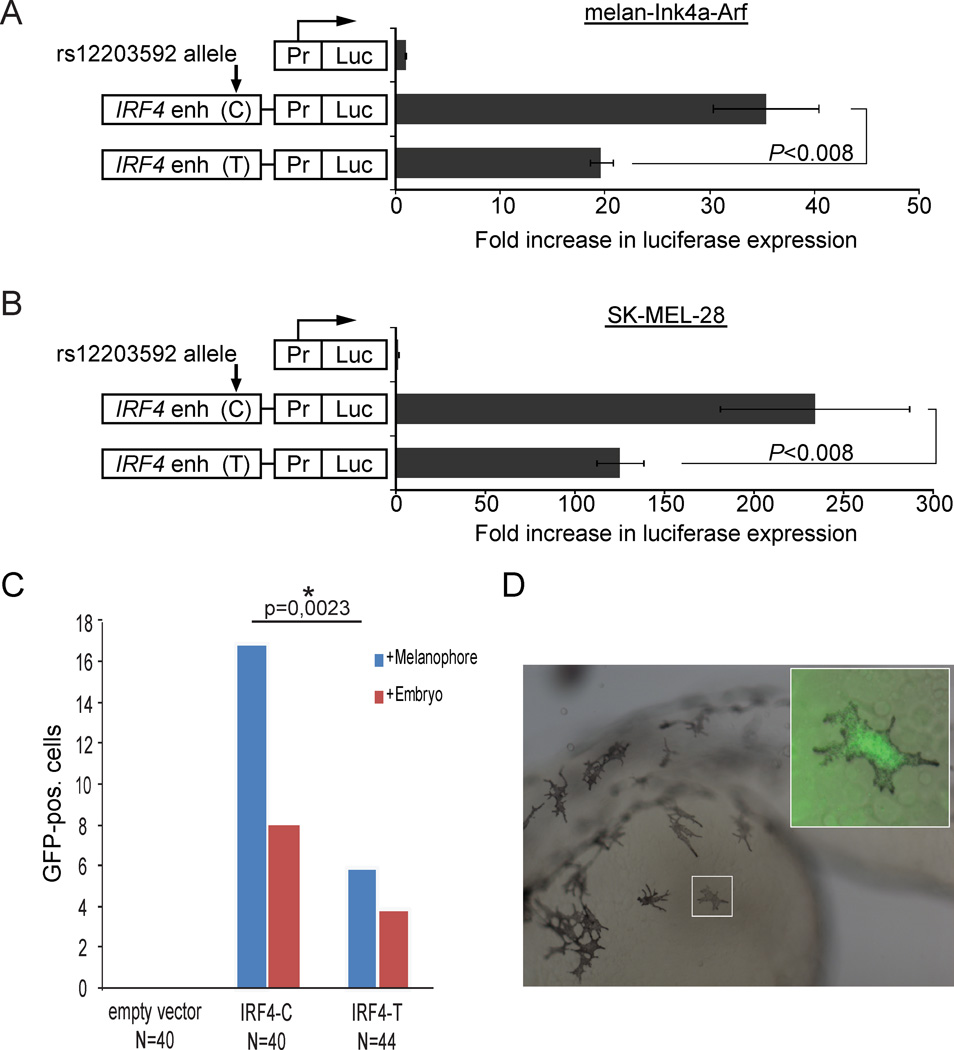
To confirm that the intronic sequence containing rs12203592 acts as a melanocyte-specific enhancer we sub-cloned a 450 bp fragment containing the rs12203592-C allele upstream of a minimal promoter driving luciferase, and assayed its activity in both the mouse melan-Ink4a-Arf and human SK-MEL-28 cell lines. The fragment showed strong enhancer activity, directing >35-fold higher luciferase expression than the minimal promoter alone in melan-Ink4a-Arf, and >200-fold in SK-MEL-28 (Figure 3a, b). We next asked whether the genotype at rs12203592 affects enhancer activity. In both melan-Ink4a-Arf and SK-MEL-28, the presence of the rs12203592-T allele significantly reduced the enhancer activity of the fragment (P<0.008) (Figure 3a, b).
Figure 3. Intron 4 of IRF4 drives expression in melanocytes.
a–b) Results of the luciferase assays in melan-Ink4a-Arf and SK-MEL-28 cells. Pr=promoter; Luc=Luciferase reporter gene; IRF4 enh= 450 bp fragment from the 4th intron of IRF4 containing rs12203592. ‘IRF4 enh’ fragments are identical except for the base at position rs12203592 as indicated (‘C’ is the ancestral allele, ‘T’ is the derived allele). The X-axis shows fold luciferase activity relative to the minimal promoter alone, which is normalized to 1. P-values calculated by Kolmogorov–Smirnov test. The result is also highly significant by other non-parametric tests (P=0.007937 by Wilcoxon Rank-Sum test for both melan-Ink4a-Arf and SK-MEL-28), and by the standard parametric t-test (P=0.001686 for melan-Ink4a-Arf; P=0.01032 for SK-MEL-28). Each bar represents the average of five biological replicates per reporter construct. Error bars represent standard deviation. c) Reporter constructs containing either the rs12203592-C version of the IRF4 intron 4 element (IRF4) or the rs12203592-T version (IRF4snp) upstream of the GFP reporter after injection into zebrafish. The numbers represent the number of melanophores (blue) and embryos (red) seen to be positive for GFP. The difference between the melanophores containing the wild-type allele of intron 4 of IRF4 compared to the mutant allele (rs12203592) is statistically significant (p=0,0023, unpaired t-test). d) Image showing a GFP positive zebrafish embryo with a GFP positive melanophore in the inset.
We further assayed the sequence containing rs12203592 in vivo using transgenic zebrafish. We engineered a vector containing the entire human IRF4 intron 4 sequence upstream of a minimal promoter and the gene encoding Green Fluorescent Protein (GFP). We created two versions of the vector, with either the T or C allele at rs12203592; as above, the remainder of the sequence was identical in both constructs. We injected these constructs, or a negative control construct lacking any human genomic sequence, into zebrafish embryos at the 2-cell stage, incubated them 48 hour post fertilization, and scored them for the presence of GFP-positive cells. In embryos injected with the negative control construct we did not detect GFP-positive melanocytes (0/40 embryos) (Figure 3c). In embryos injected with the IRF4 intron 4 reporter construct containing the ancestral rs12203592-C allele, we detected GFP-positive melanocytes in about 20% of embryos (8/40 embryos, 17 melanocytes total from the 8 embryos), consistent with the mosaicism expected in transient transgenic embryos (Figure 3c, d). By contrast, in embryos injected with the reporter containing rs12203592-T, only 9% of the embryos had detectable GFP-positive melanocytes (4/44 embryos, only 6 melanocytes total). The difference in the total number of melanocytes is statistically significant (p=0,0023, unpaired t-test). We did not detect expression in other tissues, beyond transient, scattered expression which was seen in all three constructs and has been reported by others (Bessa et al., 2009). We conclude that IRF4 intron 4 contains a melanocyte enhancer, likely directing IRF4 expression in these cells, and that the rs12203592-T allele reduces the activity of this enhancer.
MITF activates IRF4 gene expression
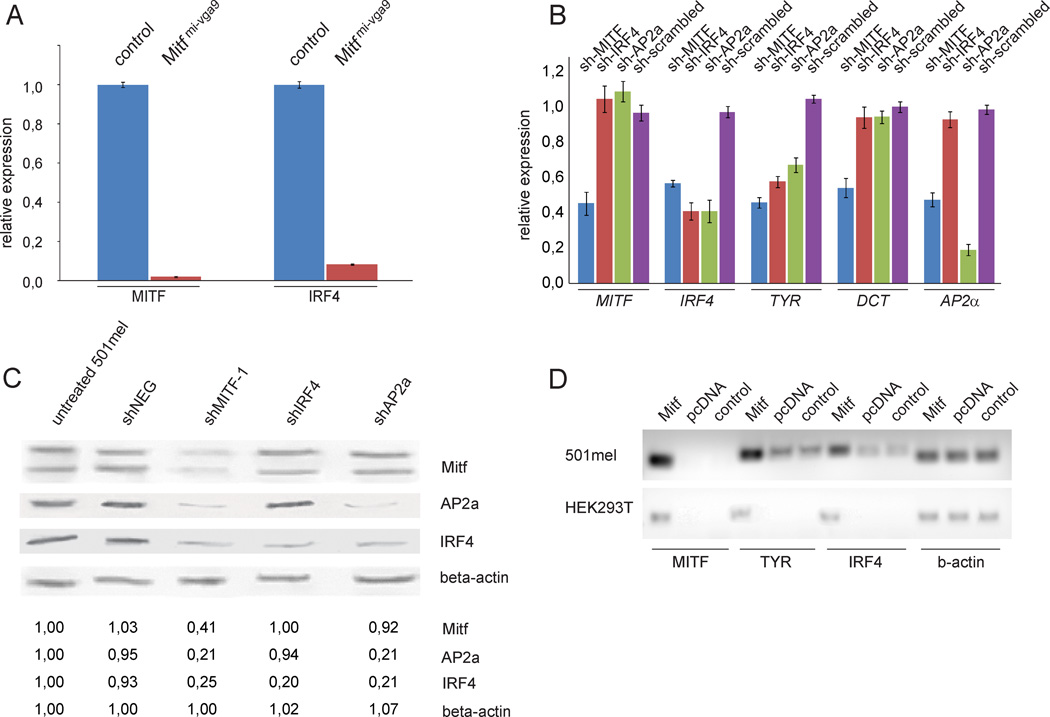
ChIP sequencing and gene expression studies have suggested that MITF may be involved in regulating IRF4 gene expression (Hoek et al., 2008, Strub et al., 2011). We analyzed Irf4 expression in mice lacking Mitf and focused on the heart because cells that normally express Mitf are still present in the heart of Mitfmi-vga9 homozygous mutants, whereas melanocytes are absent resulting in white coat color (Hodgkinson et al., 1993). Irf4 gene expression was dramatically reduced in Mitf mutants compared to wild type controls (Figure 4a) consistent with the possibility that Irf4 is regulated by MITF. To determine whether IRF4 is a target of MITF in the melanocyte lineage, we used shRNA to knock down MITF and IRF4 mRNAs in human 501mel melanoma cells which normally express these genes. Transfecting these cells with shRNA directed against MITF reduced MITF mRNA expression to 45% (Figure 4b) and MITF protein levels to 41% (Figures 4c and S2) of those seen in untreated cells or cells treated with a scrambled control shRNA. In cells treated with shMITF, the expression of IRF4 mRNA and protein were dramatically reduced (to 50% and 25%, respectively). shRNA against IRF4 resulted in reduction in IRF4 mRNA and protein expression to 40% and 20% of control levels, respectively, whereas MITF expression was unaffected (Figures 4c and S2). The expression of DCT (encoding Dopachrome tautomerase) and TYR, two known MITF target genes (Yasumoto et al., 2002; Yasumoto et al., 1994), was also reduced upon shMITF treatment whereas shIRF4 treatment only affected the expression of TYR (Figure 4b). TYR protein expression was reduced upon treatment with shMITF, shIRF4 and shAP2A, as well as with all shRNAs together (Figure S2). Knocking down TFAP2A did not affect MITF expression. We also tested this relationship in an overexpression assay. Untransfected 501mel melanoma cells express MITF but low levels of both TYR and IRF4 mRNAs whereas HEK293T cells do not express MITF endogenously and also lack expression of TYR and IRF4. In both cell lines, overexpression of mouse Mitf (mMitf) strongly induced the levels of TYR and IRF4 (Figure 4d). Analysis of the expression of MITF and IRF4 in 22 melanoma cell lines revealed a positive correlation between MITF and IRF4 (p=0.0003, Pearson correlation coefficient, Figure S3a). These results indicate that MITF directly or indirectly regulates IRF4 expression. They also suggest that TYR expression depends on IRF4.
Figure 4. MITF regulates expression of IRF4.
a) The expression of the Mitf and Irf4 mRNAs is reduced in hearts from Mitfmi-vga9 mutant mice, as determined by qPCR analysis. Data are represented as mean +/− SEM. b) Expression of the MITF, IRF4, TFAP2a, DCT and TYR genes upon treatment with shRNA against MITF, IRF4, TFAP2A and negative control in 501mel melanoma cells as determined by qPCR analysis. Data are represented as mean +/− SEM. c) Western blot showing expression of the MITF, TFAP2A, IRF4 and β-actin proteins in 501mel cells after shRNA treatment. Intensity quantification is relative to actin loading control. See also Figure S2. d) Overexpression of the mouse Mitf (mMitf) cDNA (expressed from pcDNA3.1) in 501mel melanoma cells and in HEK293T embryonic kidney cells affects expression of the TYR and IRF4 mRNAs as assayed by RT-qPCR, whereas β-actin expression is unchanged. mMitf was detected with species-specific primers which only recognize the mouse Mitf gene. See also Figure S3.
rs12203592 alters the function of a melanocyte enhancer in IRF4 through disruption of a TFAP2A binding site
The rs12203592 polymorphism is located 66 bp from 3 sites recently shown to be occupied by MITF in ChIP-sequencing studies (Figure 5a) (Strub et al., 2011). ChIP performed in 501mel and MITF-transfected 293T cells showed that MITF binds to intron 4 of IRF4 (Figure 5b). Interestingly, the rs12203592-T polymorphism occurs in a predicted binding site (GGCAAA) for TFAP2A (Do et al., 2010) which was recently shown to be involved in melanocyte differentiation in zebrafish (van Otterloo et al., 2010). To determine if TFAP2A can bind this sequence, we performed anti-TFAP2A ChIP in 501mel cells (homozygous for rs12203592-C) and showed that TFAP2A binds to the intron 4 element in the IRF4 gene (Figure 5c). Furthermore, gel shift assays showed that TFAP2A binds oligos carrying the TFAP2A motif from IRF4 intron 4 only when the oligos harbor the rs12203592-C allele. It did not bind the rs12203592-T variant, nor when the binding site was altered completely (Figure 5d). Addition of antibody resulted in a supershift only in presence of the wild-type oligo. Binding of MITF to the neighbouring MITF sites was not affected by the SNP (Figure S3b). These results show that TFAP2A binds the ancestral sequence (rs12203592-C) in intron 4 of IRF4 but fails to bind to the rs12203592-T variant.
Figure 5. MITF and TFAP2A affect IRF4 expression by binding regulatory elements in intron 4.
a) Schematic view of the IRF4 gene. The sequence shows comparison of MITF and TFAP2A binding sites in intron 4 of IRF4 between humans (H sap), gorilla (G gor), chimpanzee (P tro) and mouse (M mus). The location of the rs12203592 polymorphism is indicated. b) ChIP analysis of a GST-tagged MITF protein (a-GST AB) performed in 501mel cells. Primers specific for HSPA1B (neg. control), TYR (pos. control), and intron 4 of IRF4 are indicated. The GST-tagged Mitf only precipitated TYR and IRF4 sequences. c) ChIP analysis of TFAP2A in 501mel cells. PCR products specific for TGFA (pos. control), HSPA1B (neg. control) and intron 4 of IRF4 are shown. TFAP2A only precipitated TGFA and INT4 sequences. d) Gel shift analysis showing the binding of TFAP2A to the ancestral sequence (INT4-wt) in intron 4 of IRF4 (coordinates chr6: 396309–396333 in build GRCh37.p5 of the human genome) but not to the rs12203592-T sequence (INT4-RS) or to a completely mutated sequence (INT4-mut). e) Luciferase reporter assays performed in 501mel melanoma cells using intron 4 of IRF4 as a reporter. The ancestral IRF4 intron 4 sequence (intron 4, blue), the rs12203592-T polymorphic sequence (intron 4-rs), a sequence where all MITF binding sites were mutated (intron 4-3xE) and a sequence containing mutated MITF sites and the rs12203592-T polymorphism (intron 4-3xE+rs) were tested. The luciferase reporters were co-transfected with the wild type MITF and TFAP2A proteins and with a dominant negative version of MITF (MITFmi). Statistical analysis (p<0.0001) was done using unpaired t-test. Data are represented as mean +/− SEM. f) Cells homozygous for the ancestral allele (CC) express significantly higher levels of IRF4 than cells homozygous for the rs12203592-T polymorphism (TT). Fold expression data is represented relative to pooled mean TT expression +/− SEM. Statistical analysis was performed using analysis of variance (***, p < 0.001). g) A western blot showing expression of the IRF4 protein in melanocytes from CC and TT individuals. Intensity quantification is relative to GAPDH loading control. Note the decrease of basal IRF4 protein in the TT melanocyte cell lines. See also Figures S3–S5.
To determine if TFAP2A plays a role in the activity of the IRF4 enhancer, we performed reporter assays using intron 4 of IRF4 as regulatory element. 501mel melanoma cells were co-transfected with the reporter constructs and with plasmids encoding MITF, TFAP2A or both. Neither MITF nor TFAP2A alone elevated expression from this element above basal levels (Figure 5e). However, when both MITF and TFAP2A were expressed simultaneously, transcription was increased 6-fold (Figure 5e), suggesting that the proteins have cooperative effects on the activity of this enhancer. This is further supported by the shRNA experiments which showed that knocking down TFAP2A reduces IRF4 expression (Figure 4b and c). Removing all three MITF binding sites together (intron 4-3xE) lead to background-level expression (Figure 5e). Importantly, when the rs12203592-T variant was introduced into the reporter construct (intron 4-rs), the cooperation seen between MITF and TFAP2A was reduced to levels close to background (Figure 5e). This suggests that MITF requires TFAP2A in order to induce expression from the intron 4 element and that the rs12203592-T polymorphism abolishes the MITF-mediated activation.
To determine whether the rs12203592-T variant affects IRF4 expression in melanocytes, RNA and protein were isolated from melanocytes differentiated from human neonatal foreskin melanoblasts from Australian subjects (Cook et al., 2009). RT-qPCR showed a significantly lower (p<0,01, unpaired t-test) expression of IRF4 mRNA in cells homozygous for the rs12203592-T polymorphism than in cells homozygous for the ancestral C (Figure 5f, Figure S3c). IRF4 protein levels in cells from T/T individuals were considerably reduced compared to that of cells from C/C individuals (Figure 5g).
To determine whether the rs12203592 polymorphism affects expression of the transcript on the chromosome on which the variation resides we used an allele discrimination assay. We used RT-qPCR on human primary melanocytes heterozygous for the IRF4 3’UTR SNP rs872071-A/G. The rs872071-G allele resides 15kb distal to rs12203592 and is present on the haplotype on which the rs12203592-T allele arose, thus allowing for comparison of IRF4 allele specific expression levels, controlling for potential trans-factor variations that may contribute to expression level differences between individuals. We observed no difference in IRF4 allelic expression in rs12203592-C/C homozygous melanocytes, as represented by the rs872071-A/G allele ratio (Figure S4). In contrast, cells heterozygous for the rs12203592 C/T alleles exhibit a significantly higher rs872071-A/G allele ratio (p<0.01) consistent with reduced transcription occurring from IRF4 rs872071-G alleles in phase with rs12203592-T.
Several alternate products are made from the IRF4 gene, including transcripts which initiate in intron 4 (Figure S5a), leading to an mRNA containing internal ATGs (lacking Kozak consensus sequences) and the tentative production of a truncated protein lacking the N-terminal DNA-binding domain. To determine the major product in the melanocyte, RT-qPCR analysis with transcript specific primers was performed on RNA isolated from foreskin melanoblasts from Australian subjects. This showed that the major IRF4 product in melanocytes is the full length transcript; the smaller alternative product was barely detectable (Figure S5b). Similarly, in mouse melanoblasts isolated from E15.5 and E17.5 embryos and melanocytes from P1 and P7 neonatal pups (Debbache et al., 2012), the full-length Irf4 transcript was the only transcript detected at all developmental stages (Figure S6). We therefore conclude that the regulatory element in intron 4 affects the production of the full-length IRF4 transcript.
IRF4 and MITF cooperatively regulate TYR expression
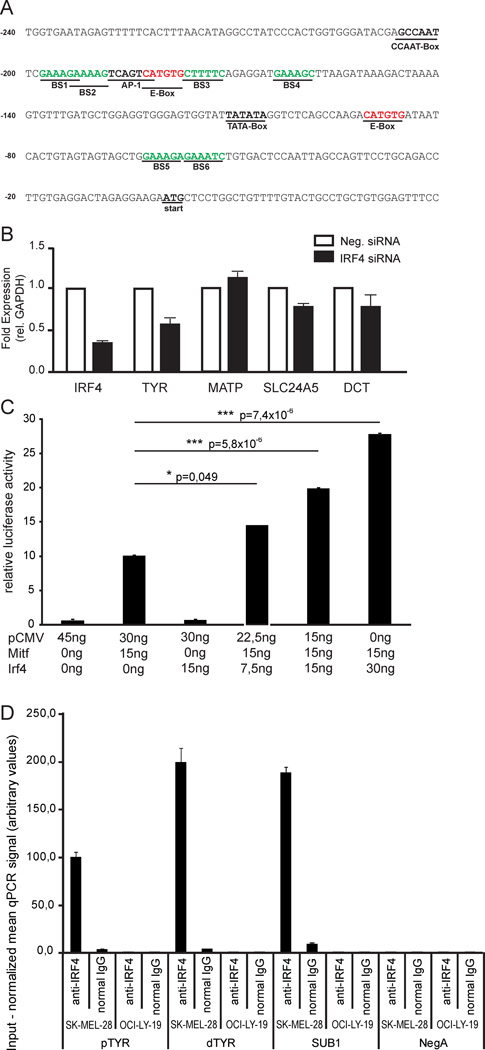
To determine how IRF4 levels might affect pigmentation we searched the promoter sequences of several pigmentation genes for the IRF4 binding motif. Several potential IRF4 binding motifs were found in the TYR promoter, on either side of known MITF binding sites previously shown to be essential for gene activation (Figure 6a) (Yasumoto et al., 1994; Bertolotto et al., 1996). To investigate the relationship between IRF4 and the expression of several pigmentation genes, two human melanocyte strains with high expression of IRF4 were independently transfected with siRNA targeting IRF4 for 48 hours, followed by total RNA extraction and RT-qPCR. These results combined show that expression of IRF4 was reduced on average to 40% of cells treated with negative control siRNA (Figure 6b). Interestingly, the expression of TYR was reduced to 50% compared to untreated control whereas the expression of the pigmentation genes SLC45A2 (MATP), SLC24A5 and DCT were largely unaffected (Figure 6b). Similarly, knockdown of IRF4 affected expression of TYR but not of DCT in 501mel melanoma cells (Figure 2b, c). This suggests that IRF4 is involved in regulating TYR expression but does not affect expression of SLC45A2 or SLC24A5. The expression of IRF4 correlates with the expression of TYR in 22 melanoma cell lines (p=0.0007, Pearson correlation coefficient, Figure S3a).
Figure 6. MITF and IRF4 regulate expression of TYR.
a) Schematic view of the human TYR promoter showing the sequence of the MITF and potential IRF4 binding sites. b) Expression of IRF4, TYR, MATP, SLC24A5 and DCT as determined by qPCR, after treatment with siIRF4 or control siRNAs. Fold expression of each gene is represented relative to cells transfected with negative siRNA (arbitrarily set to 1). Data represent the mean ± range of two experiments using two primary melanocytes (QF1438 and QF1424). c) Luciferase reporter assay using wild-type TYR promoter as reporter construct and co-transfected with constructs expressing the MITF and IRF4 proteins. Statistical analysis was performed using the t-test; data are represented as average with the standard deviation. d) IRF4 binds the TYR locus. Chromatin immunoprecipitation real time PCR (ChIP-qPCR) analysis of IRF4 binding at sites proximal (around transcription start site; pTYR) and more distal (~1800 bp upstream of transcription start site; dTYR) to TYR coding region and other control loci in SK-MEL-28 melanoma cell line and in negative control OCI-Ly-19, a germinal center type B-cell lymphoma (GCB-DLBCL) cell line previously shown to lack IRF4 expression (Yang et al., 2012). Primer pair amplifying a region upstream of the SUB1 locus was used as a positive control, since this region showed strong IRF4 binding both in multiple myeloma (Shaffer et al., 2008), and activated B-cell type B-cell lymphoma (Yang et al., 2012) cell lines. NegA is a region on chromosome 7, used as a negative control for IRF4 binding, due to lack of observable IRF4 binding in previous studies with multiple myeloma and ABC-DLBCL cell lines. Error bars depict standard error of the mean. See also Figure S7.
Co-transfection assays in HEK293T cells (which do not express MITF) showed that whereas MITF can activate expression of the TYR gene, the IRF4 protein is unable to do so on its own (Figure 6c). However, in the presence of both MITF and IRF4, cooperative effects were observed with increasing amounts of IRF4 (Figure 6c). A mutant MITF protein with defective DNA-binding ability resulted in less activation and reduced cooperativity (Figure S7a). Similar effects were observed when the E-box was mutated (Figure S7b), confirming that the cooperativity is mediated through MITF. Quantitative ChIP experiments in Skmel28 (Figure 6d) and Skmel5 (Figure S7c) melanoma cells showed that IRF4 binds the proximal TYR promoter as well as a more distant region.
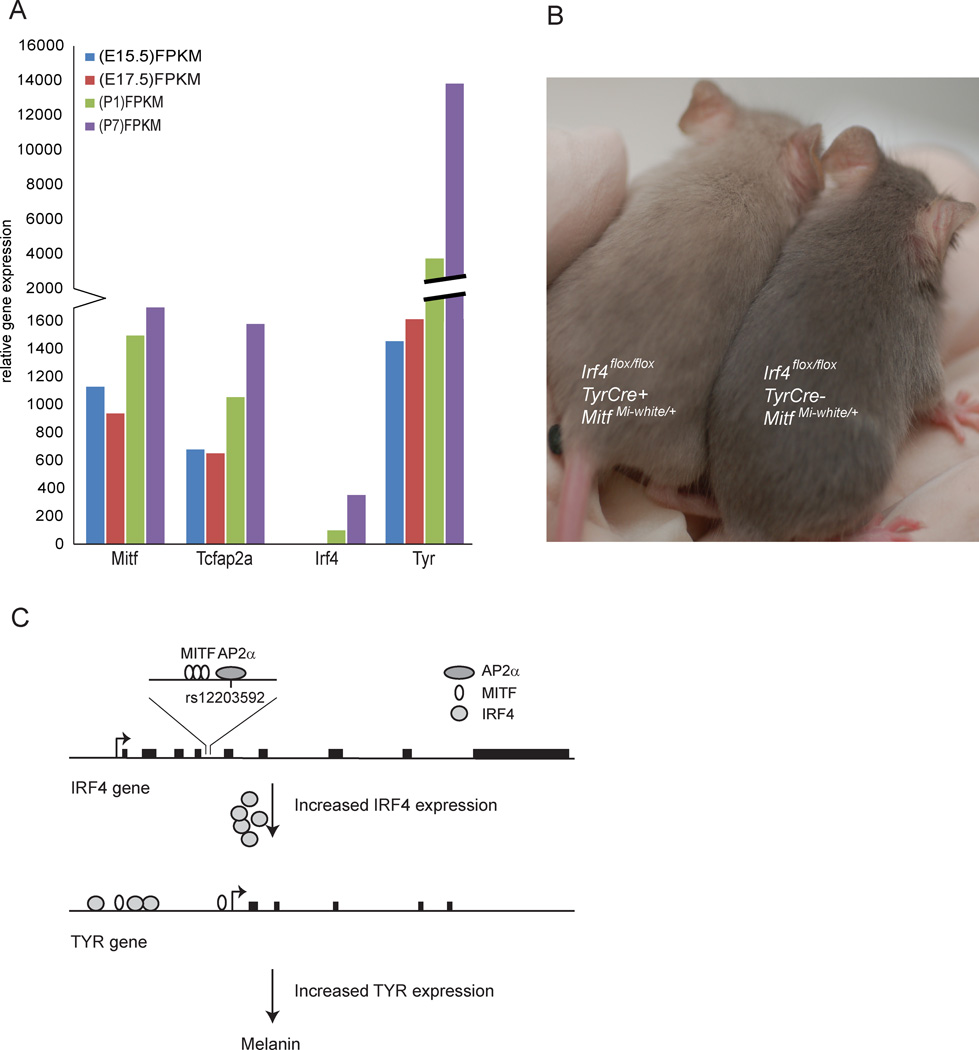
Irf4 affects pigmentation in mice
No pigment phenotypes were reported in mice with a targeted deletion of Irf4 (Mittrucker et al., 1997). We analyzed the expression of Mitf, Tfap2a, Irf4 and Tyr in melanoblasts isolated from E15.5, E17.5, P2 and P7 mouse embryos (Debbache et al., 2012). Mitf and Tfap2a are both expressed at the embryonic and postnatal stages whereas Irf4 is detected at a low level at the two postnatal stages (Figure 7a). Tyr is expressed at all stages but the level of expression increases at the postnatal stages and is ten-fold higher at P7 than at the earlier stages (Figure 7a). This expression pattern is consistent with our data that MITF and TFAP2A regulate IRF4 expression and MITF and IRF4 regulate TYR expression. Since early stages of development do not express Irf4, this also suggests that IRF4 is mostly involved in melanocyte differentiation. To determine if Irf4 plays a role in mouse melanocytes, mice carrying an Irf4-flox cassette (Irf4 f) (Klein et al., 2006) were crossed to mice expressing the Cre-recombinase under control of the Tyr promoter (Tyr::Cre) (Delmas et al., 2003). This led to the lack of Irf4 expression in Tyr expressing cells, but no apparent pigmentation phenotype, similar to the Irf4 null mice (Mittrucker et al., 1997). In order to test the relationship between Mitf and Irf4 in the mouse, we crossed the Tyr::Cre/0; Irf4 f/f mice to mice carrying the dominant negative allele MitfMi-White (MitfMi-Wh). Mice heterozygous for this allele (MitfMi-Wh/+) have a partial deficiency of Mitf and exhibit a stable, grey coat color with white belly spot. Interestingly, the Tyr::Cre/0; Irf4 f/f; MitfMi-Wh/+ mice showed distinctly lighter coat color than seen in MitfMi-Wh/+ mice which are negative for the Tyr-Cre transgene (Irf4 f/f; MitfMi-Wh/+) (Figure 7b). This confirms that IRF4 plays a role in mouse pigmentation, like in humans, and that the effects of Irf4 in the mouse also depend on Mitf. In humans, the IRF4 variant rs12203592-T is linked to brown hair color (Sulem et al., 2007) (Table S1). Thus, reduced IRF4 expression alone is sufficient for lighter hair color in humans whereas in mice MITF function also needs to be reduced to see effects on pigmentation. This suggests that there may be species-specific differences between mice and humans in terms of requirement for MITF and IRF4 in melanocytes of the hair.
Figure 7. IRF4 is involved in regulating pigmentation in mice.
a) The expression plot shows presence and clear differences in the expression of Mitf, Tcfap2a, Irf4 and Tyr at different developmental time points, E15.5, E17.5, P1 and P7, measured in FPKM. Indicated expression levels of a transcript are proportional to the number of reads sequenced from that transcript after normalizing for respective transcript’s length. RNA from E15.5 embryos is shown in blue bars, E17.5 embryos in red, P1 pups in green and P7 in violet. b) Mice where Irf4 has been conditionally knocked out in heterozygotes for the semidominant MitfMi-Wh mutation. MitfMi-Wh heterozygotes which are simultaneously lacking Irf4 show lighter coat color (left) than mice which are wild type for Irf4 (right). c) Model depicting the relationship between MITF, TFAP2A and IRF4 in melanocytes and their effects on the expression of TYR. MITF and TFAP2A bind together to the intron 4 element in the IRF4 gene and regulate the expression of IRF4 from the upstream promoter. MITF and IRF4 then bind to and activate expression of TYR, leading to normal pigmentation. The rs12203592-T polymorphism leads to reduced IRF4 activation and thus reduced TYR expression which consequently leads to sun sensitivity and blue eyes.
Discussion
Genome-wide association studies have yielded a long list of sequence variants correlated with a wide range of human phenotypes. Most of these variants are located in non-coding regions of the genome, and do not alter the amino-acid sequence of proteins or have known regulatory functions. Two steps are essential in order to characterize the biology behind such associations. First, the association signals must be fine-mapped using full sequence data. Second, the biological processes involved must be unravelled experimentally. In this study, we have taken both steps to shed light on the functional consequences of the IRF4 non-coding variant rs12203592 on pigmentation.
Our work proposes a model describing how IRF4 acts in cooperation with MITF to influence pigmentation phenotypes (Figure 7c). In melanocytes, MITF and TFAP2A cooperatively activate IRF4 expression through an enhancer element in intron 4. In turn, IRF4 and MITF cooperate to activate transcription of TYR. The rs12203592-T allele reduces binding of TFAP2A to its cognate site in intron 4 of IRF4, thereby suppressing the induction of IRF4 expression and, as a consequence, impairing the cooperative induction of TYR. This leads to the pigmentation phenotypes associated with the rs12203592-T allele. The effects of this variant must be cell-type specific since Do et al. (2010) have shown that it leads to increased IRF4 promoter activity in Burkitt lymphoma B cells (Raji), HEK293T human embryonic kidney cells and the NCI-H295R human adrenal cells. Consistent with our results, they showed that TFAP2A binds with higher affinity to the C allele than to the T allele (Do et al., 2010). Thus, the cell-type specific effects must be mediated by transcription factors other than TFAP2A.
In humans, genetic variants in different genes have particular effects on pigmentation traits. For example, MC1R and ASIP variants have major effects on freckling, sun sensitivity and red hair, whereas they have relatively little effect on eye color (Sulem et al., 2007; Sulem et al., 2008). The rs12203592-T variant of IRF4 also has major effects on freckling and sun sensitivity, but is associated with brown (over blond) hair and has a notable effect on eye color (Table S1). The results presented here suggest that IRF4, like MC1R and ASIP, exerts its major effects on pigmentation traits through the master regulator MITF. However some divergence in pathways must occur, in order to account for the differences in effects on pigmentation traits. We have shown that TYR is a target of MITF-IRF4 cooperative activation. However, TYRP1 and DCT are targets of MITF activation that do not appear to be responsive to IRF4. Clearly, the MITF-IRF4 cooperation only occurs in a subset of MITF responsive genes. A broader search of MITF target genes for potential IRF4 responsiveness might shed light on the pattern of pigmentation traits affected by IRF4.
Freckles represent clusters of concentrated melanin in the skin, without an increase in melanocyte numbers. Freckles are either light brown or red but usually become darker and more visible upon exposure to sunlight. GWAS studies have shown that variations in MC1R, ASIP, TYR, and BNC2, in addition to IRF4, are associated with freckles (Sulem et al., 2007; Sulem et al., 2008; Eriksson et al., 2010). Mutations in the MC1R gene have been highly associated with the formation of freckles (Bastiaens et al., 2001). In addition to regulating TYR and IRF4 gene expression, MITF is also known to regulate expression of MC1R (Adachi et al., 2000; Smith et al., 2001; Aoki and Moro, 2002). This suggests the possibility that, like with TYR, IRF4 cooperates with MITF in regulating expression of MC1R in melanocytes. Indeed, ChIP-seq studies show that MITF binds MC1R regulatory sequences (Strub et al., 2011).
The effects of both TFAP2A and IRF4 are mediated through MITF which is needed both for the TFAP2A-mediated activation of IRF4 and the IRF4-mediated activation of TYR. ChIP-seq studies suggest that MITF regulates TFAP2A as well as IRF4 (Strub et al., 2011). The cooperative effects of MITF and IRF4 suggest that these proteins might physically interact on the TYR promoter. TFAP2A is expressed in a variety of both mesenchymal and epithelial cell types. It is not known how MITF and TFAP2A interact in the regulation of IRF4. Since the binding sites are physically close to each other, it is possible that both proteins are part of a larger complex formed at the site. Together, these data suggest that transcription factors which are expressed in multiple cell types can form cell-type specific complexes to govern differentiation.
Several studies have shown that polymorphisms identified as susceptibility loci in GWAS studies affect the binding of particular transcription factors, in some cases leading to differences in expression of a nearby target gene (Meyer et al., 2008; Rahimov et al., 2008; Pomerantz et al., 2009; Schodel et al., 2012; Tuupanen et al., 2009). However, the effects of the gene expression differences have not been characterized and, more importantly, the biological effects on the phenotype are only implied in these studies. A recent example is provided by the rs12913832 polymorphism in intron 86 of the HERC2 gene which shows a strong association with human eye color and pigmentation. Since the pigment gene OCA2 is located 21kb downstream of HERC2 it has been speculated that this polymorphism is located in a distal regulatory element of OCA2 and may affect binding of the transcription factors HLTF, LEF1 and MITF (Visser et al., 2012). This results in the formation of a loop between the HERC2 enhancer and the OCA2 promoter, thus affecting OCA2 expression. The rs12913832-C allele prevents HLTF from binding to the element, thus reducing loop formation. The regulation of OCA2 expression may be more complex however, with separate regulatory elements responsible for eye and skin color (Beleza et al., 2013). Here we have gone one step further and shown the effects of transcription factor binding on gene expression and how this gene expression difference leads to effects on human pigmentation.
Materials and Methods
Whole genome sequence analysis
Icelandic population samples and phenotypic information
Icelandic adults were recruited as cases, family members or controls thorough a series of cardiovascular, oncology, neurology and metabolic studies conducted by deCODE Genetics. Blood samples were taken for DNA isolation upon recruitment. The studies were approved by the National Bioethics Committee of Iceland and the Icelandic Data Protection Commission. Personal identifiers associated with phenotypic information, blood samples and genotypes were encrypted using a third party encryption system. Each participant completed a questionnaire that included questions about natural eye color categories (blue/gray, green or black/brown), natural hair color categories (red/reddish, blond, dark blond/light brown or brown/black) and the presence of freckles at any time. Sun sensitivity was assessed using the Fitzpatrick skin-type score (Fitzpatrick, 1988), where the lowest score (I) represents very fair skin that is sensitive to UVR and the highest score represented in native Icelanders (IV) represents dark skin that tans rather than burns in reaction to UVR exposure. Individuals scoring I or II were classified as being sensitive to sun whereas individuals scoring III and IV were classified as not being sensitive to sun.
SNP chip genotyping
The Icelandic chip-typed samples were genotyped with Illumina Human Hap300, HumanHap CNV370, HumanHap 610, 1M, Omni1-Quad, Omni 2.5 or Omni Express bead chips at deCODE Genetics. SNPs were excluded if they had (i) yield less than 95%, (ii) minor allele frequency less than 1% in the population (iii) significant deviation from Hardy-Weinberg equilibrium in the controls (P < 0.001), (iv) if they produced and excessive inheritance error rate (over 0.001), (v) if there was a substantial difference in allele frequency between the chip types (SNPs from a single chip platform were excluded if that resolved all differences, but otherwise from all chip platforms). All samples with a call rate below 97% were excluded from the analysis. For the HumanHap series of chips 308,840 SNPs were used for long rang phasing, whereas for the Omni series of chips 642,079 SNPs were included. The final set of SNPs used for long-range phasing was composed of 785,863 SNPs.
Whole genome sequencing and imputation
Sequence data was obtained from about 2,230 Icelanders using methods described previously (Holm et al., 2011). Sequencing was carried out to an average depth of at least 10×. This resulted in the identification of approximately 38.5 million SNPs and small indels that were available for imputation. Long range phasing, imputation and association testing (by logistic regression) was done as described previously (Kong et al., 2008; Kong et al., 2009; Holm et al., 2011; Stacey et al., 2011). For conditional analysis of the IRF4 region the rs12203592-T allele count of each individual was given as a covariate in the logistic regression.
Supplementary Material
Acknowledgements
We thank Thorunn Rafnar, Magnus Karl Magnusson, Unnur Thorsteinsdottir, Colin R. Goding and Thorarinn Gudjonsson for critical comments on the manuscript. This work was supported by grants from NIH (5R01 AR043369-16), Melanoma Research Alliance, Dr. Miriam and Sheldon Adelson Medical Research Foundation, US-Israel Binational Science Foundation (to DEF), from Ligue Nationale Contre le Cancer (Equipe labellisée) and INCa (to LL), from the Icelandic Research Fund the Research Fund of the University of Iceland (to ES), from the Haskolasjodur Student Fund (ES, CP and CG), from the Jules Verne Fund (ES and LL), by the National Institute of General Medical Sciences (GM071648) and National Institute of Neurological Disease and Stroke (NS062972) to ASM, by the National Human Genome Research Institute's (NHGRI) Intramural Research Program (WJP, SL) and by an NSF Graduate Research Fellowship to DUG.
Footnotes
Declaration of Contribution
ES conceived and designed the study; SNS, AH and KS analyzed whole genome sequence data and the frequency of IRF4 variants, AMM, RSQK, AGS and RAS characterized expression of IRF4 and other genes in cultured human melanocytes, DUG, ASM, SKL, DRA and WJP identified and characterized the enhancer element in intron 4 of IRF4 and performed the allele-specific expression analysis, EVO and RC performed experiments in zebrafish, CP analyzed the effects of MITF and TFAP2A on regulation of IRF4 and other target genes and CG, MHO and KB determined the involvement of IRF4 and MITF in TYR regulation, EM, US and NCTE performed ChIP on the TYR promoter, PJM, TG, MRZ, SRD, PSM and GM analyzed expression of MITF, TFAP2A, IRF4 and TYR in mouse melanocytes and VP and LL in human melanoma cells. KCR, LL and DEF characterized the interaction between MITF and IRF4 in mouse pigmentation. CP and ES wrote the manuscript. All authors approved the final version of the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Adachi S, Morii E, Kim D, Ogihara H, Jippo T, Ito A, Lee YM, Kitamura Y. Involvement of mi-transcription factor in expression of alpha-melanocyte-stimulating hormone receptor in cultured mast cells of mice. Journal of immunology. 2000;164:855–860. doi: 10.4049/jimmunol.164.2.855. [DOI] [PubMed] [Google Scholar]
- Aoki H, Moro O. Involvement of microphthalmia-associated transcription factor (MITF) in expression of human melanocortin-1 receptor (MC1R) Life Sci. 2002;71:2171–2179. doi: 10.1016/s0024-3205(02)01996-3. [DOI] [PubMed] [Google Scholar]
- Bastiaens M, ter Huurne J, Gruis N, Bergman W, Westendorp R, Vermeer BJ, Bouwes Bavinck JN. The melanocortin-1-receptor gene is the major freckle gene. Hum Mol Genet. 2001;10:1701–1708. doi: 10.1093/hmg/10.16.1701. [DOI] [PubMed] [Google Scholar]
- Beleza S, Johnson NA, Candille SI, Absher DM, Coram MA, Lopes J, Campos J, Araujo II, Anderson TM, Vilhjalmsson BJ, et al. Genetic architecture of skin and eye color in an African-European admixed population. PLoS Genet. 2013;9:e1003372. doi: 10.1371/journal.pgen.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto C, Bille K, Ortonne JP, Ballotti R. Regulation of tyrosinase gene expression by cAMP in B16 melanoma cells involves two CATGTG motifs surrounding the TATA box: implication of the microphthalmia gene product. J Cell Biol. 1996;134:747–755. doi: 10.1083/jcb.134.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernandez-Minan A, Naranjo S, Fernandez A, Montoliu L, Akalin A, Lenhard B, Casares F, et al. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 2009;238:2409–2417. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- Cook AL, Chen W, Thurber AE, Smit DJ, Smith AG, Bladen TG, Brown DL, Duffy DL, Pastorino L, Bianchi-Scarra G, et al. Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. J Invest Dermatol. 2009;129:392–405. doi: 10.1038/jid.2008.211. [DOI] [PubMed] [Google Scholar]
- Debbache J, Zaidi MR, Davis S, Guo T, Bismuth K, Wang X, Skuntz S, Maric D, Pickel J, Meltzer P, et al. In vivo role of alternative splicing and serine phosphorylation of the microphthalmia-associated transcription factor. Genetics. 2012;191:133–144. doi: 10.1534/genetics.111.135996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Martinozzi S, Bourgeois Y, Holzenberger M, Larue L. Cre-mediated recombination in the skin melanocyte lineage. Genesis. 2003;36:73–80. doi: 10.1002/gene.10197. [DOI] [PubMed] [Google Scholar]
- Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, Sullivan K, Vijayakrishnan J, Wang Y, Pittman AM, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nature genetics. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- Do TN, Ucisik-Akkaya E, Davis CF, Morrison BA, Dorak MT. An intronic polymorphism of IRF4 gene influences gene transcription in vitro and shows a risk association with childhood acute lymphoblastic leukemia in males. Biochim Biophys Acta. 2010;1802:292–300. doi: 10.1016/j.bbadis.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, Zhernakova A, Heap GA, Adany R, Aromaa A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nature genetics. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy DL, Iles MM, Glass D, Zhu G, Barrett JH, Hoiom V, Zhao ZZ, Sturm RA, Soranzo N, Hammond C, et al. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am J Hum Genet. 2010;87:6–16. doi: 10.1016/j.ajhg.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. Transcriptional control of adipose lipid handling by IRF4. Cell metabolism. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, Saxonov S, Avey L, Wojcicki A, Pe'er I, Mountain J. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS genetics. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante CR, Yie J, Thanos D, Aggarwal AK. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB. The validity and practicality of sun-reactive skin types I through VI. Archives of dermatology. 1988;124:869–871. doi: 10.1001/archderm.124.6.869. [DOI] [PubMed] [Google Scholar]
- Gorkin DU, Lee D, Reed X, Fletez-Brant C, Bessling SL, Loftus SK, Beer MA, Pavan WJ, McCallion AS. Integration of ChIP-seq and machine learning reveals enhancers and a predictive regulatory sequence vocabulary in melanocytes. Genome research. 2012 doi: 10.1101/gr.139360.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A, Mittrucker HW, Nicholl J, Suzuki A, Chung S, Antonio L, Suggs S, Sutherland GR, Siderovski DP, Mak TW. Cloning of human lymphocyte-specific interferon regulatory factor (hLSIRF/hIRF4) and mapping of the gene to 6p23-p25. Genomics. 1996;37:229–233. doi: 10.1006/geno.1996.0547. [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, Hankinson SE, Hu FB, Duffy DL, Zhao ZZ, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS genetics. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, Rademakers R, de Silva R, Litvan I, Riley DE, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nature genetics. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, Magnusson OT, Helgason A, Saemundsdottir J, Gylfason A, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nature genetics. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S, Rao PH, Butler M, Corradini P, Boccadoro M, Klein B, Chaganti RS, Dalla-Favera R. Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nature genetics. 1997;17:226–230. doi: 10.1038/ng1097-226. [DOI] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Kong A, Masson G, Frigge ML, Gylfason A, Zusmanovich P, Thorleifsson G, Olason PI, Ingason A, Steinberg S, Rafnar T, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nature genetics. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S, Jonasdottir A, Sigurdsson A, Kristinsson KT, Jonasdottir A, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KB, Maia AT, O'Reilly M, Teschendorff AE, Chin SF, Caldas C, Ponder BA. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS biology. 2008;6:e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittrucker HW, Matsuyama T, Grossman A, Kundig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89:744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodel J, Bardella C, Sciesielski LK, Brown JM, Pugh CW, Buckle V, Tomlinson IP, Ratcliffe PJ, Mole DR. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet. 2012;44:420–425. doi: 10.1038/ng.2204. S421–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Box NF, Marks LH, Chen W, Smit DJ, Wyeth JR, Huttley GA, Easteal S, Sturm RA. The human melanocortin-1 receptor locus: analysis of transcription unit, locus polymorphism and haplotype evolution. Gene. 2001;281:81–94. doi: 10.1016/s0378-1119(01)00791-0. [DOI] [PubMed] [Google Scholar]
- Stacey SN, Sulem P, Jonasdottir A, Masson G, Gudmundsson J, Gudbjartsson DF, Magnusson OT, Gudjonsson SA, Sigurgeirsson B, Thorisdottir K, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nature genetics. 2011;43:1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the Microphthalmia Transcription Factor Network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Strub T, Giuliano S, Ye T, Bonet C, Keime C, Kobi D, Le Gras S, Cormont M, Ballotti R, Bertolotto C, et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene. 2011;30:2319–2332. doi: 10.1038/onc.2010.612. [DOI] [PubMed] [Google Scholar]
- Sturm RA. Molecular genetics of human pigmentation diversity. Human molecular genetics. 2009;18:R9–R17. doi: 10.1093/hmg/ddp003. [DOI] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Jakobsdottir M, Steinberg S, Gudjonsson SA, Palsson A, Thorleifsson G, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, Manolescu A, Karason A, Palsson A, Thorleifsson G, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- Sundram U, Harvell JD, Rouse RV, Natkunam Y. Expression of the B-cell proliferation marker MUM1 by melanocytic lesions and comparison with S100, gp100 (HMB45), and MelanA. Mod Pathol. 2003;16:802–810. doi: 10.1097/01.MP.0000081726.49886.CF. [DOI] [PubMed] [Google Scholar]
- Sviderskaya EV, Hill SP, Evans-Whipp TJ, Chin L, Orlow SJ, Easty DJ, Cheong SC, Beach D, DePinho RA, Bennett DC. p16(Ink4a) in melanocyte senescence and differentiation. Journal of the National Cancer Institute. 2002;94:446–454. doi: 10.1093/jnci/94.6.446. [DOI] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi K, Iida S, Inagaki H, Kato M, Hayami Y, Hanamura I, Miura K, Harada S, Kikuchi M, Komatsu H, et al. MUM1/IRF4 expression as a frequent event in mature lymphoid malignancies. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2000;14:449–456. doi: 10.1038/sj.leu.2401696. [DOI] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- Van Otterloo E, Li W, Bonde G, Day KM, Hsu MY, Cornell RA. Differentiation of zebrafish melanophores depends on transcription factors AP2 alpha and AP2 epsilon. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome research. 2012;22:446–455. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Shaffer AL, 3rd, Emre NC, Ceribelli M, Zhang M, Wright G, Xiao W, Powell J, Platig J, Kohlhammer H, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer cell. 2012;21:723–737. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto K, Takeda K, Saito H, Watanabe K, Takahashi K, Shibahara S. Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. Embo J. 2002;21:2703–2714. doi: 10.1093/emboj/21.11.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Molecular and cellular biology. 1994;14:8058–8070. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.