Abstract
Core 3 derived glycans, a major type of O-glycan expressed by normal epithelial cells of the gastrointestinal tract, are downregulated during malignancy, because of loss of expression of functional β3-N-acetylglucosaminyltransferase-6 (core 3 synthase). We investigated the expression of core 3 synthase in normal pancreas and pancreatic cancer and evaluated the biological effects of re-expressing core 3 synthase in pancreatic cancer cells that had lost expression. We determined that pancreatic tumors and tumor cell lines have lost expression of core 3 synthase. We therefore re-expressed in human pancreatic cancer cells (Capan-2 and FG) to investigate the contribution of core 3 glycans to malignant progression. Pancreatic cancer cells expressing core 3 synthase showed reduced in vitro cell proliferation, migration and invasion compared with vector control cells. Expression of core 3 O-glycans induced altered expression of β1 integrin, decreased activation of focal adhesion kinase, led to the down regulation of expression of several genes including REG1α and FGFR3, and altered lamellipodia formation. The addition of a GlcNAc residue by core 3 synthase leads to the extension of the tumor associated Tn structure on MUC1. Orthotopic injection of FG cells expressing core 3 synthase into the pancreas of nude mice produced significantly smaller tumors and decreased metastasis to the surrounding tissues compared to vector control FG cells. These findings indicate that expression of core 3 derived O-glycans in pancreatic cancer cells suppresses tumor growth and metastasis through modulation of glycosylation of mucins and other cell surface and extracellular matrix proteins.
Keywords: pancreatic cancer, O-glycan, core 3 synthase, MUC1, α2β1 integrin
Introduction
Oligosaccharide modifications influence the activity of proteins that mediate oncogenic, tumor suppressor, and metastatic cellular activities and thereby dramatically affect the progression and biological properties of tumors. The regulation of carbohydrate chain modification to proteins during the development and progression of cancer is in part mediated by differential expression (or epigenetic or mutational inactivation) of glycosyltransferases that create oligosaccharide modifications. Aberrant expression of carbohydrate chains, such as Tn and sialyl Tn (STn), which result from truncated synthesis of structures produced by mucin type O-glycosylation, are among the most tumor-specific alterations observed in adenocarcinomas as compared normal cells. These and other aberrant glycan modifications are believed to contribute significantly to tumor progression, though the underlying mechanisms are poorly understood. Mucin type O-glycosylation is initiated by the GALNT (UDP-GalNAc: polypeptide GalNAc transferases) family of glycosyltransferases, whose members transfer N-Acetylgalactosamine (GalNAc) to serine or threonine residues on protein backbones to form the Tn epitope (GalNAcα-Ser/Thr). 1, 2 Somatic and germline mutations in polypeptide GalNAc transferases have been implicated in cancer progression.3 These O-glycans are sequentially extended by a series of enzymatic reactions that are classified into four major groups (core 1, core 2, core 3 and core 4) based on the addition of different carbohydrate residues to GalNAcα-Ser/Thr.4 Changes in mucin-type glycosylation occur in both the terminal carbohydrate chains and the core structures of O-glycans leading to a heterogeneous group of glycan structures in cancer. Production of Tn or STn structures on glycoproteins as is observed in cancer requires the failure of extension of the GalNAc residue by either core 1 or core 3 enzymes. This can occur through loss of expression or inactivating mutations in these glycosyltransferases or associated factors, such as the Cosmc protein that is a required molecular chaperone for core 1 synthase activity.4 It is likely that other factors contribute to the creation of these truncated structures.5 Core 2 O-glycans are overexpressed in pancreatic, lung, colon, breast, and prostate cancers; however, core 3 and core 4 structures are dramatically down regulated in gastric and colorectal carcinomas.6–12 Core 3 structures and enzyme activity have not been widely investigated in pancreatic cancer, which is the subject of this report.
Core 3 synthase (β3GnT-6) adds β1-3 linked N-Acetylglucosamine (GlcNAc) to GalNAc at the reducing saccharide terminus to form core 3 structures (GlcNAcβ1-3GalNAcα-Ser/Thr) on glycoproteins. Human core 3 synthase is highly expressed in normal stomach, colon and small intestine13, but is down regulated in gastric and colon carcinoma.11, 14, 15 Ectopic expression of core 3 synthase in human fibrosarcoma cell lines significantly reduced lung tumor foci in mice after intravenous injection.11 Increased susceptibility to experimentally induced inflammatory diseases and colorectal tumors were also observed in mice lacking core 3 synthase.16 Recently, Lee et al. showed that forced expression of core 3 synthase in human prostate cancer cells suppressed tumor formation and metastasis to surrounding tissues. This suppression was mediated in part through down-regulation of α2β1 integrin, an extracellular matrix protein that plays an essential role in cell-cell adhesion by binding to collagen.17 In pancreatic cancer, α2β1 integrin expression induces a malignant phenotype through interaction with type I collagen.18 α2β1 integrin also possesses both N- and O-glycosylation sites, leading to the possibility that alterations in the O-glycan structures affect pancreatic cancer growth and metastasis.17, 19
MUC1, a type I transmembrane protein comprised primarily of a large extracellular domain and a differentially O-glycosylated variable number of tandem repeat rich in serine, threonine, and proline residues, contributes to the invasive and metastatic potential of pancreatic cancer.20 Differential glycosylation of MUC1 is an important modulator of MUC1 function and may also regulate protein stability and subcellular localization.21 Park et al. showed that differential O-glycosylation of MUC1 by GalNAc-T6 promotes mammary carcinogenesis through stabilization of MUC1 proteins.22 Recently, Wandall et al. detected auto-antibodies against Tn or sialyl Tn (STn) epitope on MUC1 in ovarian, breast and prostate cancer patient sera, suggesting these epitopes might play a critical role in carcinogenesis.23 Expression of Tn or STn epitope on MUC1 is also observed in pancreatic cancer tissues (unpublished data). These reports indicate that altered expression and glycosylation of MUC1 can promote tumor growth and metastasis; however, it is currently not known whether changes in O-glycan structure on MUC1 that result from the activity of core 3 synthase affect the capacity of MUC1 to modulate the progression of pancreatic cancer.
In this study we used two human pancreatic cell lines, Capan-2 and FG, to investigate the role of core 3 O-glycans in tumor formation and metastasis. We show that ectopic expression of core 3 synthase resulted in reduced cell proliferation and reduced in vitro migration and invasion compared to vector control cells. Increased core 3 synthase expression in FG cells resulted in the loss of Tn epitope on MUC1, deregulation of the α2β1 integrin complex, and decreased activation of focal adhesion kinase (FAK) compared to vector control cells, suggesting the addition of core 3 derived O-glycans to MUC1 and α2β1 integrin can inhibit pancreatic tumor progression to metastasis.
Materials and Methods
Cell lines and cell culture
Cell lines Capan-2, FG, Colo357, Panc1, Panc1-MUC1 and HT293A were maintained at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone, Logan, UT, USA ) containing 10% heat inactivated fetal bovine serum (HIFBS) (Valley Biomedical Inc., Winchester, VA, USA), 100 µg/mL streptomycin and 100 units/mL penicillin (Mediatech Inc., Manassas, VA, USA).
Human samples
Human normal pancreas (NP), primary pancreatic adenocarcinoma (PCa), matched liver metastatic (L.Mets) tumor tissues (from two different patients) and normal stomach tissue were acquired from the University of Nebraska Medical Center tissue bank.
Reverse transcriptase PCR
Total RNA was isolated from tissues of normal pancreas, pancreatic cancer, liver metastases and from the pancreatic cancer cell lines, Capan-2, FG, Colo357, Panc1 and Panc1-MUC1 cells using Tri Reagent (Molecular Research Center, Cincinnati, OH, USA). The cDNA was synthesized from isolated RNAs using the Verso cDNA synthesis kit according to manufacturer instruction (Thermo Fisher Scientific, Waltham, MA, USA).
PCR analysis of glycosyltransferases
The following genes were amplified using cDNA specific primers as described previously 17: C2GnT-1, C2GnT-2, C2GnT-3, β3GnT-6 (core 3 synthase), C1GalT1C1 (Cosmc), C1GalT1 (core 1 synthase) and GAPDH. PCR was carried out at 94°C for 5min followed by 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min followed by a single incubation at 72°C for 5min. PCR products were resolved by electrophoresis on a 1% agarose gel.
Real time PCR analysis
Real-time PCR analysis was performed as described previously.24 Expression of core 3 synthase and 18s rRNA was carried out on human normal pancreas, two primary pancreatic tumor and matched liver metastatic tumor tissues. Expression of Cosmc, core 1 synthase, FGFR-3 and GAPDH was analyzed in vector control and Core 3 synthase expressing pancreatic cancer cells using the following forward (F) and reverse primers (R): Core 3 synthase, F-TTCACCGTGCAAAGTGGAAACCAG and R-AGGACCCAACTAGGCCATTAGCTT; Cosmc, F-ATGCACCACCATGAGCATCATCAC and R-ACTGCAGCCCAAAGACTCACATCT; Core 1 synthase, F-CAAGATCAGAGCAGCAACCA and R-ATCCCTTTGTGCCAGAACAC; FGFR3, F-TCAGCTCCACAGCATCCC and R-GTCCTTGGGGACGGAGC; REG1α, F-GTAGGAGACCAGGGACCCA and R-CCTTTGTGGCCTCACTGATT; GAPDH, F-TCGACAGTCAGCCGCATCTTCTTT and R-ACCAAATCCGTTGACTCCGACCTT; 18s rRNA, F-CTTTCGAGGCCCTGTAATTG and R-CCTCCAATGGATCCTCGTTA.
Vector constructs and cell line transduction
The cDNA encoding core 3 synthase was amplified from normal stomach cDNA using the following primers (F-GGATCCGCCACCATGGCTTTTCCCTG and R- ATCTAGATCAGGAGACCCGGTG). The amplified core 3 synthase cDNA was cloned into pLVX-Puro lentiviral vector expression system (Clontech Laboratories Inc, Mountainview, CA, USA). HT293 A cells were transfected with pLVX-β3GnT-6-Puro and pLVX-Puro vector according to manufacturer’s instructions and viral particles were collected and used to transduce Capan-2 and FG cells. Transduced cells were selected and maintained with 3µg/mL puromycin (Invitrogen, Grand Island, NY, USA).
Western blot analysis
Total cell lysates (50µg) from FG and Capan-2 (vector control and core 3) were used for western blot analysis. Proteins were separated on 4–20% SDS-PAGE gels, transferred to PVDF membrane, and membranes were incubated overnight at 4°C with mouse polyclonal anti-core 3 synthase antibody (Abnova, Neihu District, Taipei City, Taiwan), rabbit polyclonal anti-β1 integrin antibody (Cell Signaling Technology Inc, Danvers, MA, USA), rabbit anti-α2 integrin antibody, rabbit anti-phospho FAK, rabbit anti-FAK (Millipore, Billerica, MA, USA) rabbit anti-p21 (Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA), or mouse monoclonal anti- β-actin (Sigma-Aldrich Corp., St. Louis, MO, USA) at 1:500 dilution. Membranes were washed three times in TBS-T (0.1% Tween 20) for 5 min each, followed by a 1 hr incubation with horse radish peroxidase (HRP) conjugated goat-anti rabbit and anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA, USA). Membranes were washed three times in TBS-T (0.1% Tween 20) for 5 min each and Super signal® West Pico Chemiluminescent substrate reagents (Thermo scientific, Rockford, IL, USA) were used for signal detection.
Phalloidin staining
The F-actin distribution was visualized in vector control and core 3 synthase transduced FG cells using Rhodamine-conjugated Phalloidin (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions. The staining intensity was quantified by use of the image J program (NIH).
Cell proliferation assay
Cell proliferation was evaluated by the Alamar blue reduction method as previously described.25 Pancreatic cancer cells (5,000 cells/well) were seeded in triplicate in 96-well plates and incubated for 1, 2, 3 or 4 days at 37°C. After each incubation time, alamar blue (equal to 10% volume) (AbD serotec, Kidlington, Oxford, UK) was aseptically added and incubated for 2–4 h at 37°C. Fluorescence was measured at 560 nm excitation and 590 nm emission using a Spectramax M5e (Molecular devices, Sunnyvale, CA, USA) fluorescence reader. This experiment was repeated three times and statistical analysis of vector control versus core 3 synthase was calculated by unpaired Student’s t test (p<0.05 considered statistically significant).
Cell migration assay
In vitro tumor cell migration assays were performed according to previous methods.26 Briefly, 0.5×106 cells of FG (vector control and core 3 synthase) and 0.25×106 cells of Capan-2 (vector control and core 3 synthase) were suspended in 1% FBS containing DMEM and seeded on the top chambers of polyethylene terapthalate membranes (PET) (12-well insert, pore size 8µm) (BD biosciences, Bedford, MA, USA). Following a 36 hr incubation, cells in the top chamber that did not migrate through the membrane were removed by scraping the membrane with a cotton swab. Cells that migrated through the membrane were stained with diff-Quick cell staining kit (Siemens Healthcare Diagnostic Inc., Newark, DE, USA). The number of cells that migrated were quantified in 5 different random fields at 40X magnification. The results are represented as the average number of cells/field. Three independent experiments were carried out and the data is represented as the average ± standard deviation.
Cell invasion assay
Tumor cell invasion assays were performed according to previously described methods.26 Briefly, 0.5×106 cells of FG (vector control and core 3 synthase) and 0.25×106 cells of Capan-2 (vector control and core 3 synthase) were seeded on Matrigel-coated membranes (BD biosciences, Bedford, MA, USA) and incubated for 36 hrs at 37°C. After incubation, non-invading cells on the upper surface of the filter were removed with cotton swabs. Cells that invaded through the pores onto the lower side of the filet were fixed and stained with Diff-Quick cell stain kit (Siemens Healthcare Diagnostic Inc., Newark, DE, USA). Invading cells were counted and analysed as mentioned in above procedure.
Orthotopic implantation of FG cells
Orthotopic implantation of tumor cells into the pancreas was performed according to previous methods.20 Briefly, core 3 synthase and vector control stably expressing FG cells were harvested from subconfluent cultures by standard procedures. Cells suspended in serum free DMEM medium, with > 90% viability, were used for implantations. Athymic nude mice (Crl:NU-Foxn1nu) (n=15 each group) were anesthetized with Ketamine-xylazine solution. A small left abdominal flank incision was made, and FG cells (1×106) in 30µl of serum free DMEM medium was injected in to the subcapsular region of the pancreas using a 29-gauge needle. Four weeks after injection, mice were euthanized and tumor weight was measured and metastasis to other organs was evaluated by histology. The statistical differences in tumor weight and metastasis between vector control and core 3 injected mice were calculated by two-tailed Student’s t-test and two-tailed Fisher’s exact test.
SDS-Agarose electrophoresis
SDS-Agarose gel electrophoresis for mucin and mucin glycans were carried out as described previously.27 Briefly, cell lysates (50–100 µg proteins) were resolved on SDS (0.1%) - Agarose (2%) gel electrophoresis and transferred overnight to PVDF membrane by capillary transfer. Membranes were blocked in 5% milk in TBS-T (Tris-buffered saline and 0.1% Tween 20, pH 7.4) and incubated with the following primary antibodies, 5E5 (mouse IgG against S/Tn epitope on MUC1) a kind gift from Dr. Henrik Clausen, Department of Cellular and Molecular Medicine, University of Copenhagen, Denmark and mouse anti-β-tubulin IgG (Developmental studies hybridoma bank, Iowa, USA). The membranes were washed and incubated with HRP conjugated Goat-anti mouse secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA). The signal was detected with Super signal® West Pico Chemiluminescent substrate (Thermo scientific, Rockford, IL, USA).
Confocal immunofluorescence microscopy
For confocal immunofluorescence analysis, vector control and core 3 synthase expressing FG cells were grown overnight on sterile cover slips. Cells were washed with PBS and fixed with 4% paraformaldehyde with 120mM sucrose for 15 min. Cells were permeabilized with 0.15% TritonX-100 in PBS containing 1% BSA. After blocking, cells were incubated with mouse monoclonal 5E5 antibody for 1hr at room temperature. Cells were then washed and incubated with FITC-conjugated goat anti-mouse IgG secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA). Cells were mounted with DAPI nuclear stain containing anti-fade vectashield mounting medium (Vector laboratories, Burlingame, CA, USA). Laser confocal microscopy was performed using a LSM 510 microscope at Confocal Microscopy Core facilities at UNMC.
RNA isolation and Gene microarray
Total RNA was isolated from vector control and core 3 synthase expressing FG cells (three independent samples of each cells) by using MRI TRI reagent and used for microarray analysis. Human cDNA microarray (39200 genes) analysis was performed by the Gene Microarray Core Facilities at University of Nebraska Medical Center. The data were analyzed by the Biostatistics Core facilities. Briefly, microarray data were preprocessed by background correction, within-array normalization through Loess normalization and between-array Aquantile normalization. The LIMMA (Linear Models for Microarray Analysis) method was fit on the normalized data to compare the intensities between the Core 3 synthase and vector control samples. The Benjamini Hochberg approach was used to control the false discovery rate (FDR), and the genes with false discovery rate less than 0.1 were designated as differentially expressed.
Results
Expression of glycosyltransferases in pancreatic tumor and cancer cells
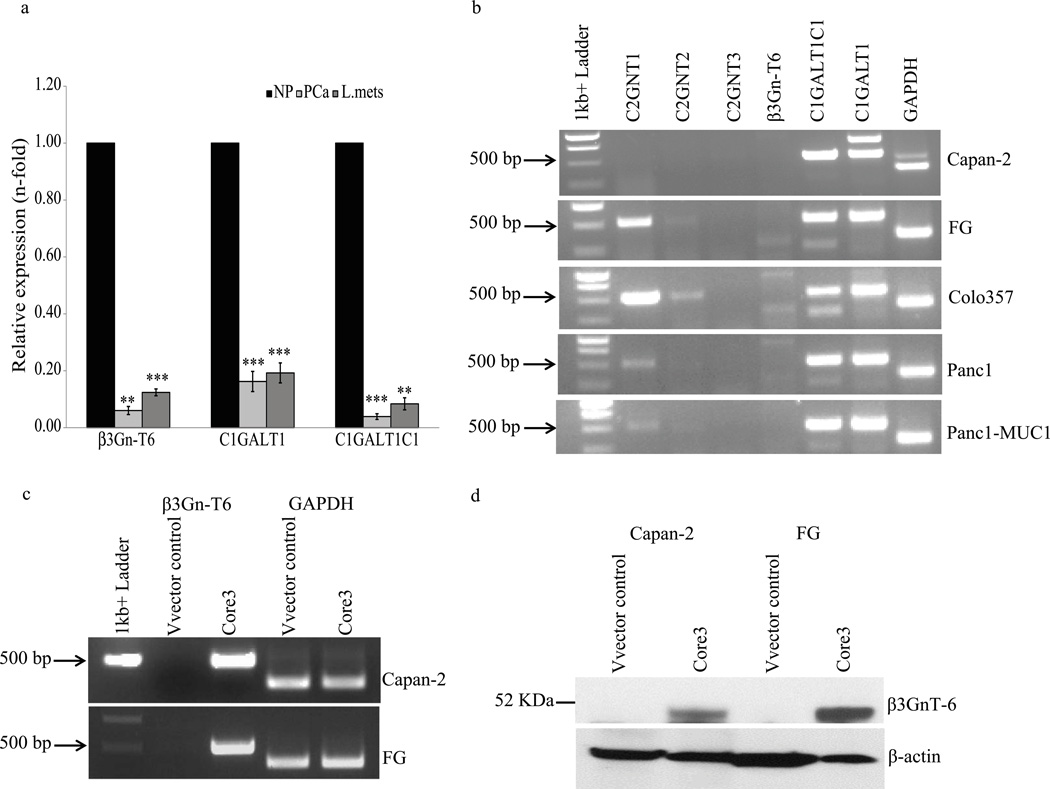
We detected low levels of core 3 synthase mRNA in normal uninvolved pancreas, and there was significantly decreased or no expression observed in primary and liver-metastatic pancreatic tumors (Fig.1a). Analysis of additional key O-glycan encoding glycosyltransferases that produce oligosaccharide core structures revealed differential mRNA expression in pancreatic cancer cell lines (Fig. 1b). Core 1 synthase and its molecular chaperone Cosmc were expressed in all pancreatic cancer cell lines examined (Capan-2, FG, Colo357, Panc1 and Panc1-MUC1). The core 2 enzyme C2GnT-1 was expressed at high levels in FG and Colo357 cells, whereas Panc1 and Panc1-MUC1 cells expressed lower amounts. C2GnT-2 expression was observed at low levels in FG and Colo357 cells, while C2GnT-3 and core 3 synthase were not detected in any of the cell lines analyzed here. Capan-2 (well differentiated) and FG (moderately differentiated) cells were selected to characterize the effect of core 3 synthase expression on pancreatic cancer cell growth and metastasis. Consistent with previous results from normal pancreas,13 we observed down regulation of core 3 synthase in pancreatic ductal adenocarcinoma patients (n=36) compared to normal pancreas (n=36) (Supplemental Fig. S1).
Figure 1.
Expression of selected glycosyltransferases in pancreas and tumors. Normal pancreas, pancreatic tumors and selected pancreatic cancer cell lines were evaluated for expression of glycosyltransferases that extend mucin type O-glycosylation. (a) Real time PCR analysis of core 3 synthase, core 1 synthase and its molecular chaperone C1GALT1C1 (Cosmc) mRNA expression in two normal pancreas (NP), two primary pancreatic adenocarcinoma (PCa) and matched tumors that were metastatic to liver (L. Mets). Each value was normalized with 18sRNA expression, and is presented as relative to expression levels of normal pancreas. The experiments were repeated three times in triplicate. The values are expressed as averages of two samples mean (2 normal pancreases, 2 PCa and 2 L.Mets) and error bars represents S.D of mean. A p-value less than 0.05 was considered to be statistically significant (***p<0.0005, **p<0.005). (b). Gene specific primers were used to amplify the following genes, C2GnT-1, C2GnT-2, C2GnT-3, β3GnT-6 (core 3 synthase), C1GALT1C1 (Cosmc), C1GALT1 (core1 synthase) and GAPDH. Low or no expression of core 3 synthase was observed in all the pancreatic cancer cell lines. Core 3 synthase coding cDNA sequence was cloned in pLVX lentiviral vector and transduced into Capan-2 and FG cells for stable expression. The expression of core 3 synthase was detected by RT PCR (c) and western blot (d) using gene specific primers and core 3 synthase antibody, respectively. Amplification of GAPDH and detection of β-actin was used as an internal control.
Stable expression of core 3 synthase in Capan-2 and FG cells was achieved through the transduction of each cell line with lentivirus containing either the vector alone or the core 3 synthase coding sequence. Cells were selected with puromycin and expression of core 3 synthase transcript and protein was confirmed by RT PCR and western blot analysis (Fig.1c and 1d). Expression of core 3 synthase did not affect the levels of either Cosmc or core 1 synthase (data not shown).
Core 3 synthase reduces cancer cell proliferation and affects actin polymerization
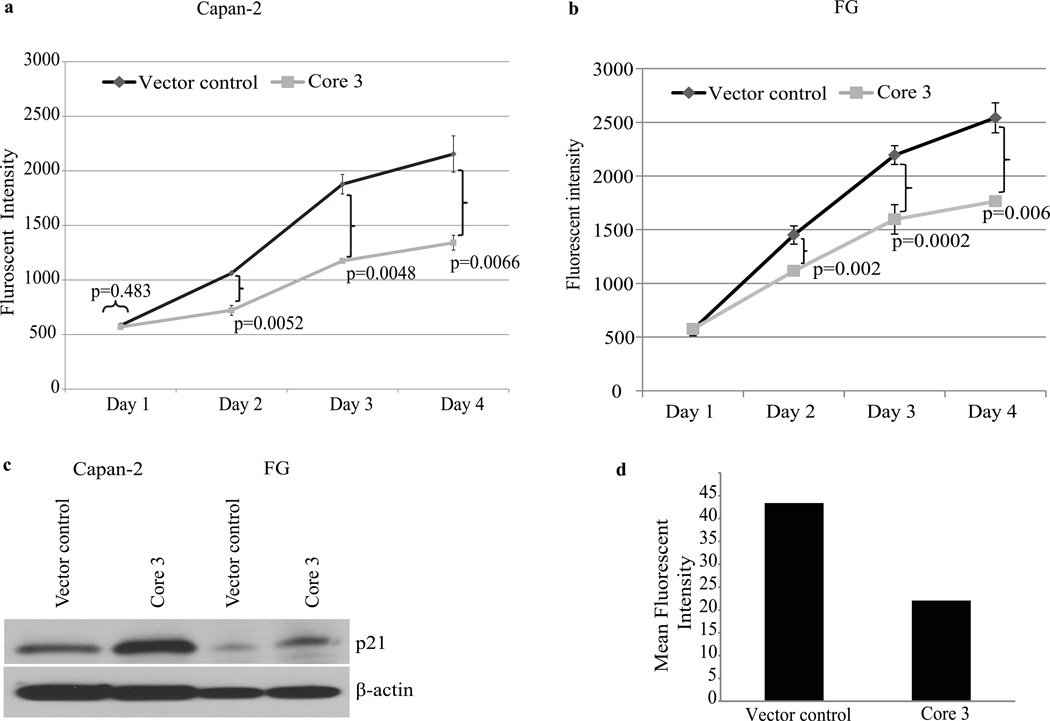
We evaluated the effect of re-expression of core 3 synthase on proliferation in vitro of Capan-2 and FG cells. Vector control and core 3 synthase expressing cells were seeded into 96 well plates and proliferation was evaluated using an alamar blue assay at 24, 48 72 and 96 hrs. Significantly reduced (p<0.05) rates of cell proliferation were observed at 48, 72 and 96 hrs in both Capan-2 (Fig. 2a) and FG cells (Fig. 2b) expressing core 3 synthase compared to vector control cells. Reduced cell proliferation was likely due in part to higher expression of cyclin dependent kinase inhibitor p21 by core 3 synthase expressing cells (Fig. 2c). Consistent with previous work in prostate cancer,17 FG cells expressing core 3 synthase showed reduced Phalloidin staining and altered cytoskeletal organization compared to vector control FG cells (Fig. 2d and Supplemental Fig. S3), suggesting that expression of core 3 O-glycans alters stress fiber formation and actin polymerization.
Figure 2.
Cell proliferation analyses and actin polymerization. Core 3 synthase and vector control cell proliferation was analysed by Alamar blue dye reduction method. Fluorescence intensity was measured at each incubation period (day1, 2, 3 and 4) for Capan-2 (a) and FG cells (b). Points represent the mean of triplicates; bars represent SE. Data are representative of three independent experiments that included triplicate samples (A p-value <0.05 is considered to be statistically significant). (c) Expression p21 on core 3 and vector control cell lysates were assessed by western blot. Detection of β-actin was used as an internal loading control. (d) Quantification of F-actin Phalloidin staining. Core 3 and vector control FG cells F-actin were stained with Rhodamine conjugated Phalloidin and the fluorescence intensity was measured by the Image J program. Core 3 expressing FG cells show reduced Phalloidin staining intensity. Mean fluorescence intensity values of duplicate experiments are presented in the graph.
Expression of core 3 O-glycans affects the malignant properties of pancreatic cancer cells
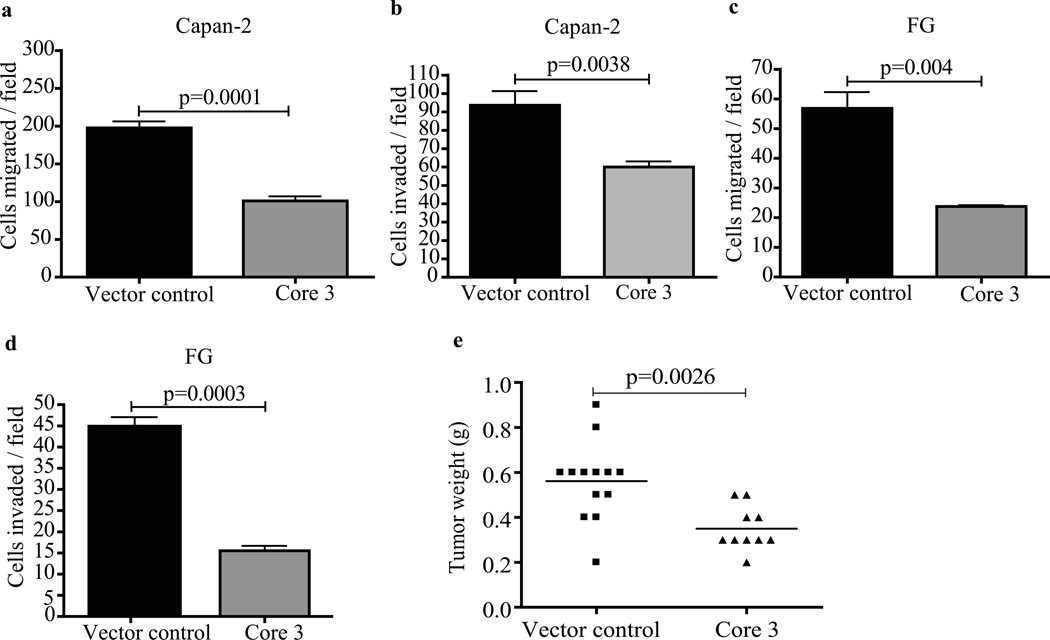
We evaluated the contribution of core 3 synthase expression to in vitro cell migration and invasion properties. Capan-2 and FG cells expressing core 3 synthase or vector controls were seeded on top chambers of polyethylene terapthalate inserts and matrigel coated membranes for 36 hrs. Cells that migrated and invaded through the membranes were fixed and stained. Core 3 synthase expression resulted in a significant reduction in numbers of cells that migrated through filters (Fig.3a–c) and invaded through matrigel (Fig.3b–d) compared to vector control cells, respectively. We evaluated the influence of core 3 derived glycans on tumor growth properties. FG-vector control and FG-core 3 synthase expressing cells were injected orthotopically into the pancreas of nude mice. After 4 weeks, the animals were sacrificed and examined for tumor growth and metastasis. Mice inoculated with FG-core 3 synthase cells showed significantly smaller pancreas tumors (p=0.0026), a low incidence of metastasis to lymph nodes (10%), and an absence of metastasis to the peritoneal cavity. In contrast, mice injected with vector control FG cells had larger tumors, and higher incidences of lymph node (30.7%) and peritoneal metastasis (69.2%) (Fig. 3e and Table 1). These results support the hypothesis that expression of core 3 derived O-glycans suppresses tumor growth and metastasis.
Figure 3.
In vitro migration and invasion assay. FG vector control and core 3 cells were seeded into upper chambers of transwell inserts and matrigel chambers; cells that migrated and invaded to the bottom side of the chambers were fixed, stained and counted. Core 3 expressing FG and Capan-2 cells shows significantly reduced migration (a–c) and invasion (b–d). The data represents the mean of three independent experiments and error bars represent S.D of the mean. A p-value less than 0.05 considered to be statistically significant. (e). Core 3 synthase suppresses tumor formation. Vector control and core 3 expressing FG cells (1×106) were orthotopically implanted into the pancreases of nude mice, animals were sacrificed after 4 weeks, and tumor weight was measured. Tumors with core 3 (n=10) show significantly reduced tumor weight (p=0.0026) as compared to tumors with vector control cells (n=13). A p-value less than 0.05 was considered to be statistically significant.
Table 1.
Incidence of primary tumor formation and metastasis
| FG cells | Pancreatic tumor | Lymph node metastasis | Peritoneal metastasis |
|---|---|---|---|
| Vector control | 13/13 (100%) | 4/13 (30.7%) | 9/13 (69.2%) |
| Core 3 | 10/10 (100%) | 1/10 (10%) | 0/10 (0%) |
| p-value | 1.00 | 0.339 | 0.0016* |
The incidence of tumor formation and metastasis was compared between the control vector and core3 cells injected nude mice by the χ2-test. A p-value of < 0.05 was considered statistically significant.
indicates the significant difference between two groups.
Core 3 synthase extends Tn structures on MUC1, deregulates α2β1integrin expression and reduces FAK phosphorylation
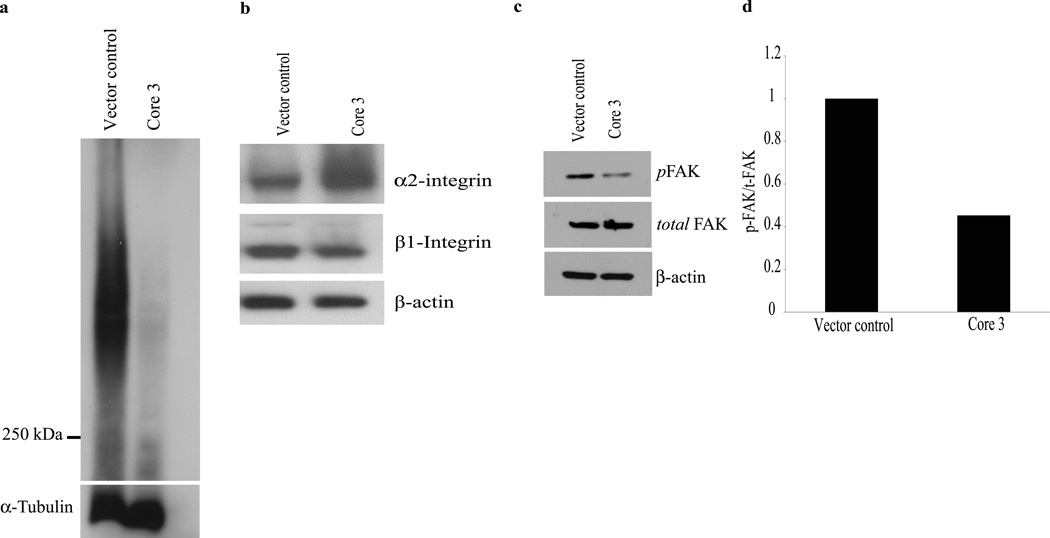
We examined the glycosylation status of the MUC1 glycoprotein, which is predicted to be affected by expression of core 3 synthase, as this oncoprotein is heavily O-glycosylated and aberrantly overexpressed in pancreatic cancer. The Tn epitope (GalNAcα1 – Thr / Ser), which contains the precursor saccharide structure that is extended by core 1 or core 3 synthases, is detected on the MUC1 tandem repeat domain (detected by 5E5 mouse monoclonal antibody) in cancer and cancer cell lines but is not seen in normal tissues. We observed substantial reduction of Tn epitope on MUC1 in core 3 synthase expressing in FG cells compared to vector control cells (Fig. 4a). Additional confirmatory evidence was provided by the fact that we also observed a reduction in T antigen expression in these cells upon expression of core 3 synthase (Supplemental Fig. S4). These data suggest that core 3 synthase activity extends the Tn epitope by adding a N-Acetylglucosamine (GlcNAc) moiety to enable the production of extended core 3 derived O-glycans (GlcNAc β1-3GalNAcα1 – Thr / Ser) on MUC1.
Figure 4.
Extension of Tn epitope by core 3 synthase. (a) Western blot analysis of Tn epitope on MUC1 in core 3 and vector control FG cells using the 5E5 antibody (Tn on MUC1 tandem repeat). Tn epitope on MUC1 is reduced as indicated by decreased band intensity (above 250 KDa) in core 3 synthase expressing cells. Detection of α-tubulin served as a loading control. Deregulation of α2β1 integrin and impaired FAK phosphorylation. (b) Core 3 synthase expression induces increased levels of α2 integrin and decreased levels of β1 integrin. (c) Immunoblot analysis of FAK shows decreased levels of phosphorylated FAK (pFAK) in core 3 synthase expressing FG cells. (d) A densitometric quantification of FAK activation (phosphorylation) carried out using the Image J program. The relative intensity of pFAK is calculated by core 3 total FAK (−) pFAK (/) vector control total FAK (−) pFAK. Experiments were repeated twice and the mean values are presented in the graph. Detection of β-actin was used as an internal loading control.
It has been previously reported that expression of core 3 synthase down regulates α2β1 integrin complex formation in prostate cancer cells.17 We sought to confirm that expression of core 3 synthase affected the α2 and β1 integrins in pancreatic cancer by examining the expression of α2 and β1 integrin by immunoblot analysis. Expression of α2 integrin was increased in core 3 synthase expressing cells, whereas there was reduced expression of β1 integrin in core 3 synthase expressing cells compared to vector control cells (Fig. 4b). These data suggest that addition of core 3 O-glycans deregulates the production and/or stability of α2 and β1 integrins in FG cells.
The deregulation of α2 and β1 integrins in FG cells expressing core 3 synthase suggests that core 3 glycan alters the function of α2β1 integrin complex. Integrin mediated cell adhesion directly activates different tyrosine kinases including FAK, which is directly activated by α2β1 integrin. We examined the phosphorylation status of FAK as a measure of α2β1 downstream signaling. FG cells expressing core 3 synthase showed reduced levels of phosphorylated FAK relative to FAK when compared to vector control cells (Fig.4c–d).
Core 3 O-glycans affect gene expression
The finding that core 3 glycosylation affected integrin signaling, suggested that other signaling pathways may be affected, given that a number of cell surface proteins that engage in signal transduction are O-glycosylated, including cell surface mucins such as MUC1 and receptor tyrosine kinases. The net result of differences in signal transduction is reflected in the transcriptional profile of the cells. Thus, we evaluated the global effects of extending O-glycans with core 3 structures on gene expression in these pancreatic cancer cells by cDNA microarray analysis. This analysis revealed that both tumor promoting and tumor suppressing genes were significantly up and down regulated in FG core 3 synthase expressing cells compared with vector control cells (listed in Table 2). Differential expression of two down regulated genes (FGFR3 and REG1α) were validated by real time PCR (Fig. S2). The expression array data was deposited in the GEO database, accession number GSE42417.
Table 2.
Differential expression of genes regulated by core 3 synthase
| Accession number | Gene name | Fold change |
|---|---|---|
| Tumor suppressor genes fold up-regulated (core 3 synthase versus vector control cells) | ||
| NM_144975 | SLFN5 – Schlafen family member 5 | 2.16 |
| NM_005100 | AKAP12 – A-kinase anchor protein 12 | 1.88 |
| NM_001030287 | ATF3 – Activating transcription factor 3 | 1.88 |
| NM_001731 | BTG1- B-cell translocation gene 1 | 1.75 |
| Tumor enhancer genes fold down regulated (core3 synthase versus vector control cells) | ||
| NM_002909 | REG1A – Regenerating islet-derived 1 alpha | 0.61 |
| NM_002160 | TNC – Tenascin C | 0.62 |
| NM_000142 | FGFR3 – Fibroblast growth factor receptor 3 | 0.70 |
Discussion
Core 3 synthase, an important glycosyltransferase that mediates synthesis of mucin-type O-glycans in the normal gastrointestinal tract, is down regulated in gastric and intestinal cancer cells. We demonstrate here that the core 3 synthase is also down regulated in pancreatic cancer. Our results support the general concept that increased expression of core 3 O-glycan structures on cell surface glycoproteins limits pancreatic tumor growth and metastasis. This supports conclusions of previous studies of core 3 synthase-deficient mice, which exhibit reduced expression of the mucin Muc2 and increased intestinal permeability, and were found to be more susceptible to experimentally induced colitis and to carcinoma, implicating mucin-type glycoproteins containing core 3 O-glycans in the suppression of tumor formation.16
We investigated the functional role of core 3 derived O-glycans by re-expressing core 3 synthase in two human pancreatic cancer cells, Capan-2 and FG. Expression of core 3 synthase in these cells significantly reduced in vitro proliferation, migration, invasion, and metastasis, which strongly supports the hypothesis that alteration of mucin type O-glycan structures affects cancer progression. These results are consistent with previous studies in other tumors, which showed that ectopic expression of core 3 synthase in prostate cancer cells reduced primary tumor formation and metastasis.17 Reduced lung tumor focus formation was also observed in human fibrosarcoma cells expressing core 3 synthase.11
The finding that induced expression of core 3 synthase in the FG pancreatic cancer cell line significantly reduced both primary pancreas tumor formation and metastatic dissemination to the surrounding tissues (lymph nodes and peritoneum) is likely explained by several factors. O-glycosylation affects a number of cell surface proteins including growth factor and cytokine receptors, molecules that configure structural and adhesive properties of cells, and molecules that otherwise engage in signal transduction. This is confirmed by the fact that core 3 synthase expressing pancreatic cancer cells showed altered expression of α2β1 integrin and decreased FAK activation, which is consistent with a previous report by Lee et al in prostate cancer.17 We extend the previous findings by demonstrating for the first time that core 3 enzyme expression eliminated expression of Tn epitope on MUC1, likely a consequence of the construction of core 3 O-glycans through addition of β1-3 linked GlcNAc residues to terminal GalNAc residues on Tn epitopes to form the core 3 O-glycan (GlcNAcβ1-3GalNAcα-Ser/Thr) and the potential for extended glycan structures. MUC1 is overexpressed in greater than 90% of pancreatic adenocarcinomas, and a significant percentage of other adenocarcinomas, and is believed to contribute to tumor progression through its influence in configuring the cell surface properties of cells28 and by oncogenic signaling effects that are modulated by its differential glycosylation.29, 30 Differential glycosylation includes the appearance of Tn structures on MUC1. For example, induced expression of Tn epitope on MUC1 has been reported to promote mammary carcinogenesis in part through stabilization of the MUC1 oncoprotein.22 The identification of Tn or STn on MUC1 specific auto-antibodies in ovarian, breast and prostate cancer patients further support the concept that Tn epitopes on MUC1 are associated with disease progression.23 Alterations in oncogenic signaling through MUC1, integrins or other cell surface proteins have downstream effects on tumor progression. Here, we observed that one of those downstream effects was reduced cell proliferation by core 3 synthase expressing pancreatic cancer cells through higher expression of cell cycle inhibitor p21, which inhibited cell cycle progression through G1 to S phase.31
Our microarray data revealed additional downstream effects of core 3 expression including up-regulation of expression of the tumor suppressor function associated genes schlafen family member 5 (SLFN5), A-kinase anchor protein 12 (AKAP12), Activating transcription factor 3 (ATF3) and B-cell translocation gene 1 (BTG1). SLFN5 is a cell cycle regulatory protein that mediates growth inhibition responses.32 AKAP12 is a kinase anchoring protein that is mainly involved in regulation of β2-adrenergic complex. Several tumors have been shown to down regulate expression of this gene by epigenetic mechanisms.33 Overexpression of AKAP12 reduces colony formation and induces apoptosis.34 ATF3 is a member of ATF/cyclic AMP response element-binding family of transcription factors that is mainly involved in stress responses. Overexpression of ATF3 inhibits tumor cell invasiveness and tumor growth.35–37 BTG1 is an anti-proliferative gene, highly expressed in G0/G1 phases of cell cycle. Reduced cell proliferation was observed in BTG1 cDNA transfected cells.38
Expression of tumor promoting genes were decreased in core 3 synthase expressing FG cells: Regenerating gene 1α (REG1 α), Fibroblast growth factor receptor 3 (FGFR3) and Tenascin C (TNC). REG1 α is an epithelial growth factor mainly expressed in pancreatic islets cells involved in carcinogenesis of numerous cancers including pancreatic cancer with diabetes.39 Overexpression of REG1α in pancreatic cancer cell lines induced cell proliferation and tumor growth both in in vitro and in vivo.39 FGFR3, an extracellular cell surface glycoprotein that regulates cellular processes including cell growth and division, is expressed by many cancers, and has been associated with increased cancer cell migration and invasion.40 TNC is an extracellular matrix glycoprotein that is up-regulated in pancreatic and other cancers.41 TNC exhibits de-adhesive properties for cell-extracellular matrix interactions. This in turn enhances cell proliferation and motility can be associated with the initiation of epithelial to mesenchymal transition (EMT) during progression of breast cancers.42
In summary the findings reported here suggest that the core 3 synthase is expressed at low levels by normal pancreas but is lost in pancreatic tumors. Expression of core 3 synthase suppresses tumor growth and metastatic potential of pancreatic cancer cells. The mechanism whereby expression is lost is not known currently but represents a novel future therapeutic target to reduce the early metastatic phenotype of pancreatic cancer. This occurs in part through altered glycosylation of MUC1, deregulated expression of α2β1 integrin and the resultant reduced phosphorylation of FAK, and alterations in downstream signaling through MUC1, integrins, and other cell surface proteins that are affected by differential glycosylation, and which in turn result in altered expression and activity of tumor suppressing and oncogenic proteins. Further studies should be undertaken to elucidate the molecular mechanisms whereby the addition of core 3 structures to cell surface proteins suppresses tumor growth and metastasis.
Supplementary Material
What’s new: Core 3 derived glycans are downregulated during malignancy, because of loss of expression of functional β3-N-acetylglucosaminyltransferase-6 (core 3 synthase). We show that pancreatic cancer cells have lost expression of core 3 synthase and that re-expression of core 3 derived O-glycans in pancreatic cancer cells suppresses tumor growth and metastasis through modulation of glycosylation of mucins and other cell surface and extracellular matrix proteins
Acknowledgements
This work was supported in part by the following grants from the NCI:
Grant sponsor: NIH R01 CA57362, NIH P50 CA127297 SPORE in Pancreatic Cancer, NCI P30 CA036727-2452 Cancer Center Support Grant, NIH U01 CA111294 Early Detection Research Network, NIH U01 CA128437 Alliance of Glycobiologists for Detection of Cancer and Cancer Risk, NIH U54 CA163120 Tumor Microenvironment Network
References
- 1.Ten Hagen KG, Tetaert D, Hagen FK, Richet C, Beres TM, Gagnon J, Balys MM, VanWuyckhuyse B, Bedi GS, Degand P, Tabak LA. Characterization of a UDP-GalNAc:Polypeptide N-acetylgalactosaminyltransferase that displays glycopeptide N-acetylgalactosaminyltransferase activity. J Biol Chem. 1999;274:27867–27874. doi: 10.1074/jbc.274.39.27867. [DOI] [PubMed] [Google Scholar]
- 2.Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guda K, Moinova H, He J, Jamison O, Ravi L, Natale L, Lutterbaugh J, Lawrence E, Lewis S, Willson JK, Lowe JB, Wiesner GL, et al. Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc Natl Acad Sci U S A. 2009;106:12921–12925. doi: 10.1073/pnas.0901454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng PW, Radhakrishnan P. Mucin O-glycan branching enzymes: Structure, function, and gene regulation. Adv Exp Med Biol. 2011;705:465–492. doi: 10.1007/978-1-4419-7877-6_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcos NT, Bennett EP, Gomes J, Magalhaes A, Gomes C, David L, Dar I, Jeanneau C, DeFrees S, Krustrup D, Vogel LK, Kure EH, et al. ST6GalNAc-I controls expression of sialyl-tn antigen in gastrointestinal tissues. Front Biosci (Elite Ed) 2011;3:1443–1455. doi: 10.2741/e345. [DOI] [PubMed] [Google Scholar]
- 6.Nagaraj S, Neumann J, Winzen B, Frank S, Ziske C, Sievers E, Koch N, Schmidt-Wolf IG. Pancreas carcinoma antigen fused to invariant chain elicits T-cell response and tumor growth inhibition. Pancreas. 2008;37:321–327. doi: 10.1097/MPA.0b013e318166722e7. [DOI] [PubMed] [Google Scholar]
- 7.Machida E, Nakayama J, Amano J, Fukuda M. Clinicopathological significance of core 2 beta1,6-N-acetylglucosaminyltransferase messenger RNA expressed in the pulmonary adenocarcinoma determined by in situ hybridization. Cancer Res. 2001;61:2226–2231. [PubMed] [Google Scholar]
- 8.Shimodaira K, Nakayama J, Nakamura N, Hasebe O, Katsuyama T, Fukuda M. Carcinoma-associated expression of core 2 beta-1,6-N-acetylglucosaminyltransferase gene in human colorectal cancer: Role of O-glycans in tumor progression. Cancer Res. 1997;57:5201–5206. [PubMed] [Google Scholar]
- 9.Yousefi S, Higgins E, Daoling Z, Pollex-Kruger A, Hindsgaul O, Dennis JW. Increased UDP-GlcNAc:Gal beta 1-3GaLNAc-R (GlcNAc to GaLNAc) beta-1, 6-N-acetylglucosaminyltransferase activity in metastatic murine tumor cell lines control of polylactosamine synthesis. J Biol Chem. 1991;266:1772–1782. [PubMed] [Google Scholar]
- 10.Hagisawa S, Ohyama C, Takahashi T, Endoh M, Moriya T, Nakayama J, Arai Y, Fukuda M. Expression of core 2 beta1,6-N-acetylglucosaminyltransferase facilitates prostate cancer progression. Glycobiology. 2005;15:1016–1024. doi: 10.1093/glycob/cwi086. [DOI] [PubMed] [Google Scholar]
- 11.Iwai T, Kudo T, Kawamoto R, Kubota T, Togayachi A, Hiruma T, Okada T, Kawamoto T, Morozumi K, Narimatsu H. Core 3 synthase is down-regulated in colon carcinoma and profoundly suppresses the metastatic potential of carcinoma cells. Proc Natl Acad Sci U S A. 2005;102:4572–4577. doi: 10.1073/pnas.0407983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang MC, Chen HY, Huang HC, Huang J, Liang JT, Shen TL, Lin NY, Ho CC, Cho IM, Hsu SM. C2GnT-M is downregulated in colorectal cancer and its re-expression causes growth inhibition of colon cancer cells. Oncogene. 2006;25:3267–3276. doi: 10.1038/sj.onc.1209350. [DOI] [PubMed] [Google Scholar]
- 13.Iwai T, Inaba N, Naundorf A, Zhang Y, Gotoh M, Iwasaki H, Kudo T, Togayachi A, Ishizuka Y, Nakanishi H, Narimatsu H. Molecular cloning and characterization of a novel UDP-GlcNAc:GalNAc-peptide beta1,3-N-acetylglucosaminyltransferase (beta 3Gn-T6), an enzyme synthesizing the core 3 structure of O-glycans. J Biol Chem. 2002;277:12802–12809. doi: 10.1074/jbc.M112457200. [DOI] [PubMed] [Google Scholar]
- 14.Vavasseur F, Dole K, Yang J, Matta KL, Myerscough N, Corfield A, Paraskeva C, Brockhausen I. O-glycan biosynthesis in human colorectal adenoma cells during progression to cancer. Eur J Biochem. 1994;222:415–424. doi: 10.1111/j.1432-1033.1994.tb18880.x. [DOI] [PubMed] [Google Scholar]
- 15.Vavasseur F, Yang JM, Dole K, Paulsen H, Brockhausen I. Synthesis of O-glycan core 3: Characterization of UDP-GlcNAc: GalNAc-R beta 3-N-acetyl-glucosaminyltransferase activity from colonic mucosal tissues and lack of the activity in human cancer cell lines. Glycobiology. 1995;5:351–357. doi: 10.1093/glycob/5.3.351. [DOI] [PubMed] [Google Scholar]
- 16.An G, Wei B, Xia B, McDaniel JM, Ju T, Cummings RD, Braun J, Xia L. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J Exp Med. 2007;204:1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SH, Hatakeyama S, Yu SY, Bao X, Ohyama C, Khoo KH, Fukuda MN, Fukuda M. Core3 O-glycan synthase suppresses tumor formation and metastasis of prostate carcinoma PC3 and LNCaP cells through down-regulation of alpha2beta1 integrin complex. J Biol Chem. 2009;284:17157–17169. doi: 10.1074/jbc.M109.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grzesiak JJ, Bouvet M. The alpha2beta1 integrin mediates the malignant phenotype on type I collagen in pancreatic cancer cell lines. Br J Cancer. 2006;94:1311–1319. doi: 10.1038/sj.bjc.6603088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clement M, Rocher J, Loirand G, Le Pendu J. Expression of sialyl-tn epitopes on beta1 integrin alters epithelial cell phenotype, proliferation and haptotaxis. J Cell Sci. 2004;117:5059–5069. doi: 10.1242/jcs.01350. [DOI] [PubMed] [Google Scholar]
- 20.Tsutsumida H, Swanson BJ, Singh PK, Caffrey TC, Kitajima S, Goto M, Yonezawa S, Hollingsworth MA. RNA interference suppression of MUC1 reduces the growth rate and metastatic phenotype of human pancreatic cancer cells. Clin Cancer Res. 2006;12:2976–2987. doi: 10.1158/1078-0432.CCR-05-1197. [DOI] [PubMed] [Google Scholar]
- 21.Altschuler Y, Kinlough CL, Poland PA, Bruns JB, Apodaca G, Weisz OA, Hughey RP. Clathrin-mediated endocytosis of MUC1 is modulated by its glycosylation state. Mol Biol Cell. 2000;11:819–831. doi: 10.1091/mbc.11.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JH, Nishidate T, Kijima K, Ohashi T, Takegawa K, Fujikane T, Hirata K, Nakamura Y, Katagiri T. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 2010;70:2759–2769. doi: 10.1158/0008-5472.CAN-09-3911. [DOI] [PubMed] [Google Scholar]
- 23.Wandall HH, Blixt O, Tarp MA, Pedersen JW, Bennett EP, Mandel U, Ragupathi G, Livingston PO, Hollingsworth MA, Taylor-Papadimitriou J, Burchell J, Clausen H. Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 2010;70:1306–1313. doi: 10.1158/0008-5472.CAN-09-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behrens ME, Grandgenett PM, Bailey JM, Singh PK, Yi CH, Yu F, Hollingsworth MA. The reactive tumor microenvironment: MUC1 signaling directly reprograms transcription of CTGF. Oncogene. 2010;29:5667–5677. doi: 10.1038/onc.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voytik-Harbin SL, Brightman AO, Waisner B, Lamar CH, Badylak SF. Application and evaluation of the alamarBlue assay for cell growth and survival of fibroblasts. In Vitro Cell Dev Biol Anim. 1998;34:239–246. doi: 10.1007/s11626-998-0130-x. [DOI] [PubMed] [Google Scholar]
- 26.Radhakrishnan P, Lin MF, Cheng PW. Elevated expression of L-selectin ligand in lymph node-derived human prostate cancer cells correlates with increased tumorigenicity. Glycoconj J. 2009;26:75–81. doi: 10.1007/s10719-008-9167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabriel M, Zentner A. Sodium dodecyl sulfate agarose gel electropheresis and electroelution of high molecular weight human salivary mucin. Clin Oral Investig. 2005;9:284–286. doi: 10.1007/s00784-005-0007-2. [DOI] [PubMed] [Google Scholar]
- 28.Hollingsworth MA, Swanson BJ. Mucins in cancer: Protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 29.Brockhausen I, Yang JM, Burchell J, Whitehouse C, Taylor-Papadimitriou J. Mechanisms underlying aberrant glycosylation of MUC1 mucin in breast cancer cells. Eur J Biochem. 1995;233:607–617. doi: 10.1111/j.1432-1033.1995.607_2.x. [DOI] [PubMed] [Google Scholar]
- 30.Mungul A, Cooper L, Brockhausen I, Ryder K, Mandel U, Clausen H, Rughetti A, Miles DW, Taylor-Papadimitriou J, Burchell JM. Sialylated core 1 based O-linked glycans enhance the growth rate of mammary carcinoma cells in MUC1 transgenic mice. Int J Oncol. 2004;25:937–943. [PubMed] [Google Scholar]
- 31.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 32.Katsoulidis E, Mavrommatis E, Woodard J, Shields MA, Sassano A, Carayol N, Sawicki KT, Munshi HG, Platanias LC. Role of interferon {alpha} (IFN{alpha})-inducible schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J Biol Chem. 2010;285:40333–40341. doi: 10.1074/jbc.M110.151076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, Gong J, Hu J, Hu T, Sun Y, Du J, Sun C, Guan M, Jiang H, Lu Y. Quantitative assessment of AKAP12 promoter methylation in human prostate cancer using methylation-sensitive high-resolution melting: Correlation with gleason score. Urology. 2011;77 doi: 10.1016/j.urology.2010.12.010. 1006.e1,1006.e7. [DOI] [PubMed] [Google Scholar]
- 34.Choi MC, Jong HS, Kim TY, Song SH, Lee DS, Lee JW, Kim TY, Kim NK, Bang YJ. AKAP12/Gravin is inactivated by epigenetic mechanism in human gastric carcinoma and shows growth suppressor activity. Oncogene. 2004;23:7095–7103. doi: 10.1038/sj.onc.1207932. [DOI] [PubMed] [Google Scholar]
- 35.Hackl C, Lang SA, Moser C, Mori A, Fichtner-Feigl S, Hellerbrand C, Dietmeier W, Schlitt HJ, Geissler EK, Stoeltzing O. Activating transcription factor-3 (ATF3) functions as a tumor suppressor in colon cancer and is up-regulated upon heat-shock protein 90 (Hsp90) inhibition. BMC Cancer. 2010;10:668. doi: 10.1186/1471-2407-10-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bottone FG, Jr, Moon Y, Kim JS, Alston-Mills B, Ishibashi M, Eling TE. The anti-invasive activity of cyclooxygenase inhibitors is regulated by the transcription factor ATF3 (activating transcription factor 3) Mol Cancer Ther. 2005;4:693–703. doi: 10.1158/1535-7163.MCT-04-0337. [DOI] [PubMed] [Google Scholar]
- 37.Lu D, Wolfgang CD, Hai T. Activating transcription factor 3, a stress-inducible gene, suppresses ras-stimulated tumorigenesis. J Biol Chem. 2006;281:10473–10481. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- 38.Rouault JP, Rimokh R, Tessa C, Paranhos G, Ffrench M, Duret L, Garoccio M, Germain D, Samarut J, Magaud JP. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–1670. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou L, Zhang R, Wang L, Shen S, Okamoto H, Sugawara A, Xia L, Wang X, Noguchi N, Yoshikawa T, Uruno A, Yao W, et al. Upregulation of REG ialpha accelerates tumor progression in pancreatic cancer with diabetes. Int J Cancer. 2010;127:1795–1803. doi: 10.1002/ijc.25188. [DOI] [PubMed] [Google Scholar]
- 40.Henriksson ML, Edin S, Dahlin AM, Oldenborg PA, Oberg A, Van Guelpen B, Rutegard J, Stenling R, Palmqvist R. Colorectal cancer cells activate adjacent fibroblasts resulting in FGF1/FGFR3 signaling and increased invasion. Am J Pathol. 2011;178:1387–1394. doi: 10.1016/j.ajpath.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Juuti A, Nordling S, Louhimo J, Lundin J, Haglund C. Tenascin C expression is upregulated in pancreatic cancer and correlates with differentiation. J Clin Pathol. 2004;57:1151–1155. doi: 10.1136/jcp.2003.015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagaharu K, Zhang X, Yoshida T, Katoh D, Hanamura N, Kozuka Y, Ogawa T, Shiraishi T, Imanaka-Yoshida K. Tenascin C induces epithelial-mesenchymal transition-like change accompanied by SRC activation and focal adhesion kinase phosphorylation in human breast cancer cells. Am J Pathol. 2011;178:754–763. doi: 10.1016/j.ajpath.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.