Abstract
We have used statistical models based on Fourier transform-infrared spectra to differentiate between the DNA structure of normal granulocytes and those obtained from patients with myelodysplastic syndrome (MDS). The substantial degree of discrimination achieved between the two DNA groups is attributed to differences in the nucleotide base and backbone structures. These structural differences allowed for the development of a discriminant analysis model that predicted, with high sensitivity and specificity, which DNA came from normal granulocytes vs. granulocytes from MDS patients. The findings are a promising basis for developing a blood test to diagnose and predict the occurrence of MDS, for which there is currently a paucity of molecular markers.
Keywords: clonal blood disease, blood test, disease diagnosis, prediction, Fourier transform-infrared spectroscopy
Myelodysplastic syndrome (MDS) is a clonal hematopoietic stem cell disorder characterized by peripheral blood cytopenias caused by ineffective hematopoiesis that can lead to bone marrow failure (1). It is estimated that 15,000 to 20,000 cases of MDS are diagnosed each year in the United States, primarily in patients over 60 years of age (2, 3), most of whom die of infection or bleeding. Moreover, up to 40% of cases transform to acute myelogenous leukemia (2, 4). Only half of MDS cases are characterized by nonrandom chromosomal abnormalities, and for the rest diagnosis is largely dependent on the presence of blood count suppression in association with morphologic abnormalities of both mature and immature myeloid cells (5, 6). As a consequence of this phenotypic mimicry, the distinction between MDS and nonclonal disorders causing bone marrow failure is often difficult.
Although the molecular basis of MDS remains to be clarified, it is recognized that aberrant DNA hypermethylation is one event implicated in the transformation of MDS to acute myelogenous leukemia (7-9). This reaction, which is catalyzed by DNA methyltransferase enzymes (10), has been shown to favor promoter-associated CpG-rich regions (CpG islands) of the DNA (8, 11-13). The methylation of cytosine and certain other epigenetic events (e.g., reactions of oxygen free radicals; ref. 14) leading to base modifications would be expected to interfere with the normal vertical base stacking properties of DNA, which, in turn, would induce conformational changes in the phosphodiester-deoxyribose backbone (15, 16). These structural alterations are likely to affect the fidelity of DNA replication and transcription (17, 18).
We previously have demonstrated that highly sensitive statistical models of Fourier transform-infrared (FT-IR) spectra are capable of identifying subtle differences in the base and backbone structures of DNA in normal and abnormal tissues. This technology has been used successfully to discriminate between DNA structures of the normal and cancerous breast (19-23) and prostate (24, 25). The structural differences were the basis for constructing statistical models that predict the probability of cancer vs. noncancer with a high sensitivity and specificity (21, 23-25).
In this article, we tested the hypothesis that statistical models of FT-IR spectra, adjusted for age and batch effects, would discriminate between granulocyte DNA from MDS patients and patients lacking this clonal disease. We describe the application of FT-IR statistical models to a human blood disorder and show that normal granulocyte DNA is readily distinguishable from the granulocyte DNA of MDS patients. Moreover, a discriminant analysis model classified with high sensitivity and specificity which DNA samples were from normal granulocytes and which were from patients with MDS, thus setting the stage for future clinical applications.
Materials and Methods
Granulocyte Purification. With approval of the Johns Hopkins Medical Institution's Institutional Review Board, excess whole blood from specimens obtained in EDTA anticoagulant for diagnostic testing (one specimen per patient) were used as the source for both MDS granulocytes and control granulocytes. The study was double-blinded and included 21 control and 12 MDS samples. There was a significant difference (P = 0.03) in the mean ages of the control (55.4 ± 18.3 years) and MDS (69.8 ± 14.2 years) groups. These differences were taken into account in the development of statistical models. Gender composition did not differ significantly between groups (100% male in MDS group and 70% male in control group; P = 0.07) and thus was not included in any models. The diagnosis of MDS was based on the French-American-British (FAB) classification (5) together, when possible, with cytogenetic analysis after exclusion of benign causes for the blood and marrow abnormalities. The International Prognostic Scoring System (IPSS) classifications (26) for the MDS samples were distributed as follows: low (four samples), intermediate-1 (five samples), intermediate-2 (one sample), and “no cytogenetics” (two samples). The World Health Organization classifications (6) were as follows: RARS (six samples), RAEB (one sample), MDSU (two samples), and RCMD (three samples). The 21 control samples included 9 from patients with no known blood disorders and 12 from patients with nonclonal blood disorders (e.g., hemochromatosis, erythrocytosis, and anemia). (One patient assigned to the control group was subsequently diagnosed with chronic myeloid leukemia.)
The blood samples (≈5 ml) were obtained over two separate time periods (batches) from patients attending the general hematology and MDS clinics at the Johns Hopkins Medical Institution. The blood specimens were diluted with 2 vol of phosphate-buffered normal saline (pH 7.4) containing 0.05% BSA and 0.6% sodium citrate. The diluted blood was layered over 15 ml of ficoll-hypaque (Ficoll-Paque Plus; Amersham Pharmacia Biosciences) and centrifuged at 835 × g for 30 min at room temperature. After removal of the supernatant, the cell pellet was resuspended in 15 ml of 0.83% ammonium chloride and incubated with gentle mixing for 15 min at room temperature to lyse the erythrocytes. The lysed cell preparation was centrifuged at 470 × g for 10 min at room temperature, and, if necessary, the lysis procedure was repeated to remove residual erythrocytes. The cell pellet was washed twice with phosphate-buffered normal saline and inspected for granulocyte purity by light microscopy with Wright-stained cytocentrifuge preparations. Granulocyte purity was always >98%. The granulocytes were centrifuged at 210 × g for 10 min at room temperature and then frozen and stored at -80°C.
DNA Extraction. DNA (≈25 μg) was obtained from the granulocytes with Qiagen 100/G Genomic-tips (Qiagen, Chatsworth, CA) by using a modification of the Qiagen extraction procedure (24). The DNA was washed three times with ice-cold 70% ethanol and subsequently dissolved in 10-40 μl (depending on the size of the pellet) of optima grade distilled water (Fisher Scientific). The Qiagen procedure is an ion-exchange system and does not constitute a source for artifactual oxidation of DNA during extraction.
FT-IR Spectral Analysis. DNA analysis by FT-IR spectroscopy was conducted as described (24), and as with the DNA extraction all samples were randomized to mitigate batch effects between control and MDS samples. Briefly, aliquots (0.2 μl) of DNA solution were spotted on a BaF2 plate and allowed to spread, forming an outer ring containing the DNA. Two separate spots (splits) were created for each sample and allowed to dry. Spotting was repeated until the ring was at least 100 μm wide, the width of the aperture of the FT-IR microscope spectrometer (System 2000, Perkin-Elmer). The plate was placed in a lyophilizer for 1 hr to completely dry the DNA. Ten spectral determinations were made around each of the two rings per sample, and the percent transmittance was Fourier-transformed into absorbance values. Each spectrum was baselined and then normalized to have a mean of 1.0 in the range of 1,750 to 1,550 cm-1 (27). Baselining and normalization adjusted for the optical characteristics of each sample (e.g., related to film thickness). The mean absorbance of the 20 spectra per sample was determined at each integer wavenumber between 1,750 and 700 cm-1.
Statistical Analyses. Generally, these analyses were performed as described (19, 24). Principal components analysis was used to integrate different properties of the spectra (e.g., varying peak heights, peak locations, and various combinations thereof). This analysis yields summary variables that capture most of the variation in the data set. Principal components analysis was undertaken on the data set composed of the mean spectra for each sample. The first 10 principal component (PC) scores were retained for comparison between the two DNA groups (28).
Histograms and scatter plots of combinations of the 10 PC scores suggested the presence of outliers, so a formal test for outliers was carried out by using the method of Hadi (29). Three outliers were identified, and these were significantly different from the balance of the samples, with P values between 0.01 and 0.001. Each outlier was from 3.9 to 5.9 standard deviation units different from the mean of the nonoutlier samples for at least 1 of the 10 PC scores. One of the outliers was a control sample and two were MDS samples. After removal of outliers, FT-IR spectral analyses were carried out on the DNA of the remaining 20 control and 10 MDS samples. It should be noted, however, that statistical analyses before the removal of outliers yielded good discrimination with high sensitivity and specificity.
The t test was used to compare mean PC scores between the control and MDS samples, between male and female patients, and between the two laboratory batches of specimens. Discriminant analysis was used to build a model to classify samples (19). A Receiver Operating Characteristic (ROC) curve was constructed by using probabilities of group membership determined from the discriminant analysis. Variables used in the discriminant analysis were drawn from the collection of PC scores, age, and a dichotomous indicator of batch. Because some PC scores differed significantly between batches, the discriminant analysis models were fit separately in each batch. The model provides the predicted probability of membership in the MDS group for each DNA sample.
In contrast to the discriminant analysis model using the PC scores, an additional extensive series of discriminant analyses were carried out (one analysis per integer wavenumber). The resulting model predicts group membership from the normalized absorbances (at the specific wavenumber), along with the other independent variables of batch, an absorbance-batch interaction term, and age. This series of analyses was carried out in the R statistical language (30) and yielded a total of 1,051 discriminant analysis models. The P value of interest from these models reflects the contribution of spectral absorbance to the prediction of group membership. Formally, the P value at a given wavenumber results from a test of the null hypothesis that absorbance at the wavenumber and the absorbance-batch interaction do not incrementally improve the prediction of group membership when they are added to a model that includes age and batch. A plot of these P values vs. wavenumbers is useful in identifying regions of the spectrum where the absorbance differs between the two groups. Regions of the spectrum with P ≤ 0.05 are likely to reflect real structural differences between the DNA groups when they account for >5% of the spectral range (17). In addition, a predicted probability and risk score (logit of predicted probability) for MDS group membership vs. control was obtained by means of discriminant analysis with age, batch, and PC scores. The resulting model allowed us to estimate the probability that a DNA sample was derived from an MDS patient.
Results and Discussion
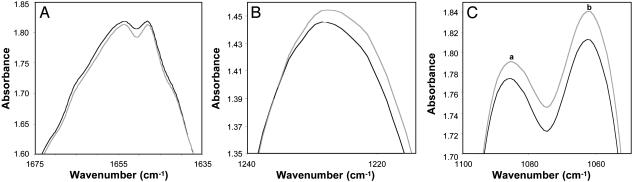
A comparison of the mean DNA spectra of the control (n = 20) and MDS (n = 10) samples revealed statistically significant differences (P ≤ 0.05) over 43.4% of the total spectral range (1,750-700 cm-1). This result is considerably more than the 5% that would be expected by chance. Four spectral peaks are shown in Fig. 1 in which comparisons of strong peak amplitudes indicate significant structural differences in DNA between the control and MDS groups. These differences reflect in-plane ring and C═N stretching vibrations of cytosine at 1,651 cm-1 (P = 0.02; Fig. 1A) (31, 32), antisymmetric stretching vibrations of the PO2- structure at 1,230 cm-1 (P = 0.046; Fig. 1B), symmetric stretching vibrations of the PO2- structure at 1,084 cm-1 (P = 0.001; Fig. 1C, peak a), and C─O stretching vibrations assigned to ribose/deoxyribose at 1,062 cm-1 (P = 0.0001; Fig. 1C, peak b). In addition to these important differences shown in Fig. 1, a significant difference was found in the weak peak at 1,481 cm-1 (P = 0.048), representing in-plane vibrations of base residues for N─H and C─H deformation modes (data not shown). Differences in several weak peaks also were found between ≈950 and 760 cm-1. These differences represent various ribose-phosphate main-chain vibrations (31). The significant spectral differences found over a substantial portion of the spectral range clearly demonstrate that the transformation of the control granulocyte DNA to the MDS structure involves readily discernable alterations in both the nucleotide bases and backbone. The differences in the cytosine peaks for the control and MDS samples at 1,651 cm-1 may well arise from methylation reactions (2, 12). Because patients were not on medications known to affect DNA methylation, it is reasonable to speculate that the observed differences in cytosine vibrations between the groups may be a consequence of this reaction (7-9), although other factors cannot be ruled out. In addition, the significant backbone difference found between the two DNA groups (represented by the strong peak at 1,062 cm-1) may have at least partly arisen (i.e., by means of induction) from the disruption in vertical base stacking interactions (31) resulting from cytosine methylation (15, 16), or even from alterations occurring in other base structures. This finding seems plausible given that the introduction of a single oxygen atom on an adenine in a 25-base oligonucleotide strand induced conformational changes in the backbone (17).
Fig. 1.
Mean DNA spectra of the control (gray line; n = 20) and MDS (black line; n = 10) groups are compared. Three spectral areas are shown, each containing strong peaks representing significant (P < 0.05) differences in peak amplitudes. (A) Cytosine at 1,651 cm-1. (B) The antisymmetric stretching vibration of the PO2- at 1,230 cm-1.(C) Peak a shows the symmetric stretching vibration of the PO2- at 1,084 cm-1, and peak b shows the C─O stretching vibration of ribose/deoxyribose at 1,062 cm-1 (31).
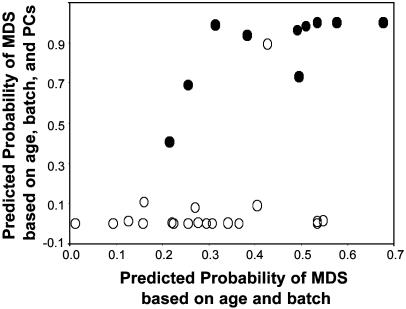
The differences between the mean spectra are major factors influencing the results subsequently obtained by principal components analysis, which involves ≈106 correlations between absorbance values relating to diverse spectral properties, such as peak heights and locations. Analysis of PC scores showed significant differences between the control and MDS samples. Among the 10 PC scores used, 3 PCs added significantly to the prediction of group membership in models including age and calculated separately by batch (with P ≤ 0.01 used conservatively to designate significance in this analysis): PC3 in the first batch (P = 0.002) and PC4 (P = 0.01) and PC7 (P = 0.003) in the second batch. By using discriminant analysis, 10 PC scores were compared between control and MDS groups in each of the two batches, controlling for age. Of the 20 tests of PC scores, 5 yielded P values between 0.002 and 0.05, whereas only 1 with P < 0.05 would be expected by chance and only 2 of 1,000 tests would be expected to yield P ≤ 0.002. The gain in discrimination between control and MDS samples by use of the FT-IR data is illustrated in Fig. 2, which includes two sets of predicted probabilities of MDS group membership from discriminant analysis. The horizontal axis is the predicted probability based only on age and batch, and the vertical axis is the predicted probability based on age, batch, and the PC scores. Horizontally, there is considerable overlap of controls and MDS cases, but vertically there is almost complete separation, with only one sample outside its correct group. This finding indicates that the discrimination is caused by real differences in DNA structure, as reflected by PC scores, rather than age or batch alone.
Fig. 2.
Predicted probability of MDS (•; n = 10) group membership vs. control (○; n = 20) from discriminant analysis models based on (i) age and batch alone (horizontal axis) vs. (ii) age, batch, and PC scores (vertical axis) from FT-IR spectra. Note the superior discrimination between the two groups in the vertical direction (incorporating spectral information) compared with the horizontal direction (ignoring spectral information). See text for details.
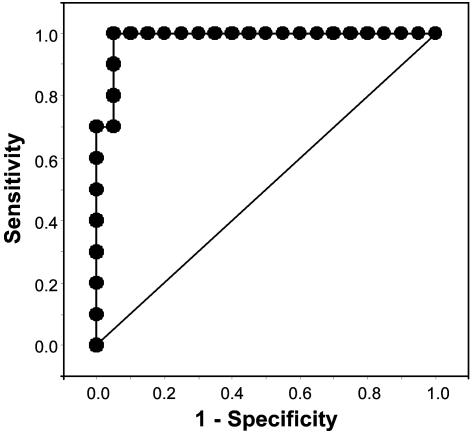
The ROC curve for the full model incorporating the spectral, age, and batch data is presented in Fig. 3. This model has an area under the curve of 0.985 of a possible maximum of 1.0, which indicates almost perfect discrimination. The model based on age and batch alone had an area of 0.758. The diagonal reference line (Fig. 3) represents an ROC curve for a hypothetical diagnostic marker unrelated to the disease status and, therefore, has an area under the curve of 0.5 (33). By using a cut point ≥0.4 for predicted probability of a sample having the characteristics of MDS, the sensitivity was 100% and the specificity was 95%. The predictive model with age and batch alone had a sensitivity and specificity of 70% and 75%, respectively.
Fig. 3.
ROC curve based on predicted probability of a granulocyte sample being derived from a patient with MDS, using 20 control and 10 MDS samples. The more closely the curve follows the top and left-hand border of the ROC space, the more accurate the test. Area under the curve = 0.985.
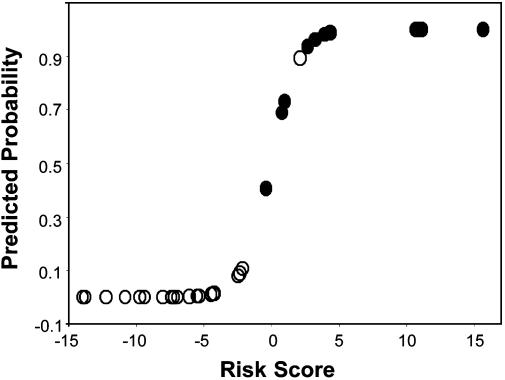
Discriminant analysis of age, batch, and PC data provided a highly promising model for estimating the probability that a particular DNA sample was derived from a patient with MDS vs. a patient free of the disease (Fig. 4). It is noteworthy that 19 of the 20 control samples are clustered at the bottom end of the curve at or below ≈0.14 predicted probability of MDS. The one anomalous control sample is clustered with the MDS samples at ≈0.9 predicted probability of MDS (the same sample is clustered with the MDS samples in Fig. 2). This model has an almost perfect sensitivity and specificity, as illustrated by the ROC curve in Fig. 3. It is interesting to speculate that the one anomalous control may well be a DNA sample whose structure has changed, reflecting an early stage in the development of MDS, although collaborative evidence was not obtained at the cellular level.
Fig. 4.
Predicted probability and risk score (logit of predicted probability) of MDS (•; n = 10) group membership vs. control (○; n = 20) from discriminant analysis models based on age, batch, and PC scores from FT-IR spectra.
MDS comprises a spectrum of clonal hematopoietic stem cell disorders of unknown etiology. In the absence of widely applicable clonal markers, the identification of MDS currently relies on morphologic features of blood and bone marrow cells. A number of classification schemes have been proposed for this disease, based on morphologic characteristics, for purposes of risk stratification (5, 6); however, no one classification has met with universal approval (34, 35). Moreover, when there are no cytogenetic abnormalities present, these schemes are not always helpful in distinguishing MDS from nonclonal forms of bone marrow failure for which there are effective therapies. Because <50% of MDS patients (particularly those in the early stages of the disorder) will have a nonrandom chromosomal abnormality (26), the development of a test that can distinguish MDS from the disorders that it mimics would be of great clinical utility.
Conclusions
The results of this initial, double-blinded study indicate unequivocally that MDS granulocyte DNA is distinguishable from normal granulocyte DNA by using the FT-IR statistical models (taking age and possible batch effects into account). The underlying basis for this capability was the structural differences found between the mean DNA spectra of the control and MDS groups (Fig. 1).
This is an initial molecular test for MDS, other than cytogenetic analysis, and one that is likely based on one or more biochemical abnormalities, such as aberrant DNA methylation that is reported to be associated with the evolution of this disorder (7-9, 36). In this study, which involved only a small number of patients, no attempt was made to correlate the French-American-British (FAB) classification or the International Prognostic Scoring System (IPSS) score with the FT-IR spectral data. However, we have established the ability of the spectral models to credibly identify DNA structural alterations in MDS granulocytes and used these data to construct statistical models for classifying, with high sensitivity and specificity, whether a patient has MDS. Correlation of the predictive model data with findings from various morphologic and prognostic classification schemes currently used clinically will be of great interest and importance and is anticipated to lead to a highly discriminating blood test.
Acknowledgments
We thank Drs. Karl Erik Hellström, Kandace Williams, Tom K. Hei, and Peter J. Quesenberry for helpful comments, Blazej Neradilek for assistance with the statistical evaluations, and Mary Ann Isaacs for preparing the granulocytes.
Abbreviations: MDS, myelodysplastic syndrome; FT-IR, Fourier transform-infrared; PC, principal component; ROC, Receiver Operating Characteristic.
References
- 1.Heaney, M. L. & Golde, D. W. (1999) N. Engl. J. Med. 340, 1649-1660. [DOI] [PubMed] [Google Scholar]
- 2.Silverman, L. R. (2001) Oncologist 6, Suppl. 5, 8-14. [DOI] [PubMed] [Google Scholar]
- 3.Aul, C., Germing, U., Gattermann, N. & Minning, H. (1998) Leuk. Res. 22, 93-100. [DOI] [PubMed] [Google Scholar]
- 4.Silverman, L. R. (2000) in Cancer Medicine e.5, eds. Bast, R. C., Kufe, D. W., Pollock, R. E., Weischselbaum, R. R., Holland, J. F. & Frei, E. (B. C. Decker, Hamilton, Canada), pp. 1931-1946.
- 5.Bennett, J. M., Catovsky, D., Daniel, M. T., Flandrin, G., Galton, D. A., Gralnick, H. R. & Sultan, C. (1982) Br. J. Haematol. 51, 189-199. [PubMed] [Google Scholar]
- 6.Vardiman, J. W., Harris, N. L. & Brunning, R. D. (2002) Blood 100, 2292-2302. [DOI] [PubMed] [Google Scholar]
- 7.Tien, H. F., Tang, J. L., Tsay, W., Liu, M. C., Lee, F. Y., Wang, C. H., Chen, Y. C. & Shen, M. C. (2001) Br. J. Haematol. 112, 148-154. [DOI] [PubMed] [Google Scholar]
- 8.Melki, J. R. & Clark, S. J. (2002) Semin. Cancer Biol. 12, 347-357. [DOI] [PubMed] [Google Scholar]
- 9.Voso, M. T., Scardocci, A., Guidi, F., Zini, G., Di Mario, A., Pagano, L., Hohaus, S. & Leone, G. (2004) Blood 103, 698-700. [DOI] [PubMed] [Google Scholar]
- 10.Bestor, T. H. (2000) Hum. Mol. Genet. 9, 2395-2402. [DOI] [PubMed] [Google Scholar]
- 11.Wachsman, J. T. (1997) Mutat. Res. 375, 1-8. [DOI] [PubMed] [Google Scholar]
- 12.Toyota, M., Kopecky, K. J., Toyota, M. O., Jair, K. W., Willman, C. L. & Issa, J. P. J. (2001) Blood 97, 2823-2829. [DOI] [PubMed] [Google Scholar]
- 13.Parker, B. S., Cutts, S. M. & Phillips, D. R. (2001) J. Biol. Chem. 276, 15953-15960. [DOI] [PubMed] [Google Scholar]
- 14.Aul, C., Bowen, D. T. & Yoshida, Y. (1998) Haematologica 83, 71-86. [PubMed] [Google Scholar]
- 15.Forde, G., Gorb, L., Shiskin, O., Flood, A., Hubbard, C., Hill, G. & Leszczynski, J. (2003) J. Biomol. Struct. Dyn. 20, 819-828. [DOI] [PubMed] [Google Scholar]
- 16.Banyay, M. & Graslund, A. (2002) J. Mol. Biol. 324, 667-676. [DOI] [PubMed] [Google Scholar]
- 17.Malins, D. C., Polissar, N. L., Ostrander, G. K. & Vinson, M. A. (2000) Proc. Natl. Acad. Sci. USA 97, 12442-12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner, B. M. (2001) Chromatin and Gene Regulation: Mechanisms in Epigenetics (Blackwell Scientific, Oxford).
- 19.Garcia-Closas, M., Hankinson, S. E., Ho, S., Malins, D. C., Polissar, N. L., Schaefer, S. N., Su, Y. & Vinson, M. A. (2000) J. Natl. Cancer Inst. Monogr. 27, 147-156. [DOI] [PubMed] [Google Scholar]
- 20.Malins, D. C., Polissar, N. L., Schaefer, S., Su, Y. & Vinson, M. (1998) Proc. Natl. Acad. Sci. USA 95, 7637-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malins, D. C., Polissar, N. L., Su, Y., Gardner, H. S. & Gunselman, S. J. (1997) Nat. Med. 3, 927-930. [DOI] [PubMed] [Google Scholar]
- 22.Malins, D. C., Polissar, N. L. & Gunselman, S. J. (1996) Proc. Natl. Acad. Sci. USA 93, 14047-14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malins, D. C., Polissar, N. L., Nishikida, K., Holmes, E. H., Gardner, H. S. & Gunselman, S. J. (1995) Cancer 75, 503-517. [DOI] [PubMed] [Google Scholar]
- 24.Malins, D. C., Johnson, P. M., Barker, E. A., Polissar, N. L., Wheeler, T. M. & Anderson, K. M. (2003) Proc. Natl. Acad. Sci. USA 100, 5401-5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malins, D. C., Polissar, N. L. & Gunselman, S. J. (1997) Proc. Natl. Acad. Sci. USA 94, 259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenberg, P., Cox, C., LeBeau, M. M., Fenaux, P., Morel, P., Sanz, G., Sanz, M., Vallespi, T., Hamblin, T., Oscier, D., et al. (1997) Blood 89, 2079-2088. [PubMed] [Google Scholar]
- 27.Malins, D. C., Hellström, K. E., Anderson, K. M., Johnson, P. M. & Vinson, M. A. (2002) Proc. Natl. Acad. Sci. USA 99, 5937-5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malins, D. C., Holmes, E. H., Polissar, N. L. & Gunselman, S. J. (1993) Cancer 71, 3036-3043. [DOI] [PubMed] [Google Scholar]
- 29.Hadi, A. S. (1994) J. R. Stat. Soc. B 56, 393-396. [Google Scholar]
- 30.Maindonald, J. H. & Braun, J. (2003) Data Analysis and Graphics Usingr: An Example-Based Approach (Cambridge Univ. Press, Cambridge).
- 31.Tsuboi, M. (1969) Appl. Spectrosc. Rev. 3, 45-90. [Google Scholar]
- 32.Brewer, S. H., Anthireya, S. J., Lappi, S. E., Drapcho, D. L. & Franzen, S. (2002) Langmuir 18, 4460-4464. [Google Scholar]
- 33.Pepe, M. S. (2003) The Statistical Evaluation of Medical Tests for Classification and Prediction (Oxford Univ. Press, Oxford).
- 34.Nösslinger, T., Reisner, R., Koller, E., Grüner, H., Tüchler, H., Nowotny, H., Pittermann, E. & Pfeilstöcker, M. (2001) Blood 98, 2935-2941. [DOI] [PubMed] [Google Scholar]
- 35.Germing, U., Gattermann, N., Strupp, C., Aivado, M. & Aul, C. (2000) Leuk. Res. 24, 983-992. [DOI] [PubMed] [Google Scholar]
- 36.Leone, G., Teofili, L., Voso, M. T. & Lübbert, M. (2002) Haematologica 87, 1324-1341. [PubMed] [Google Scholar]