Abstract
Sinorhizobium meliloti, a legume symbiont, and Brucella abortus, a phylogenetically related mammalian pathogen, both require the bacterial-encoded BacA protein to establish chronic intracellular infections in their respective hosts. We found that the bacterial BacA proteins share sequence similarity with a family of eukaryotic peroxisomal-membrane proteins, including the human adrenoleukodystrophy protein, required for the efficient transport of very-long-chain fatty acids out of the cytoplasm. This insight, along with the increased sensitivity of BacA-deficient mutants to detergents and cell envelope-disrupting agents, led us to discover that BacA affects the very-long-chain fatty acid (27-OHC28:0 and 29-OHC30:0) content of both Sinorhizobium and Brucella lipid A. We discuss models for how BacA function affects the lipid-A fatty-acid content and why this activity could be important for the establishment of chronic intracellular infections.
Sinorhizobia and brucellae are Gram-negative α-proteobacteria that live intracellularly within their respective hosts. Sinorhizobia form a beneficial symbiosis with agriculturally important legumes that results in the conversion of N2 to NH3 (1). In contrast, brucellae are highly infectious pathogens that cause abortions and infertility in domestic and wild mammals and a severe and debilitating zoonotic disease in humans (2). Brucella melitensis, Brucella suis, and Brucella abortus are potential biological warfare agents, and they are a serious concern because there is presently no human vaccine (3). Despite the strikingly different outcomes that sinorhizobia and brucellae eventually have on their hosts, commonalties exist in the chronic-infection process because both are endocytosed into host cells, where they adapt and survive for extensive periods of time within acidic, membrane-bound compartments (1, 2, 4, 5). More importantly, the close phylogenetic relatedness of the sinorhizobia and the brucellae that was revealed initially by RNA homology studies has been confirmed recently by determination of the complete genome sequences of Sinorhizobium meliloti, B. melitensis, and B. suis (6-8).
The BacA protein, initially found to be essential for S. meliloti to form a long-term infection within alfalfa-plant cells (9), was also shown subsequently to be essential for the establishment and maintenance of chronic spleen and liver infections by B. abortus in BALB/c mice (10). BacA is predicted to span the inner membrane of S. meliloti and B. abortus seven times, and it is homologous to the SbmA protein of Escherichia coli, a putative transporter of peptide antibiotics (9, 11). Although an S. meliloti bacA null mutant displays altered sensitivity to peptide antibiotics (11), the increased sensitivity of this mutant to detergents and cell envelope-disrupting agents supports an alternative model wherein the function of BacA affects the integrity of the bacterial cell envelope (12). Recently, an S. meliloti lpsB mutant, altered dramatically in its lipopolysaccharide (LPS) carbohydrate content, was found to be defective at the same stage as a bacA mutant in establishing a chronic infection of alfalfa nodule cells (13). Unlike the S. meliloti lpsB mutant, however, the S. meliloti bacA mutant does not have a dramatic alteration in the carbohydrate composition of its LPS (12). Here, we report our findings that the S. meliloti and B. abortus BacA proteins, essential for the chronic-infection process in plants and animals, respectively, are related distantly to a family of peroxisomal-membrane proteins. This insight, combined with our physiology data for S. meliloti and B. abortus bacA null mutants that suggested that they had altered cell envelopes, led us to discover that BacA affects the LPS fatty-acid content.
Materials and Methods
Bacterial Growth and LPS Extraction. The sequenced S. meliloti strain Rm1021 (wild type) and an isogenic Rm1021 bacA null mutant were used in this study (11, 12). The plasmids pBacA D198G, pBacA R284G, pBacA Q193G, and pBacA R389G containing site-directed mutations in the S. meliloti bacA gene were constructed (14). Expression of the site-directed mutant BacA proteins was determined by a dominant-negative test in the presence of the wild-type BacA protein. For this study, the bacA mutant plasmids were transferred from E. coli into the Rm1021 bacA null mutant by triparental mating by using an E. coli strain containing the helper plasmid pRK600. As described in the Rm8002 bacA null background (14), all four mutant BacA proteins produced from the plasmids in the Rm1021 bacA null background were unable to function in the alfalfa symbiosis (data not shown). As controls, the plasmid pRK404 alone with no insert (pControl) and the plasmid pJG51A (9) containing a wild-type S. meliloti bacA gene (pBacAWT) were also transferred to Rm1021 bacA null mutant, and they produced the expected responses on alfalfa plants (11).
For the LPS extractions, cultures of the appropriate S. meliloti strain were grown to stationary phase in Luria-Bertani (LB) media supplemented with 2.5 mM MgS04/2.5 mM CaCl2/500 μg·ml-1 streptomycin (Sm500). The cultures were then centrifuged, and cell pellets were washed with LB, resuspended to OD600 ≈0.2 in 4 liters of LB/Sm500 and grown until midexponential phase (OD600 ≈1-2). The bacteria were then harvested by centrifugation and resuspended in 150 ml of EDTA solution (0.05 M Na2HPO4/5 mM EDTA), and the LPS was extracted into the aqueous phase by using a hot phenol/water extraction procedure (15). LPS was extracted from LB-grown cultures because it has been shown (12) that divalent cations increase the stress resistance of S. meliloti and mask the phenotypes of an S. meliloti bacA null mutant. When S. meliloti-containing plasmids were grown, the media was also supplemented with 5 μg·ml-1 tetracycline.
B. abortus is a biosafety level 3 pathogen that is highly infectious by means of the aerosol route in a laboratory setting. To avoid potential aerosol exposure associated with the processing of large-scale liquid cultures, B. abortus strains were cultivated on solid growth medium. B. abortus 2308 and the bacA null mutant KL7 (10) were streaked from -80°C stocks onto Schaedler agar (Becton Dickinson) supplemented with 5% defibrinated bovine blood and incubated at 37°C for 48-72 h. The colonies from five plates were pooled, and the brucellae were heat-killed after a standard procedure (16). After testing for loss of viability, the LPS was extracted under BL2 conditions from at least four combined pools by using the hot phenol/water procedure (15). The LPS from B. abortus partitions into the phenol phase, as described (17). Deoxycholate gradient plates were prepared as described, except that Brucella broth agar (Becton Dickinson) replaced LB agar (12).
Lipid-A Isolation. Lipid A was isolated from the crude LPS extractions by mild-acid hydrolysis (18). LPS (≈5 mg) was dissolved in 1 ml of 1% SDS in 10 mM sodium acetate (pH adjusted to 4.5 with 4 M HCl) and dissolved by incubation in an ultrasound bath. The sample was then heated 100°C for 1 h and freeze-dried, and the SDS was removed by washing the dried residue with 100 μl of distilled water and 500 μl of acidified ethanol (prepared by combining 100 μl of 4 M HCl with 20 ml of 95% ethanol), followed by centrifugation (2,000 × g, 20 min). The sample was then washed with 500 μl of nonacidified 95% ethanol and centrifuged. The centrifugation steps and washing steps were then repeated. Finally, the sample was lyophilized to give purified lipid A.
Matrix-Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) MS of Purified Lipid A. Lipid A samples were analyzed by MALDI-TOF MS by using an LD-TOF system (Hewlett-Packard). Lipid A samples were prepared to a concentration of 2 μg·ml-1 in 2:1 CHCl3/CH3OH, and 1 μl was mixed with the matrix (trihydroxyacetone in methanol) for analysis. These conditions were essential to solubilize lipid A with and without very-long-chain fatty acid (VLCFA) modifications. The proposed composition of the major identified lipid A species were determined based on the GC-MS fatty-acid composition data and the reported structures of the Sinorhizobium NGR234 (19) and B. abortus (20) lipid A species.
Lipid A Fatty-Acid Quantification by GC-MS. The fatty acids were identified and quantitated by combined GC-MS of their trimethylsilyl (TMS) ether (because these are hydroxy fatty acids) methyl esters. Each sample was treated with methanolic 1 M HCl at 80°C for 18 h. The methanolic HCl was then evaporated by using a stream of nitrogen, and the TMS derivatives were prepared by the addition of Tri·Sil reagent (Pierce) and heating at 80°C for 25 min. The solvent was removed by using a stream of nitrogen, and the resulting TMS fatty-acid methyl esters were dissolved in hexane and analyzed by GC-MS using a 30-m DB-5 capillary column (J & W Scientific, Folsom, CA).
Results
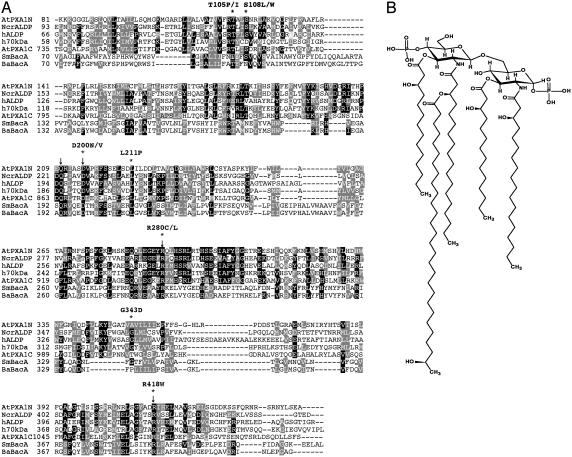
BacA Shares Sequence Similarity with Peroxisomal-Membrane Proteins. A possible function for BacA was suggested by our observation that its sequence shares similarity with a family of peroxisomal-membrane proteins that include the human adrenoleukodystrophy protein (hALDP) (Fig. 1A), consistent with the idea that BacA is a distant member of this family. This finding was intriguing because several members of this family are thought to be involved in the efficient transport of either VLCFAs or long-chain fatty acids out of the cytoplasm into peroxisome, where they can then be degraded (21-24). One member of this family, hALDP, is affected in patients with X-linked adrenoleukodystrophy (25), an X-linked disorder that is secondary to a mutation in the ABCD1 gene (which encodes hALDP). Genetic defects in hALDP result in defects in peroxisomal β-oxidation (21) and the accumulation of VLCFAs in all tissues of the body (26). Our analysis has revealed that several identified (25) missense mutations that alter hALDP function affect amino acids conserved with S. meliloti BacA (Fig. 1 A). We also determined by using information from a genetic study (14) of S. meliloti bacA missense mutants that changes in four different amino acids in BacA, which prevent BacA from functioning effectively in the symbiosis with alfalfa plants, are conserved with amino acids in hALDP (Fig. 1 A).
Fig. 1.
Alignment of BacA proteins with proteins affecting long-chain fatty acid and VLCFA transport and structure of major lipid A in S. meliloti showing VLCFA modification. By using position-specific iterative blast (NCBI), we identified that S. meliloti BacA appears to be distantly related to the central domain of a family of peroxisomal-membrane proteins (position-specific iterative blast results showed that S. meliloti BacA residues 63-418 share 32% similarity and 12% identity with residues 98-453 of the Neurospora crassa related to adrenoleukodystrophy protein). Standard blast (NCBI), as had been used in our earlier sequence comparisons of S. meliloti BacA (14), does not detect this similarity. (A) Clustal T-Coffee 1.41 (available at http://ch.EMBnet.org) (see ref. 45) was then used to align bacterial BacA proteins with several members of the peroxisomal-membrane protein family. The following protein sequences were aligned: S. meliloti BacA (SmBacA; GenBank accession no. CAC49525), B. abortus BacA (BaBacA; GenBank accession no. AAF76873), N. crassa related to hALDP (NcrALDP; GenBank accession no. CAB91246), hALDP (GenBank accession no. P33897), human 70-kDa peroxisomal-membrane protein (h70Kda; GenBank accession no. CAA4146), and Arabidopsis thaliana PXA1 (AtPXA1, also known as COMATOSE, NP568072), both N- and C-terminal domains. Identical and similar residues throughout the alignment are shown in black and gray, respectively, by using the parameters of box shade 3.21 (available at http://ch.EMBnet.org). Human ALDP residues altered in X-linked adrenoleukodystrophy patients that affect residues conserved with S. meliloti BacA residues are indicated (*); the precise hALDP residues changes are shown. Arrows indicate the four residues altered in the S. meliloti bacA missense mutants (pBacA Q193G, pBacA D198G, pBacA R284G, and R389G) (14) that change residues conserved with hALDP. (B) Major lipid-A species in free-living S. meliloti, as determined by our composition analysis (see Fig. 2) and the mass spectra of the published lipid-A species of Sinorhizobium NGR234 (19). The lipid-A molecule shown is modified by 27-OHC28:0, which can be replaced by 27-O (βOMeC4:0) C28:0 (forming another major species) or 29-OHC30:0 (forming a minor species).
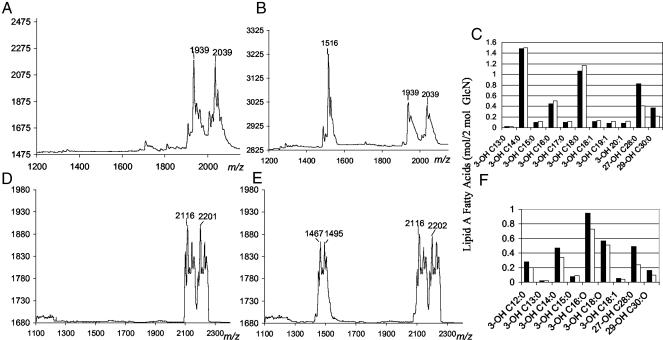
Sinorhizobium and Brucella BacA Affect Lipid A Fatty Acids. The involvement of hALDP in a process involving VLCFAs was of particular interest because Sinorhizobium and Brucella LPS each contain a saturated VLCFA of 27-OHC28:0, 27-O (βOMeC4:0) C28:0, or 29-OHC30:0 (27). VLCFAs are attached to the lipid-A component of Sinorhizobium LPS (Fig. 1B), and each lipid-A molecule of free-living wild-type sinorhizobia contains one VLCFA (Fig. 2A and Table 1); lipid A lacking VLCFAs was not detected in wild-type S. meliloti. Given the relationship between BacA and hALDP, it seemed to be possible that BacA might affect the VLCFA content of S. meliloti LPS. We found that more than one half of the lipid-A molecules isolated from the S. meliloti bacA null mutant, in contrast to lipid A from wild-type sinorhizobia, lacked either a 27-OHC28:0 or 27-O (βOMeC4:0) C28:0 modification (Fig. 2B and Table 1). Furthermore, compositional analysis of the lipid-A fatty acids revealed that the S. meliloti bacA null mutant had a significant decrease in both the 27-OHC28:0 and 29-OHC30:0 levels compared with wild-type S. meliloti, whereas the levels of 3-OHC20:1 or shorter were not reduced (Fig. 2C). The remaining VLCFA modification of the lipid A that occurred in the bacA null mutant was not due to residual BacA activity because the entire promoter and ≈70% of the bacA coding sequence had been replaced with a spectinomycin cassette during the creation of the S. meliloti bacA null mutant (11).
Fig. 2.
BacA affects the lipid-A fatty-acid modifications of both S. meliloti and B. abortus. (A) MALDI-TOF MS of S. meliloti wild-type Rm1021 lipid A; major species [M - H]- 2039 and [M - H]- 1939. (B) Same as A, except S. meliloti Rm1021 bacA null mutant; major species [M - H]- 1516 with lesser amounts of [M - H]- 2039 and [M - H]- 1939. (C) Fatty-acid compositions of S. meliloti wild-type Rm1021 (filled bars) and its bacA null mutant (open bars) lipid A as determined by GC-MS. (D) MALDI-TOF MS of B. abortus wild-type 2308 lipid A; major species [M - H]- 2201 and [M - H]- 2116. (E) Same as D, except B. abortus 2308 bacA null mutant; major species [M - H]- 1495, [M - H]- 1467, [M - H]- 2202 and [M - H]- 2116. (F) The fatty-acid composition of B. abortus wild type (filled bars) and bacA null mutant (open bars) lipid A, as determined by GC-MS. For the MALDI-TOF MS experiments, the proposed composition of the major lipid-A species identified are shown in Table 1. The remaining ions of lesser intensity, some of which represent lipid A where the 27-OHC28:0 modification has been replaced with 29-OHC30:0, are due to variation in the chain length of the fatty-acyl substituents.
Table 1. Major lipid A species identified by MALDI-TOF MS.
| Observed [M - H]− | Calculated molecular weight | Proposed lipid A composition |
|---|---|---|
| 2,039 | 2,040 | GlcN2P23-OHC14:O23-OHC18:02 27-O(βOMeC4:0)C28:01 |
| 1,939 | 1,940 | GlcN2P23-OHC14:023-OH C18:02 27-OHC28:01 |
| 1,515/1,516* | 1,517 | GlcN2P23-OHC14:023-OHC18:02 |
| 2,201/2,202 | 2,203 | GlcN2P23-OHC12:013-OHC14:01 3-OHC16:023-OHC18:01C18:01 27-O(βOMeC4:0)C28:01 |
| 2,116 | 2,117 | GlcN2P23-OHC12:013-OHC14:01 3-OHC16:023-OHC18:01C18:01 27-OHC28:01 |
| 1,495* | 1,496 | GlcN2P23-OHC16:023-OHC18:01 C18:013-OHC14:01 |
| 1,467* | 1,468 | GlcN2P23-OHC16:023OHC18:01 C18:013-OHC12:01 |
Species lacking VLCFA only observed in bacA mutant strains.
Because the B. abortus bacA null mutant is defective also at long-term survival within the host cell (10), we rationalized that it could have an alteration resembling that of the S. meliloti bacA null mutant in its LPS. Similar to S. meliloti, we found that each molecule of lipid A from wild-type B. abortus contains a VLCFA modification (Fig. 2D and Table 1), whereas a significant fraction of the lipid A extracted from the B. abortus bacA null mutant lacked either a 27-OHC28:0 or 27-O (βOMeC4:0) C28:0 modification (Fig. 2E and Table 1). However, unlike the S. meliloti bacA null mutant lipid A, the brucellae lipid-A molecules lacking VLCFAs also lacked either a 3-OHC14:0 or 3-OHC12:0 modification (Fig. 2E and Table 1). The lipid-A fatty-acid composition data showed that the B. abortus bacA null mutant had the greatest reduction in 27-OHC28:0 and 29-OHC30:0 levels compared with wild-type B. abortus, with a lesser reduction in some of the shorter-chain fatty acids (Fig. 2F). Consistent with an alteration in the cell envelope, we observed that the B. abortus bacA mutant also displayed an increased sensitivity to detergents (data not shown), as has been described for the S. meliloti bacA null mutant (12).
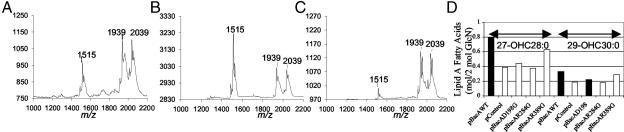
Changes in BacA Residues Conserved with hALDP Affect Lipid-A Fatty Acids. As described, we discovered that changes in four different amino acids in the S. meliloti BacA protein that are conserved with amino acids in hALDP (Fig. 1 A) prevent S. meliloti from establishing a chronic infection in alfalfa-plant cells. For three of these mutants, changes in the corresponding conserved residue in hALDP have been detected in X-linked adrenoleukodystrophy patients (Fig. 1 A). Analysis of the lipid A from these four S. meliloti bacA missense mutants revealed that, unlike the wild-type strain, they all produced a lipid-A species lacking a VLCFA (Fig. 3 and Table 1). However, the proportion of the different lipid-A species present in each mutant varied. Intriguingly, the S. meliloti bacA missense mutant R389G (14) produced only a relatively small fraction (≈20%) of its total lipid-A molecules lacking VLCFAs compared with the bacA null mutant (Fig. 3 C and D), yet it was completely defective in forming a chronic infection in alfalfa. As with the S. meliloti bacA null mutant, the bacA missense mutants did not affect the shorter-chain lipid-A fatty acids (data not shown).
Fig. 3.
Changing S. meliloti BacA residues identical to hALDP affects the amount of lipid A-containing VLCFAs. MALDI-TOF MS of lipid-A molecules from the S. meliloti bacA null mutant carrying plasmids encoding different mutant BacA proteins (14), pBacA Q193G (A), pBacA D198G or pBacA R284G (B), and pBacA R389G (C). The lipid-A molecules from the mutants were affected differently, resulting in an increase of [M - H]- 1515 (which lacks a VLCFA) at the expense of [M - H]- 1939 and [M - H]- 2039. The MALDI-TOF MS of lipid A from the S. meliloti bacA null mutant carrying either the plasmid pRK404 with no insert (pControl) or the plasmid pJG51A carrying a wild-type copy of S. meliloti bacA (pBacAWT) were virtually indistinguishable from the MALDI-TOF MS of the bacA null mutant alone (Fig. 2B) and wild-type S. meliloti (see Fig. 2 A), respectively (data not shown). (D) Fatty-acid composition of lipid A from the different mutants, as determined by GC-MS. For the MALDI-TOF MS experiments, the proposed composition of the major lipid-A species identified are shown in Table 1.
Discussion
Our data show that both S. meliloti and B. abortus BacA proteins have striking effects on the modification of at least one cell-envelope component, the LPS, by specific fatty acids. This is clear evidence that BacA, an integral membrane protein originally postulated to be a peptide transporter (9), affects specific fatty acids rather than peptides. The fact that BacA affects the degree of modification of the LPS by VLCFAs (27-OHC28:0 and 29-OHC30:0), taken together with the sequence similarity of BacA to the adrenoleukodystrophy family of proteins [some of which affect transport of activated long (C18) and VLCFAs (>C22) (21, 22-24)], suggests that the molecular mechanism of BacA action is related to that of human ALDP and other peroxisomal-membrane proteins. As described above, BacA function is critically required for S. meliloti symbiosis and B. abortus chronic pathogenesis (9, 10). Thus, the fact that BacA affects the VLCFA modification of a cell-envelope component, the LPS, both in S. meliloti and B. abortus, is consistent with the hypothesis that a BacA-mediated change in the VLCFA modification of the LPS and/or some other cell-envelope component is critically important for the chronic intracellular infections that underlie S. meliloti symbiosis and B. abortus chronic pathogenesis.
There are outstanding questions to be resolved from our analyses. (i) Is the lipid-A modification in the bacA mutants the proximal cause of the symbiotic/chronic pathogenesis defects and, if so, what is the mechanism for this? (ii) What is the molecular mechanism of the BacA protein? And (iii) why do the bacA mutants still produce some lipid A with a VLCFA modification? We will now address each of these questions in turn. For acute intracellular pathogens such as Salmonella enterica serovar Typhimurium, changes in the lipid-A structure are known to have a profound effect on virulence and resistance against vertebrate antimicrobial peptides (28-30). In contrast, the role of lipid A in the establishment of chronic intracellular infections is poorly understood. Thus, an attractive model is that the observed alterations in the fatty-acid modification of the lipid A in S. meliloti and B. abortus bacA mutants are responsible, at least in part, for their inability to establish chronic intracellular infections. However, before considering why lipid-A alterations could be important, we will discuss two observations that, at least on the surface, might seem to be inconsistent with such a model.
First, it has been shown recently that the LPS from an S. meliloti acpXL mutant (unable to synthesize a particular acyl carrier protein involved with VLCFA synthesis) (31) lacks 27-OHC28:0 under free-living conditions, yet this mutant is able to form an effective symbiosis with alfalfa (32) although substantially less efficiently than a wild-type S. meliloti (32). These observations have been used to argue that 27-OHC28:0-containing LPS is beneficial but not essential for the symbiosis. However, many acyl carrier proteins exist in S. meliloti (33, 34). Thus, it is possible that one of these is able to substitute for AcpXL during the nodulation process such that LPS of the S. meliloti acpXL mutant may contain 27-OHC28:0 modifications in planta. This seems to be a real possibility because preliminary studies of the analogous acpXL mutant in the closely related bacterium Rhizobium leguminosarum have shown that its LPS in the symbiotic state contains some 27-OHC28:0 (R.W.C., unpublished data), whereas, just as observed with the S. meliloti acpXL mutant, the LPS in the free-living state lacks 27-OHC28:0 (35). Thus, for this reason, we do not think that the free-living properties of the S. meliloti acpXL mutant rule out a critical role for BacA in symbiosis by it affecting the degree of lipid-A modification by VLCFAs.
The second issue is raised by our own data because a model hypothesizing the importance of correct lipid-A structure for chronic intracellular infection has to rationalize why reductions in the VLCFA modifications of the LPS, ranging from ≈50% for the S. meliloti and B. abortus bacA null mutants to ≈20% in the S. meliloti R389G bacA mutant, could prevent successful chronic intracellular infection. Because we were unable to detect any lipid A lacking a VLCFA modification in our LPS preparations from wild-type S. meliloti and wild-type B. abortus, it is possible that even a small fraction of LPS lacking a VLCFA modification may be sufficient to prevent a chronic infection. Furthermore, if the VLCFA content of LPS increases during S. meliloti/alfalfa symbiosis, as it does during R. leguminosarum/pea symbiosis (36), the differences in the VLCFA content of the LPS of the wild-type strains and respective bacA mutants could be even more pronounced in the host.
One possible consequence of the reduction of the VLCFA on lipid A is that it might make the LPS more easily recognized by the defense systems of the host. Lipid A is a powerful stimulator of innate immune responses in mammals, and recognition of this molecule by Toll-like receptors plays a critical role in this process (37). Toll-like receptors have been identified in plants (38), where they also contribute to innate immunity. It is well documented that rhizobial and brucellae lipid-A molecules exhibit attenuated biological activity and very low endotoxicity compared with enterobacterial lipid-A molecules (39, 40). This low level of activity of brucellae lipid A is thought to serve as a mechanism for immune evasion during the initial stages of host infection. Thus, an interesting model is that the reduced VLCFA modification of the lipid A in the S. meliloti and B. abortus bacA mutants renders the LPS more easily detectable by host Toll-like receptors, thereby inducing a robust innate immune response and accelerated clearance of the bacA mutants from their hosts. An alternative or additional consequence of the reduction of the VLCFA on lipid A is that it could compromise the integrity of the outer membrane. Unlike shorter-chain LPS fatty acids, the VLCFAs present in Sinorhizobium and Brucella LPS have the potential to span the whole outer membrane. Thus, the reduction in the amount of VLCFAs present in the lipid A of the bacA mutants could increase their sensitivity to host-derived stresses, such as exposure to low pH, reactive oxygen intermediates, and antimicrobial peptides.
What is the molecular mechanism of the BacA protein? As mentioned earlier, based not only on the sequence similarity between BacA and the ALDP family of peroxisomal-membrane proteins but also on the fact that all of these proteins affect processes involving long-chain fatty acids and VLCFAs, it seems to be likely that BacA could have a related function. It is known (22, 23, 41) that members of the ALDP family affect the transport of activated long-chain fatty acids and VLCFAs out of the cytoplasm, but their precise mechanistic role is controversial. They could either act by directly transporting the activated fatty acid out of the cytoplasm across the peroxisomal membrane or by facilitating fatty-acid transport out of the cytoplasm by some other peroxisomal-membrane protein. Similar considerations apply to the BacA proteins, which could either be involved directly in the transport of activated fatty acids out of the cytoplasm across the inner membrane or could facilitate the transport of fatty acids by another inner-membrane protein. The simplest model is that BacA is involved directly in the transport of activated fatty acids. Because the BacA protein is highly expressed throughout the symbiosis of S. meliloti with alfalfa (9), this mechanism would permit the bacteria to modify existing cell-envelope components by fatty-acid addition rather than having to resynthesize them in the modified form. Because the B. abortus BacA protein also affects the shorter-chain lipid-A fatty acids, the B. abortus protein may resemble other members of the adrenoleukodystrophy family (22) and have a broader transport specificity compared with the S. meliloti BacA protein. If this model is correct, then S. meliloti and B. abortus would need one or more proteins outside of the cytoplasm that could transfer the activated fatty acids onto the LPS and possibly other cell-envelope components. Salmonella PagP, an outer-membrane protein that transfers a fatty acid to existing LPS, is an example of this general class of protein (42). It is known that S. meliloti produces a class of nodulation factors that are acylated with ω-1-hydroxylated fatty acids ranging from C18 to C26 (43). However, the occurrence of VLCFAs on other cellular components has yet to be determined. Intriguingly, the gene encoding the S. typhimurium BacA homologue SbmA is only 14 bp away from yaiW, a gene encoding a putative outer-membrane lipoprotein. Although the S. meliloti and B. abortus bacA genes are not organized in operons and these bacteria lack yaiw, they do possess many outer-membrane lipoproteins. Thus, the S. meliloti and B. abortus BacA proteins may also affect acylation of an outer-membrane lipoprotein.
Why do the bacA mutants still produce some lipid A with a VLCFA modification? If our model presented above is correct, then in the free-living state, S. meliloti and B. abortus must have another mechanism by which lipid A can be modified with a VLCFA in the absence of BacA. However, the identity of this system remains to be determined. Although less attractive based on our sequence comparison data, we cannot rule out alternative models by which BacA could affect the lipid-A VLCFA content. In E. coli in which knowledge of the process of LPS assembly and transport is the most advanced, the lipid A is assembled on the cytoplasmic face of the inner membrane where MsbA is involved in a “flip-flop” mechanism whereby the rough LPS is moved to the periplasmic face of the inner membrane (37, 44). Many MsbA-like proteins are encoded in the S. meliloti genome. Thus, BacA could potentially influence the activity of one of the MsbA-like proteins such that, in the absence of BacA, the “flipping” of rough LPS-containing VLCFAs is less efficient. If the sole function of BacA were to modify the LPS, then this MsbA-facilitator model would provide a means by which BacA could fulfill this function. However, if BacA affects fatty-acid modification of additional cell-envelope components then the activated-fatty-acid transport mechanism would be favored.
It is also worth considering that, in addition to the cell-envelope alteration, the reduction and/or absence of BacA activity in our bacA mutants could also be detrimental to the bacterial cell within the host. Our group observed previously that plant cells infected with the S. meliloti bacA null mutant appear to accumulate aggregates of lipids that are not present during infection with wild-type S. meliloti (9). When hALDP is defective in X-linked adrenoleukodystrophy patients, VLCFAs accumulate within tissues, and this accumulation ultimately leads to their misincorporation into the myelin sheath (25, 26). Thus, if BacA affects the transport of either activated VLCFAs or rough LPS-containing VLCFA out of the cytoplasm, then in our bacA mutants in which BacA is either defective or absent, activated VLCFAs or rough LPS-containing VLCFA could accumulate, leading to adverse effects on cellular processes. Under free-living conditions, we observed that the bacA mutants do not have substantial growth defects. However, if the VLCFA level increases within the host (36), this problem could be exacerbated, contributing, at least in part, to the death of the bacA mutants in planta.
In summary, these findings present clear evidence that BacA affects a fatty-acid modification of at least one component of the bacterial cell envelope, the LPS. Although the precise role that this LPS alteration plays in the establishment of chronic infections is unclear, it is likely to be important. We have noted (14) that BacA and BacA-related proteins are present in other bacteria, including the chronic intracellular pathogen Mycobacterium tuberculosis. Because Gram-positive bacteria lack LPS, research into BacA and BacA-related proteins could better define the role of these proteins in cell-envelope modifications and the effect that such modifications have on chronic bacteria-host interactions. In addition, these studies may also yield important insights regarding the function of the ALDP family of proteins in eukaryotes.
Acknowledgments
We thank David Housman, Chris Burge, Nancy Hopkins, Harvey Lodish, Phil Reeves, and all of the members of the laboratory of G.C.W. for their critical insights. G.P.F. was funded by National Institutes of Health Grant GM31030 (to G.C.W.). G.C.W. was also supported by an American Cancer Society Research Professorship and a Howard Hughes Medical Institute Professorship. This work was also funded by Department of Energy Grant DE/FG02/93ER20097 (to the Complex Carbohydrate Research Center) and National Institutes of Health Grant GM39583 (to R.W.C.). Work in the laboratory of R.M.R. was funded by United States Army Medical Research and Material Command Contract DAMD17-98-C-8045.
Abbreviations: LPS, lipopolysaccharide; LB, Luria-Bertani; MALDI-TOF, matrix-assisted laser desorption ionization-time of flight; VLCFA, very-long-chain fatty acid; hALDP, human adrenoleukodystrophy protein.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Niner, B. M. & Hirsch, A. M. (1998) Symbiosis 24, 51-102. [Google Scholar]
- 2.Corbel, M. J. (1997) Emerg. Infect. Dis. 3, 213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franz, D. R., Jahrling, P. B., Friedlander, A. M., McClain, D. J., Hoover, D. L., Bryne, W. R., Pavlin, J. A., Christopher, G. W. & Eitzen, E. M., Jr. (1997) J. Am. Med. Assoc. 278, 399-411. [DOI] [PubMed] [Google Scholar]
- 4.Mellor, R. B. (1989) J. Exp. Bot. 40, 831-839. [Google Scholar]
- 5.Porte, F., Liautard, J. P. & Kohler, S. (1999) Infect. Immun. 67, 4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paulsen, I. T., Seshadri, R., Nelson, K. E., Eisen, J. A., Heidelberg, J. F., Read, T. D., Dodson, R. J., Umayam, L., Brinkac, L. M., Beanan, M. J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galibert, F., Finan, T. M., Long, S. R., Pühler, A., Abola, P., Ampe, F., Barloy-Hubler, F., Barnett, M. J., Becker, A., Boistard, P., et al. (2001) Science 293, 668-672. [DOI] [PubMed] [Google Scholar]
- 8.DelVecchio, V. G., Kapatral, V., Elzer, P., Patra, G. & Mujer, C. V. (2002) Vet. Microbiol. 90, 587-592. [DOI] [PubMed] [Google Scholar]
- 9.Glazebrook, J., Ichige, A. & Walker, G. C. (1993) Genes Dev. 7, 1485-1497. [DOI] [PubMed] [Google Scholar]
- 10.LeVier, K., Phillips, R. W., Grippe, V. K., Roop, R. M., Jr. & Walker, G. C. (2000) Science 287, 2492-2493. [DOI] [PubMed] [Google Scholar]
- 11.Ichige, A. & Walker, G. C. (1997) J. Bacteriol. 179, 209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson, G. P., Roop, R. M., Jr. & Walker, G. C. (2002) J. Bacteriol. 184, 5625-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell, G. R., Reuhs, B. L. & Walker, G. C. (2002) Proc. Natl. Acad. Sci. USA 99, 3938-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeVier, K. & Walker, G. C. (2001) J. Bacteriol. 183, 6444-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuhs, B. L., Kim, J. S., Badgett, A. & Carlson, R. W. (1994) Mol. Plant Microbe Interact. 7, 240-247. [DOI] [PubMed] [Google Scholar]
- 16.Alton, G. G., Jones, L. M., Angus, R. D. & Verger, J. M. (1988) Techniques for the Brucellosis Laboratory (Institut National de la Recherche, Paris).
- 17.Moreno, E., Pitt, M. W., Jones, L. M., Schurig, G. G. & Berman, D. T. (1979) J. Bacteriol. 138, 361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caroff, M., Tacken, A. & Szabo, L. (1988) Carbohydr. Res. 175, 273-282. [DOI] [PubMed] [Google Scholar]
- 19.Gudlavalleti, S. K. & Forsberg, L. S. (2003) J. Biol. Chem. 278, 3957-3968. [DOI] [PubMed] [Google Scholar]
- 20.Qureshi, N., Takayama, K., Seydel, U., Wang, R., Cotter, R. J., Agrawal, P. K., Bush, C. A., Kurtz, R. & Berman, D. T. (1994) J. Endotoxin Res. 1, 137-148. [Google Scholar]
- 21.Braiterman, L. T., Watkins, P. A., Moser, A. B. & Smith, K. D. (1999) Mol. Genet. Metab. 66, 91-99. [DOI] [PubMed] [Google Scholar]
- 22.Footitt, S., Slocombe, S. P., Larner, V., Kurup, S., Wu, Y., Larson, T., Graham, I., Baker, A. & Holdsworth, M. (2002) EMBO J. 21, 2912-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verleur, N., Hettema, E. H., van Roermund, C. W., Tabak, H. F. & Wanders, R. J. (1997) Eur. J. Biochem. 249, 657-661. [DOI] [PubMed] [Google Scholar]
- 24.Zolman, B. K., Silva, I. D. & Bartel, B. (2001) Plant Physiol. 127, 1266-1278. [PMC free article] [PubMed] [Google Scholar]
- 25.Kemp, S., Pujol, A., Waterham, H. R., van Geel, B. M., Boehm, C. D., Raymond, G. V., Cutting, G. R., Wanders, R. J. & Moser, H. W. (2001) Hum. Mutat. 18, 499-515. [DOI] [PubMed] [Google Scholar]
- 26.Valianpour, F., Selhorst, J. J., van Lint, L. E., van Gennip, A. H., Wanders, R. J. & Kemp, S. (2003) Mol. Genet. Metab. 79, 189-196. [DOI] [PubMed] [Google Scholar]
- 27.Bhat, U. R., Carlson, R. W., Busch, M. & Mayer, H. (1991) Int. J. Syst. Bacteriol. 41, 213-217. [DOI] [PubMed] [Google Scholar]
- 28.Guo, L., Lim, K. B., Poduje, C. M., Daniel, M., Gunn, J. S., Hackett, M. & Miller, S. I. (1998) Cell 95, 189-198. [DOI] [PubMed] [Google Scholar]
- 29.Jones, B. D., Nichols, W. A., Gibson, B. W., Sunshine, M. G. & Apicella, M. A. (1997) Infect. Immun. 65, 4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan, S. A., Everest, P., Servos, S., Foxwell, N., Zahringer, U., Brade, H., Rietschel, E. T., Dougan, G., Charles, I. G. & Maskell, D. J. (1998) Mol. Microbiol. 29, 571-579. [DOI] [PubMed] [Google Scholar]
- 31.Brozek, K. A., Carlson, R. W. & Raetz, C. R. (1996) J. Biol. Chem. 271, 32126-32136. [DOI] [PubMed] [Google Scholar]
- 32.Sharypova, L. A., Niehaus, K., Scheidle, H., Holst, O. & Becker, A. (2003) J. Biol. Chem. 278, 12946-12954. [DOI] [PubMed] [Google Scholar]
- 33.Geiger, O. & Lopez-Lara, I. M. (2002) FEMS Microbiol. Lett. 208, 153-162. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Lara, I. M. & Geiger, O. (2000) Microbiology 146, 839-849. [DOI] [PubMed] [Google Scholar]
- 35.Vedam, V., Kannenberg, E. L., Haynes, J. G., Sherrier, D. J., Datta, A. & Carlson, R. W. (2003) J. Bacteriol. 185, 1841-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kannenberg, E. L. & Carlson, R. W. (2001) Mol. Microbiol. 39, 379-391. [DOI] [PubMed] [Google Scholar]
- 37.Raetz, C. R. & Whitfield, C. (2002) Annu. Rev. Biochem. 71, 635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeda, K. & Akira, S. (2003) Cell. Microbiol. 5, 143-153. [DOI] [PubMed] [Google Scholar]
- 39.Rasool, O., Nnalue, N. A. & Jarstrand, C. (1992) Clin. Exp. Immunol. 90, 63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandenplas, M. L., Carlson, R. W., Jeyaretnam, B. S., McNeill, B., Barton, M. H., Norton, N., Murray, T. F. & Moore, J. N. (2002) J. Biol. Chem. 277, 41811-41816. [DOI] [PubMed] [Google Scholar]
- 41.Aubourg, P. & Dubois-Dalcq, M. (2000) Glia 29, 186-190. [DOI] [PubMed] [Google Scholar]
- 42.Bishop, R. E., Gibbons, H. S., Guina, T., Trent, M. S., Miller, S. I. & Raetz, C. R. (2000) EMBO J. 19, 5071-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demont, N., Ardourel, M., Maillet, F., Prome, D., Ferro, M., Prome, J. C. & Denarie, J. (1994) EMBO J. 13, 2139-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang, G. & Roth, C. B. (2001) Science 293, 1793-1800. [DOI] [PubMed] [Google Scholar]
- 45.Notredame, C., Higgins, D. G. & Heringa, J. (2000) J. Mol. Biol. 302, 205-217. [DOI] [PubMed] [Google Scholar]