Abstract
Context
Lynch syndrome is the most common form of hereditary colorectal cancer (CRC) and is caused by germline mutations in DNA mismatch repair (MMR) genes. Identification of gene carriers currently relies on germline analysis in patients with MMR-deficient tumors, but criteria to select individuals in whom tumor MMR testing should be performed are unclear.
Objective
To establish a highly sensitive and efficient strategy for the identification of MMR gene mutation carriers among CRC probands.
Design, Setting, and Patients
Pooled-data analysis of 4 large cohorts of newly diagnosed CRC probands recruited between 1994 and 2010 (n = 10 206) from the Colon Cancer Family Registry, the EPICOLON project, the Ohio State University, and the University of Helsinki examining personal, tumor-related, and family characteristics, as well as microsatellite instability, tumor MMR immunostaining, and germline MMR mutational status data.
Main Outcome Measures
Performance characteristics of selected strategies (Bethesda guidelines, Jerusalem recommendations, and those derived from a bivariate/multivariate analysis of variables associated with Lynch syndrome) were compared with tumor MMR testing of all CRC patients (universal screening).
Results
Of 10 206 informative, unrelated CRC probands, 312 (3.1%) were MMR gene mutation carriers. In the population-based cohorts (n=3671 probands), the universal screening approach (sensitivity, 100%;95% CI, 99.3%–100%; specificity, 93.0%; 95% CI, 92.0%–93.7%; diagnostic yield, 2.2%; 95% CI, 1.7%–2.7%) was superior to the use of Bethesda guidelines (sensitivity, 87.8%; 95% CI, 78.9%–93.2%; specificity, 97.5%; 95% CI, 96.9%–98.0%; diagnostic yield, 2.0%; 95% CI, 1.5%–2.4%; P <.001), Jerusalem recommendations (sensitivity, 85.4%; 95% CI, 77.1%–93.6%; specificity, 96.7%; 95% CI, 96.0%–97.2%; diagnostic yield, 1.9%; 95% CI, 1.4%–2.3%; P < .001), and a selective strategy based on tumor MMR testing of cases with CRC diagnosed at age 70 years or younger and in older patients fulfilling the Bethesda guidelines (sensitivity, 95.1%; 95% CI, 89.8%–99.0%; specificity, 95.5%; 95% CI, 94.7%–96.1%; diagnostic yield, 2.1%; 95% CI, 1.6%–2.6%; P <.001). This selective strategy missed 4.9% of Lynch syndrome cases but resulted in 34.8% fewer cases requiring tumor MMR testing and 28.6% fewer cases undergoing germline mutational analysis than the universal approach.
Conclusion
Universal tumor MMR testing among CRC probands had a greater sensitivity for the identification of Lynch syndrome compared with multiple alternative strategies, although the increase in the diagnostic yield was modest.
Colorectal cancer (CRC) is the third most common cancer worldwide and the second leading cause of cancer-related death.1 Lynch syndrome, also known as hereditary nonpolyposis colorectal cancer (HNPCC), is the most common form of hereditary CRC, accounting for 1% to 3% of all these tumors. It is an autosomaldominant disorder caused by germline mutations in DNA mismatch repair (MMR) genes (ie, MSH2, MLH1, MSH6, and PMS2).2 The abnormal function of these genes leads to accumulation of errors during DNA replication, especially in repetitive sequences known as microsatellites. As a result, tumors of patients with Lynch syndrome characteristically demonstrate MMR deficiency, defined as the presence of microsatellite instability (MSI) or loss of the MMR protein expression, which is the hallmark of this disorder.3,4
Identification of patients with Lynch syndrome needs to be improved because, unless there is strong clinical suspicion, the majority of cases remain undetected, leading to the lack of implementation of highly effective preventive measures. Indeed, intensive CRC screening by colonoscopy and prophylactic gynecological surgery have been demonstrated to reduce both the incidence and mortality of these tumors.5
In 1991, the International Collaborative Group on HNPCC proposed the Amsterdam criteria and subsequently the extended Amsterdam II criteria,6 the first clinical definition of the syndrome and as a means to identify the genes responsible. However, these criteria were limited in clinical practice because of their low sensitivity. Consequently, the National Cancer Institute proposed the Bethesda guidelines, and more recently the revised Bethesda guidelines,7 for identifying those individuals who should undergo tumor MSI testing. Although this strategy has been demonstrated to be both effective and cost-effective,8 it is not fully accepted because some MMR gene mutation carriers do not fulfill these criteria and because they are difficult to apply in clinical practice.9 Virtually all Lynch syndrome-associated CRC display MMR deficiency, so universal tumor MMR screening has been proposed using MSI testing or immunostaining of all CRC patients.2,4 Recently, it was suggested that tumor MMR screening should be performed in, at a minimum, all CRC occurring in individuals younger than 70 years (ie, Jerusalem recommendations).10 Nevertheless, while this strategy overcomes the limitations of using any selection based on clinical criteria, it might not represent the most effective approach.
The controversy reflects that, at present, tumor MMR testing is the cornerstone for identification of Lynch syndrome. However, it is still under debate which CRC patients should undergo these analyses. Most sets of recommendations are not empirically based6,7 or derived from series in which patients were selected on the basis of their personal or family history.11–14 To overcome these limitations, a pooled data analysis of population-based series with fully integrated, comprehensive, and reliable data seems the most appropriate approach to outline a highly sensitive, efficient, and widely accepted strategy for the identification of MMR gene mutation carriers among CRC probands.
METHODS
The study sample came from the Colon Cancer Family Registry (CFR), the EPICOLON project,8 the Clinical Cancer Genetics Program of the Ohio State University,4,9 and the Department of Medical Genetics of the University of Helsinki, Finland15,16 (Figure 1 and Figure 2). Overall, cases were recruited between 1994 and 2010. The Colon CFR, an international resource for studies on the etiology of CRC described in detail elsewhere,17 recruited families through 6 administrative centers.18 The EPICOLON, Ohio, and Helsinki cohorts are population based and represent the core of the comparative analyses of diagnostic strategies for identification of Lynch syndrome (Figure 2). The Colon CFR recruited from both population-based cancer registries and through cancer family and high-risk clinics and used an upper age limit of 75 years (except for the Australian site, which did not recruit participants older than 60 years).17 Therefore, Colon CFR probands were used only in the analysis of variables associated with the presence of germline MMR gene mutations and not in ascertaining the performance characteristics of selected strategies for Lynch syndrome identification.
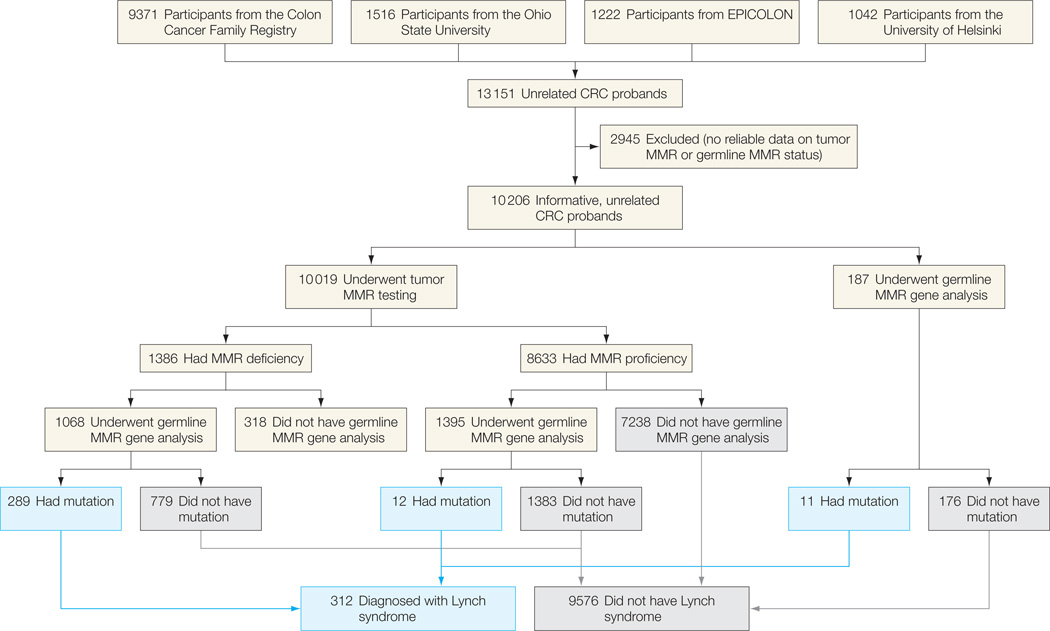
Figure 1.
Flowchart of the Study for the Overall Series
A patient is assumed to not have Lynch syndrome if the tumor is mismatch repair (MMR) proficient; germline MMR gene analysis was not performed for most of these individuals. CRC indicates colorectal cancer.
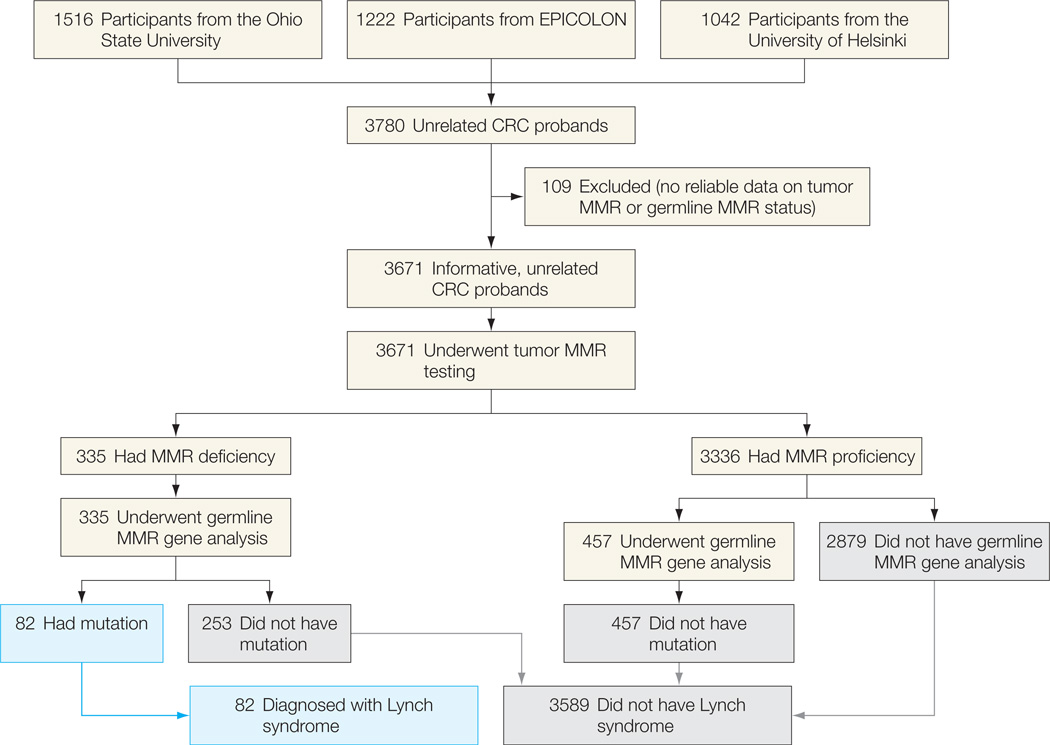
Figure 2.
Flowchart of the Study for the Population-Based Cohorts
A patient is assumed to not have Lynch syndrome if the tumor is mismatch repair (MMR) proficient; germline MMR gene analysis was not performed for most of these individuals. CRC indicates colorectal cancer.
Exclusion criteria were polyposis syndromes and personal history of inflammatory bowel disease. Written informed consent was obtained from all study participants, and the study protocol was approved at each participating center.
Personal, tumor-related, and familial characteristics of probands were pooled from each series. Tumor MSI testing and immunostaining for the 4 MMR proteins were performed as previously described.4,8,9,15–17,19 MSI testing was done at each center using different panels of microsatellite markers, and patients were classified as MSI high or MSI-low/microsatellite stable according to previously described criteria.20 Overall, tumors were deemed MSI-high if instability was seen at 30% or more markers or instability was present at monomorphic mononucleotide markers. Tumors were considered MMR deficient if they were MSI high, exhibited loss of MMR protein expression, or both.
Germline MMR gene testing was performed by both multiple ligation probe amplification analysis and direct sequencing at each participating center. Whereas MSH2, MLH1, and MSH6 genes were evaluated in all cohorts, evaluation of the PMS2 gene was not included in the study design of the cohorts of EPICOLON, Helsinki, and the University of Southern California Consortium (part of the Colon CFR). Deletions, insertions, duplications, nonsense, and frame shift mutations were considered deleterious; missense mutations were considered deleterious based on published data and existing mutation databases. Tumor MMR status was not used to classify any variant of unknown significance. Germline MMR mutational analysis was usually driven by demonstration of tumor MMR deficiency, although in a subset of 187 patients (1.8%), direct germline MMR gene testing was performed without assessment of MMR status (Figure 1). These patients were used in the analysis of variables associated with presence of MMR gene mutation but not in ascertaining the performance characteristics of selected strategies for Lynch syndrome identification. Similarly, in a subset of 1395 Colon CFR probands, germline gene testing was done although they had an MMR-proficient tumor. On the other hand, germline MMR gene testing was not performed in 318 patients (3.1%) in spite of having an MMR deficient tumor (Figure 1), and consequently, they were excluded from all analyses.
Statistical Analysis
The focus of the analysis was to establish, primarily, the most sensitive strategy and, secondarily, the most efficient one for identification of MMR gene mutation carriers among CRC patients. The presence of a germline mutation was considered the gold standard. Efficiency was defined as the capacity to detect a germline mutation with the minimum amount of diagnostic resources (ie, tumor MMR testing and germline MMR gene analysis).
Age at diagnosis was treated as a continuous variable. In probands diagnosed with the same type of cancer more than once, the age at diagnosis of cancer at which they were first identified as having Lynch syndrome was considered. In relatives diagnosed with the same type of cancer more than once, the age at diagnosis was defined as the earliest one. The number of relatives with CRC or other Lynch syndrome–related tumors were also treated as continuous variables. All other evaluated variables were considered dichotomous.
Logistic regression analysis was performed, adjusted by age, sex, and participating center, to identify individual variables associated with the presence of germline MMR gene mutations. Multivariate models based on regression tree analysis were explored to establish the most discriminative combination of variables to identify MMR gene mutation carriers. Recursive partitioning programs build classification or regression models of a very general structure using a 2-stage procedure; the resulting models can be represented as binary trees. Because the proportion of carriers was low, a high cost for misclassification was used to prime sensitivity over specificity. These analyses were limited to probands with information on the mutational status of MMR genes. Results were expressed as odds ratios (ORs) with 95% CI.
Performance characteristics (ie, sensitivity, specificity, positive and negative predictive values, positive and negative likelihood ratio, diagnostic yield, and false-positive yield) of selected strategies for Lynch syndrome identification were calculated with respect to the presence of germline MMR gene mutations in the population-based cohorts. Selected strategies included germline testing of probands with an MMR deficient lesion (1) after tumor testing of any CRC (ie, universal screening strategy); (2) after tumor testing of patients fulfilling the revised Bethesda guidelines7; (3) after tumor testing of patients fulfilling the Jerusalem recommendations10; or (4) after tumor testing of patients fulfilling the model derived from the multivariate analysis. These analyses were performed overall (ie, mutation in any MMR gene) and for each specific MMR gene. Comparison among strategies was made using the Matthews correlation coefficient and its 95% CI, which appropriately weights sensitivity and specificity values, as a measure of the quality of binary classifications.21
All calculations were performed with SPSS version 18.0 (SPSS) and R package rpart version 3.1–50.22 All tests were 2-sided, and a P value of less than .05 was considered statistically significant.
RESULTS
A total of 13 151 unrelated CRC probands from the 4 cohorts were included (Figure 1). Of these, 2945 cases were excluded due to lack of reliable information on tumor MMR or germline MMR mutational status. Therefore, 10 206 informative, unrelated CRC probands constituted the basis of this pooled-data analysis. Demographic, clinical, and tumor-related characteristics of the patients are summarized in Table 1.
Table 1.
Characteristics of Probands From Each Colorectal Cancer Cohort Included in the Pooled-Data Analysis
| Characteristics | No. (%) | Total | |||
|---|---|---|---|---|---|
| Colon CFR17 | OSU4,9 | EPICOLON8 | University of Helsinki15,16 | ||
| Probands included, No. | 6535 | 1516 | 1222 | 933 | 10 206 |
| Age, mean (SD), y | 53.7 (11.4) | 63.2 (13.0) | 69.9 (11.3) | 67.4 (11.8) | 59.1 (13.4) |
| Male sex | 3305 (50.5) | 830 (54.9) | 731 (59.8) | 528 (56.6) | 5394 (52.8) |
| First-degree relatives with CRC | 1105 (16.9) | 331 (21.9) | 156 (12.8) | 142 (15.2) | 1736 (17.0) |
| Revised Bethesda guidelines | 3056 (46.8) | 561 (37.1) | 287 (23.5) | 156 (16.7) | 4060 (39.8) |
| Amsterdam criteria I |
85 (1.3) | 15 (1.0) | 18 (1.5) | 2 (0.2) | 120 (1.2) |
| II | 156 (2.4) | 35 (2.3) | 22 (1.8) | 31 (3.3) | 246 (2.5) |
| Tumor MMR deficiencya | 948 (14.5) | 214 (14.2) | 90 (7.4) | 133 (14.2) | 1386 (13.8) |
| Germline MMR gene mutationb | 227 (3.5) | 44 (2.9) | 11 (0.9) | 30 (3.2) | 312 (3.1) |
Abbreviations: CFR, Cancer Family Registry; CRC, colorectal cancer; MMR, mismatch repair; OSU, Ohio State University.
Tumor MMR testing was performed in 10 019 patients. MMR deficiency was defined as loss of MMR protein expression (MLH1, MSH2, MSH6, and/or PMS2) and/or microsatellite instability-high.
Germline MMR gene mutational analysis was done in 2650 probands.
Tumor MMR testing was performed in 10 019 probands (98.1%), whereas in 187 patients (1.8%), germline MMR gene analysis was done without previous tumor MMR testing (Figure 1). The number of cases that were tested by MSI only was 2150; by immunostaining only, 2278; and by both MSI and immunostaining, 5591. In this latter group, concordance between MSI and immunostaining was 97.5% (94 cases [1.7%] showed MSI with retained protein expression and 49 [0.8%] exhibited loss of expression with microsatellite stability). A total of 1386 cases (13.8%) exhibited tumor MMR deficiency. Germline MMR mutational analysis was completed in 2650 probands and identified 312 gene mutation carriers in MSH2 (n=129), MLH1 (n = 114), MSH6 (n = 40), or PMS2 (n=29), representing 3.1% of the whole series (individual data available from the authors on request).
Among the 312 probands diagnosed with Lynch syndrome, mean (SD) age at CRC diagnosis was 48.1 (2.9) years; 131 (42.5%) had 1 or more first-degree relatives with CRC; 41 (14.0%) and 85 (27.2%) fulfilled Amsterdam I and II criteria, respectively; and 214 (68.6%) fulfilled at least 1 criterion of the revised Bethesda guidelines (eTable 1, available at http://www.jama.com). Moreover, 289 probands (92.6%) exhibited tumor MMR deficiency, whereas 12 (3.8%) (ie, 5 MLH1, 3 MSH2, 3 MSH6, and 1 PMS2 gene carriers) showed MMR proficiency. Of those, 5 cases had a tumor retaining protein expression (MSI analysis not performed), 4 cases exhibited microsatellite stability (immunostaining not performed), and 3 cases retained protein expression and showed microsatellite stability. In the remaining 11 probands (3.5%), tumor MMR testing was not performed (eTable 1).
MMR Gene Mutation Carriers
To identify variables associated with Lynch syndrome, a bivariate analysis was performed in those probands with information regarding germline MMR mutational status (n=2650) (Table 2). This analysis identified CRC diagnosed at age 70 years or younger (OR, 4.0; 95% CI, 2.2–7.1) and fulfillment of at least 1 criterion of the revised Bethesda guidelines (OR, 7.3; 95% CI, 4.6–11.0) as the variables with the highest sensitivity (94.2% and 88.1%, respectively) and negative predictive value (97.0% and 97.3%, respectively). All other evaluated characteristics showed sensitivities lower than 70% and negative predictive values lower than 95% (Table 2). Distribution of germline MMR gene mutations according to the age at CRC diagnosis is shown in eTable 2.
Table 2.
Analyses of Variables Associated With Presence of a Germline Mismatch Repair Gene Mutation (Bivariate Analysis)a
| Variable | Evaluable Probands, No.b |
MMR Gene Carriers Fulfilling the Condition, No./Total No.c |
OR (95% CI)d | % (95% CI) | |||
|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | ||||
| Proximal CRCe | 1644 | 130/208 | 2.5 (1.7–3.7) | 62.3 (55.6–69.3) | 51.4 (48.7–53.9) | 15.6 (13.1–18.2) | 90.4 (88.3–92.5) |
| Mucinous CRC | 1492 | 52/201 | 1.7 (1.2–2.4) | 25.9 (19.5–32.1) | 84.0 (82.0–86.0) | 20.2 (15.0–25.2) | 87.9 (86.0–89.7) |
| Poorly differentiated CRC | 745 | 24/104 | 1.3 (0.8–2.1) | 23.1 (14.5–31.6) | 81.1 (78.0–84.2) | 16.6 (10.1–22.9) | 86.7 (83.8–89.4) |
| Crohn-like lymphocytic reaction | 300 | 27/55 | 2.0 (1.1–3.7) | 49.1 (34.9–63.2) | 67.8 (61.7–73.8) | 25.5 (16.7–34.2) | 85.6 (80.3–90.7) |
| Tumor infiltrating lymphocytes | 367 | 44/64 | 3.2 (1.7–5.9) | 68.8 (56.6–80.8) | 61.7 (56.0–67.3) | 27.5 (20.2–34.7) | 90.3 (86.0–94.6) |
| Synchronous or metachronous CRC |
2160 | 50/244 | 5.0 (3.3–7.8) | 20.7 (15.4–26.0) | 94.3 (93.2–95.3) | 31.4 (23.9–38.9) | 90.5 (89.1–91.7) |
| Metachronous Lynch syndrome– related tumorf |
2650 | 53/311 | 5.1 (3.4–7.7) | 17.0 (12.7–21.3) | 95.7 (94.8–96.5) | 34.6 (26.7–42.5) | 89.6 (88.4–90.8) |
| CRC excluded | 2650 | 21/311 | 4.7 (2.6–8.4) | 6.7 (3.8–9.7) | 97.6 (97.0–98.2) | 27.6 (16.9–38.3) | 88.7 (87.4–89.9) |
| Diagnosed ≤50 y | 2649 | 36/310 | 6.2 (3.7–10.4) | 11.6 (7.8–15.3) | 98.4 (97.8–98.9) | 48.6 (36.5–60.7) | 89.3 (88.1–90.5) |
| CRC excluded (diagnosed ≤50 y) |
2650 | 12/311 | 4.2 (1.9–9.2) | 3.8 (1.5–6.1) | 99.1 (98.7–99.5) | 37.5 (19.1–55.8) | 88.5 (87.5–89.8) |
| FDR with CRC ≥1 |
2644 | 131/308 | 3.0 (2.3–4.1) | 42.5 (37.0–48.4) | 81.9 (80.3–83.5) | 23.7 (20.1–27.4) | 91.5 (90.3–92.7) |
| ≥2 | 2644 | 56/306 | 5.0 (3.3–7.6) | 18.3 (13.8–22.8) | 96.0 (95.1–96.8) | 37.3 (29.2–45.4) | 90.0 (88.7–91.1) |
| FDR with CRC diagnosed ≤50 y ≥1 |
2610 | 79/289 | 8.0 (5.5–11.8) | 27.3 (22.2–32.9) | 96.3 (95.4–97.0) | 47.6 (40.0–55.7) | 91.5 (90.2–92.5) |
| ≥2 | 2633 | 22/297 | 11.9 (5.4–26.0) | 7.4 (4.2–10.5) | 99.5 (99.1–99.8) | 64.7 (47.1–82.2) | 89.4 (88.2–90.6) |
| FDR with Lynch syndrome– related tumorf ≥1 |
2640 | 154/303 | 2.8 (2.1–3.7) | 50.8 (45.0–56.6) | 74.0 (72.1–75.7) | 20.2 (17.2–23.1) | 92.1 (90.8–93.3) |
| ≥2 | 2648 | 88/310 | 4.5 (3.1–6.4) | 28.4 (23.1–33.5) | 93.2 (92.1–94.2) | 35.5 (29.3–41.6) | 90.8 (89.5–91.9) |
| FDR with Lynch syndrome– related tumor diagnosed ≥50yf ≥1 |
2589 | 95/281 | 5.7 (4.1–8.0) | 33.8 (28.1–39.5) | 92.9 (91.8–94.0) | 36.8 (30.7–42.9) | 92.0 (90.9–93.1) |
| ≥2 | 2619 | 32/283 | 12.4 (6.6–23.0) | 11.3 (7.9–15.8) | 99.1 (98.7–99.5) | 61.5 (49.1–76.7) | 90.3 (89.0–91.3) |
| CRC diagnosed ≤70 y (Jerusalem recommendations) |
2112 | 226/240 | 4.0 (2.2–7.1) | 94.2 (90.4–97.0) | 26.3 (24.4–28.3) | 13.4 (11.7–15.0) | 97.0 (94.7–98.3) |
| Fulfillment of Amsterdam criteria I |
2627 | 41/291 | 9.6 (5.7–16.2) | 13.7 (9.5–17.7) | 98.6 (98.1–99.1) | 55.6 (43.3–67.7) | 90.2 (88.9–91.2) |
| II | 2650 | 85/312 | 11.4 (7.3–17.7) | 27.2 (22.1–32.3) | 97.9 (97.3–98.5) | 63.4 (54.9–71.9) | 91.0 (89.7–92.0) |
| Fulfillment of ≥1 criterion of revised Bethesda guidelines |
2128 | 214/243 | 7.3 (4.6–11.0) | 88.1 (83.7–92.3) | 54.4 (52.1–56.6) | 19.9 (17.4–22.3) | 97.3 (96.2–98.2) |
Abbreviations: CRC, colorectal cancer; FDR, first-degree relative; MMR, mismatch repair; NPV, negative predictive value; OR, odds ratio; PPV, positive predictive value.
This analysis was limited to patients with information on the germline mutational status of MMR genes and without considering the result of tumor MMR testing.
Probands in whom the corresponding variable could be assessed.
MMR gene carriers fulfilling the condition with respect to those MMR gene carriers in whom the corresponding variable could be evaluated.
Adjusted by age, sex, and participating center.
With respect to the splenic flexure.
Lynch syndrome–related tumors: colorectal, endometrial, ovarian, gastric, hepatobiliary, small bowel, urinary tract, pancreatic, and brain cancer.
In the multivariate analysis, based on regression trees, the highest discrimination was achieved when MMR testing was done for probands with any of the following characteristics: CRC diagnosed at 60 years or younger, presence of at least 1 first-degree relative with CRC diagnosed at 50 years or younger, or personal history of metachronous Lynch syndrome–related tumors diagnosed at 50 years or younger (OR, 11.3; 95% CI, 6.70–19.0). The sensitivity of this model was 90.1%, with a negative predictive value of 97.5%.
Performance of Selected Strategies
Strategies based on tumor MMR testing of probands fulfilling at least 1 criterion of the revised Bethesda guidelines, Jerusalem recommendations, the model resulting from the most sensitive variables in the bivariate analysis (ie, CRC diagnosis at ≤70 years and fulfillment of at least 1 criterion of the revised Bethesda guidelines, henceforth “selective strategy”), or the model resulting from the multivariate analysis, followed by germline MMR testing of individuals with an MMR-deficient tumor, were compared with the universal screening approach in which tumor MMR testing was performed in all CRC patients (Table 3 and Table 4). As expected, only the universal screening strategy achieved 100% sensitivity (95% CI, 99.3%–100%) and negative predictive value (95% CI, 99.9%–100%) in the identification of patients with Lynch syndrome, when the analysis was limited to population-based cohorts (n=3671) (Figure 2).
Table 3.
Performance Characteristics of Selected Strategies for the Identification of Patients With Lynch Syndromea
| Tumor MMR Testing | No.(%) | Germline MMR Gene Mutation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Probands Requiring Tumor MMR Testingb |
Probands Requiring Germline MMR Gene Analysisc |
Sensitivity | Specificity | Positive Predictive Value |
Negative Predictive Value |
|||||
| No./ Total No |
% (95% CI) |
No./ Total No. |
% (95% CI) | No./ Total No. |
% (95% CI) |
No./ Total No. |
% (95% CI) |
|||
| CRC patients fulfilling a condition Multivariate modeld |
1158 (31.5) | 128 (3.5) | 67/82 | 81.7 (72.7–90.6) |
3528/3589 | 98.3 (97.8–98.7) |
67/128 | 52.3 (43.3–61.3) |
3528/3543 | 99.6 (99.3–99.8) |
| Jerusalem recommendationse |
2125 (57.8) | 189 (5.1) | 70/82 | 85.4 (77.1–93.6) |
3470/3589 | 96.7 (96.0–97.2) |
70/189 | 37.0 (29.9–44.2) |
3470/3482 | 99.7 (99.4–99.8) |
| ≥1 Criterion of revised Bethesda guidelines |
992 (27.0) | 162 (4.4) | 72/82 | 87.8 (78.9–93.2) |
3499/3589 | 97.5 (96.9–98.0) |
72/162 | 44.4 (36.4–52.4) |
3499/3509 | 99.7 (99.5–99.9) |
| Jerusalem recommendationse or ≥1 criterion of revised Bethesda guidelines |
2394 (65.2) | 239 (6.5) | 78/82 | 95.1 (89.8–99.0) |
3428/3589 | 95.5 (94.7–96.1) |
78/239 | 32.6 (26.4–38.7) |
3428/3432 | 99.9 (99.7–100) |
| Any CRC patient (universal strategy) | 3671 (100) | 335 (9.1) | 82/82 | 100 (99.3–100) |
3336/3589 | 93.0 (92.0–93.7) |
82/335 | 24.5 (19.7–29.2) |
3336/3336 | 100 (99.9–100) |
Abbreviations: CRC, colorectal cancer; MMR, mismatch repair.
This analysis was limited to population-based cohorts (n=3671 probands).
Probands requiring tumor MMR testing in each strategy, with respect to those in whom it could be assessed.
Probands requiring germline MMR gene analysis because of the demonstration of tumor MMR deficiency in each strategy, with respect to those in whom it could be assessed.
Defined as fulfillment of ≥1 of the following characteristics: CRC diagnosed at ≤60 years, ≥1 first-degree relative with CRC diagnosed at ≤50 years, or personal history of metachronous Lynch syndrome-related tumors diagnosed at ≤50 years.
Age at CRC diagnosis ≤70 years.
Table 4.
Diagnostic and False-Positive Yields of Selected Strategies for the Identification of Patients With Lynch Syndromea
| Tumor MMR Testing | Germline MMR Gene Mutation | |||||
|---|---|---|---|---|---|---|
| Diagnostic Yieldb | P Valuec | Incremental Diagnostic Yield, %d |
False–Positive Yielde | |||
| No./Total No. | % (95% CI) | No./Total No. | % (95% CI) | |||
| CRC patients fulfilling a condition Multivariate modelf |
67/3671 | 1.8 (1.3–2.2) | <.001 | 61/3671 | 1.7 (1.2–2.1) | |
| Jerusalem recommendationsg | 70/3671 | 1.9 (1.4–2.3) | <.001 | 0.08 | 119/3671 | 3.2 (2.6–3.8) |
| ≥1 Criterion of revised Bethesda guidelines |
72/3671 | 2.0 (1.5–2.4) | <.001 | 0.05 | 90/3671 | 2.5 (2.0–2.9) |
| Jerusalem recommendationsg or ≥1 criterion of revised Bethesda guidelines |
78/3671 | 2.1 (1.6–2.6) | <.001 | 0.16 | 161/3671 | 4.4 (3.7–5.0) |
| Any CRC patient (universal strategy) | 82/3671 | 2.2 (1.7–2.7) | [Reference] | 0.11 | 253/3671 | 6.9 (6.0–7.7) |
Abbreviations: CRC, colorectal cancer; MMR, mismatch repair.
This analysis was limited to population-based cohorts (n=3671 probands).
Diagnostic yield refers to probands requiring germline MMR gene analysis in whom a mutation was found.
Matthews correlation coefficient comparison of diagnostic yield with respect to the universal strategy.
Compared with the next least intensive strategy.
False-positive yield refers to probands requiring germline MMR gene analysis in whom no mutation was found.
Defined as fulfillment of ≥1 of the following characteristics: CRC diagnosed at ≤60 years, ≥1 first-degree relative with CRC diagnosed at ≤50 years, or personal history of metachronous Lynch syndrome-related tumors diagnosed at ≤50 years.
Age at CRC diagnosis ≤70 years.
Universal tumor testing (sensitivity, 100%; 95% CI, 99.3%–100%; specificity, 93.0%; 95% CI, 92.0%–93.7%; diagnostic yield, 2.2%; 95% CI, 1.7%–2.7%) was superior to the selective strategy (sensitivity, 95.1%; 95% CI, 89.8%–99.0%; specificity, 95.5%; 95% CI, 94.7%–96.1%; diagnostic yield, 2.1%; 95% CI, 1.6%–2.6%; Matthews correlation coefficient, 0.54; P<.001), Bethesda guidelines (sensitivity, 87.8%; 95% CI, 78.9%–93.2%; specificity, 97.5%; 95% CI, 96.9%–98.0%; diagnostic yield, 2.0%; 95% CI, 1.5%–2.4%; Matthews correlation coefficient, 0.61; P <.001), and Jerusalem recommendations (sensitivity, 85.4%; 95% CI, 77.1%–93.6%; specificity, 96.7%; 95% CI, 96.0%–97.2%; diagnostic yield, 1.9%; 95% CI, 1.4%–2.3%; Matthews correlation coefficient, 0.55; P<.001) (Table 3 and Table 4). However, differences in diagnostic yield from the universal approach were small, with a difference between universal screening and the next less intensive strategy (ie, selective strategy) of only 0.11% (Table 3 and Table 4) and accompanied by an increase in false-positive yield of 2.5%. Indeed, the selective strategy resulted in a 34.8% fewer CRC patients requiring tumor MMR testing and an additional 28.6% fewer cases undergoing germline MMR mutational analysis in comparison with universal screening (Table 3 and Table 4). All these results were similar to those obtained in the overall series (eTable 3).
When the analysis was conducted for each specific MMR gene, the selective strategy resulted in identical sensitivity and negative predictive value to those achieved with the universal tumor MMR testing approach but only for the identification of MLH1 and MSH2 gene carriers, in a similar manner as the fulfillment of Bethesda guidelines for the identification of MLH1 gene carriers (Table 5). Again, these results were similar to those obtained in the whole series (eTable 4).
Table 5.
Performance Characteristics of Selected Strategies for the Identification of Patients With Lynch Syndrome, According to the Mismatch Repair Gene Mutateda
| Tumor MMR Testing | Germline MMR Gene Mutation | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Positive Predictive Value |
Negative Predictive Value |
||||||
| No./ Total No. |
% (95% CI) |
No./ Total No. |
% (95% CI) |
No./ Total No. |
% (95% CI) |
No./ Total No. |
% (95% CI) |
P Valueb | |
|
MLH1 (n = 3589)c CRC patients fulfilling a condition ≥1 Criterion of revised Bethesda guidelines |
34/34 | 100 (98.5–100) |
3481/3555 | 97.9 (97.4–98.4) |
34/108 | 31.5 (22.2–40.7) |
3481/3481 | 100 (99.9–100) |
<.001 |
| Jerusalem recommendationsd | 32/34 | 94.1 (84.7–100) |
3464/3555 | 97.4 (96.9–97.9) |
32/123 | 26.0 (17.8–34.1) |
3464/3466 | 99.9 (99.8–100) |
<.001 |
| Jerusalem recommendationsd or ≥1 criterion of revised Bethesda guidelines |
34/34 | 100 (98.5–100) |
3424/3555 | 96.3 (95.6–96.8) |
34/165 | 20.6 (14.1–27.0) |
3424/3424 | 100 (99.9–100) |
<.001 |
| Multivariate modele | 28/34 | 82.4 (68–96.6) |
3513/3555 | 98.8 (98.4–99.1) |
28/70 | 40.0 (27.8–52.1) |
3513/3519 | 99.8 (99.6–99.9) |
<.001 |
| Any CRC patient (universal strategy) | 34/34 | 100 (98.5–100) |
3336/3555 | 93.8 (93.0–94.6) |
34/253 | 13.4 (9.0–17.8) |
3336/3336 | 100 (99.9–100) |
[Reference] |
|
MSH2 (n = 3422)c CRC patients fulfilling a condition ≥1 Criterion of revised Bethesda guidelines |
31/33 | 93.9 (84.2–100) |
3366/3389 | 99.3 (99.0–99.6) |
31/54 | 57.4 (43.2–71.5) |
3366/3368 | 99.9 (99.8–100) |
<.001 |
| Jerusalem recommendationsd | 29/33 | 87.9 (75.2–100) |
3361/3389 | 99.2 (98.8–99.4) |
29/57 | 50.9 (37.0–64.7) |
3361/3365 | 99.9 (99.7–100) |
<.001 |
| Jerusalem recommendationsd or ≥1 criterion of revised Bethesda guidelines |
33/33 | 100 (98.4–100) |
3353/3389 | 98.9 (98.5–99.3) |
33/69 | 47.8 (35.3–60.3) |
3353/3353 | 100 (99.9–100) |
<.001 |
| Multivariate modele | 31/33 | 93.9 (84.2–100) |
3370/3389 | 99.4 (99.1–99.6) |
31/50 | 62.0 (47.5–76.4) |
3370/3372 | 99.9 (99.8–100) |
<.001 |
| Any CRC patient | 33/33 | 100 (98.4–100) |
3336/3389 | 98.4 (98.0–98.8) |
33/86 | 38.4 (27.5–49.2) |
3336/3336 | 100 (99.9–100) |
[Reference] |
|
MSH6 (n = 3391)c CRC patients fulfilling a condition ≥1 criterion of revised Bethesda guidelines |
3/9 | 33.3 (0–69.6) |
3362/3382 | 99.4 (99.1–99.6) |
3/23 | 13.0 (0–28.9) |
3362/3368 | 99.8 (99.6–99.9) |
<.001 |
| Jerusalem recommendationsd | 6/9 | 66.7 (30.3–100) |
3359/3382 | 99.3 (99.0–99.6) |
6/29 | 20.7 (4.22–37.1) |
3359/3362 | 99.9 (99.7–100) |
.11 |
| Jerusalem recommendationsd or ≥1 criterion of revised Bethesda guidelines |
7/9 | 77.8 (45.0–100) |
3350/3382 | 99.1 (98.6–99.3) |
7/39 | 17.9 (4.62–31.2) |
3350/3352 | 99.9 (99.8–100) |
.13 |
| Multivariate modele | 5/9 | 55.6 (17.5–93.5) |
3365/3382 | 99.5 (99.2–99.7) |
5/22 | 22.7 (2.94–42.5) |
3365/3369 | 99.9 (99.7–100) |
.02 |
| Any CRC patient | 9/9 | 100 (94.4–100) |
3336/3382 | 98.6 (98.2–99.0) |
9/55 | 16.4 (5.6–27.0) |
3336/3336 | 100 (99.9–100) |
[Reference] |
|
PMS2 (n = 3351)c CRC patients fulfilling a condition ≥1 criterion of revised Bethesda guidelines |
4/6 | 66.7 (20.6–100) |
3342/3345 | 99.9 (99.7–99.9) |
4/7 | 57.1 (13.3–100) |
3342/3344 | 99.9 (99.8–100) |
.30 |
| Jerusalem recommendationsd | 3/6 | 50.0 (1.6–98.3) |
3340/3345 | 99.9 (99.7–100) |
3/8 | 37.5 (0–77.3) |
3340/3343 | 99.9 (99.7–100) |
<.001 |
| Jerusalem recommendationsd or ≥1 criterion of revised Bethesda guidelines |
4/6 | 66.7 (20.6–100) |
3339/3345 | 99.8 (99.6–99.9) |
4/10 | 40.0 (4.64–75.3) |
3339/3341 | 99.9 (99.8–100) |
<.001 |
| Multivariate modele | 3/6 | 50.0 (1.6–98.3) |
3341/3345 | 99.9 (99.7–100) |
3/7 | 42.9 (0–86.6) |
3341/3344 | 99.9 (99.7–100) |
<.001 |
| Any CRC patient | 6/6 | 100 (91.6–100) |
3336/3345 | 99.7 (99.5–99.9) |
6/15 | 40.0 (11.8–68.1) |
3336/3336 | 100 (99.9–100) |
[Reference] |
Abbreviations: CRC, colorectal cancer; MMR, mismatch repair.
This analysis was limited to population-based cohorts.
With respect to universal strategy (Mathews correlation coefficient comparison).
Probands in whom strategies for the identification of germline mutations for each specific MMR gene could be assessed.
Age at CRC diagnosis ≤70 years.
Defined as fulfillment of ≥1 of the following characteristics: CRC diagnosed at ≤60 years, ≥1 first-degree relative with CRC diagnosed at ≤50 years, or personal history of metachronous Lynch syndrome-related tumors diagnosed at ≤50 years.
COMMENT
Results of this international, multicenter, pooled-data analysis demonstrate that unless a universal screening approach consisting of tumor MMR testing in all CRC patients is performed, a clinically meaningful proportion of MMR gene mutation carriers will remain undiagnosed. Specifically, use of the revised Bethesda guidelines will miss approximately 12%, use of the Jerusalem recommendations will miss approximately 15%, and use of a selective criteria (performing tumor MMR testing of CRC probands diagnosed at 70 years or younger or fulfilling ≥ 1 criterion of the revised Bethesda guidelines) will miss approximately 5%. Conversely, the specificity for these strategies ranged from 93.0% for the universal tumor MMR testing approach to 97.5% for the Bethesda guidelines. These data may be useful to more empirically inform discussions on the most efficient approaches for the identification of Lynch syndrome among CRC probands.
This study has several strengths. First, this is the largest series published so far in which fully characterized CRC patients were evaluated to ascertain the most effective and efficient strategy for the identification of Lynch syndrome, using personal and family history, tumor MMR testing, and germline MMR mutational data. This comprehensive approach overcomes previous attempts—Amsterdam criteria,6,23 Bethesda guidelines,7 and Jerusalem recommendations10—in which strategies were not empirical or were based on expert consensus. Second, this analysis was based on population-based cohorts,4,8,9,15,16 its results being applicable to an unremarkable newly diagnosed CRC patient rather than in the subset of individuals usually referred to genetic counseling because of a high suspicion of an inherited disorder. Third, the methodological approach, which included an exploratory analysis of the most discriminative variables associated with presence of germline MMR mutations and evaluation of the performance characteristics of comprehensive strategies, allowed us not only to establish their accuracy for the identification of Lynch syndrome, but also to estimate the molecular resources needed.
We are aware of some limitations of the study. First, the results of this investigation have not been replicated in an independent set of CRC patients because the prevalence of Lynch syndrome is relatively low and, accordingly, it is difficult to find 2 database sets adequate for such analyses. Second, all probands were diagnosed with CRC, thus precluding our ability to extrapolate our results to patients presenting with other Lynch syndrome–related tumors. Nevertheless, CRC represents the most prevalent neoplasm in such patients, and in fact, it is the most common “red flag” to drive the subsequent molecular confirmation.24,25 Third, germline MMR mutational analysis was not performed in all probands, although it was done in the vast majority of patients with MMR-deficient tumors and also in a notable proportion of those with proficient lesions. In that sense, it is important to note that 12 mutation carriers had an MMR-proficient neoplasm, thus indicating that a reduced number of patients with Lynch syndrome will remain undiagnosed if screening relies on MMR tumor testing. To overcome this limitation, sequencing all genes of concern in all CRC patients would represent the most sensitive approach. When high-throughput technology becomes more affordable, cost-effectiveness analysis of this approach will be warranted.
Fourth, no information was available regarding either tumor BRAF V600E mutation or tumor MLH1 gene promoter methylation. Both molecular techniques are helpful in excluding epigenetically driven inactivation of the MLH1 gene among patients with MLH1-deficient tumors.26 This fact may explain the lower specificity of all evaluated strategies for the identification of MLH1 gene carriers with respect to the other 3 MMR genes, but it does not affect their sensitivity, which is the main goal of our analysis. Finally, in contrast to the other 3 MMR genes, the PMS2 gene was not systematically analyzed in all evaluated cohorts. This limitation, however, has been addressed by analyzing the results separately for each specific gene.
Our analysis demonstrates that, although the revised Bethesda guidelines have been considered as the mainstay for selecting patients to undergo tumor MMR testing so far,7,8,27,28 they have a low sensitivity for the identification of Lynch syndrome. The lack of sensitivity is mainly due to its poor performance in identifying MSH6 gene carriers29 and, to less extent, PMS2 and MSH2 gene carriers. On the other hand, the use of age at CRC diagnoses as a criterion to select patients requiring tumor MMR testing, as was suggested in the Lynch syndrome conference held in Jerusalem,10 is also limited by a low sensitivity, because 15% of patients were older than 70 years at the diagnosis of Lynch syndrome.
Universal tumor screening has, as expected, the highest sensitivity. Although it is not sufficient to just consider sensitivity when comparing different strategies, this is the most important parameter clinically (ie, to minimize the number of patients with undiagnosed Lynch syndrome). Indeed, it is accepted that the whole Lynch syndrome screening process is cost-effective when the benefits to immediate relatives of identified patients are considered30; accordingly, the more patients who are diagnosed, the more atrisk relatives can undergo genetic evaluation and receive appropriate cancer surveillance and other preventive interventions.
On the other hand, any policy recommendation needs to consider the economic and psychosocial harms of false-positive results obtained in each strategy. It is notable that the selective strategy of performing tumor MMR testing of CRC probands diagnosed at 70 years or younger, and in older probands fulfilling at least 1 criterion of the revised Bethesda guidelines, achieved a similar diagnostic yield to the universal strategy, while reducing by about 35% and 30% the number of patients requiring tumor and germline MMR testing, respectively. Therefore, if resources are limited, this selective strategy may represent an alternative approach to universal tumor screening for the identification of Lynch syndrome, although it remains to be demonstrated that this strategy can be implemented consistently in a clinical setting. Whereas recent data suggest that universal tumor testing may yield substantial benefits at acceptable costs,31 further studies assessing cost-effectiveness of those strategies evaluated in this study are still needed.
In addition to the pragmatic approach proposed in this study, a more precise characterization of probands exhibiting MLH1-deficient tumors is needed. Because tumor MMR deficiency in the vast majority of such patients is due to epigenetic MLH1 inactivation, performance of tumor BRAF V600E mutation analysis,32,33 or even better, methylation analysis of MLH1 gene promoter,34,35 may contribute to increasing the specificity of this strategy for the identification of MLH1 gene carriers and consequently to further decreasing the cost associated with germline testing.
The strategies evaluated in this study rely heavily on tumor testing. However, they should not be in conflict with available mathematical algorithms to predict MMR gene mutation carriers based on personal and family history.11–14,36–38 Indeed, both approaches must be viewed as complementary because it is not always feasible to obtain tumor tissue.36,39 More importantly, these models may also encompass individuals affected by other non-CRC Lynch syndrome–related tumors.11–14,36–38 In addition, it would be interesting to explore whether the use of predictive algorithms may contribute to identifying gene mutation carriers among patients with MMR-proficient tumors.
Finally, it is important to note that significant differences were observed among the 3 population-based cohorts evaluated in this study. Indeed, in the EPICOLON cohort, the prevalence of Lynch syndrome, as well as of tumor MMR deficiency, was roughly half that observed in the Ohio and Helsinki series. This finding, rather than being considered as a drawback of our analysis, should be regarded as an opportunity to generalize its results broadly. The geographical variation in Lynch syndrome genotypic and phenotypic characteristics4,8,9,15–17 may reflect some specific gene-environment interactions and therefore deserves further investigation.
In conclusion, identification of patients with Lynch syndrome is critical to drive presymptomatic diagnosis of relatives at risk, as well as subsequent preventive measures for decreasing morbidity and mortality. Universal tumor MMR testing followed by germline testing offers the highest sensitivity and a somewhat lower specificity than alternative screening strategies for this purpose, although the increase in the diagnostic yield is modest. The empirical data from this large multinational study may help inform clinical recommendations for individuals diagnosed with CRC.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by grants from the Ministerio de Economía y Competividad (SAF2010-19273), Agència de Gestió d'Ajuts Universitaris i de Recerca (2009 SGR 849), Asociación Española contra el Cáncer (Fundación Científica and Junta de Barcelona), Hospital Clínic of Barcelona (Josep Font grant), and Instituto de Salud Carlos III (PI10/00384). CIBEREHD and CIBERESP are funded by the Instituto de Salud Carlos III. The research at the Ohio State University Comprehensive Cancer Center was supported by National Cancer Institute grants CA67941 and CA16058. The research at the University of Pennsylvania was supported by the National Institutes of Health under 5R01DK056645, the National Colon Cancer Research Alliance, and the Hansen Foundation. This work was also supported by the National Cancer Institute, by the National Institutes of Health under RFA CA-95-011, and through cooperative agreements with members of the Colon Cancer Family Registry and Principal Investigators. Collaborating centers include Australasian Colorectal Cancer Family Registry (U01 CA097735), Familial Colorectal Neoplasia Collaborative Group (U01 CA074799) (University of Southern California), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783), Seattle Colorectal Cancer Family Registry (U01 CA074794), and University of Hawaii Colorectal Cancer Family Registry (U01 CA074806). The work was carried out in part at the Esther Koplowitz Center, Barcelona.
Role of the Sponsor: The funding sources had no role in the design and conduct of the study; in the collection, analysis, and interpretation of the data; or in the preparation, review, or approval of the manuscript
Additional Contributions: We thank all study participants of the Colon Cancer Family Registry, the Ohio State University, the University of Helsinki, and the EPICOLON consortium and corresponding staff for their contributions to this project. None of them were compensated for their contributions.
Footnotes
Author Contributions:
Drs Moreira and Castells had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Moreira, Balaguer, Lindor, de la Chapelle, Aaltonen, Hopper, Gallinger, Jenkins, Andreu, Castells.
Acquisition of data: Moreira, Lindor, Hampel, Aaltonen, Hopper, Le Marchand, Gallinger, Newcomb, Haile, Thibodeau, Gunawardena, Jenkins, Buchanan, Potter, Ahnen, Andreu, Ponz de Leon, Castells.
Analysis and interpretation of data: Moreira, Balaguer, Hopper, Haile, Jenkins, Baron, Ahnen, Moreno, Rustgi, Castells.
Drafting of the manuscript: Moreira, Balaguer, Lindor, Hopper, Jenkins, Rustgi, Castells.
Critical revision of the manuscript for important intellectual content: Moreira, Balaguer, Lindor, de la Chapelle, Hampel, Aaltonen, Hopper, Le Marchand, Gallinger, Newcomb, Thibodeau, Gunawardena, Jenkins, Buchanan, Potter, Baron, Ahnen, Moreno, Andreu, Ponz de Leon, Rustgi.
Statistical analysis: Moreira, Balaguer, Moreno, Castells.
Obtained funding: Hopper, Gallinger, Newcomb, Haile, Jenkins, Buchanan, Rustgi.
Administrative, technical, or material support: Lindor, Hampel, Aaltonen, Hopper, Le Marchand, Gallinger, Newcomb, Haile, Jenkins, Buchanan, Potter.
Study supervision: Lindor, de la Chapelle, Hopper, Thibodeau, Jenkins, Buchanan, Ponz de Leon, Castells.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest and none were reported.
Disclaimer: The content of this article does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Colon Cancer Family Registry.
Online-Only Material: The eTables are available at http://www.jama.com.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Vasen HF, Boland CR. Progress in genetic testing, classification, and identification of Lynch syndrome. JAMA. 2005;293(16):2028–2030. doi: 10.1001/jama.293.16.2028. [DOI] [PubMed] [Google Scholar]
- 3.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348(10):919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 4.Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26(35):5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoffel EM, Chittenden A. Genetic testing for hereditary colorectal cancer: challenges in identifying, counseling, and managing high-risk patients. Gastroenterology. 2010;139(5):1436–1441. doi: 10.1053/j.gastro.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116(6):1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 7.Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piñol V, Castells A, Andreu M, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293(16):1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 9.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352(18):1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 10.Boland CR, Shike M. Report from the Jerusalem workshop on Lynch syndrome-hereditary nonpolyposis colorectal cancer. Gastroenterology. 2010;138(7):2197. doi: 10.1053/j.gastro.2010.04.024. e1-e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balmaña J, Stockwell DH, Steyerberg EW, et al. Prediction of MLH1 and MSH2 mutations in Lynch syndrome. JAMA. 2006;296(12):1469–1478. doi: 10.1001/jama.296.12.1469. [DOI] [PubMed] [Google Scholar]
- 12.Barnetson RA, Tenesa A, Farrington SM, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006;354(26):2751–2763. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 13.Chen S, Wang W, Lee S, et al. Colon Cancer Family Registry. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA. 2006;296(12):1479–1487. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wijnen JT, Vasen HF, Khan PM, et al. Clinical findings with implications for genetic testing in families with clustering of colorectal cancer. N Engl J Med. 1998;339(8):511–518. doi: 10.1056/NEJM199808203390804. [DOI] [PubMed] [Google Scholar]
- 15.Aaltonen LA, Salovaara R, Kristo P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338(21):1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 16.Salovaara R, Loukola A, Kristo P, et al. Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2000;18(11):2193–2200. doi: 10.1200/JCO.2000.18.11.2193. [DOI] [PubMed] [Google Scholar]
- 17.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 18.Breast and colon cancer family registries. National Cancer Institute; [Accessed September 25, 2012]. http://epi.grants.cancer.gov/CFR/. [Google Scholar]
- 19.Cicek MS, Lindor NM, Gallinger S, et al. Quality assessment and correlation of microsatellite instability and immunohistochemical markers among population- and clinic-based colorectal tumors results from the Colon Cancer Family Registry. J Mol Diagn. 2011;13(3):271–281. doi: 10.1016/j.jmoldx.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–5257. [PubMed] [Google Scholar]
- 21.Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16(5):412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 22.Therneau TM, Atkinson B. [Accessed September 21, 2012];R port. http://CRAN.R-project.org/package=rpart.
- 23.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34(5):424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 24.Aarnio M, Mecklin JP, Aaltonen LA, Nyström-Lahti M, Järvinen HJ. Life-time risk of different cancers in hereditary non-polyposis colorectal cancer (HNPCC) syndrome. Int J Cancer. 1995;64(6):430–433. doi: 10.1002/ijc.2910640613. [DOI] [PubMed] [Google Scholar]
- 25.Chung DC, Rustgi AK. The hereditary nonpolyposis colorectal cancer syndrome: genetics and clinical implications. Ann Intern Med. 2003;138(7):560–570. doi: 10.7326/0003-4819-138-7-200304010-00012. [DOI] [PubMed] [Google Scholar]
- 26.Loughrey MB, Waring PM, Tan A, et al. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary nonpolyposis colorectal cancer. Fam Cancer. 2007;6(3):301–310. doi: 10.1007/s10689-007-9124-1. [DOI] [PubMed] [Google Scholar]
- 27.Rodríguez-Moranta F, Castells A, Andreu M, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Clinical performance of original and revised Bethesda guidelines for the identification of MSH2/MLH1 gene carriers in patients with newly diagnosed colorectal cancer: proposal of a new and simpler set of recommendations. Am J Gastroenterol. 2006;101(5):1104–1111. doi: 10.1111/j.1572-0241.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 28.Lindor NM, Petersen GM, Hadley DW, et al. Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: a systematic review. JAMA. 2006;296(12):1507–1517. doi: 10.1001/jama.296.12.1507. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y, Berends MJ, Mensink RG, et al. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet. 1999;65(5):1291–1298. doi: 10.1086/302612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsey SD, Clarke L, Etzioni R, Higashi M, Berry K, Urban N. Cost-effectiveness of microsatellite instability screening as a method for detecting hereditary nonpolyposis colorectal cancer. Ann Intern Med. 2001;135(8 pt 1):577–588. doi: 10.7326/0003-4819-135-8_part_1-200110160-00008. [DOI] [PubMed] [Google Scholar]
- 31.Ladabaum U, Wang G, Terdiman J, et al. Strategies to identify the Lynch syndrome among patients with colorectal cancer: a cost-effectiveness analysis. Ann Intern Med. 2011;155(2):69–79. doi: 10.7326/0003-4819-155-2-201107190-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bessa X, Ballesteé B, Andreu M, et al. Gastrointestinal Oncology Group of Spanish Gastroenterological Association. A prospective, multicenter, population-based study of BRAF mutational analysis for Lynch syndrome screening. Clin Gastroenterol Hepatol. 2008;6(2):206–214. doi: 10.1016/j.cgh.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Domingo E, Niessen RC, Oliveira C, et al. BRAF-V600E is not involved in the colorectal tumorigenesis of HNPCC in patients with functional MLH1 and MSH2 genes. Oncogene. 2005;24(24):3995–3998. doi: 10.1038/sj.onc.1208569. [DOI] [PubMed] [Google Scholar]
- 34.Payá A, Alenda C, Pérez-Carbonell L, et al. Utility of p16 immunohistochemistry for the identification of Lynch syndrome. Clin Cancer Res. 2009;15(9):3156–3162. doi: 10.1158/1078-0432.CCR-08-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49(3):151–157. doi: 10.1136/jmedgenet-2011-100714. [DOI] [PubMed] [Google Scholar]
- 36.Balaguer F, Balmaña J, Castellví-Bel S, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Validation and extension of the PREMM1,2 model in a population-based cohort of colorectal cancer patients. Gastroenterology. 2008;134(1):39–46. doi: 10.1053/j.gastro.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kastrinos F, Steyerberg EW, Mercado R, et al. The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology. 2011;140(1):73–81. doi: 10.1053/j.gastro.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kastrinos F, Syngal S. Inherited colorectal cancer syndromes. Cancer J. 2011;17(6):405–415. doi: 10.1097/PPO.0b013e318237e408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balmaña J, Balaguer F, Castellví-Bel S, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Comparison of predictive models, clinical criteria and molecular tumour screening for the identification of patients with Lynch syndrome in a population-based cohort of colorectal cancer patients. J Med Genet. 2008;45(9):557–563. doi: 10.1136/jmg.2008.059311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.