Abstract
The feasibility of using a stochastic form of Pennes bioheat model within a 3D finite element based Kalman filter (KF) algorithm is critically evaluated for the ability to provide temperature field estimates in the event of magnetic resonance temperature imaging (MRTI) data loss during laser induced thermal therapy (LITT). The ability to recover missing MRTI data was analyzed by systematically removing spatiotemporal information from a clinical MR-guided LITT procedure in human brain and comparing predictions in these regions to the original measurements. Performance was quantitatively evaluated in terms of a dimensionless L2 (RMS) norm of the temperature error weighted by acquisition uncertainty. During periods of no data corruption, observed error histories demonstrate that the Kalman algorithm does not alter the high quality temperature measurement provided by MR thermal imaging. The KF-MRTI implementation considered is seen to predict the bioheat transfer with RMS error < 4 for a short period of time, Δt < 10sec, until the data corruption subsides. In its present form, the KF-MRTI method currently fails to compensate for consecutive for consecutive time periods of data loss Δt > 10sec.
Index Terms: Bioheat Transfer, MR Temperature Imaging, Kalman Filtering, Finite Element Modeling
I. Introduction
Magnetic resonance temperature imaging (MRTI) using the temperature sensitivity of the proton resonance frequency (PRF) shift is used in the guidance of clinical thermal therapy procedures, such as laser ablation in the brain [1]. Acquisition of the spatiotemporal temperature history during delivery aids in monitoring tissue temperature changes in real-time. This feedback can be used to control the therapy, allowing the user to reduce power or stop delivery if safety thresholds are exceeded [2]. Biological models of damage may also be used to predict outcomes in real-time and potentially enhance the efficacy of treatment. Unfortunately, like any measurement device, there are conditions under which data can be corrupted or lost during acquisition. This could be from low signal or signal loss due to heating or tissue motion. While a substantial amount of work on the acquisition and processing of PRF images has been performed to aid in mitigating these errors in a clinical environment, there remain instances of failure that lead to lost data [3], [4].
Proliferation of novel image guided thermal therapy procedures in the clinical setting has generated substantial interest in the use of high performance computing for computer-assisted prospective treatment planning as well as real-time monitoring and control of the procedures. Given trends of increasingly powerful computational and visualization resources, the role of the computational sciences is likely to continue to escalate in image-guided thermal therapy procedures. Computer simulation of the bioheat equation has been used in the past to provide a methodology for more optimal pre-treatment planning [5] as well as being incorporated into the process of automated control [6]. However, embedding a real-time MRTI data driven bioheat transfer model based simulation into the procedure may enable a new form of monitoring which can provide a robust estimate of the state of the procedure in the presence of lost information due to motion, low signal-to-noise-ratio (SNR), excessive heating, catheter induced signal voids, and other data corruption. The ability to perform realtime model based extrapolation in the spatial or temporal dimension can also enhance real-time control algorithms for efficacy as well as provide a means for more accurately monitoring high temperatures for safety [7], [8], [9], [10].
Kalman filter (KF) theory [11], [12] provides a precise mathematical framework to estimate the state of the induced temperature field during therapy delivery given a computer model of the bioheat transfer, all available temperature measurement data from MRTI, and estimated uncertainties in both the model and measurement data. Within the Kalman framework, the model of the temperature field is viewed in a statistical sense. The mean and variance of the predicted temperature, conditioned on the MRTI data may be interpreted as confidence in the treatment time predicted temperature values. This may subsequently be used to embed valuable information within the confidence levels of the lethal thermal dose. The state estimation provided by the KF will be important for the emerging adaptive image guided controllers [13], [14], [15], [6], [16] to allow the best estimate of the state and subsequent control decisions in the presence of modeling and image guided measurement uncertainties. Moreover, computer modeling may be further incorporated to extrapolate predictions into regions where MRTI measurements are not available [17], thereby extending the range of prediction when monitoring capabilities are limited. The technology could potentially facilitate confident completion of procedures that would otherwise have been considered too difficult to perform or possibly unsafe due to unusable or limited data.
In this study, we evaluated the potential performance of a Pennes bioheat transfer based Kalman filtered-MR thermal imaging (KF-MRTI) algorithm for predicting temperature during MR-guided laser induced thermal therapy (MRgLITT) delivery in the presence of noise or missing data. We consider the case where data corruption has occurred for the entire temperature imaging region of interest (ROI). Here we focus on demonstrating the feasibility of the idea that the filter can provide reasonably accurate temperature estimates comparable with the MRTI measurements. The particular stochastic differential equation (SDE) form of the bioheat model inherent to the Kalman algorithm is used with tabulated constitutive data [18], [19], [20]. The overall predictability of a deterministic form of the bioheat transfer model has been shown to be substantially increased through bioheat transfer constrained calibration algorithms [21], [22]. The patient-specific calibration algorithms are not coupled to the Kalman temperature estimates in this work. Real-time performance of the bioheat transfer algorithms have been studied previously [23], [21] and is not critically evaluated within the scope of the presented work.
A characterization study was performed to evaluate the ability of the implementation to provide accurate estimations of procedure progress in the presence of a range of simulated signal loss for a representative data set obtained from a clinical MRgLITT procedure. An online form of the Kalman algorithm is considered as the simulated signal loss manifests through acquisition time measurement uncertainty. The effect of the amount of data lost and error covariance used were evaluated in terms of a L2 metric (RMS) between the temperature imaging and KF prediction. The study was designed with two primary metrics in mind for monitoring: the maximum estimated temperature [2] (safety) and the estimated extent of tissue damage as predicted by an Arrhenius model of damage [24] (efficacy).
II. Materials and Methods
A. Kalman Algorithm
The goal of the Kalman filtered MRTI is to recursively estimate the temperature distribution throughout the tissue domain, U, given a computational model of the bioheat transfer and estimated temperature from MRTI. A linear stochastic differential form of the Pennes [25] bioheat transfer equation provides the underlying model for temperature field.
where the tissue specific heat, cp, tissue density, ρ, thermal conductivity, κ, perfusion, ω, blood specific heat, cblood, laser source, qlaser, and arterial blood temperature, ua, are assumed deterministic values. The primary difference between this approach and the classical deterministic Pennes model is that the unknown temperature field, u(·), is assumed to be a probabilistic random variable mapping a probability space, Ω, to an appropriate function space,
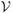 , that contains well-defined temperature solutions, u(·) : Ω →
, that contains well-defined temperature solutions, u(·) : Ω →
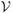 . In addition to the conduction, perfusion, and laser terms, the temperature change, ∂u(·), is also assumed to be driven by a Brownian field, β(·) : Ω →
. In addition to the conduction, perfusion, and laser terms, the temperature change, ∂u(·), is also assumed to be driven by a Brownian field, β(·) : Ω →
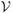 . A solution to these equations may be considered as a Galerkin expansion of the temperature field and Brownian field chosen such that the spatial basis
is deterministic and stochasticity is considered only in the time variable [12].
. A solution to these equations may be considered as a Galerkin expansion of the temperature field and Brownian field chosen such that the spatial basis
is deterministic and stochasticity is considered only in the time variable [12].
| (1) |
Here, the conventional summation notation over the index is implied for the number of the degrees of freedom, Ndof, in the Galerkin expansion. Statistics of the zero mean Brownian motion are assumed known. The diffusion matrix, Q, is a measure of the rate of divergence of the mean squared value from its initial value. Initial conditions for the temperature are given by P0. Both Q and P0 are assumed known symmetric positive definite Ndof × Ndof matrices.
The laser source, qlaser, is assumed a deterministic function of the applied power, p(t), optical scattering, μs, optical absorption, μa, anisotropy factor, g, and distance, ‖x − ξ ‖, from the source, Utip.
Active cooling of the applicator is modeled by holding the temperature in this region fixed through Dirichlet boundary conditions at the ambient temperature [26].
Proceeding formally, substituting the Galerkin discretization (1) into a variational form of our stochastic differential equation (SDE) with a deterministic test function yields the typical system dynamics matrix vector form.
where F and B are the known time invariant system dynamics matrix (Ndof × Ndof) and deterministic input matrix (Ndof × 1), respectively. In context of the time discretized form of these equations implemented to simulate laser ablations on the computer, the vector valued Brownian process, β⃗, may be considered representative of the spatial and temporal discretization errors as well as any modeling errors including inaccurate model parameters, system inputs, and constitutive nonlinearities in the thermal conductivity, perfusion, and optical absorption and scattering.
MRTI temperature estimates, z⃗(ti, ·) : Ω → ℝMdof i = 1, 2, 3, … are uniformly spaced at discrete time intervals, Δt, and are assumed to be a linear transformation of the state vector, u⃗(ti, ·), corrupted by white Gaussian noise, v⃗(ti, ·).
| (2) |
In this work, there are more state degrees of freedom, Ndof, than measurements, Mdof, such that the 3D image measurements are acquired on a subset of the 3D finite element simulation domain, Ndof > Mdof. The measurement matrix, H, is used to transform the 3D model prediction into the average value over the slice thickness of the MR thermal imaging. This provides a direct comparison between predicted and measured temperature.
Under Kalman theory assumptions, the nice algebraic properties when manipulating linear combinations and independent products of Gaussian distributions maintain the Gaussian structure throughout the time evolution of the state variable, u⃗(ti, ·). Hence, the state of the system at time, ti, is completely characterized by the mean and covariance conditioned on the available data. Following, standard conventions, the time immediately before the measured data is incorporated is denoted and the time immediately after the measured data is incorporated is denoted Step one of the two step Kalman algorithm begins with propagating the state from to [12]. This utilizes the stochastic form of the bioheat transfer model to make a prediction of the temperature field before data is available at time .
| (3) |
Here, and denote the mean and covariance of the state conditioned on all data up to the previous time step, , respectively. For this time invariant system, the state transition matrix, Φ(t,t0), is given by the matrix exponential of the system dynamics matrix, Φ(t, t0) = eF(t−t0). Intuitively, the mean, , provides an estimate of the temperature field given potential modeling inaccuracies and the covariance, , provides an estimate of the pixel-wise correlation expected in the temperature imaging. For example, the temperature of two pixels i and j near a laser fiber are expected to be positively correlated, Pij > 0, as their temperature is expected to increase and decrease together.
In step two of the algorithm, predictions are updated based on measurements at time ti and the Kalman gain, K(ti), to obtain the mean, , and covariance, , of the state conditioned on all data up to the current time step.
| (4) |
The updated temperature estimate, , is essentially a weighted sum of the measured and predicted temperature with weights given by the relative uncertainties in the modeling predictions and measurement accuracy. Offline implementations of the Kalman filter in which the error covariance is precomputed [27], [16] are not considered. The measurement model used in this work assumes that the acquisition time measurement uncertainty, R(ti) is not known a priori. The reader is referred to [12] for further details of the Kalman algorithm.
a) Covariance Propagation in Time
The high computational intensity of propagating the covariance matrix, P(t) in time is well known [28], [29], [30], [31], [32], [33], [34]. According to the Kalman algorithm (3) (4), any initial assumed sparsity in the covariance matrix is quickly lost as the error is propagated and becomes spatially correlated. The resulting dense linear algebra becomes computationally expensive very quickly and requires dense linear system solves, , with multiple right hand sides at each time step.
Localization approximations [35] provide a technique to approximate the dense covariance matrix with a sparse matrix, thus reducing computational storage, computational complexity, and runtime. Localization techniques will be needed in future efforts to achieve real-time performance of the implemented algorithm but are not considered in this work. Parallel computing techniques are used to solve the Kalman algorithm directly. The magnitude of the floating point operations required for the dense linear algebra, , is not feasible to achieve real-time performance, but is still important, within the context of MRgLITT, as it provides a baseline for future localization studies in which approximations to the Kalman algorithm can be evaluated for real-time performance and accuracy. A Crank Nicolson scheme was implemented to approximate the covariance prediction from to , see Appendix.
B. MR-guided Laser Ablation
A schematic of the MR-guided laser ablation procedure in brain from which the MRTI data is obtained is illustrated in Figure 1. Briefly, laser ablation was carried out in a dedicated neurosurgical MRI suite (BrainSUITE®, BrainLAB, AG, Feldkirchen, Germany) on a patient with a recurrent glioblastoma amenable to the procedure. The patient was under general endotracheal anesthesia and on a ventilator. The patient was positioned within a 1.5T clinical whole body scanner (MAGNETOM Espree, Siemens Medical Solutions, Erlangen, Germany) with an 8-channel, receive-only head coil (NORAS MRI products GmbH, Germany). A 1.5mm outer diameter water-cooled laser applicator with 1 cm diffusing fiber (VCLAS, Visualase, Inc, Houston, TX) was positioned into the right cerebellar lesion with MRI driven cranial guidance (Vector Vision Sky®, BrainLAB, AG, Feldkirchen, Germany). Laser ablation was carried out under MR-guidance using a 980 nm solid state laser source with a real-time visualization and control workstation (Visualase System, Visualase, Inc., Houston, TX). Exposures of 4W for less than 30s were used under MRTI guidance to verify the location of the fiber and 10W for up to 140s were used for therapy. Exposures were monitored in real-time using the temperature-sensitive proton resonance frequency (PRF) shift technique acquired with a 2D spoiled gradient-echo to generate temperature measurements, z⃗(ti, ·), every Δt =5 seconds (TR/TE/FA = 38 ms/20 ms/30°, frequency × phase = 256 × 128, FOV = 26 cm2, BW = 100Hz/pixel, slice thickness 5mm). For the purposes of this research, the magnitude and phase data was processed offline to investigate the feasibility of real-time KF-MRTI.
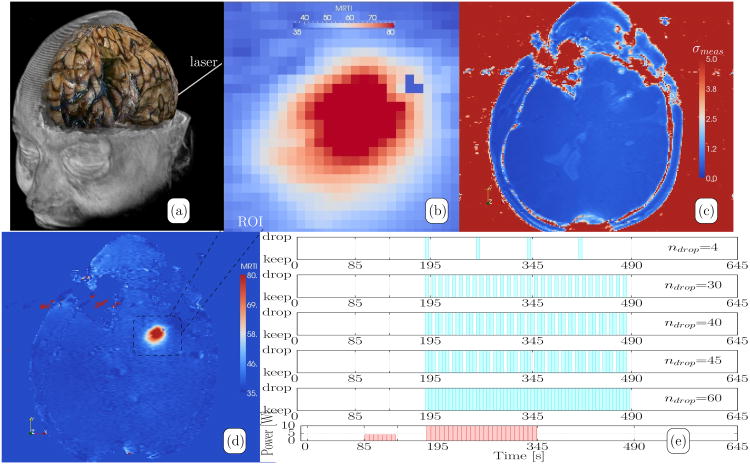
Fig. 1.
Summary of MRgLITT Data. (a) A schematic diagram of the in vivo MR-guided LITT procedure in brain is shown. A catheter is delivered to the target under MR guidance. (b) The ROI of the temperature imaging data considered in the Kalman algorithm is shown. Collectively, these pixels represent the Mdof = 26 × 26 = 676 measured temperature values of the measurement model. The full (c) uncertainty map, σ(x,t) shown in equation (5), and (d) MR thermal image are provided as a reference and shown in °C. (e) To simulate data loss, thermal images were dropped uniformly over the heating and cooling time period. The plot is intended to represent the number of dropped images, ndrop, as well as the time instances in which images were dropped in this study. Within the Kalman framework, data loss may be simulated by manually setting the measurement uncertainty to a large value, R(tdrop) → ∞, at the intended time instances. As a reference, the laser exposure history of the procedure is provided as power as a function of time at the bottom of (e). The 4 Watt location finding pulse is followed by the 10 Watt delivery.
The SNR is assumed sufficiently large (SNR > 10) such that the noise model for the PRF-based MR thermal image measurements is approximated as Gaussian. An uncorrelated Gaussian noise model for the measurements is assumed a function of the acquisition time SNR. The measurement model noise covariance is not available before the procedure.
| (5) |
A temperature sensitivity coefficient of α = −0.0097 was used [36]. The gyromagnetic ratio is denoted γ.
To characterize the performance of the implemented Kalman filter in the presence of incorrect or even incomplete data, data loss was systematically simulated through artificial removal of an entire thermal imaging data set at varying time instances. The total number of time instances of data loss considered was ndrop = 4, 30, 40, 45, 60. Data loss was considered uniform over the active laser exposure and tissue cooling period, 60 image acquisitions (300 [s]). Data loss of ndrop = 4 is sparse; two time instances during heating and cooling are dropped. Data loss of ndrop = 30, 40, 45 repeatedly drops data of 1,2,3 consecutive time points, respectively, over the 60 image data set considered. Full data loss ndrop = 60 is considered as a reference for the underlying bioheat transfer model used. The data loss provides the opportunity to investigate the ability of the Kalman filter to provide temperature estimates both near the applicator, in the case of heat induced signal loss due to large spin-lattice relaxation times, and at the boundaries of the treatment if they extended into regions that were not monitored. Data drop was implemented by setting the uncertainty to a large value, σ → ∞, at the time instance of the data loss. A summary of the simulated data artifacts is illustrated in Figure 1.
The original dropped MRTI temperature estimates were compared with the Kalman prediction as a metric of the Kalman filter performance. The metric of comparison, ∈(t), for comparing the temperature estimates is a weighted L2 norm within the time interval of interest, [0, T], and normalized by the volume of the ROI, U ⊂ ℝ3
| (6) |
The metric is normalized by the time varying and spatially dependent maps of the temperature image noise, σ(x,t).
The final thermal dose outcome was also quantitatively evaluated to compare the spatial overlap between the binary images of the Kalman filtered thermal dose estimate and the originally acquired MR temperature measurements. A Dice similarity coefficient [37] (DSC) was used to calculate the overlap of two thermal dose volumes A,B ⊂ ℝ3. The DSC has been recommended as a good validation metric for spatial overlap [38]. The DSC is defined as:
The possible values of DSC range from 0 (no overlap) to 1 (complete overlap). An Arrhenius rate process model [39] was used to evaluate the thermal dose resulting from the time-temperature history of the laser exposure.
In this Arrhenius thermal dose model, the frequency factor, A, and the activation energy, EA, are known experimentally determined kinetic parameters. The values for A and EA were and ; respectively, and have been used in previous studies [1], [40], [2]. R is the universal gas constant. The thermal dose was assumed to be lethal at doses ≥ 1 as seen in previous reports [1], [2].
C. Constitutive Data
Table I summarizes the constitutive data used. The model and initial temperature covariance matrices are assumed un-correlated. Within the context of the discretized equations, the model covariance, Q, may be considered representative of discretization errors as well modeling errors including inaccurate model parameters, system inputs, and constitutive nonlinearities in the thermal conductivity, perfusion, and optical absorption and scattering. A single value for the model covariance is difficult to select. A range of model covariance values, , was considered that was the order of magnitude of the measurement covariance, Figure 1(c). The Kalman temperature estimate is essentially a weighted sum of the measured and predicted temperature with weights given by Q and R. These values were chosen to investigate the behavior of the filter for a range of values where the final temperature state would weigh the measurement and model more heavily at the two extremes. Errors incurred due to time stepping inaccuracies for the Δt= 5 sec time steps of the Crank Nicolson algorithm implemented are also assumed to be included in the range considered. Initial computations using the current implementation of the Kalman filter showed little sensitivity to the range of acceptable state covariance values, .
Table I. Constitutive Data [18], [19], [20].
|
|
|
g |
|
|
|
|
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.527 | 9.0 | 0.88 | 140.0 | 5.0 | 1045 | 3840 | 3600 |
III. Results
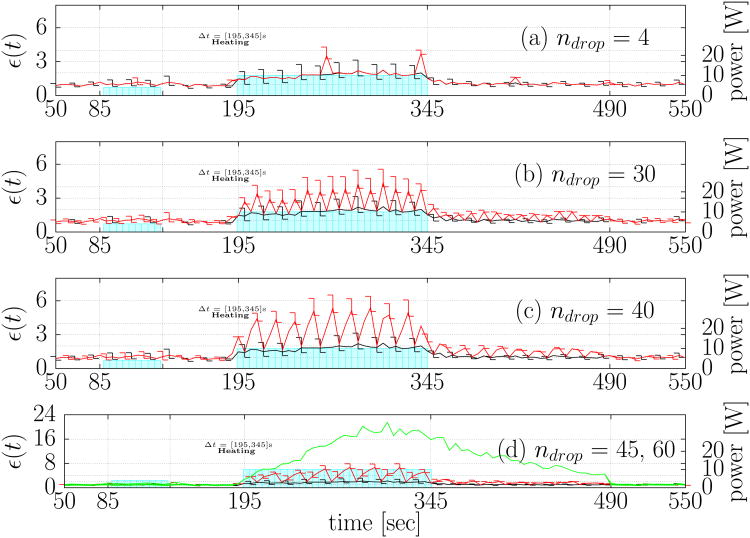
In total, 15 permutations of the simulated corruption of the original data combined with the different degrees of assumed model covariance were considered. A comprehensive summary of the time history of the L2 error (6) between the Kalman predicted temperature estimates and original data for each permutation of data loss is presented in Figure 2. The power profile is plotted against the right axis as a reference for the time points of the data drop. The non damaging location finding test pulse is seen at t ≈ 85[s], and the data drop is seen during the main thermal dose delivery t ∈ [190,490]. The error bars plot the variation over the model variances, σModel, considered. As a reference, the Kalman prediction in the absence of any data loss is provided as a reference in Figure 2(a)-(d). The error in the prediction is seen to increase with the number of consecutive data losses at a given time instance. The Kalman estimates returns to the baseline value when the data loss subsides.
Fig. 2.
Error history summary. Error histories of the L2 difference (6) between the original data and the Kalman prediction under the simulated data loss for (a) ndrop = 4 (b) ndrop = 30 (c) ndrop = 40 (d) ndrop = 45 are plotted as a 2D line graph. The time points of the simulated data loss are illustrated in Figure 1 (e). In each plot the Kalman estimate under no data loss, ndrop = 0, is shown in black. The line plots are shown for The lower and upper error bars represent the error values (6) seen at these time points for . The lower and upper bounds of the error bar provides concise presentation of the range of modeling error covariance considered. The power history is plotted against the right axis in Watts as a reference. The error history for (a) represents the error in prediction under sparsely dropped data. The error history for (b), (c), and (d) shows the error history under repeated data loss of 1,2,3 consecutive time points, respectively, over the time period of interest. The model prediction error history without any data updates, ndrop = 60, is provided as a reference as the green line in (d); the Kalman prediction is not sensitive to the model covariance in this situation and no error bars are necessary.
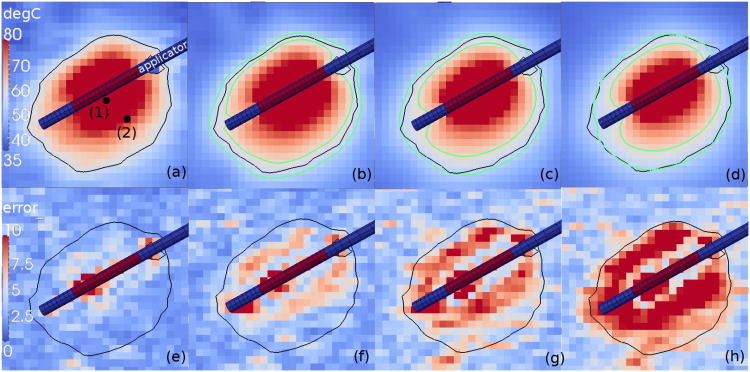
Figure 3(a)-(d) provides an illustration comparing the original MRTI data to the Kalman temperature predictions under the simulated data loss for (b) ndrop = 4 (c) ndrop = 30 (d) ndrop = 40 at the same time point, t = 340s, near peak heating. The measured 57°C isotherm is shown relative to the 95% confidence interval in each case. Maximum pixel-wise error is shown in Figure 3(e)-(h). As a reference, the maximum error for the Kalman prediction is provided without any simulated data loss in Figure 3(e). Maximum error under the simulated data loss for ndrop = 4, 30, 40 is shown in Figure 3 (f),(g),(h); respectively.
Fig. 3.
Kalman temperature prediction summary. The (a) original MRTI and the Kalman temperature estimate under the simulated data loss for (b) ndrop = 4 (c) ndrop = 30 (d) ndrop = 40 is shown at t = 340s for comparison. The time points of the simulated data loss are illustrated in Figure 1 (e). The same 57°C isotherm is shown for the MRTI data in black for (a)-(d). Green isotherms mark the 95% confidence interval for the predicted 57°C isotherm in (b)-(d). The temperature scale provided is 35-80°C. The corresponding measurement uncertainty is shown in °C. The applicator orientation with respect to the imaging is shown. The applicator diameter is 1.5mm. between the Kalman prediction and the MRTI is plotted in (e)-(h). (e) Max error for the Kalman prediction is provided without any simulated data loss as a reference. Maximum error under the simulated data loss for (f) ndrop = 4 (g) ndrop = 30 (h) ndrop = 40 is shown. All bioheat transfer predictions were done in 3D on a finite element mesh of the same resolution as the imaging data. The 3D finite element simulation data is projected onto the imaging data through the measurement model (2).
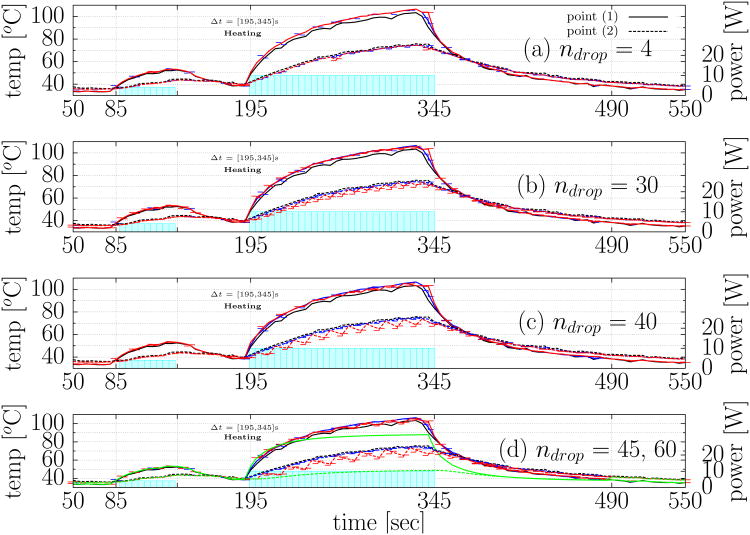
As a reference, temperature is plotted as function of time at the two spatial locations labeled in Figure 3(a). Temporal profiles at the selected spatial locations are shown in Figure 4. A comprehensive summary of the temperature history is shown for the the Kalman temperature estimates and original data for each permutation of data loss. As in Figure 3, the power profile is plotted against the right axis as a reference for the time points of the data drop. The non damaging location finding test pulse is seen at t ≈ 85[s], and the data drop is seen during the main thermal dose delivery t ∈ [190,490]. The error bars plot the variation over the model variances, σModel, considered. As a reference, the Kalman prediction in the absence of any data loss as well as original MRTI data is provided as a reference at both points in Figure 4(a)-(d). The error in the prediction is again seen to increase with the number of consecutive data loss at a given time instance.
Fig. 4.
Temporal profiles at selected spatial locations. The temperature is plotted as function of time at the two spatial locations labeled in Figure 3(a). The Kalman prediction under the simulated data loss for (a) ndrop = 4 (b) ndrop = 30 (c) ndrop = 40(d) ndrop = 45 are plotted as a 2D line graph. Solid-lines and dotted-lines represent the spatial locations labeled “(1)” and “(2)” in Figure 3(a); respectively. The time points of the simulated data loss are illustrated in Figure 1 (e). In each plot the Kalman estimate under no data loss, ndrop = 0, is shown in blue. The original MRTI data is shown in black. Similar to Figure 2, the line plots are shown for . The lower and upper error bars represent the error values (6) seen at these time points for . The lower and upper bounds of the error bar provides concise presentation of the range of modeling error covariance considered. The model prediction error history without any data updates, ndrop = 60, is representative of the uncalibrated bioheat transfer prediction using tabulated data [18], [19], [20] and is provided as a reference as the green line in (d); the uncalibrated bioheat transfer model is seen to systematically underestimate the temperature.
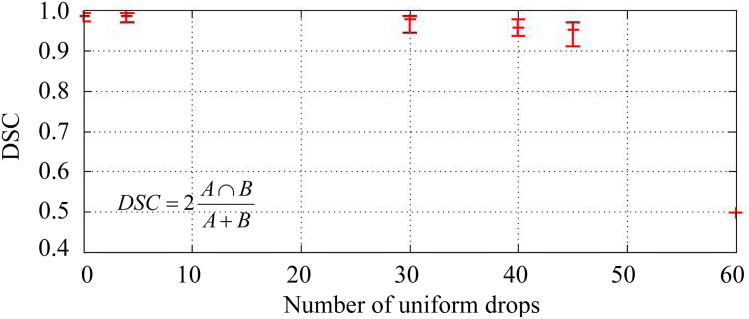
DSC values as a function of simulated data loss is shown in Figure 5. The data is shown for . The range in DSC values when using is represented by the lower and upper error bars. The DSC value is seen to decrease as a function of the assumed data loss.
Fig. 5.
DSC values as a function of simulated data loss. The DSC value provides a quantitative metric of spatial overlap between the binary images of the Kalman filtered thermal dose estimate and the originally acquired MR temperature measurements. The data is shown for . Error bars represent the range in DSC values when using .
IV. Discussion
The current work attempts to demonstrate the feasibility as well as to characterize the performance of a real-time Kalman filtered MRTI algorithm for laser ablation which uses an uncalibrated linear bioheat transfer model with homogeneous coefficients. Our primary goal in this instance was to demonstrate that, under limited circumstances, KF-MRTI can act as a reasonable surrogate for the MRTI data when MRTI data is not available. For this reason, the KF-MRTI data is compared to the MRTI data itself. Our results indicate that embedding this real-time predictive simulation within MRTI may facilitate more robust monitoring of laser ablation procedures. The low error seen in Figure 2 during periods of no data corruption indicates that the Kalman framework provides a rigorous methodology that matches well with realtime MRTI.
As observed by the errors in the model comparison to the MR temperature imaging data, Figure 2 (ndrop = 60), the underlying uncalibrated bioheat transfer model outside the KF framework does not accurately reproduce the MRTI. The uncalibrated bioheat transfer model used is seen to systematically underestimate the temperature, Figure 4(ndrop = 60). As expected, the period of time for which the model can provide a reasonable prediction of the bioheat transfer is determined by the accuracy of the model parameters. Incorporating Pennes bioheat transfer constrained calibration techniques [21], [22] into the underlying model predictions is one potential option that has been shown capable of dramatically increasing the accuracy of the computer model prediction. Alternatively, the Kalman algorithm uses the MR temperature image as an initial condition at each time step, and the available data may be thought of as continually calibrating the model [41]. The stochastic representation of the temperature field provides an opportunity to exploit the uncertainty in the model parameters and still provide predictive simulations without model calibration of the parameters in the conventional sense. As shown in Figure 3, temperature estimates are provided ± the standard deviation. The measured temperature values at the 57°C isotherm are seen to lie within the 95% confidence interval of the model predictions for ndrop = 4, 30. The Kalman algorithm essentially re-calibrates the temperature estimates as reliable data is obtained. High quality measured data essentially provides an initial condition from which the misconfigured model predictions are accurate for a short period of time. Poor predictions ∊ > 4 are seen for consecutive time periods of data loss Δt > 10sec. Note that since our MRTI was sampled every 5sec, one limitation of this study is that we do not investigate the impact of MRTI sampling on performance. As observed in the error plots shown in Figure 2 (b) and Figure 3 (b),(f) the un-calibrated model may be used to accurately predict, ∊ < 4, the bioheat transfer for a short period of time until the data corruption subsides. This potentially provides a methodology to provide predictive simulations without the added complexity of model calibration. Naturally, future efforts that exploit the predictive capabilities of the calibrated bioheat transfer models [21] may be expected to increase the tolerance of the Kalman algorithm to substantially large amounts of data corruption in both space and time and could dramatically reduce the aging of the underlying model of the bioheat transfer.
During rapid laser ablation in tissue, the best estimate for damage and corresponding uncertainty is valuable information for the safe and effective delivery of treatment. The ability to provide confidence intervals for isotherms, Figure 3, is a very powerful feature of the underlying uncertainty quantification framework provided by the Kalman algorithm. This can be transformed into a quantitative confidence level in the lethal thermal dose that can be readily conveyed to the physician and used to adjust treatment parameters to better conform to treatment goals and achieve the desired region of tissue damage [42]. Error metrics that mathematically characterize the desired clinical outcome, including the Dice similarity coefficients [37] shown in Figure 5 and metrics that accurately quantify the variance in the dose, will be a key interest in future work.
Figure 2 implies that the implemented algorithm, , seems to introduce essentially no bias during periods in which the laser power is zero and a bias of approximately two standard deviations near peak heating. Figure 3 (e) shows that the majority of this error is due to modeling in accuracies in the model predictions near the applicator. For underlying uncalibrated bioheat transfer model, the source of the inaccuracies near the applicator is difficult to distinguish between the cooling model used or the laser source term and will need further investigation in future efforts. Future efforts would also incorporate these errors as an increased spatially dependent model error covariance σModel(x). This capability is not present in the current study. Inaccuracies near the applicator could be exacerbated by the SNR based measurement model. The SNR loss with temperature causes a higher degree of weighting towards the computer prediction nearer the applicator. There is much future work possible in developing and validating measurement models that detect the data corruption outside the realm of the SNR based approximations of the presented work. More realistic measurement models that provide a temperature based estimate of measurement uncertainty from the predicted MR-relaxation times [43] are possible. Further work could also incorporate a thermal sensor internal to the applicator; the additional temperature information could potentially improve the prediction accuracy.
An Ensemble Kalman filter [35] (EnKF) framework and appropriate nonlinear SDE model [44] will be needed to address nonlinearities resulting from directly propagating uncertainties in the model parameters, i.e. perfusion, thermal conductivity, etc. Additionally, further work is required to study the optimal level of additional computational complexity inherent to the Pennes bioheat transfer constrained calibration and localization framework that would benefit the currently developed Kalman algorithms. Square root filters [12] may be needed to improve the numerical stability of these higher complexity algorithms. Higher order modeling of the effect of the flow of the cooling fluid [45] within the laser applicator may also have additional benefit in better predicting the maximum temperature near the applicator for safety purposes. These techniques must be critically evaluated in terms of the desired clinical result and computational cost.
Finally, in addition to the online case where only treatment time information is utilized, Kalman like algorithms may be utilized within the context of optimal smoothing [46] to retrospectively provide the best representation of the temperature state using all available information. The underlying finite element models may be used for prediction outside the region of data acquisition, making it much more powerful and robust than a mere filtering or interpolation of the MRTI data. The resulting map of the predicted tissue damage may operate as a surrogate for post-treatment imaging verification and even provide an improved assessment of the outcome of the procedure.
V. Conclusion
In summary, the work presented here provides evidence that the bioheat model driven KF-MRTI may be used to provide a robust model based estimate of the temperature state during thermal therapy delivery when MRTI data is lost. When using MRTI as the initial condition, the embedded linear Pennes bioheat transfer model with homogeneous parameters is seen to accurately predict the measured temperature for a limited range of data corrupting artifacts and may be beneficial in the case where MR artifacts temporarily prevent accurate temperature measurements to be used in real-time MR-guided treatment monitoring. However, the overall algorithm would significantly benefit from a calibration of the bioheat transfer model parameters [21]. The algorithm is not intrusive on the measured MR temperature imaging state when the signal is acceptable.
Acknowledgments
The research in this paper was supported in part through NIH 5T32CA119930-03, 1R21EB010196-01 and CA79282 as well as the Cancer Center Support Grant CA016672. The authors would like to thank Roger McNichols, PhD and Ashok Gowda, PhD for their assistance in clinical laser ablation as well as the ITK [48], Paraview [49], PETSc [50], libMesh [51], and CUBIT [52] communities for providing enabling software for scientific computation and visualization. Parameter studies were performed using allocations at the Texas Advanced Computing Center.
Appendix A. Prediction Approximations
A Crank Nicolson scheme and uncorrelated error propagation algorithm were considered as approximations of the covariance prediction from to . For the time invariant system dynamics matrix, F, the state transition matrix is represented as a matrix exponential [12]. A Taylor series approximation of the state transition matrix, Φ(t,t0), was used to obtain analytical expressions of the discrete time covariance predictions.
| (7) |
Anticipating implementation on future stream processing computing architectures, we considered a Crank Nicolson scheme that allows for a perfectly parallel column-wise update of the covariance matrix, .
| (8) |
Recall that and Q are symmetric but in general, and F are not. To maintain symmetry, the average of the column wise computed covariance, , and its transpose are taken as the covariance prediction.
Adding (8) and its transpose yields a Crank Nicolson form of the prediction covariance, .
The additional terms in this Crank Nicolson approximation are proportional to the lack of symmetry of . As noted in [44] the column wise solution to (8) has the intuitive interpretation that the pixel wise error, ∊, propagates and diffuses as the original bioheat equation with the source given by the assumed modeling error, σModel.
The bioheat equation imposes a natural correlation length that decays to zero away from the error source term and can be exploited in future localization studies of the covariance matrix to speed up the computations.
Appendix B. Covariance Perturbation Effect on MR Temperature Imaging
The effect of a correlation change due to propagation of the state covariance matrix on the final Kalman estimate is a key interest in the present study. In general, it is difficult to infer the effect of the off-diagonal covariance entries on the Kalman estimate and resulting error (6), however, intuition may be built through simplified assumptions. Consider the state update equation (4) with H = I and an arbitrary perturbation to the state covariance matrix, δP. The change in the Kalman state, , is given as the difference between the original Kalman prediction, , and the solution to the Kalman equations under the covariance perturbation, . The linear algebra of the resulting perturbation in the Kalman state, , may be interpreted as a matrix perturbation to a linear system of equations [47].
Intuition for the effect of a perturbation of a single entry may be understood by considering the block matrix inversion of the full n × n system of equations. Under the assumptions of the MRTI data, it is reasonable to assume an initially uncorrelated temperature covariance with matrix, S11 ∈ ℝ(n− 1)×(n− 1), and scalar variance, s22 ∈ ℝ, becomes correlated through a perturbation in the correlation vector δp12 ∈ ℝn−1.
The linear algebra for this system can be written very succinctly in terms of the difference between the measure and predicted value, ,
| (9) |
In general, the inverse of a sparse matrix is potentially dense. This implies that a single non-zero entry, , and a single correlation, δp12 = (0,…,0, δp, 0,…,0), could perturb the entire Kalman state estimate. Of course, the given sparsity of S11 would affect the relative magnitude of the perturbation for each state entry. Due to the potentially dense nature of the inverse of a sparse matrix the degree of perturbation is difficult to rigorously quantify. However, under further assumptions of diagonal matrix S11, further intuition can be gathered for specific scenarios of interest. Also, notice the potential singularity at that could cause numerical instabilities.
Contributor Information
D. Fuentes, Email: dtfuentes@mdanderson.org.
R. J. Stafford, Email: jstafford@mdanderson.org.
References
- 1.Carpentier A, McNichols R, Stafford R, Itzcovitz J, Guichard J, Reizine D, Delaloge S, Vicaut E, Payen D, Gowda A, et al. Real-time magnetic resonance-guided laser thermal therapy for focal metastatic brain tumors. Neurosurgery. 2008;6363, no. 1(1):8. doi: 10.1227/01.neu.0000335007.07381.df. [DOI] [PubMed] [Google Scholar]
- 2.McNichols R, Gowda A, Kangasniemi M, Bankson J, Price R, Hazle J. MR thermometry-based feedback control of laser interstitial thermal therapy at 980 nm. Lasers in Surgery and Medicine. 2004;3463, no. 1:48–55. doi: 10.1002/lsm.10243. [DOI] [PubMed] [Google Scholar]
- 3.Roujol S, Ries M, Quesson B, Moonen C, Denis de Senneville B. Real-time MR-thermometry and dosimetry for interventional guidance on abdominal organs. Magnetic Resonance in Medicine. 2010;63, no. 4:1080–1087. doi: 10.1002/mrm.22309. [DOI] [PubMed] [Google Scholar]
- 4.Rieke V, Butts Pauly K. MR thermometry. Journal of Magnetic Resonance Imaging. 2008;27, no. 2:376–390. doi: 10.1002/jmri.21265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Miga M, Galloway R., Jr Optimizing electrode placement using finite-element models in radiofrequency ablation treatment planning. IEEE Trans Biomed Eng. 2009;56:237–245. doi: 10.1109/TBME.2008.2010383. [DOI] [PubMed] [Google Scholar]
- 6.Mougenot C, Quesson B, de Senneville B, de Oliveira P, Sprinkhuizen S, Palussière J, Grenier N, Moonen C. Three-dimensional spatial and temporal temperature control with MR thermometry-guided focused ultrasound (MRgHIFU) Magnetic Resonance in Medicine. 2009;61, no. 3:603–614. doi: 10.1002/mrm.21887. [DOI] [PubMed] [Google Scholar]
- 7.Potocki J, Tharp H. Concurrent hyperthermia estimation schemes based on extended kalman filtering and reduced-order modelling. International journal of hyperthermia. 1993;9, no. 6:849–865. doi: 10.3109/02656739309034987. [DOI] [PubMed] [Google Scholar]
- 8.Todd N, Payne A, Parker D. Model predictive filtering for improved temporal resolution in MRI Temperature imaging. Magnetic Resonance in Medicine. 2010;63, no. 5:1269–1279. doi: 10.1002/mrm.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuentes D. Kalman Filtered MR Temperature Imaging. ISMRM 2011 Annual Meeting. Track: MR Guided Focused Ultrasound, Thermotherapy & Thermometry; Montreal, Canada. May, 2011. [Google Scholar]
- 10.Roujol S, Denis S, Hey S, Moonen C, Ries M. Robust adaptive extended Kalman filtering for real time MR-thermometry guided HIFU interventions. IEEE transactions on medical imaging. 2011 doi: 10.1109/TMI.2011.2171772. [DOI] [PubMed] [Google Scholar]
- 11.Kalman R. A new approach to linear filtering and prediction problems. Journal of basic Engineering. 1960;8263, no. 1:35–45. [Google Scholar]
- 12.Maybeck PS. ser Mathematics in Science and Engineering. Vol. 141 Academic Press; 1979. Stochastic models, estimation, and control. [Google Scholar]
- 13.Cheng K, Dewhirst M, Stauffer P, Das S. Effective learning strategies for real-time image-guided adaptive control of multiple-source hyperthermia applicators. Medical physics. 2010;37:1285. doi: 10.1118/1.3302829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arora D, Skliar M, Roemer R. Model-predictive control of hyperthermia treatments. IEEE Transactions on Biomedical Engineering. 2002;49, no. 7:629–639. doi: 10.1109/TBME.2002.1010846. [DOI] [PubMed] [Google Scholar]
- 15.Hutchinson E, Dahleh M, Hynynen K. The feasibility of mri feedback control for intracavitary phased array hyperthermia treatments. International Journal of Hyperthermia. 1998;1463, no. 1:39–56. doi: 10.3109/02656739809018213. [DOI] [PubMed] [Google Scholar]
- 16.Malinen M, Duncan S, Huttunen T, Kaipio J. Feedforward and feedback control of ultrasound surgery. Applied Numerical Mathematics. 2006;5663, no. 1:55–79. [Google Scholar]
- 17.Arora D, Skliar M, Roemer R. Minimum-time thermal dose control of thermal therapies. IEEE Transactions on Biomedical Engineering. 2005;52, no. 2:191–200. doi: 10.1109/TBME.2004.840471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diller KR, Valvano JW, Pearce JA. Bioheat transfer. In: Kreith F, Goswami Y, editors. The CRC handbook of mechanical engineering. 2nd. CRC Press; Boca Raton: 2005. pp. 4–278.pp. 4–357. [Google Scholar]
- 19.Welch AJ, van Gemert MJC. Optical-thermal response of laser-irradiated tissue. New York: Plenum Press; 1995. [Google Scholar]
- 20.Duck F. Physical properties of tissue: a comprehensive reference book. Academic Press; 1990. [Google Scholar]
- 21.Fuentes D, Feng Y, Elliott A, Shetty A, McNichols RJ, Oden JT, Stafford RJ. Adaptive real-time bioheat transfer models for computer driven MR-guided laser induced thermal therapy. IEEE Trans Biomed Eng. 2010;57, no. 5 doi: 10.1109/TBME.2009.2037733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diller KR, Oden JT, Bajaj C, Browne JC, Hazle J, Babuška I, Bass J, Bidaut L, Demkowicz L, Elliott A, Feng Y, Fuentes D, Goswami S, Hawkins A, Khoshnevis S, Kwon B, Prudhomme S, Stafford RJ. Advances in Numerical Heat Transfer. Vol. 3. Taylor & Francis Group; 2008. Numerical Implementation of Bioheat Models and Equations, ch. 9: Computational Infrastructure for the Real-Time Patient-Specific Treatment of Cancer. [Google Scholar]
- 23.Oden JT, Diller KR, Bajaj C, Browne JC, Hazle J, Babuška I, Bass J, Demkowicz L, Feng Y, Fuentes D, Prudhomme S, Rylander MN, Stafford RJ, Zhang Y. Dynamic data-driven finite element models for laser treatment of prostate cancer. Num Meth PDE. 2007;23, no. 4:904–922. doi: 10.1002/num.20251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yung J, Shetty A, Elliott A, Weinberg J, McNichols R, Gowda A, Hazle J, Stafford R. Quantitative comparison of thermal dose models in normal canine brain. Medical physics. 2010;37:5313. doi: 10.1118/1.3490085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pennes HH. Analysis of tissue and arterial blood temperatures in the resting forearm. J Appl Physiol. 1948;1:93–122. doi: 10.1152/jappl.1948.1.2.93. [DOI] [PubMed] [Google Scholar]
- 26.Fuentes D, Walker C, Elliott A, Shetty A, Hazle J, Stafford RJ. MR temperature imaging validation of a bioheat transfer model for LITT. International Journal of Hyperthermia. 2011;27, no. 5:453–464. doi: 10.3109/02656736.2011.557028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stengel R. Optimal control and estimation. Dover Pubns; 1994. [Google Scholar]
- 28.Todling R, Cohn S. Suboptimal schemes for atmospheric data assimilation based on the Kalman filter. Monthly Weather Review. 1994;122, no. 11:2530–2557. [Google Scholar]
- 29.Gauthier P, Courtier P, Moll P. Assimilation of simulated wind lidar data with a Kalman filter. Monthly Weather Review. 1993;121, no. 6:1803–1820. [Google Scholar]
- 30.Evensen G. Sequential data assimilation with a nonlinear quasi-geostrophic model using Monte Carlo methods to forecast error statistics. Journal of geophysical research. 1994;99, no. C5:10143. [Google Scholar]
- 31.Bouttier F. The dynamics of error covariances in a barotropic model. Tellus A. 1993;45, no. 5:408–423. [Google Scholar]
- 32.Rabier F, Courtier P, Ehrendorfer M. Four-dimensional data assimilation: comparison of variational and sequential algorithms. Quarterly Journal of the Royal Meteorological Society. 1992;118, no. 506:673–713. [Google Scholar]
- 33.Cohn S, Parrish D. The behavior of forecast error covariances for a Kalman filter in two dimensions. Monthly Weather Review. 1991;119, no. 8:1757–1785. [Google Scholar]
- 34.Cohn S. Dynamics of short-term univariate forecast error covariances. Monthly Weather Review. 1993;121, no. 11:3123–3149. [Google Scholar]
- 35.Ott E, Hunt B, Szunyogh I, Zimin A, Kostelich E, Corazza M, Kalnay E, Patil D, Yorke J. A local ensemble Kalman filter for atmospheric data assimilation. Tellus A. 2004;56, no. 5:415–428. [Google Scholar]
- 36.Peters R, Hinks R, Henkelman R. Ex vivo tissue-type independence in proton-resonance frequency shift MR thermometry. Magnetic resonance in medicine. 1998;40, no. 3:454–459. doi: 10.1002/mrm.1910400316. [DOI] [PubMed] [Google Scholar]
- 37.Dice L. Measures of the amount of ecologic association between species. Ecology. 1945;26, no. 3:297–302. [Google Scholar]
- 38.Zou K, Wells W, III, Kikinis R, Warfield S. Three validation metrics for automated probabilistic image segmentation of brain tumours. Statistics in medicine. 2004;23, no. 8:1259–1282. doi: 10.1002/sim.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearce J, Thomsen S. Rate process analysis of thermal damage. Optical-thermal response of laser-irradiated tissue. 1995:561–606. [Google Scholar]
- 40.Schwarzmaier H, Yaroslavsky I, Yaroslavsky A, Fiedler V, Ulrich F, Kahn T. Treatment planning for MRI-guided laser-induced interstitial thermotherapy of brain tumorsThe role of blood perfusion. Journal of Magnetic Resonance Imaging. 1998;863, no. 1:121–127. doi: 10.1002/jmri.1880080124. [DOI] [PubMed] [Google Scholar]
- 41.Aidala V. Parameter estimation via the Kalman filter. Automatic Control, IEEE Transactions on. 2002;22, no. 3:471–472. [Google Scholar]
- 42.Arora D, Skliar M, Cooley D, Roemer R. Constrained predictive control of thermal therapies for minimum-time delivery of thermal dose. Control Systems Technology, IEEE Transactions on. 2007;15, no. 6:1030–1037. [Google Scholar]
- 43.Moser E, Winklmayr E, Holzmuller P, Krssak M. Temperature-and pH-dependence of proton relaxation rates in rat liver tissue. Magnetic resonance imaging. 1995;13, no. 3:429–440. doi: 10.1016/0730-725x(94)00135-p. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Xiu D. On numerical properties of the ensemble Kalman filter for data assimilation. Computer Methods in Applied Mechanics and Engineering. 2008;197, no. 43-44:3574–3583. [Google Scholar]
- 45.Fasano A, Hömberg D, Naumov D. On a mathematical model for laser-induced thermotherapy. Applied Mathematical Modelling. 2010 [Google Scholar]
- 46.Simon D. Optimal state estimation: Kalman, H [infinity] and nonlinear approaches. John Wiley and Sons; 2006. [Google Scholar]
- 47.Demmel J. Society for Industrial and Applied Mathematics Philadelphia. 1997. Applied numerical linear algebra. [Google Scholar]
- 48.Ibanez L, Schroeder W, Ng L, Cates J. The ITK Software Guide. 2nd. Kitware, Inc; 2005. ISBN 1-930934-15-7, http://www.itk.org/ItkSoftwareGuide.pdf. [Google Scholar]
- 49.Henderson A, Ahrens J. The ParaView Guide. Kitware; 2004. [Google Scholar]
- 50.Balay S, Gropp W, McInnes L, Smith B. PETSc Users Manual. Argonne National Laboratory, Tech Rep ANL-95/11 - Revision 2.1.5. 2003 [Google Scholar]
- 51.Kirk B, Peterson J. libMesh-a C++ Finite Element Library. CFDLab. 2003. URL http://libmesh.sourceforge.net.
- 52.Blacker T, et al. Cubit Users Manual. 2008. http://cubit.sandia.gov/documentation .