Abstract
Objective
Meniscus tears are associated with a heightened risk for osteoarthritis. We aimed to advance our understanding of the metabolic state of human injured meniscus at the time of arthroscopic partial meniscectomy through transcriptome-wide analysis of gene expression in relation to patient age and degree of cartilage chondrosis.
Methods
The degree of chondrosis of knee cartilage was recorded at the time of meniscectomy in symptomatic patients without radiographic osteoarthritis. RNA preparations from resected menisci (N=12) were subjected to transcriptome-wide microarray and QuantiGene Plex analyses. The relative changes in gene expression variation with age and chondrosis were analyzed and integrated biological processes were investigated computationally.
Results
We identified a set of genes in torn meniscus that were differentially expressed with age and chondrosis. There were 866 genes differentially regulated (≥1.5-fold; P<0.05) with age and 49 with chondrosis. In older patients, genes associated with cartilage and skeletal development and extracellular matrix synthesis were repressed while those involved in immune response, inflammation, cell cycle, and cellular proliferation were stimulated. With chondrosis, genes representing cell catabolism (cAMP catabolic process) and tissue and endothelial cell development were repressed and those involved in T cell differentiation and apoptosis were elevated.
Conclusion
Differences in age-related gene expression suggest that in older adults, meniscal cells might de-differentiate and initiate a proliferative phenotype. Conversely, meniscal cells in younger patients appear to respond to injury, but maintain the differentiated phenotype. Definitive molecular signatures identified in damaged meniscus could be segregated largely with age and, to a lesser extent, with chondrosis.
Keywords: Meniscus, Transcriptome analysis, Gene expression, Aging, Articular cartilage, Chondrosis
The meniscus is an essential component of the tibiofemoral articulation that contributes to the complex biomechanics of the knee joint, protecting the underlying articular cartilage. The role of the meniscus in the initiation and progression of osteoarthritis (OA) is thought to be minimal unless it is injured (1, 2). The location of the meniscus and its role in shock absorption and load transmission predispose the meniscus to traumatic and degenerative injuries, thus, meniscus tears are one of the most common intra-articular injuries of the knee (3, 4). There is an age bias in terms of mechanisms of meniscal injury (5): younger people are more likely to have acute tears due to trauma whereas older people are more likely to have tears due to degeneration (6-9). Since the majority of meniscus tears are unsuitable for repair (3, 10, 11), partial meniscectomy is used to redress most meniscus tears (7, 12).
A large body of information suggests that damage to or loss of meniscus is associated with changes in cartilage (13-16). It has also been reported that following partial meniscectomy, there is an elevated risk for functional deterioration of cartilage (11, 17, 18), often resulting in the development of OA and irreparable joint damage (2, 10, 14, 17, 19-21). Despite the fact that meniscus and knee articular cartilage have several characteristics in common and the mechanisms of meniscus and knee cartilage degeneration have been demonstrated (22, 23), the molecular pathogenesis of traumatic and degenerative meniscus tears and their association with degenerative changes in the articular cartilage are not known.
We have previously shown, through a candidate gene approach, that the expression of various OA- and obesity-related genes diverged with age (24, 25) and to some extent with body mass index (BMI) (25) in the injured meniscus. As these studies provided information on only a handful of genes (known for their specific role in OA and obesity), we sought to determine the transcriptome-wide gene expression profile in meniscus tears in patients undergoing arthroscopic partial meniscectomy to identify a comprehensive set of targets for future studies. The goal is to identify changes in the gene expression profile of the torn meniscus that may be relevant to age-associated meniscal and cartilage degeneration. We believe that this information will allow for a better understanding of why and how degenerative changes occur in the meniscus and how injury to the meniscus, whether traumatic or degenerative, contributes to the initiation and progression of OA in the knee.
Materials and Methods
Patients and meniscus samples
The study protocol was approved by Washington University Institutional Review Board and complied with the Health Insurance Portability and Accountability Act. All subjects furnished a written informed consent to allow use of their tissues for research purposes. The dysfunctional fragment of the human injured meniscus resected from patients with symptomatic, non-repairable tears during arthroscopic partial meniscectomy was used for gene expression analysis.
Characteristics of the study population
The characteristics of the study population are presented in Table 1. Human meniscal tissues were collected from 12 patients (6 ≤40 years, 6 >40 years and 5 with chondrosis; 7 without chondrosis) at the time of arthroscopic meniscal resection. The average age of patients in ≤40 (young) group was 31.3 years (range 16–40 years) and in >40 (old) group was 50.0 years (range 43–55 years). Most injuries were complex tears of the posterior horn of the medial meniscus with surgery an estimated 1–12 months after injury. Only one patient had a concomitant ligament injury, which was a chronic anterior cruciate ligament tear. In patients without chondrosis, no cartilage changes (grade 0) were observed in any of the three knee compartments. On the other hand, in patients with chondrosis, cartilage damage (grade ≥2) was observed in at least one compartment of the knee. The mean age of patients with chondrosis was 48.40 years (range 40–55 years) and for those without chondrosis was 35.14 years (range 16–48).
Table 1. Clinical description of meniscus tears, chondrosis and RNA quality.
| Meniscus | Type of tear | Location of tear | Time from injury (estimated) | Concomitant injury | Age (years) | Chondrosis | RIN |
|---|---|---|---|---|---|---|---|
| Medial | Complex | Posterior horn | 2 months | ACL | 28 | No | 6.3 |
| Medial | Parrot beak | Posterior horn | 2.5 months | None | 40 | Yes | 6.7 |
| Medial | Bucket handle | Posterior horn | 1 month | None | 16 | No | 7.7 |
| Medial | Complex | Posterior horn | 3.5 months | None | 55 | Yes | 6.9 |
| Lateral | Complex | Posterior horn | 10 months | None | 28 | No | 6.8 |
| Lateral | Complex | Posterior horn/body | 4 months | None | 53 | Yes | 7.3 |
| Medial | Complex | Posterior horn | 3 months | None | 36 | No | 7.2 |
| Medial | Parrot beak | Posterior horn | 12 months | None | 40 | Yes | 7.5 |
| Medial | Complex | Posterior horn | 9 months | None | 48 | No | 6.9 |
| Medial | Complex | Posterior horn | 4 months | None | 43 | No | 7.1 |
| Medial | Complex | Posterior horn | 1.5 months | None | 55 | Yes | 6.9 |
| Lateral | Complex | Posterior horn | 4 months | None | 47 | No | 6.9 |
ACL = Anterior cruciate ligament; RIN = RNA integrity number
Arthroscopic chondral changes and grading
At the time of arthroscopy, changes in the cartilage were recorded by the operating surgeon (fellowship trained academic orthopedic sports medicine) in three compartments of the knee: same compartment from which injured meniscus was resected, the other weight-bearing compartment and the patellofemoral compartment. The chondrosis was recorded using a modified Outerbridge scoring system (26). For the purpose of this analysis, we simplified chondrosis to a binomial variable with a value of 1 if any compartment had grade 2 or greater chondrosis and a value of 0 if no compartment had grade 2 or greater chondrosis.
RNA preparation from resected meniscus
Meniscus tissues received in the laboratory were processed and used for RNA isolation as has been described previously (25). Total RNA concentration and purity were determined by use of Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) according to manufacturer's instructions. RNA integrity number for each sample is shown in Table 1. RNA samples were sent to Washington University Genome Technology Access Center (GTAC) for microarray and QuantiGene Plex assays.
Transcriptome analysis
Transcriptome analysis was done according to standard protocol by the GTAC. Briefly, 100 ng of RNA transcripts were amplified by T7 linear amplification using ABI-Ambion MessageAmp TotalPrep amplification kit (Life Technologies, Grand Island, NY). A total of 750 μg of amplified RNA was applied to HumanHT-12-v4 Expression BeadChips (Illumina Inc., San Diego, CA) and hybridized at 58°C for 16–20 h at high humidity. Immobilized, biotinylated amplified RNAs were then detected by staining with Cy3 Streptavidin (1 μg Cy3-SA per 1 ml of Illumina Block E1) for 10 min at room temperature. Arrays were washed, dried and scanned on an Illumina BeadArray Reader according to recommended protocols. Images were quantitated by Illumina BeadScan-v3 and the quantitated data was imported into Illumina GenomeStudio data analysis software (Illumina Inc., San Diego, CA). Finally, on-slide spot replicates were averaged by GenomeStudio and an individual spot probe was reported.
Comparisons and calculations of results
The gene expression differences were compared between young (≤40 years) and old (>40 years) patients and patients with and without concomitant chondrosis. We used the age cutoff of 40 years as per our previous studies where we found significant differences in gene expression between the age groups of >40 and ≤40 (24, 25).
Statistical analysis
To identify differentially expressed genes in injured menisci, one-way Analysis of Variance (ANOVA) model was performed on the probe sets using Partek Genomics Suite v6.6 (Partek Inc., St. Louis, MO). To control the false-positive response rate, a false discovery rate (FDR) was used to adjust the P value. Genes were considered differentially expressed whose probes passed these tests: minimum expression, FDR and fold change. An FDR of 0.05 and a statistical significance of a P value <0.05 was used. An arbitrary cutoff of absolute fold-change ≥1.5 was applied to narrow down the number of differentially regulated genes.
Hierarchical clustering and Venn diagrams
Hierarchical clustering was done in Partek Genomics Suite v6.6 using the full-featured hierarchical clustering option on genes significantly up- or down-regulated with age as well as with the presence or absence of chondrosis. This analysis allows for a dual clustering of genes and samples, interactive branch flipping, and other advanced features for clustering and coloring the resulting dendrograms and heat maps. Similarly, Venn diagrams were created by the Partek list manager to show number of genes differentially up- or down-regulated with age and chondrosis and their potential overlap.
Validation of selected genes by QuantiGene Plex assay
To confirm the consistency and reliability of microarray (transcriptome) data, we selected 45 target genes and 3 housekeeping genes for validation. First, the top 15 genes up-regulated and 15 genes down-regulated with age were selected. Additionally, we handpicked another 15 genes known to be important to cartilage homeostasis and OA. The validation of gene expression was done by QuantiGene Plex assay (Panomics Inc., Fremont, CA), which has proven to be a reliable tool to validate microarray data (27). This is an unbiased assay, which is based on branched-chain DNA technology and provides a novel approach for gene expression analysis by analyzing the reporter gene (signal amplification) rather than a target amplification as achieved by polymerase chain reaction (28, 29). A list of genes validated by QuantiGene Plex assay with some of their features is provided in Supplementary Table 1. The quantification of RNA was carried out in biological and technical replicates using the QuantiGene Plex 2.0 assay kit. Panomics, Inc. developed individual bead-based oligonucleotide probe sets for each target (Plex set # 312184, http://www.panomics.com). All steps were performed according to supplied protocols by GTAC and as has been described elsewhere (30).
Biological processes and functional network analysis for differentially expressed genes
To determine the biological significance of differentially expressed genes, we analyzed gene sets by GeneGo MetaCore (GeneGo Inc., Carlsbad, CA) pathway analysis software to analyze the gene ontology (GO). The altered cellular and biological processes (GO distribution) were ranked based upon significant −log(P Value). GeneGo MetaCore was also used to investigate functional and molecular networks over represented by the differentially expressed genes. The gene expression data was used to generate functional and molecular networks also by utilizing GeneGo MetaCore pathways analysis software. Genes differentially expressed with age and chondrosis were subjected to gene networking to see the association of genes with each other in canonical biological pathways.
Microarray data deposition
The raw microarray data have been deposited in the Gene Expression Omnibus (GEO) with the accession number GSE45233 (http:www.ncbi.nlm.nih.gov/projects/geo).
Results
Transcriptome analysis
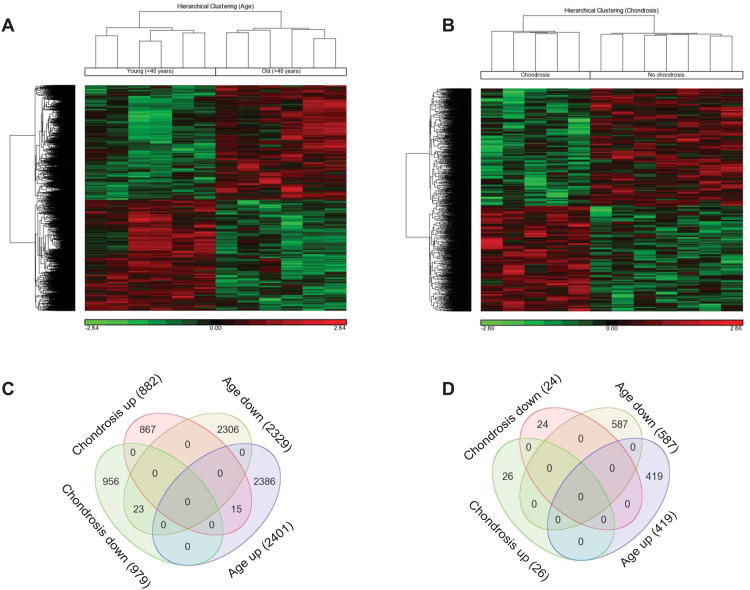
We analyzed gene expression changes in transcriptome-wide gene expression profiles in the meniscus tears. A principal component analysis was performed to see how the samples segregated in a 3-dimensional space based on patients' age and on the presence or absence of chondrosis (not shown). The samples from younger patients clustered together and the samples from older patients clustered together. Likewise, samples also segregated with the presence or absence of chondrosis. This segregation suggested that there would be molecular differences between the samples based on age and chondrosis. Then we identified differentially expressed genes by one-way ANOVA analysis. Of the 47,432 transcripts detectable by Illumina HumanHT-12-v4 BeadChips Kit, 4730 (∼10%) transcripts were significantly (P<0.05) and differentially up- or down-regulated with age and 1861 (4%) with chondrosis. Hierarchical clustering analyses of significantly differentially regulated genes, in the torn meniscus between young and old patients (Fig. 1A) or in patients with and without chondrosis (Fig. 1B), are shown. Hierarchical clustering also indicated the molecular differences between meniscal samples based on age or chondrosis.
Fig. 1. Hierarchical clustering and Venn diagram of differentially expressed genes.
Transcriptome analysis of human injured meniscus from young (≤40 years) and old (>40 years) patients with and without chondrosis was performed. Hierarchical clustering representing the transcripts that were significantly (P<0.05) and differentially up- and down-regulated are shown for age (A) and chondrosis (B). Each vertical row represents sample and each horizontal line represents a single gene. The numbers of significantly differentially expressed genes are shown in Venn diagrams without (C) and with (D) a fold-change cutoff of ≥1.5. The numbers of differentially expressed genes are shown in parenthesis for all four categories (young, old, chondrosis and no chondrosis). The numbers shown in overlapping areas depict the number of genes common to two or more categories. Color codes for hierarchical clustering: down-regulated genes are shown in green while up-regulated genes are shown in red.
Analysis of differentially expressed genes varying with age
We identified a total of 4730 genes in injured meniscus that were significantly differentially expressed between patients ≤40 years and >40 years of age. Among these, 2401 genes were significantly up-regulated while 2329 genes were down-regulated with age. Because of the large number of genes, we set an arbitrary cutoff of ≥1.5-fold to narrow down the number of differentially expressed genes. This filtering criterion allowed us to limit the number of differentially expressed genes to 1006; following removal of the non-annotated and duplicate genes, the number of differentially expressed genes dropped to 866. Among these 866 genes, 373 genes (Supplementary Table 2) were up-regulated while 493 genes (Supplementary Table 3) were down-regulated with age. Some of the important differentially expressed genes (top 15 up- or down-regulated plus 15 other genes selected for their significance in meniscus and cartilage metabolism and OA), and their respective QuantiGene Plex fold-change validation, are shown in Table 2. QuantiGene Plex assay showed a similar fold-change for genes down-regulated and a somewhat lower fold-change for genes up-regulated with age, however, the overall trend remained the same between microarray and QuantiGene Plex assays.
Table 2. Genes differentially regulated with age and with chondrosis.
| Symbol | Gene Name | Microarray | QGP assay | |
|---|---|---|---|---|
| Fold-change | P | Fold-change | ||
| Genes down-regulated with age (top 15) | ||||
| COL2A1 | Collagen, type II, alpha 1 | -10.38 | 0.018 | -14.66 |
| CHAD | Chondroadherin | -6.73 | 0.014 | -6.57 |
| COL11A2 | Collagen, type XI, alpha 2 | -6.14 | 0.006 | -10.59 |
| COL9A2 | Collagen, type IX, alpha 2 | -5.99 | 0.003 | -5.98 |
| CLEC3A | C-type lectin domain family 3, member A | -5.54 | 0.023 | -5.87 |
| SCIN | Scinderin | -4.88 | 0.005 | -7.41 |
| FGFR3 | Fibroblast growth factor receptor 3 | -4.65 | 0.007 | -4.36 |
| SHC4 | SHC (Src homology 2 domain containing) family, member 4 | -4.55 | 0.016 | -7.94 |
| CILP2 | Cartilage intermediate layer protein 2 | -4.32 | 0.030 | -3.04 |
| SOX8 | SRY (sex determining region Y)-box 8 | -4.23 | 0.015 | -1.12 |
| SBSPON | RPE-Spondin | -4.14 | 0.038 | -3.37 |
| S100A1 | S100 calcium binding protein A1 | -4.08 | 0.012 | -12.47 |
| Additional selected candidate genes | ||||
| GLDN | Gliomedin | -4.06 | 0.031 | -4.94 |
| MT1E | Metallothionein 1E | -3.86 | 0.003 | -1.33 |
| S100A13 | S100 calcium binding protein A13 | -3.82 | 0.014 | -1.04 |
| HAPLN1 | Hyaluronan and proteoglycan link protein 1 | -3.70 | 0.008 | -4.69 |
| LEPREL1 | Leprecan-like 1 | -3.36 | 0.016 | -4.14 |
| SOX9 | SRY (sex determining region Y)-box 9 | -2.98 | 0.042 | -2.53 |
| VEGFA | Vascular endothelial growth factor A | -2.86 | 0.029 | -1.71 |
| ACAN | Aggrecan | -2.52 | 0.032 | -3.37 |
| SPRY4 | Sprouty homolog 4 | -2.09 | 0.038 | -1.51 |
| SULF2 | Sulfatase 2 | -2.07 | 0.031 | -2.16 |
| FGF18 | Fibroblast growth factor 18 | -1.88 | 0.033 | -1.58 |
| WNT16 | Wingless-type MMTV integration site family, member 16 | -1.67 | 0.022 | -3.45 |
| Genes up-regulated with age (top 15) | ||||
| HLA-DRB5 | Major histocompatibility complex, class II, DR beta 5 | 15.73 | 0.011 | 3.01 |
| HLA-DRB1 | Major histocompatibility complex, class II, DR beta 1 | 15.01 | 0.002 | 3.70 |
| HBA2 | Hemoglobin, alpha 2 | 5.92 | 0.040 | 2.50 |
| ACTG2 | Actin, gamma 2, smooth muscle, enteric | 5.18 | 0.016 | 1.24 |
| HBB | Hemoglobin, beta | 5.12 | 0.045 | 2.44 |
| RPL14 | Ribosomal protein L14 | 4.55 | 0.006 | 1.02 |
| FCER1A | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | 4.15 | 0.000 | 3.00 |
| IFI27 | Interferon, alpha-inducible protein 27 | 3.21 | 0.000 | 1.75 |
| CD36 | CD36 molecule (thrombospondin receptor) | 3.21 | 0.003 | 4.33 |
| DHRS9 | Dehydrogenase/reductase (SDR family) member 9 | 3.16 | 0.017 | 2.98 |
| DARC | Duffy blood group, chemokine receptor | 3.07 | 0.022 | 1.11 |
| SCARA5 | Scavenger receptor class A, member 5 (putative) | 2.95 | 0.031 | 1.96 |
| LXN | Latexin | 2.87 | 0.002 | 2.17 |
| IL7R | Interleukin 7 receptor | 2.83 | 0.000 | 1.31 |
| OAS2 | 2′-5′-oligoadenylate synthetase 2, 69/71kDa | 2.80 | 0.000 | 1.99 |
| Additional selected candidate genes | ||||
| WISP2 | WNT1 inducible signaling pathway protein 2 | 2.59 | 0.039 | 1.63 |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 | 2.28 | 0.022 | 2.86 |
| CDC25C | Cell division cycle 25 homolog C | 1.61 | 0.024 | 1.40 |
| GDNF | Glial cell derived neurotrophic factor | 1.57 | 0.011 | 1.27 |
| CCL8 | Chemokine (C-C motif) ligand 8 | 1.56 | 0.031 | 1.24 |
| CCNF | Cyclin F | 1.55 | 0.021 | 1.32 |
| Genes down-regulated with chondrosis* | ||||
| AGC1 | Aggrecan | -2.10 | 0.043 | – |
| KCTD4 | Potassium channel tetramerisation domain containing 4 | -1.95 | 0.035 | – |
| MMP13 | Matrix metallopeptidase 13 (collagenase 3) | -1.91 | 0.038 | – |
| PDE4B | Phosphodiesterase 4B, cAMP-specific | -1.83 | 0.003 | – |
| HIST1H4A | Histone cluster 1, H4a | -1.75 | 0.003 | – |
| PRAMEF10 | PRAME family member 10 | -1.69 | 0.006 | – |
| LOC100133180 | N.A. | -1.67 | 0.009 | – |
| ARHGAP8 | Rho GTPase activating protein 8 | -1.64 | 0.013 | – |
| C21orf100 | N.A. | -1.62 | 0.013 | – |
| DACT2 | Dapper, antagonist of beta-catenin, homolog 2 (Xenopus laevis) | -1.61 | 0.026 | – |
| TM4SF5 | Transmembrane 4 L six family member 5 | -1.61 | 0.032 | – |
| RRAD | Ras-related associated with diabetes | -1.60 | 0.030 | – |
| LAMB3 | Laminin, beta 3 | -1.59 | 0.007 | – |
| OLA1 | Obg-like ATPase | -1.58 | 0.017 | – |
| FLJ42953 | Breakpoint cluster region pseudogene 2 | -1.57 | 0.016 | – |
| SLC22A10 | Solute carrier family 22, member 10 | -1.57 | 0.039 | – |
| FLG | Filaggrin | -1.55 | 0.021 | – |
| LOC652710 | N.A. | -1.54 | 0.002 | – |
| KLF14 | Kruppel-like factor 14 | -1.54 | 0.015 | – |
| BIRC5 | Baculoviral IAP repeat containing 5 | -1.54 | 0.002 | – |
| MEOX1 | Mesenchyme homeobox 1 | -1.53 | 0.022 | – |
| TULP2 | Tubby like protein 2 | -1.53 | 0.006 | – |
| PER2 | Period homolog 2 (Drosophila) | -1.52 | 0.041 | – |
| LOC645960 | ribosomal protein L10 pseudogene | -1.52 | 0.003 | – |
| GPC6 | glypican 6 | -1.51 | 0.005 | – |
| Genes up-regulated with chondrosis* | ||||
| TBX5 | T-box 5 | 1.52 | 0.006 | – |
| OR52I2 | Olfactory receptor, family 52, subfamily I, member 2 | 1.53 | 0.008 | – |
| MAL | Mal, T-cell differentiation protein | 1.53 | 0.031 | – |
| SNORD89 | Small nucleolar RNA, C/D box 89 | 1.54 | 0.004 | – |
| OR7C1 | Olfactory receptor, family 7, subfamily C, member 1 | 1.56 | 0.004 | – |
| ETV3L | Ets variant 3-like | 1.56 | 0.024 | – |
| GOLGA6B | Golgin A6 family, member B | 1.56 | 0.040 | – |
| DDX11 | DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 | 1.58 | 0.024 | – |
| SLC5A12 | Solute carrier family 5 (sodium/glucose cotransporter), member 12 | 1.59 | 0.032 | – |
| LOC440558 | Uncharacterized LOC440558 | 1.60 | 0.033 | – |
| ITGB1BP1 | Integrin beta 1 binding protein 1 | 1.61 | 0.030 | – |
| TCF2 | HNF1 homeobox B | 1.62 | 0.025 | – |
| ERCC-00156 | N.A. | 1.63 | 0.004 | – |
| S1PR5 | Sphingosine-1-phosphate receptor 5 | 1.65 | 0.010 | – |
| LOC100132761 | N.A. | 1.70 | 0.017 | – |
| IFNA4 | Interferon, alpha 4 | 1.70 | 0.001 | – |
| MYL4 | Myosin, light chain 4, alkali; atrial, embryonic | 1.72 | 0.037 | – |
| TUBB1 | Tubulin, beta 1 class VI | 1.79 | 0.045 | – |
| NKD2 | Naked cuticle homolog 2 (Drosophila) | 1.81 | 0.044 | – |
| RGPD5 | RANBP2-like and GRIP domain containing 5 | 1.81 | 0.030 | – |
| IKZF3 | IKAROS family zinc finger 3 (Aiolos) | 2.04 | 0.017 | – |
| FBN2 | Fibrillin 2 | 2.14 | 0.043 | – |
| CA1 | Carbonic anhydrase I | 2.76 | 0.019 | – |
Genes up- or down-regulated with chondrosis were not validated by QGP (QuantiGene Plex) assay
Biological processes and pathway analysis
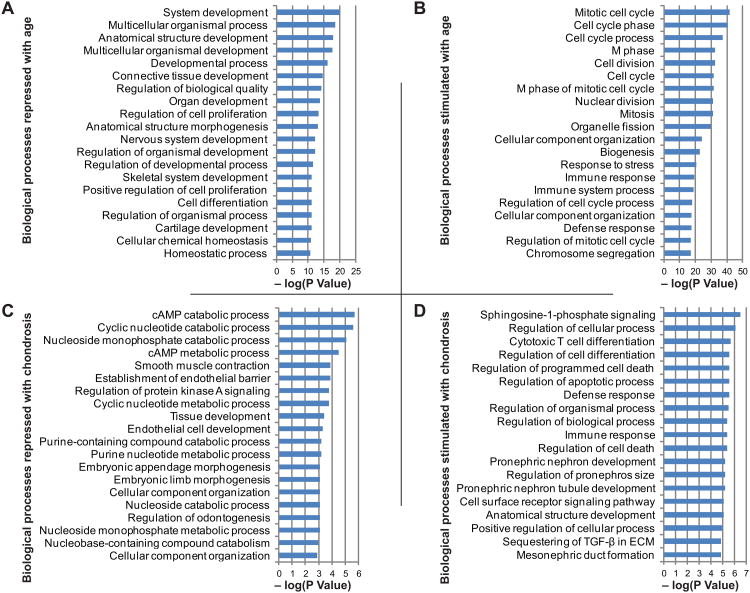
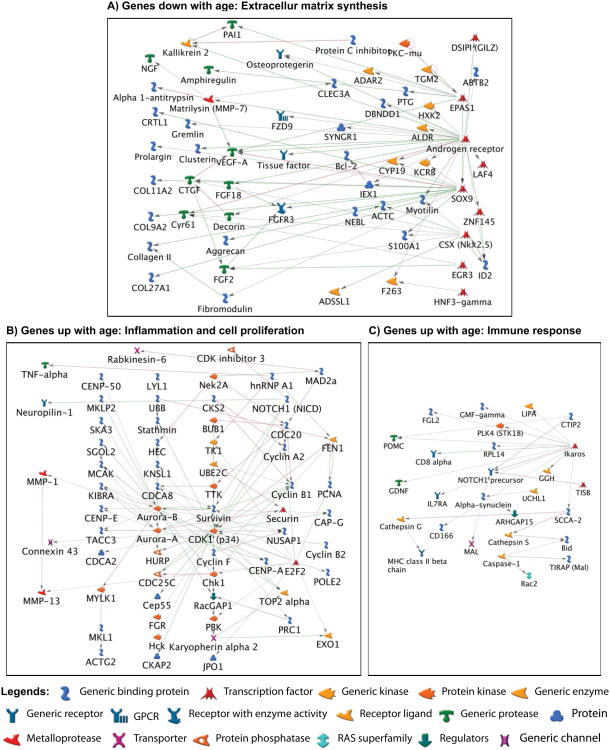
The genes up-regulated (≥1.5-fold) or down-regulated (≥1.5-fold) with age were subjected to GO analysis to determine the biological processes they represent. Fig. 2A-B show the top 20 ontologies related to biological processes based on significant –log(P Value) for genes up- or down-regulated with age. The biological processes that appeared to be repressed with age included skeletal development, cartilage development and cartilage extracellular matrix synthesis (Fig. 2A). Genes representing these processes mainly included COL2A1, CHAD, COL11A2, COL9A2, CLEC3A, SCIN, FGFR3, SHC4, CILP2, SOX8, SBSPON, S100A1, GLDN, MT1E, S100A13, HAPLN1, LEPREL1, SOX9, VEGFA, ACAN, SPRY4, SULF2, FGF18 and WNT16. Conversely, the biological processes that were elevated (Fig. 2B) with age included cell cycle and cell division (PCNA, DLGAP5, CDC25C), and immune response and inflammation (HLA-DRB5, HLA-DRB1, IL-7R, CXCR1, CCL8) pathways. Pathway analysis showed that genes down-regulated with age demonstrated significant interactions with each other, suggesting that these interactions act in concert to down-regulate the extracellular matrix genes (Fig. 3A). In addition, the genes up-regulated with age were also inter-correlated but for the sake of clarity, the processes up-regulated with age have been divided into two components: one component mainly depicts networking and inter-correlation of genes associated with cellular proliferation and inflammation (Fig. 3B) and the other component depicts interaction between genes that lead to the up-regulation of the immune response (Fig. 3C).
Fig. 2. Biological processes associated with age and chondrosis.
Based on statistical significance –log(P Value) as identified by GeneGo MetaCore, the top 20 GO biological processes down-regulated (A) or up-regulated (B) with age, and down-regulated (C) or up-regulated (D) with chondrosis are shown.
Fig. 3. Molecular networks generated from the genes differentially regulated with age.
The molecular networks generated from genes differentially expressed in the injured meniscus with age using GeneGo MetaCore are shown. Pathway analysis showed that genes down-regulated with age represent extracellular matrix development pathways and demonstrate significant interactions with each other (A). In addition, the genes up-regulated with age were also inter-correlated and were associated with either cellular proliferation and inflammation (B) or immune response (C) pathways. Green arrows indicate activation and red arrows indicate inhibition.
These results suggest that the differential regulation of these genes is not a random occurrence, but could have important biological consequences. For instance, genes down-regulated with age ranged from transcription factors (SOX9) in the nucleus with direct links to genes involved in extracellular matrix development (COL2A1, ACAN etc.) that were also down-regulated implicating an interlinked degenerative mechanism and loss of potential repair genes in meniscal cells.
Analysis of differentially expressed genes varying with chondrosis
A total of 1861 genes, out of 47432 transcripts examined, showed significant differential expression (P<0.05) with chondrosis. As stated above, using a minimal fold-change of ≥1.5 as an additional criterion, only 50 genes (49 after removing one non-annotated gene) were identified to have significant differential expression with chondrosis. Among these 49 genes, 26 genes were up-regulated in patients with chondrosis and 23 were up-regulated in patients without chondrosis. All differentially regulated genes along with their fold-changes are listed in Table 2.
Biological processes
GO analysis demonstrated that with chondrosis cellular catabolism including cAMP and cyclic nucleotide catabolic processes were repressed and biological processes representing cell differentiation and cell apoptosis were stimulated. The top 20 biological processes in each category are shown in Fig. 2C-D. While the pathway analysis showed no direct gene interactions suggesting that the genes belonged to disparate pathways that did not overlap, the variance in patient age and degree of chondrosis also implies that overlap would have been unlikely.
Genes commonly regulated with age and chondrosis
As cartilage damage is correlated with age (31, 32), we sought to determine if we could identify genes that changed with both age and chondrosis. As shown in the Venn diagram (Fig. 1C), it was found that there were 38 genes common between age and chondrosis: 15 genes were up-regulated and 23 genes were down-regulated by the combined effect of age and chondrosis (Supplementary Table 4). However, there were no common genes between age and chondrosis at ≥1.5-fold difference (Fig. 1D). These findings suggest that separate sets of genes and mechanisms are involved in age- and chondrosis-related molecular changes in meniscal tissues.
Discussion
In this study, we identified a larger number of differentially expressed genes, from human injured meniscus tissues, that diverged mainly with age and to some extent with the presence of chondrosis of the articular cartilage. This divergence was observed for both individual genes, as well as sets of coordinately expressed genes, which provides a number of possible pathways of interest in meniscus tissues from an aging population. Since aging affects both meniscus tears and OA (9, 33-35), we and others have shown age-related differences in gene expression in OA for diverse orthopedic tissues including cartilage (34, 36), injured meniscus (24, 25), cartilage and meniscus combined (31) and all knee joint tissues (32). These studies together have demonstrated that the degenerative processes in joint tissues are accelerated with age. In addition to gene expression differences, severity of OA (defined by the degree of chondrosis) also increases with age (4, 37). It has been reported that meniscus tears are associated with changes in cartilage (13, 14) which are thought to accelerate OA progression (2, 10, 14, 17, 19-21, 33). Our study shows that the torn meniscus metabolic state is mainly associated with age and less with the degree of chondrosis characterized by arthroscopic examination (i.e. early degenerative changes in cartilage).
To the best of our knowledge, there is no study to-date reporting unbiased microarray analysis for global gene expression patterns in human meniscus tears. There are, however, two studies that have shown gene expression changes in harvested meniscal cells (i.e. not meniscal tissues) through microarray analysis. In one study (38), the authors compared gene expression signatures from normal meniscal cells to OA meniscal cells and found that genes associated with inflammation and cytokine production were up-regulated in OA meniscal cells while genes representing DNA repair processes were down-regulated in OA meniscal cells. In the second study (39), the investigators compared a handful of genes to determine gene expression association with pathological deposition of calcium in the meniscus. Our study clearly demonstrates an elevation of gene expression related to immune response, inflammation, cell cycle and cellular proliferation in the injured menisci from older patients (>40 years). In addition, this study also shows that biological processes such as skeletal development, cartilage development and cartilage extracellular matrix development are repressed in older patients (>40 years). The loss of extracellular matrix genes indicates decreased ability to repair and increased degenerative changes in the injured meniscus. Both of these findings indicate that the torn meniscus is metabolically active in young and old patients but with an age-dependent molecular degenerative state. Therefore, a combination of both the metabolic activity of the injured meniscus and loss of extracellular matrix synthesis could drive meniscus-associated cartilage damage, as the end result of all these molecular changes may lead to early cartilage damage and the initiation of OA (2, 10, 14, 17, 19-21, 33).
Our findings on down-regulation of extracellular matrix genes and up-regulation of inflammation and proliferation genes with age are very relevant to the clinical picture seen in arthritic tissues, where loss of extracellular matrix gene expression is observed on cartilage degeneration (35, 40). These findings suggest that there are decelerated anabolic and accelerated catabolic (molecular) activities taking place in the injured meniscus with age. As with age, there were important biological processes detected with chondrosis, which included cellular catabolism, cell differentiation and apoptosis, although the number of genes was much lower and the pathways affected were more diverse. The variation in gene expression by age and chondrosis taken together indicates a dysregulation of cell cycle and cell metabolism with age.
A recent study (41) has shown a zonally dependent degenerative phenotype (characterized by collagenolysis and aggrecanolysis) of the meniscus after exogenous induction of proinflammatory cytokines. Thus, we also suspect that decreased expression of extracellular matrix genes in the injured meniscus as seen in this study might contribute to meniscal degeneration. Furthermore, the finding that inflammation-related genes were up-regulated with age is consistent with studies that have documented the role of intermittent inflammation in OA (42-46). Our current study also demonstrates the loss of extracellular matrix gene expression and the increased involvement of the immune system and chronic low-grade proinflammatory state in the injured meniscus with age. These two aspects of the aging meniscus tears are very conducive to exacerbating degenerative-type meniscus tears. Past studies have demonstrated a positive link between severity of cartilage degeneration and degree of meniscus degeneration (1, 38, 47, 48) and few studies have reported that meniscus degeneration precedes cartilage degeneration (15, 16). In light of these studies, it is puzzling that our study did not show any overlap between genes differentially regulated when using age as a variable as opposed to genes when using chondrosis (presence or absence) as a variable. One reason for the lack of this overlap could be that the transcriptome analysis is a molecular analysis of the injured meniscus tissues and not of the cartilage on which the observation of chondrosis is based. Furthermore, the torn meniscus tissues were harvested at the time of meniscectomy, which are definitely a pre-radiographic phase of OA and perhaps an early phase of cartilage degeneration. A molecular analysis of the cartilage specimens taken at the time of meniscectomy or a follow-up study on the chondrosis in the same patients may allow for more molecular overlaps between meniscus tears associated with aging and presence or absence of chondrosis. Finally, in our study only three patients (out of 12) who did not have chondrosis were over 40 years, which did not allow for statistically significant associations between aging and chondrosis in relation to meniscus tears.
Considering our findings on age-related gene expression differences in injured meniscus, we propose that the molecular events in the injured meniscus could be critical indicators of future degenerative changes in cartilage. Age-related loss of extracellular matrix genes and gain in inflammation-related genes synergistically make the meniscus susceptible to degenerative tears and the knee cartilage more susceptible to damage from altered joint mechanics due to an impaired ability of chondrocytes to synthesize matrix (49). Furthermore, the degenerative meniscus itself likely results in altered joint mechanics. In our previous studies (24, 25), we have observed that the expression of various cytokines, chemokines and enzyme mediators was suppressed and that of matrix genes was stimulated with age. However, except for MMP1, none of the genes was found to show a statistically significant divergence from our current findings. While it is not possible to explain these differences in MMP1 expression at this point, it is however, believed that its expression is decreased with an advanced stage of OA and higher degree of cartilage damage (50).
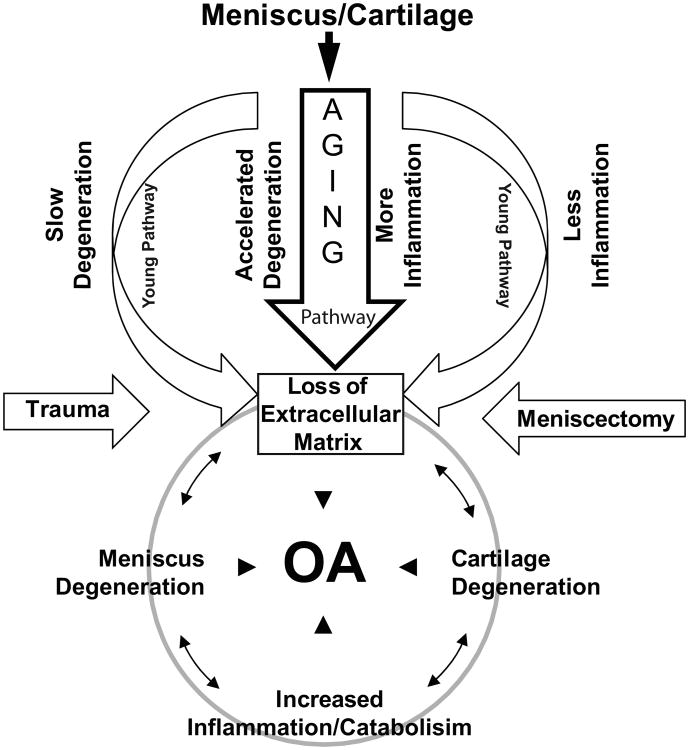
Based on our current findings and those gleaned from Pennock et al. (49), we propose a working model (Fig. 4) that highlights how aging accelerates meniscal and cartilage degeneration and metabolic dysregulation with elevated levels of inflammation-related genes resulting in OA phenotype of the joint. In addition, the decreased expression of extracellular matrix genes with age indicates failure of repair potential. These findings suggest that in older patients there is an increased risk of meniscal degeneration and tearing, which fuels the cartilage matrix degradation, accelerates the dedifferentiated phenotype, and in conjunction with inflammation leads to development of OA. On the other hand in younger individuals matrix synthesis is not compromised, which likely maintains the cartilage phenotype. This model further warrants the investigation of complex molecular events in the meniscus and cartilage as they relate in the overall joint health.
Fig. 4. A working model of molecular events in articular cartilage and meniscus degeneration and knee OA.
This model is modified from Pennock et al. (49) and is based on the age-related gene expression changes and potential links between the meniscus degeneration and articular cartilage damage leading to OA. The transcriptome analysis showed that with age there is a decreased expression of extracellular matrix genes and an increased expression of inflammation-related genes in the injured meniscus. These findings suggest that in older population (compared to young individuals) there is a risk of meniscal degeneration and meniscus tear that may accelerate meniscus and cartilage matrix degradation with progressive inflammation ultimately leading to development of OA. The meniscus tears as well as the subsequent meniscectomy aid to degenerative processes and to OA development.
A limitation associated with our study is that we lack data on gene expression in normal (uninjured) meniscus tissues for comparison with injured meniscus tissues and their potential relationship to age and chondrosis. While it would be informational to have data on gene expression from normal tissues, such information is not essential in our analysis for a couple of reasons: i) current study solely focuses on meniscus tears with no intention to compare with uninjured meniscus but rather to identify age- and chondrosis-related differences in gene expression of injured menisci. We are interested in the individuals with meniscus tears to monitor the impact of age and chondrosis on the gene expression as it is more relevant clinically in terms of the prognosis for these patients, ii) given the heightened risk of developing OA in patients with meniscus tears, our analysis could help to stratify this risk independently of the potential relationship of gene expression in torn meniscus to gene expression in normal meniscus, iii) age and chondrosis may have similar or dissimilar effects on gene expression in the injured meniscus compared with the uninjured and that question is not the focus of our present investigation. Considering that measuring gene expression in normal meniscus is neither paramount nor practical, the relationship, if any, between gene expression of normal meniscus tissue and torn meniscus tissue is interesting but not integral to the current investigation. Furthermore, it would be of great value in the future to have histology samples of the injured meniscus to correlate morphology with gene expression.
Conclusion
In summary, our work identified definitive molecular signatures in damaged meniscus that could be segregated based on age and, to a lesser extent, on the degree of chondrosis in the knee. Although this is a complex interaction, a substantial number of genes identified to be differentially down-regulated are known to have critical roles in cartilage extracellular matrix synthesis and maintenance, and genes that are differentially up-regulated are known for their roles in inflammation and proliferation. These findings suggest an emerging paradigm that the metabolic state of the injured meniscus is dependent on age but not mild chondrosis of the articular cartilage. Taken, together, our transcriptome analysis reveals that a torn meniscus has molecular footprint that resembles OA in individuals >40 years and a healing potential in individuals ≤40 years. These studies may provide a molecular rationale for the cause or frequency of degenerative meniscus tears in the aging population and the initiation and propagation of degenerative changes in the cartilage. Therefore, further investigation of metabolic activity in the injured meniscus and its relationship to cartilage degeneration and the early development of knee OA may identify new therapies to prevent or limit the contribution of meniscal degeneration to the initiation and progression of OA.
Supplementary Material
Supplementary Table 1: Characteristics of selected genes validated by QGP (QuantiGene Plex) assay
Supplementary Table 2: List of genes up-regulated with age with fold difference of ≥1.5-fold and P<0.05.
Supplementary Table 3: List of genes down-regulated with age with fold difference of ≥1.5-fold and P<0.05.
Supplementary Table 4: List of genes common to both age and chondrosis with P<0.05.
Acknowledgments
We thank Washington University Genome Technology Access Center (GTAC) for help with transcriptome and QuantiGene Plex assays.
Financial support: This study was supported by an Orthopaedic Research and Education Foundation (OREF) grant to Dr. Brophy, National Institute of Arthritis and Musculoskeletal and Skin Diseases grants # R01-AR050847 and R01-AR045550 to Dr. Sandell and Musculoskeletal Research Center, grant # P30-AR057235. Dr. Rai is supported by Ruth L. Kirschstein National Research Service Award Fellowship from National Institutes of Health through grant # T32-AR060719. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Arthritis, Musculoskeletal and Skin Diseases or the National Institutes of Health.
Footnotes
Conflict of interest: Dr. Sandell owns stock options in ISTO Technologies and receives royalties from Merck/Millipore for a type IIA collagen N-propeptide enzyme-linked immunosorbent assay. Drs. Rai, Patra and Brophy have no conflict of interest to disclose.
Author contributions: All authors were involved in the drafting and revision of the manuscript and all authors approved the final version to be published. Dr. Brophy had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design: Sandell, Brophy, Rai.
Acquisition of data: Brophy, Rai, Patra.
Analysis and interpretation of data: Rai, Patra, Sandell, Brophy.
Contributor Information
Muhammad Farooq Rai, Email: raim@wudosis.wustl.edu.
Debabrata Patra, Email: patrad@wudosis.wustl.edu.
Linda J. Sandell, Email: sandelll@wustl.edu.
Robert H. Brophy, Email: brophyr@wudosis.wustl.edu.
References
- 1.Hunter DJ, Zhang YQ, Niu JB, Tu X, Amin S, Clancy M, et al. The association of meniscal pathologic changes with cartilage loss in symptomatic knee osteoarthritis. Arthritis Rheum. 2006;54(3):795–801. doi: 10.1002/art.21724. [DOI] [PubMed] [Google Scholar]
- 2.Lohmander LS, Englund PM, Dahl LL, Roos EM. The long-term consequence of anterior cruciate ligament and meniscus injuries: osteoarthritis. Am J Sports Med. 2007;35(10):1756–69. doi: 10.1177/0363546507307396. [DOI] [PubMed] [Google Scholar]
- 3.Morgan CD, Wojtys EM, Casscells CD, Casscells SW. Arthroscopic meniscal repair evaluated by second-look arthroscopy. Am J Sports Med. 1991;19(6):632–7. 7–8. doi: 10.1177/036354659101900614. [DOI] [PubMed] [Google Scholar]
- 4.Christoforakis J, Pradhan R, Sanchez-Ballester J, Hunt N, Strachan RK. Is there an association between articular cartilage changes and degenerative meniscus tears? Arthroscopy. 2005;21(11):1366–9. doi: 10.1016/j.arthro.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 5.Tandogan RN, Taser O, Kayaalp A, Taskiran E, Pinar H, Alparslan B, et al. Analysis of meniscal and chondral lesions accompanying anterior cruciate ligament tears: relationship with age, time from injury, and level of sport. Knee Surg Sports Traumatol Arthrosc. 2004;12(4):262–70. doi: 10.1007/s00167-003-0398-z. [DOI] [PubMed] [Google Scholar]
- 6.Englund M, Guermazi A, Lohmander SL. The role of the meniscus in knee osteoarthritis: a cause or consequence? Radiologic clinics of North America. 2009;47(4):703–12. doi: 10.1016/j.rcl.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Englund M, Roos EM, Roos HP, Lohmander LS. Patient-relevant outcomes fourteen years after meniscectomy: influence of type of meniscal tear and size of resection. Rheumatology (Oxford) 2001;40(6):631–9. doi: 10.1093/rheumatology/40.6.631. [DOI] [PubMed] [Google Scholar]
- 8.Roos H, Adalberth T, Dahlberg L, Lohmander LS. Osteoarthritis of the knee after injury to the anterior cruciate ligament or meniscus: the influence of time and age. Osteoarthritis Cartilage. 1995;3(4):261–7. doi: 10.1016/s1063-4584(05)80017-2. [DOI] [PubMed] [Google Scholar]
- 9.Pauli C, Grogan SP, Patil S, Otsuki S, Hasegawa A, Koziol J, et al. Macroscopic and histopathologic analysis of human knee menisci in aging and osteoarthritis. Osteoarthritis Cartilage. 2011;19(9):1132–41. doi: 10.1016/j.joca.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodkey WG, Steadman JR, Li ST. A clinical study of collagen meniscus implants to restore the injured meniscus. Clin Orthop Relat Res. 1999;(367 Suppl):S281–92. doi: 10.1097/00003086-199910001-00027. [DOI] [PubMed] [Google Scholar]
- 11.Baratz ME, Fu FH, Mengato R. Meniscal tears: the effect of meniscectomy and of repair on intraarticular contact areas and stress in the human knee. A preliminary report. Am J Sports Med. 1986;14(4):270–5. doi: 10.1177/036354658601400405. [DOI] [PubMed] [Google Scholar]
- 12.Rath E, Richmond JC. The menisci: basic science and advances in treatment. British journal of sports medicine. 2000;34(4):252–7. doi: 10.1136/bjsm.34.4.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper C, McAlindon T, Snow S, Vines K, Young P, Kirwan J, et al. Mechanical and constitutional risk factors for symptomatic knee osteoarthritis: differences between medial tibiofemoral and patellofemoral disease. J Rheumatol. 1994;21(2):307–13. [PubMed] [Google Scholar]
- 14.Bennett LD, Buckland-Wright JC. Meniscal and articular cartilage changes in knee osteoarthritis: a cross-sectional double-contrast macroradiographic study. Rheumatology (Oxford) 2002;41(8):917–23. doi: 10.1093/rheumatology/41.8.917. [DOI] [PubMed] [Google Scholar]
- 15.Englund M, Roos EM, Lohmander LS. Impact of type of meniscal tear on radiographic and symptomatic knee osteoarthritis: a sixteen-year followup of meniscectomy with matched controls. Arthritis Rheum. 2003;48(8):2178–87. doi: 10.1002/art.11088. [DOI] [PubMed] [Google Scholar]
- 16.Berthiaume MJ, Raynauld JP, Martel-Pelletier J, Labonte F, Beaudoin G, Bloch DA, et al. Meniscal tear and extrusion are strongly associated with progression of symptomatic knee osteoarthritis as assessed by quantitative magnetic resonance imaging. Ann Rheum Dis. 2005;64(4):556–63. doi: 10.1136/ard.2004.023796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDermott ID, Amis AA. The consequences of meniscectomy. J Bone Joint Surg Br. 2006;88(12):1549–56. doi: 10.1302/0301-620X.88B12.18140. [DOI] [PubMed] [Google Scholar]
- 18.Fairbank TJ. Knee joint changes after meniscectomy. J Bone Joint Surg Br. 1948;30B(4):664–70. [PubMed] [Google Scholar]
- 19.Englund M, Lohmander LS. Patellofemoral osteoarthritis coexistent with tibiofemoral osteoarthritis in a meniscectomy population. Ann Rheum Dis. 2005;64(12):1721–6. doi: 10.1136/ard.2005.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz JN, Martin SD. Meniscus--friend or foe: epidemiologic observations and surgical implications. Arthritis Rheum. 2009;60(3):633–5. doi: 10.1002/art.24363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roos H, Lauren M, Adalberth T, Roos EM, Jonsson K, Lohmander LS. Knee osteoarthritis after meniscectomy: prevalence of radiographic changes after twenty-one years, compared with matched controls. Arthritis Rheum. 1998;41(4):687–93. doi: 10.1002/1529-0131(199804)41:4<687::AID-ART16>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Shin SJ, Fermor B, Weinberg JB, Pisetsky DS, Guilak F. Regulation of matrix turnover in meniscal explants: role of mechanical stress, interleukin-1, and nitric oxide. J Appl Physiol. 2003;95(1):308–13. doi: 10.1152/japplphysiol.00131.2003. [DOI] [PubMed] [Google Scholar]
- 23.Kisiday JD, Vanderploeg EJ, McIlwraith CW, Grodzinsky AJ, Frisbie DD. Mechanical injury of explants from the articulating surface of the inner meniscus. Arch Biochem Biophys. 2010;494(2):138–44. doi: 10.1016/j.abb.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 24.Brophy RH, Rai MF, Zhang Z, Torgomyan A, Sandell LJ. Molecular analysis of age and sex-related gene expression in meniscal tears with and without a concomitant anterior cruciate ligament tear. J Bone Joint Surg Am. 2012;94(5):385–93. doi: 10.2106/JBJS.K.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rai MF, Sandell LJ, Cheverud JM, Brophy RH. Relationship of age and body mass index to the expression of obesity and osteoarthritis-related genes in human meniscus. Int J Obes (Lond) 2013 doi: 10.1038/ijo.2012.221. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Outerbridge RE. The etiology of chondromalacia patellae. J Bone Joint Surg Br. 1961;43-B:752–7. doi: 10.1302/0301-620X.43B4.752. [DOI] [PubMed] [Google Scholar]
- 27.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24(9):1115–22. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 28.Yang W, Maqsodi B, Ma Y, Bui S, Crawford KL, McMaster GK, et al. Direct quantification of gene expression in homogenates of formalin-fixed, paraffin-embedded tissues. BioTechniques. 2006;40(4):481–6. doi: 10.2144/000112133. [DOI] [PubMed] [Google Scholar]
- 29.Urdea MS, Horn T, Fultz TJ, Anderson M, Running JA, Hamren S, et al. Branched DNA amplification multimers for the sensitive, direct detection of human hepatitis viruses. Nucleic acids symposium series. 1991;(24):197–200. [PubMed] [Google Scholar]
- 30.Aleksunes LM, Yeager RL, Wen X, Cui JY, Klaassen CD. Repression of hepatobiliary transporters and differential regulation of classic and alternative bile Acid pathways in mice during pregnancy. Toxicological sciences : an official journal of the Society of Toxicology. 2012;130(2):257–68. doi: 10.1093/toxsci/kfs248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAlinden A, Dudhia J, Bolton MC, Lorenzo P, Heinegard D, Bayliss MT. Age-related changes in the synthesis and mRNA expression of decorin and aggrecan in human meniscus and articular cartilage. Osteoarthritis Cartilage. 2001;9(1):33–41. doi: 10.1053/joca.2000.0347. [DOI] [PubMed] [Google Scholar]
- 32.Loeser RF, Olex AL, McNulty MA, Carlson CS, Callahan MF, Ferguson CM, et al. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis Rheum. 2012;64(3):705–17. doi: 10.1002/art.33388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan WP, Lang P, Stevens MP, Sack K, Majumdar S, Stoller DW, et al. Osteoarthritis of the knee: comparison of radiography, CT, and MR imaging to assess extent and severity. AJR Am J Roentgenol. 1991;157(4):799–806. doi: 10.2214/ajr.157.4.1892040. [DOI] [PubMed] [Google Scholar]
- 34.Martin JA, Buckwalter JA. Roles of articular cartilage aging and chondrocyte senescence in the pathogenesis of osteoarthritis. Iowa Orthop J. 2001;21:1–7. [PMC free article] [PubMed] [Google Scholar]
- 35.Squires GR, Okouneff S, Ionescu M, Poole AR. The pathobiology of focal lesion development in aging human articular cartilage and molecular matrix changes characteristic of osteoarthritis. Arthritis Rheum. 2003;48(5):1261–70. doi: 10.1002/art.10976. [DOI] [PubMed] [Google Scholar]
- 36.Todd Allen R, Robertson CM, Harwood FL, Sasho T, Williams SK, Pomerleau AC, et al. Characterization of mature vs aged rabbit articular cartilage: analysis of cell density, apoptosis-related gene expression and mechanisms controlling chondrocyte apoptosis. Osteoarthritis Cartilage. 2004;12(11):917–23. doi: 10.1016/j.joca.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Noble J, Hamblen DL. The pathology of the degenerate meniscus lesion. J Bone Joint Surg Br. 1975;57(2):180–6. [PubMed] [Google Scholar]
- 38.Sun Y, Mauerhan DR, Honeycutt PR, Kneisl JS, Norton JH, Hanley EN, Jr, et al. Analysis of meniscal degeneration and meniscal gene expression. BMC Musculoskelet Disord. 2010;11:19. doi: 10.1186/1471-2474-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Mauerhan DR, Honeycutt PR, Kneisl JS, Norton HJ, Zinchenko N, et al. Calcium deposition in osteoarthritic meniscus and meniscal cell culture. Arthritis Res Ther. 2010;12(2):R56. doi: 10.1186/ar2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldring SR, Goldring MB. The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin Orthop Relat Res. 2004;(427 Suppl):S27–36. doi: 10.1097/01.blo.0000144854.66565.8f. [DOI] [PubMed] [Google Scholar]
- 41.Fuller ES, Smith MM, Little CB, Melrose J. Zonal differences in meniscus matrix turnover and cytokine response. Osteoarthritis Cartilage. 2012;20(1):49–59. doi: 10.1016/j.joca.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Rai MF, Sandell LJ. Inflammatory mediators: tracing links between obesity and osteoarthritis. Critical reviews in eukaryotic gene expression. 2011;21(2):131–42. doi: 10.1615/critreveukargeneexpr.v21.i2.30. [DOI] [PubMed] [Google Scholar]
- 43.Pelletier JP, Martel-Pelletier J, Abramson SB. Osteoarthritis, an inflammatory disease: potential implication for the selection of new therapeutic targets. Arthritis Rheum. 2001;44(6):1237–47. doi: 10.1002/1529-0131(200106)44:6<1237::AID-ART214>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 44.Hedbom E, Hauselmann HJ. Molecular aspects of pathogenesis in osteoarthritis: the role of inflammation. Cell Mol Life Sci. 2002;59(1):45–53. doi: 10.1007/s00018-002-8404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelletier JP, DiBattista JA, Roughley P, McCollum R, Martel-Pelletier J. Cytokines and inflammation in cartilage degradation. Rheum Dis Clin North Am. 1993;19(3):545–68. [PubMed] [Google Scholar]
- 46.Abramson SB, Attur M. Developments in the scientific understanding of osteoarthritis. Arthritis Res Ther. 2009;11(3):227. doi: 10.1186/ar2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crema MD, Guermazi A, Li L, Nogueira-Barbosa MH, Marra MD, Roemer FW, et al. The association of prevalent medial meniscal pathology with cartilage loss in the medial tibiofemoral compartment over a 2-year period. Osteoarthritis Cartilage. 2010;18(3):336–43. doi: 10.1016/j.joca.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 48.Melrose J, Fuller ES, Roughley PJ, Smith MM, Kerr B, Hughes CE, et al. Fragmentation of decorin, biglycan, lumican and keratocan is elevated in degenerate human meniscus, knee and hip articular cartilages compared with age-matched macroscopically normal and control tissues. Arthritis Res Ther. 2008;10(4):R79. doi: 10.1186/ar2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pennock AT, Robertson CM, Emmerson BC, Harwood FL, Amiel D. Role of apoptotic and matrix-degrading genes in articular cartilage and meniscus of mature and aged rabbits during development of osteoarthritis. Arthritis Rheum. 2007;56(5):1529–36. doi: 10.1002/art.22523. [DOI] [PubMed] [Google Scholar]
- 50.Rubenhagen R, Schuttrumpf JP, Sturmer KM, Frosch KH. Interleukin-7 levels in synovial fluid increase with age and MMP-1 levels decrease with progression of osteoarthritis. Acta Orthop. 2012;83(1):59–64. doi: 10.3109/17453674.2011.645195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Characteristics of selected genes validated by QGP (QuantiGene Plex) assay
Supplementary Table 2: List of genes up-regulated with age with fold difference of ≥1.5-fold and P<0.05.
Supplementary Table 3: List of genes down-regulated with age with fold difference of ≥1.5-fold and P<0.05.
Supplementary Table 4: List of genes common to both age and chondrosis with P<0.05.