Abstract
Aims
Epidemiological and animal data suggest that the development of adult chronic conditions is influenced by early-life exposure-induced changes to the epigenome. This study investigates the effects of perinatal lead (Pb) exposure on DNA methylation and bodyweight in weanling mice.
Materials & methods
Viable yellow agouti (Avy) mouse dams were exposed to 0, 2.1, 16 and 32 ppm Pb acetate before conception through weaning. Epigenetic effects were evaluated by scoring coat color of Avy/a offspring and quantitative bisulfite sequencing of two retrotransposon-driven (Avy and CDK5 activator-binding protein intracisternal A particle element) and two imprinted (Igf2 and Igf2r) loci in tail DNA.
Results
Maternal blood Pb levels were below the limit of detection in controls, and 4.1, 25.1 and 32.1 μg/dl for each dose, respectively. Pb exposure was associated with a trend of increased wean bodyweight in males (p = 0.03) and altered coat color in Avy/a offspring. DNA methylation at Avy and the CDK5 activator-binding protein intracisternal A-particle element was significantly different from controls following a cubic trend (p = 0.04; p = 0.01), with male-specific effects at the Avy locus. Imprinted genes did not shift in methylation across exposures.
Conclusion
Dose- and sex-specific responses in bodyweight and DNA methylation indicate that Pb acts on the epigenome in a locus-specific fashion, dependent on the genomic feature hosting the CpG site of interest, and that sex is a factor in epigenetic response.
Keywords: developmental origins of health and disease, DNA methylation, environmental epigenomics, epigenetics, lead, metastable epiallele, plasticity, viable yellow agouti
The ‘early origins’ hypothesis postulates that nutrition and other environmental factors during prenatal and early postnatal development influence developmental plasticity, thereby, altering susceptibility to adult chronic diseases, including obesity [1,2]. However, relatively little research has considered the mechanisms by which perinatal environmental exposures may influence physical growth and development during sensitive periods in childhood. For example, animal and human data have established that exposure to lead (Pb) in utero affects birth size, neurodevelopment and the development of adult chronic conditions including high blood pressure and cardiovascular disease [3–7]. Despite these associations, no studies to date have robustly assessed the role of environmentally induced epigenetic gene alterations as a mechanism linking in utero Pb exposure to later in life disease risk.
Pb is commonly found throughout the environment as it is used in the manufacture of automotive batteries, paints, glazes, ammunition and piping, and in some parts of the world as an additive to gasoline. Thus, air, soil, water, old paint and food are all avenues for Pb uptake via ingestion, inhalation, and dermal absorption [8]. Since 1991, the US CDC have established a blood Pb level (BLL) of 10 μg/dl as the level of concern and recently lowered this to 5 μg/dl, yet research continues to find adverse health effects at even lower doses [9]. Transfer of Pb from mother to offspring is more efficient transplacentally and lactationaly than by oral ingestion in adults [10]. In mice, early-life Pb exposure acts as an obesogen, and even very low in utero doses can result in early puberty [11,12]. Mechanisms of health effects following early-life exposure are not well established, and studies of epigenetic modifications induced by developmental exposure to Pb are limited.
Epigenetic patterns in offspring, including DNA methylation and histone modifications, are known to be influenced by maternal nutrition, behavior, stress and toxins, causing changes that can persist into adulthood long after the acute developmental exposure has ceased [13,14]. Recent findings have confirmed this hypothesis following exposure to several organic toxicants [15], and here we explore the effects of the heavy metal Pb. Previously, our group has shown global and gene-specific changes in weanling DNA methylation induced by fetal exposure to bisphenol A [15]. Similarly, exposure to heavy metals has been shown to correlate with gene specific and global epigenetic effects for several metals including arsenic, nickel, chromium, cadmium and mercury, in combination [16]. To address the long-term stable epigenetic alterations resulting from early Pb exposure, we used the viable yellow agouti (Avy) mouse model as a biosensor to expose mice to multiple physiologically relevant doses of Pb in utero and in early life.
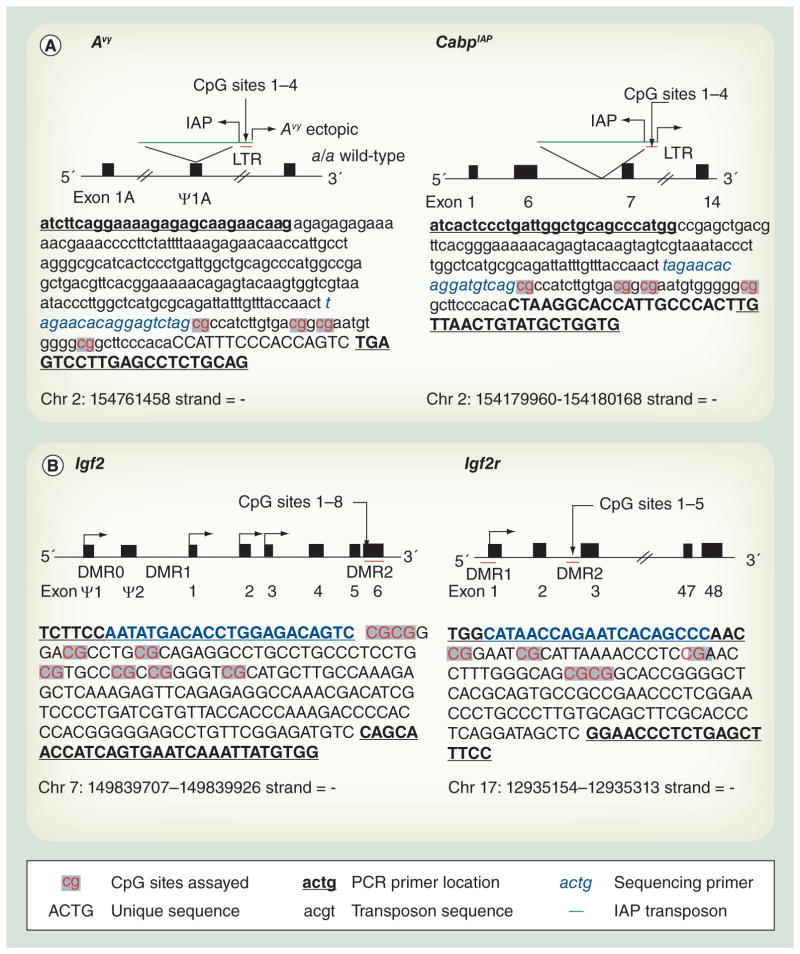
The epigenetics of the Avy mouse model are well established [17]. Interindividual variation in DNA methylation at this locus is visually reflected by the coat color of carrier mice, where increased DNA methylation levels yield darker coat colors in contrast to decreased DNA methylation levels yielding lighter yellow coat colors. The average level of DNA methylation, and consequently coat color, among a litter of mice is somewhat normally distributed; however, the distribution can be shifted towards more or less DNA methylation both nutritionally and toxicologically [15,18,19]. Avy’s DNA methylation status is stochastically established in development, and remains fixed thereafter, so it is considered a metastable epiallele – a locus whose DNA methylation status varies among genetically identical individuals. In addition to Avy the CDK5 activator-binding protein intracisternal A particle element (IAP; CabpIAP) metastable epiallele has been shown to be sensitive to environmental exposures, making them ideal for evaluating the effect of perinatal environmental exposures on the epigenome [20]. Both the Avy and CabpIAP metastable epialleles are the result of an inserted IAP retrotransposon repetitive element, and the variable methylation of CpG sites within their long terminal repeats drives the variable expression of nearby genes (Figure 1A). By contrast, imprinted genes harbor epigenetic marks based upon the sex of the parent transmitting each copy [21]. Because methylation profiles at imprinted genes are allele specific and are often well characterized due to their importance in growth and development, they have frequently served as biomarkers of environmental exposure [22–28]. The Igf2 and its receptor (Igf2r) are imprinted in mice and contain differentially methylated regions (Figure 1B). Both have previously shown varying or stable responses to environmental perturbations and are measured here as potential biomarkers.
Figure 1. Metastable and imprinted genic regions.
(A) Metastable epiallele loci, Avy and CabpIAP, are caused by the insertion of an IAP element (green lines) containing variably methylated CpGs (sites labeled 1–4) in the LTR region (red lines). (B) Imprinted loci, Igf2 and Igf2r, contain multiple differentially methylated regions (red lines) whose methylation status is dependent on the parent of origin. Assayed CpG sites are in red text. Primer locations are underlined; see Table 1 for primer sequences for bisulfite converted DNA. The sequencing primer location is in blue text.
Avy: Viable yellow agouti; CabpIAP: CDK5 activator-binding protein intracisternal A particle element; Chr: Chromosome; DMR: Differentially methylated region; IAP: Intracisternal A particle; LTR: Long terminal repeat.
Here, using a well-established mouse model of perinatal environmental exposures, we evaluate the effects of three physiologically relevant concentrations of Pb, 2.1, 16 and 32 ppm, on bodyweight and epigenetic patterning by observing weaning weight, coat color shift and concurrent change in DNA methylation at the Avy locus. We also evaluate DNA methylation at a second IAP-driven metastable epiallele CabpIAP, and at two imprinted genes, Igf2 and Igf2r. To our knowledge, this is the first study of early-life Pb exposure and its effects on DNA methylation and weaning weight in the Avy mouse model.
Methods
Animals & diet
Avy mice were obtained from a colony that has been maintained with sibling mating and forced heterozygosity of the Avy allele through the male line for over 220 generations, resulting in a genetically invariant background [29]. To minimize the effects of parity and age, virgin a/a (wild-type) dams, of 6–8 weeks of age, were randomly assigned to one of four Pb treatment groups and fed a phytoestrogen-free AIN-93G diet (diet 95092 with 7% corn oil substituted for 7% soybean oil; Harlan Teklad, WI, USA). Pb (II) acetate trihydrate (Sigma-Aldrich, MO, USA) was dissolved in a single batch for each concentration level in distilled water, and the concentration was verified by inductively coupled plasma mass spectrometry (ICPMS; NSF International, MI, USA). Prior to study commencement, the distilled diluent water was confirmed as Pb free by ICPMS, and the same batch dilution of Pb-acetate-treated water was used throughout the study for all mice and administered using standard 8 oz gravity bottles that were changed at least weekly. Thus, exposure groups were as follows: distilled water (n = 11 litters, 78 total surviving offspring, 39 surviving Avy/a offspring); distilled water supplemented with 2.1 ppm Pb acetate (n = 12 litters, 75 total surviving offspring, 41 surviving Avy/a offspring); distilled water supplemented with 16 ppm Pb acetate (n = 12 litters, 86 total surviving offspring, 48 surviving Avy/a offspring); distilled water supplemented with 32 ppm Pb acetate (n = 14 litters, 75 total surviving offspring, 42 surviving Avy/a offspring). Blood was collected in a subset of dams at weaning by cardiac puncture and tested at the Michigan Department of Community Health (MI, USA), via ICPMS, with a limit of detection of 1.3 μg/dl, as summarized in Figure 2.
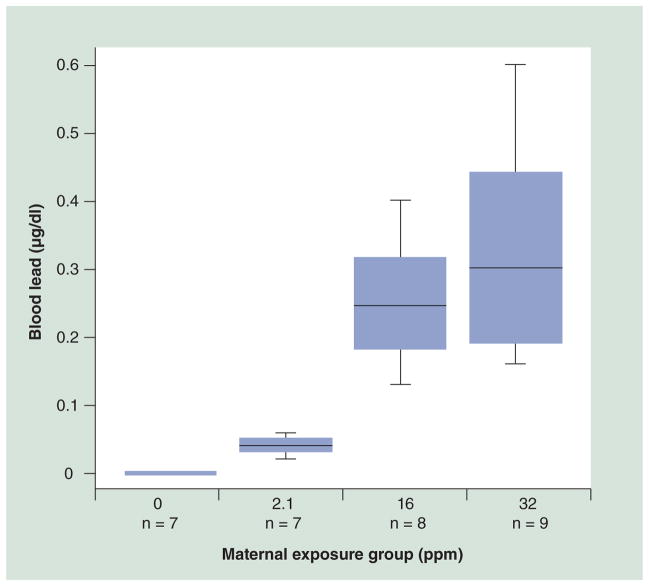
Figure 2. Maternal blood lead levels at 8 weeks of exposure.
At weaning (day 22), blood lead levels were measured for 7–9 days from each exposure group. All controls (0 ppm) measured below the detection limit (1.3 μg/dl), the 2.1 ppm group averaged 4.1 μg/dl (± 1.3), the 16 ppm group averaged 25 μg/dl (± 7.3) and the 32 ppm group averaged 32 μg/dl (± 11.4).
Following 2 weeks on their respective Pb-acetate water, dams were mated with Avy/a males of 7–10 weeks of age. The animals were provided with access to diet and water ad libitum. Pb-acetate water was provided to dams throughout pregnancy and lactation. All offspring were weaned, tail tipped and weighed at postnatal day 22 (d22). Furthermore, Avy/a offspring were scored for phenotype as follows: a single observer visually allocated all Avy/a mice into five categories depending on the proportion of brown fur: yellow (<5% brown fur), slightly mottled (between 5 and 40% brown), mottled (~50% brown), heavily mottled (between 60 and 95% brown) and pseudoagouti (>95% brown).
The mice in this study were monitored by the University of Michigan Unit for Laboratory Animal Medicine (MI, USA) and treated humanely during the study and sacrifice. The guidelines for the care and use of laboratory animals were followed when maintaining the mice. The protocol for the study was reviewed and approved by the University of Michigan Committee on Use and Care of Animals.
DNA isolation & methylation analyses
Using a standard phenol–chloroform–isoamyl alcohol protocol, total genomic DNA was isolated from d22 tail tissue (<3 mm) of all a/a and Avy/a offspring [30]. The Qiagen Epitect kit (Qiagen, Venlo, The Netherlands) automated on the Qiagen QIAcube® purification system, was utilized for bisulfite conversion. Sodium bisulfite was added to approximately 1 μg of genomic DNA, thereby converting unmethylated cytosines to uracil, which are then replaced by thymine during PCR; methylated cytosines remained unchanged [31]. After bisulfite conversion, amplification of candidate gene regions of interest was performed using HotStarTaq master mix (Qiagen), forward primer (50 pmol), and reverse primer (50 pmol) in a 30 μl PCR and subsequently resolved by gel electrophoresis. The reverse primer was biotinylated in all assays. The PCR conditions, forward, reverse and sequencing primers and biotin labeling for all assays are shown in Table 1.
Table 1.
Primers (5′ to 3′) and sequences to analyze DNA methylation quantification via pyrosequencing for candidate genes.
| Primer/sequence to analyze | Avy assay | CabpIAP assay | Igf2 assay | Igf2r assay |
|---|---|---|---|---|
| Strand | Chr2: 154761458 strand = reverse | Chr2: 154179960–154180168 strand = reverse | Chr7: 149839707–149839926 strand = reverse | Chr17: 12935154–12935313 strand = reverse |
| Forward PCR primer | ATTTTTAGGAAAAGAGAGTAAGAAGTAAG | ATTATTTTTTGATTGGTTGTAGTTTATGG | TTTTTTAATATGATATTTGGAGATAGTT | TGGTATAATTAGAATTATAGTTTAAT |
| Reverse PCR primer | Biotin-CTACAAAAACTCAAAAACTCA | Biotin-CACCAACATACAATTAACA | Biotin-CCACATAATTTAATTCACTAATAATTACTA | Biotin-AAAAAACTCAAAAAATTCC |
| Sequencing primer | TAGAATATAGGATGTTAG | TAGAATATAGGATGTTAG | AATATGATATTTGGCGATAGTT | ATAATTAGAATTATAGTTTA |
| Sequence to analyze | YGTTATTTTGTGAYGGYGAATGTGGGGGYGGTT | YGTTATTTTGTGAYGGYGAATGTGGGGGYGGTT | YGYGGGAYGTTTGYGTAGAGGTTTGTTTGTTTTTTTGYGTGTTYGTYGGGGTYGT | ATYGGAATYGTATTAAAATTTTTYGAATTTTTGGGTAGYG |
| Amplicon length (bp) | 294 | 209 | 220 | 127 |
| Temperature (°C) | 53 | 47 | 56 | 52 |
| Cycles (n) | 42 | 40 | 50 | 50 |
Avy: Viable yellow agouti; CabpIAP: CDK5 activator-binding protein intracisternal A particle element; Chr: Chromosome.
Pyrosequencing technology using PyroMark MD (Qiagen) was used to quantify DNA methylation of CpG sites of interest. To determine the percentage of methylation, PyroMark software calculates the fraction of 5-methylated cytosines among the total sum of methylated and unmethylated cytosines. Duplicate runs of all samples were performed, and these duplicates were averaged to determine the mean CpG site methylation to be used in statistical analysis. Figure 1 lists the mm9 chromosomal position, primer location, CpG sites, and sequences to analyze for pyrosequencing runs. The four CpG sites considered at the Avy allele can be found at nucleotide positions 306, 319, 322 and 334 of GenBank accession number AF540972.1 and the insertion exists at mm9 genomic position chromosome (chr)2:154776947 (complementary strand) [29]. The four CpG sites considered at the CabpIAP allele can be found at nucleotide positions 44, 57, 60 and 72 of GenBank accession number BB842254 or mm9 genomic position chr2:154179960–154180168 (complementary strand) [32]. The eight CpG sites considered at the Igf2 allele can be found at nucleotide positions 1227, 1229, 1234, 1240, 1264, 1270, 1273 and 1279 of GenBank accession number AY849922 or mm9 genomic position chr7:149839707–149839926 (complementary strand) [33]. The five CpG sites considered at the Igf2r allele can be found at nucleotide positions 1070, 1076, 1091, 1106 and 1108 of GenBank accession number L06446 or mm9 genomic position chr17: 12935154–12935313 (complementary strand) [34].
Statistical analysis
The influence of perinatal Pb exposure on sex ratio, genotypic ratio and pup survival rate significance were determined by Fisher’s exact test comparing exposure groups with control. Litter size variation across exposure groups was tested via analysis of variance with a Tukey’s honestly significant difference post hoc adjustment to determine intergroup significance. Coat color distribution variation across exposure levels was performed using the χ2 goodness-of-fit test with the control coat color distribution representing the expected distribution. The margin of error for maternal BLL was calculated at the 95% confidence level. Weaning weight and candidate gene methylation analyses were carried out by fitting to linear mixed models in the R statistical package, version 2.13.2 (package lme4) with weaning weight or gene methylation and exposure as fixed effects in each model and a random effect component to account for within-litter effects. P-values were calculated by Markov Chain Monte Carlo resampling (obtained by the pvals.fnc function). Pb exposure was an ordered factor, therefore linear, quadratic and cubic trends were fitted. Statistical significance for all analyses was defined as p < 0.05.
Results
Maternal BLLs for the control group on distilled water were all below the analytical limit of detection (1.3 μg/dl). While levels for the 2.1 ppm group ranged from 2.0 to 5.8 μg/dl (mean: 4.1), levels for the 16 ppm group ranged from 13 to 40 μg/dl (mean: 25.1), and levels for the 32 ppm group ranged from 16 to 60 μg/dl (mean: 32.1) (Figure 2). In comparison with control offspring, perinatal exposure to Pb acetate at 2.1, 16 or 32 ppm in drinking water did not significantly alter litter size, sex ratio or genotype ratio of a/a to Avy/a offspring (Table 2). Sex ratio, as compared with control, reached near significance in the 16 ppm group (p = 0.06), with 64% male offspring. Survival rate to weaning was significantly altered between the control and 2.1 ppm concentration (p = 0.01), and was the result of poor maternal care in several litters in this group, causing a disproportionate loss of pups. The control group had a 96% survival rate, while the 2.1, 16 and 32 ppm groups had rates of 82, 96 and 90%, respectively (Table 2).
Table 2.
Litter parameters: offspring litter size, survival rate, genotype and sex ratio across exposure groups.
| Exposure (ppm) | Litter (n) | Pups (n) | Mean pups (n) | Pup survival rate | a/a offspring (%) | Male offspring (%) |
|---|---|---|---|---|---|---|
| Control | 11 | 81 | 7.4 | 0.96 | 50 | 49 |
| 2.1 | 12 | 91 | 7.6 | 0.82* | 45 | 56 |
| 16 | 12 | 90 | 7.5 | 0.96 | 44 | 64** |
| 32 | 14 | 83 | 5.9 | 0.90 | 44 | 57 |
| Total | – | 345 | – | – | – | – |
| Survived | – | 314 | – | – | – | – |
p < 0.05 compared with control exposure group.
p < 0.10 compared with control exposure group.
a/a: Wild-type.
Bodyweight
Pb exposure did not result in a significant linear trend of altered wean bodyweight by exposure concentration for all animals (p = 0.15; n = 314); however, when females were excluded, we found a male-specific increase in the trend (p = 0.03; n = 177) (Table 3). When weaning weight analysis was restricted to a/a males, the significance was still observed (p = 0.04; n = 80) (Figure 3). In Avy/a males, weaning weight was higher and showed a similar trend with increasing exposure, but did not reach significance across exposure groups. Female mice did not trend towards weight increase overall (p = 0.93) or in any exposure or genotype subgroup (p > 0.79).
Table 3.
Offspring weaning weight from maternal lead exposure.
| Exposure (ppm) | All live pups; g (SD) | All male; g (SD) | All female; g (SD) | a/a pups; g (SD) | a/a males; g (SD) | a/a females; g (SD) | Avy/a pups; g (SD) | Avy/a males; g (SD) | Avy/a females; g (SD) |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 8.68 (1.4) | 8.73 (1.4) | 8.64 (1.4) | 8.57 (1.3) | 8.55 (1.1) | 8.6 (1.5) | 8.8 (1.5) | 8.97 (1.7) | 8.68 (1.3) |
| 2.1 | 8.78 (1.5) | 8.93 (1.6) | 8.55 (1.4) | 8.97 (1.1) | 9.14 (1.1) | 8.74 (1.0) | 8.61 (1.8) | 8.76 (1.9) | 8.37 (1.7) |
| 16 | 9.02 (1.4) | 9.37 (1.3) | 8.41 (1.4) | 8.93 (1.4) | 9.51 (1.2) | 8.35 (1.4) | 9.09 (1.4) | 9.29 (1.3) | 8.5 (1.6) |
| 32 | 9.11 (1.5) | 9.39 (1.3) | 8.72 (1.6) | 8.99 (1.3) | 9.22 (1.2) | 8.63 (1.5) | 9.2 (1.6) | 9.54 (1.5) | 8.78 (1.7) |
| Linear trend p-value | 0.15 | 0.03* | 0.89 | 0.30 | 0.04* | 0.83 | 0.16 | 0.15 | 0.79 |
p < 0.05 compared with control exposure group.
a/a: Wild-type; Avy: Viable yellow agouti; SD: Standard deviation.
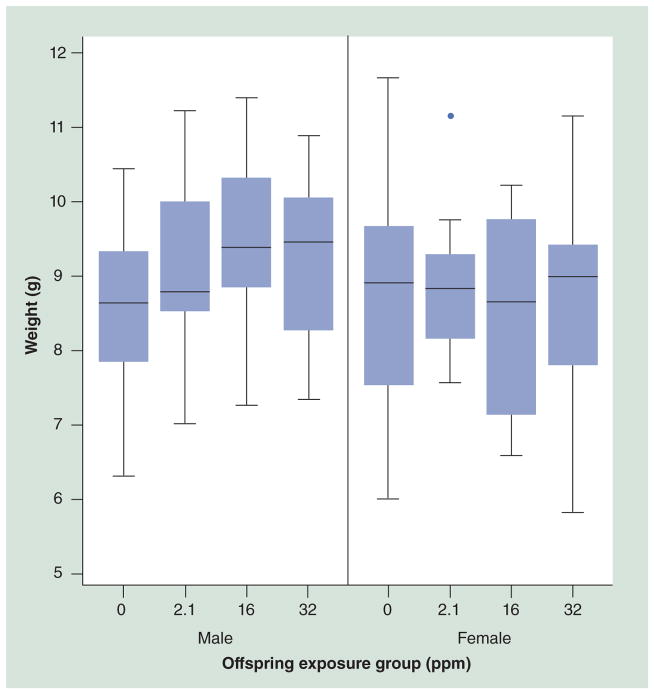
Figure 3. Day 22 weight in a/a (wild-type) animals across exposure groups.
A linear increase in mean weight with increasing lead exposure was found in males (p = 0.04) but not females (p = 0.83) in a/a (wild-type) animals (n = 144). Among males, the mean weight for 0 ppm was 8.55 g (1.10 standard deviation [SD]), the 2.1 ppm group measured 9.14 g (1.15 SD), the 16 ppm group measured 9.51 g (1.22 SD) and the 32 ppm group measured 9.22 g (1.16 SD). Among females, the mean weight for 0 ppm was 8.60 g (1.48 SD), the 2.1 ppm group measured 8.74 g (0.99 SD), the 16 ppm group measured 8.35 g (1.37 SD) and the 32 ppm group measured 8.63 g (1.45 SD). The dot represents an outlier.
Coat color effects
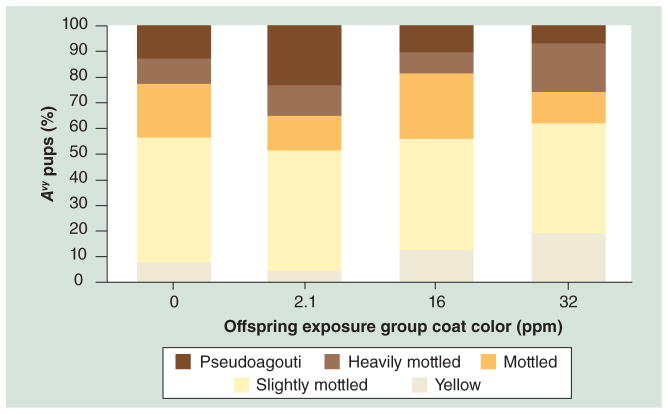
Avy/a mice (n = 170; 54% of the total offspring) were evaluated at weaning for coat color classification, a preliminary visual indication of DNA methylation status at the Avy metastable epiallele. Perinatal exposure to Pb acetate in drinking water shifted the coat color distribution of the Avy/a offspring in a dose-dependent fashion (Figure 4). Maternal exposure to 32 ppm of Pb acetate shifted offspring coat color towards yellow (19% of control offspring characterized as yellow compared with 7% of control offspring; p = 0.01). The 16 ppm group did not significantly differ in color distribution from the control (p = 0.64). However, given the similarity in maternal BLLs among the 16 and 32 ppm groups, shown in Figure 2, we repeated the coat color classification analysis with these groups combined and found a significant shift in coat color from control (p = 0.04; n = 90). Interestingly, the 2.1 ppm exposure level resulted in a small decrease in yellow mice compared with control and a relatively large increase in pseudoagouti mice, 12% in control and 23% in the 2.1 ppm exposure group, suggesting a non-monotonic effect at this locus between low-level and the higher levels of exposure.
Figure 4. Coat color distribution by exposure.
The percentage of animals with each coat color across five categories for all Avy/a animals (n = 170) separated by lead exposure group is shown. A significant increase in yellow offspring between control and 32 ppm exposure (p = 0.01) reflects a decrease in methylation of the Avy allele. At the 2.1 ppm concentration, the percentage of pseudoagouti mice increased from 12 to 23%, indicating an increased frequency of highly methylated Avy offspring in this group and suggesting a non-monotonic effect at this locus with low lead exposure.
Avy: Viable yellow agouti.
IAP-associated DNA methylation
As expected in this mouse strain, DNA methylation levels at the locus underlying the coat color, Avy, corresponded with coat color with yellow mice displaying low levels of methylation compared with higher levels in mottled and pseudoagouti animals (data not shown). The levels of methylation at Avy in Pb-exposed offspring were significantly different from control offspring following a cubic trend (p = 0.04; n = 145). The 2.1 ppm exposure group displayed a 10.9% increase in Avy methylation, while minimal changes or a slight decrease in average DNA methylation were noted in the two higher exposure groups, compared with controls (Figure 5). Sex-specific analysis indicated that this trend was driven by male offspring (p = 0.02; n = 83) (Table 4). Similarly, DNA methylation patterns at a second murine metastable epiallele, CabpIAP, were significantly different in all exposed animals as compared with control animals following a cubic trend (p = 0.01; n = 285). At this locus, both males and females displayed significant differences (p = 0.05 and 0.05; n = 166 and 119, respectively) (Table 4). When divided by genotype subgroups, the a/a offspring had a significant cubic trend (p = 0.01; n = 132), driven by the females (p = 0.01; n = 58), which is opposite to the results seen at the Avy locus. Neither male nor female Avy/a animals showed a significant trend in CabpIAP methylation differences compared with the exposure group.
Figure 5. DNA methylation levels at metastable and imprinted loci.
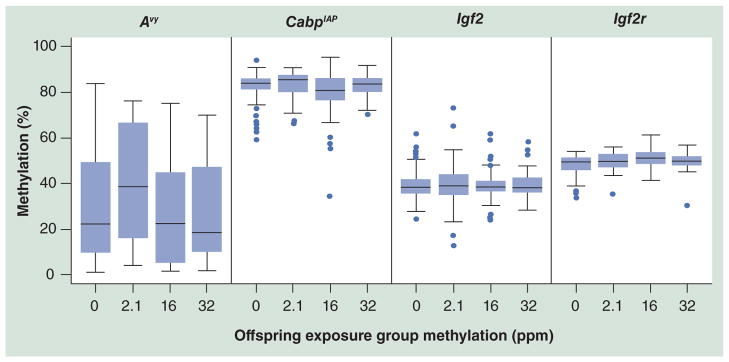
Day 22 tail tissue methylation assayed via pyrosequencing reveals a cubic trend of methylation in Avy (p = 0.04; all Avy animals) and CabpIAP (p = 0.01; all Avy/a and a/a [wild-type] animals) loci, resulting in an increased methylation level at the 2.1 ppm level and decreased methylation at higher levels. The imprinted loci, Igf2 and Igf2r, do not show a significant shift in methylation levels across dosage in either linear or cubic trends for any subgroup of animals, suggesting that methylation response is dependent on the genetic features at each locus. Avy naturally exhibits a wide range of variation from near 0 to 85% in this population, and the range is slightly reduced as dosage increases. By contrast, CabpIAP has a smaller range and higher median methylation. The dots represent outliers.
Avy: Viable yellow agouti; CabpIAP: CDK5 activator-binding protein intracisternal A particle element.
Table 4.
Percentage methylation summary for Avy and CabpIAP loci.
| Exposure (ppm) | All live pups; % (SD) | All male; % (SD) | All female; % (SD) | a/a pups; % (SD) | a/a males; % (SD) | a/a females; % (SD) | Avy/a pups; % (SD) | Avy/a males; % (SD) | Avy/a females; % (SD) |
|---|---|---|---|---|---|---|---|---|---|
| CabpIAP (Avy/a and a/a offspring) mean methylation across four CpG sites | |||||||||
| 0 | 82.2 (6.7) | 82.4 (7.1) | 82 (6.3) | 82.7 (5.4) | 84.2 (4.8) | 80.6 (5.7) | 81.7 (7.7) | 80 (8.9) | 82.9 (6.7) |
| 2.1 | 83.5 (5.7) | 82.8 (6.5) | 84.3 (4.3) | 83.9 (6.7) | 83.3 (8.1) | 84.7 (4.6) | 83 (4.5) | 82.3 (4.7) | 83.9 (4.1) |
| 16 | 80.1 (9.4) | 79.7 (10.3) | 80.9 (7.6) | 79.2 (7.6) | 79.6 (7.7) | 78.8 (7.6) | 80.7 (10.7) | 79.7 (11.6) | 83.7 (6.9) |
| 32 | 82.7 (4.6) | 83.2 (4.8) | 82 (4.4) | 81.2 (4.2) | 81.4 (4.5) | 80.9 (3.9) | 83.9 (4.7) | 84.5 (4.6) | 83 (4.8) |
| Cubic trend p-value | 0.01* | 0.05* | 0.05* | 0.01* | 0.26 | 0.01* | 0.11 | 0.12 | 0.96 |
| Avy (Avy/a offspring) mean methylation across four CpG sites | |||||||||
| 0 | – | – | – | – | – | – | 29.5 (24.8) | 24.1 (26.1) | 33 (23.9) |
| 2.1 | – | – | – | – | – | – | 40.4 (24.8) | 41.3 (21.2) | 39.2 (29.4) |
| 16 | – | – | – | – | – | – | 27.1 (23.1) | 24.8 (23.1) | 34 (23.1) |
| 32 | – | – | – | – | – | – | 26.9 (21.8) | 29.4 (24.1) | 23.4 (18.5) |
| Cubic trend p-value | – | – | – | – | – | – | 0.04* | 0.02* | 0.85 |
p < 0.05 for the cubic trend.
a/a: Wild-type; Avy: Viable yellow agouti; CabpIAP: CDK5 activator-binding protein intracisternal A particle element; SD: Standard deviation.
Imprinted gene DNA methylation
Two murine imprinted genes were assessed for methylation, Igf2 and Igf2r. Unlike the IAP-associated loci, neither imprinted gene revealed consistent trends in methylation shifts across exposures or genotype groups. For Igf2, only a/a males reached near significance (p = 0.07; n = 173) in cubic trend. No subgroup of female or Avy/a pups was significantly altered in methylation across exposures. In Igf2r, neither the group of mice as a whole, nor any subgroup of animals responded with a statistically significant cubic trend (Table 5) or linearly (data not shown).
Table 5.
Percentage methylation summary for Igf2 and Igf2r imprinted loci.
| Exposure (ppm) | All live pups; % (SD) | All male; % (SD) | All female; % (SD) | a/a pups; % (SD) | a/a males; % (SD) | a/a females; % (SD) | Avy/a pups; % (SD) | Avy/a males; % (SD) | Avy/a females; % (SD) |
|---|---|---|---|---|---|---|---|---|---|
| Igf2 (Avy/a and a/a offspring) mean methylation across eight CpG sites | |||||||||
| 0 | 39.3 (6.9) | 38.9 (7.4) | 39.7 (6.3) | 40.6 (7.1) | 40.9 (7.5) | 40.2 (6.5) | 37.9 (6.5) | 36 (6.4) | 39.3 (6.3) |
| 2.1 | 39.8 (9.8) | 40.4 (10.0) | 39.1 (9.6) | 41.1 (9.0) | 42.9 (8.1) | 38.9 (9.8) | 38.6 (10.5) | 38.1 (11.2) | 39.2 (9.8) |
| 16 | 39.2 (5.9) | 39.1 (6.5) | 39.3 (4.8) | 39.6 (4.7) | 38.4 (4.5) | 40.9 (4.6) | 38.9 (6.8) | 39.6 (7.5) | 36.8 (4.0) |
| 32 | 39.2 (5.1) | 39.6 (5.7) | 38.6 (4.2) | 39.9 (4.8) | 40.3 (5.5) | 39.4 (3.7) | 38.6 (5.4) | 39.1 (6.0) | 38.1 (4.5) |
| Cubic trend p-value | 0.51 | 0.37 | 0.76 | 0.50 | 0.07* | 0.38 | 0.97 | 0.85 | 0.45 |
| Igf2r (Avy/a and a/a offspring) mean methylation across five CpG sites | |||||||||
| 0 | 48.0 (5.1) | 48.2 (5.9) | 47.9 (4.0) | 49.1 (4.2) | 49.2 (5.0) | 49 (2.8) | 46.8 (5.9) | 46.8 (7.1) | 46.7 (4.9) |
| 2.1 | 49.4 (4.5) | 49.1 (5.3) | 49.6 (3.8) | 49.9 (3.7) | 52.6 (NA*) | 49.4 (3.9) | 49.2 (4.8) | 48.8 (5.4) | 49.8 (4.0) |
| 16 | 51.0 (4.3) | 51.7 (4.5) | 49.2 (3.3) | 50.9 (4.8) | 52.1 (5.1) | 48.9 (4.2) | 51.1 (4.1) | 51.6 (4.4) | 49.5 (2.6) |
| 32 | 49.6 (4.4) | 48.9 (2.5) | 50.3 (5.6) | 50.2 (3.4) | 48.2 (2.4) | 52.2 (3.1) | 49.2 (5.0) | 49.3 (2.6) | 49.1 (6.6) |
| Cubic trend p-value | 0.62 | 0.19 | 0.50 | 0.80 | 0.96 | 0.52 | 0.59 | 0.37 | 0.56 |
p < 0.10 for the cubic trend.
a/a: Wild-type; Avy: Viable yellow agouti; CabpIAP: CDK5 activator-binding protein intracisternal A particle element; NA: Not applicable; SD: Standard deviation.
Discussion
In the current study, we report a dose- and sex-dependent response in bodyweight and tail DNA methylation levels in weanling mice following perinatal exposure to three levels of Pb acetate in drinking water. First, we observed a significant linear trend increase in bodyweight at weaning with sex-specific effects. When segregated by sex, males exposed to 2.1, 16 and 32 ppm of Pb acetate exhibited increased weight compared with control offspring; however, effects on wean bodyweight were not observed for females. This association was stronger when analysis was restricted to a/a males and is not evident in a/a females. The Avy/a males exhibited higher weaning weight trending with exposure, but did not reach significance across exposure groups, likely due to the confounding effect of the varying levels of methylation at the Avy locus, which is known to affect bodyweight [35]. Weight gain in mice exposed to moderate levels of Pb during gestation has also been associated with male-specific increases in bodyweight at 1 year of age [11]. No other obvious health effects were evident in mice at this age, however, persistent effects throughout the life course should be determined. Rodent studies suggest Pb’s effect on bodyweight may have a dual site of action: at the level of the hypothalamic pituitary unit, and directly at the level of gonadal steroid biosynthesis [36]. Among animals, Pb is believed to act on the hypothalamic–pituitary–gonadal axis by blocking the release of GnRH, thus decreasing puberty-related hormones such as LH, IGF-1 and estradiol [37–41]. At the gonadal steroid biosynthesis level, Pb has been shown to impair Leydig cell and Sertoli cell functions [42, 43]. Our measurement here was limited to overall weanling bodyweight rather than in changes in fat and lean body mass. Such measurements, particularly taken over the life course after early-life Pb exposure, will be important in defining human health relevance and toxicant influences on obesity.
Second, we observed that perinatal exposure to Pb acetate in drinking water shifted the coat color distribution of the Avy/a offspring, a preliminary visual indication of altered DNA methylation status, in a dose-dependent fashion. When candidate IAP-associated and imprinted genes were assayed by quantitative bisulfite sequencing, IAP loci were associated with altered DNA methylation with exposure, following a cubic trend, while candidate imprinted genes were not associated with altered DNA methylation. The two IAP-associated metastable epialleles studied here, Avy and CabpIAP, both vary in methylation at the site of an inserted transposable IAP long terminal repeat element. While IAP elements do not exist in human genome and are not directly relevant to human health, their frequency of occurrence and activity in the mouse genome serves as a good proxy for changes in similar elements in humans. Although these insertions share 98.5% sequence identity [44], their shifts in methylation in response to Pb are subtly different. CabpIAP is more highly methylated on average, and has a smaller methylation range. While both CabpIAP and Avy show a significant cubic trend with increased methylation levels in the 2.1 ppm group, the results for Avy are sex-specific, driven by male offspring, and the results for CabpIAP are significant among both male and female offspring. Sex-specific epigenetic effects have been observed following diverse environmental exposures, including in Avy mice from radiation [45] and in mice exposed to altered nutrition [28,46], bisphenol A exposure [47], and with stress in rats [48]. Furthermore, in rats, multidose Pb exposure at two time points prior to weaning showed significant changes in Dnmt1, Dnmt3a and MeCP2 expression, with differences seen even at the lowest concentration correlated to both sex and developmental window of exposure [49]. Interestingly, the hormetic response reported in Bernal et al. to low-dose radiation is similar to our low-level Pb exposure (2.1 ppm) increase in both coat color and directly tested DNA methylation levels at the IAP-associated loci [45].
Imprinted genes have been targeted as potentially more environmentally labile than other genomic regions [50,51]. Here, we studied two murine imprinted genes, Igf2 and Igf2r, which have been previously shown to exhibit altered methylation upon various environmental exposures. Both are associated with differences in growth rate and present good targets for any correlation between weight and DNA methylation shifts [27,33,52]. In contrast, we found both genes to be stable in methylation after exposure to Pb in utero and during lactation. In our study, we assayed mouse tail tissue at d22, collected at weaning, whereas previous studies finding alterations have examined other tissues during different life stages, such as whole embryo, pancreas, fetal germ cells, muscle and liver. Similar to our results, several studies have also found stable methylation at these loci in tissues such as fetal gut, embryo and sperm. We consider the tail as a useful proxy tissue for imprinting methylation analyses since methylation changes early in development can be expected to propagate throughout the body. A literature survey of mouse exposure studies where our candidate genes were also subject to methylation analysis is summarized in Table 6. In light of these other studies, our data reinforce the notion that loci can react differently to various environmental exposures, suggesting tailored epigenetic reactions rather than a universal response. Pb has also been shown to act as both a gene-specific as well as global hypomethylation agent in rat pheochromocytoma cells, increasing the expression of amyloid precursor protein, suggesting a link between DNA methylation changes and disease etiology [53]. Together, these data suggest that Pb acts in a locus-specific fashion, potentially dependent on the genomic feature hosting the CpG site of interest (i.e., transposon, promoter or differentially methylated region), and that sex is a factor in the epigenetic response to Pb.
Table 6.
Literature review of imprinted and metastable gene methylation shifts in mice.
| Study (year) | Gene | Exposure | Methylation status | Tissue | Disease/condition | Ref. |
|---|---|---|---|---|---|---|
| McKay et al. (2011) | Igf2 | Low maternal folate | Stable | Fetal gut | – | [23] |
| Downing et al. (2011) | In utero alcohol | Stable | Embryonic | Teratogenesis | [22] | |
| Ding et al. (2012) | – | Hypermethylation | Pancreatic | Gestational diabetes mellitus | [24] | |
| Susiarjo et al. (2013) | In utero BPA | Hypermethylation | Embryonic | Abnormal placentation | [27] | |
|
| ||||||
| Zhang et al. (2012) | Igf2r | In utero BPA | Hypermethylation | Fetal germ cells | – | [25] |
| Somm et al. (2013) | In utero TCDD | Stable Hypermethylation |
Sperm Muscle and liver |
Decreased sperm count | [26] | |
| Gallou-Kabani et al. (2010) | High-fat maternal diet | Sexual dimorphism | Placental | Altered nutrient transfer from the placenta to the fetuses | [28] | |
|
| ||||||
| Dolinoy et al. (2006) | CabpIAP | In utero moderate-dose BPA | Hypomethylation | Liver | – | [19] |
| Anderson et al. (2012) | In utero low-dose BPA | Hypermethylation | Tail | Decreased weaning weight | [15] | |
|
| ||||||
| Dolinoy et al. (2006) | Avy |
In utero moderate-dose BPA In utero BPA followed by genistein |
Hypomethylation Stable |
Tail | – | [19] |
| Anderson et al. (2012) | In utero low-dose BPA | Hypomethylation | Tail | Decreased weaning weight | [15] | |
| Kaminen-Ahola et al. (2010) | In utero ethanol | Hypermethylation | Tail | Skull deformation | [60] | |
| Bernal et al. (2013) | In utero radiation | Hypermethylation | Liver | – | [45] | |
| Waterland et al. (2006) | In utero methyl donor diet | Hypermethylation | Tail | – | [18] | |
| Dolinoy et al. (2006) | In utero genistein | Hypermethylation | Tail | – | [61] | |
Avy: Viable yellow agouti; BPA: Bisphenol A; CabpIAP: CDK5 activator-binding protein intracisternal A particle element; TCDD: Tetrachlorodibenzo-p-dioxin.
The toxic effects of Pb, especially in relation to neuropathology and cardiovascular disease risk have been studied for decades, resulting in direct human benefits across a range of outcomes (i.e., removal of Pb from gasoline and paint) [54,55]. Despite these efforts, Pb is still present in the environment and in consumer products, and with a half-life of years to decades, can be mobilized from bone during pregnancy and lactation. Thus, although the epigenetic effects of early-life Pb exposure are subtle, they may serve as an underlying mechanism linking exposure to later-in-life disease risk. Here we measured both the actual Pb content of the drinking water supplied to all mice in the study from a single dilution and the resulting BLLs in dams at weaning. Previous mouse studies involving Pb acetate exposure in drinking water range from exposures of 0.02 [12] to 4000 ppm [56] with widely varying resulting BLLs. Many factors may account for the differences in resulting BLLs between studies, including impurities in the water source, Pb acetate source and pH, among others. Thus, it is crucial to report both the diluted Pb water supply and the resulting BLLs in future studies. At the 2.1 ppm level, we found significant epigenetic effects, from a blood level of approximately 4.1 μg/dl, corresponding to human relevant doses. The impact of in utero Pb exposure on the epigenome and downstream health implications for today’s youth is relatively unknown but could be substantial. In a Mexican cohort recruited in the 1990s, over 75% of new mothers had a BLL greater than 5 μg/dl [57]. In the US population from 1988–1994, 5.8% of women of reproductive age had BLLs greater than 5 μg/dl while 25.6% of children between 1 and 5 years of age had a BLL above this level [58,59]. Therefore, low-level Pb exposure, particularly during the epigenetically sensitive developmental period, impacts a significant percentage of the human population and understanding its epigenetic consequences may provide direct health benefits.
Future perspective
An increasing understanding of environmental impacts on the epigenome is resulting in the recognition that the context and makeup of genes may govern their epigenetic response to toxic exposures. Likewise, more sensitive studies are revealing that effects of early-life exposures can reverberate throughout the life course. The continuing global Pb exposure risk combined with internal Pb sequestration in bone and subsequent release into blood, including during pregnancy, warrants an understanding of epigenetic effects established in early life that may have direct bearing on disease risk in later life.
Executive summary.
Background
Chronic health conditions have been associated with early-life exposures to lead (Pb).
Epigenetic mechanisms such as DNA methylation at CpG sites may mediate the connection between early-life exposure and later health effects.
Materials & methods
The viable yellow agouti mouse (Avy) varies in coat color according to DNA methylation at a single locus, and was exposed to Pb at concentrations of 0, 2.1, 16 and 32 ppm.
Weaning weight was measured in both Avy allele carriers (Avy/a) and noncarriers (a/a [wild-type]).
DNA methylation was assayed via pyrosequencing at two transposon-associated genes, Avy and CabpIAP, and at two imprinted genes, Igf2 and Igf2r.
Results
Pb exposure was associated with increased weaning weight in a dose-dependent fashion in males only (p = 0.03).
Coat color distribution in agouti offspring was shifted towards the hypomethylated yellow phenotype, and found to be significant between the control and highest dose group (32 ppm).
In three dose groups, methylation at the transposon-associated genes Avy and CabpIAP was significantly shifted in a cubic trend, compared with controls (p = 0.04 and p = 0.01, respectively), with male-specific effects predominating at the Avy locus.
Imprinted gene methylation at Igf2 and Igf2r was not significantly affected by Pb exposure.
Discussion
Our findings suggest that the epigenome is affected by Pb in a dose- and sex-specific manner, and that sex is a predictor of physiological and epigenetic response to exposure.
The epigenetic effects of Pb are specific and suggestive of a toxicant-specific response in development that can have long-term consequences for adult health.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
This work was supported by the University of Michigan (UM) NIEHS/EPA Children’s Environmental Health Formative Center (P20 ES018171/RD834800), as well as the UM NIEHS Core Center (P30 ES017885). C Faulk was supported by the UM NIEHS Core Center (T32 ES007062). JM Goodrich was supported by the UM NIH National Center for Advancing Translational Sciences grant (2UL1TR000433). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
The authors would like to thank BN Sanchez, Z Zhang and KE Peterson for their technical and intellectual assistance with this project.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Barker D. Intrauterine programming of coronary heart disease and stroke. Acta Paediatra Suppl. 1997;423:178–182. doi: 10.1111/j.1651-2227.1997.tb18408.x. [DOI] [PubMed] [Google Scholar]
- 2▪.Barker D. Programming the baby. In: Barker D, editor. Mothers, Babies, and Disease in Later Life. BMJ Publishing; London, UK: 1994. Developmental origins of adult disease are clearly presented and human health implications laid forth. [Google Scholar]
- 3.Hu H, Tellez-Rojo MM, Bellinger D, et al. Fetal lead exposure at each stage of pregnancy as a predictor of infant mental development. Environ Health Perspect. 2006;114(11):1730–1735. doi: 10.1289/ehp.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellinger DC. Teratogen update: lead and pregnancy. Birth Defects Res A Clin Mol Teratol. 2005;73(6):409–420. doi: 10.1002/bdra.20127. [DOI] [PubMed] [Google Scholar]
- 5.Lanphear BP, Hornung R, Khoury J, et al. Low-level environmental lead exposure and children’s intellectual function: an international pooled analysis. Environ Health Perspect. 2005;113(7):894–899. doi: 10.1289/ehp.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navas-Acien A, Guallar E, Silbergeld EK, Rothenberg SJ. Lead exposure and cardiovascular disease – a systematic review. Environ Health Perspect. 2007;115(3):472–482. doi: 10.1289/ehp.9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong GP, Ng TL, Martin TR, Farquharson DF. Effects of low-level lead exposure in utero. Obstet Gynecol Surv. 1992;47(5):285–289. doi: 10.1097/00006254-199205000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Agency for Toxic Substances and Disease Registry. Toxicological Profile for Lead. Agency for Toxic Substances and Disease Registry; GA, USA: 2007. [PubMed] [Google Scholar]
- 9.Gilbert SG, Weiss B. A rationale for lowering the blood lead action level from 10 to 2 microg/dl. Neurotoxicology. 2006;27(5):693–701. doi: 10.1016/j.neuro.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder JE, Filipov NM, Parsons PJ, Lawrence DA. The efficiency of maternal transfer of lead and its influence on plasma IgE and splenic cellularity of mice. Toxicol Sci. 2000;57(1):87–94. doi: 10.1093/toxsci/57.1.87. [DOI] [PubMed] [Google Scholar]
- 11▪▪.Leasure JL, Giddabasappa A, Chaney S, et al. Low-level human equivalent gestational lead exposure produces sex-specific motor and coordination abnormalities and late-onset obesity in year-old mice. Environ Health Perspect. 2008;116(3):355–361. doi: 10.1289/ehp.10862. Notable for its careful study of low levels of gestational lead exposure in mice and the effects in male and female offspring at 1 year of life. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iavicoli I, Carelli G, Stanek EJ, Castellino N, Li Z, Calabrese EJ. Low doses of dietary lead are associated with a profound reduction in the time to the onset of puberty in female mice. Reprod Toxicol. 2006;22(4):586–590. doi: 10.1016/j.reprotox.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Faulk C, Dolinoy DC. Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics. 2011;6(7):791–797. doi: 10.4161/epi.6.7.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol. 2012;863:359–376. doi: 10.1007/978-1-61779-612-8_23. [DOI] [PubMed] [Google Scholar]
- 15.Anderson OS, Nahar MS, Faulk C, et al. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ Mol Mutagen. 2012;53(5):334–342. doi: 10.1002/em.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koedrith P, Kim H, Weon JI, Seo YR. Toxicogenomic approaches for understanding molecular mechanisms of heavy metal mutagenicity and carcinogenicity. Int J Hyg Environ Health. 2013;216(5):587–598. doi: 10.1016/j.ijheh.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Dolinoy DC. The agouti mouse model: an epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutr Rev. 2008;66(Suppl 1):S7–S11. doi: 10.1111/j.1753-4887.2008.00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation at axin fused. Genesis. 2006;44(9):401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- 19.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114(4):567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18(7):348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 21.Hackett JA, Surani MA. DNA methylation dynamics during the mammalian life cycle. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20110328. doi: 10.1098/rstb.2011.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Downing C, Johnson TE, Larson C, et al. Subtle decreases in DNA methylation and gene expression at the mouse Igf2 locus following prenatal alcohol exposure: effects of a methyl-supplemented diet. Alcohol. 2011;45(1):65–71. doi: 10.1016/j.alcohol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKay JA, Williams EA, Mathers JC. Effect of maternal and post-weaning folate supply on gene-specific DNA methylation in the small intestine of weaning and adult apc and wild type mice. Front Genet. 2011;2:23. doi: 10.3389/fgene.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding GL, Wang FF, Shu J, et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61(5):1133–1142. doi: 10.2337/db11-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang XF, Zhang LJ, Feng YN, et al. Bisphenol A exposure modifies DNA methylation of imprint genes in mouse fetal germ cells. Mol Biol Rep. 2012;39(9):8621–8628. doi: 10.1007/s11033-012-1716-7. [DOI] [PubMed] [Google Scholar]
- 26.Somm E, Stouder C, Paoloni-Giacobino A. Effect of developmental dioxin exposure on methylation and expression of specific imprinted genes in mice. Reprod Toxicol. 2013;35:150–155. doi: 10.1016/j.reprotox.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol A exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9(4):e1003401. doi: 10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallou-Kabani C, Gabory A, Tost J, et al. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One. 2010;5(12):e14398. doi: 10.1371/journal.pone.0014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23(15):5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. 4. Cold Spring Harbor Laboratory Press; NY, USA: 2012. [Google Scholar]
- 31.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29(13):E65–E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Druker R. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32(19):5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15(5):705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 34.Fauque P, Ripoche MA, Tost J, et al. Modulation of imprinted gene network in placenta results in normal development of in vitro manipulated mouse embryos. Hum Mol Genet. 2010;19(9):1779–1790. doi: 10.1093/hmg/ddq059. [DOI] [PubMed] [Google Scholar]
- 35.Miltenberger RJ, Mynatt RL, Wilkinson JE, Woychik RP. The role of the agouti gene in the yellow obese syndrome. J Nutr. 1997;127(9):1902S–1907S. doi: 10.1093/jn/127.9.1902S. [DOI] [PubMed] [Google Scholar]
- 36.Rossi-George A, Virgolini MB, Weston D, Cory-Slechta DA. Alterations in glucocorticoid negative feedback following maternal Pb, prenatal stress and the combination: a potential biological unifying mechanism for their corresponding disease profiles. Toxicol Appl Pharmacol. 2009;234(1):117–127. doi: 10.1016/j.taap.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronis MJ, Badger TM, Shema SJ, et al. Endocrine mechanisms underlying the growth effects of developmental lead exposure in the rat. J Toxicol Environ Health A. 1998;54(2):101–120. doi: 10.1080/009841098158944. [DOI] [PubMed] [Google Scholar]
- 38.Pine M, Hiney J, Dearth R, Bratton J, Les Dees WL. IGF-1 administration to prepubertal female rats can overcome delayed puberty caused by maternal Pb exposure. Reprod Toxicol. 2006;21:104–109. doi: 10.1016/j.reprotox.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Dearth RK, Hiney JK, Srivastava V, Burdick SB, Bratton GR, Dees WL. Effects of lead (Pb) exposure during gestation and lactation on female pubertal development in the rat. Reprod Toxicol. 2002;16(4):343–352. doi: 10.1016/s0890-6238(02)00037-0. [DOI] [PubMed] [Google Scholar]
- 40.McGivern RF, Sokol RZ, Berman NG. Prenatal lead exposure in the rat during the third week of gestation: long-term behavioral, physiological, and anatomical effects associated with reproduction. Toxicol Appl Pharmacol. 1991;110(2):206–215. doi: 10.1016/s0041-008x(05)80003-1. [DOI] [PubMed] [Google Scholar]
- 41.Petrusz P, Weaver CM, Grant LD, Mushak P, Krigman MR. Lead poisoning and reproduction: effects on pituitary and serum gonadotropins in neonatal rats. Environ Res. 1979;19(2):383–391. doi: 10.1016/0013-9351(79)90063-x. [DOI] [PubMed] [Google Scholar]
- 42.Gorbel F, Boujelbene M, Makni-Ayadi F, et al. Cytotoxic effects of lead on the endocrine and exocrine sexual function of pubescent male and female rats. Demonstration of apoptotic activity. C R Biol. 2002;325(9):927–940. doi: 10.1016/s1631-0691(02)01492-0. [DOI] [PubMed] [Google Scholar]
- 43.Ronis MJ, Gandy J, Badger T. Endocrine mechanisms underlying reproductive toxicity in the developing rat chronically exposed to dietary lead. J Toxicol Environ Health A. 1998;54(2):77–99. doi: 10.1080/009841098158935. [DOI] [PubMed] [Google Scholar]
- 44.Faulk C, Barks A, Dolinoy DC. Phylogenetic and DNA methylation analysis reveal novel regions of variable methylation in the mouse IAP class of transposons. BMC Genomics. 2013;14(1):48. doi: 10.1186/1471-2164-14-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45▪.Bernal AJ, Dolinoy DC, Huang D, Skaar DA, Weinhouse C, Jirtle RL. Adaptive radiation-induced epigenetic alterations mitigated by antioxidants. FASEB J. 2013;27(2):665–671. doi: 10.1096/fj.12-220350. Hormetic response was observed following low-dose gestational radiation exposure, correlating with increased DNA methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li G, Kohorst JJ, Zhang W, et al. Early postnatal nutrition determines adult physical activity and energy expenditure in female mice. Diabetes. 2013;62(8):2773–2783. doi: 10.2337/db12-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel BB, Raad M, Sebag IA, Chalifour LE. Lifelong exposure to bisphenol A alters cardiac structure/function, protein expression, and DNA methylation in adult mice. Toxicol Sci. 2013;133(1):174–185. doi: 10.1093/toxsci/kft026. [DOI] [PubMed] [Google Scholar]
- 48.Sterrenburg L, Gaszner B, Boerrigter J, et al. Chronic stress induces sex-specific alterations in methylation and expression of corticotropin-releasing factor gene in the rat. PLoS One. 2011;6(11):e28128. doi: 10.1371/journal.pone.0028128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider JS, Kidd S, Anderson DW. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol Lett. 2012;217(1):75–81. doi: 10.1016/j.toxlet.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson SL, Konfortova G, Gregory RI, Reik W, Dean W, Feil R. Environmental effects on genomic imprinting in mammals. Toxicol Lett. 2001;120(1–3):143–150. doi: 10.1016/s0378-4274(01)00292-2. [DOI] [PubMed] [Google Scholar]
- 51.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts CT, Owens JA, Sferruzzi-Perri AN. Distinct actions of insulin-like growth factors (IGFs) on placental development and fetal growth: lessons from mice and guinea pigs. Placenta. 2008;29(Suppl A):S42–S47. doi: 10.1016/j.placenta.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 53▪▪.Li YY, Chen T, Wan Y, Xu SQ. Lead exposure in pheochromocytoma cells induces persistent changes in amyloid precursor protein gene methylation patterns. Environ Toxicol. 2012;27(8):495–502. doi: 10.1002/tox.20666. Gene-specific and global DNA hypomethylation were observed in this study, and may be associated with lead-induced neurodegeneration. [DOI] [PubMed] [Google Scholar]
- 54.Park SK, Elmarsafawy S, Mukherjee B, et al. Cumulative lead exposure and age-related hearing loss: the VA normative aging study. Hear Res. 2010;269(1–2):48–55. doi: 10.1016/j.heares.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz BS, Hu H. Adult lead exposure: time for change. Environ Health Perspect. 2007;115(3):451–454. doi: 10.1289/ehp.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tokar EJ, Diwan BA, Waalkes MP. Early life inorganic lead exposure induces testicular teratoma and renal and urinary bladder preneoplasia in adult metallothionein-knockout mice but not in wild type mice. Toxicology. 2010;276(1):5–10. doi: 10.1016/j.tox.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez-Cossio T, Peterson KE, Sanin LH, et al. Decrease in birth weight in relation to maternal bone-lead burden. Pediatrics. 1997;100(5):856–862. doi: 10.1542/peds.100.5.856. [DOI] [PubMed] [Google Scholar]
- 58.Lee MG, Chun OK, Song WO. Determinants of the blood lead level of US women of reproductive age. J Am Coll Nutr. 2005;24(1):1–9. doi: 10.1080/07315724.2005.10719436. [DOI] [PubMed] [Google Scholar]
- 59.Bernard SM, McGeehin MA. Prevalence of blood lead levels > or = 5 micro g/dl among US children 1 to 5 years of age and socioeconomic and demographic factors associated with blood of lead levels 5 to 10 micro g/dl, Third National Health and Nutrition Examination Survey, 1988–1994. Pediatrics. 2003;112(6 Pt 1):1308–1313. doi: 10.1542/peds.112.6.1308. [DOI] [PubMed] [Google Scholar]
- 60.Kaminen-Ahola N, Ahola A, Flatscher-Bader T, et al. Postnatal growth restriction and gene expression changes in a mouse model of fetal alcohol syndrome. Birth Defects Res Part A Clin Mol Teratol. 2010;88(10):818–826. doi: 10.1002/bdra.20729. [DOI] [PubMed] [Google Scholar]
- 61.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114(4):567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]