Abstract
Adsorption and genome ejection are fundamental to the bacteriophage life cycle, yet their molecular mechanisms are not well understood. We used cryo–electron tomography to capture T7 virions at successive stages of infection of Escherichia coli minicells at ~4-nm resolution. The six phage tail fibers were folded against the capsid, extending and orienting symmetrically only after productive adsorption to the host cell surface. Receptor binding by the tail triggered conformational changes resulting in the insertion of an extended tail, which functions as the DNA ejection conduit into the cell cytoplasm. After ejection, the extended phage tail collapsed or disassembled, which allowed resealing of the infected cell membrane. These structural studies provide a detailed series of intermediates during phage infection.
After encountering a cell, a bacteriophage delivers its genetic material into the host cytoplasm via an orchestrated series of conformational changes (1). T7 is a member of the Podoviridae, having short, noncontractile tails that are too short to span a bacterial cell envelope (2). The icosahedral 2-nm-thick T7 capsid is 60 to 61 nm in diameter, and inside, coaxial to the dodecameric head-tail connector, is a 26- by 21-nm internal core, which is thought to consist of three proteins: gp14, gp15, and gp16 (2, 3). The core is essential for both virion morphogenesis and ejection of the 40-kb genome. Near the head-proximal end of the 23-nm tail are six fibers, trimers of gp17. Each fiber has an N-terminal domain that attaches the fiber to the tail, followed by proximal and distal half-fibers. The latter interact with the cell surface (4, 5). After adsorption, the core proteins are ejected through the portal-tail complex into the infected cell, and it has been hypothesized that they extend the tail, creating a trans-envelope channel used for DNA transport (6-8). Here, we used cryo–electron tomography (cryo-ET) to provide three-dimensional (3D) structures of infected cell complexes in near-native, frozen-hydrated states at ~4-nm resolution (fig. S1).
We infected small (≤0.3 μm in diameter) Escherichia coli minicells (9) to achieve maximum resolution. The T7-infected minicells produced infective centers at normal efficiencies, and the parent skinny cells were good hosts for the phage, which suggested that our structures reflected the normal infection pathway (10). The 3D reconstructions revealed adsorbed virions at three stages of infection (Fig. 1 and movie S1) and supported the hypothesis (6-8) that T7 internal core proteins form a trans-envelope channel (Fig. 1, B and E). Dual-axis tomography confirmed the presence of various conformations (movie S2). In order to obtain better structural details, we used subvolume averaging to determine the 3D structures during infection (10).
Fig. 1.
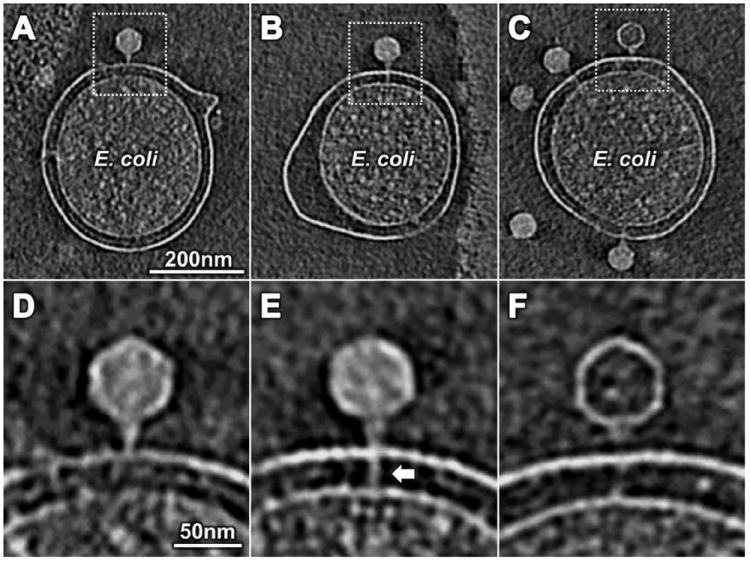
Three stages of T7 infection. (A and D) T7 adsorbed to the outer membrane. The density spanning the cell envelope at 270° in (A) was derived from fiducial gold markers (shown explicitly in movie S1 as a series of slices across the cell in (A). (B and E) An extended tail (arrow) spanned the cell envelope. (C and F) After DNA ejection, the extended tail was not apparent.
Asymmetric reconstructions of a fiberless gene 17 mutant (Fig. 2, A and D) and wild-type (Fig. 2, B and E) virions were derived from 1945 and 6116 subvolumes, respectively. The overall structure of wild-type T7 (Fig. 2B) is similar to that reported (3), but our tomographic reconstruction resolved the tail fibers (Fig. 2, B, C, E, and F). Placing a known structure of part of the distal half-fiber (5) into the cryo-ET density revealed a good fit (Fig. 2F). Each fiber was folded back onto the capsid with the proximal half-fiber rising across the face of a capsomer. The ~90° kink between proximal and distal half-fibers was visible in 3D surface renderings (Fig. 2, E and F). Fibers retained a conformation similar to that of isolated tail complexes (fig. S2) (4).
Fig. 2.
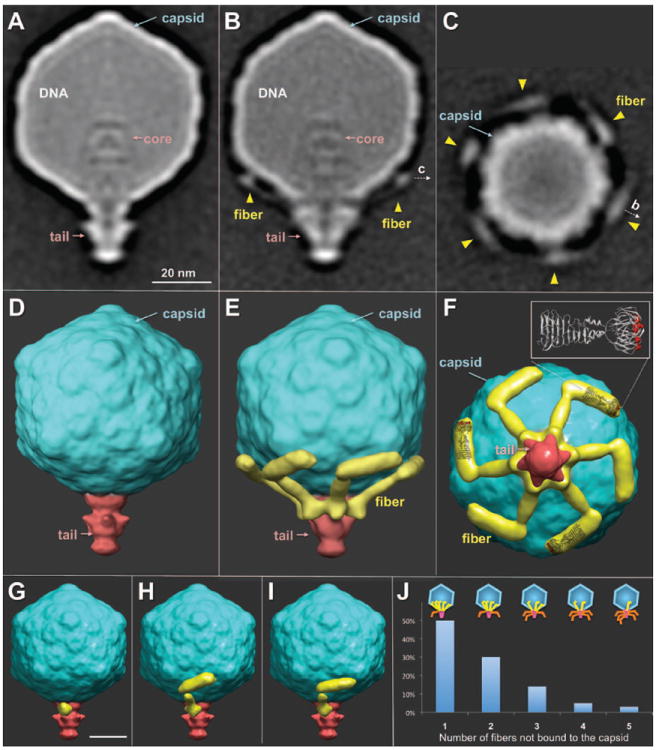
Asymmetric reconstructions of virions. (A) A central slice of a fiberless virion revealed the tail, capsid, and internal core. Fibers were visible in a central slice (B) and a cross section of wild-type virions (C). Surface rendering of fiberless (D) and wild-type virions in side (E) and bottom (F) views. A crystal structure of the receptor-binding domain of T7 fibers (5) was placed into three fiber densities. Residues A518, D520, and V544, known to affect phage host range (5), are highlighted in red (F). Classification revealed different conformations of fibers not bound (G) or bound to the capsid (H and I). (J) Distribution of the number of bound fibers on free virions; few had either six or zero fibers bound.
The T7 tail exhibited 6-fold symmetry, but the distal half-fibers were bound to the capsid, which is 5-fold symmetrical at the connector. To better understand the symmetry mismatch and the fiber-capsid interactions, we classified each fiber independently by correspondence analysis (figs. S3 and S4). Class I fibers exhibited only a short fragment corresponding to the N-terminal domain that is bound to the tail (4), which indicated that neither half-fiber was bound to the capsid. Proximal and distal half-fibers were visible in class II and were subdivided into subclasses by their different orientations on the capsid (Fig. 2, H and I). Projecting all distal half-fiber conformations onto the same surface showed that there is no specific binding site (fig. S4).
Phage tail fibers are normally depicted extending away from the capsid, poised for adsorption. However, we found that ~50% of mature T7 virions had five fibers bound to the capsid and ~30% had four; very few had no bound fibers (Fig. 2J). Most published electron microscopy (EM) images of T7 do not resolve fibers; they were visible in a tomogram of negatively stained T7 particles only as short projections from the capsid (fig. S2). More than 95% of the wild-type T7 prepared for this study made infective centers in exponentially growing cells within 2 to 3 min, which suggested that extended fibers were not required for rapid adsorption (10). Proximal half-fibers of the T7-related phage P-SSP7 also appear bound in mature virions but are extended in spontaneously emptied free particles (11). Furthermore, cryo-EM reconstructions of T4 virions show fibers wrapped around the tail sheath, not pointing away from the capsid (12). Maintaining tail fibers on the body of free virions may thus be typical of many phages.
Adsorbed T7 virions were generally oriented perpendicular to the cell surface with the tail slightly indenting the outer membrane (Fig. 3, A and B; fig. S5; and movie S3), similar to adsorbed phage P-SSP7 (11). Proximal half-fibers extended horizontally, whereas distal half-fibers were vertical; bound fibers exhibited the same 6-fold symmetry as the tail, covering ~3000 nm2 of the cell surface. During adsorption, fibers underwent a large rotation, which was probably due to the N-terminal domain pivoting on its connection to the tail (fig. S6). Two-thirds (3352) of adsorbed phages analyzed were captured at this intermediate stage, with the internal core remaining intact within the capsid, which suggests that recognition of the cell surface by all six fibers is insufficient to trigger later steps of infection.
Fig. 3.

Adsorption structures. (A) Central slice and (B) 3D surface view of a subvolume average derived from 3352 virions. All six fibers were bound to the cell, and the tail made a small indentation in the outer membrane (OM); the internal core and DNA were still in the capsid. (C) Central slice and (D) 3D surface view of a subvolume average derived from 1886 virions. The internal core had been ejected from the virion, forming an extended tail that penetrated the cell cytoplasm. PG, peptidoglycan cell wall; IM, inner membrane.
About one-third (1886) of adsorbed particles displayed a tubelike extension of the tail that penetrated both cell membranes (Fig. 3, C and D; figs. S5 and S7; and movie S3). A functional extension of the T7 tail is essential for genome delivery into the host (2). The energy required for this dramatic change in structure must be stored in the virion, because cellular energy sources will not yet be accessible. The interaction between the tail and the outer membrane also changed, which presumably reflects the conformational changes that signal ejection of the internal core, which was no longer visible (Fig. 3, A and C). However, the head still appeared full, which suggested that the encap-sidated DNA had expanded to occupy the volume originally occupied by the core.
The extended tail is probably composed of ejected core proteins, which have been found in the infected cell envelope (7, 8). Gp14 localizes to the infected cell outer membrane, whereas gp15 and gp16 span the periplasm and cytoplasmic membrane. Various mutant gp16-containing virions change the kinetics of genome ejection or prevent DNA from entering the cytoplasm; the latter defect can be suppressed by virions also harboring a mutant gp15 (8, 13-16). After infection by 16E37F mutant virions, which are defective in cell wall hydrolysis and which exhibit delayed DNA entry into the cytoplasm (14, 17), the extended tail took ~20 times as long to form (fig. S8). The estimated volume of the internal core (3) is sufficient to form the entire extension. The overall tail extension was ~45 nm, and the tube had an outer diameter of ≥8 nm, sufficient to accommodate double-stranded DNA in its lumen. As the tube approached the cytoplasmic membrane, density broadened (Fig. 3, C and D), probably because the extended tail was flexible (movie S4). Density associated with the tube was less distinct inside the cytoplasm, but ~12 nm below the center of the membrane a ~30 by 6 nm toroid with a ~4-nm central cavity was clearly present (fig. S9 and movie S4). The toroid may be part of the molecular motor that has been hypothesized to pull the leading genome end into the cell (6, 8, 16).
About 3% (162) of adsorbed phages had completely ejected their genome. Virion structure at this stage of infection was thus determined only at relatively low resolution (fig. S10 and movie S3). The cytoplasmic toroid was not visible, and the tube traversing the periplasm had collapsed or disassembled, which presumably prevented dissipation of the membrane potential, which would inhibit phage development. We tested whether an energized membrane was required for channel formation by treating minicells with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) before infection. Of the 244 virions observed, 14% did not adsorb, 55% adsorbed without tail extension, 31% formed a trans-envelope structure, but none ejected their genome (fig. S11). An energized membrane was thus not necessary for adsorption or ejection of internal proteins, but, as expected (16, 18), it is essential for genome translocation.
The combination of technical advances in cryo-ET and a reduction in E. coli cell size has allowed us to describe detailed features of the mature T7 virion and structural changes during infection. Tail fibers, usually depicted extending away from the virion, were mostly bound to the capsid. Our data suggest a dynamic equilibrium between bound and unbound states. Maintaining fibers in a folded-back configuration would allow a higher rate of virion diffusion, whereas transient extension could still permit the phage to explore a large volume in search of a cell. Binding of fibers to bacterial receptors is weak, but transient binding could reduce the dimensional space for finding a suitable site for infection from three to two (19). Weak binding could potentially achieve the necessary specificity of adsorption by cooperative binding among fibers or by a second component, i.e., the tail, binding with high specificity (2, 7, 20). We propose that the transition to a stably adsorbed state occurs following a random walk after an initial interaction between a fiber and the cell; e.g., bound fiber no. 1 is directly replaced by binding of fiber no. 2 without virion release (Fig. 4 and movie S5). Alternatively, in a quasi-2D diffusion process, fiber no. 1 may dissociate, followed rapidly by fiber no. 2 detaching from the capsid and binding to the cell. Note that the common lab strain of phage λ, which lacks side tail fibers, has been suggested to move across the cell surface in a quasi-1D process (21). At a preferred site of infection, the tail interacts specifically with its receptor, which prevents further lateral movement and allows all fibers to bind. The fibers of phage T7 may thus function primarily to facilitate interaction of the tail with its specific receptor. After stable adsorption, infection is triggered by ejection of the internal core proteins into the cell envelope and the formation of an extended tail, which may protect the entering genome from periplasmic nucleases.
Fig. 4.

Schematic model of T7 infection. The insert tomograms show the different orientations of tail fibers. Fibers bound to the cell are highlighted with yellow arrows, unbound fibers with green arrows. After “walking” across the cell surface to find the receptor for the tail, all fibers rotate downward to contact the outer membrane. Commitment to infection occurs after internal core proteins are ejected from the virion and the extended tail (red arrow) forms.
Supplementary Material
Acknowledgments
We thank P. Leiman, S. Norris, and Y. W. Yin for comments. B.H. and J.L. were supported in part by grants R01AI087946 from the National Institute on Allergy and Infection, NIH, and AU-1714 from the Welch Foundation. W.M. was supported by NIH grant R01GM61074 and a grant from the Human Frontier Science Program. EM maps have been deposited in the EM Data Bank (www.ebi.ac.uk/pdbe/emdb/) with accession nos. EMD-5534, 5535, 5536, and 5537. J.L. and I.J.M. designed research; B.H. and W.M. constructed the E. coli mutants; B.H. and J.L. collected and analyzed data; and B.H., W.M., J.L., and I.J.M. wrote the paper.
Footnotes
Supplementary Materials
www.sciencemag.org/cgi/content/full/science.1231887/DC1
Materials and Methods
Figs. S1 to S11
Table S1
References (22–30)
Movies S1 to S5
References and Notes
- 1.Johnson JE, Chiu W. Curr Opin Struct Biol. 2007;17:237. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Casjens SR, Molineux IJ. Adv Exp Med Biol. 2012;726:143. doi: 10.1007/978-1-4614-0980-9_7. [DOI] [PubMed] [Google Scholar]
- 3.Agirrezabala X, et al. EMBO J. 2005;24:3820. doi: 10.1038/sj.emboj.7600840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steven AC, et al. J Mol Biol. 1988;200:351. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Doval C, van Raaij MJ. Proc Natl Acad Sci U S A. 2012;109:9390. doi: 10.1073/pnas.1119719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molineux IJ. Mol Microbiol. 2001;40:1. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 7.Kemp P, Garcia LR, Molineux IJ. Virology. 2005;340:307. doi: 10.1016/j.virol.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 8.Chang CY, Kemp P, Molineux IJ. Virology. 2010;398:176. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, et al. Proc Natl Acad Sci U S A. 2012;109:E1481. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.See supplementary materials on Science Online.
- 11.Liu X, et al. Nat Struct Mol Biol. 2010;17:830. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leiman PG, et al. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García LR, Molineux IJ. J Bacteriol. 1996;178:6921. doi: 10.1128/jb.178.23.6921-6929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moak M, Molineux IJ. Mol Microbiol. 2000;37:345. doi: 10.1046/j.1365-2958.2000.01995.x. [DOI] [PubMed] [Google Scholar]
- 15.Struthers-Schlinke JS, Robins WP, Kemp P, Molineux IJ. J Mol Biol. 2000;301:35. doi: 10.1006/jmbi.2000.3940. [DOI] [PubMed] [Google Scholar]
- 16.Kemp P, Gupta M, Molineux IJ. Mol Microbiol. 2004;53:1251. doi: 10.1111/j.1365-2958.2004.04204.x. [DOI] [PubMed] [Google Scholar]
- 17.Moak M, Molineux IJ. Mol Microbiol. 2004;51:1169. doi: 10.1046/j.1365-2958.2003.03894.x. [DOI] [PubMed] [Google Scholar]
- 18.Zimkus AZ, Zavriev SK, Grinius LL. Mol Biol (Mosk) 1986;20:185. [PubMed] [Google Scholar]
- 19.Adam G, Delbrück M. In: Structural Chemistry and Molecular Biology. Rich R, Davidson N, editors. Freeman; San Francisco: 1968. pp. 198–215. [Google Scholar]
- 20.Qimron U, Marintcheva B, Tabor S, Richardson CC. Proc Natl Acad Sci U S A. 2006;103:19039. doi: 10.1073/pnas.0609428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothenberg E, et al. Biophys J. 2011;100:2875. doi: 10.1016/j.bpj.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


