Abstract
Pseudo-nitzschia-specific PCR primers (PnAll F/R) were designed to amplify a polymorphic region of the internal transcribed spacer 1 (ITS1) from at least 11 Pseudo-nitzschia species. The primers were used to generate environmental clone libraries from Puget Sound, Washington, and Vancouver Island, British Columbia, to confirm that the primers were specific for Pseudo-nitzschia and to determine the extent of ITS1 sequence diversity within individual species. All environmental ITS1 sequences generated with PnAll primers displayed the greatest similarity to known Pseudo-nitzschia ITS1 sequences. The length of cloned ITS1 fragments differed among species but was conserved within a species. Intraspecific genotypes exhibited <3% sequence divergence for seven of the 10 species detected in clone libraries. Several ITS1 genotypes unique to the Pacific Northwest were identified in environmental samples, and other genotypes were more broadly distributed. The Pseudo-nitzschia primers were also used to develop an automated ribosomal intergenic spacer analysis (ARISA) to rapidly identify Pseudo-nitzschia species in environmental samples based on species-specific variation in the length of the targeted ITS1 region. The ARISA peaks were then associated with the environmental clone sequences for Pseudo-nitzschia species. Surveying the genetic composition of communities at both the inter- and intraspecific levels will enhance our understanding of Pseudo-nitzschia bloom dynamics.
Key index words: ARISA, Bacillariophyceae, diatoms, diversity, environmental clone libraries, genotypes, ITS, Pseudo-nitzschia, Puget Sound, Vancouver Island
Diatoms are considered one of the more species-rich groups of eukaryotes, even though current species classifications of diatoms are relatively coarse and probably underestimate the true species number (Mann and Droop 1996). The genus Pseudo-nitzschia is composed of at least 30 species and is common to diatom assemblages in all ocean basins, both in coastal and offshore waters. In recent years, increased focus has been directed toward identifying Pseudo-nitzschia species for two important reasons. First, some species of Pseudo-nitzschia produce the neurotoxin domoic acid, which can result in the death or illness of vertebrates (including humans) if seafood contaminated with domoic acid is consumed. Second, Pseudo-nitzschia has been identified as one of the main genera to increase in cell numbers during iron fertilization experiments in high nitrate low chl (HNLC) regions of the Southern Ocean, the subarctic Pacific, and the east equatorial Pacific (de Baar et al. 2005).
Despite concerted efforts, the taxonomy of Pseudo-nitzschia is still largely unresolved, and morphological species complexes and/or cryptic species are increasingly described for the genus (Hasle 1995, Manhart et al. 1995, Villac and Fryxell 1998, Lundholm et al. 2002, 2003, 2006, Orsini et al. 2004, Hasle and Lundholm 2005). Links between physiology and environment remain elusive for Pseudo-nitzschia, in part due to the difficulty in identifying when and where different species and strains occur. Multiple toxigenic Pseudo-nitzschia species frequently coexist in the environment (Bates 1998, Cho et al. 2002, Trainer et al. 2002), even during blooms that appear to be dominated by a single species of Pseudo-nitzschia (Horner et al. 1996, Parsons et al. 1999, Bill 2005, Bill et al. 2006). Isolates of Pseudo-nitzschia species that bloom in open iron fertilization experiments do not produce detectable levels of domoic acid (Marchetti et al. 2008), but most coastal species can produce the toxin (Bates 1998, Baugh et al. 2006), although to different extents depending on environmental conditions and perhaps strain differences (Bates 1998). Onshore and offshore distributions of coastal and open-ocean species and strains remain unknown.
The most routinely used technique for identifying Pseudo-nitzschia species is SEM or TEM, even though EM does not always provide the necessary resolution required for identification of all Pseudo-nitzschia species (Lundholm et al. 2003, 2006, Hasle and Lundholm 2005). Molecular techniques are increasingly being used to rapidly and/or more precisely identify Pseudo-nitzschia species and include sequencing the LSU rRNA gene (Miller and Scholin 1996, Lundholm et al. 2002, Orsini et al. 2004) or the internal transcribed spacer (ITS) regions (Manhart et al. 1995, Orsini et al. 2004), utilizing hybridization probes for the LSU of rRNA (Miller and Scholin 1996, Parsons et al. 1999), and DNA fingerprinting with PCR (Bornet et al. 2005). Microsatellites have been developed for two Pseudo-nitzschia species to examine population structure, or intraspecific diversity, in field populations (Evans and Hayes 2004, Evans et al. 2004). Most current molecular approaches require isolation and at least short-term cultivation of Pseudo-nitzschia cells, which adds an inherent bias to these studies.
Here, we describe the development and use of a community-based approach to assess the diversity of Pseudo-nitzschia populations at both the inter- and intraspecific levels. Primers for PCR were designed to specifically amplify the ITS1 of the genus Pseudo-nitzschia. This region of the genome was targeted because ITS1 sequence and fragment length can vary among different species and different morphotypes of Pseudo-nitzschia (Orsini et al. 2004). An automated ribosomal intergenic spacer analysis (ARISA) approach (Fisher and Triplett 1999) was developed to identify Pseudo-nitzschia species because this approach does not require isolation or culturing of individual cells. Environmental clone libraries and ITS1 sequences were generated to ground-truth the ARISA approach and to investigate intraspecific diversity. The ability to specifically and routinely identify species and intraspecific variability in Pseudo-nitzschia communities will lead to better understanding of how this genus interacts with the environment and may help clarify the role of diversity in phytoplankton populations.
MATERIALS AND METHODS
Isolate collection and species identification
Pseudo-nitzschia cells were isolated from surface water samples collected within Puget Sound or along the coast of Washington State (USA) by pipetting single cells or chains of cells into 12-well culture plates containing 1 mL sterile seawater amended with f/20 nutrients (Sigma, St. Louis, MO, USA; Guillard 1975). Five unialgal isolates were successfully obtained and transferred into 20 mL sterile seawater amended with f/10 nutrients, and each was subsequently maintained in sterile seawater amended with f/2 nutrients at 13°C on a 12:12 light:dark (L:D) cycle of 50 µmol photons · m−2 · s−1 cool-white light. None of the isolates was axenic. Four isolates were identified to species using a combination of SEM and ITS1 sequencing (see below). Cells for SEM were acid cleaned (Hasle and Fryxell 1970), filtered, and dried onto 1 µm pore-size, 13 mm diameter polycarbonate filters (Whatman Inc., Florharm Park, NJ, USA). Filters were attached to aluminum stubs using graphite bonding material, coated with gold palladium, and examined with a JEOL JSM-6360 LV scanning electron microscope (JEOL Ltd., Tokyo, Japan). Morphometric characteristics were used to identify two P. pungens (Grunow ex Cleve) Hasle isolates, designated PNWH2O 101WB and PNWH2O 105; one P. multiseries (Hasle) Hasle isolate, designated PNWH2O A4; and one P. seriata (Cleve) H. Perag. isolate, designated PNWH2O 109 (Table S1 in the supplementary material). A third P. pungens isolate, designated PNWH2O C1, was identified to species based on ITS1 sequence alone (Table S4 in the supplementary material).
DNA extractions and DNA sequencing of the ITS1
Eight isolates of Pseudo-nitzschia provided by collaborators (Table S4) and five isolates of Pseudo-nitzschia collected for this study (see above) were maintained in f/2 media as described above. Cells from 20 mL cultures in late exponential phase were concentrated onto 0.45 µm pore-size, 47 mm diameter mixed cellulose filters (Millipore, Billerica, MA, USA) for the 13 isolates of four species of Pseudo-nitzschia (Table S4). Genomic DNA was extracted immediately following filtration with the DNeasy Plant Mini Kit (Qiagen Inc., Valencia, CA, USA) according to manufacturer’s instructions. Extracted DNA was quantified with PicoGreen (Invitrogen, Carlsbad, CA, USA) and a SpectraMax M2 microplate reader (Molecular Devices Corp., Sunnyvale, CA, USA).
PCR primers 18SF-euk and 5.8SR-euk were designed to amplify the full-length ITS1 sequence of Pseudo-nitzschia species (Table 1), based on 18S-ITS1-5.8S sequences of P. pungens and P. multiseries (Manhart et al. 1995). PCRs contained 2–6 ng of genomic DNA, 0.4 mM dNTP’s, 0.4 µM of each primer, 0.375 U of Promega taq polymerase, 2.5 mM of MgCl2, and 1× buffer (Promega, Madison, WI, USA). Amplifications consisted of an initial denaturation at 94°C for 2 min, followed by 30 cycles of 95°C for 30 s, 67°C for 30 s, and 72°C for 60 s. PCR products were purified with the QIAquick PCR purification kit (Qiagen). Cycle sequencing was conducted with the DYEnamic ET dye terminator kit (Amersham Biosciences Corp., Piscataway, NY, USA), and the products were analyzed on a MegaBACE 1000 automated sequencer (Amersham Biosciences). Both strands of the ITS1 were sequenced to completion. ITS1 sequences were deposited in GenBank under accession numbers DQ996016–DQ996028.
Table 1.
Nucleotide sequence, locus, and specificity of 18SF-euk, 5.8SR-euk, and PnAllF/R primers designed in this study.
| Name | Locus | Sequence | Specificity |
|---|---|---|---|
| 18SF-euk | 18S rDNA | CTTATCATTTAGAGGAAGGTGAAGTCG | Eukaryotesa |
| 5.8SR-euk | 5.8S rDNA | CTGCGTTCTTCATCGTTGTGG | Eukaryotesb |
| PnAllF | ITS1 | TCTTCATTGTGAATCTGA | Pseudo-nitzschia |
| PnAllR | ITS1 | CTTTAGGTCATTTGGTT | Pseudo-nitzschia |
ITS, internal transcribed spacer.
Eukaryotes amplified by ITS1F include representatives from the fungi, diatoms, unclassified stramenopiles, ciliates, and streptophytes.
Eukaryotes amplified by ITS1R include representatives of brown algae, diatoms, raphidophytes, and ciliates.
In silico development of Pseudo-nitzschia specific primers
A sequence database was compiled in ARB (Ludwig et al. 2004) that consisted of the 13 ITS1 sequences generated in this study; 161 Pseudo-nitzschia ITS1 sequences (without ambiguous base calls) available in GenBank (Table 2) as of April 15, 2007; and 186 ITS1 sequences for 12 different genera of other diatoms available in GenBank (Table S5 in the supplementary material) as of August 1, 2006. “Pseudo-nitzschia-specific” primers, PnAllF and PnAllR (Table 2), were designed to amplify an ITS1 fragment from a maximum number of Pseudo-nitzschia species with no mismatches in either the forward or reverse primers, and no exact matches to non-Pseudo-nitzschia species. Primer specificity was examined in silico against the Ribosomal Database Project (RDP) with the Probe Match function (Cole et al. 2005); against the GenBank nucleotide (nr/nt) database with the blastn function; and against the GenBank nr, wgs, and env_nt databases on September 9, 2007, using the “search for short, nearly exact matches” function within blastn (Altschul et al. 1990).
Table 2.
Summary of Pseudo-nitzschia ITS1 sequences used for in silico development of Pseudo-nitzschia-specific PCR primers and predicted length (including primer sequence) of the resulting ITS1 PnAll fragments.
| Species/typea | Predicted ARISA length (bp) |
No. sequences | Origin | GenBank accession no. |
|---|---|---|---|---|
| P. pungens type 1 | 142 | 1 | China | AY544769b |
| 1 | Mexico | AY257846c | ||
| 1 | Portugal | AY257845c | ||
| 1 | Vietnam | DQ166533b | ||
| P. pungens type 2 | 143 | 1 | Vietnam | DQ062665 |
| P. multiseries | 144 | 1 | Monterey Bay, California | AY257844c |
| 1 | Chesapeake Bay | DQ445651b | ||
| 1 | Ofunato Bay, Japan | DQ062664 | ||
| P. australis | 150 | 2 | Scotland | AY452527–528 |
| 1 | Monterey Bay | AY559850b | ||
| 1 | Portugal | AY257842c | ||
| P. seriata type 1 | 151 | 2 | Scotland | AY452523c–24c |
| 3 | Denmark | AY257841c, DQ062666c, DQ062663c | ||
| P. seriata f. obtusa | 147 | 1 | Tromsø, Norway | DQ062667c |
| P. delicatissima type 1 | 222 | 40 | Gulf of Naples, Italy | AY519282, AY519288–296, AY519302, AY519304, AY519305–06, AY519308, AY519310–11, AY519314, AY519316, AY519320, AY519322, AY519325–26, AY519328, AY519331–3, AY519334c, AY519335c, AY519336c, AY519338–347 |
| 1 | Boca Piccola, Italy | DQ336150c | ||
| 1 | Casttellamare, Italy | DQ329210c | ||
| P. delicatissima type 2 | 220 | 3 | Gulf of Naples, Italy | AY519303, AY519307, AY519337c |
| P. delicatissima type 3 | 223 | 8 | Gulf of Naples, Italy | AY519285–286, AY519299, AY519309, AY519315, AY519317, AY519323, AY519330 |
| P. delicatissima type 4 | 221 | 10 | Gulf of Naples | AY519287, AY519297, AY519300, AY519312–313, AY519318–319, AY519324, AY519327, AY519329 |
| P. delicatissima type 5 | 219 | 2 | Gulf of Naples | AY519301, AY519321 |
| P. delicatissima type 6 | 231 | 1 | Gulf of Naples | AY519283c |
| P. delicatissima type 7 | 165 | 2 | Gulf of Naples | AY519280–281 |
| P. delicatissima type 8 | 164 | 1 | Gulf of Naples | AY519279 |
| P. delicatissima type 9 | 210 | 1 | Gulf of Naples | AY519274 |
| P. delicatissima type 10 | 207 | 1 | Tasmania | AY257848c |
| P. delicatissima type 11 | 168 | 2 | Denmark | AY257849c, DQ329206c |
| 1 | Portugal | DQ329207c | ||
| P. delicatissima type 12 | 192 | 1 | Tuxpam, Mexico | DQ329211c |
| P. delicatissima type 13 | 212 | 1 | Ofunato Bay, Japan | DQ329208c |
| P. cuspidata type 1 | 230 | 1 | Canary Islands | AY257853c |
| 1 | Australia | AY257862c | ||
| P. cuspidata type 2 | 216 | 1 | Mexico | AY257852c |
| P. galaxiae type 1 | 151 | 1 | Mexico | AY257850c |
| P. galaxiae type 2 | 143 | 1 | Sydney, Australia | DQ336158c |
| P. pseudodelicatissima | 230 | 1 | Portugal | AY257854c |
| P. subpacifica | 195 | 1 | Limens, Spain | AY257859c |
| 1 | Costa Nova, Portugal | AY257858c | ||
| P. micropora type 1 | 146 | 1 | Vietnam | AY257847c |
| P. micropora type 2 | 143 | 1 | Phuket, Thailand | DQ329209c |
| P. inflatula | 156 | 1 | Phuket, Thailand | DQ329204c |
| P. spp. | 226 | 1 | Tasmania | AY257851c |
No symbol represents unknown identification method, but sequence is associated with published research.
ITS, internal transcribed spacer.
When multiple PnAll fragment lengths were predicted for a single species, each unique length was designated as a type (e.g., type 1, type 2, etc.).
Unknown identification method due to unpublished research.
Organisms identified using EM based on published research.
Primer specificity and ARISA identification of Pseudo-nitzschia isolates
PnAllF and PnAllR specificity was tested with DNA isolated from P. pungens (strain PNWH2O 101WB), P. multiseries (strain PNWH2O A4), P. australis Freng. (strain 04063_3C), and P. seriata (strain PNWH2O 109), in addition to DNA isolated from the diatoms Lithodesmium undulatum Ehrenb. (CCMP 472), Skeletonema costatum (Grev.) Cleve (CCMP780), Phaeodactylum tricornutum Bohlin (CCMP 632), Chaetoceros socialis Lauder (CCMP 205), Fragilariopsis cylindrus (Grunow) Willi Krieger (CCMP 1102); the cyanobacterium Synechococcus sp. strain UW76; the haptophyte Phaeocystis globosa Scherff. (CCMP 629); and the dinoflagellate Gyrodinium sp. (CCMP 1737). Two types of PCRs were conducted with the PnAllF and PnAllR primers. For ARISA, the PnAllR primer was modified with a 5′ fluorescent FAM label (Qiagen); all other PCRs used an unlabeled version of PnAllR. All PCRs with PnAllF and PnAllR used 4–12 ng DNA and reagents described above for ITS1 PCR amplification. Cycling parameters were 94°C for 2 min, 32 cycles of 95°C for 30 s, 50.6°C for 30 s, and 72°C for 60 s, with a final 72°C extension for 10 min. Unlabeled PCR products were visualized on a 6% nondenaturing polyacrylamide gel stained with SYBR® green (Invitrogen). For ARISA, triplicate PCR products were purified using MultiScreen PCRμ96 filter plates or Montage™ PCR96 filter plates (Millipore). Products were analyzed on a MegaBACE 1000 automated sequencer with the fluorescently labeled internal size standard ET-Rox 400 (Amersham Biosciences). Electropherograms were analyzed using DAx (van Mierlo Software, Eindhoven, the Netherlands); fragment sizes of peaks exceeding 0.2% of the total area and 30 times the noise signal of the electropherogram curve were determined.
Collection of environmental samples and DNA extraction
Surface water samples were collected with Niskin bottles from Tofino Inlet, Vancouver Island, British Columbia (49°10′ N, 125°38′ W), on August 28, 2004; from Port Madison in the main basin of Puget Sound, Washington (47°43′ N, 122°32′ W), on November 2, 2004; and from outside Esperanza Inlet, Vancouver Island, British Columbia (49°47′ N, 127°02′ W), on August 25, 2005 (Fig. 1). Each sample was examined using LM to confirm the presence of Pseudo-nitzschia cells. For each sample, 500 mL was filtered through 0.45 µm pore-size, 47 mm diameter mixed cellulose filters (Millipore), and the filters were stored at −80°C prior to analysis. DNA was extracted with the Dneasy Plant Mini Kit (Qiagen) according to manufacturer instructions, except that each filter was shredded manually in lysis buffer using scissors sterilized with bleach and UV radiation.
Fig. 1.
Map of Pacific Northwest, with sampling sites designated by stars.
Isolate and environmental clone libraries
An iCycler iQ Real-Time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) was used for quantitative PCRs (Q-PCR) with PnAllF and PnAllR on all environmental samples, and on the isolate P. pungens strain PNWH20 101WB, to determine the cycle numbers for exponential amplification. Q-PCR samples were run in triplicate in 20 µL amplifications with 5–20 ng of isolate or environmental DNA, 0.8 µM of primers, and iQ SYBR Green Supermix (Bio-Rad Laboratories). Q-PCR cycling parameters were 95°C for 3 min followed by 50 cycles of 95°C for 10 s, 50.6°C for 50 s, and 72°C for 50 s. PCRs used to generate product for clone library construction were conducted with unlabelled PnAll primers using the cycle number determined by Q-PCR as early exponential. To minimize cloning artifacts, a “reconditioning” step was implemented as described by Thompson et al. (2002).
Two PnAll clone libraries were generated for a single strain of P. pungens, PNWH2O WB101 (Table S4). One clone library was generated from DNA extracted from PNWH2O WB101 approximately 4 months after cell isolation (February 2005), and a second from DNA extracted approximately 2.5 years after cell isolation (June 2007). One clone library was generated for each environmental sample.
PCR products were cloned into the TOPO TA Cloning Kit for Sequencing (Invitrogen) and used to transform the Top 10 strain of Escherichia coli. Thirteen clones from the February 2005 extract and five clones from the June 2007 extract of P. pungens PNWH2O WB101 were randomly chosen for sequencing. From the environmental samples, 40 clones from Puget Sound, 79 clones from Esperanza Inlet, and 19 clones from Tofino were randomly chosen for sequencing.
DNA template for sequencing was prepared with the TempliPhi DNA Amplification Kit (Amersham Biosciences). Sequencing was performed with the DYEnamic ET dye terminator kit (Amersham Biosciences) and analyzed with a MegaBACE 1000 automated sequencer (Amersham Biosciences). Both strands of the ITS1 fragment were sequenced to completion using universal M13 vector primers. Sequence data was checked and aligned using Sequencher Version 4.5 (Gene Codes, Ann Arbor, MI, USA). Environmental sequences were deposited into GenBank under accession numbers EF014758–EF014895. Environmental clone sequences were affiliated with Pseudo-nitzschia species in GenBank using blast (Altschul et al. 1990). Nucleotide divergence (see below) and sequence length of clone sequences were compared to known species of Pseudo-nitzschia to determine sequence similarity of clone sequences and GenBank sequences.
Minimum spanning networks
Pseudo-nitzschia ITS1 sequences generated in this study, including isolate and environmental clone sequences, were aligned to sequences of the most similar Pseudo-nitzschia species from GenBank with clustalw (Chenna et al. 2003) and compiled in ARB. Unique genotypes were identified based on nucleotide sequence differences, insertions, and/or deletions in the DNA sequences. Divergence was calculated based on the number of unique characters relative to the number of total characters (Page and Holmes 1998). Two different divergence indices were used. The first counted each nucleotide polymorphism and each base pair (bp) insertion or deletion (in/dels) as a unique character, while the second counted only nucleotide polymorphisms as unique characters and neglected in/dels; in both cases, PnAll primer regions (35 bp total) were excluded from analysis. Minimum spanning networks were constructed manually for each species detected in clone libraries based on the methods of Excoffier and Smouse (1994).
Optimization and use of ARISA with environmental samples
ARISA profiles for environmental samples were generated by amplifying ITS1 fragments with PnAllF and the fluorescently labeled PnAllR primer, and the resulting fragment lengths were determined as described above. PCR cycle numbers and peak identification were optimized by testing a range of cycle numbers (20, 22, 24, 26, 28, 32, 34, 40, 45), signal-to-noise cutoff values (0, 3.6×, 12×, 30×), and percent area cutoff values (0.15%, 1%, 2%, 5%). At least one blank with nanopure water rather than DNA template was used to establish a baseline value for noise. The statistics package PRIMER (Clarke and Warwick 2001) was used to conduct nonparametric multivariate analyses on ARISA profiles for each environmental sample. Similarities between ARISA profiles were calculated by generating resemblance matrices based on the presence or absence of peaks. Cluster analysis was conducted on the matrices to evaluate the significance of PCR cycle number and reproducibility among triplicate PCRs, and SIMPROF was used to determine which clusters were statistically significant (P < 0.05).
RESULTS
Development of Pseudo-nitzschia-specific primers and identification of ITS1 fragment length polymorphisms
Full-length ITS1 sequence data and SEM results were congruent for 12 Pseudo-nitzschia isolates (identified in this study) that corresponded to four different species (Table S4). Both EM-based identification and ITS1 sequence were available for at least one representative of 19 Pseudo-nitzschia species (Table 2 and Tables S2, S3, and S4 in the supplementary material). Over half (90) of the 161 Pseudo-nitzschia ITS1 sequences in GenBank corresponded to P. delicatissima (Cleve) Heiden, 76 of which were from a single study (Orsini et al. 2004); only 13 of the 90 sequences were identified with SEM.
Primers PnAllF and PnAllR were designed to PCR amplify (without mismatches in either primer) a variable region from 11 of the 19 Pseudo-nitzschia species (106 of 161 sequences) for which ITS1 sequence was available (Table 2). The primers did not recognize the ITS1 of 12 other diatom genera examined (Table S5). Species of Pseudo-nitzschia not predicted to be amplified by PnAllF/R exhibited a total of between one and four mismatches to both primers (Tables S2 and S3), and the other diatom genera examined in silico exhibited more mismatches. Interestingly, none of the polar isolates of Pseudo-nitzschia was recognized by the primers.
Species predicted to be amplified by PnAllF/R are listed in Table 2, and represented Pseudo-nitzschia isolates originating from waters surrounding each continent except Antarctica. The primers recognized multiple isolates of a single species (e.g., P. pungens) originating from different oceans (e.g., Mexico, Portugal, Vietnam, the North Sea, and the U.S. Pacific Northwest). A subset of sequences corresponding to some P. delicatissima, P. seriata, and P. subpacifica (Hasle) Hasle isolates in GenBank were not specifically recognized by the primers. Based on in silico analyses, the PnAll primers did not recognize ribosomal RNA sequence of bacteria in the RDP database, nor protists in the GenBank nr/nt database, and no contiguous non-Pseudo-nitzschia sequences containing both PnAll primers were detected in the nr, wgs, and env_nt GenBank databases.
Twenty-five distinct ITS1 fragment lengths (142–231 bp) were predicted for the PnAll amplicons associated with the 11 Pseudo-nitzschia species recognized by the primers (Table 2). A single sequence each was available in GenBank, and therefore a single fragment length was predicted for three known species and one unidentified species (Table 2). Two species displayed a single fragment length for all isolates of that species (five and seven sequences, respectively, Table 2). Six species each displayed two ITS1 fragment lengths; different isolates of the same species that display different fragment lengths are here referred to as different species types. Some species types differed by a single in/del, while other species types consisted of insertions, deletions, and sequence polymorphisms. The species types of P. pungens, P. seriata, and P. micropora Priisholm, Moestrup et Lundholm exhibited <3% sequence divergence over the PnAll amplicon. Other species types exhibited greater levels of divergence: P. galaxiae Lundholm et Moestrup types (143 and 151 bp) differed by 16.4% (9.5% without in/dels), and P. cuspidata (Hasle) Hasle types (216 and 230 bp) differed by 13.3% (5.13% without in/dels).
The greatest ITS1 fragment length variation was observed in isolates identified as P. delicatissima. Thirteen different fragments lengths, ranging from 164 to 231 bp, were detected for the 76 P. delicatissima sequences predicted to be amplified by the PnAll primers (Table 2). Sequence variability between the different P. delicatissima types was similar to variability observed between different Pseudo-nitzschia species; small differences distinguished some types, and large differences in fragment lengths and sequence content distinguished others. Sequences of the ITS1 fragment from P. delicatissima types 1–5 (fragment lengths of 223, 222, 221, 220, and 219 bp, respectively), from types 7 and 8 (164 and 165 bp), and from types 10 and 13 (207 and 212 bp) were easily aligned, with sequence differences resulting in <3% sequence divergence between similar types. Sequence alignment of the ITS1 fragments was not possible across the different P. delicatissima sequence groups. For example, none of the 219–223 bp types could be aligned with any of the 164–168 bp types, and divergence among isolate sequences from the less easily aligned types was >13% (6.8% without in/dels).
Fragment lengths of 143, 151, and 230 bp were common to more than one species of Pseudo-nitzschia (Table 2). One P. cuspidata (AY257853) sequence was identical to the P. pseudodelicatissima (Hasle) Hasle (AY257854) sequence (230 bp), suggesting either that one isolate may have been misidentified or that these species share the same ITS1 sequence. A second isolate of P. cuspidata (AY257862) was also expected to generate a 230 bp amplicon but exhibited high sequence divergence (12.3% with in/dels, 5.1% without) from the first P. cuspidata isolate. For all species except P. pseudodelicatissima and P. cuspidata, shared fragment lengths were easily distinguished from one another based on sequence content, with divergence ranging from 12.3% to 59.3% (5.1%–50% without in/dels).
Primer specificity and identification of environmental clones
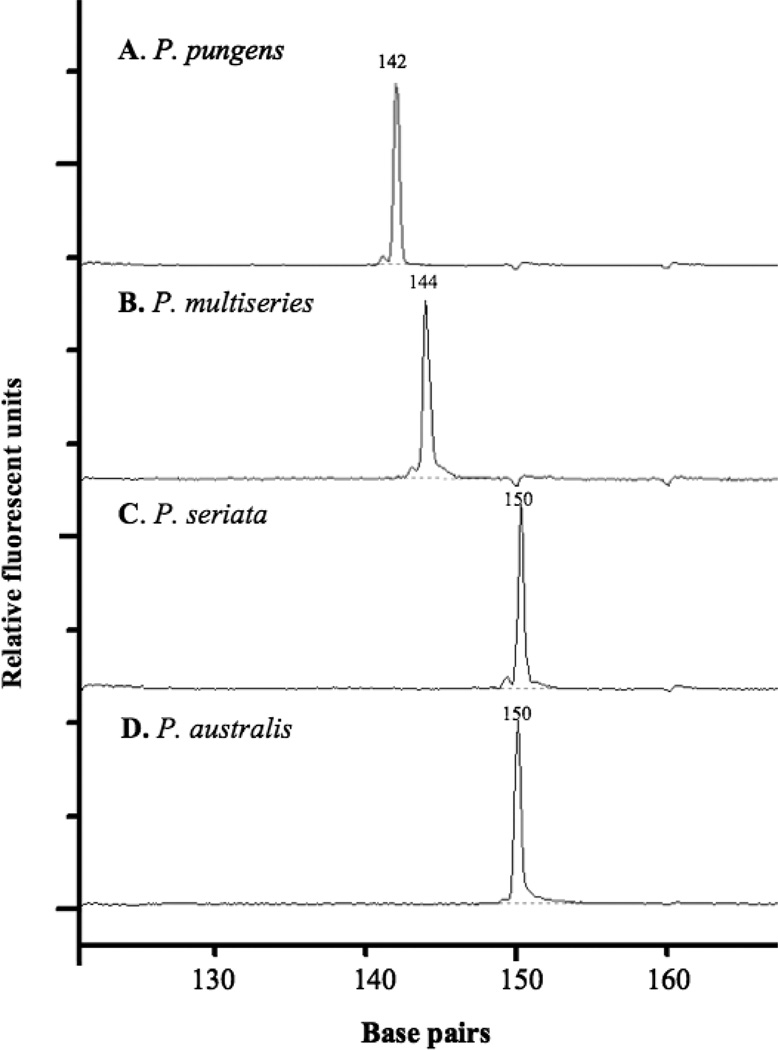
The PnAllF/R primers were tested experimentally, and a single fragment corresponding to the predicted size was detected for one isolate each of P. seriata, P. pungens, P. australis, and P. multiseries (Fig. 2). The P. pungens fragment of 142 bp was easily distinguished from the P. multiseries fragment of 144 bp, whereas fragment sizes for P. seriata type 1 and P. australis were both 150 bp (Fig. 2). None of the tested isolates was axenic, and yet only a single peak of the expected size was obtained for each Pseudo-nitzschia isolate, confirming that contaminant bacterial DNA was not amplified by the primers. No amplification was detected when Synechococcus (cyanobacterium), Phaeocystis (haptophyte), or Gyrodinium (dinoflagellate) DNA was used as template or when DNA from diatoms of genera other than Pseudo-nitzschia was used, with the single exception of the closely related polar diatom Fragilariopsis cylindrus, which produced a 213 bp amplicon and exhibited a 2 bp mismatch to the PnAllF primer (data not shown).
Fig. 2.
Automated rRNA intergenic spacer analysis (ARISA) profiles for Pseudo-nitzschia isolates: (A) P. pungens, (B) P. multiseries, (C) P. seriata, and (D) P. australis.
The specificity of the PnAll primers was also tested with DNA isolated from whole seawater samples collected from Tofino and Esperanza inlets on Vancouver Island, Canada, and Port Madison in Puget Sound, Washington. All environmental PnAll-generated ITS1 sequences displayed the greatest sequence similarity to known Pseudo-nitzschia ITS1 sequences rather than sequences from different diatom genera or bacteria.
A total of nine distinct fragment lengths were detected for sequences from the clone libraries (Table 3); eight fragment lengths were associated with nine known or putative Pseudo-nitzschia species (P. australis and P. seriata shared the same fragment length). Seven species were identified based both on predicted lengths (Table 2) and intraspecific sequence divergence of <4%: P. pungens (142 bp), P. multiseries (144 bp), P. australis (150 bp), P. seriata (150 bp), P. delicatissima (168 bp), and P. subpacifica (195 bp). Interestingly, the sequence of one ITS1 fragment displayed 1.2% divergence when compared to P. fraudulenta (Cleve) Hasle (202 bp), which had not been predicted to be amplified because of a single mismatch to the PnAllF primer. Three ITS1 fragments of 233, 209, and 196 bp were detected in the environmental clone library sequences but differed in both sequence and length compared to any known ITS1 sequence. These fragments were assumed to correspond to uncultivated Pseudo-nitzschia species or types and are referred to as P. sp. 233 (233 bp), P. sp. 209 (209 bp), and P. sp. 196 (196 bp). P. sp. 233 exhibited 1.5% divergence from P. pseudodelicatissima/P. cuspidata type 1 (230 bp), with the only sequence difference consisting of a 3 bp insertion. The ITS1 sequence for P. sp. 209 exhibited 9% divergence (6.7% without in/dels) when compared to P. decipiens Lundholm et Moestrup, which was not predicted to be amplified due to a single mismatch to PnallF. P. sp. 196 was highly divergent from all Pseudo-nitzschia sequences in GenBank and was >50% diverged from its closest BLAST match, P. pungens.
Table 3.
Comparison of intraspecific diversity in PnAll-generated sequences from clone libraries with sequences from known Pseudo-nitzschia species.
| Clone GenBank accession nos. | PnAll size (bp) w/primers (w/o primers) |
Puget sound | Tofino Inlet | Esperanza Inlet | DPNW | Closest blast match | DGB | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS | NG | DPS | NS | NG | DTI | NS | NG | DEI | |||||
| EF014838–58, EF014760–96 | 142 (107) | 21 | 5 | 0.0374 | – | – | – | 37 | 8 | 0.0280 | 0.0467 | P. pungens | 0.084 |
| EF014859–76, EF014879 | 144 (109) | 18 | 6 | 0.0183 | 1 | 1 | n/a | – | – | – | 0.0183 | P. multiseries | 0.0183 |
| EF014822–29 | 150 (115) | – | – | – | – | – | – | 8 | 5 | 0.0261 | 0.0261 | P. australis | 0.0174 |
| EF014809–13 | 150 (115) | – | – | – | – | – | – | 5 | 2 | 0.0087 | 0.0087 | P. seriata | 0.0261 |
| EF014894–95, EF014758–59 | 168 (133) | – | – | – | 2 | 1 | 0 | 2 | 0 | 0 | 0 | P. delicatissima | 0 |
| EF014814–21 | 195 (160) | – | – | – | – | – | – | 8 | 4 | 0.0313 | 0.0313 | P. subpacifica | 0.0313 |
| EF014837 | 196 (161) | 1 | 1 | n/a | – | – | – | – | – | – | n/a | P. pungensa | 0.640 |
| EF014830–36 | 202 (167) | – | – | – | – | – | – | 7 | 4 | 0.0120 | 0.0120 | P. fraudulenta | 0.0180 |
| EF014877–88 | 209 (174) | – | – | – | 2 | 2 | 0.0057 | – | – | – | 0.0057 | P. decipiens (DQ336156) | 0.0899 |
| EF014880–93, EF014797–808 | 233 (198) | – | – | – | 14 | 5 | 0.0101 | 12 | 7 | 0.0152 | 0.0152 | P. pseudodelicatissima (AY257854)/ | 0.0253 |
| P. cuspidata (AY257853) | |||||||||||||
NS, number of sequences; NG, number of genotypes; DPS, DTI, and DEI, total nucleotide divergence among clones within the respective clone libraries; DPNW, total nucleotide divergence among all Pacific Northwest clones; DGB, total nucleotide divergence between the closest blast match from GenBank and the most divergent Pacific North-west clone.
Bold text is used to identify clones that exhibit >50% divergence from any known Pseudo-nitzschia species but are still more closely related to Pseudo-nitzschia than other genera in GenBank.
Microdiversity in Pseudo-nitzschia species
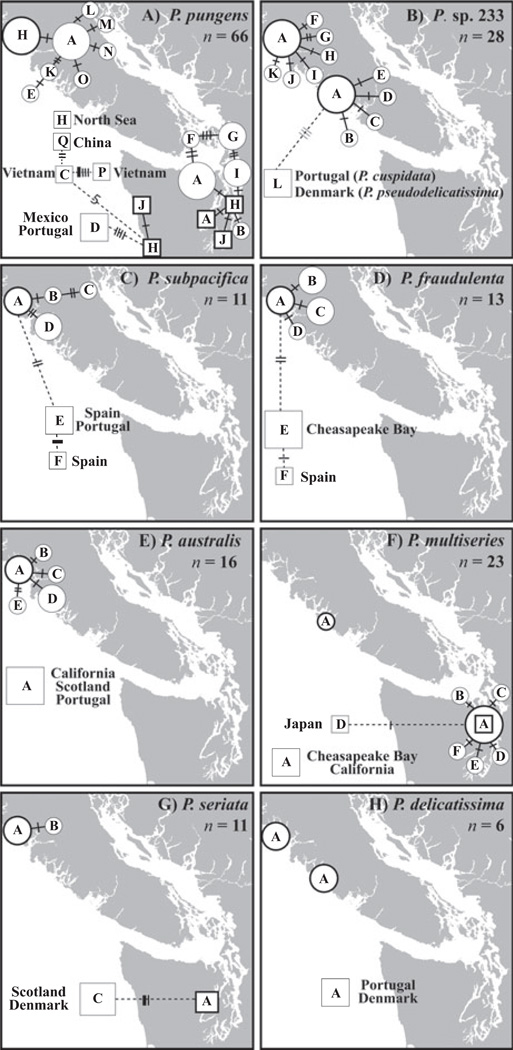
Each species detected in the clone libraries displayed a single fragment length variant, and all 18 clones of a single Pseudo-nitzschia pungens isolate (PNWH2O 101WB) were identical to the original ITS1 sequence obtained for that strain (GenBank accession DQ996020). Most sequence fragments detected more than once in the environmental clone libraries displayed sequence variation that consisted of polymorphisms at between one and five sites, here referred to as different genotypes. The only species type that did not display intraspecific variation was P. delicatissima type 11, as all four detected sequences were identical (Table 3). The most frequently detected species in the clone libraries, P. pungens, also had the greatest number of distinct genotypes (12) and the greatest nucleotide divergence among genotypes compared to other species, with high amounts of intraspecific diversity detected regardless of whether the P. pungens isolates originated from Puget Sound (3.74%) or from Esperanza Inlet (2.80%). High levels of intraspecific diversity were also detected for P. australis (2.61%) and for P. subpacifica (3.13%) despite the fact that far fewer sequences (eight each) were compared for these two species (Table 3). Two species, P. fraudulenta (1.20%) and P. sp. 233 (1.52%), each displayed a nucleotide divergence about half that of the more diverse species despite the fact that seven and 26 sequences, respectively, were generated for these species.
The overall pattern for most species was that a single genotype was most common numerically (i.e., number of clones sequenced) in clone libraries, and that other closely related genotypes were detected less frequently. Minimum spanning networks were employed to visualize the relations and distributions of the different genotypes for each species except P. sp. 196 (one recovered sequence) and P. sp. 209 (two recovered sequences). For example, a single genotype (genotype A) of P. pungens was the most commonly detected genotype in the clone libraries generated for the Esperanza Inlet and Puget Sound samples (Fig. 3a). Five additional P. pungens genotypes differed from genotype A by single nucleotide polymorphisms at different nucleotide positions and were detected less frequently; nine other genotypes that differed by an additional 1 or 2 bp were detected even more rarely (Fig. 3a). Similarly high levels of microdiversity were detected for P. sp. 233, with genotype A found at both Esperanza Inlet and Tofino Inlet, and 11 additional genotypes identified that differed from this genotype by only 1 or 2 bp (Fig. 3b). Less microdiversity was detected for P. subpacifica, P. fraudulenta, P. australis, P. multiseries, and P. seriata (Fig. 3c–g).
Fig. 3.
Network of polymorphic genotypes for eight Pseudo-nitzschia species/species types: (A) P. pungens; (B) P. sp. 233; (C) P. subpacifica; (D) P. fraudulenta; (E) P. australis; (F) P. multiseries; (G) P. seriata; and (H) P. delicatissima type 11. ITS sequences from clone libraries (represented by circles) generated in this study and from isolates from GenBank and this study (represented by squares) were used. The size of the circle or square represents the number of sequences; the smallest size represents a single sequence, the medium size represents two to four sequences, and the largest size represents five or more sequences. Unique genotypes are distinguished by letters located inside circles or squares; thickly outlined circles and squares, or “hubs,” are placed in the actual location where the sample or isolate originated. Solid branches are used to connect genotypes from the Pacific Northwest; dotted branches are used to connect genotypes from other regions. Crossbars represent differences between genotypes; thin bars represent deletions relative to the hub sequence, medium bars represent a polymorphism, and thick bars represent insertions relative to the hub sequence. ITS, internal transcribed spacer.
To further enhance this analysis, the genotypes of distantly distributed isolates available in GenBank were included in the networks. Remarkably, P. pungens genotype H (Fig. 3a), P. australis genotype A (Fig. 3e), P. multiseries genotype A (Fig. 3f), and P. delicatissima type 11 genotype A (Fig. 3h) were detected both in local and distant waters. Again, only a single, well-distributed genotype was detected for all sequences of P. delicatissima type 11, including isolates from the Pacific Northwest, Portugal, and Denmark (Fig. 3h). In contrast, some distant isolates of P. pungens were differentiated from Pacific Northwest populations; for example, genotype D was detected in both Mexico and Portugal, but not in any of the Pacific Northwest samples (Fig. 3a).
Community diversity of environmental samples detected with ARISA
The observation that sequence polymorphisms were more common than length polymorphisms for a given species suggested that the PnAll primers could be used for ARISA to identify different Pseudo-nitzschia species within environmental samples. Consistently and accurately identifying ARISA peaks was essential to developing this potentially high throughput approach. ITS1 fragments were therefore amplified from the same environmental samples used for the clone libraries with different PCR cycle numbers, and subsequent ARISA profiles were analyzed using a range of thresholds for peak calling (see Materials and Methods for details). To minimize variability, the results from individual replicates for a single sample were combined in silico. Samples that either failed (e.g., no fluorescent signal was detected) or contained <50% of the peaks detected in replicates were not considered for analysis. Cluster analysis of the resulting ARISA profiles indicated that the most consistent results (profiles were not significantly different from one another, P > 0.05) were obtained when a cycle number was used that corresponded to four or more additional cycles than that determined by Q-PCR as the early exponential phase of amplification. In addition, variability was minimized when peaks were detected using a signal-to-noise ratio of 30 and a peak threshold of 0.15% area.
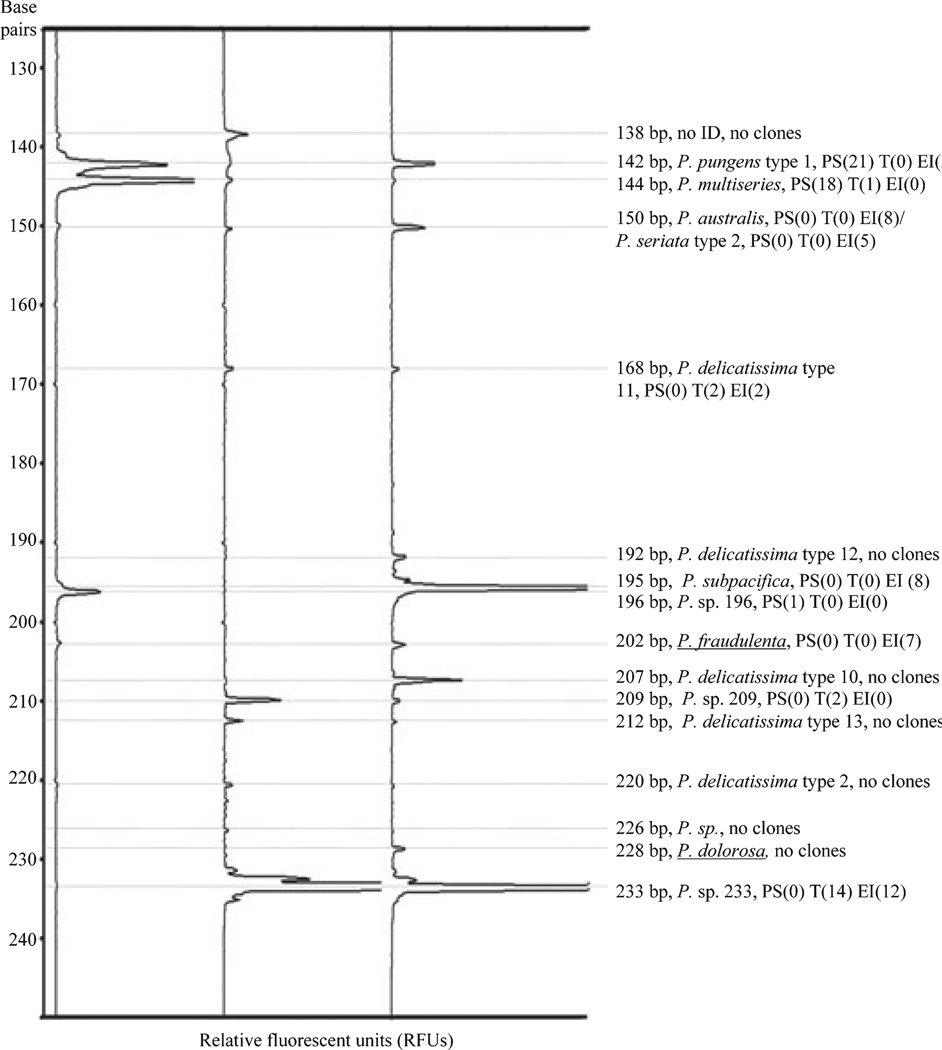
Multiple, distinct peaks were detected for each environmental sample (Fig. 4), and individual peaks, even those 1 bp apart, were easily distinguished from one another. The ARISA peaks corresponded to clone sequences for P. pungens type 1 (142 bp), P. multiseries (144 bp), P. australis/P. seriata type 2 (150 bp), P. delicatissima type 11 (168 bp), P. subpacifica (195 bp), P. sp. 196 (196 bp), P. fraudulenta (202 bp), P. sp. 209 (209 bp), and P. sp. 233 (233 bp). Seven peaks detected with ARISA (138, 192, 207, 212, 220, 226, and 228 bp) did not correspond to clone library sequences. However, five of these fragment lengths corresponded to species in GenBank—P. delicatissima type 12 (192 bp), P. delicatissima type 10 (207 bp), P. delicatissima type 13 (212 bp), P. delicatissima type 2 (220 bp), and P. sp. from Tasmania (226 bp)—and may correspond to species or types not predicted to be amplified by PnAll primers, such as one P. delicatissima type from the Gulf of Naples (226 bp) and P. dolorosa Lundholm et Moestrup (228 bp). The ARISA peak of 138 bp did not share a fragment length with any known Pseudo-nitzschia species.
Fig. 4.
Automated ribosomal intergenic spacer analysis (ARISA) profiles of environmental samples from the main basin of Puget Sound; outside Esperanza Inlet, Vancouver Island; and Tofino Inlet, Vancouver Island. Species identifications for peaks are listed to the right of the profiles, with the number of representative sequences detected in each library. Putative identifications for peaks were based on fragment length when no sequences of that fragment length were detected in the libraries. Underlined identifications represent peaks not predicted to be amplified with the PnAll primers.
Greater levels of interspecific diversity were detected with ARISA than with the clone libraries. Ten species or species types were detected by environmental sequencing of all three samples combined (Table 3). In contrast, 16 ARISA peaks were detected in all three environmental samples, suggesting that 16 or more putative Pseudo-nitzschia species or species types were present, given that some species may share a single peak. Only 19 clones (compared with 79 and 40 clones from Esperanza Inlet and Puget Sound, respectively) were sequenced from Tofino Inlet, because many of the peaks in that sample were identified in the Esperanza Inlet clone library. Three fragment lengths were detected in all three samples: 142 bp (P. pungens), 144 bp (P. multiseries), and 150 bp (P. australis/P. seriata). All other fragment lengths showed more restricted distributions (Fig. 4). For example, the 233 bp (P. sp. 233) fragment was found only in Vancouver Island stations.
The magnitude of an ARISA peak was not always representative of the number of clones detected in the corresponding clone library (Fig. 4). In the Esperanza Inlet sample, no clone sequences were obtained for ARISA fragment length 207 bp, despite the fact that the peak fluorescence signal was at least four times stronger than the peaks for 168 bp (P. delicatissima type 11) and 202 bp (P. fraudulenta; Fig. 4), both of which were represented in the clone libraries. As a corollary, the peak for 168 bp was relatively small in the Tofino Inlet and Esperanza Inlet samples, and yet sequences corresponding to P. delicatissima type 11 were detected in both samples.
DISCUSSION
PCR primers specific to the ITS1 region of about half the described Pseudo-nitzschia species were successfully designed and employed with field samples to study variation with the genus Pseudo-nitzschia. An ARISA for Pseudo-nitzschia was designed based on protocols for bacterial ARISA (Fisher and Triplett 1999, Hewson and Fuhrman 2004), and clone libraries were used to verify the identification of ARISA peaks. The ITS1 is a highly variable marker at both inter- and intraspecific levels for the Pseudo-nitzschia species, and ITS may be the best molecular marker thus far for elucidating Pseudo-nitzschia taxonomy (Manhart et al. 1995, Amato et al. 2007).
Intraspecific variation
ITS1 target sequences from EM-identified isolates fell into two categories distinguished by the presence or absence of insertions and deletions. When in/dels were excluded from analysis, sequence variation among isolates of a single species was relatively restricted and ranged from ~1% to 3%, although P. pungens displayed intraspecific variation as high as 8.4%. No within-strain sequence variability was detected in P. pungens PNWH2O 101WB over the course of 2.5 years in culture, suggesting that the detected variation in field samples is likely due to the presence of multiple strains rather than variability within a single strain. Thus, each unique sequence was treated as a unique genotype. Furthermore, potential PCR bias and error was minimized by terminating PCR prior to saturation and by using a reconditioning step (Thompson et al. 2002, Acinas et al. 2005).
Based on sequence comparisons, isolates with the same ITS1 fragment length and sequence divergence of <3% were defined as different genotypes of a single species. Isolates with different ITS1 fragment lengths but whose sequence identity (excluding in/dels) was also <3% were defined as different species types. For comparison, closely related species, such as P. australis and P. seriata, or P. pungens and P. multiseries, displayed interspecific divergence that ranged from 7.8% to 28.4%, respectively. These interspecific divergences should be considered lower estimates, as sequences of different species cannot easily be aligned.
The extent of variation detected within a single species appeared to be unrelated to geographical distance between isolation sites. Different species types could originate from geographically distinct locations or from the same sampling site. Amato et al. (2007) detected two ITS1 types for P. pseudodelicatissima in the Gulf of Naples, distinguished by a 13 bp insertion and 2.1% divergence (without in/dels). The two P. pseudodelicatissima species types were capable of reproduction, did not exhibit significant morphological variability, and thus satisfied the biological species concept (Amato et al. 2007). The available ITS1 sequence data from GenBank and results from this study and the interbreeding study (Amato et al. 2007) suggested that sequences exhibiting 3% or less variation for the PnAll sequence fragment were representative of a single species. This value is low compared to ITS1 variability within other pennate diatom species (see Vanormelingen et al. 2007), although the entire ITS1 region rather than just a fragment was considered for these other species.
Sequence divergence was used to determine the relationships of Pacific Northwest environmental ITS1 sequences to previously generated ITS1 sequences from described species; both species types and genotypes were represented in the environmental clone sequences. Variants of P. multiseries, P. australis, P. seriata, P. subpacifica, P. fraudulenta, P. delicatissima type 11, and P. sp. 233 most likely each represent a single species, since they exhibited less genetic variability than the P. pseudodelicatissima ITS1 types from the Gulf of Naples (Amato et al. 2007). Distinct genotypes and/or species types for P. sp. 233, P. subpacifica, P. fraudulenta, P. seriata, and P. sp. 209 were found only in the Pacific Northwest (Fig. 3, b–d, and g). Sequencing additional isolates and environmental DNA will be useful to determine if these Pseudo-nitzschia genotypes or species types are endemic to the Pacific Northwest, or if they will exhibit broader distribution ranges, like the genotypes of P. australis, P. multiseries, and P. delicatissima type 11 (Fig. 3, e, f, and h).
The term “pseudo-cryptic” has been used to describe highly similar species with genetic variability and distinct but overlapping morphological characteristics (Amato et al. 2007). Examples of apparently pseudo-cryptic species were also detected in the clone libraries, including P. sp. 209 (most similar to P. decipiens; 9% with in/dels, 6.7% without) and P. pungens. P. pungens exhibited higher apparent intraspecific variation than any other species (8.4%; no in/dels detected) suggesting that this morphotype may encompass at least two pseudo-cryptic species. A single fixed polymorphism split sequences into three groups, referred to here as P. pungens-A, P. pungens-T, and P. pungens-C for the nucleotide at position 77 of 142 bp. Only a single clone sequence of P. pungens-C was detected (genotype G, Fig. 3a). Other sequences were distributed between P. pungens-A (genotypes A, C–F, and J–Q; Fig. 3a) and P. pungens-T (genotypes B, H, and I; Fig. 3a). When intraspecific variation for these two groups was considered separately, the variation was <2% for the P. pungens-T (16 sequences) and <7.5% for P. pungens-A (46 sequences). Further support for the possibility of cryptic diversity in P. pungens comes from a recent microsatellite-based study documenting two distinct, coexisting populations of P. pungens in the Pacific Northwest (Adams 2006). No evidence for a pseudo-cryptic species of P. pungens was detected for North Sea isolates (Evans et al. 2005). Additional experiments should be conducted to test the species concept for P. pungens.
Interspecific variation
Environmental samples for ARISA and clone libraries were standardized based on volume of water filtered (500 mL) and by using Q-PCR to determine the optimal PCR cycle number, to minimize PCR error and to account for differences in Pseudo-nitzschia concentrations among environmental samples. Pseudo-nitzschia ARISA peaks ranged from 138 to 233 bp, had 1 bp resolution, and were appropriately sized when compared to clone sequences. Precision in bacterial ARISA tends to decrease as amplicon lengths increase, but amplicons <700 bp (Brown et al. 2005) are generally called precisely; thus, it was not surprising that Pseudo-nitzschia peaks were straightforward to identify in ARISA profiles. The amplification efficiencies associated with each Pseudo-nitzschia species in a mixed sample are likely variable, especially if some species (like P. fraudulenta from the Esperanza Inlet sample) have mismatches to the primer set. For this reason, and because ARISA peak strength and the number of clones detected for a species were not always correlated, we did not attempt to correlate abundance with either ARISA peak strength or frequency of clones detected. The number of species detected with ARISA was greater than the number detected with clone libraries for each environmental sample; this may reflect differences between techniques (e.g., cycle numbers for ARISA, reconditioning for clone libraries, potential artifacts) or between a fingerprinting community approach like ARISA and sequencing a discrete number of clones.
It was remarkable that 13 ARISA peaks were detected in the Esperanza Inlet, Vancouver Island, sample, as only seven species were detected in a multiyear survey of the Washington coast (Stehr et al. 2002). Several of these ARISA types remain to be characterized by additional sequencing. All sequences in the environmental clone libraries were most similar to Pseudo-nitzschia sequences in Gen- Bank, further confirming the specificity of the PnAll primers for these environmental samples. The amplification of Fragilariopsis cylindrus isolate DNA with the PnAll primer set and the identification of species variants with distinct fragment lengths in this study highlight the importance of generating clone libraries to identify ARISA peaks prior to beginning a study in a new region.
The genus Pseudo-nitzschia displays a “continuum of diversity” (Soininen 2006), including genotypes (intraspecific variation), species types (intraspecific variation), cryptic and pseudo-cryptic species (intra- or interspecific variation), species, and the genus itself. The combination of ARISA with sequences from isolates and environmental clones allows resolution across this continuum of diversity. The process of isolation and culturing might select for certain variants, which in combination with the misidentification of genetic variants, likely biases the current view of worldwide distributions. ARISA is a robust way to identify Pseudo-nitzschia species and species types in the environment and may be applicable to other similarly cryptic genera of phytoplankton. The ability to process many samples easily and quickly allows for rapid identification of species in samples, which is necessary for routine monitoring of toxigenic algae, and the ability to identify genotypes may be important for understanding why and under what conditions this diatom blooms and produces domoic acid.
Supplementary Material
Acknowledgments
We thank Brian Bill, Katherine Evans, and Jason Smith for providing Pseudo-nitzschia cultures, and Ben Gilmore and Ellen Ostlund Lin for helping with DNA clone libraries. Leon Delwiche generously collected samples from Vancouver Island. We also thank Julie Koester, Adrian Marchetti, Michele Wrabel, and Micaela Parker for helpful comments and discussions. This work is supported by the Pacific Northwest Center for Human Health and Ocean Sciences (National Institute of Environmental Health: P50 ES012762 and National Science Foundation: OCE-0434087) and by a Gordon and Betty Moore Foundation Marine Microbiology Investigator Award to E. V. Armbrust.
Abbreviations
- ARISA
automated ribosomal intergenic spacer analysis
- bp
base pairs
- in/del
insertion/deletion
- ITS1
internal transcribed spacer 1
Footnotes
Supplementary Material
The following supplementary material is available for this article:
Table S1. Morphometric characters of Pacific Northwest Pseudo-nitzschia isolates.
Table S2. Sequence mismatches to PnAllF for Pseudo-nitzschia species not predicted to be amplified (in silico) by PnAll primer set.
Table S3. Sequence mismatches to PnAllR for Pseudo-nitzschia species not predicted to be amplified (in silico) by PnAll primer set.
Table S4. Summary of Pacific Northwest and North Sea Pseudo-nitzschia isolates used to generate ITS1 sequences.
Table S5. List of diatoms used as negative controls for in silico design of Pseudo-nitzschia-specific PCR primers.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1529-8817.2008.00518.x.
(This link will take you to the article abstract.)
Please note: Blackwell Publishing are not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. PCR-induced sequence artifacts and bias: insights form comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams NG. Master’s thesis. Seattle: University of Washington; 2006. Genetic differentiation of Pseudo-nitzschia pungens from the Pacific Northwest and the North Sea; p. 112. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amato A, Kooistra WHCF, Ghiron HL, Mann DG, Proschold T, Montresor M. Reproductive isolation among sympatric cryptic species in marine diatoms. Protist. 2007;158:193–207. doi: 10.1016/j.protis.2006.10.001. [DOI] [PubMed] [Google Scholar]
- de Baar HJW, Boyd PW, Coale KH, Landry MR, Tsuda A, Assmy P, Bakker DCE, et al. Synthesis of iron fertilization experiments: from the iron age in the age of enlightenment. J. Geophys. Res. Oceans. 2005;110:1–24. [Google Scholar]
- Bates SS. Ecophysiology and metabolism of ASP toxin production. In: Anderson DM, Cembella AD, Hallegraeff GM, editors. Physiological Ecology of Harmful Algal Blooms. Heidelberg, Germany: Springer-Verlag; 1998. pp. 405–426. [Google Scholar]
- Baugh KA, Bush JM, Bill BD, Lefebvre KA, Trainer VL. Estimates of specific toxicity in several Pseudo-nitzschia species from the Washington coast, based on culture and field studies. Afr. J. Mar. Sci. 2006;28:403–407. [Google Scholar]
- Bill BD. Domoic acid in Pseudo-nitzschia cuspidata from Washington State coastal waters. Third Symposium on Harmful Algae in the U.S; Monterey Bay; California. 2005. (Abstract) [Google Scholar]
- Bill BD, Cox FH, Horner RA, Borchert JA, Trainer VL. The first closure of shellfish harvesting due to domoic acid in Puget Sound, Washington, USA. Afr. J. Mar. Sci. 2006;28:437–442. [Google Scholar]
- Bornet B, Antoine E, Francoise S, Marcaillou-Le C. Development of sequence characterized amplified region markers from intersimple sequence repeat fingerprints for the molecular detection of toxic phytoplankton Alexandrium catenella (Dinophyceae) and Pseudo-nitzschia pseudodelicatissima (Bacillariophyceae) from French coastal waters. J. Phycol. 2005;41:704–711. [Google Scholar]
- Brown MV, Schwalbach MS, Hewson I, Fuhrman JA. Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ. Microbiol. 2005;7:1466–1479. doi: 10.1111/j.1462-2920.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho ES, Hur HJ, Byun HS, Lee SG, Rhodes LL, Jeong CS, Park JG. Monthly monitoring of domoic acid producer Pseudo-nitzschia multiseries (Hasle) Hasle using species-specific DNA probes and WGA lectins and abundance of Pseudo-nitzschia species (Bacillariophyceae) from Chinhae Bay, Korea. Bot. Mar. 2002;45:364–372. [Google Scholar]
- Clarke KR, Warwick RM. Changes in Marine Communities: An Approach to Statistical Analysis and Interpretation. 2nd ed. Plymouth, UK: PRIMER-E; 2001. p. 172. [Google Scholar]
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KM, Bates SS, Medlin LK, Hayes PK. Microsatellite marker development and genetic variation in the toxic marine diatom Pseudo-nitzschia multiseries (Bacillariophyceae) J. Phycol. 2004;40:911–920. [Google Scholar]
- Evans KM, Hayes PK. Microsatellite markers for the cosmopolitan marine diatom Pseudo-nitzschia pungens. Mol. Ecol. Notes. 2004;4:125–126. [Google Scholar]
- Evans KM, Kühn SF, Hayes PK. High levels of genetic diversity and low levels of genetic differentiation in North Sea Pseudo-nitzschia pungens (Bacillariophyceae) populations. J. Phycol. 2005;41:506–514. [Google Scholar]
- Excoffier L, Smouse PE. Using allele frequencies and geographic subdivision to reconstruct gene trees within a species – molecular variance parsimony. Genetics. 1994;136:343–359. doi: 10.1093/genetics/136.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MM, Triplett EW. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 1999;65:4630–4636. doi: 10.1128/aem.65.10.4630-4636.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of Marine Invertebrate Animals. New York: Plenum Press; 1975. pp. 26–60. [Google Scholar]
- Hasle GR. Pseudo-nitzschia pungens and P. multiseries (Bacillariophyceae) – nomenclatural history, morphology, and distribution. J. Phycol. 1995;31:428–435. [Google Scholar]
- Hasle GR, Fryxell GA. Diatoms – cleaning and mounting for light and electron microscopy. Trans. Am. Microsc. Soc. 1970;89:469–474. [Google Scholar]
- Hasle GR, Lundholm N. Pseudo-nitzschia seriata f. obtusa (Bacillariophyceae) raised in rank based on morphological, phylogenetic and distributional data. Phycologia. 2005;44:608–619. [Google Scholar]
- Hewson I, Fuhrman JA. Richness and diversity of bacterioplankton species along an estuarine gradient in Moreton Bay, Australia. Appl. Environ. Microbiol. 2004;70:3425–3433. doi: 10.1128/AEM.70.6.3425-3433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner RA, Hanson L, Hatfield CL, Newton JA. Domoic acid in Hood Canal, Washington, USA. In: Yasumoto T, Oshima Y, Fukuyo Y, editors. Harmful and Toxic Algal Blooms. Paris: Intergovernmental Oceanographic Commission of UNESCO; 1996. pp. 127–129. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhu K, Buchner A, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundholm N, Hasle GR, Fryxell GA, Hargraves PE. Morphology, phylogeny and taxonomy of species within the Pseudo-nitzschia americana complex (Bacillariophyceae) with descriptions of two new species, Pseudo-nitzschia brasiliana and Pseudo-nitzschia linea. Phycologia. 2002;41:480–497. [Google Scholar]
- Lundholm N, Moestrup Ø, Hasle GR, Hoef-Emden K. A study of the Pseudo-nitzschia pseudodelicatissima/cuspidata complex (Bacillariophyceae): what is P. pseudodelicatissima? J. Phycol. 2003;39:797–813. [Google Scholar]
- Lundholm N, Moestrup Ø, Kotaki Y, Hoef-Emden K, Scholin C, Miller P. Inter- and intraspecific variation of the Pseudo-nitzschia delicatissima complex (Bacillariophyceae) illustrated by rRNA probes, morphological data and phylogenetic analyses. J. Phycol. 2006;42:464–481. [Google Scholar]
- Manhart JR, Fryxell GA, Villac MC, Segura LY. Pseudo-nitzschia pungens and P. multiseries (Bacillariophyceae): nuclear ribosomal DNA’s and species differences. J. Phycol. 1995;31:421–427. [Google Scholar]
- Mann DG, Droop SJM. Biodiversity, biogeography and conservation of diatoms. Hydrobiologia. 1996;336:19–32. [Google Scholar]
- Marchetti A, Lundholm N, Kotaki Y, Hubbard K, Harrison PJ, Armbrust EV. Identification and assessment of domoic acid production in oceanic Pseudo-nitzschia (Bacillariophyceae) from iron-limited waters in the northeast subarctic Pacific. J. Phycol. 2008;44 doi: 10.1111/j.1529-8817.2008.00526.x. (in press) [DOI] [PubMed] [Google Scholar]
- Miller PE, Scholin CA. Identification of cultured Pseudonitzschia (Bacillariophyceae) using species-specific LSU rRNA-targeted fluorescent probes. J. Phycol. 1996;32:646–655. [Google Scholar]
- Orsini L, Procaccini G, Sarno D, Montresor M. Multiple rDNA ITS-types within the diatom Pseudo-nitzschia delicatissima (Bacillariophyceae) and their relative abundances across a spring bloom in the Gulf of Naples. Mar. Ecol. Prog. Ser. 2004;271:87–98. [Google Scholar]
- Page RDM, Holmes EC. Molecular Evolution: A Phylogenetic Approach. Oxford, UK: Blackwell Science; 1998. p. 359. [Google Scholar]
- Parsons ML, Scholin CA, Miller PE, Doucette GJ, Powell CL, Fryxell GA, Dortch Q, Soniat TM. Pseudonitzschia species (Bacillariophyceae) in Louisiana coastal waters: molecular probe field trials, genetic variability, and domoic acid analyses. J. Phycol. 1999;35:1368–1378. [Google Scholar]
- Soininen J. Local and regional coexistence of diatoms – on the mechanisms promoting high local diatom species richness. Diatom Res. 2006;21:217–223. [Google Scholar]
- Stehr CM, Connell L, Baugh KA, Bill BD, Adams NG, Trainer VL. Morphological, toxicological, and genetic differences among Pseudo-nitzschia (Bacillariophyceae) species in inland embayments and outer coastal waters of Washington state, USA. J. Phycol. 2002;38:55–65. [Google Scholar]
- Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainer VL, Hickey BM, Horner RA. Biological and physical dynamics of domoic acid production off the Washington coast. Limnol. Oceanogr. 2002;47:1438–1446. [Google Scholar]
- Vanormelingen P, Chepurnov VA, Mann DG, Cousin S, Vyverman W. Congruence of morphological, reproductive and ITS rDNA sequence data in some Australasian Eunotia bilunaris (Bacillariophyta) Eur. J. Phycol. 2007;42:61–79. [Google Scholar]
- Villac MC, Fryxell GA. Pseudo-nitzschia pungens var. cingulata var. nov. (Bacillariophyceae) based on field and culture observations. Phycologia. 1998;37:269–274. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.