Abstract
Microarrays do not yield direct evidence for functional connections between genes. However, transcription factors (TFs) and their binding sites (TFBSs) in promoters are important for inducing and coordinating changes in RNA levels and thus represent the first layer of functional interaction. Similar to genes TFs act only in context, which is why a TF/TFBS-based promoter analysis of genes needs to be done in the form of gene(TF)-gene networks, not individual TFs or TFBSs. In addition, integration of literature and various databases (e.g. GO, MeSH, etc) allows adding genes relevant for the functional context of the data even if they were initially missed by the microarray as their RNA levels did not change significantly. Here we outline a TF-TFBSs network-based strategy to assess involvement of transcription factors in agonist signaling and demonstrate its utility in deciphering the response of human microvascular endothelial cells (HMEC-1) to leukemia inhibitory factor (LIF). Our strategy identified a central core of eight TFs, of which only STAT3 had previously been definitively linked to LIF in endothelial cells. We also found potential molecular mechanisms of gene regulation in HMEC-1 upon stimulation with LIF that allows for the prediction of changes of genes not used in the analysis. Our approach, which is readily applicable to a wide variety of expression microarray and next generation sequencing RNA-seq results, illustrates the power of a TF-gene networking approach for elucidation of the underlying biology.
Keywords: Microarray data analysis, high-throughput (HT) approaches, transcription factor-gene networking, transcription factor binding sites, transcription factors
Introduction
Microarrays record a snapshot of transcriptional changes caused by the administration of drugs or agonists to cells and define all changes, as far as the genome is covered by the microarray design, regardless of whether they have relevance to the functional actions of the drug or agonist (1). They provide long lists of genes that show changes in steady-state RNA levels, but do not yield direct evidence for functional connections between genes and miss even important genes if their steady state RNA levels are not significantly changed. However, as recently demonstrated by results of the ENCODE project (2), functional interactions of genes depend on a variety of functional genomic elements with transcription factors (TFs) and their binding sites (TFBSs) in promoters and enhancers being important for inducing and coordinating changes in RNA levels. Moreover, multiple databases and the scientific literature provide huge amounts of functional information on genes and their interactions including TFs. Therefore, an approach based on elucidation of TF/TFBS interactions (i.e. networks) by promoter analysis of genes with significantly changed transcripts is very well suited to elucidate functional connections between significantly changed genes in microarray data sets that might be missed in any individual gene or factor oriented analysis.
Attempts to include additional data frequently make use of pathways, GeneOntology (GO)-terms, or molecular features such as transcription factor binding sites (TFBSs) in the vicinity of genes, e.g., an approach focusing on transcriptional regulation by transcription factor binding was recently described (3). However, with the exception of pathways all these approaches just produce more lists still missing a structured biological context. Another clear-cut lesson from ENCODE as well as many previous smaller scale studies is that neither genes nor TFs or their corresponding TFBSs act in isolation, but are highly interconnected usually in the form of gene-gene networks. Biological functionality only becomes apparent on the network level (pathways representing small networks themselves). Moreover, integration of additional functional connections as taken from the literature and various databases (e.g. GO, MeSH, etc.) allows for inclusion of genes relevant to functional context of the data even if they were initially missed as their RNA levels do not change significantly. An integrative approach has the additional advantage to compensate for the intrinsic weaknesses of individual methods; enrichment analyses are necessarily biased by uneven distribution of knowledge, co-citation literature networks face the same challenge and in addition inevitably contain variable numbers of false positive connections. However, by bringing several lines of evidence together outliers due to erroneous results of one method are readily identified and discarded. This rationale is based on “biological consistency”, i.e. every finding in one area of analysis must be reflected in the results of other lines of evidence also in order to be accepted as real.
We developed a widely applicable strategy entirely focusing on TF and TFBSs-centered networks complemented by literature and knowledge mining for expression profiling. Other approaches report as the final results GO-terms, pathways, associated TFBSs. These are only “stepping stones” in our strictly context/network-oriented approach. One of the most important principles of this strategy is to complement findings from expression data with conclusions drawn from our network approaches (biological consistency between data and knowledge based analyses). We applied this strategy to elucidate the potential involvement of transcription factors in the regulation of genes in response to leukemia inhibitory factor (LIF) in human microvascular endothelial cells (HMEC-1). We were able to identify a central core of eight TFs based on multiple lines of evidence, most likely involved in the regulatory network of LIF-induced gene expression changes in HMEC-1 cells, although initially almost 100 TFs showed significant expression changes (one line of evidence). We also found potential molecular mechanisms of gene regulation in HMEC-1 cells upon stimulation with LIF that allowed prediction of changes of genes observed on the microarray but not used in the analysis. This demonstrates clearly the power of a TF-gene networking approach for elucidation of the underlying biology. Our approach is widely applicable to high-throughput analyses of transcriptional changes such as all expression microarrays as well as all pertinent next generation sequencing (NGS) applications (ChIP-seq, RNA-seq, bisulfite-resequencing), where the capability to reduce the amount of data to a biologically-linked small network is especially important.
Materials and Methods
Materials
Cell culture reagents were obtained from Invitrogen (Carlsbad, CA, USA). Epidermal growth factor was from BD Biosciences (Franklin Lakes, NJ USA), hydrocortisone from Sigma-Aldrich (St. Louis, MO, USA), recombinant human LIF from Millipore (Billerica, MA, USA), and fetal bovine serum (SH30070.03) from Thermo Fisher Scientific (Waltham, MA, USA).
Experimental design
HMEC-1 cells were obtained from the Centers for Disease Control and Prevention (CDC), and grown in MCDB 131 with 15% fetal bovine serum (FBS), 10 ng/mL epidermal growth factor, 1 μg/mL hydrocortisone, 10 mM glutamine, and antibiotic-antimycotic. Cells were grown in 100 mm dishes to near confluency and incubated in medium with 0.5% FBS 12–15 hours before being used in experiments. Cells were dosed with vehicle or 2 ng/mL LIF for 90 min at 37°C, placed on ice, and washed 2x with 10 mL ice cold Hanks’ buffered saline solution.
Microarray analysis
RNA was isolated using the RNAquous-4PCR Kit from Applied Biosystems (Foster City, CA, USA). RNA quality was established using the NanoDrop 3300 Fluorospectrometer (Thermo Scientific) and Agilent 2100 Bioanalyser. Only samples with a 260/280 ratio close to 2 and RNA Integrity Number (RIN) value > 9 were processed for microarray analysis. Microarray processing was performed by the core facility of the University Of Mississippi School Of Medicine using Agilent technology and whole human genome slides. Cy3 and Cy5 dye swap and background correction were applied. Genes were considered downregulated with treatment to control ratios < 0.5 and upregulated with treatment to control ratios > 2. Image processing was done using ImaGene (version 8.0.1) and statistical analysis performed using the R statistical program (version 2.10.1). Array signals for 6 replicates (channel median values) were calculated by first subtracting the local background mean followed by normalization using loess (within array) and quantile (between arrays) algorithms. P values for differential expression were determined using the R/Bioconductor package limma, which incorporates both Bayesian and linear modeling methods and is routinely used in microarray data analyses (4). In the calculation of signal values for each probe there was a subtraction of the local background, which is the recommended procedure to remove bias (e.g., one array or part of an array was not washed as well after hybridization). This is thought to represent somewhat of a trade-off with reduced bias and lower variability for highly expressed genes but with higher variability for genes with low expression. For that reason, we used an unadjusted p-value < 0.05 as significance threshold. Annotation for the probe sets on the array was obtained from the Gene Expression Omnibus (GEO) at the NCBI using accession number GPL4133 and from the Agilent internet site (http://www.chem.agilent.com/cag/bsp/gene_lists.asp).
Regulatory network analysis
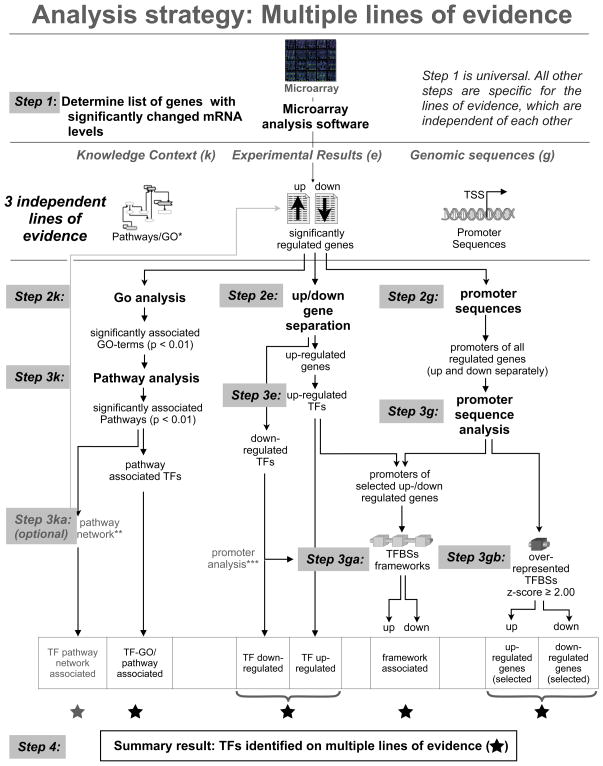
Figure 1 summarizes the strategies used for the analysis of the significantly regulated genes. We separated up-and down-regulated genes by GO and pathway-analysis in order to find TFs specifically associated with up or down-regulation. The whole strategy is a combination of five results originating from three independent lines of evidence: a) mRNA values and their relative changes, b) literature and pathway analysis, c) sequence-based promoter analysis (Figure 1 top “lines of evidence”). The only experiment-specific data used were the list of significantly regulated genes and their expression values. We posed our main focus onto the analysis of TF genes and their potential targets in order to understand the transcriptional effects of LIF treatment.
Figure 1. Analysis strategy and summary of results.
The upper part of the figure indicates the three major lines of evidence used in the subsequent analysis. Three parallel threads of analysis were carried out from the associated lines of evidence, each using results from all lines of evidence to focus and restrict the next analysis step. This is indicated by the cross-connections. The whole strategy focused onto transcription factors (TFs) throughout and collected all positive evidence for involvement of a TF. *In case no pathways are available GO categories can be used in the same way. **The pathway network essentially produces a reduced initial list which can be treated exactly the same way as the initial list.***Promoter analysis for the down-regulated genes is carried out exactly as for the up-regulated, if a down-regulated TF is thought to be responsible for the down-regulation.
Analysis downstream of the significant microarray signals was carried out using the standard integrated analysis package Genomatix Software Suite (Genomatix Software GmbH, Munich, Germany) and the various databases and software tools within this package, including all of the following: Gene-ontology (GO)-analysis was carried out by the program GeneRanker using default parameters recommended by the supplier. All literature-based analyses were carried out using the Genomatix Pathways System (GePS), which combines co-citation analysis from the whole PubMed database with canonical pathway analysis. GePS was used with the default parameters recommended by the supplier. Promoters used for TFBSs analysis were all extracted from the ElDorado genome database (Release 12/2010) using the program Gene2Promoter. The various promoter collections were then analyzed using the program RegionMiner, which contains precompiled databases of TFBSs match numbers for whole genomes and whole-genome promoter collections, and for which over-representations and p-values are automatically calculated. We refer to whole-genome promoter collections as the relevant background throughout this study.
Promoter context is defined as sets of TFBSs that show a specific organization within sequences: The individual TFBSs (e.g. TFBSs A, B, and C) and their relative order is conserved (A-B-C only, A-C-B rejected), and a flexible but limited distance range is allowed between the individual TFBSs, and which also must have a conserved strand-orientation. In this way a complete framework of three TFBSs would have the annotation A(+) - distance range 1 - B(−) - distance range 2 - C(+) where + and − symbolize the strand orientation of the individual TFBSs. Such a framework needs to be found conserved in a minimum number of sequences (sequence quorum) which can be set as a user parameter. Throughout this study we used the following parameters: Minimum number of TFBSs in a framework 3, variation of distance range 20 (in case no results were found, this was increased to 30), minimum distance 10, maximum distance 200 (between TFBSs). The sequence quorum was set high initially (no results) and then stepwise reduced till frameworks of three elements were found or the minimum quorum was reached without finding frameworks. For each search single TFBSs identified as important in previous analyses were set as mandatory elements and all frameworks found with the described settings were collected as framework sets and the sets were the evaluated.
Evaluation for association of the frameworks with the respective promoter sets was carried out using the program ModelInspector as follows: Each set was searched for matches in the promoters of the specific set the frameworks were derived from, various larger subsets from the significantly regulated genes (such as network genes, 3-and higher up-regulated genes, etc.). This was compared to match results obtained either from all microarray-derived promoters or all promoters from the human genome (automatically carried out by ModelInspector). The over-representation of the framework sets in the specific promoter sets as compared to random sampling of the genome was calculated. These are the results shown in the tables. For more detailed description of the methodologies see (5).
Results
Differentially expressed genes
Steady-state mRNA levels of HMEC-1 cells were analyzed by microarray assays for genes significantly changing in mRNA level in response to LIF treatment. LIF-treated cells were compared to untreated control cells. Microarray files were analyzed as described in Methods using the Bioconductor package limma in order to find the significantly regulated genes. We found a total of 1,171 genes significantly regulated between the LIF-treated cells and the control: 589 genes were up-regulated and 582 genes were found to be down-regulated. Out of the 1,171 genes 1,107 were annotated allowing Gene Ontology (GO) and pathway analysis, which were the first steps in our data analysis.
GO-term and pathway analysis
We had a total of 368 GO-terms from the significantly (p-value ≤ e−03) associated biological processes. Table 1 shows the top ten GO-terms according to their p-value. There is a clear preference for kinase-cascade signaling in GO-terms, which is a hallmark of multiple signal transduction pathways. Therefore, we went on to pathway analysis as the third step using the GenomatixPathwaySystem (GePS, Genomatix Software, Munich) database/tool. Table 2 shows the six pathways that were significantly associated with the 1,107 regulated (and annotated) genes. Again, JAK-STAT regulation is evident (IL7 signaling pathway). However, several other signaling pathways are also found. There were 7 transcription factor families, i.e., TFs that are very similar and bind to the same motifs, directly implicated by the six pathways (AP1, ETS, STAT, HNF, CREB, CEBP, DDIT3). This step concluded the analysis of the knowledge–based GO- and pathway-based line of evidence.
Table 1.
Top 10 GO categories “biological process” associated with genes differentially regulated by LIF treatment
| GO process | GO-ID | p-value | Go | Ge | Gt |
|---|---|---|---|---|---|
| enzyme linked receptor protein signaling pathway | GO:0007167 | 8.58e-08 | 57 | 27.22 | 472 |
| transmembrane receptor protein tyrosine kinase signaling pathway | GO:0007169 | 1.56e-07 | 41 | 17.07 | 296 |
| prostate gland growth | GO:0060736 | 2.13e-07 | 7 | 0.58 | 10 |
| phosphate metabolic process | GO:0006796 | 3.98e-07 | 117 | 74.45 | 1291 |
| phosphorus metabolic process | GO:0006793 | 3.98e-07 | 117 | 74.45 | 1291 |
| MAPKKK cascade | GO:0000165 | 4.31e-07 | 39 | 16.44 | 285 |
| phosphorylation | GO:0016310 | 7.32e-07 | 104 | 64.82 | 1124 |
| regulation of cellular component movement | GO:0051270 | 8.72e-07 | 34 | 13.73 | 238 |
| regulation of MAPKKK cascade | GO:0043408 | 9.07e-07 | 26 | 8.99 | 156 |
| regulation of phosphorus metabolic process | GO:0051174 | 9.46e-07 | 59 | 30.68 | 532 |
All associated GO-processes were ranked by their p-value. Go = Number of genes observed in the significantly regulated genes belonging to the respective biological process, Ge = Number of genes expected in the significantly regulated genes belonging to the respective biological process by a random pick of the same size, Gt = total number of genes belonging to the respective biological process.
Table 2.
Six pathways associated with the differentially regulated genes
| Pathway | p-value | input genes in pathway | gene IDs |
|---|---|---|---|
| PDGFR-alpha signaling pathway | 1.39E-03 | ITGAV, IFNG, SHF, JUN, CSNK2A1, PDGFRA, CAV1 | 3685, 3458, 90525, 3725, 1457, 5156, 857 |
| pertussis toxin-insensitive ccr5 signaling in macrophage | 2.42E-03 | CCL2, CCR5, JUN, CXCL12 | 6347, 1234, 3725, 6387 |
| E-cadherin signaling events | 5.25E-03 | EPHA2, EXOC3, AKT1, HGF, IGF1, IGF1R, EFNA1 | 1969, 11336, 207, 3082, 3479, 3480, 1942 |
| IL-7 signaling pathway(JAK1 JAK3 STAT5) | 6.74E-03 | IL7, RIPK3, AKT1, SYK, ZAP70, MAPK13, KIT, BRAF, LCK, FGFR2, IRAK4, PRKCD, PIK3CD, FLT4, IGF1R, PAK2, CSNK1A1, CAMK2G, AKT2, PDGFRA, MAP3K2, ITK | 3574, 11035, 207, 6850, 7535, 5603, 3815, 673, 3932, 2263, 51135, 5580, 5293, 2324, 3480, 5062, 1452, 818, 208, 5156, 10746, 3702 |
| ATF-2 transcription factor network | 6.94E-03 | IFNG, POU2F1, SOCS3, JUN, CCND1, DUSP8, PDGFRA, BCL2, NOS2 | 3458, 5451, 9021, 3725, 595, 1850, 5156, 596, 4843 |
| TCR signaling in naive CD4+ T cells | 8.93E-03 | VAV1, AKT1, ZAP70, LAT, FYB, LCK, LCP2, PTPRC, DBNL, PTEN, ITK | 7409, 207, 7535, 27040, 2533, 3932, 3937, 5788, 28988, 5728, 3702 |
All associated pathways were ranked by their p-value as determined by the program GePS/GeneRanker (Genomatix Software, Munich). Input genes in pathways: these genes were part of the list of regulated genes as well as the pathway.
TF-regulation analysis
This is another line of evidence independent from the literature-based analyses shown above, except for the literature-derived TF-gene annotation. The only common starting point is the list of significantly changed genes. GePS is also able to identify genes for TFs and we used this feature to evaluate the number of TF genes that show altered expression. We found 50 TF genes to be up-regulated among the 1,107 genes and 45 TF genes down-regulated. Merging results from pathway and TF-regulation analysis showed that from the pathway-associated TF genes ETS and CEBP factors were up-regulated, while AP1 and Jun (a CREB family factor) were down-regulated, yielding a total of 4 differentially expressed TFs so far supported by two lines of evidence (expression data and pathway analysis). However, as many more TFs were regulated we also looked for additional evidence for association of these factors with regulated genes. This step concluded the analysis of the konwledge–based lines of evidence.
Statistical promoter analysis for TFBSs
Sequence-based analyses have the advantage to be largely independent of the above mentioned heavily knowledge-dependent methods. The genomic sequence (and thus the promoters) is universal, entirely independent of literature and the detection of TFBSs is based on sequence patterns derived by sequence analysis. The only part where knowledge comes into play is the completeness of the library, i.e. TF identification. TFs may act directly or indirectly on genes and some may change transcriptional activity without any apparent change in their own mRNA levels. In order to estimate direct regulation by TFs we decided to look at the other end of TF-mediated transcriptional regulation namely the TFBSs in the promoters of differentially regulated genes. If any particular TF is directly involved in regulation of a set of genes, then those genes should contain at least one TFBS for such TFs. Thus TFBSs for factors prominently involved in mediating transcriptional signaling might be statistically enriched in the regulated promoters. Lack of overrepresentation does not preclude a functional connection but a positive result is an additional evidence for inclusion. We extracted all 5,371 promoters associated with the 1,107 regulated genes using the Gene2Promoter tool (Genomatix Software GmbH, Munich) and analyzed them for statistical overrepresentation of TFBSs from the MatBase Matrix Family Library (Version 8.3, Genomatix Software GmbH, Munich). A total of 53 TFBSs families were found to be overrepresented (as compared to a random sampling from all promoters in the human genome, using a cutoff threshold of a z-score of 2.00), 47 TFBSs families were in those promoters that were upregulated and 6 TFBSs families were associated with up-regulated TF genes (HOMF (HMX1), FKHD (FOXD1), BCDF (OTX1), CEBP (CEBPD), IRFF (IRF1, IRF8), DMRT (DMRTB1).
In promoters from down-regulated genes 35 TFBSs were found to be significantly associated, 6 of which were also associated with down-regulated TF genes FKHD (FoxP4, FOXJ2), PARF (HLF), VTBP (TBP), NKXH (NKX2-2, NKX2-3), HOXF (HOXD8), OCT1 (POU2F1). It became evident that different factors belonging to the same TF family (e.g. forkhead, FKHD) and their respective TFBSs were associated with up- and down-regulated genes. It also became evident that 8 transcription factor families showed up in at least 2 of 3 analyses (Table 3). Of the 3 that were not associated with a differentially expressed TF gene (STAT, HOMF, HOXF) only STAT was directly associated with one of the six associated pathways as well as being co-cited with LIF in the context of vascular endothelium (6), resulting in a short list of 6 TFs: FKHD, IRF, OCT1, CEBP, BCDF, and STAT (Table 3).
Table 3.
TFs prominently associated with significantly regulated genes
| TFBS family | TF/up (+) or down (−) regulated | pathway association | z-score all regulated promoters | z-score up-regulated promoters | z-score down-regulated promoters |
|---|---|---|---|---|---|
| OCT1 | POUF2 + | + | 5.5 | 5.07 | 2.55 |
| FKHD | FOXD1 + FOXP4 − FOXJ2 − |
− | 7.4 | 5.31 | 4.92 |
| IRF | IRF1 + IRF8 + |
− | 5.59 | 3.32 | 4.9 |
| CEBP | CEBPD + | + | 3.65 | 4.67 | - |
| BCDF | OTX1 + | − | 3.18 | 4.85 | - |
| STAT | - | + | 3.59 | 3.57 | - |
| HOMF | - | − | 8.03 | 7.3 | 4.33 |
| HOXF | - | − | 5.75 | 5.64 | 2.63 |
Column 1 shows the TFBS family of which the individual TFs shown in column 2 are members. Column 3 indicates whether the TF was directly implicated by an associated pathway and columns 4 to 6 indicate the statistical over-representation of the respective TFBS family as compared to all promoters in the human genome. Only factors that show at positive values in at least three columns are shown.
So far the selection was based on a combination of classical analyses essentially focusing on individual TFs. Next we focused on functional connections between TFs not necessarily restricted to these 8 TFs in Table 3 but using them as a starting set.
Promoter context analysis of TFBSs (frameworks)
Presence of TFBSs is a physical phenomenon while the organization of TFBSs into clearly defined groups (frameworks) is connected to transcriptional function. Thus frameworks establish another line of evidence on top of the TFBSs presence. Thus we extended our analysis to find such TFBSs networks in regulated promoters. Table 3 shows three forkhead factors one of which was up-regulated transcriptionally (FOXD1) while two (FOXP4 and FOXJ2) were down-regulated. As all three factors are able to bind to the same FKHD binding sites (MatBase, Matrix Family Library Version 8.3, Genomatix Software GmbH) this suggests that the transcription factors most likely act in different contexts with other factors. Such context can be specifically addressed and elucidated by promoter analysis for conserved TFBSs frameworks (strand, order and distance correlated sets of TFBSs) (5). However, as there are 2,744 promoters associated with the up-regulated genes (Gene2Promoter, Genomatix Software GmbH, Munich) systematic analysis of all up-regulated promoters could not be carried out due to technical limitations of the software (limit is 1000 promoters due to the combinatorial explosion of possible TFBSs combinations). Therefore, we decided to select the subset of 764 promoters of three-fold or more up-regulated genes.
We analyzed these 764 promoters for frameworks of at least three TFBSs (essentially representing regulatory networks with one molecular mechanism), where one of TFBS was mandatory (exhaustively for all six TFBSs families corresponding to the six most important TFs identified in this study). Table 4 summarizes the results of these context searches. Most framework sets show a modest association with the selected promoter set (Z-score cutoff 2.00, promoters of three-fold or more up-regulated genes) except for one FKHD-group (3.13) and the STAT-group, which has the highest association (> 8 fold overrepresented). However, none show an association with all regulated microarray promoters (the STAT group being borderline with 2.03). However, restriction to one model that contained also a second associated TFBS (CEBP) resulted in more selective results (Table 4, last row). Interestingly, the two TFBSs families HOMF and HOXF originally found but discarded based on few lines of evidence, showed up numerous times in context of the significant factors. Thus, all six previously selected TFs, OCT1, FKHD, IRF, CEBP, BCDF, and STAT were also supported by associated TFBSs framework context (3-fold or more up-regulated promoters).
Table 4.
Framework analysis of the six associated TFBSs families
| Framework set mandatory TFBSs in bold | 3 and more upregulated promoters (764) | all microarray promoters (5371) | all genome promoters (82703) | 3 up overrepresentation | all microarray overrepresentation |
|---|---|---|---|---|---|
| DMRT-HOMF-OCT1 | 36 | 153 | 1990 | 1.96 | 1.2 |
| PDX1-OCT1-HOXF MYT-OCT1-HOXF |
39 | 129 | 1655 | 2.55 | 1.2 |
| CDXF-HOMF-FKHD | 25 | 76 | 870 | 3.13 | 1.26 |
| IRFF-HOMF-BRNF | 23 | 84 | 897 | 2.80 | 1.44 |
| X-CEBP-FKHD-X | 144 | 531 | 6316 | 2.46 | 1.29 |
| BCFD-OCT-FKHD | 119 | 457 | 5855 | 2.33 | 1.26 |
| STAT-set | 36 | 60 | 454 | 8.78 | 2.03 |
| CEBP-BRNF-STAT | 9 | 11 (allup) | 120 | na | 2.76 |
Column 1 shows the main TFBSs determining the Framework sets as automatically determined by FrameWorker (Genomatix Software GmbH, Munich). Columns 2 to 4 show the number of promoters matched by the whole sets of frameworks in the 3 respective promoter collections, and columns 5 and 6 show the respective over-representation with respect to all human promoters
Functional context analysis (TFBSs-frameworks) already linked several TFBSs even when based only on a statistical selection (≥ 3-fold up regulated). Therefore, we expected an approach based on a subset based on biologically linked genes to confirm the results and maybe be even more successful.
The following analysis is currently only possible using the Genomatix solution, which is commercial. However, as also indicated in figure 1 this analysis is optional and essentially supports the findings achieved without it, albeit in a much faster time with much less interactive steps.
Pathway network analysis
We used another selection method that is more biology-oriented. Based on the initially associated pathways and the regulated genes the new pathway-network tool determines a subset of genes that link those pathways into a network with optimal co-citation connectivity, i.e. the network of genes has the highest number of co-citation based edges (normalized for gene count). This is motivated by best-knowledge based biological connections bypassing any fold-change based criteria and should be more biologically correlated to LIF action than the 3-fold or higher sub-section as expression values represent only one of three selection criteria (pathways, co-citations, and expression changes). The network method is entirely data-driven, and requires no more input than the complete list of all regulated genes (Hahn et al in preparation). A network of 335 genes was defined (as detailed in Methods) by this method 190 of which were up-regulated, connecting all six significantly associated pathways into one network. We then applied the exact same strategy as for the unselected and the 3-fold-up-regulated genes to the analysis of the network-selected genes.
GO-term analysis comparison
All together the network was significantly associated with 988 GO-terms (as compared to 368 for all regulated genes). Table 5 shows that several GO/Medical Subject Heading (MeSH) terms significantly associated with both gene groups (all regulated and network-selected genes) show a dramatic lower p-value in the network genes than in all regulated genes suggesting a sharper focus on the corresponding biology by the network selection.
Table 5.
GO/MeSH term comparison all regulated genes/network genes
| GO-term | p-value 1107 regulated genes | p-value 335 network genes |
|---|---|---|
| top ranked GO term | e-8 | e-29 |
| MapKKK cascade | e-7 | 1.32 e-15 |
| signal transmission via phosphorylation event | 1.11 e-6 | 2.80 e-19 |
| inflammation (MeSH disease) | 1.93 e-11 | 2.75 e-64 |
Selected GO-processes were compared by their p-value. All 3 selected individual GO-terms (rows 2 to 4) showed a much lower p-value for the network association than for all of the regulated genes.
Pathway analysis
The 190 up-regulated genes of the network were significantly associated with 10 pathways (Table 6). These 10 pathways are related/overlap as can be seen from the fact that there were six genes shared by 5 out of 10 pathways (SOCS3 ZAP70, ITK, PDGFRA, PRKCD, SYK). Promoter modeling of this set of 6 genes most common to the 10 pathways revealed also a strong association with STAT and FKHD TFBSs (data not shown).
Table 6.
Ten pathways associated with the 190 up-regulated genes contained in the network
| Pathway | p-value | input genes in pathway |
|---|---|---|
| Cytokine receptor degradation signaling | 2.84E-04 | ILA1, MAP3K2, IRAK4, IL4R, AKT2, IGF1R, FLT4, ITK, IL7, IL1B, PDGFRA, IFNG, SOCS3, BRAF, PRLR, PRKCD, SYK, FGFR2, ZAP70 |
| IL-7 signaling pathway(JAK1 JAK3 STAT5) | 5.82E-04 | MAP3K2, IRAK4, PIK3CD, AKT2, IGFR1, FLT4, ITK, IL7, PDGFRA, BRAF, PRKCD, SYK, FGFR2, ZAP70 |
| pertussis toxin-insensitive ccr5 signaling in macrophage | 1.86E-03 | CCL2, CCR5, JUN, CXCL12 |
| AKT(PKB)-Bad signaling | 1.95E-03 | MAP3K2, IRAK4, PIK3CD, AKT2, IGFR1, FLT4, ITK, PDGFRA, BRAF, PRKCD, SYK, FGFR2, TZAP70 |
| Migration | 2.15E-03 | MAP3K2, IRAK4, PIK3CD, AKT2, IGFR1, FLT4, ITK, PDGFRA, BRAF, PRKCD, SYK, FGFR2, ZAP70 |
| ATF-2 transcription factor network | 2.89E-03 | DUSP8, I BCL2, NOS2, PDGFRA, IFNG, SOCS3 |
| Signaling events mediated by PTP1B | 3.92E-03 | ITGB3, LAT, LYN, SOCS3, PRLR, CSF1R |
| IL23-mediated signaling events | 4.23E-03 | CCL2, NOS2, IL1B, IFNG, SOCS3 |
| Class I PI3K signaling events | 8.96E-03 | ITK, LYN, VAV1, SYK, ZAP70 |
| IL-6-mediated signaling events | 9.85E-03 | CEBPD, IRF1, VAV1, SOCS3, PRKCD |
All associated pathways were ranked by their p-value as determined by the program GePS/GeneRanker (Genomatix Software, Munich). Input genes in pathways: these genes were part of the list of regulated genes as well as the pathway.
Statistical promoter analysis for TFBSs
The 190 up-regulated genes in the network were associated with 18 TFBSs (data not shown) and although there were only 49 down-regulated genes, they were associated with 17 TFBSs (data not shown). As shown in Table 7 the network analysis so far identified 8 TFs supported by at least 2 of four lines of evidence (TF mRNA regulation, network pathway association, TFBSs association with up and/or down-regulated network promoters). Notably, there is an overlap of 5 factors (in bold) already identified by the same approach in all regulated genes. Joining all lines of evidence including the network analysis, all together a list of 8 TFs emerged, confirming the initially detected OCT1 and adding SP1 to the list (Table 8).
Table 7.
7 associated with genes in the network of LIF-associated pathways
| TFBS family | TF/up + or down − regulated | network pathway association | z-score up-regulated network promoters | z-score down-regulated network promoters |
|---|---|---|---|---|
| SP1 | KLF11 + | + | 3.89 | 2.97 |
| CEBP | CEBPD + | + | 2.79 | - |
| FKHD | FOXD1 + FOXP4 − FOXJ2 − |
+ | 2.38 | - |
| IRF | IRF1 + IRF8 + |
− | - | 2.54 |
| STAT | - | + | 2.54 | - |
| ETS | SPI1 + PBRM1 + |
- | - | |
| ZBP | ZNF219 + | − | 5.22 | - |
| BCDF | OTX1 + | − | 2.00 | - |
Column 1 shows the TFBS family of which the individual TFs shown in column 2 are members of. Column 3 indicates whether the TF was directly implicated by an associated pathway, and columns 4 and 5 indicate the statistical over-representation of the respective TFBS family in network promoters as compared to all promoters in the human genome.
Table 8.
Final results: Core set of TFs involved in response to LIF
| TF | Matrix family | pathway network | associated Pathway | TF regulated (+)/(−) | TF Framework associated | TFBS over-represented (+)/(−) |
|---|---|---|---|---|---|---|
| FOXD1 | FKHD | + | − | + | − | |
| FOXP4 - FOXJ2 | FKHD | + | + | + | + | |
| STAT 1/3/4/5a | STAT | + | + | + | + | |
| CEBPD | CEBP | + | + | + | + | |
| SP1 | + | + | + | + | ||
| IRF1 IRF8 |
IRF | + | + | + | − | |
| CREB | + | + | + | |||
| POU2F1 | OCT1 | − | + | − | ||
| OTX1 | BCDF | + | + | + |
The table summarizes the results from five analyses (pathway network, pathway association, TF gene up/down regulation, framework association, and TFBSs overrepresentation in promoters of up/down-regulated genes) derived from three independent lines of evidence: generic knowledge databases, experimental measurements, and promoter sequence analysis. Final selection was made with a cutoff of 3/5, i.e. only factors supported by at least 3 of the five analyses are shown. (+) and (−) indicate association with up (+) or down (−) regulated genes and are for the purpose of sum scores treated as equivalent.
Promoter context analysis of TFBSs (frameworks)
An analogous approach as described for the 3-fold or more up-regulated promoters based on network-derived up-regulated promoters yielded framework sets that also were associated with the up-regulated network promoters as well as with the 3 and more up-regulated promoters (data not shown).
TFBS-frameworks in promoters are associated with transcriptional regulation of the corresponding genes and can be located by computational search in promoters of genes not involved in the detection of those frameworks. Hence, they are also suitable to predict transcriptional up-regulation for genes that contain such frameworks in their promoters.
Framework-predicted gene regulation is confirmed by microarray data
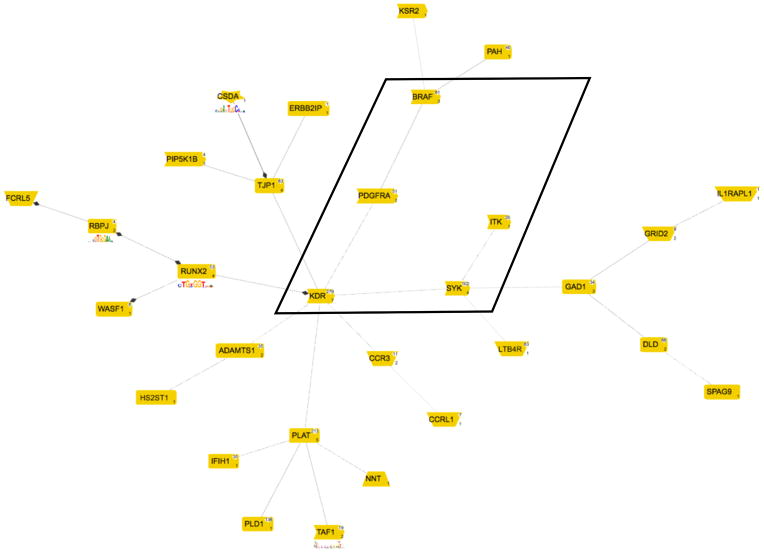
We selected the FKHD-CREB-SORY framework (defined from promoters of ITK, PDGFRA, SYK) as it is associating 2 relevant TFBSs (FKHD and CREB) with the central genes of the gene-interaction network-derived pathways. All promoters of up-regulated genes on the whole microarray were searched with this framework. Any matching promoter is supposed to be associated with an up-regulated transcript, which in turn can be verified using the microarray data for these genes. It is important to note, that none of these microarray results have been used at any time to generate the framework, which makes them independent data. The framework was overrepresented in the promoters of the up-regulated genes on the microarray (6.41 fold enriched) matching just 11 promoters (Table 9). The only down-regulated gene was skipped as it was not annotated and thus not suitable for further evaluation. We then used GePS to construct a co-citation linked network from the 206 genome.-wide matches. A central AREA connected five genes including the three input genes and consisting of: ITK-SYK-KDR (Vascular endothelial growth factor receptor 2 VEGFR2)-PDGFRA-BRAF (Figure 2). BRAF was also associated with 4 of the 10 network-up-regulated genes associated pathways.
Table 9.
FKHD-CREB-SORY containing promoters are all up-regulated with one exception.
| all microarray promoters (5371) | all upregulated promoters (764) | all genome promoters (101233) | all microarray promoters | all upregulated promoters |
|---|---|---|---|---|
| matches | matches | matches | overrepresentation | overrepresentation |
| 11 | 10 | 206 | 1.01 | 6.41 |
Overrepresentation analysis was carried out in the same way as for the data in table 4.
Figure 2. Literature-derived co-citation network based on the 206 genes selected by genome-wide search for the FKHD-CREB-SORY promoter TFBS-framework.
This network represents the largest contiguous network detectable in the set of 206 genes. The central area containing the framework-founding genes ITK, SYK and PDGFRA is boxed.
Discussion
We applied a predominantly data-driven and strictly network-focused strategy to the analysis of microarray data - in our case HMEC-1 cells treated with LIF. Several attempts have already been published employing more data-driven strategies, such as identification of co-expression of transcription factors and their putative target genes (7), which worked best in yeast. A more recent approach aimed at the identification of functionally coordinated TF-clusters also in human and Arabidopsis microarray data (8). These and many other approaches are truly data-driven analyses but focus on expression data only while our approach was designed to include as many sources of information as possible in a data-driven and network-focused analysis. Even the simplest analysis of the ENCODE data as published recently in Nature (Nature 489, 2012) provided overwhelming evidence how strongly network-oriented gene regulation is.
The actions of LIF include several mRNA independent steps such as kinase cascades which can never be observed directly in microarray data (6). However, we were not only able to identify STAT as a central factor in LIF action solely by data analysis but could also determine a short list of eight TFs most of which were not known to be important for LIF action (Table 8). IRF8, STAT3, SP1, IRF from that list are significantly associated with myeloid leukemia (p = 1.21e−10) yielding further support for the validity of the TF selection.
The most compelling part of the regulatory network-oriented analyses is the ability to actually predict RNA changes of other genes not used in the definition of the TFBSs frameworks defining regulatory networks. We ran the prediction using a network-associated framework containing two of the best associated TF/TFBSs (FKHD-CREB-SORY), found 11 promoters of genes interrogated on the microarray and 10 of these matched the prediction derived from the framework analysis. At this point verification by other experimental methods such as RT-PCR, NGS or the like would be required to turn most likely candidates into verified transcriptional regulators or transcriptional targets (by ChIP-seq, ChIP-on-chip, siRNA or vector-driven over-expression approaches), but this is clearly beyond the scope of this study that focused on strategies for the computational data analysis. However, supporting evidence can also be collected from existing knowledge: Four of the core TFs are part of the androgen receptor pathway (STAT3, SP1, POU2F1, and ATF2) and three are part of the IL-6 and the c-Myc signaling pathways respectively (STAT3, CEPBD, IRF1, and CEBPD, IRF8, SP1). This may allow selective inhibition of such pathway-oriented downstream reactions, which might even enable differentiating inflammatory responses from others such as angiogenesis.
Our strategy focused early onto TFs, their TFBSs and the potential functional network-context by combining knowledge-based measures (GO-terms, pathways, co-citations) with experimental data (expression changes) and genomics-based sequence analysis (TFBSs and promoters) as outlined before (9). The almost perfect agreement of framework-derived predictions with the actual microarray readings on genes is another line of supporting evidence. We used specific prior knowledge solely to judge our results not to generate them, e.g. we used the knowledge about STAT and SOCS3 involvement to qualify our results as valid but both factors were identified without explicit use of this knowledge.
A TF involved in the regulation should bind to its target genes and would naturally act together with other factors in this context, which is modeled by the framework approach (5, 10). Each line of evidence basically provides quantitative results of some kind (scores, expression values etc.). But it is almost impossible to normalize knowledge-based (11) and genomics based data in any way that would allow a quantitative comparison. Therefore, we count a line of evidence as supportive (i.e. associated significantly with the data) or not without any internal ranking or order. This safeguarded against the bias of “more” evidence (e.g. from literature) available for particularly popular factors and premature filtering. For example, STAT factors turned out to be among the most important TFs in the end despite the fact, that we did not observe a significant mRNA regulation in the microarray data as STAT is finally activated by phosphorylation even if transcriptionally upregulated (12). The collection of multiple lines of evidence made the results robust with respect to missing lines of evidence as long as enough lines remained supportive.
We have successfully used a highly systematic network-focused approach, which can be applied to almost all high-throughput data sets such as microarrays, NGS-based experiments (e.g. RNA-Seq, and ChIP-seq) as well as protein-interaction maps with very few adaptations. The general process contains steps with quantitative limitation requiring some pre-selections by the scientist, that cannot always be strictly motivated from the data, as in our case the selection of 3-fold or higher induced genes. Here, a best guess approach is required, but it is possible to test a few alternatives. This is one of the reasons why we also used a novel pathway-network oriented approach that does not suffer from such limitations and essentially confirmed results obtained on the arbitrarily selected gene subset. The network tool can take an unlimited number of pathways and genes and always results in a single network, optimal in terms of co-citation based connectivity. The biggest advantage is that the network is constructed in a fully automatic process within less than a minute requiring no user-defined parameters. The results appeared to be more focused on the LIF-relevant biology as indicated by the much lower p-values of pertinent GO-terms. SOCS3 featured prominently as a central gene in the network associated pathways, and is already known to be involved in the actions of LIF (13). All in all, we hope that this strategy can contribute another building block for standardized data analysis of experimental high-throughput methods aiming at rapid selection of subsets of data relevant for the experimental question.
Supplementary Material
Acknowledgments
We would like to thank Ruth Brack-Werner for helpful comments and discussion on this manuscript. This work was supported by grants from the National Heart, Lung, and Blood Institute to G. W. Booz (R01HL088101-06 and R01HL088101-02S1) and by grant 01EX1021L (M4 Personalized Medicine Ring funding project) to Genomatix (TW and SD)
References
- 1.Altman RB, Raychaudhuri S. Whole-genome expression analysis: challenges beyond clustering. Curr Opin Struct Biol. 2001;11:340–347. doi: 10.1016/s0959-440x(00)00212-8. [DOI] [PubMed] [Google Scholar]
- 2.Dunham I, Kundaje A, Aldred SF, et al. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrera LO, Ren B. The transcriptional regulatory code of eukaryotic cells--insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr Opin Cell Biol. 2006;18:291–298. doi: 10.1016/j.ceb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 5.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 6.Kubota Y, Hirashima M, Kishi K, Stewart CL, Suda T. Leukemia inhibitory factor regulates microvessel density by modulating oxygen-dependent VEGF expression in mice. J Clin Invest. 2008;118:2393–2403. doi: 10.1172/JCI34882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Z, Pilpel Y, Church GM. Computational identification of transcription factor binding sites via a transcription-factor-centric clustering (TFCC) algorithm. J Mol Biol. 2002;318:71–81. doi: 10.1016/S0022-2836(02)00026-8. [DOI] [PubMed] [Google Scholar]
- 8.Nie J, Stewart R, Ruan F, Thomson JA, Zhang H, Cui X, Wei H. TF-Cluster: A Pipeline For Identifying Functionally Coordinated Transcription Factors Via Network Decomposition of the Shared Coexpression Connectivity Matrix (SCCM) BMC Syst Biol. 2011;5:53. doi: 10.1186/1752-0509-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werner T. Bioinformatics applications for pathway analysis of microarray data. Curr Opin Biotechnol. 2008;19:50–54. doi: 10.1016/j.copbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Werner T, Fessele S, Maier H, Nelson PJ. Computer modeling of promoter organization as a tool to study transcriptional coregulation. Faseb J. 2003;17:1228–1237. doi: 10.1096/fj.02-0955rev. [DOI] [PubMed] [Google Scholar]
- 11.Scherf M, Epple A, Werner T. The next generation of literature analysis: integration of genomic analysis into text mining. Brief Bioinform. 2005;6:287–297. doi: 10.1093/bib/6.3.287. [DOI] [PubMed] [Google Scholar]
- 12.Kurdi M, Booz GW. JAK redux: a second look at the regulation and role of JAKs in the heart. Am J Physiol Heart Circ Physiol. 2009;297:H1545–H1556. doi: 10.1152/ajpheart.00032.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrai A, Boyle K, Hart AH, Hartley L, Rakar S, Willson TA, Simpson KM, Roberts AW, Alexander WS, Voss AK, Robb L. Absence of suppressor of cytokine signalling 3 reduces self-renewal and promotes differentiation in murine embryonic stem cells. Stem Cells. 2006;24:604–614. doi: 10.1634/stemcells.2005-0323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.