Abstract
DNA transfer across membranes is a fundamental life process. The structure of part of a protein channel that performs this task offers insight into the mechanism of DNA passage through bacterial cell envelopes.
Many bacterial species transfer DNA to neighbouring cells in a process called conjugation. Over evolutionary time, conjugation has played a dominant part in shaping bacterial genomes. On a more immediate timescale, conjugation poses an enormous public-health problem as a mechanism underlying the rapid dissemination of antibiotic-resistance genes and other virulence traits among pathogens. The question of how bacteria move DNA from one cell to the next has been investigated for more than 50 years, but only recently have structural details emerged. On page 1011 of this issue, a group led by Gabriel Waksman1 reports the first crystal structure of a large portion of a conjugation ‘machine’. The researchers’ findings represent a great leap in our understanding of how these transfer systems function at the molecular level.
Conjugation systems are a subfamily of type IV secretion (T4S) systems, whose range of functions includes DNA exchange with the environment and delivery of DNA and protein effectors to bacterial, fungal, plant and human target cells2. A few T4S systems in Gram-negative bacteria have been extensively studied, and one of these, the VirB/VirD4 T4S system from Agrobacterium tumefaciens, serves as a point of reference for this superfamily of translocation systems. In A. tumefaciens, 11 VirB sub units assemble together with a VirD4 subunit to form two structures: a gated channel for substrate transfer and a conjugative pilus (a hairlike appendage) for establishing contact with target cells2.
During the past decade, Waksman and colleagues have solved the structures of several subunits of T4S systems3–8, adding crucial details to an emerging picture of channel architecture. Earlier this year, these investigators presented a structure9 of a portion of a conjugation channel, known as the core complex, from Escherichia coli, solved at a resolution of about 15 Å using cryo-electron microscopy (cryo-EM). The structure showed that 14 copies of the complex’s TraN, TraO and TraF subunits — which are functionally equivalent to VirB7, VirB9 and VirB10 in A. tumefaciens — assemble as a large chamber, 185 Å wide and high, that spans the bacterial cell envelope (Fig. 1). The authors designated the portions of the chamber that are close to the inner and outer membranes of the envelope as inner (I) and outer (O) layers, respectively.
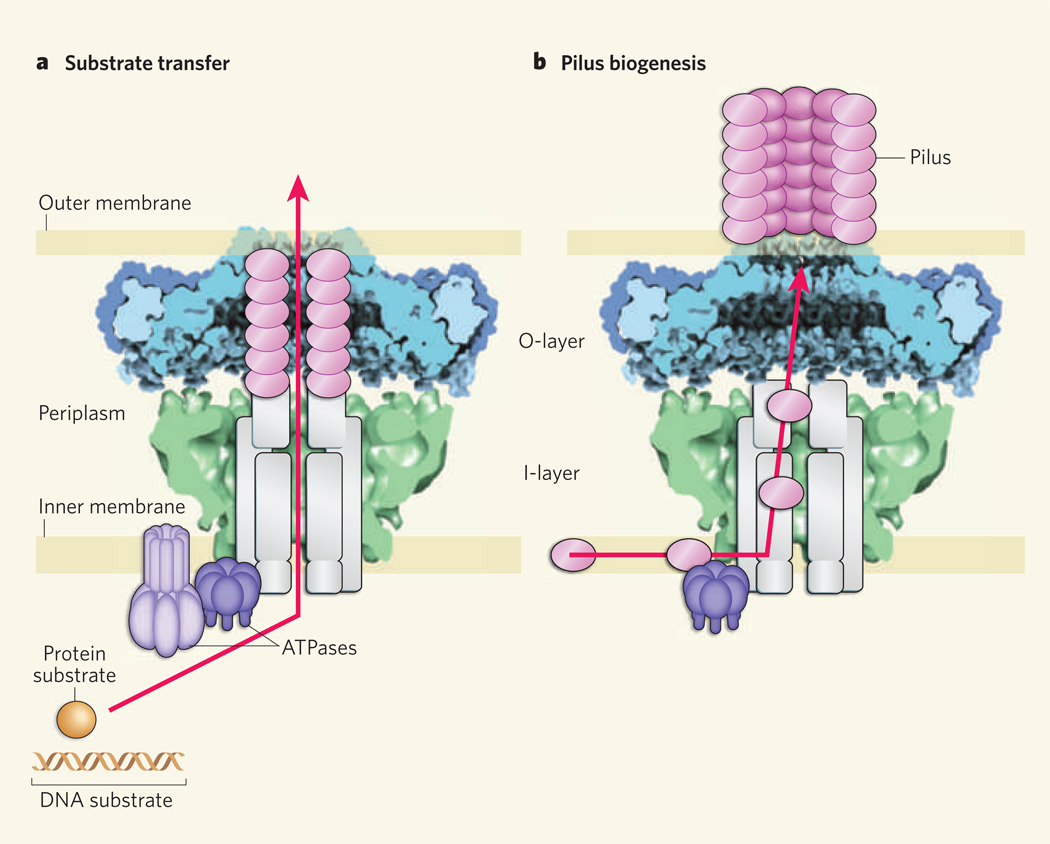
Figure 1. Architecture and possible functions of a type IV secretion system.
Bacteria transfer DNA and protein substrates to each other and to eukaryotic cells, such as those of plants and animals, using type IV secretion (T4S) systems. These systems span the periplasm region between the outer and inner membranes of bacterial cell envelopes. T4S systems contain a core complex that consists of two layers, the O-layer and the I-layer. The X-ray crystal structure of an O-layer (blue) reported by Waksman and colleagues1 is shown here with a cryoelectron-microscope image9 (green) of the same core complex. The remaining subunits of the system are depicted in schematic form. Waksman and colleagues propose two possible functions for the core complex. a, It might serve as a structural scaffold that houses the translocation channel through which substrates pass. Several channel subunits, including pilin proteins (magenta ovals), form a conduit encased within the core chamber, and ATPase subunits bind the substrates ready for translocation. b, The core complex might also act as a scaffold and assembly platform for the conjugative pilus — the hair-like appendage of the T4S system that is used to make contact with other bacteria (or eukaryotic cells) prior to substrate transfer. Pilin proteins in the inner membrane would be passed through the core chamber to the bacterial cell surface to build the pilus.
Waksman and colleagues1 now provide the first high-resolution (2.6 Å) three-dimensional crystal structure of a portion of the core complex from E. coli (Fig. 1). The structure, which corresponds to the O-layer of the cryo-EM structure9, is a tour de force in structural biology. It is the first X-ray structure of an outer-membrane pore that consists of more than one type of subunit; all other known outer-membrane pores or channels consist of variable numbers of a single subunit. It is also one of the largest membrane-protein complexes solved to date.
Surprisingly, the structure reveals that the outer-membrane pore, called the cap, is composed not of TraO (VirB9), as previously predicted9, but of a domain of TraF (VirB10) known as the antennae projection6. This projection consists of two α-helices and an intervening loop; 14 copies of these two-helix bundles arrange to form the membrane-spanning pore. This is a strikingly unusual structure — almost all other bacterial outer-membrane proteins assemble as β-barrels in the membrane, rather than as α-helices.
Even more intriguingly, the new structure1, taken together with earlier findings10, establishes that TraF (VirB10) spans the cell envelope. This is unprecedented among bacterial membrane proteins, and is especially fascinating because VirB10 is structurally dynamic11. Specifically, VirB10 senses conformational changes that accompany either the binding or hydrolysis of ATP (an energy source) by protein subunits situated at the conjugation channel’s entrance. VirB10 responds to these changes by undergoing its own structural transition, which is required for the conjugation system to transfer a substrate. In the context of Waksman and colleagues’ new structure1, this ATP-sensing mechanism could serve to modulate gating of the α-helical pore to permit substrate passage. Indeed, the authors suggest that structural differences detected in their cryo-EM9 and X-ray structures1 might reflect different energy states of the complex.
So what is the large chamber in the core complex for? In A. tumefaciens, besides the VirB7–VirB9–VirB10 core complex, at least seven other subunits are required for substrate transfer12. These subunits might assemble within the core chamber, the dimensions of which are sufficiently large to house an intact translocation channel (Fig. 1a). Additionally (or alternatively), the core chamber could serve as a staging arena for pilus biogenesis (Fig. 1b).
The idea that the core chamber acts as a structural scaffold for channel and/or pilus assembly is gratifyingly compatible with most biochemical data. But such a model needs to be reconciled with at least three experimental observations. First, it is known that deletion of at least half of the amino acids in the VirB10 antennae projections (which come together to form the outer-membrane pore) selectively abolishes pilus biogenesis without disrupting substrate transfer10. If substrates pass through the core chamber (Fig. 1a), how can they transit through the outer membrane without an intact pore?
Second, although most T4S-system substrates engage with their T4S systems at the channel entrance in the cytoplasm, some substrates — such as the pertussis toxin, a penta meric protein complex — first assemble in the periplasm (the space between the inner and outer membranes of the bacterial cell envelope) and then engage with the T4S system for transfer across the outer membrane13. But Waksman and colleagues’ X-ray structure1 reveals an extensive network of interactions among the core subunits of T4S systems, and it is not evident how multisubunit substrates could gain access to the core’s chamber for translocation.
Finally, the diameter of the α-helical pore in the new crystal structure1 is only 32 Å, a size that could accommodate DNA and unfolded protein substrates, but not a multisubunit protein substrate or, for that matter, the conjugative pilus (80–120 Å in diameter)14 without inducing gross structural rearrangements.
These and other issues await further study, but in the meantime Waksman and colleagues are to be credited for a quantum leap in our understanding of macromolecular translocation in bacteria. On the structural front, the next task is to resolve even larger subassemblies of T4S systems and, ultimately, the entire translocation machine. Defining how the core complex physically associates with the conjugative pilus will also be a formidable technical challenge, but this is an essential goal if we are to understand the role, if any, of the pilus in substrate transfer. Equally necessary are functional studies to establish that the structures solved in vitro exist and have biological activity in vivo. Besides stimulating these and other avenues of research, Waksman and colleagues’ findings1 move us a step closer to developing drugs that target T4S systems, for controlling the spread of anti biotic resistance and mitigating the proliferation of medically important pathogens.
References
- 1.Chandran V, et al. Nature. 2009;462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Martinez CE, Christie PJ. Microbiol. Mol. Biol. Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeo H-J, Savvides SN, Herr AB, Lanka E, Waksman G. Mol. Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 4.Savvides SN, et al. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeo H-J, Yuan Q, Beck MR, Baron C, Waksman G. Proc. Natl Acad. Sci. USA. 2003;100:15947–15952. doi: 10.1073/pnas.2535211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terradot L, et al. Proc. Natl Acad. Sci. USA. 2005;102:4596–4601. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hare S, Bayliss R, Baron C, Waksman G. J. Mol. Biol. 2006;360:56–66. doi: 10.1016/j.jmb.2006.04.060. [DOI] [PubMed] [Google Scholar]
- 8.Bayliss R, et al. Proc. Natl Acad. Sci. USA. 2007;104:1673–1678. doi: 10.1073/pnas.0609535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fronzes R, et al. Science. 2009;323:266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakubowski SJ, et al. Mol. Microbiol. 2009;71:779–794. doi: 10.1111/j.1365-2958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascales E, Christie PJ. Proc. Natl Acad. Sci. USA. 2004;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascales E, Christie PJ. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns DL. Curr. Opin. Microbiol. 2003;6:29–34. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 14.Wang YA, Yu X, Silverman PM, Harris RL, Egelman EH. J. Mol. Biol. 2009;385:22–29. doi: 10.1016/j.jmb.2008.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]