Abstract
Previous studies have identified multiple blood pressure and renal disease quantitative trait loci located on rat chromosome 12. In the present study, we narrowed blood pressure loci using a series of overlapping SS-12BN congenic lines. We found that transferring 6.1Mb of SS chromosome 12 (13.4-19.5Mb) onto the consomic SS-12BN background significantly elevated blood pressure on 1% NaCl (146±6 vs. 127±1 mmHg, P<0.01) and 8% NaCl diets (178±7 vs. 144±2 mmHg, P<0.05). Compared with the SS-12BN consomic, these animals also had significantly elevated albumin (218±31 vs. 104±8 mg/day, P<0.01) and protein excretion (347±41 vs. 195±12 mg/day, P<0.01) on 1% NaCl diet. Elevated blood pressure, albuminuria, and proteinuria coincided with greater renal and cardiac damage, demonstrating that SS allele(s) within the 6.1Mb congenic interval are associated with strong cardiovascular disease phenotypes. Sequence analysis of the 6.1Mb congenic region revealed 12,675 single nucleotide polymorphisms between SS and BN. Of these polymorphisms, 295 lie within coding regions and 20 resulted in nonsynonymous changes in conserved genes, of which 5 were predicted to be potentially damaging to protein function. Syntenic regions in human chromosome 7 have also been identified in multiple linkage and association studies of cardiovascular disease, suggesting that genetic variants underlying cardiovascular phenotypes in this congenic strain can likely be translated to a better understanding of human hypertension.
Keywords: Hypertension, Genetic, Dahl Salt-Sensitive Rat, Brown Norway, Congenic
Introduction
Hypertension is the most prevalent cardiovascular disease (CVD) worldwide and is a major risk factor for stroke, atherosclerosis, renal failure, and heart disease.1 In most cases, a single cause of hypertension cannot be found (i.e., essential hypertension), but rather multiple environmental and genetic factors contribute to overall disease susceptibility.1 This, combined with complex gene interaction,2 poses significant challenges in sifting through the many variants that could potentially contribute to hypertension.
One strategy to characterize regions of the genome that affect CVD traits is through chromosome substitution (i.e., consomics). Mattson and colleagues3 used this approach to characterize blood pressure (BP) and renal damage by transferring chromosomes from the normotensive Brown Norway (BN) strain onto the hypertensive Dahl Salt-Sensitive/Mcwi (SS) background using marker-assisted selection.4 The SS model exhibits CVD traits5 and end organ damage that are highly similar to human hypertension.6-10 This comprehensive study identified multiple chromosomes from the normotensive BN strain that attenuate salt-induced hypertension and renal damage or had no effects; whereas, a few BN chromosomes (e.g., 12 and 17) surprisingly increased susceptibility to CVD.3 Several additional mapping studies have confirmed that RNO12 harbors quantitative trait loci (QTL) for BP and renal damage,5, 11-13 suggesting collectively that BP QTL are located on RNO12 and may be derived from normotensive as well as hypertensive strains.
In this study we narrowed regions by introgressing SS chromosome 12 back onto the SS-12BN consomic background to generate overlapping congenic strains that were phenotyped for BP, renal function, and cardiac damage. These congenics revealed that transferring a 6.1Mb region of SS chromosome 12 (13.4-19.5Mb) onto the consomic SS-12BN genetic background significantly elevated BP by 20-40 mmHg. This BP phenotype coincided with ~2-fold increase in albuminuria and proteinuria and a substantial increase in kidney and heart damage. Importantly, this region overlaps with multiple human and mouse quantitative trait loci (QTL) for BP and renal damage.14-18 This synteny suggests that renal and cardiovascular genetic elements in this region might be shared across species, thus, the SS-12BN congenic model may provide insight into genetic susceptibility to cardiovascular and renal disease in humans.
Materials and Methods
Generation of SS-12BN Congenic Rats
All rats were provided food and water ad libitum and were housed at the Medical College of Wisconsin (MCW) Animal Resource Center. All procedures were approved by the MCW IACUC committee. The SS-12BN/Mcwi consomic was made by selectively breeding the SS/JrHsD/Mcwi (SS) and BN/SsN/Mcwi (BN) strains19, and SS-12BN congenic strains were subsequently generated by backcrossing SS-12BN/Mcwi to SS/JrHsD/Mcwi.
Blood Pressure Measurement
Mean arterial pressure was measured by telemetry transmitter implantation with a catheter inserted into the abdominal aorta (TA11PA-C40, Data Sciences, St. Paul, MN) as previously described.20
Measurement of Protein and Albumin Excretion
On post-surgical day 3, rats were acclimated for 24 hours in metabolic cages, followed by 24 hour urine collection. Urinary protein and albumin were measured as described previously3, 12
Echocardiography
Echocardiography was performed using a GE Vivid 7 ultrasound machine (GE Healthcare, Waukesha, WI) equipped with a 10MHz transducer as described previously.21
Histology
Histological analysis of kidneys and hearts was performed as described previously.21
Genomic Sequencing and Analysis
SS/JrHsD/Mcwi genomic DNA was sequenced using an Illumina HiSeq 2000 and analyzed by CASAVA v1.8.1 (Illumina, San Diego, CA). Variants were analyzed by Variant Effect Predictor22 and Polyphen23. Analysis of the SS/JrHsD/Mcwi genome can be accessed on the RGD website (http://rgd.mcw.edu).
Statistical analysis
Statistical analyses were performed using Sigma Plot 11.0 software. Data are presented as mean ± SEM. Interventricular septum thickness, interstitial fibrosis, and capillary density were analyzed by unpaired Student’s t test. All other data were analyzed by one-way ANOVA followed by the Holm-Sidak multiple comparison test. Because albumin measurements failed the equal variance test, the data were log-transformed prior to one-way ANOVA followed by the Holm-Sidak multiple comparison test.
Expanded Materials and Methods are provided in the supplement.
Results
Blood Pressure
To narrow the regions on RNO12 that modify BP, we generated a series of SS-12BN congenic lines as depicted in Figure 1. At 9 weeks-of-age, SS-12BN consomic and congenic male rats fed a 1% NaCl diet were implanted with radiotelemeters and MAP was recorded over 3 consecutive days. On 1% NaCl diet, the MAP of line C (144±2 mmHg, n=11) was significantly higher than SS-12BN consomic (127±1 mmHg, P<0.001, n=10), line A (114±2 mmHg, P<0.001, n=8), and line B (119±1 mmHg, P<0.001, n=8). Likewise, after an 8% NaCl challenge for 7 days, the MAP of line C rose to 178±7 mmHg, which was significantly higher than the SS-12BN consomic (144±2 mmHg, P<0.001), line A (136±5 mmHg, P<0.001), and line B (139±2 mmHg, P<0.001) (Figure 1). Because line C is genetically identical to the SS-12BN consomic except for 6.1Mb (13.4-19.5Mb), these data indicate that allele(s) within this region strongly regulate BP.
Figure 1.
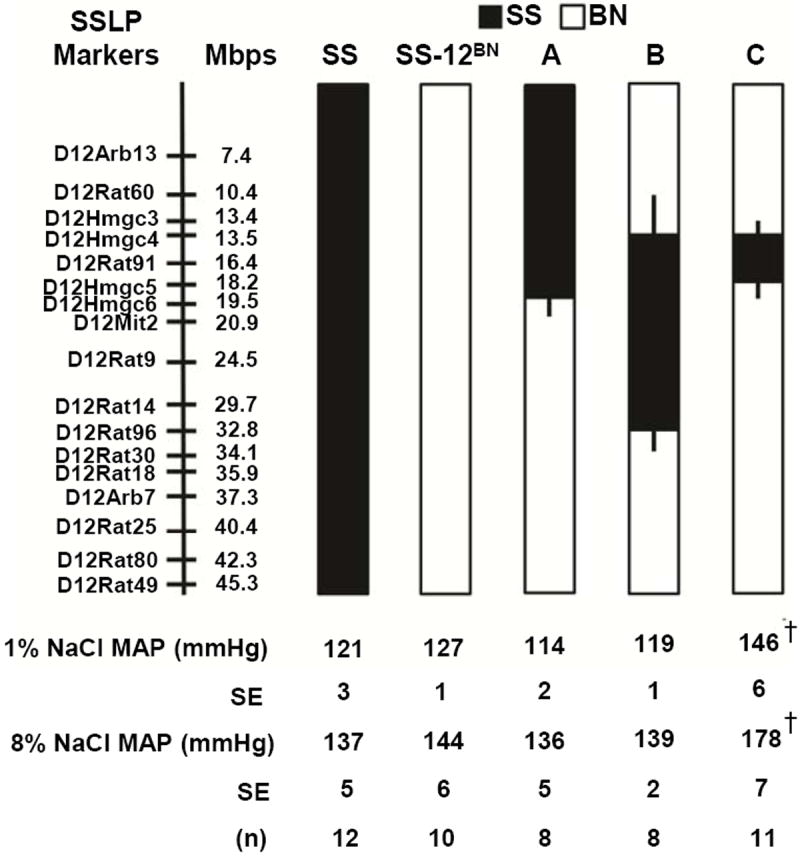
Schematic representation of the overlapping SS-12BN congenic strains that were generated by introgressing segments of the SS chromosome 12 (black) into the genetic background of the SS-12BN consomic rat (white) by marker assisted breeding. Thin black bars represent chromosomal regions that could be BN or SS. MAP of SS-12BN consomic and congenic rats on 1% and 8% NaCl diets. Data represent MAP ± SEM from 8-12 animals per group. ***P<0.001 as determined by a one-way ANOVA followed by a Holm-Sidak post hoc test.
Renal Damage and Histology
To assess renal damage, urine was collected from 9-week old SS-12BN consomic and congenic rats on 1% NaCl Purina diet and total albumin and protein excretion was quantified. In agreement with BP results, line C had significantly higher albumin (218±29 mg, P<0.001) and protein (347±38 mg, P<0.001) excretion compared with the SS-12BN consomic, line A, and line B, with the values collectively ranging from 85-125 mg and 195-236 mg for total albumin and protein excretion, respectively (Figure 2A and 2B).
Figure 2.
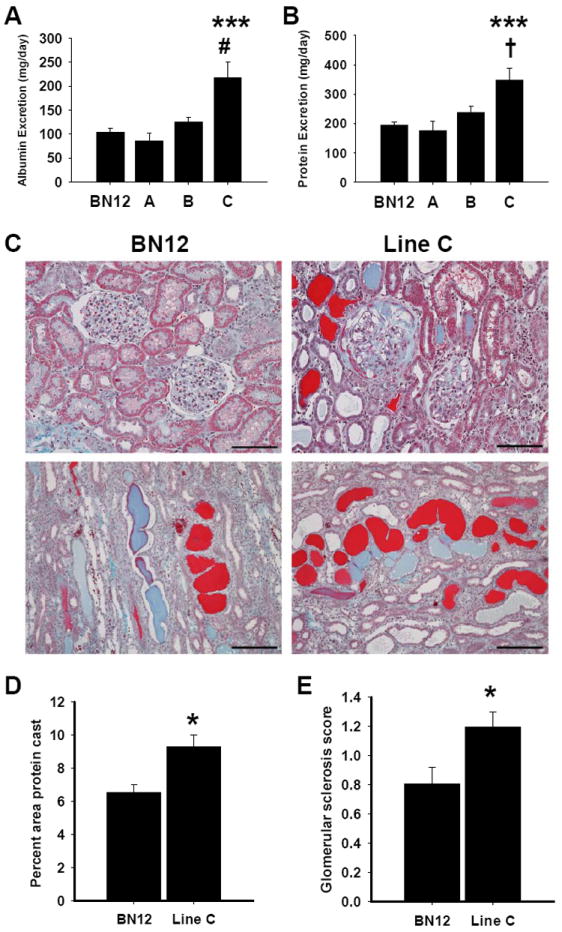
Urinary albumin and protein excretion of SS-12BN consomic and congenic rats on a 1% NaCl diet. The rats were placed in metabolic cages overnight to acclimate, followed by a 24 hour urine collection. Data are presented as mean albumin (A) or protein excretion (B) ± SEM from 8-12 animals per group. Data were not normally distributed, so each data point was log transformed before statistical analysis. ***P<0.001 vs. all groups, #P<0.01 vs. line B, and †P<0.01 vs. line B, as determined by a one-way ANOVA followed by a Holm-Sidak post hoc test. (C) Histology of trichrome-stained SS-12BN consomic and line C kidneys. (D) Quantification of percent area of protein casting in the outer stripe of the medulla (n=4 animals per group). Data are presented as mean percent area of protein casting ± SEM. (E) Average sclerosis score for SS-12BN consomic and line C kidneys (n=4 animals per group). Data are presented as mean sclerosis score ± SEM. For (D) and (E) *P<0.05 as determined by Student’s unpaired t-test.
The extent of renal damage was assessed histologically using trichrome-stained kidney sections of 12-week old SS-12BN consomic and line C rats placed on 8% NaCl. Compared with SS-12BN consomics, the morphology of line C kidneys appeared to have significantly more tubular and glomerular damage (Figure 2C). Quantitative analysis of the percent area of protein casting showed significantly higher tubular casting in the outer medulla of line C (9.3±0.7%) as compared with SS-12BN consomic kidneys (6.5±0.5%, P<0.05; Figure 2D). Kidneys from line C rats also showed considerably more glomerular sclerosis and basement membrane thickening as well as increased interstitial fibrosis compared to the SS-12BN consomic. Glomerular sclerosis was significantly higher in line C compared to SS-12BN (1.2±0.1 versus 0.8±0.1, P<0.05), as quantified by glomerular sclerosis scoring (Figure 2E).
Heart Function and Histology
To determine whether elevated BP in line C also coincided with increased cardiac hypertrophy and remodeling, the hearts of 4-month old line C and SS-12BN consomic rats on 1% NaCl Purina diet were measured by echocardiography and then collected for histological analysis. Compared with SS-12BN consomics, morphological analysis of line C hearts showed notable increase in LV wall thickness (Figure 3A) and a 24±0.1% (P < 0.01) increase in interstitial fibrosis (Figure 3B and 3C). Cardiac hypertrophy in line C was confirmed by a 31±6% (P < 0.05) increase in septal wall thickness compared with SS-12BN consomics, as determined by echocardiography (Figure 3D).
Figure 3.
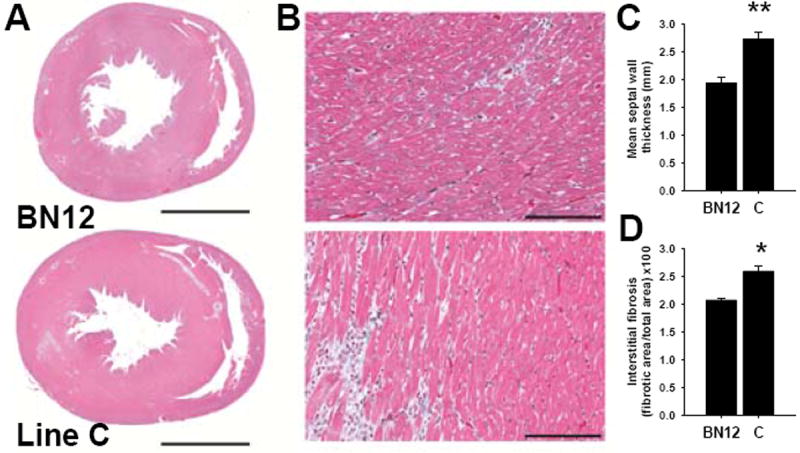
Left ventricular hypertrophy (LVH) and cardiac remodeling in line C compared with the SS-12BN consomic. (A) Trichrome-stained mid-ventricular sections from SS-12BN consomics (top) and line C (bottom) show greater LVH in line C compared with age-matched SS-12BN consomics. (B) Higher magnification images (400X) of trichrome-stained mid-ventricular sections from SS-12BN consomic (top) and line C (bottom) show increased interstitial fibrosis in line C as compared with the SS-12BN consomic. (C) The percent fibrotic area was quantified from images of trichrome-stained mid-ventricular sections acquired at 200X magnification. Data are presented as the average % fibrotic tissue area ± SEM (n=50 images per rat; 4-5 rats per group). (D) Septal wall thickness of SS-12BN and line C hearts was assessed by echocardiography. Data represent the mean septal wall thickness ± SEM from 3-5 rats per group. For (C) and (D), *P<0.05 and **P<0.01 as determined by Student’s unpaired t-test. Scale bars represent 3mm (A) and 150μm (B).
Comparative Genomics
Our data demonstrate that genetic element(s) within the line C congenic region strongly affect BP, renal function, and cardiac remodeling (Figures 1-3). This 6.1Mb region contains 133 genes and is syntenic to two regions on human chromosome 7 and one region on mouse chromosome 5 (Figure 4). A 3.9Mb region toward the p terminal of line C (13.4-17.3Mb) is homologous to the p end of human chromosome 7 (0.2-4.1Mb) in the reverse orientation (p to q) compared with the rat. The human homolog to line C from 17.2 to 17.9Mb is also located on human chromosome 7 spanning from 99.2 to 100.3Mb in the same orientation (p to q) as the rat. The remaining 1.1Mb of the line C congenic region (16.8-17.3Mb) aligned to the human X and Y chromosomes from 1.5 to 2.1Mb and 155.0 to 155.1Mb (X chromosome) and from 1.4 to 2.1Mb and 59.1 to 59.3Mb (Y chromosome). In comparison, line C is syntenic to a contiguous region on mouse chromosome 5 (137.8-142.7Mb) in the reverse orientation (q to p) compared with the rat.
Figure 4.
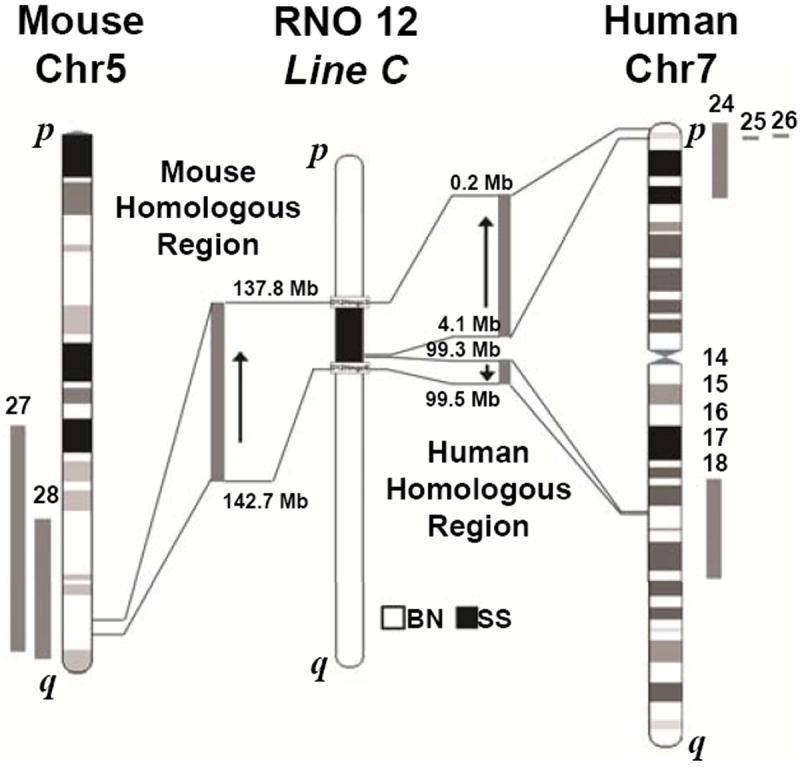
Schematic representation of the flanking markers on RNO12 for line C and the orthologous regions in human and mouse. The flanking markers on RNO12 for the line C congenic region are D12-Hmgc3 (RGD ID: 5683878) and D12-Hmgc6 (RGD ID: 5683884). References for previous studies showing overlapping QTLs or associations are indication in the figure.
A total of 6 human QTLs14-18, 24, 2 human SNP association studies25, 26 and 2 mouse QTLs27, 28 for BP or renal function have been previously mapped to regions that are syntenic to the line C congenic region (Figure 4). Of these studies, two significantly associated the 3.9Mb toward the p end of human chromosome 7 with elevated BP25 and diabetic nephropathy24, while five studies linked the marker D7S1799 (chr12:103982240) in multiple populations with elevated MAP, diastolic BP, and systolic BP.14-18 Two additional linkage studies in the mouse identified concordant BP QTLs on mouse chromosome 5 spanning 89-148Mb27 and 126-162Mb.28 On average the QTLs identified in mouse and human linkage studies spanned 30-60Mb and contained approximately 600-700 genes. In comparison, the number of candidates in the line C congenic region is considerably less at 133 genes, of which 56 are validated, 62 are predicted, and 15 belong to the nonconserved family of vomeronasal receptors (V2R)29 (Supplementary Table S1). Thus, if the same genetic elements influence BP and renal damage across species, then we can use species conservation to preliminarily narrow our list to 118 candidate genes that overlap in human, mouse and rat QTLs.
Identification of Sequence Variants within the Candidate Region
The SS genome was aligned across the 6.1Mb line C congenic region using Illumina sequencing reads. We aligned 5,283,594bp (86.9% of non-gap reference) with an average depth of 20X. Compared with the BN reference, we identified a total of 12,675 variants by two or more reads that constitute at least 30% of total reads detected at that loci (Supplementary Table S2). Of the total variants, 295 lie within coding regions of known and predicted genes and 141 are expected to cause nonsynonomous amino acid changes. Of the 141 nonsynonymous variants, 20 reside in conserved validated genes and 5 of these were predicted to be damaging by PolyPhen (http://genetics.bwh.harvard.edu/pph/) (Table 1). Four miRNA were also predicted within this region using miRBase (http://www.mirbase.org/), but we found no sequence variations within these predicted miRNAs (data not shown). Likewise, using the TargetScan tool (http://www.targetscan.org/) to predict miRNA-targeted genes within the line C congenic interval revealed 18 putative miRNA target sites; however, no sequence variants were found in any of these sites.
Table 1.
Nonsynonomous variants in conserved genes within the SS-12BN line C congenic region
| Gene Symbol | Gene ID* | Position | Ref† nt | Var† nt | AA Change | Polyphen Prediction |
|---|---|---|---|---|---|---|
| Ttyh3 | ENSRNOT00000038773 | 14,480,874 | A | C | N331T | Benign |
| Elfn1 | ENSRNOT00000033551 | 15,139,510 | C | T | M287I | Damaging |
| Micall2 | ENSRNOT00000036744 | 15,395,661 | T | C | V181A | Benign |
| Micall2 | ENSRNOT00000036744 | 15,396,513 | A | G | T280A | Benign |
| Adap1 | ENSRNOT00000038777 | 15,844,453 | C | T | P23L | Benign |
| Zfp68 | ENSRNOT00000001784 | 16,545,529 | C | T | L168F | Benign |
| MGC112692 | ENSRNOT00000060155 | 16,635,046 | A | C | V332G | Benign |
| MGC112692 | ENSRNOT00000060155 | 16,635,542 | C | T | V167I | Benign |
| Dhrsx | ENSRNOT00000029485 | 16,809,701 | G | T | V78F | Benign |
| Akap17a | ENSRNOT00000037829 | 16,818,972 | A | G | C476R | Unknown |
| Il3ra | ENSRNOT00000001792 | 16,828,844 | C | T | V103I | Damaging |
| Cyp3a9 | ENSRNOT00000001863 | 17,176,498 | A | G | I207V | Benign |
| Cyp3a9 | ENSRNOT00000001863 | 17,203,077 | G | A | V454I | Benign |
| Spry3 | ENSRNOT00000011128 | 17,343,558 | C | A | G123V | Damaging |
| Azgp1 | ENSRNOT00000001801 | 17,494,146 | T | C | L40P | Damaging |
| Zscan21 | ENSRNOT00000001810 | 17,567,622 | A | G | R51G | Benign |
| Mcm7 | ENSRNOT00000001825 | 17,612,322 | G | A | S74L | Benign |
| Mblac1 | ENSRNOT00000001831 | 17,636,671 | A | G | Q36R | Benign |
| Mblac1 | ENSRNOT00000001831 | 17,636,674 | G | A | G37E | Benign |
| Mblac1 | ENSRNOT00000001831 | 17,636,772 | G | A | G70S | Damaging |
Ref nt, reference nucleotide; Var nt, variant nucleotide; AA, amino acid
Designated by Ensembl gene ID
Reference (BN/SsN/Mcwi) and variant (SS/JrHsD/Mcwi) were confirmed by Sanger sequencing
Discussion
Previous chromosome substitution strains3 and linkage analyses5, 12, 13, 30 have demonstrated that RNO12 impacts BP and renal damage, but the causative variants underlying these large QTLs have not yet been identified. Our goal was to narrow cardiovascular QTLs on RNO12 by screening overlapping SS-12BN congenic strains for BP, renal, and cardiac phenotypes. Analysis of the line C interval showed that transferring 6.1Mb of RNO12 (13.4-19.5Mb) from the SS strain onto the SS-12BN background significantly elevated BP by 20-40 mmHg (Figure 1), which coincided with extensive renal and cardiac damage compared with the SS-12BN consomic (Figures 2-3). Our observation that line C develops significant hypertension, proteinuria, albuminuria, and cardiac remodeling (Figures 1-3) on 1% NaCl Purina diet demonstrates that variants within the line C congenic region have strong phenotypic effects. These data also demonstrate that we have reduced the overlapping rat BP QTLs on RNO12 at 1.8-46.6Mb,11 11.4-29.0Mb5 and renal function QTLs at 9.6-33.2Mb12 and 10.7-31.4Mb13 by ~85%, which provides a more manageable region to screen for potentially damaging variants.
Genetic Modifiers of BP and Renal Function on RNO12
Our data provide evidence for genetic elements surrounding the 6.1Mb region (13.4-19.5Mb) that likely modulate blood pressure. The flanking SS genome in line A (1-13.4Mb) and line B (19.5-36.2Mb) significantly attenuated BP and renal phenotypes (Figures 2 and 3) compared to the line C region alone. This observation suggests that either SS alleles in these regions decrease blood pressure or BN alleles increase blood pressure. Several studies have previously demonstrated that interacting “+BP QTLs” and “−BP QTLs” can be derived from either hypertensive or normotensive strains2, 31 and the effect of these genes is only manifested under certain genetic combinations. Furthermore, it is possible that genes within the different regions (lines A, B, and C) interact with one another (i.e., epistasis), as has been demonstrated for BP in other regions of the rat genome (e.g., RNO3,31 RNO8,32 RNO10,33 and RNO1320). However, further experimentation would be required to test this hypothesis. Nevertheless, our data highlight the power of using congenic strains to map complex genetic traits that rely on allelic combinations, which may not be apparent from the parental strains.
Comparative Analysis of Line C Sequence Variants
The line C congenic region is syntenic with two regions in human chromosome 7 that have been linked with BP15-18, 25 and renal disease14, 34 by linkage and association studies (Figure 4), suggesting that common genetic elements might play a similar role in rat and human. In the line C region we identified 20 nonsynonomous amino acid changes in conserved validated genes between SS and BN, of which Mblac1, Il3ra, Spry3, Elfn1, and Azgp1 were predicted by PolyPhen to be damaging variants (Table 1). Mblac1 is a poorly characterized metal-binding enzyme35 with no reported role in BP or renal function. Il3ra and Spr3 are involved in hematopoiesis36 and neurogenesis,37 respectively, but also have no reported role in BP or renal function. Elfn1, also known in humans as PPP1R28, is an inhibiting regulatory subunit of the ubiquitous protein phosphatase, PP1.38 Although the function of PPP1R28 is largely unknown, the PP1 signaling pathway has been linked in humans with hypertension39, diabetes40, and heart failure.41 This human data makes Elfn1 an interesting candidate for our congenic model, as its associated pathway may regulate cardiovascular phenotypes across species. Azgp1 is a lipid mobilizing adipokine that has been associated with serum lipid levels,42, 43 multiple inflammatory phenotypes,44 and is also implicated in preeclampsia45 and renal function45, 46 suggesting that it might mediate disease-associated remodeling pathways.
The larger of the two syntenic regions in humans (0.2-4.1Mb of human chr7) has been linked with BP and renal disease by multiple studies.24-26 Two studies associated single nucleotide polymorphisms (SNPs) in the SDK1 (sidekick-1) locus with elevated BP in Nigerian25 and Japanese26 populations. The predicted rat homologue of SDK1 (RGD1560686) partially overlaps the line C congenic region, suggesting that SDK1 or variant(s) in close proximity to this gene might contribute to elevated BP across species. None of the SDK1 SNPs that were associated with elevated BP in humans25, 26 caused amino acid changes our line C congenic region (Supplementary Table S2), which leads us to believe that mutations of SDK1 itself are probably not causing BP changes in our model. However, we cannot rule out potential variants in non-coding regulatory regions that might affect expression levels of SDK1. Multiple studies have shown that elevation of SDK1 in podocytes has been shown to increase glomerular sclerosis in both animal models47 and HIV-associated nephropathy,48, 49 making this gene an interesting candidate for complex renal disease.
Perspectives
We narrowed rat BP QTLs on RNO12 by ~85% to a 6.1Mb congenic region that contains 118 conserved genes and is syntenic with BP QTLs from multiple human and mouse studies. Further congenic mapping will be needed to narrow the loci and elucidate the specific variant(s) and mechanisms that drive increased BP and renal damage in the SS-12BN strain. Additionally, with the advent of rat knockout models developed by our group50 and others,51 it is now possible to specifically target and test functional alleles in the rat. Meanwhile, these genes serve as candidates to be tested in subsets GWAS and other species or strains.
Supplementary Material
Novelty and Significance.
1. What Is New?
For the first time we show that the SS-12BN congenic region (13.4-19.5Mb) drives a 20-40 mmHg change in MAP and is associated with extensive renal and cardiac damage.
We identified 12,675 SNPs, of which 295 lie within known and predicted genes and 5 are anticipated to be potentially damaging.
2. What Is Relevant?
The SS-12BN congenic region overlaps with 8 human and 2 mouse CVD QTLs, offering new insight to common genetic elements mediate susceptibility to CVD.
3. Summary
Variant(s) within the SS-12BN congenic region (13.4-19.5Mb) strongly regulate BP that increases susceptibility to end-stage renal and cardiac disease.
Acknowledgments
We thank Becky Schilling, Angela Lemke, Jesse Meyer, Samantha Richter, Allison Zappa, and the RGD team for excellent technical support.
Sources of Funding
This study was supported by NHLBI-5R01HL089930 to HJJ and a NHLBI training grant (5T32HL007792) to MJF.
Footnotes
Conflict(s) of Interest/Disclosure(s) Statement - None
References
- 1.Cowley AW., Jr The genetic dissection of essential hypertension. Nat Rev Genet. 2006;7:829–840. doi: 10.1038/nrg1967. [DOI] [PubMed] [Google Scholar]
- 2.Deng AY. Genetic basis of polygenic hypertension. Hum Mol Genet. 2007;16(Spec No. 2):R195–202. doi: 10.1093/hmg/ddm126. [DOI] [PubMed] [Google Scholar]
- 3.Mattson DL, Dwinell MR, Greene AS, Kwitek AE, Roman RJ, Jacob HJ, Cowley AW., Jr Chromosome substitution reveals the genetic basis of dahl salt-sensitive hypertension and renal disease. Am J Physiol Renal Physiol. 2008;295:F837–842. doi: 10.1152/ajprenal.90341.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markel P, Shu P, Ebeling C, Carlson GA, Nagle DL, Smutko JS, Moore KJ. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nature genetics. 1997;17:280–284. doi: 10.1038/ng1197-280. [DOI] [PubMed] [Google Scholar]
- 5.Stoll M, Cowley AW, Jr, Tonellato PJ, Greene AS, Kaldunski ML, Roman RJ, Dumas P, Schork NJ, Wang Z, Jacob HJ. A genomic-systems biology map for cardiovascular function. Science. 2001;294:1723–1726. doi: 10.1126/science.1062117. [DOI] [PubMed] [Google Scholar]
- 6.Campese VM. Salt sensitivity in hypertension. Renal and cardiovascular implications. Hypertension. 1994;23:531–550. doi: 10.1161/01.hyp.23.4.531. [DOI] [PubMed] [Google Scholar]
- 7.Cowley AW, Jr, Roman RJ. The role of the kidney in hypertension. JAMA. 1996;275:1581–1589. [PubMed] [Google Scholar]
- 8.Feldman HI, Klag MJ, Chiapella AP, Whelton PK. End-stage renal disease in us minority groups. Am J Kidney Dis. 1992;19:397–410. doi: 10.1016/s0272-6386(12)80945-0. [DOI] [PubMed] [Google Scholar]
- 9.Grim CE, Wilson TW, Nicholson GD, Hassell TA, Fraser HS, Grim CM, Wilson DM. Blood pressure in blacks. Twin studies in barbados. Hypertension. 1990;15:803–809. doi: 10.1161/01.hyp.15.6.803. [DOI] [PubMed] [Google Scholar]
- 10.Lackland DT, Keil JE. Epidemiology of hypertension in african americans. Semin Nephrol. 1996;16:63–70. [PubMed] [Google Scholar]
- 11.Ramos A, Moisan MP, Chaouloff F, Mormede C, Mormede P. Identification of female-specific qtls affecting an emotionality-related behavior in rats. Mol Psychiatry. 1999;4:453–462. doi: 10.1038/sj.mp.4000546. [DOI] [PubMed] [Google Scholar]
- 12.Moreno C, Dumas P, Kaldunski ML, Tonellato PJ, Greene AS, Roman RJ, Cheng Q, Wang Z, Jacob HJ, Cowley AW., Jr Genomic map of cardiovascular phenotypes of hypertension in female dahl s rats. Physiol Genomics. 2003;15:243–257. doi: 10.1152/physiolgenomics.00105.2003. [DOI] [PubMed] [Google Scholar]
- 13.Schulz A, Litfin A, Kossmehl P, Kreutz R. Genetic dissection of increased urinary albumin excretion in the munich wistar fromter rat. J Am Soc Nephrol. 2002;13:2706–2714. doi: 10.1097/01.asn.0000031803.55613.86. [DOI] [PubMed] [Google Scholar]
- 14.McDonough CW, Palmer ND, Hicks PJ, Roh BH, An SS, Cooke JN, Hester JM, Wing MR, Bostrom MA, Rudock ME, Lewis JP, Talbert ME, Blevins RA, Lu L, Ng MC, Sale MM, Divers J, Langefeld CD, Freedman BI, Bowden DW. A genome-wide association study for diabetic nephropathy genes in african americans. Kidney Int. 2011;79:563–572. doi: 10.1038/ki.2010.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atwood LD, Samollow PB, Hixson JE, Stern MP, MacCluer JW. Genome-wide linkage analysis of pulse pressure in mexican americans. Hypertension. 2001;37:425–428. doi: 10.1161/01.hyp.37.2.425. [DOI] [PubMed] [Google Scholar]
- 16.Atwood LD, Samollow PB, Hixson JE, Stern MP, MacCluer JW. Genome-wide linkage analysis of blood pressure in mexican americans. Genet Epidemiol. 2001;20:373–382. doi: 10.1002/gepi.7. [DOI] [PubMed] [Google Scholar]
- 17.Hunt SC, Ellison RC, Atwood LD, Pankow JS, Province MA, Leppert MF. Genome scans for blood pressure and hypertension: The national heart, lung, and blood institute family heart study. Hypertension. 2002;40:1–6. doi: 10.1161/01.hyp.0000022660.28915.b1. [DOI] [PubMed] [Google Scholar]
- 18.Cheng LS, Davis RC, Raffel LJ, Xiang AH, Wang N, Quinones M, Wen PZ, Toscano E, Diaz J, Pressman S, Henderson PC, Azen SP, Hsueh WA, Buchanan TA, Rotter JI. Coincident linkage of fasting plasma insulin and blood pressure to chromosome 7q in hypertensive hispanic families. Circulation. 2001;104:1255–1260. doi: 10.1161/hc3601.096729. [DOI] [PubMed] [Google Scholar]
- 19.Cowley AW, Jr, Stoll M, Greene AS, Kaldunski ML, Roman RJ, Tonellato PJ, Schork NJ, Dumas P, Jacob HJ. Genetically defined risk of salt sensitivity in an intercross of brown norway and dahl s rats. Physiol Genomics. 2000;2:107–115. doi: 10.1152/physiolgenomics.2000.2.3.107. [DOI] [PubMed] [Google Scholar]
- 20.Moreno C, Kaldunski ML, Wang T, Roman RJ, Greene AS, Lazar J, Jacob HJ, Cowley AW., Jr Multiple blood pressure loci on rat chromosome 13 attenuate development of hypertension in the dahl s hypertensive rat. Physiol Genomics. 2007;31:228–235. doi: 10.1152/physiolgenomics.00280.2006. [DOI] [PubMed] [Google Scholar]
- 21.Kriegel AJ, Greene AS. Substitution of brown norway chromosome 16 preserves cardiac function with aging in a salt-sensitive dahl consomic rat. Physiol Genomics. 2008;36:35–42. doi: 10.1152/physiolgenomics.00054.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the ensembl api and snp effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramensky V, Bork P, Sunyaev S. Human non-synonymous snps: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowden DW, Colicigno CJ, Langefeld CD, Sale MM, Williams A, Anderson PJ, Rich SS, Freedman BI. A genome scan for diabetic nephropathy in african americans. Kidney Int. 2004;66:1517–1526. doi: 10.1111/j.1523-1755.2004.00915.x. [DOI] [PubMed] [Google Scholar]
- 25.Tayo BO, Luke A, Zhu X, Adeyemo A, Cooper RS. Association of regions on chromosomes 6 and 7 with blood pressure in nigerian families. Circ Cardiovasc Genet. 2009;2:38–45. doi: 10.1161/CIRCGENETICS.108.817064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oguri M, Kato K, Yokoi K, Yoshida T, Watanabe S, Metoki N, Yoshida H, Satoh K, Aoyagi Y, Nozawa Y, Yamada Y. Assessment of a polymorphism of sdk1 with hypertension in japanese individuals. Am J Hypertens. 2010;23:70–77. doi: 10.1038/ajh.2009.190. [DOI] [PubMed] [Google Scholar]
- 27.Feng M, Deerhake ME, Keating R, Thaisz J, Xu L, Tsaih SW, Smith R, Ishige T, Sugiyama F, Churchill GA, DiPetrillo K. Genetic analysis of blood pressure in 8 mouse intercross populations. Hypertension. 2009;54:802–809. doi: 10.1161/HYPERTENSIONAHA.109.134569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugiyama F, Churchill GA, Higgins DC, Johns C, Makaritsis KP, Gavras H, Paigen B. Concordance of murine quantitative trait loci for salt-induced hypertension with rat and human loci. Genomics. 2001;71:70–77. doi: 10.1006/geno.2000.6401. [DOI] [PubMed] [Google Scholar]
- 29.Young JM, Trask BJ. V2r gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet. 2007;23:212–215. doi: 10.1016/j.tig.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Iwai N, Yasui N, Naraba H, Tago N, Yamawaki H, Sumiya H. Klk1 as one of the genes contributing to hypertension in dahl salt-sensitive rat. Hypertension. 2005;45:947–953. doi: 10.1161/01.HYP.0000161969.65767.0d. [DOI] [PubMed] [Google Scholar]
- 31.Palijan A, Dutil J, Deng AY. Quantitative trait loci with opposing blood pressure effects demonstrating epistasis on dahl rat chromosome 3. Physiol Genomics. 2003;15:1–8. doi: 10.1152/physiolgenomics.00084.2003. [DOI] [PubMed] [Google Scholar]
- 32.Ariyarajah A, Palijan A, Dutil J, Prithiviraj K, Deng Y, Deng AY. Dissecting quantitative trait loci into opposite blood pressure effects on dahl rat chromosome 8 by congenic strains. J Hypertens. 2004;22:1495–1502. doi: 10.1097/01.hjh.0000133720.94075.6f. [DOI] [PubMed] [Google Scholar]
- 33.Charron S, Duong C, Menard A, Roy J, Eliopoulos V, Lambert R, Deng AY. Epistasis, not numbers, regulates functions of clustered dahl rat quantitative trait loci applicable to human hypertension. Hypertension. 2005;46:1300–1308. doi: 10.1161/01.HYP.0000192024.72367.c3. [DOI] [PubMed] [Google Scholar]
- 34.Turner ST, Kardia SL, Mosley TH, Rule AD, Boerwinkle E, de Andrade M. Influence of genomic loci on measures of chronic kidney disease in hypertensive sibships. J Am Soc Nephrol. 2006;17:2048–2055. doi: 10.1681/ASN.2005121254. [DOI] [PubMed] [Google Scholar]
- 35.Hu Z, Spadafora LJ, Hajdin CE, Bennett B, Crowder MW. Structure and mechanism of copper- and nickel-substituted analogues of metallo-beta-lactamase l1. Biochemistry. 2009;48:2981–2989. doi: 10.1021/bi802295z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Testa U, Riccioni R, Diverio D, Rossini A, Lo Coco F, Peschle C. Interleukin-3 receptor in acute leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U K. 2004;18:219–226. doi: 10.1038/sj.leu.2403224. [DOI] [PubMed] [Google Scholar]
- 37.Panagiotaki N, Dajas-Bailador F, Amaya E, Papalopulu N, Dorey K. Characterisation of a new regulator of bdnf signalling, sprouty3, involved in axonal morphogenesis in vivo. Development. 2010;137:4005–4015. doi: 10.1242/dev.053173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Shen GQ, Ikegami H, Fujisawa T, Hamada Y, Kamide K, Rakugi H, Higaki J, Murakami H, Shimamoto K, Ogihara T. Asp905tyr polymorphism of protein phosphatase 1 g subunit gene in hypertension. Hypertension. 1997;30:236–239. doi: 10.1161/01.hyp.30.2.236. [DOI] [PubMed] [Google Scholar]
- 40.Delibegovic M, Armstrong CG, Dobbie L, Watt PW, Smith AJ, Cohen PT. Disruption of the striated muscle glycogen targeting subunit ppp1r3a of protein phosphatase 1 leads to increased weight gain, fat deposition, and development of insulin resistance. Diabetes. 2003;52:596–604. doi: 10.2337/diabetes.52.3.596. [DOI] [PubMed] [Google Scholar]
- 41.Gupta RC, Mishra S, Rastogi S, Imai M, Habib O, Sabbah HN. Cardiac sr-coupled pp1 activity and expression are increased and inhibitor 1 protein expression is decreased in failing hearts. Am J Physiol Heart Circ Physiol. 2003;285:H2373–2381. doi: 10.1152/ajpheart.00442.2003. [DOI] [PubMed] [Google Scholar]
- 42.Olofsson LE, Olsson B, Lystig T, Jacobson P, Jernas M, Sjoholm K, Gummesson A, Sjostrom L, Eriksson P, Hamsten A, Hale LP, Thelle DS, Carlsson B, Carlsson LM. Preliminary report: Zn-alpha2-glycoprotein genotype and serum levels are associated with serum lipids. Metabolism. 2010;59:1316–1318. doi: 10.1016/j.metabol.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 43.Yeung DC, Lam KS, Wang Y, Tso AW, Xu A. Serum zinc-alpha2-glycoprotein correlates with adiposity, triglycerides, and the key components of the metabolic syndrome in chinese subjects. J Clin Endocrinol Metab. 2009;94:2531–2536. doi: 10.1210/jc.2009-0058. [DOI] [PubMed] [Google Scholar]
- 44.Gao D, Trayhurn P, Bing C. Macrophage-secreted factors inhibit zag expression and secretion by human adipocytes. Mol Cell Endocrinol. 2010;325:135–142. doi: 10.1016/j.mce.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 45.Stepan H, Philipp A, Roth I, Kralisch S, Jank A, Schaarschmidt W, Lossner U, Kratzsch J, Bluher M, Stumvoll M, Fasshauer M. Serum levels of the adipokine zinc-alpha2-glycoprotein are increased in preeclampsia. J Endocrinol Invest. 2011 doi: 10.3275/7877. In Press. [DOI] [PubMed] [Google Scholar]
- 46.Philipp A, Kralisch S, Bachmann A, Lossner U, Kratzsch J, Bluher M, Stumvoll M, Fasshauer M. Serum levels of the adipokine zinc-alpha2-glycoprotein are increased in chronic hemodialysis. Metabolism. 2011;60:669–672. doi: 10.1016/j.metabol.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 47.Kaufman L, Potla U, Coleman S, Dikiy S, Hata Y, Kurihara H, He JC, D’Agati VD, Klotman PE. Up-regulation of the homophilic adhesion molecule sidekick-1 in podocytes contributes to glomerulosclerosis. J Biol Chem. 2010;285:25677–25685. doi: 10.1074/jbc.M110.133959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaufman L, Yang G, Hayashi K, Ashby JR, Huang L, Ross MJ, Klotman ME, Klotman PE. The homophilic adhesion molecule sidekick-1 contributes to augmented podocyte aggregation in hiv-associated nephropathy. FASEB J. 2007;21:1367–1375. doi: 10.1096/fj.06-7191com. [DOI] [PubMed] [Google Scholar]
- 49.Kaufman L, Hayashi K, Ross MJ, Ross MD, Klotman PE. Sidekick-1 is upregulated in glomeruli in hiv-associated nephropathy. J Am Soc Nephrol. 2004;15:1721–1730. doi: 10.1097/01.asn.0000128975.28958.c2. [DOI] [PubMed] [Google Scholar]
- 50.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Menoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong C, Li P, Wu NL, Yan Y, Ying QL. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature. 2010;467:211–213. doi: 10.1038/nature09368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


