Summary
Almost all aspects of protein traffic in bacteria were covered at the ASM-FEMS meeting on the topic in Iraklio, Crete in May 2006. The studies presented ranged from mechanistic analysis of specific events leading proteins to their final destinations to the physiological roles of the targeted proteins. Among the highlights from the meeting that are reviewed here are the molecular dynamics of SecA protein, membrane protein insertion, type III secretion needles and chaperones, type IV secretion, the two partner and autosecretion systems, the ‘secretion competent state’, and the recently discovered type VI secretion system.
Who is counting?
Protein translocation is one area of bacterial molecular biology that impinges substantially on eukaryotic molecular biology. Although specific details of protein translocation mechanisms might differ considerably in bacteria and eukaryotes, there are numerous commonalities (e.g. ATP-driven protein unfolding and translocation motors) and even shared systems (the SRP-Sec systems for endoplasmic reticulum and bacterial plasma membrane protein traffic, for example, or the TAT systems in bacteria and chloroplasts and the YidC system in bacteria, chloroplasts and mitochondria). As a result, bacterial protein translocation systems feature prominently at meetings on protein traffic. In organizing an ASM-FEMS meeting specifically devoted to protein traffic in prokaryotes on the island of Crete in May 2006, the authors of this report intended to give prominence to mechanistic aspects of as many prokaryotic protein translocation systems as possible with the aim of bringing to the forefront the many and varied pathways that have been discovered. Indeed, bacteria have evolved so many protein translocation systems that they might outnumber those in eukaryotes.
Bacterial protein translocation can be divided into two or even three steps, dealing, respectively, with the plasma membrane (inner membrane in Gram-negative bacteria), the outer membrane and the plasma membrane of target cell. Gram-negative bacterial protein translocation systems have evolved sophisticated solutions to deal with the second barrier. In some cases, an appendage has been grafted onto the inner membrane translocation systems such that the entire process is divided into two physically separable steps. A second successful solution involved the evolution of entirely new systems that bypass the conventional or broad specificity inner membrane systems altogether. In all cases where specific, non-destructive translocation of bacterial proteins (effectors) occurs across a eukaryotic membrane, the latter is negotiated by an additional appendage that forms an integral part of a system that bypasses the general export pathways in the inner membrane. The following sections of this report deal with some of the major issues raised at the meeting.
Typing errors
According to common usage, ‘export’ defines the localization of non-cytoplasmic proteins to within the cell envelope, whereas ‘secretion’ applies to extracellular proteins that are entirely outside of the outer-most lipid bilayer, including soluble (free) and surface-associated proteins and surface appendages. Confusion arises, for example, in the use of the term Sec to define (components of) the general export pathway in Gram-negative bacteria, which directs proteins into the inner membrane or periplasm but cannot ensure their secretion sensu strictu, which is mediated by specific pathways for transport across the outer membrane. This confusion does not arise in Gram-positive bacteria or Archaea, where soluble proteins reach the outside medium directly via the Sec system.
Initial interest in the mechanisms of bacterial protein translocation focused on the Sec system, largely in Escherichia coli. Characterization of other translocation systems began a little later (around 20 years ago) and often focused more on the physiological function of the system (usually in relation to pathogenesis) than on mechanistic aspects. When George Salmond introduced the numerical classification of protein secretion systems (Salmond and Reeves, 1993), there were only three: two Sec-bypass pathways [(the type I system (T1SS) composed of three components, and the more complex type III system (T3SS or injectisome)] and the type II secretion system (T2SS or secreton), which takes proteins exported by the Sec pathway (or, rarely, the TAT pathway) and translocates them in an folded state across the outer membrane (Table 1). Even at this early stage, however, the numerical classification system excluded the chaperone-usher pilus assembly pathway and the flagellum assembly pathway. The latter was subsequently shown to be a variant (possibly an evolutionary progenitor) of the T3SS, while one of the most common pilus biogenesis pathways, the type IV pilus system (T4PS), was more recently shown to be almost identical to the T2SS. The latter was nicely illustrated at the meeting on a poster presented by Julian Rood from Monash University, Melbourne, who showed that Dichelobacter nodosus uses the T4PS to secrete a protease. Another example is the T4PS of Francisella, which secretes several proteins (Hager et al., 2006). The type IV secretion system (T4SS) was discovered later and, understandably, is often confused with the T4PS, from which it is completely different. The T4SSs are ancestrally related to conjugation machines. They are composed of a translocation channel and surface structures such as conjugative pili (Gram-negative bacteria) or adhesive surface proteins (Gram-positive bacteria) to mediate attachment to target cells. Although the autotransporter system was originally called type IV, it was later renamed as type V (T5SS). The superficially similar but structurally quite distinct two-partner secretion system (TPSS) was never given a number (and we will refrain from doing so here), but a novel system recently discovered in several Gram-negative pathogenic bacteria was named type VI (T6SS) (see below).
Table 1.
Protein export, assembly and secretion systems in bacteria.
| Name | Process | Direct Sec (Tat) dependencea | Gram− | Gram+ | Comments |
|---|---|---|---|---|---|
| Sec (SRP-Sec) | Export | nab | Yes | Yes | |
| YidC | Export and assembly | Variable | Yes | Yes | Inner membrane protein assembly |
| Tat | Export | na | Yes | Yes | |
| YaeT (Omp85) | Assembly | Yes | Yes | No | Outer membrane protein assembly |
| T1SS | Secretion | No | Yes | (Yes) | Generally small substrates in Gram-positives |
| T2SS | Secretion | Yes | Yes | No | |
| T3SS | Secretion | No | Yes | No | |
| Flagellum | Secretion | No | Yes | Yes | Equivalent of T3SS |
| T4SS | Secretion | Variable | Yes | Yes | Conjugation system |
| T4PS | Secretion | Yes | Yes | Yes | Equivalent to T2SS |
| Autosecretion (T5SS) | Secretion | Yes | Yes | No | |
| T6SS | Secretion | No | Yes | No | |
| Chaperone-usher | Secretion | Yes | Yes | No | Pili |
| Esc | Secretion | No (see text) | No | Yes | |
| TPSS | Secretion | Yes | Yes | No | |
| Sortase | Secretion | Yes | No | Yes | Pili |
For translocated protein; assembly of machinery requires Sec.
na, not applicable.
With the exception of the bacterial flagellum system and the T1SS and T4SS, these secretion systems are restricted to Gram-negative bacteria and have evolved varying degrees of sophistication to overcome the bacterial outer membrane barrier and, in the T3SS and T4SS, the plasma membrane of the cell into which the secreted proteins are delivered. Although, as noted by Caparon’s group (Madden et al., 2001), secreted proteins from Gram-positive bacteria can also find their way into target cells, the procedure used is completely different and in no way justifies the name ‘type III-like’. However, many Gram-positive bacteria and the Gram-variable Actinomycetes have at least one novel Sec-bypass pathway for which the name Esc is generally used. Mycobacterium tuberculosis has several such systems encoded at different locations in the genome. As illustrated by Roland Brosch from the Institut Pasteur, Paris, primary interest focuses on the role of this system in pathogenicity (attenuated strains lack the best characterized of these genetic loci), but mechanistic studies of substrate recognition, energization and translocation through the two lipid bilayers in the M. tuberculosis cell envelope are now beginning (Brodin et al., 2006). There are even some indirect indications that this secretion pathway might use components of the universal general export pathway, and in particular the signal recognition particle targeting pathway (Singh et al., 2006).
How many does it take to tango?
Several studies on the venerable Sec system were presented at the meeting. In a way, these studies define the shape of things to come in the analysis of the other, so far less extensively characterized protein translocation machines. A complete set of structures of the molecules that are involved in the Sec pathway is now in hand: the SRP/ribosome complex presented by Irmi Sinning (University of Heidelberg) (Halic et al., 2006), the SecB chaperone (Zhou and Xu, 2005), the SecA motor (Hunt et al., 2002), the SecYEG protein-conducting channel alone (Osborne et al., 2005) and in complex with a ribosome (Mitra et al., 2005) and the signal (leader) peptidase (Paetzel et al., 1998). Structural biology is the most recent member of the powerful trinity of tools (the others being biochemistry and genetics) that are beginning to lead to at least limited understanding of the mechanics involved in Sec-mediated export. The exiting preprotein chain appears to be enclosed within a channel formed by SecY (Joly and Wickner, 1993; Cannon et al., 2005). Outward movement of a periplasmic SecY ‘plug’ region might facilitate (Van den Berg et al., 2004; Cannon et al., 2005; Tam et al., 2005), but is not essential (Junne et al., 2006) for translocation. In agreement with previous studies (Arkowitz et al., 1993), data presented by Nico Nouwen (University of Groningen) from experiments with a OmpA-titin chimera suggested that long preproteins might be actively unfolded by the translocase during the export process. However, the bore of the SecY channel is too narrow to accommodate even a single extended polypeptide chain (Van den Berg et al., 2004) and Tom Rapoport (Harvard University, Boston) proposed that it must dilate to accommodate the exiting chain.
However, if a single SecY channel is sufficient to form the protein export conduit, why are two or more ‘channels’ seen bound to the ribosome (Mitra et al., 2005; Osborne et al., 2005) or to SecA (Manting et al., 2000; Scheuring et al., 2005). A complex thought to contain a single SecY with a single SecA and a trapped preprotein has been identified (Duong, 2003) but it is unclear whether this detergent-solubilized extract represents a functional, physiological state, and later studies by the same authors revealed all possible numerical combinations of SecY/SecA complexes (Tziatzios et al., 2004).
The quaternary organization of the SecA motor raised heated debate. Tom Rapoport suggested that the SecA ATPase motor functions as a monomer (Or et al., 2002; 2005), a concept refuted by Arnold Driessen (University of Groningen), who reported that cross-linked SecA dimers are functional (de Keyzer et al., 2005), supporting his earlier FRET studies (Driessen, 1993). The disagreement in the field extends to the surfaces involved in SecA dimerization, because two published (Hunt et al., 2002; Sharma et al., 2003) and three new structures of SecA presented at the meeting from Bacillus subtilis (Tom Rapoport), Thermus thermophilus (Hiroyuki Mori, Kyoto University) and E. coli (Tassos Economou, IMBB and University of Crete) all show different dimerization interfaces. What is more, there are conflicting data on the importance of an amino-terminal decapeptide of SecA, which has been reported to affect (Or et al., 2005) and to be unrelated to (Karamanou et al., 2005) dimer formation.
Clearly, cocrystallized complexes of an active translocase holoenzyme with and without preproteins will be required to address many of these issues. Until we have them, we can only speculate. One open possibility is that many (or all) of the presented models of translocase and its subunits are correct. The Sec translocase might assemble on demand into quaternary complexes of different functional specificities. It is conceivable, for example, that the quaternary organization of SecYEG that catalyses membrane protein insertion (see below) is different from that of SecYEGA that catalyses protein export.
Membrane insertion
Historically, considerably more attention has been devoted to how proteins get across membranes than to how they insert into them. In part, this has been due to the experimental intractability of membrane insertion; unlike protein translocation, one cannot simply look in a supernatant fraction to see how much protein has appeared there. The subject of membrane insertion was given a boost, however, with the discovery that bacteria possess an essential protein, YidC (Samuelson et al., 2000), that is similar to Oxa1, a protein involved in the insertion of mitochondrial inner membrane proteins. As explained by Ross Dalbey from Ohio State University, Columbus, YidC depletion turns out to have quite pleiotropic effects on protein traffic in bacteria. Some of these effects are direct, while others are due to defects in the insertion of YidC-dependent proteins involved in energy transduction and translocation (Yi et al., 2003). Interestingly, YidC seems to operate both in conjunction with (Houben et al., 2002) and without (van der Laan et al., 2004) the SRP-Sec system. Exactly how YidC operates remains a mystery.
The translocation role of the Sec machinery is now well established and the crystal structure of one such machinery has helped conceptualize models for how it works. Transmembrane segments of integral inner membrane proteins that use the Sec machinery in E. coli must leave the translocon and slip laterally into the membrane. This process presumably occurs through sideways opening of the translocation channel to the lipid bilayer (Osborne et al., 2005). The orientation of successive transmembrane segments is determined by charged amino acids on either side of these hydrophobic anchors and, as explained by Mikhail Bogdanov from the University of Texas, Houston, by charged phospholipids. Incorrect topology results from phosphatidylethanolamine (PE) depletion in vivo (usually, one transmembrane segment loops out of the membrane and subsequent segments are in the wrong order). This can be mimicked by reconstituting membrane proteins into phospholipid vesicles in vitro. Furthermore, the incorrect topology can be corrected by adding back the missing PE, implying that, contrary to established dogma, topology is not irrevocably fixed in time and space (Zhang et al., 2003).
Despite substantial effort by a number of groups, the mechanisms of outer membrane protein (OMP) insertion remained only vaguely understood until the breakthrough discovery of the role played by the essential Omp85/YaeT protein (Voulhoux et al., 2003; Wu et al., 2005). In the talks by Tom Silhavy (Princeton University) and Jan Tommassen (University of Utrecht), we learned that Omp85/YaeT is required for the insertion of many, and possibly all β-barrel OMPs and, in E. coli at least, is associated with several lipoproteins, some of which are also essential cell components (Malinverni et al., 2006). Jan Tommassen demonstrated that, in artificial lipid bilayers, E. coli YaeT forms channels whose activity can be modulated by the C-terminal amphipathic β strand from an outer membrane porin. This is a very significant finding because Tommassen’s previous work showed the critical importance of this segment of OMPs for their insertion (Struyve et al., 1991). Why YaeT should form a channel is not immediately clear, and neither is the way it promotes insertion. One intriguing possibility is that the POTRA repeat domains (Sanchez-Pulido et al., 2003), of which there are five in the large predicted periplasmic domain of YaeT, interact with successive amphipathic β strands as they arrive at the outer membrane. As all OMPs characterized to date have less than 20 transmembrane segments, there would have to be four YaeT monomers per complex to accommodate a single OMP. According to Tommassen, Omp85/YaeT does appear to be tetrameric, although the number of protomers per complex might vary to accommodate proteins with different numbers of transmembrane segments.
Thus, the POTRA domains could form a periplasmic cavity in which the OMP β strands are correctly organized (note that, in contrast to membrane proteins with α-helical transmembrane segments, which are often arranged in non-linear fashion, transmembrane segments in OMPs are organized in numerical order around the walls of the barrel) prior to insertion. Once the complete barrel has assembled, it might slide perpendicularly into the membrane within the Omp85/YaeT superstructure, which would then dissociate to release the OMP into the membrane, where multimerization could occur.
Outer membrane blebs
By definition, the term secretion implies an active, selective and non-destructive process. The (usually genetic) identification of cellular components specifically devoted to the process is one of the cornerstones of studies into the molecular mechanisms underlying protein secretion. If no such cellular components can be identified, then the system cannot be designated as bona fide secretion. Many Gram-negative bacteria release outer membrane vesicles that contain secreted proteins, apparently without the intervention of any known ‘secretion’ factor but also independently of membrane perturbation (McBroom et al., 2006; Whiteley and Mashburn-Warren, 2006). Vesicle blebbing increases when the load of secreted protein increases, as illustrated by Tomoko Yamamoto from Chiba University for PagC release by Salmonella enterica. Although there is ample evidence that membrane blebs might play important roles in the delivery of toxins and other compounds to target cells (Whiteley and Mashburn-Warren, 2006), the jury is still out on whether the process is dedicated to this function and whether it can be considered as active and selective.
Crossing the outer membrane from the periplasm
The translocation-competent state that secreted proteins adopt in the periplasm prior to and during secretion by two-step systems remains elusive. In the case of autotransporters (T5SS), dogma states that the N-terminal passenger domain of these modular proteins traverses linearly as a hairpin in an unfolded conformation through a β-barrel channel formed by the C-terminal domain. This model has come under increasing fire (Veiga et al., 1999; Oomen et al., 2004; Skillman et al., 2005). The small size of the C-terminal domain channel is incompatible with accommodating folded passengers whose structures are known, such as pertactin (Emsley et al., 1996) and the haemoglobin protease, Hbp (Otto et al., 2005), yet passengers can fold in the periplasm (Veiga et al., 1999; Brandon and Goldberg, 2001). Indeed, EspP (Skillman et al., 2005) and IcsA (Brandon and Goldberg, 2001), two passengers with neighbouring cysteines and that fold in the periplasm, are efficiently secreted. However, there appears to be a limit to the extent that folding can be tolerated. Wouter Jong from Joen Luirink’s lab at the Vrije Universiteit in Amsterdam showed that translocation of the folded Hbp passenger engineered with strategically located cysteine pairs was hampered significantly when the cysteine pairs were well separated but not when they were close.
If native passengers can fold in the periplasm, how do they traverse the outer membrane? Rachel Fernandez from the University of British Columbia in Vancouver presented further characterization of a region at the C-terminus of autotransporter passengers called the ‘junction’. She showed that the junction is subdivided into a moiety that nucleates passenger folding and a highly conserved region that appears to promote efficient secretion (Velarde and Nataro, 2004). Deletion of this region significantly reduced the surface presentation of BrkA but did not affect folding of the passenger domain. How this region functions to potentiate secretion and whether or not Omp85/YaeT (Oomen et al., 2004) or other proteins are involved in this process are not known. Stephanie Pommier from Kim Hardie’s lab at the University of Nottingham presented a poster further characterizing six candidates derived from a screen of transposon mutants unable to secrete EspC. Whether one or more of these candidates contributes to the folded state and translocation of the passenger remains to be determined.
The folded state of the 250-residue secretion domain that targets filamentous haemagglutinin (FHA) to its cognate channel of its dedicated TPSS, FhaC was discussed by Françoise Jacob-Dubuisson (Institut Pasteur, Lille). Using overlay, and pull-down assays, she showed that a denatured (i.e. non-native) derivative of FHA was better able to bind FhaC than its native counterpart (Hodak et al., 2006). Jacob-Dubuisson suggested that FHA attains its final conformation only after it is secreted. Interestingly, the secretion domain of FHA seems to interact with the periplasmic domain of FhaC, a member of the Omp85/YaeT superfamily. Thus, FhaC might be an Omp85/YaeT-like protein that has evolved to permit specific translocation of a large soluble protein across the outer membrane.
Embrace me till I fold
In the T2SS, secretory proteins are first taken to the periplasm by the Sec machinery (or, in a few cases, by the TAT machinery) and then fold before becoming secreted through large ‘channels’ formed in the outer membrane by proteins termed secretins (Bayan et al., 2006). Lipases that use the T2SS for secretion are very particular about acquiring their proper native state in the periplasm, because they could easily adopt a non-native secretion-incompetent state, akin to a molten globule, that would not be recognized by the secretion machinery. Proper folding requires assistance from a ‘lipase foldase’ (Lif), a ‘steric chaperone’. In fact, Lif can be added exogenously to resurrect the lipase from an inactive state to its properly folded form. Lif is anchored in the inner membrane, with its large ‘foldase’ domain in the periplasm. Patrick Van Gelder from the Flanders Interuniversity Institute for Biotechnology and Free University of Brussels presented the crystal structure of recombinant Lif devoid of its membrane anchor and complexed with its lipase substrate (Pauwels et al., 2006) (Fig. 1). The structure reveals the rather mundane lipase structure, ‘properly’ folded (blue) and practically indistinguishable from a structure of the lipase crystallized alone. Lif (green) on the other hand presents a novel, extensively α-helical elongated scaffold that wraps around the lipase, making numerous contacts. Based on these and other findings, Van Gelder proposed that the malleable Lif embraces the lipase, making several simultaneous contacts that promote proper folding.
Fig. 1.
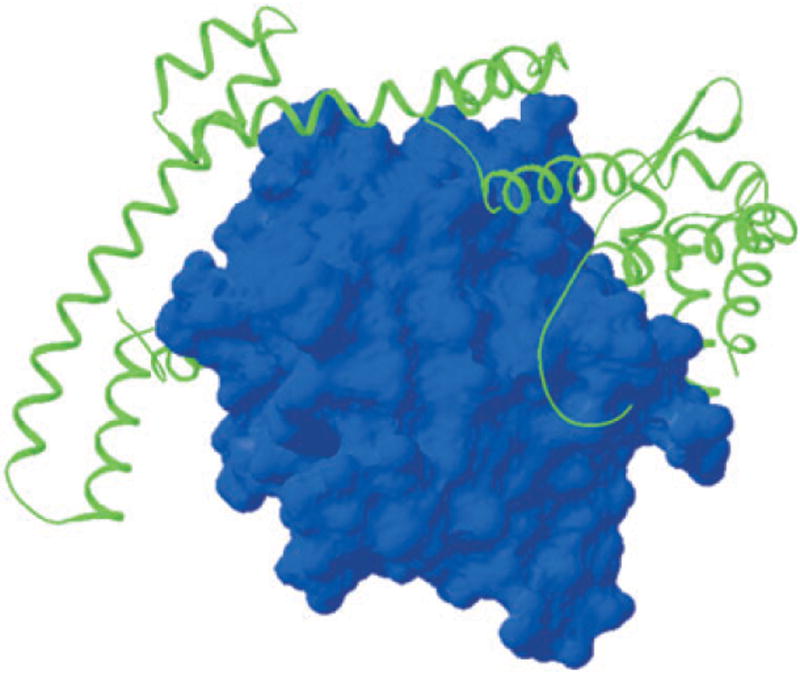
Lipase of Burholderia glumae (blue) with its specific chaperone (Lif) wrapped around it to form a large folding platform (Pauwels et al., 2006).
Type III secretion chaperones
T3SS substrates for this system fall into two classes: components of the machinery itself (the filamentous surface appendage that bridges the gap between bacterial and target cell components that integrate into the plasma membrane of the target cell) and late substrates (effectors) that are translocated into the target cell. Effectors sometimes come in waves according to the way their production is synchronized by complex, interwoven regulatory pathways (Mattoo et al., 2004; Parsot et al., 2005; Yahr and Wolfgang, 2006). Most effectors have a cognate chaperone (Page and Parsot, 2002) that always binds to the unfolded substrate as a dimer and is often encoded in an operon with the effector. Not all effectors have known cognate chaperones, however. One such effector was YopO of Yersinia enterocolitica. Guy Cornelis and colleagues at the University of Basel noted that the product of a gene located in the same operon as yopO on the Y. enterocolitica virulence plasmid has features shared by effector chaperones. Using pull-down assays, the Cornelis group found that, in common with most chaperones, SycO binds around amino acids 20–77 of its substrate (YopO) (Letzelter et al., 2006). Previous studies with a sycO knock-out strain failed to identify this chaperone function because SycO does not have any influence on the long-term (cotranslational) secretion of YopO in vitro; i.e. in the absence of target cells and under artificial secretion-activating conditions. However, SycO is clearly required in vivo, where pre-synthesized effectors are secreted in a sudden burst during infection.
Cornelis and colleagues went on to explore the function of T3SS chaperones; their conclusion might alter the way we view the function of these proteins. Surprisingly, deletion of the YopO chaperone-binding domain actually increased rather than decreased its secretion in vitro. Moreover, the chaperone-binding domain of YopO caused the protein to aggregate in the bacterial cytoplasm unless SycO was present. Thus, the sole function of SycO is to prevent YopO from aggregating in the producing cell, and the only reason that YopO has a chaperone-binding domain is to bind its chaperone. Cornelis reasoned that this interpretation must be too naive and that the SycO-binding domain in YopO must play a critical role within the eukaryotic cell. This is indeed the case: the SycO binding site targets YopO, a serine-threonine protein kinase, to the target cell plasma membrane. Subsequent work with other translocated Yops led Cornelis to propose that the role of many T3SS chaperones is to mask essential but aggregation-prone domains in effectors prior to their secretion.
Nikhil Thomas from Dalhousie University in Halifax, reported on the hierarchical delivery of effector proteins in enteropathogenic E. coli (EPEC). In EPEC, the type III secretion apparatus and effector proteins are encoded on the LEE (Locus of Enterocyte Effacement). One of the major effectors in EPEC is Tir, which inserts into the host cell membrane and serves as a receptor for the EPEC OMP intimin (Hartland et al., 1999). The chaperone CesT, which is essential for Tir secretion (Abe et al., 1999), is structurally similar to other effector chaperones. Thomas showed that immobilized CesT binds to all of the different effector proteins encoded on the LEE, indicating that it probably acts as a multivalent chaperone (Thomas et al., 2005). Alignment of the sequences of the multiple effectors allowed the identification of a degenerate CesT binding domain. Deletion of this domain from Tir, or from another effector protein, NleH, abolished CesT binding and effector secretion. Furthermore, replacement of the CesT binding domain of Tir by that of NleH produced a chimera that was secreted, indicating the modular nature of the chaperone binding domains. Interestingly, Thomas reported that the co-ordinated interaction of CesT with Tir was necessary for the efficient secretion of other effectors, suggesting yet another way that the hierarchy of effector secretion might be established.
Effector translocation by the T3SS is usually stimulated by contact with the host cell. In Yersinia pestis this can be mimicked by growing cells at 37°C in a medium lacking calcium, whereupon effectors are secreted into the growth medium. Secretion of Y. pestis Yop effectors is prevented in the presence of calcium by a quaternary protein complex located in the bacterial cytoplasm and comprising YopN (a secreted effector), SycN-YscB (the heterodimeric secretion chaperone for YopN) and TyeA (a small protein that binds to the C-terminus of YopN). David Waugh from the National Cancer Institute, Frederick, Maryland presented a model for the structure of this quaternary complex, deduced from the X-ray structures of two overlapping complexes (YopN-SycN-YscB and YopN-TyeA).
The interaction of TyeA with YopN prevents the chaperone-assisted secretion of YopN. Upon dissociation of TyeA, YopN is secreted but TyeA remains inside the bacterium. Curiously, the YopN-TyeA homologues in some bacteria are encoded by a single gene and are produced as a fused protein [for example SepL from EPEC (O’Connell et al., 2004)]. Greg Plano from the University of Miami presented evidence that, in Y. pestis, a proportion of the secreted YopN protein is fused to TyeA, despite the fact that separate but overlapping genes encode these two proteins. Plano demonstrated that this protein fusion arose as a result of a +1 translation frameshift close to the end of the yopN open reading frame. Engineering a frameshift in the yopN-tyeA coding region produced a chimera that was almost fully able to maintain the calcium-dependent block in secretion. It is not clear whether there is any selective advantage to producing these regulatory proteins as separate- or as fused polypeptides. Indeed, frameshifting seems to be an increasingly common feature of T3SSs, but its raison d’être remains elusive (Parsot et al., 2005; Penno et al., 2005).
Pins and needles
T3SS machineries span both the inner and outer membranes and are topped by a hook/filament structure (flagellar T3SSs) or a needle-like structure (virulence-associated T3SSs). The flagellar hook and filament function as a protein secretion channel during hook/filament assembly and effector secretion, and as a rotary propeller for bacterial motility. The external needle of virulence-associated T3SSs is required for the secretion of T3SS substrates and for the injection of effector proteins into eukaryotic cells. The crystal structure of the bacterial filament protein flagellin has been determined (Samatey et al., 2001) but relatively little is known about the structure and function of the T3SS needle.
Janet Deane in Susan Lea’s lab at the University of Oxford reported the long-awaited crystal structure of the Shigella flexneri needle subunit protein MxiH. A MxiH protein lacking the C-terminal five residues, which are required for MxiH polymerization, was crystallized as a monomer. Analysis of the MxiH-Δ5C structures revealed two distinct structural conformations. The basic structure consists of N- and C-terminal helices connected by a loop formed by a conserved P-X-X-P motif. A distinct kink in the C-terminal helix of one MxiH structure differentiates it from the other and suggests the presence of a molecular hinge. The extreme N-terminus of MxiH was disordered and not seen in the crystal structure (Deane et al., 2006).
A three-dimensional electron microscopy (EM) reconstruction of the assembled MxiH needle has been presented previously (Cordes et al., 2003). The needle is approximately 7 nm in diameter with a 2–3 nm wide central channel. A model of the assembled T3SS needle was generated by docking the MxiH crystal structure into the EM reconstruction. The model has the C-terminal helix of MxiH positioned on the outside of the needle and the N-terminal helix forming the inside surface of the needle. This arrangement is opposite to that reported for the assembled flagellin subunits (Samatey et al., 2001). Interestingly, MxiH residues where substitution causes constitutive secretion (Kenjale et al., 2005) mapped primarily to the interface between MxiH subunits, suggesting that alterations in intersubunit interactions could be involved in triggering secretion upon contact with a host cell. Deane showed that the dimensions of the central channel of the needle are only sufficient to accommodate partially unfolded translocation substrates (Deane et al., 2006) (Fig. 2), which therefore must occupy a considerable length of the needle at any given moment during their movement through the needle.
Fig. 2.
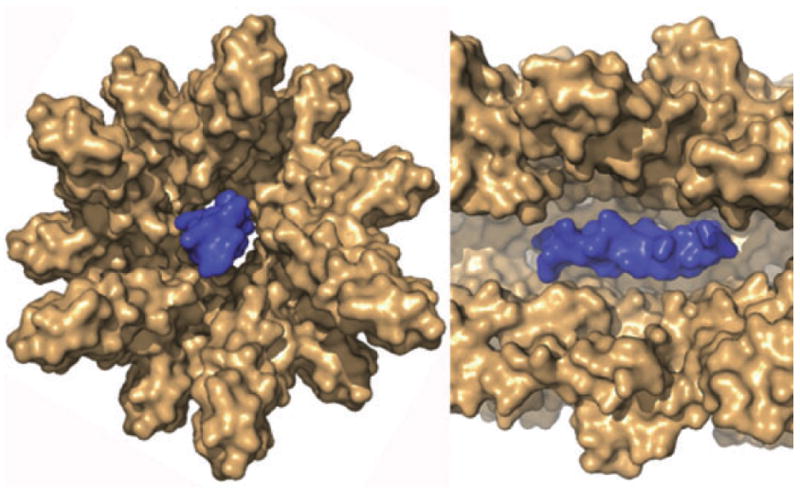
Cross sections of the proposed structure of the T3SS needle complex (brown), based on the crystal structure of MxiH modelled into a three-dimensional EM reconstruction of the needle with an helix (blue) modelled at its centre to show that the dimensions of the central channel cannot accommodate more than simple secondary structures in proteins (needle subunits, translocator proteins, effectors) that move through it. (Figure generously supplied by Janet Deane).
Effector secretion depends totally on the presence of the T3SS needle subunit protein. As already mentioned, assembled T3SS needles also play an as yet undefined role in regulating T3SS activity (Kenjale et al., 2005; Torruellas et al., 2005). A direct role of the needle in effector translocation across the eukaryotic membrane has also been suggested (Torruellas et al., 2005) but direct evidence of this activity is lacking. The recent discovery of a tip complex composed of the secreted LcrV protein that is required for translocon assembly suggests the existence of a bridge between the needle and the translocation effectors inserted into the membrane of the target cell (Mueller et al., 2005).
Alison Davis in Joan Mecsas’ lab at Tufts University, Boston, presented evidence that the Yersinia pseudotuberculosis YscF needle plays an essential role in effector translocation. An elegant two-step screen was devised to identify YscF mutants that assembled secretion-competent but translocation-defective T3SS needles. In other words, these mutants could secrete proteins across the bacterial membranes but could not translocate them across a eukaryotic membrane. Error-prone PCR was used to generate a library of YscF expression plasmids. A Congo red dye plate-based screen identified YscF mutants that remained competent for secretion. Finally, a HEP-2 cell-based cytotoxicity assay identified secretion-competent strains that were unable to inject effectors. Approximately 25 YscF mutants showed significant secretion of all effector and translocator proteins but severely reduced effector translocation. The amino acid substitutions concerned were spread throughout the 87-residue YscF protein. Several of these YscF mutants also exhibited defects in the calcium- and cell contact-dependent regulation of the T3SS. Most importantly, at least five YscF variants showed normal calcium-dependent T3SS regulation but essentially no effector protein translocation. Most of these variants were unable to form T3SS-dependent pores in target cells, which could be explained by a defective YscF–LcrV interaction. Overall, these studies suggest that the T3SS needle is much more than a static channel for protein secretion but, instead is a multifunctional appendage that plays a role in effector secretion and in regulating secretion and effector translocation across the eukaryotic membrane.
Type IV secretion
The T4SSs of Gram-negative bacteria are composed of a substrate receptor, an envelope-spanning translocation channel and an extracellular pilus or surface filament. These systems translocate DNA and protein substrates, generally by a contact-dependent mechanism, though some T4SS machines release protein and DNA to the milieu and take up DNA (transformation). As with the T3SSs, the two major areas of investigation have addressed the T4SS machine structures and functions, and the roles of effector protein and DNA substrates upon translocation to target cells. The findings reported in Crete focused mainly on the former topic, with the exciting exception of the report by Christoph Dehio (University of Basel) that Bartonella henselae can deliver the MobA relaxase together with covalently bound transfer DNA into human cells. Evidence was presented for random integration of the transfer DNA in the chromosome and expression of a neomycin transferase gene. The mechanism of integration appears to resemble illegitimate recombination, however, in contrast to the well studied mechanism of T4SS-mediated T-DNA transfer from Agrobacterium tumefaciens into plant, yeast and mammalian cells, the illegitimate integration event does not appear to depend on the covalently attached protein.
Structural studies of T4SS components have been at the forefront of efforts to define its architecture. Pete Christie from the University of Texas Medical School, Houston reported results from collaborative studies with Gabriel Waksman on the bitopic energy sensor protein VirB10. In A. tumefaciens, VirB10 senses ATP energy use by two inner membrane-associated proteins, VirD4 and VirB11 and, in turn, undergoes a conformational change required for stable complex formation with the outer membrane-associated VirB9 protein (Cascales and Christie, 2004). Crystallography of a soluble, periplasmic domain of a VirB10 homologue revealed two alpha-helical extensions projecting from the overall globular structure (Terradot et al., 2005). Mutagenesis studies revealed that the alpha helical domain 1 contributes to both VirB10 energy sensing and VirB9 complex formation. The model based on these findings proposes that VirB10 senses ATP energy use through TM contacts with VirD4 and/or VirB11 and then undergoes a conformational change enabling a stable interaction to form between its periplasmic globular domain and VirB9 (V. Krishnamoorthy and P.J. Christie, submitted).
What is the physiological role of energy sensing? The VirB9 homologues of T4SSs have been proposed to assemble as outer membrane channels through which substrates and conjugative pili protrude. Beyond weak sequence similarities with secretins, however, there is no experimental evidence to support this model. The Waksman group has now resolved an NMR structure of a complex of the C-terminal domain of pKM101 TraO (VirB9 homologue) bound to full-length TraN (VirB7 lipoprotein homologue). VirB9CT forms a β-sandwich around which VirB7 winds. On the opposite side of the VirB7-binding site, VirB9CT contains a protruding three-stranded β-appendix. Immunofluorescence microscopy and cysteine accessibility studies showed that this structure is surface-exposed. Thus, VirB9 protrudes across the outer membrane, raising the intriguing possibility that VirB10 energy sensing might regulate VirB9 insertion and β-pore formation in the outer membrane (R. Bayliss et al., submitted).
Anath Das from the University of Minnesota, Minneapolis updated earlier findings that the VirB/D4 T4SS machine assembles predominantly at one cell pole of A. tumefaciens. Mutagenesis studies revealed that the periplasmic domain of VirD4, the substrate receptor (also termed the coupling protein), is necessary for polar positioning. Moreover, using GFP fusions, Das and colleagues showed that the N-terminal third of VirD4 contains a transmembrane loop that is necessary and sufficient for polar localization. Last, alanine-scanning mutagenesis identified a single amino acid substitution in the periplasmic domain that abolished polar targeting and DNA transfer function. These approaches were extended to study VirB8, a bitopic protein whose polar positioning is a prerequisite for recruiting several other VirB proteins. The data establish that the periplasmic region of VirB8 is both necessary and sufficient for polar positioning as well as for VirB8 to function as an assembly factor for the VirB/D4 machine (Judd et al., 2005). Pete Christie reported that VirC1, a ParA-like protein required for processing at T-DNA borders, also assembles at A. tumefaciens cell poles but independently of the VirD4 and VirB proteins. Besides its role in conjugative DNA metabolism, VirC1 recruits the VirD2 relaxase-T-DNA transfer intermediate to the cell poles. Through its interaction with VirD4, VirC1 could function as a spatial determinant for the T-DNA substrate – VirD4 receptor docking reaction (K. Atmakuri and P.J. Christie, submitted).
Hiroshi Nagai from Osaka University reported intriguing results on how T4SS substrates are recognized by the Legionella pneumophila Dot/Icm T4SS. Nagai used a computational screen that focused on candidate effectors bearing a specific C-terminal secretion motif. He calculated the number of effectors to be around 150, and tested candidates using an adenylate cyclase-based assay (Nagai et al., 2005) used previously to identify the first ~30 substrates. Nagai has now added two dozen more effector candidates to this list, making this one of the most prolific ‘specific’ translocation systems known. It will now be necessary to determine whether this secretion motif is both necessary and sufficient for translocation of the native protein. Such further studies are especially important in view of emerging evidence that membrane-bound substrate binding proteins play crucial roles in spatial and/or temporal regulation of substrate presentation to cognate T4SS machines. In addition to the role of VirC1 as a spatial determinant in the A. tumefaciens system, talks by Marc Couturier from Markus Stein’s laboratory at the University of Alberta in Edmonton and Wolfgang Fischer from the Max von Pettenkofer Institute in Munich reported that a membrane-associated protein CagF interacts with the CagA substrate of the Helicobacter pylori Cag T4SS. This interaction is required for CagA translocation and, thus, CagF could function as a chaperone required for early recognition and configuration (possibly unfolding) of CagA for translocation. In line with its proposed chaperone activity, CagF binds the N-terminal region and probably also a C-terminal domain of CagA. Fischer also reported that deletions of the extreme C-terminus (not including the CagF-binding domain) render CagA translocation-incompetent. Together, these data suggest that a combination of CagF binding to distinct regions of CagA plus a C-terminal region composed of a cluster of positive charges is necessary for translocation (Couturier et al., 2006; Hohlfeld et al., 2006). In these properties, CagA resembles A. tumefaciens VirE2, which possesses distinct N- and C-terminal VirE1-chaperone binding domains as well as a charged C-terminal motif for translocation by the VirB/D4 T4SS apparatus (Zhao et al., 2001; Atmakuri et al., 2004). Finally, adding to the evidence that T4SS substrates carry multiple secretion/translocation signals, Christoph Dehio has shown that the Bartonella-translocated effector proteins (Beps) require both a short C-terminal charged tail sequence and at least one other domain (Bep intracellular delivery or BID) domain for translocation by the Bartonella VirB/D4 system. BID domains might serve as binding targets for proteins involved in spatio-temporal control of substrate presentation to the cognate T4SS machine (Schulein et al., 2005).
Until recently, mechanistic studies have focused mainly on the T4SS machines of Gram-negative bacteria but Elisabeth Grohmann from the Technische Universität in Berlin has begun to explore the mechanics of T4SS in Gram-positive bacteria. She is investigating the Tra system of plasmid pIP501, which is capable of mobilizing pIP501 transfer among a wide range of Gram-positive bacteria and even to Gram-negative bacteria. This system is composed of ~15 Tra proteins, of which several are homologues of the A. tumefaciens VirB/D4 transfer system. Homologues include TraA relaxase, whose catalytic action at the pIP501 origin of transfer sequence resembles that of VirD2 and other relaxases operating in Gram-negative bacteria. Among the T4SS machine components, Orf5 is related to the VirB4 ATPase, Orf7 to the VirB1 transglycosylase, and Orf10 to the VirD4 substrate receptor. In collaboration with Günther Koraimann at the Karl-Franzens-Universität, Graz, Grohmann demonstrated Orf7 transglycosylase activity as well as evidence for an Orf7 self-association and interaction with Orf5. These important pioneering studies are setting the stage for detailed mechanistic comparisons between the T4SS machines of Gram-negative and Gram-positive bacteria (Grohmann et al., 2003; Kurenbach et al., 2006).
Counting up to 6
Joseph Mougous from Harvard Medical School presented details of his work on the T6SS of Pseudomonas aeruginosa. The first detailed description of this system, from the same lab, reported its requirement for Vibrio cholerae virulence on Dictyostelium (Pukatzki et al., 2006). In Pseudomonas, the system contributes to pathogenicity in patients with cystic fibrosis (Mougous et al., 2006). Hallmarks of this system include the presence of an orthologue of IcmF, a cytoplasmic membrane protein that confers stability to the Legionella T4SS (Sexton et al., 2004) and a AAA+ ATPase. The proteins secreted by this system lack a Sec signal peptide, but information on where proteins accumulate when the system is genetically blocked, the identity of the signal that mediates their interaction with the secretion machinery and, indeed, the identity, location and functions of most of the T6SS components remain unknown. One interesting substrate is Hcp1, which forms a homohexameric ring with a large internal diameter. It was speculated that Hcp1 monomers oligomerize following secretion, but the precise secretion mechanism remains to be explored. Hcp1 seems to be actively secreted in lungs of CF patients with long-term P. aeruginosa infections. Mougous further reported that one component of the system is a hexameric AAA+ ClpB-like ATPase that locates at specific sites in the cell envelope when the machinery is active, and presumably provides the driving force for direct translocation through the cell envelope. The fact that the T6SS was discovered only this year suggests that the field of bacterial protein translocation might well be enriched by more newly discovered systems in the next few years.
References
- Abe A, de Grado M, Pfuetzner RA, Sanchez-Sanmartin C, Devinney R, Puente JL, et al. Enteropathogenic Escherichia coli translocated intimin receptor, Tir, requires a specific chaperone for stable secretion. Mol Microbiol. 1999;33:1162–1175. doi: 10.1046/j.1365-2958.1999.01558.x. [DOI] [PubMed] [Google Scholar]
- Arkowitz RA, Joly JC, Wickner W. Translocation can drive the unfolding of a preprotein domain. EMBO J. 1993;12:243–253. doi: 10.1002/j.1460-2075.1993.tb05650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayan N, Guilvout I, Pugsley AP. Secretins take shape. Mol Microbiol. 2006;60:1–4. doi: 10.1111/j.1365-2958.2006.05084.x. [DOI] [PubMed] [Google Scholar]
- Brandon LD, Goldberg MB. Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. J Bacteriol. 2001;183:951–958. doi: 10.1128/JB.183.3.951-958.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, et al. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun. 2006;74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon KS, Or E, Clemons WM, Jr, Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA. 2004;101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes FS, Komoriya K, Larquet E, Yang S, Egelman EH, Blocker A, Lea SM. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- Couturier M, Tasca E, Montecucco C, Stein M. Interaction with CagF is required for translocation of CagA into the host via the Helicobacter pylori type IV secretion system. Infect Immun. 2006;74:271–283. doi: 10.1128/IAI.74.1.273-281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane J, Roversi P, Cordes F, Johnson S, Kenjale R, Daniell S, et al. Molecular model of a Type Three Secretion System needle: implications for host-cell sensing. Proc Natl Acad Sci USA. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen AJ. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry. 1993;32:13190–13197. doi: 10.1021/bi00211a030. [DOI] [PubMed] [Google Scholar]
- Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22:4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Charles IG, Fairweather NF, Isaacs NW. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature. 1996;381:90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. Table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A, Bolton D, Pelletier M, Brittnacher M, Gallagher L, Kaul R, et al. Type IV pili-mediated secretion modulates Francisella virulence. Mol Microbiol. 2006;62:227–237. doi: 10.1111/j.1365-2958.2006.05365.x. [DOI] [PubMed] [Google Scholar]
- Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- Hartland EL, Batchelor M, Delahay RM, Hale C, Matthews S, Dougan G, et al. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol Microbiol. 1999;32:151–158. doi: 10.1046/j.1365-2958.1999.01338.x. [DOI] [PubMed] [Google Scholar]
- Hodak H, Clantin B, Willery E, Villeret V, Locht C, Jacob-Dubuisson F. Secretion signal of the filamentous haemagglutinin, a model two-partner secretion substrate. Mol Microbiol. 2006;61:368–382. doi: 10.1111/j.1365-2958.2006.05242.x. [DOI] [PubMed] [Google Scholar]
- Hohlfeld S, Pattis I, Puls J, Plano G, Haas R, Fischer W. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol Microbiol. 2006;59:1624–1637. doi: 10.1111/j.1365-2958.2006.05050.x. [DOI] [PubMed] [Google Scholar]
- Houben EN, Urbanus ML, Van Der Laan M, Ten Hagen-Jongman CM, Driessen AJ, Brunner J, et al. YidC and SecY mediate membrane insertion of a Type I transmembrane domain. J Biol Chem. 2002;277:35880–35886. doi: 10.1074/jbc.M205556200. [DOI] [PubMed] [Google Scholar]
- Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- Joly JC, Wickner W. The SecA and SecY subunits of translocase are the nearest neighbors of a translocating preprotein, shielding it from phospholipids. EMBO J. 1993;12:255–263. doi: 10.1002/j.1460-2075.1993.tb05651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd PK, Kumar RB, Das A. Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc Natl Acad Sci USA. 2005;102:11498–11503. doi: 10.1073/pnas.0505290102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junne T, Schwede T, Goder V, Spiess M. The plug domain of yeast Sec61p is important for efficient protein translocation but is not essential for cell viability. Mol Biol Cell. 2006 doi: 10.1091/mbc.E06-03-0200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanou S, Sianidis G, Gouridis G, Pozidis C, Papanikolau Y, Papanikou E, Economou A. Escherichia coli SecA truncated at its termini is functional and dimeric. FEBS Lett. 2005;579:1267–1271. doi: 10.1016/j.febslet.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Kenjale R, Wilson J, Zenk SF, Saurya S, Picking WL, Picking WD, Blocker A. The needle component of the type III secretion of Shigella regulates the activity of the secretion apparatus. J Biol Chem. 2005;280:42929–42937. doi: 10.1074/jbc.M508377200. [DOI] [PubMed] [Google Scholar]
- de Keyzer J, van der Sluis EO, Spelbrink RE, Nijstad N, de Kruijff B, Nouwen N, et al. Covalently dimerized SecA is functional in protein translocation. J Biol Chem. 2005;280:35255–35260. doi: 10.1074/jbc.M506157200. [DOI] [PubMed] [Google Scholar]
- Kurenbach B, Kopec J, Magdefrau M, Andreas K, Keller W, Bohn C, et al. The TraA relaxase autoregulates the putative type IV secretion-like system encoded by the broad-host-range Streptococcus agalactiae plasmid pIP501. Microbiology. 2006;152:637–645. doi: 10.1099/mic.0.28468-0. [DOI] [PubMed] [Google Scholar]
- van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJ. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J Cell Biol. 2004;165:213–222. doi: 10.1083/jcb.200402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzelter M, Sorg I, Mota LJ, Meyer S, Stalder J, Feldman M, et al. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 2006;25:3223–3233. doi: 10.1038/sj.emboj.7601202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. Outer membrane vesicle production by Escherichia coli Is independent of membrane instability. J Bacteriol. 2006;188:5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden JC, Ruiz N, Caparon M. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell. 2001;104:143–152. doi: 10.1016/s0092-8674(01)00198-2. [DOI] [PubMed] [Google Scholar]
- Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- Manting EH, van Der Does C, Remigy H, Engel A, Driessen AJ. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 2000;19:852–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Whiteley M. Special delivery: vesicle trafficking in prokaryotes. Mol Microbiol. 2006;61:839–846. doi: 10.1111/j.1365-2958.2006.05272.x. [DOI] [PubMed] [Google Scholar]
- Mattoo S, Yuk MH, Huang LL, Miller JF. Regulation of type III secretion in Bordetella. Mol Microbiol. 2004;52:1201–1214. doi: 10.1111/j.1365-2958.2004.04053.x. [DOI] [PubMed] [Google Scholar]
- Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CA, Broz P, Muller SA, Ringler P, Erne-Brand F, Sorg I, et al. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci USA. 2005;102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell CB, Creasey EA, Knutton S, Elliott S, Crowther LJ, Luo W, et al. SepL, a protein required for enteropathogenic Escherichia coli type III translocation, interacts with secretion component SepD. Mol Microbiol. 2004;52:1613–1625. doi: 10.1111/j.1365-2958.2004.04101.x. [DOI] [PubMed] [Google Scholar]
- Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J, Gros P. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23:1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E, Navon A, Rapoport T. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21:4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Or E, Boyd D, Gon S, Beckwith J, Rapoport T. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem. 2005;280:9097–9105. doi: 10.1074/jbc.M413947200. [DOI] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Otto BR, Sijbrandi R, Luirink J, Oudega B, Heddle JG, Mizutani K, et al. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J Biol Chem. 2005;280:17339–17345. doi: 10.1074/jbc.M412885200. [DOI] [PubMed] [Google Scholar]
- Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- Page AL, Parsot C. Chaperones of the type III secretion pathway: jacks of all trades. Mol Microbiol. 2002;46:1–11. doi: 10.1046/j.1365-2958.2002.03138.x. [DOI] [PubMed] [Google Scholar]
- Parsot C, Ageron E, Penno C, Mavris M, Jamoussi K, d’Hauteville H, et al. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol Microbiol. 2005;56:1627–1635. doi: 10.1111/j.1365-2958.2005.04645.x. [DOI] [PubMed] [Google Scholar]
- Pauwels K, Lustig A, Wyns L, Tommassen J, Savvides SN, Van Gelder P. Structure of a membrane-based steric chaperone in complex with its lipase substrate. Nat Struct Mol Biol. 2006;13:374–375. doi: 10.1038/nsmb1065. [DOI] [PubMed] [Google Scholar]
- Penno C, Sansonetti P, Parsot C. Frameshifting by transcriptional slippage is involved in production of MxiE, the transcription activator regulated by the activity of the type III secretion apparatus in Shigella flexneri. Mol Microbiol. 2005;56:204–214. doi: 10.1111/j.1365-2958.2004.04530.x. [DOI] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmond GP, Reeves PJ. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem Sci. 1993;18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- Samatey FA, Imada K, Nagashima S, Vonderviszt F, Kumasaka T, Yamamoto M, Namba K. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature. 2001;410:331–337. doi: 10.1038/35066504. [DOI] [PubMed] [Google Scholar]
- Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, et al. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem Sci. 2003;28:523–526. doi: 10.1016/j.tibs.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Scheuring J, Braun N, Nothdurft L, Stumpf M, Veenendaal AK, Kol S, et al. The oligomeric distribution of SecYEG is altered by SecA and translocation ligands. J Mol Biol. 2005;354:258–271. doi: 10.1016/j.jmb.2005.09.058. [DOI] [PubMed] [Google Scholar]
- Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, Vergunst AC, et al. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci USA. 2005;102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton JA, Miller JL, Yoneda A, Kehl-Fie TE, Vogel JP. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect Immun. 2004;72:5983–5992. doi: 10.1128/IAI.72.10.5983-5992.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, et al. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci USA. 2003;100:2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Mai D, Kumar A, Steyn AJ. Dissecting virulence pathways of Mycobacterium tuberculosis through protein–protein association. Proc Natl Acad Sci USA. 2006;103:11346–11351. doi: 10.1073/pnas.0602817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skillman KM, Barnard TJ, Peterson JH, Ghirlando R, Bernstein HD. Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Mol Microbiol. 2005;58:945–958. doi: 10.1111/j.1365-2958.2005.04885.x. [DOI] [PubMed] [Google Scholar]
- Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- Tam PC, Maillard AP, Chan KK, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradot L, Bayliss R, Oomen C, Leonard GA, Baron C, Waksman G. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102:4596–4601. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NA, Deng W, Puente JL, Frey EA, Yip CK, Strynadka NC, Finlay BB. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol Microbiol. 2005;57:1762–1779. doi: 10.1111/j.1365-2958.2005.04802.x. [DOI] [PubMed] [Google Scholar]
- Torruellas J, Jackson MW, Pennock JW, Plano GV. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol Microbiol. 2005;57:1719–1733. doi: 10.1111/j.1365-2958.2005.04790.x. [DOI] [PubMed] [Google Scholar]
- Tziatzios C, Schubert D, Lotz M, Gundogan D, Betz H, Schagger H, et al. The bacterial protein-translocation complex: SecYEG dimers associate with one or two SecA molecules. J Mol Biol. 2004;340:513–524. doi: 10.1016/j.jmb.2004.04.076. [DOI] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Veiga E, de Lorenzo V, Fernandez LA. Probing secretion and translocation of a beta-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol Microbiol. 1999;33:1232–1243. doi: 10.1046/j.1365-2958.1999.01571.x. [DOI] [PubMed] [Google Scholar]
- Velarde JJ, Nataro JP. Hydrophobic residues of the autotransporter EspP linker domain are important for outer membrane translocation of its passenger. J Biol Chem. 2004;279:31495–31504. doi: 10.1074/jbc.M404424200. [DOI] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Yahr T, Wolfgang M. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2006 doi: 10.1111/j.1365-2958.2006.05412.x/. [DOI] [PubMed] [Google Scholar]
- Yi L, Jiang F, Chen M, Cain B, Bolhuis A, Dalbey RE. YidC is strictly required for membrane insertion of subunits a and c of the F(1)F(0)ATP synthase and SecE of the SecYEG translocase. Biochemistry. 2003;42:10537–10544. doi: 10.1021/bi034309h. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Sagulenko E, Ding Z, Christie PJ. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J Bacteriol. 2001;183:3855–3865. doi: 10.1128/JB.183.13.3855-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xu Z. The structural view of bacterial translocation-specific chaperone SecB: implications for function. Mol Microbiol. 2005;58:349–357. doi: 10.1111/j.1365-2958.2005.04842.x. [DOI] [PubMed] [Google Scholar]


