Abstract
Human immunodeficiency virus type 1 (HIV-1) is pandemic, but its contemporary global transmission network has not been characterized. A better understanding of the properties and dynamics of this network is essential for surveillance, prevention, and eventual eradication of HIV. Here, we apply a simple and computationally efficient network-based approach to all publicly available HIV polymerase sequences in the global database, revealing a contemporary picture of the spread of HIV-1 within and between countries. This approach automatically recovered well-characterized transmission clusters and extended other clusters thought to be contained within a single country across international borders. In addition, previously undescribed transmission clusters were discovered. Together, these clusters represent all known modes of HIV transmission. The extent of international linkage revealed by our comprehensive approach demonstrates the need to consider the global diversity of HIV, even when describing local epidemics. Finally, the speed of this method allows for near-real-time surveillance of the pandemic's progression.
Keywords: human immunodeficiency virus, transmission network, molecular epidemiology
(See the editorial commentary by Pennings et al on pages 180–2.)
The origin and geographic expansion of human immunodeficiency virus type 1 (HIV-1) have been well characterized using phylogenetic approaches [1], but these methods are suboptimal for describing recent HIV transmission. Phylogenies are well suited for differentiating distinct viral lineages but not for identifying transmission partners; it is generally accepted that phylogenetic analysis is most powerful at excluding potential transmission partners, rather than establishing linkage [2–6]. Conversely, transmission networks focus on similarity and directly link genetically similar viruses. Although network methods have previously been used to study HIV [7, 8], partly because recombination in HIV complicates phylogenetic inference, they have not been adopted in the context of global epidemiological patterns.
Previous studies investigating HIV transmission clusters typically begin by inferring a phylogeny and then identifying those clades (ie, subtrees) that have appropriate statistical support. This identification alone, however, is insufficient for epidemiological purposes, because such an analysis lacks the concept of recency; for example, individual HIV-1 subtypes will form well-resolved clades. Hence, the isolates within supported clades are often designated as a transmission cluster if the mean [9], median [10], or maximum [11, 12] genetic diversity (a proxy for time) within these clades falls below a given cutoff. The branching structure within the phylogeny is irrelevant after these clades have been identified. A conceptual problem arises as these clades are not easily resolved into transmission pairs and clusters [11, 13, 14], and, within a transmission cluster, all individuals are treated as equally related/connected; the inferred branching structure within these clusters is again discarded (but see Leigh Brown et al [12] for an alternative approach), wasting the considerable computational effort expended to infer the complete phylogeny. Furthermore, the relationship between statistical support for a clade and population dynamics may be conceptually ambiguous [15]. High statistical support (eg, bootstrap) for any specific clade indicates that there is no close relative to the clade in question, not that the members of the clade itself are necessarily closely related to each other [16]. A final problem with using phylogenetic methods to infer transmission clusters is that often only a single geographic region is considered, and the data must be subsampled for computational tractability [9, 11, 12, 15, 17–19]. Both of these simplifications can seriously bias the interpretation due to the limited scope of the analysis, as closely related or relevant sequences may be inadvertently excluded during sequence selection.
We applied a network approach to analyze global HIV-1 transmission patterns by constructing HIV-1 transmission clusters using only close genetic links to identify potential transmission partners. In contrast with phylogenetic approaches, inclusion in our transmission clusters required only a shared connection with another member of the cluster. We identified potential transmission partners—HIV-positive individuals whose viral pol gene sequences were genetically similar (≤1% nucleotide genetic distance)—and defined transmission clusters as maximal connected groups of potential transmission partners. Focusing on only potential transmission partners (1) maintains the internal structure of the transmission clusters, (2) obviates the reliance on a phylogeny to identify highly supported clades, and (3) eliminates the need to subsample the dataset, which allows for consideration of the global diversity of HIV-1 when inferring transmission clusters.
METHODS
Sequence Dataset
All HIV-1 group M protease and reverse transcriptase gene sequences (pol, HXB2 coordinates: 2253–3554) at least 500 nucleotides long were downloaded from the Los Alamos National Laboratory (LANL) HIV sequence database on 1 February 2012. Only 1 sequence per individual, based on classification in the LANL database, was retained. These sequences represent all known subtypes, 47 circulating recombinant forms, and many unique recombinant forms. Clonal and “problematic” sequences were removed, leaving 84 757 sequences. Sequences were codon-aligned to HXB2 using an extension of the Smith-Waterman algorithm [20], which uses amino acid homology to directly align nucleotide sequences (more suitable for aligning isolates from different divergent subtypes) and corrects out-of-frame insertions and deletions (likely sequencing errors). Given the evolutionary conservation of this genomic region, codon insertions relative to HXB2 were filtered from downstream analyses.
As a conservative screen for contamination, we inspected clusters that included sequences from the 1980s, as many of them are used as laboratory controls. Eight clusters, containing a total of 230 sequences, were removed as suspected contaminants because they included sequences from both the 1980s and the late 1990s/2000s; a small genetic distance between such heterochronous sequences is implausible, and we attributed it to laboratory contamination or sample mislabeling. The largest of these clusters contained the HIV reference sequence, HXB2, and 188 other sequences sampled between 1983 and 2010 representing 25 countries and regions, with more than half the isolates coming from the United States, China, France, and Italy.
The final curated pol dataset, after removing these potential contaminants, contained 84 527 sequences.
Network Construction
Genetic distance (Tamura-Nei 93, or TN93 [21]) was determined for all pairs of pol sequences (approximately 3.5 billion comparisons). TN93 distances, which model a single nucleotide transversion rate and 2 nucleotide transition rates, were used because they are the most complex genetic distances that can be represented by a closed-form solution, allowing for rapid distance calculations. The network was constructed by identifying pairs of sequences (nodes) that overlapped for a minimum of 500 nucleotides and whose divergence was ≤1% (0.01 expected substitutions per site); these sequences were connected (an undirected edge placed between 2 nodes) in our network. Due to the low divergence cutoff used to identify potential transmission partners, network construction is not expected to be noticeably affected by evolutionary model selection [22]; hence, TN93 was chosen as a balance between computational tractability and biological realism.
Network construction was neither a computationally nor memory-intensive process, because sequences were analyzed in a pairwise fashion, and only those pairs whose divergence was below a cutoff were stored in memory (≤0.002% of total). Nucleotide ambiguities were averaged during distance calculations (eg, the distance between R:A is the mean of the distances between A:A and G:A) to prevent spurious connections between sequences of poor quality or multiple infections. This analysis (https://github.com/veg/HIVClustering; https://github.com/veg/TN93; https://github.com/veg/BioExt), including alignment, distance calculation, and network construction, can be performed in <30 minutes on a small (128 CPUs) computer cluster and in <8 hours on a standard laptop computer (2.53 GHz Intel Core 2 Duo MacBook Pro), making it an extremely efficient tool for rapidly reconstructing the recent history of HIV-1. The addition of a sequence to an existing network has linear complexity in the size of the current network and, in practical terms, can be accomplished nearly instantaneously, making the method very attractive for an online implementation.
Bootstrap Datasets
To test the overall stability of our network inference, network replicates were constructed from 1000 bootstrapped codon alignments. We calculated the frequency of edges from the global network in the bootstrap datasets and explored the variance in the genetic distance these edges represent.
RESULTS
Network Inference
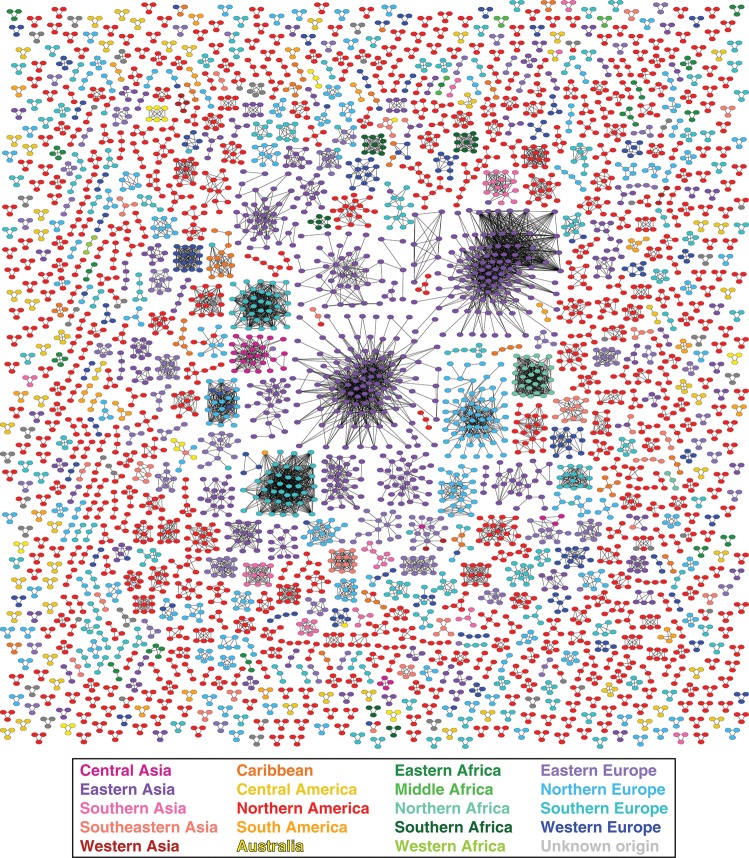
The global transmission network was built from every publicly available HIV-1 group M pol sequence (n = 84 527, 1 sequence per individual), representing 141 countries/regions (Table 1). The network contained 4342 connected components with ≥2 nodes (clusters) comprising 13 295 nodes (individual sequences) and 51 182 edges (undirected, potential transmission links) (Figures 1 and 2 and Supplementary Figure 1). The average degree (number of edges per node) was 3.84.
Table 1.
Los Alamos National Laboratory pol Sequences in Global Network Analysis and Their Propensity for Clusteringa
| Geographic Regionb | pol Sequences in LANL | Sampling Yearsc | Observed Clustered/Expected Clustered | χ2 Test |
|---|---|---|---|---|
| Central Asia | 273 | 1998–2009 | 5.43 | *** |
| Eastern Asia | 4916 | 1991–2010 | 2.38 | *** |
| Eastern Europe | 2885 | 1986–2011 | 2.02 | *** |
| Northern America | 17 836 | 1982–2010 | 1.82 | *** |
| Central America | 1124 | 2001–2010 | 1.78 | *** |
| Western Asia | 333 | 1994–2009 | 1.58 | *** |
| Northern Europe | 4330 | 1986–2009 | 1.55 | *** |
| Southern Europe | 7187 | 1991–2011 | 1.00 | NS |
| Northern Africa | 306 | 1998–2010 | 0.97 | NS |
| Southern Asia | 1748 | 1993–2010 | 0.94 | NS |
| Southeastern Asia | 4080 | 1990–2011 | 0.92 | NS |
| Caribbean | 1505 | 1996–2010 | 0.89 | NS |
| Western Europe | 5640 | 1986–2009 | 0.70 | *** |
| Eastern Africa | 5323 | 1985–2010 | 0.57 | *** |
| Australiad | 819 | 1987–2004 | 0.49 | *** |
| Middle Africa | 1967 | 1983–2009 | 0.42 | *** |
| South America | 8685 | 1989–2010 | 0.40 | *** |
| Southern Africa | 3206 | 1984–2010 | 0.34 | *** |
| Unknown origine | 9861 | 2001–2003 | 0.25 | *** |
| Western Africa | 2503 | 1990–2010 | 0.14 | *** |
Abbreviations: LANL, Los Alamos National Laboratory; NS, not significant.
a Propensity for clustering was not uniform across geographic regions (χ2 test, P < .0001, df = 19). Data are shown from most overrepresented to most underrepresented.
b Based on the United Nations geographic region and composition (http://unstats.un.org/unsd/methods/m49/m49regin.htm).
c Twenty percent of sequences in LANL do not have associated sampling years.
d Includes 1 sequence from Fiji.
e Sequences without associated geographic sampling information in LANL.
***Bonferroni-corrected P value < .001 (df = 1).
Figure 1.
Partial human immunodeficiency virus type 1 global transmission network inferred from pol locus. All clusters containing between 3 and 125 nodes are depicted (n = 1038). Edge lengths are optimized for visual presentation and do not represent genetic distances between potential transmission partners. All nodes are of equal size. Colors correspond to geographic regions.
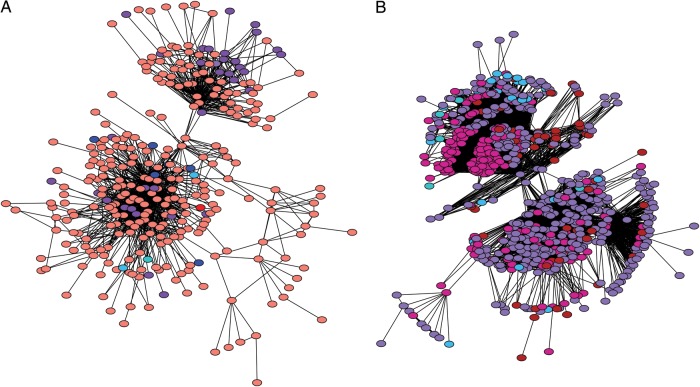
Figure 2.
Large international transmission clusters. A, Southeast Asian transmission cluster, CRF01_AE, primarily from Thailand and Vietnam. Additional isolates from Belgium, Cambodia, China, Czech Republic, France, Greece, Indonesia, Japan, Luxembourg, Malaysia, Singapore, Sweden, Taiwan, United Kingdom, and United States. B, Former Soviet Union transmission cluster, subtype A1, primarily from the former Soviet Union. Isolates from Azerbaijan, Belarus, Cyprus, Czech Republic, Denmark, Georgia, Israel, Kazakhstan, Latvia, Moldova, Portugal, Russia, Slovenia, Spain, Sweden, Ukraine, United Kingdom, and Uzbekistan. Edge lengths are optimized for visual presentation and do not represent genetic distances between potential transmission partners. All nodes are of equal size, and colors correspond to geographic regions in Figure 1.
Like smaller, local HIV-1 networks [12, 23] and sexual networks in general [24], the inferred global network appears to be scale-free. A classical mechanism for generating scale-free networks is the preferential attachment process, whereby a new member of the network connects to an existing node with probability proportional to the latter's degree (eg, a connection to an existing node with degree 10 is 5 times more likely than to a node with degree 2). We compared the fit of 4 different degree distributions (negative binomial [not scale free], Pareto, Waring, and Yule [all scale free]) using Bayesian information criterion [12]. The Waring distribution, a preferential-attachment model with a proportion of connections formed randomly [25], provided the best fit to the inferred degree distribution. The characteristic exponent of the Waring distribution (ρ) for the global transmission network is 1.74; a smaller ρ indicates a higher propensity for high-degree nodes.
The network analysis is performed without explicit regard for HIV-1 subtype. Misclassification of subtype or recombinant provenance (a common problem [26]) will not affect the inference of the global network, because no sequences were excluded based on prior classification. Moreover, distance-based approaches are able to handle the inclusion of recombinant sequences, as no single underlying phylogeny is assumed. However, the oversampling of certain subtypes and circulating recombinant forms (eg, B and CRF01_AE) could bias the network toward more high-degree nodes.
International Dimension of HIV Transmission
A global analysis including all published HIV-1 sequences recovered previously characterized transmission clusters, expanded known transmission clusters by incorporating additional isolates and/or smaller clusters, and identified previously undescribed transmission clusters (for representative examples and references, see Figure 3 and Table 2). For example, both UK clusters (shown in Figure 3) were partially characterized in the literature, but these transmission clusters had not been considered in the global context, and hence the international links had been missed. The global network analysis also discovered 2 large international transmission clusters: (1) a 333-node cluster representing 17 countries, primarily comprising heterosexuals and injection drug users in Southeast Asia, and (2) a 674-node cluster representing 18 countries, primarily comprising injection drug users and men who have sex with men in the former Soviet Union (Figure 2). Portions of both of these networks have been previously described in the literature (Supplementary Data); however, the extent of international transmission had been vastly underestimated, because these previous studies narrowly focused on single countries or regions. We argue that a global-scale analysis is a prerequisite for capturing the scope of these transmission clusters and should become commonplace in the field.
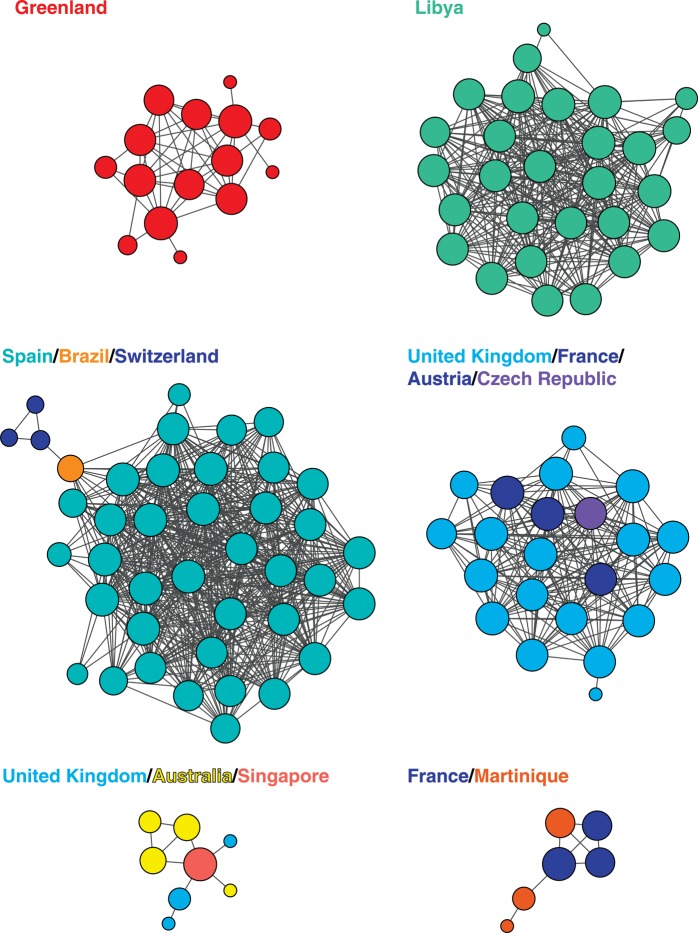
Figure 3.
Examples of transmission clusters inferred from the global network shown in Figure 1. Node size is proportional to relative degree (number of edges per node). Edge lengths are optimized for visual presentation and do not represent genetic distances between potential transmission partners. Colors correspond to geographic regions in Figure 1.
Table 2.
Examples of Transmission Clusters in the Global Network
| Countries/Regions | No. of Isolates | Sampling Year(s) | Subtype | Prior Description | Risk Factor(s) | References |
|---|---|---|---|---|---|---|
| Greenland | 15 | 1999–2006 | B | Yes | Heterosexual | [27, 28] |
| Libya | 28 | 1998 | CRF02_AG | Yes | Nosocomial | [29] |
| Spain/Switzerland/Brazil | 41 | 2005–2011 | F1 | Yes | MSM/heterosexual | [18, 30–32] |
| United Kingdom/France/Austria/Czech Republic | 16 | 2000–2006 | B | Partial | MSM | [9, 12, 33, 34] |
| United Kingdom/Australia/Singapore | 8 | 2005–2007 | B | Partial | MSM | [12, 35, 36] |
| France/Martinique | 6 | 2006 | B | None | Unknown | NA |
Abbreviations: MSM, men who have sex with men; NA, not applicable.
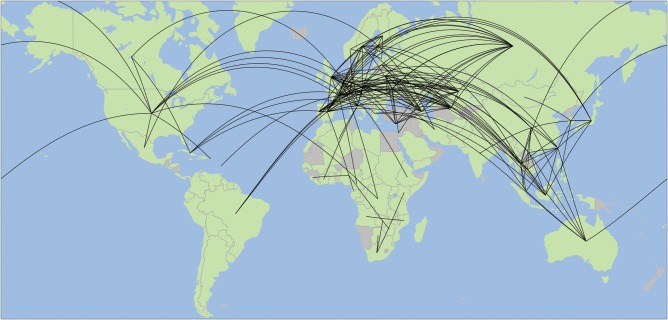
Our analysis revealed the contemporary pattern of HIV-1 transmission across international borders (Figure 4). Of the 106 countries or regions represented in transmission clusters, individuals from 72 (68%) of these countries/regions had a potential transmission partner from another country/region (see Supplementary Table 1 for a list of the most connected countries). Moreover, 22.6% of potential transmission partners were from different countries/regions, likely due to the high connectedness of larger, international transmission networks (Figure 2), and 3.6% of transmission clusters included potential transmission partners in multiple countries/regions. Of course, substantial sampling and publication biases exist within public sequence databases, particularly regarding the undersampling of isolates from sub-Saharan Africa. However, the international nature of inferred transmission clusters is all the more striking because of the biases toward densely sampling local epidemics.
Figure 4.
Global transmission patterns of human immunodeficiency virus type 1. There are 211 putative transmission links between countries/regions. Green countries/regions are sampled in the network; gray indicates absence of sampling. Connections for each country/region originate in the centroid of its map region.
Transmission Clusters
Inclusion in our transmission clusters required only 1 shared edge with another member of the cluster. Therefore, one would expect to find more and larger transmission clusters in regions with young epidemics and extensive sampling, and indeed the propensity for clustering in our network varied across geographic regions (χ2 test, P < .0001; Table 1). Isolates from recent epidemics in Central and East Asia clustered more frequently than isolates from elsewhere in the world, and isolates from older and more diverse African epidemics were less likely to be found in clusters. Furthermore, the 3 largest transmission clusters predominantly from a single country/region were found in younger epidemics from Pakistan, Taiwan, and China (Supplementary Figure 1).
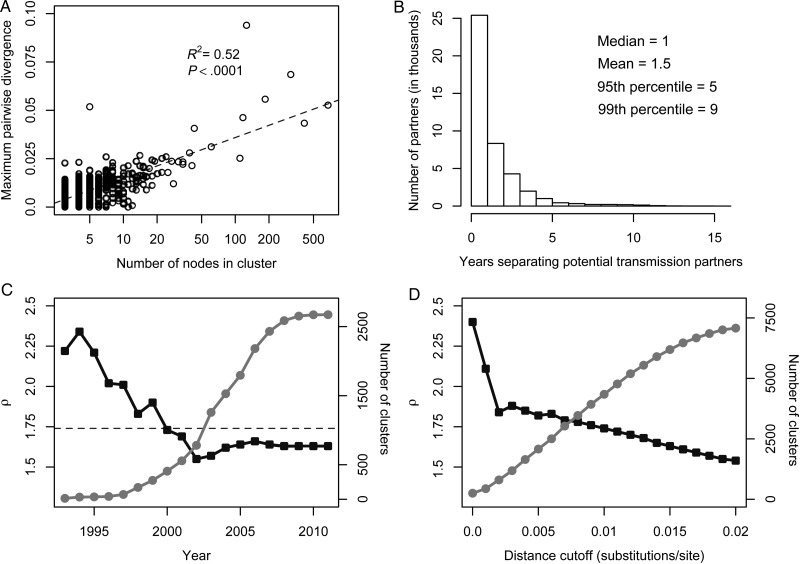
Overall, inferred transmission clusters were densely connected, suggesting that most isolates were linked to multiple other isolates in their cluster. The mean genetic distance between nodes was ≤1% for 96% of the clusters comprising ≥3 nodes, and the mean distance for the largest clusters (Figure 2 and Supplementary Figure 1) did not exceed 2%. Thus, many of these clusters could be identified using approaches and thresholds adopted in earlier studies, albeit at a much steeper computational cost and only if the relevant sequences had been considered for inclusion a priori. Notably, our network approach identifies clusters that include all potential transmission partners without excluding peripherally linked partners; these potential partners may have been excluded by previous methods that rely on mean, median, or maximum genetic distance within a cluster. As expected in a scale-free, densely connected network [37], we observed a log-linear relationship between cluster size and the maximum pairwise divergence within a cluster (Figure 5A).
Figure 5.
Properties of the human immunodeficiency virus type 1 global transmission network. A, Relationship between cluster size and maximum pairwise divergence within a cluster. B, Number of years separating viral sequence isolation from potential transmission partners in the global transmission network. C, Network characteristic exponent ρ (black squares) stabilizes as number of clusters (gray circles) increases. Dashed line shows ρ inferred from the complete network. D, Effect of distance cutoff for potential transmission partners on network characteristic exponent ρ (black squares) and the number of clusters (gray circles).
Most viruses from potential transmission partners were sampled within a narrow timeframe of each other (Figure 5B); importantly, the actual number of years of evolution separating these sequences is greater than these times, except when 1 virus is the direct descendant of the other [38]. Viral isolates from 60% of the potential transmission partners were obtained within 1 year of each other. Surprisingly, it appears as if our network approach was able to capture at least some transmission events involving chronic infections: the 95th percentile of years separating potential transmission partners extended to 5 years. Furthermore, we found that nearly 1% of potential transmission partners were separated by at least a decade. This unusually long time span separating viruses with <1% genetic divergence may be explained by (1) transmission of latent virus due to reemergence after antiretroviral therapy [39], (2) transmission of a slowly evolving virus from individuals with low viral load (ie, elite controllers) [40], (3) possible preferential transmission of ancestral variants [41, 42] (although see [43]), and/or (4) incorrectly recorded year of sampling or contamination.
Network Stability
The global transmission network inference appears robust to a number of model parameters and assumptions. More than 99% of edges were supported in a majority-rule bootstrap consensus network (Supplementary Figure 2); this analysis is a network analog of the phylogenetic bootstrap.
To determine the evolution of network properties through time, we reanalyzed the sequences, censoring the most recent year in a stepwise fashion. The current network structure appears to have been largely established by the early 2000s, when there were only about 500 inferred clusters (Figure 5C). In our dataset, 17 280 sequences (20%) lacked an associated year of sampling, and excluding these sequences resulted in slightly biased estimates of the network characteristic exponent (ρ). This bias is due to the higher frequency with which isolates without known sampling years were found in clusters (χ2 test, P < .0001). To confirm the source of this bias, we reconstructed 100 replicate networks with 17 280 random sequences removed; the network characteristic exponent ρ (95% confidence interval, 1.72–1.75) was indistinguishable from that (ρ = 1.74) in the original network.
A 1% distance cutoff (0.01 substitutions per site) for potential transmission partners was selected because it is a conservative approximation of the level of intrahost diversity early in infection and has been used in previous transmission network analyses in a cohort of recently infected individuals [19, 44]. This strict cutoff favors the detection of recent transmission partners and will likely miss older transmissions between partners with long-term chronic infections. Nevertheless, we found that the overall network structure is stable with respect to the distance cutoff used to determine putative transmission partners (Figure 5D). For reasonable levels of intrahost pol diversity (0.002–0.02 substitutions per site), the network characteristic exponent is constrained to a relatively narrow range (ρ = 1.54–1.84).
Drug Resistance
The structure of the global network supports a hypothesized fitness trade-off between drug resistance–associated mutations (DRAMs) and viral transmission. We observed an inverse relationship between the degree of the node (a proxy for transmissibility) and the number of DRAMs at that node. This analysis was performed on both the original network (ordinal logistic regression, P < .0001) and the network constructed after removing the DRAM sites (see below, P < .01), adjusting for year and region of sampling. In the absence of antiviral drug treatment, resistance mutations should incur a fitness cost that could result in decreased viral transmission [45]. Nodes with high degrees, an indicator of more frequent transmission events, were less likely to harbor DRAMs. This pattern suggests that in the absence of antiretroviral therapy, resistance mutations should dissipate from the viral population. However, most of the viral isolates analyzed here lacked information about the patient's drug treatment status. Future studies of transmission networks constructed from higher quality datasets (in which infection stage and drug treatment status are known) will be able to better address the issue of DRAM fitness costs across populations.
To determine whether DRAMs influence network construction, we removed 36 codons associated with major drug resistance mutations (http://hivdb.stanford.edu/pages/download/resistanceMutations_handout.pdf) and repeated the analysis. In the absence of these sites, the network was found to have a slightly higher characteristic exponent: ρ = 1.84 (ie, fewer high-degree nodes). To confirm the source of this shift, we reconstructed 100 replicate networks with 36 random codons removed in which ρ (mean, 1.75 [95% confidence interval, 1.72–1.78]) was indistinguishable from ρ in the original network (ρ = 1.74).
The gag Transmission Network
Although the available dataset is much smaller, similar patterns were seen in the global transmission network inferred using gag sequences. The same network inference procedure was repeated for gag (p24) (HXB2 coordinates: 1186–1879), yielding 9426 sequences. There were 592 distinct clusters in the gag network (after excluding 6 clusters, containing 36 sequences, as potentially contaminated), representing 43 putative cross-country transmission events. Using isolates whose sequence spanned both gag and pol regions, 72 clusters were linked, including the 2 large, international pol clusters (Figure 2). Therefore, pol is not unique among HIV genes in its usefulness in reconstructing transmission history, and multiple loci can be used to confirm or supplement network inference.
DISCUSSION
Analysis of the global HIV-1 transmission network provides a comprehensive and current picture of the spread of HIV-1 within and between countries; it reveals that “local” epidemics often include international transmission links and can be fully understood only in the context of global HIV diversity. This finding is in agreement with an analysis of transmission partners within a single town in the United Kingdom, which found that only 30% of the transmission events occurred locally [46], as well as with earlier studies in Uganda [47]. The large international clusters (Figure 2), along with the expanded UK clusters, which were not previously known to extend beyond that country's borders (Figure 3), explicitly demonstrate the risk incurred by arbitrarily restricting phylogenetic or network analyses to single countries or regions: international transmission links are lost. Furthermore, these and other large transmission clusters, in which each isolate is a potential transmission partner of at least 1 other isolate, could have been missed or broken up by clustering methods that begin with a phylogenetic tree and then identify highly supported clades with low levels of divergence. Even high-resolution phylogenetic analyses of local epidemics would benefit from including relevant isolates identified by a method that is global in scope.
Although edges in the global network are not synonymous with actual transmission events, they likely link individuals in the same epidemiological transmission cluster. The scope and size of many of the clusters presented here expand with a less conservative distance cutoff (Figure 5D), but the network structure remains relatively unchanged. Moreover, the absence of connectedness between countries and clusters should not be interpreted as lack of direct transmission; rather, it reflects incomplete and nonuniform sampling due to biases in publication of HIV-1 sequences. Thus, historical viral migration events in the spread of HIV-1 subtypes around the world (eg, subtype B from Haiti to the United States, subtype F1 from Angola to Brazil and Romania, and subtype C from sub-Saharan Africa to India and China [48–50]) are absent from our network (Figure 4).
Transmission network approaches are well suited for the analysis of HIV surveillance data, which are less susceptible to the effects of sampling and publication bias (eg, oversampling of geographic regions or risk groups) and would produce more robust analysis. Last, our global-network approach will allow researchers to determine, nearly instantaneously, if newly isolated HIV-1 sequences fall within known transmission clusters, enabling near-real-time surveillance of recent and growing local epidemics and patterns of international transmission. As more sequences are deposited in public databases, the accuracy and resolution of this approach will continue to improve.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the National Institutes of Health (grants AI43638, AI47745, AI093163, AI07384, GM093939, MH083552, AI74621, AI100665, DA034978); the University of California, San Diego Center for AIDS Research (grant NIH-AI36214); the University of California Laboratory Fees Research Program (grant 12-LR-236617), and the Department of Veterans Affairs.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Tebit DM, Arts EJ. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis. 2011;11:45. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 2.Hillis DM, Huelsenbeck JP. Support for dental HIV transmission. Nature. 1994;369:24. doi: 10.1038/369024a0. [DOI] [PubMed] [Google Scholar]
- 3.Holmes EC, Brown AJ, Simmonds P. Sequence data as evidence. Nature. 1993;364:766. doi: 10.1038/364766b0. [DOI] [PubMed] [Google Scholar]
- 4.Metzker ML, Mindell DP, Liu XM, et al. Molecular evidence of HIV-1 transmission in a criminal case. Proc Natl Acad Sci U S A. 2002;99:14292. doi: 10.1073/pnas.222522599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillay D, Rambaut A, Geretti AM, et al. HIV phylogenetics. BMJ. 2007;335:460. doi: 10.1136/bmj.39315.398843.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scaduto DI, Brown JM, Haaland WC, et al. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc Natl Acad Sci U S A. 2010;107:21242. doi: 10.1073/pnas.1015673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strimmer K, Moulton V. Likelihood analysis of phylogenetic networks using directed graphical models. Mol Biol Evol. 2000;17:875. doi: 10.1093/oxfordjournals.molbev.a026367. [DOI] [PubMed] [Google Scholar]
- 8.Fitch WM. Networks and viral evolution. J Mol Evol. 1997;44(suppl 1):S65. doi: 10.1007/pl00000059. [DOI] [PubMed] [Google Scholar]
- 9.Hue S, Gifford RJ, Dunn D, et al. Demonstration of sustained drug-resistant human immunodeficiency virus type 1 lineages circulating among treatment-naive individuals. J Virol. 2009;83:2645. doi: 10.1128/JVI.01556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prosperi MC, Ciccozzi M, Fanti I, et al. A novel methodology for large-scale phylogeny partition. Nat Commun. 2011;2:321. doi: 10.1038/ncomms1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes GJ, Fearnhill E, Dunn D, et al. Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog. 2009;5:e1000590. doi: 10.1371/journal.ppat.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leigh Brown AJ, Lycett SJ, Weinert L, et al. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis. 2011;204:1463. doi: 10.1093/infdis/jir550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouyos RD, von Wyl V, Yerly S, et al. Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J Infect Dis. 2010;201:1488. doi: 10.1086/651951. [DOI] [PubMed] [Google Scholar]
- 14.Lewis F, Hughes GJ, Rambaut A, et al. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volz EM, Koopman JS, Ward MJ, et al. Simple epidemiological dynamics explain phylogenetic clustering of HIV from patients with recent infection. PLoS Comput Biol. 2012;8:e1002552. doi: 10.1371/journal.pcbi.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;38:783. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Brenner B, Moodie EEM. HIV sexual networks: the Montreal experience. Stat Communiciations Infect Dis. 2012;4 doi:10.1515/1948-4690.1039. [Google Scholar]
- 18.Castro E, Khonkarly M, Ciuffreda D, et al. HIV-1 drug resistance transmission networks in southwest Switzerland. AIDS Res Hum Retroviruses. 2010;26:1233. doi: 10.1089/aid.2010.0083. [DOI] [PubMed] [Google Scholar]
- 19.Smith DM, May SJ, Tweeten S, et al. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS. 2009;23:225. doi: 10.1097/QAD.0b013e32831d2a81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 22.Wertheim JO, Kosakovsky Pond SL. Purifying selection can obscure the ancient age of viral lineages. Mol Biol Evol. 2011;28:3355. doi: 10.1093/molbev/msr170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones JH, Handcock MS. An assessment of preferential attachment as a mechanism for human sexual network formation. Proc Biol Sci. 2003;270:1123. doi: 10.1098/rspb.2003.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liljeros F, Edling CR, Amaral LA, et al. The web of human sexual contacts. Nature. 2001;411:907. doi: 10.1038/35082140. [DOI] [PubMed] [Google Scholar]
- 25.Irwin JO. The place of mathematics in medical and biological statistics. J R Stat Soc Ser A. 1963;126:1. [Google Scholar]
- 26.Kosakovsky Pond SL, Frost SD. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol Biol Evol. 2005;22:478. doi: 10.1093/molbev/msi031. [DOI] [PubMed] [Google Scholar]
- 27.Madsen TV, Leitner T, Lohse N, et al. Introduction of HIV type 1 into an isolated population: molecular epidemiologic study from Greenland. AIDS Res Hum Retroviruses. 2007;23:675. doi: 10.1089/aid.2007.0235. [DOI] [PubMed] [Google Scholar]
- 28.Madsen TV, Lohse N, Jensen ES, et al. Short communication: high prevalence of drug-resistant human immunodeficiency virus type 1 in treatment-naive patients in Greenland. AIDS Res Hum Retroviruses. 2008;24:1073. doi: 10.1089/aid.2008.0049. [DOI] [PubMed] [Google Scholar]
- 29.Yerly S, Quadri R, Negro F, et al. Nosocomial outbreak of multiple bloodborne viral infections. J Infect Dis. 2001;184:369. doi: 10.1086/322036. [DOI] [PubMed] [Google Scholar]
- 30.Cardoso LP, Queiroz BB, Stefani MM. HIV-1 pol phylogenetic diversity and antiretroviral resistance mutations in treatment naive patients from Central West Brazil. J Clin Virol. 2009;46:134. doi: 10.1016/j.jcv.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Cuevas M, Fernandez-Garcia A, Sanchez-Garcia A, et al. Incidence of non-B subtypes of HIV-1 in Galicia, Spain: high frequency and diversity of HIV-1 among men who have sex with men. Euro Surveill. 2009;14 doi: 10.2807/ese.14.47.19413-en. [DOI] [PubMed] [Google Scholar]
- 32.Thomson MM, Fernandez-Garcia A, Delgado E, et al. Rapid expansion of a HIV-1 subtype F cluster of recent origin among men who have sex with men in Galicia, Spain. J Acquir Immune Defic Syndr. 2012;59:e49. doi: 10.1097/QAI.0b013e3182400fc4. [DOI] [PubMed] [Google Scholar]
- 33.Brown AE, Gifford RJ, Clewley JP, et al. Phylogenetic reconstruction of transmission events from individuals with acute HIV infection: toward more-rigorous epidemiological definitions. J Infect Dis. 2009;199:427. doi: 10.1086/596049. [DOI] [PubMed] [Google Scholar]
- 34.Vercauteren J, Wensing AM, van de Vijver DA, et al. Transmission of drug-resistant HIV-1 is stabilizing in Europe. J Infect Dis. 2009;200:1503. doi: 10.1086/644505. [DOI] [PubMed] [Google Scholar]
- 35.Chibo D, Kaye M, Birch C. HIV transmissions during seroconversion contribute significantly to new infections in men who have sex with men in Australia. AIDS Res Hum Retroviruses. 2011;28:460–4. doi: 10.1089/AID.2011.0137. [DOI] [PubMed] [Google Scholar]
- 36.Lee CC, Sun YJ, Barkham T, et al. Primary drug resistance and transmission analysis of HIV-1 in acute and recent drug-naive seroconverters in Singapore. HIV Med. 2009;10:370. doi: 10.1111/j.1468-1293.2009.00698.x. [DOI] [PubMed] [Google Scholar]
- 37.Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167. [Google Scholar]
- 38.Drummond A, Pybus OG, Rambaut A. Inference of viral evolutionary rates from molecular sequences. Adv Parasitol. 2003;54:331. doi: 10.1016/s0065-308x(03)54008-8. [DOI] [PubMed] [Google Scholar]
- 39.Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 40.Buckheit RW, III, Salgado M, Martins KO, et al. The implications of viral reservoirs on the elite control of HIV-1 infection. Cell Mol Life Sci. 2012;70:1009–19. doi: 10.1007/s00018-012-1101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herbeck JT, Nickle DC, Learn GH, et al. Human immunodeficiency virus type 1 env evolves toward ancestral states upon transmission to a new host. J Virol. 2006;80:1637. doi: 10.1128/JVI.80.4.1637-1644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lythgoe KA, Fraser C. New insights into the evolutionary rate of HIV-1 at the within-host and epidemiological levels. Proc Biol Sci. 2012;279:3367. doi: 10.1098/rspb.2012.0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wertheim JO, Scheffler K, Choi JY, et al. Phylogenetic relatedness of HIV-1 donor and recipient populations. J Infect Dis. 2013;207:1181. doi: 10.1093/infdis/jit021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wertheim JO, Kosakovsky Pond SL, Little SJ, et al. Using HIV transmission networks to investigate community effects in HIV prevention trials. PLoS One. 2011;6:e27775. doi: 10.1371/journal.pone.0027775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leigh Brown AJ, Frost SD, Mathews WC, et al. Transmission fitness of drug-resistant human immunodeficiency virus and the prevalence of resistance in the antiretroviral-treated population. J Infect Dis. 2003;187:683. doi: 10.1086/367989. [DOI] [PubMed] [Google Scholar]
- 46.Fisher M, Pao D, Brown AE, et al. Determinants of HIV-1 transmission in men who have sex with men: a combined clinical, epidemiological and phylogenetic approach. AIDS. 2010;24:1739. doi: 10.1097/QAD.0b013e32833ac9e6. [DOI] [PubMed] [Google Scholar]
- 47.Yirrell DL, Pickering H, Palmarini G, et al. Molecular epidemiological analysis of HIV in sexual networks in Uganda. AIDS. 1998;12:285. doi: 10.1097/00002030-199803000-00006. [DOI] [PubMed] [Google Scholar]
- 48.Gilbert MT, Rambaut A, Wlasiuk G, et al. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci U S A. 2007;104:18566. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehta SR, Wertheim JO, Delport W, et al. Using phylogeography to characterize the origins of the HIV-1 subtype F epidemic in Romania. Infect Genet Evol. 2011;11:975. doi: 10.1016/j.meegid.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fontella R, Soares MA, Schrago CG. On the origin of HIV-1 subtype C in South America. AIDS. 2008;22:2001. doi: 10.1097/QAD.0b013e3283108f69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.