Abstract
The protease HAUSP is a critical component of the p53-Mdm2 pathway and acts as a specific deubiquitinase for both p53 and Mdm2 and thus is important for p53 regulation. In knock-down and knock-out cellular systems it was observed that ablation of HAUSP induces profound stabilization of p53 due to enhanced degradation of Mdm2. Thus, inhibiting HAUSP by small compound interference has been proposed as a rational therapeutic strategy to activate p53 in p53 wild type tumors. However, HAUSP-mediated effects in the p53-Mdm2 axis are highly complex and non-linear and to date the role of HAUSP in tumor suppression in vivo remains unexplored.
Here we investigate the effect of HAUSP up and downregulation on cell proliferation, apoptosis and tumor growth in vitro and in a xenograft model in vivo, using an inducible isogenic human colon carcinoma cell system. Importantly, in the absence of stress, both HAUSP up and downregulation inhibit cell proliferation in vitro and tumor growth in vivo due to constitutively elevated p53 levels. Moreover, tumors with HAUSP up and downregulation respond to radiotherapy with further growth inhibition. However, HAUSP downregulation causes resistance to Camptothecin- and irradiation-induced apoptosis, which correlates with suppressed mitochondrial translocation of p53. Our data suggest that changes in HAUSP modulate tumor growth and apoptotic sensitivity in vivo.
Keywords: HAUSP, p53, ubiquitination, Mdm2, deubiquitination, mitochondrial translocation
Introduction
p53, one of the most important tumor suppressor proteins, plays an essential role in regulating cell cycle, senescence and apoptosis in response to many types of stress. The level of p53 protein is critical for normal cellular homeostasis, and is subtly regulated by ubiquitination and deubiquitination systems. In unstressed cells, p53 levels are very low due to rapid degradation via the ubiquitin-proteasome pathway. Mdm2 (= Hdm2 in humans, however for simplicity referred to here as Mdm2), the major p53 E3 ligase, is itself a p53 target gene, thus constituting a negative feedback loop.1–3 p53 ubiquitination is reversible by the p53 deubiquitinating enzyme HAUSP (herpesvirus-associated ubiquitin-specific protease; USP7). Ectopic HAUSP expression results in p53 stabilization, induction of growth arrest and apoptosis.4 However, subsequent studies revealed that the stability of Mdm2 is also regulated by HAUSP.5–7 Thus, HAUSP targets both p53 and Mdm2 as substrates and, in concert with Mdm2 plays a dynamic role in p53 regulation.
Of note, p53 demonstrates a differential response to changes in HAUSP levels and/or the onset of stress, whose nature and significance are not well understood but suggests the existence of a HAUSP-regulated substrate switch of Mdm2 from ubiquitination of p53 in the absence of stress to ubiquitination of Mdm2 (self) in the presence of stress.8 This switch also suggests a potential role of HAUSP as a downstream target of stress signals.8 HAUSP antagonizes Mdm2-mediated p53 degradation through direct and indirect mechanisms. Structural and biochemical studies revealed that p53 and Mdm2 compete for the same direct binding site in the N-terminal TRAF-like domain of HAUSP. Mdm2 binds to HAUSP with a several fold higher affinity than p53, implying that in normal cells growing under homeostasis Mdm2 is the preferred substrate of HAUSP.9,10 However, upon DNA damage, ATM-dependent phosphorylation of Mdm2 and Mdmx lower their affinity for HAUSP, which reduces their stability and results in stress-induced degradation of Mdm2 and Mdmx.7,11 This creates a pool of liberated HAUSP. Thus, under stress conditions p53 becomes the main substrate of HAUSP, quickly stabilizing p53 and causing unbound Mdm2 to be ubiquitinated and degraded. Recently, it was shown that HAUSP can also function in trans on p53 by using Mdm2 as a bridge, and a subpopulation of such a triple p53-Mdm2-HAUSP complex exists in transfected cells and might exist in vivo.12
HAUSP may also impact on cell proliferation and apoptosis in a p53-independent manner. HAUSP was negatively implicated in the functional regulation and nuclear distribution of the transcription factor FOXO4, important in cellular metabolism, cell cycle progression and death.13 Moreover, HAUSP might directly or indirectly affect the levels of many other proteins. A recent proteomics study detected alterations in 36 proteins after downregulation of HAUSP.14 Proteins whose levels dropped after HAUSP knockdown belonged to broad functional categories involved in cell cycle, DNA replication, transcription, translation, metabolism and most interestingly, apoptosis.
Given the mounting evidence that HAUSP activity is critical to control cell death and proliferation, several groups recently proposed HAUSP interference as a therapeutic target in wild type p53 expressing tumors to activate p53.12,15 A clinically important group of this kind are hematopoietic tumors, which are characterized by decreased levels of wtp53 protein (<30% incidence) rather than p53 mutations (only around 10%).16 However, so far, all published studies on HAUSP are based on in vitro results and the role of HAUSP manipulation in tumor suppression in vivo remains completely unexplored.
To start filling this void, we explore here the in vitro and in vivo effect of HAUSP up and downregulation on cell proliferation, apoptosis and tumor growth, using an inducible isogenic human colon carcinoma system in cell culture and xenograft models. Importantly, in the absence of DNA damage, both HAUSP up and downregulation suppress cell proliferation and retard tumor growth due to stabilization of p53. Moreover, irradiation further inhibits the growth of tumors with both HAUSP up and downregulation, despite the fact that HAUSP downregulation causes resistance to stress-induced apoptosis. Thus, HAUSP modulation and irradiation have syner-gistic effects on suppressing tumor growth. Our results suggest that manipulation of HAUSP might be a rational new anticancer target to enhance p53-dependent and -independent tumor suppressor activity in established tumors.
Results
HAUSP up and downregulation both stabilize endogenous p53 levels
Based on the striking observation that knock-down or deletion of HAUSP induces profound stabilization of p53,5–7 HAUSP interference has been proposed as a rational therapeutic strategy to activate wild type p53 in tumors.12,15,16 However, the complex and non-linear interplay between p53 and its regulatory proteins Mdm2, Mdmx and HAUSP, as well as effects by other direct or indirect targets of HAUSP involved in cell proliferation and cell death, prompted us to test systematically how manipulations of HAUSP levels might affect tumor progression in vitro and in vivo. To this end, we used an isogenic human carcinoma cell system, where the level of HAUSP protein and therefore its activity could be regulated. The wild type p53 colon carcinoma line LS174TR1 expressing a tetracycline repressor (here called ‘Par’ for parental) was used to derive a HAUSP-upregulated subline that stably expresses Doxycycline (Dox)-inducible HAUSP (Myc-epitope tagged, called LS89; ‘Hup’).7 Conversely, a HAUSP-downregulated subline was used that stably expresses a Dox-inducible HAUSP RNAi (called LS126; ‘Hdown’).7
As expected, Dox treatment significantly increased HAUSP levels in the Hup line and conversely decreased HAUSP levels in the Hdown line (Fig. 1A). In contrast, Dox did not alter HAUSP, p53 or Mdm2 levels in the parental line. In complete agreement with previous reports, HAUSP upregulation resulted in strong protection of p53 and Mdm2 from their ubiquitin-mediated degradation, with subsequent stabilization of p53 and Mdm2 (Fig. 1A, lanes 3 and 4).4,7 The fact that HAUSP stabilizes and activates p53 in the face of Mdm2 accumulation is a consistent finding, but is at least partially explained by recent data showing that HAUSP can enzymatically function to deubiquitinate p53 in trans by utilizing MDM2 as a structural bridge.12 The net effect is a functional dominance of p53.
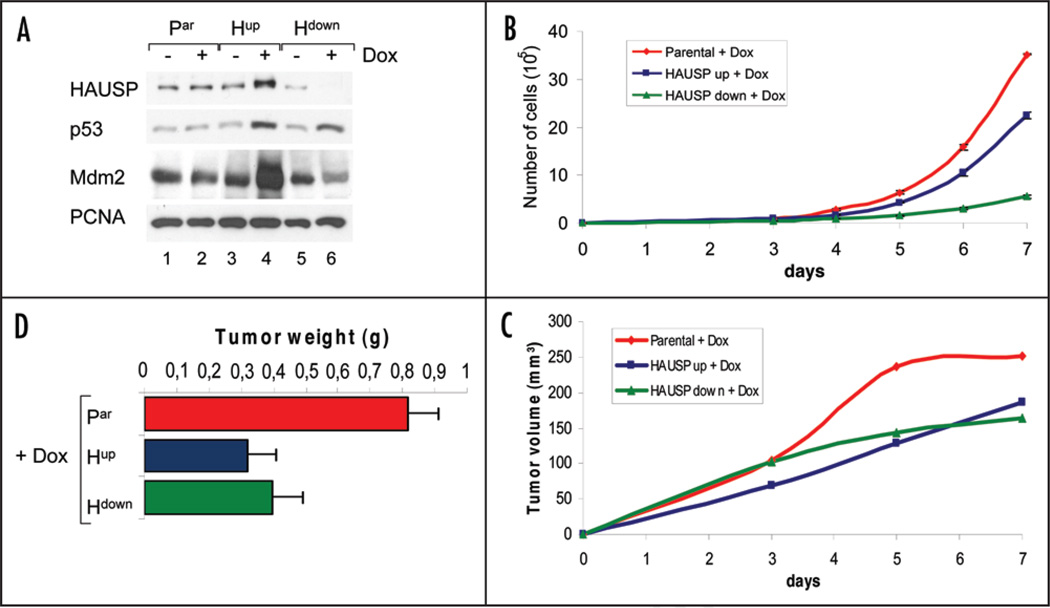
Figure 1.
Both HAUSP up and downregulation stabilize and activate p53 and induce growth suppression in the absence of stress. (A) Both HAUSP up and downregulation by Doxycycline stabilize p53 levels. Immunoblot analysis as indicated. PCNA as loading control. (B) HAUSP upregulation and especially downregulation suppress cell proliferation in vitro compared to parental cells. All cells were induced with Doxycycline for 48 hrs before seeding and kept under Dox. Each data point represents the average cell count of triplicate wells; whiskers indicate standard error. (C) Both HAUSP up and downregulation suppress tumor growth in vivo. Cells were induced for 72 hrs with Doxycycline and then injected subcutaneously into nude mice (106 cells per tumor; 6 paravertebral tumors per mouse). All mice received Doxycycline with their drinking water (2 mg/ml) ad libitum. Tumor size was measured every other day. Data from two independent experiments were pooled (a total of 24 tumors from 4 mice per time point per cell line). Growth curves represent the average tumor volume at the indicated time points. (D) Average weight of non-irradiated front tumors at endpoint day 20 (n = 2 per treatment). Consistently, front tumors in all mice were bigger than tumors located in the mid and lower back, due to uncontrollable extraneous factors in the latter areas. Thus, to exclude extraneous limitations, front tumors were chosen to determine differences in the actual growth properties in response to HAUSP modulation. Bars represent mean +/− standard error. The middle and posterior tumors showed the same trend.
Similarly, HAUSP downregulation by Dox-inducible RNAi expression also stabilized p53 protein levels, albeit to a lesser degree, due to downregulation of Mdm2 (Fig. 1A, lanes 5 and 6). Again, this is a consistent finding described previously.5,6 When HAUSP is disrupted, both p53 and Mdm2 fail to be deubiquitinated. This accelerates the degradation of ubiquitinated Mdm2. Consequently, due to the very low levels of Mdm2, p53 ubiquitination is suppressed, leading to p53 stabilization and activation. However, in contrast to HAUSP upregulation, which invariably leads to very robust p53 stabilization,4 the effect of HAUSP downregulation on p53 stabilization is highly dynamic, seems to depend on the level of HAUSP downregulation and can be paradoxical: partial reduction of endogenous HAUSP destabilizes p53. However, near complete (via siRNA) or complete ablation (via homologous recombination by a null allele) of HAUSP increases ubiquitination of Mdm2 so much so that p53 is indirectly stabilized.5,6 Together, this indicates that conditions of HAUSP downregulation exhibit a more complex p53-Mdm2-HAUSP interplay than conditions of HAUSP upregulation.
HAUSP up and downregulation inhibit cell proliferation in culture and in xenografts
Next, to investigate the effect of altered HAUSP levels and subsequent p53 stabilization on cell growth, we performed cell proliferation assays. Indeed, consistent with p53 activation, HAUSP up and downregulation both inhibited cell proliferation in vitro (Fig. 1B). This observation is in complete agreement with previous tissue culture studies. For example, HAUSP overexpression stabilized p53 and strongly inhibited the growth of wild type p53-expressing H460 human lung carcinoma cells and of p53+/+ mouse embryo fibroblasts (MEFs), but had no effect on p53-null H1299 cells nor on p53−/− MEFs, suggesting that inhibition of cell growth by HAUSP upregulation is p53-dependent.4 Likewise, HAUSP overexpression induced growth retardation in HeLa human cervical carcinoma cells in vitro.17,18 Furthermore, deletion of HAUSP in HCT116 human colon carcinoma cells and telomerase-immortalized, normal retinal-pigment epithelial cells (RPE) also stabilized p53 and inhibited cell growth.5 Interestingly, we found that in culture HAUSP downregulation inhibited cell growth more strongly than HAUSP upregulation (Fig. 1B), although p53 stabilization in Hdown cells was slightly lower than in Hup cells (Fig. 1A, compare lanes 4 and 6). Thus, p53-independent factors also appear to contribute to the growth suppression seen by HAUSP downregulation, whereas the anti-proliferative effect of HAUSP upregulation is largely p53-dependent, as shown previously.4
Importantly, both decreasing or increasing HAUSP levels also suppressed tumor cell growth in vivo, as seen in nude mouse xenograft assays (Fig. 1C and D) and thus confirmed our in vitro results (Fig. 1B). Cells pretreated with Dox were subcutaneously injected into nude mice (106 cells per tumor; 6 paravertebral tumors per mouse). All mice continuously received Dox in their drinking water. Tumor size was measured every second day. Dox alone did not affect tumor size of parental cells (data not shown). However, both tumor volume and weight from injected Hup and Hdown cells were suppressed, compared to parental cells (Fig. 1C and D).
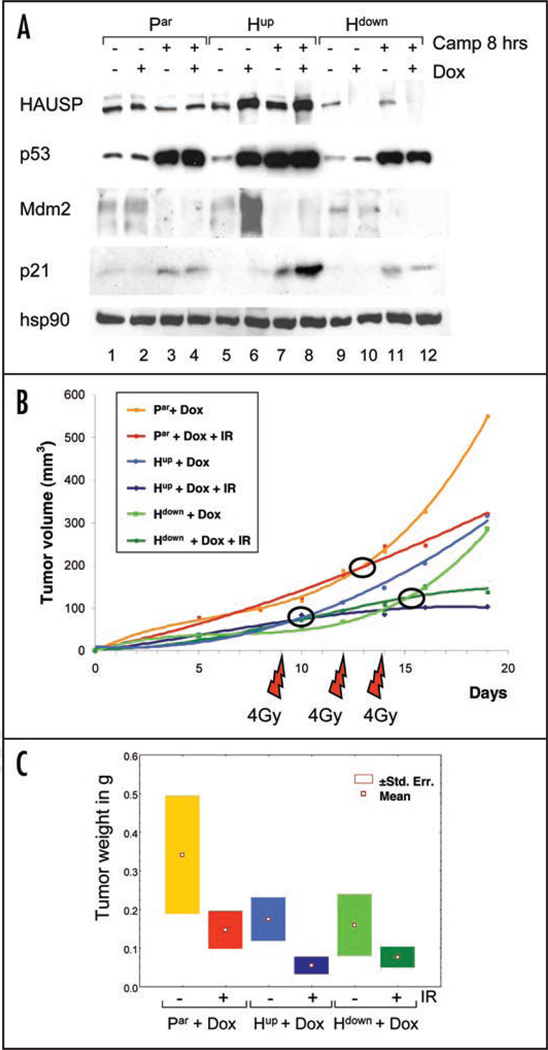
Next, we analyzed the response to DNA damage under conditions of HAUSP up and down-regulation. To this end, cells in culture were pretreated with Dox or left untreated, followed by treatment with Camptothecin (Camp) (Fig. 2A). Levels of HAUSP itself did not significantly change upon Camptothecin treatment, consistent with our previous results that HAUSP levels are not stress-sensitive.19 Moreover, independent of Dox-induced changes in HAUSP levels, DNA damage strongly stabilized total cellular p53, albeit the degree of p53 stabilization under conditions of HAUSP downregulation was somewhat lower than that under HAUSP upregulation (Fig. 2A). Of note, in response to DNA damage cells with HAUSP upregulation exhibited the highest induction of p21 protein, while cells with HAUSP downregulation did not exceed the parental level of p21 induction (Fig. 2A). To investigate how manipulation of HAUSP levels might affect tumor growth in vivo, we performed xenograft assays in response to fractionated γ-irradiation, which is typically used in clinical therapeutic applications. Half the mice were subjected to fractionated γ-irradiation on days 9, 12 and 14 receiving 4 Gy per fraction, while the other half was mock-treated. Animals were sacrificed on day 20 for evaluation. Of note, parental and tumor xenografts with HAUSP up or downregulation, which already showed growth suppression in the absence of stress, responded to γ-irradiation with further inhibition of tumor growth compared to their respective unirradiated tumors, as indicated by tumor volume and weight (Fig. 2B and C). This radiation response approached but did not reach statistical significance in any of the three cell lines (Fig. 2C). Nevertheless, at least in this system the growth retardation obtained by HAUSP manipulations alone appears to add to therapeutic efficacy. This notion deserves more extensive testing in a future study.
Figure 2.
Effect of HAUSP up and downregulation on tumor growth in response to stress. (A) Immunoblot of cells with and without pretreatment with Doxycycline, followed by treatment with 10 µM Camptothecin for 8 hrs where indicated. Hsp90 as loading control. (B and C) Response of xenograft tumors to fractionated γ-irradiation. (B) Mice were injected as in Figure 1D. Half of the mice per group (n = 6) were subjected to fractionated irradiation on day 9, 12 and 14 (4 Gy per fraction) and all animals were sacrificed on day 20. Lines represent the average tumor volumes (n = 6 per time point). (C) Boxplots depicting the mean +/− standard error of tumor weights at endpoint day 20 (n = 6 per treatment).
HAUSP downregulation is associated with resistance to apoptosis
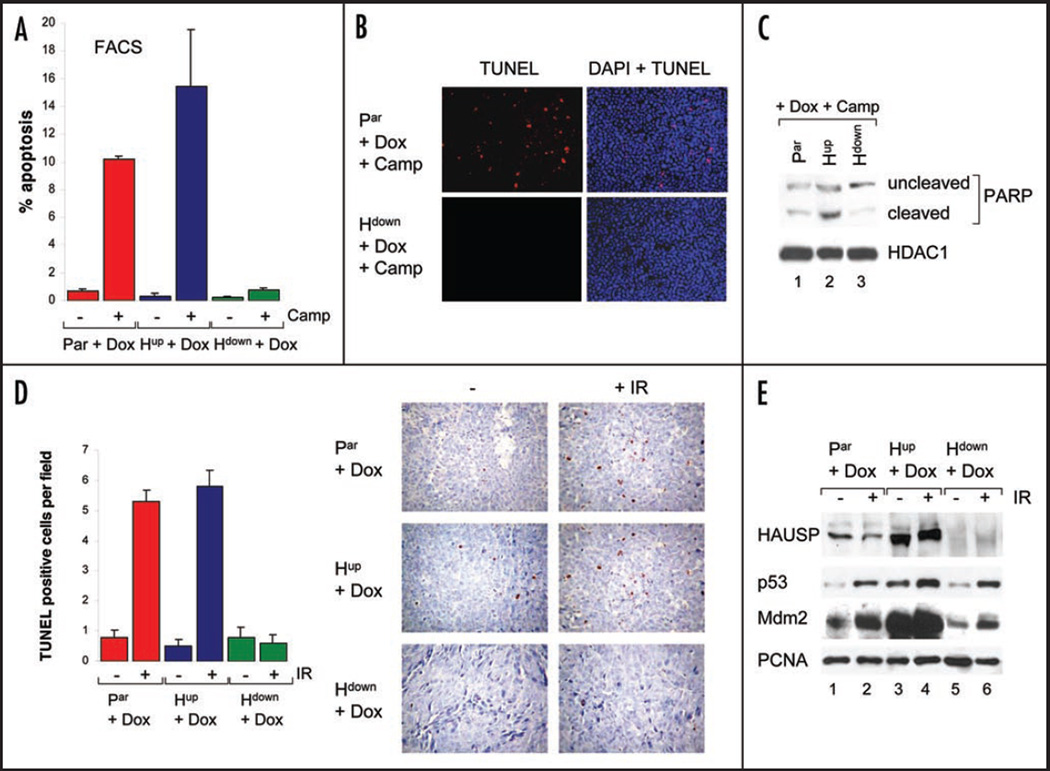
Next, we assessed the effect of HAUSP level modulation on the ability of tumor cells to undergo apoptosis. To this end, cells grown in culture and induced with Dox for 48 hrs were stressed with Camp for an additional 16 hrs and then subjected to FACS analysis and TUNEL staining. Background apoptosis in all three lines was very low (Fig. 3A). Interestingly, HAUSP upregulation slightly increased the apoptotic response to the chemotherapeutic drug compared to parental cells (Fig. 3A), consistent with some increase in cleaved PARP levels (Fig. 3C) and upregulation of the pro-apoptotic genes Noxa and Puma (Fig. 4B). More surprisingly, however, HAUSP downregulation resulted in resistance to Camp-induced apoptosis in vitro, as judged by FACS analysis (Fig. 3A), TUNEL staining (Fig. 3B) and PARP cleavage (Fig. 3C).
Figure 3.
HAUSP downregulation is associated with resistance to stress-induced apoptosis. (A) FACS analysis of sub-G1 fractions of cultured cells induced with Doxycycline and either treated with 10 µM Camptothecin for 16 hrs or left unstressed. ModFit analysis; average of two independent experiments, whiskers indicate standard error. (B) TUNEL staining of cultured parental and Hdown cells induced with Doxycycline, followed by treatment with Camptothecin for 16 hrs. (C) PARP immunoblot of Dox-induced cells after 7 hrs of Camptothecin stress. HDAC1as loading control. (D) Acute irradiation response of xenografts. Mice were either left untreated or irradiated with 7.5 Gy γIR on day 13 after inoculation and sacrificed 24 hrs later. Right, TUNEL staining of tumors (20x magnification). Hematoxylin counterstain. Left, Corresponding quantitation. Average TUNEL-positive cells per field +/− standard error (10 random fields counted from 6 tumors per group at 40x magnification). (E) Basal and stress-induced levels of HAUSP, p53 and Mdm2 in tumor xenografts. Immunoblot of pooled tumor lysates either unirradiated or 24 hrs after 7.5 Gy. PCNA as loading control.
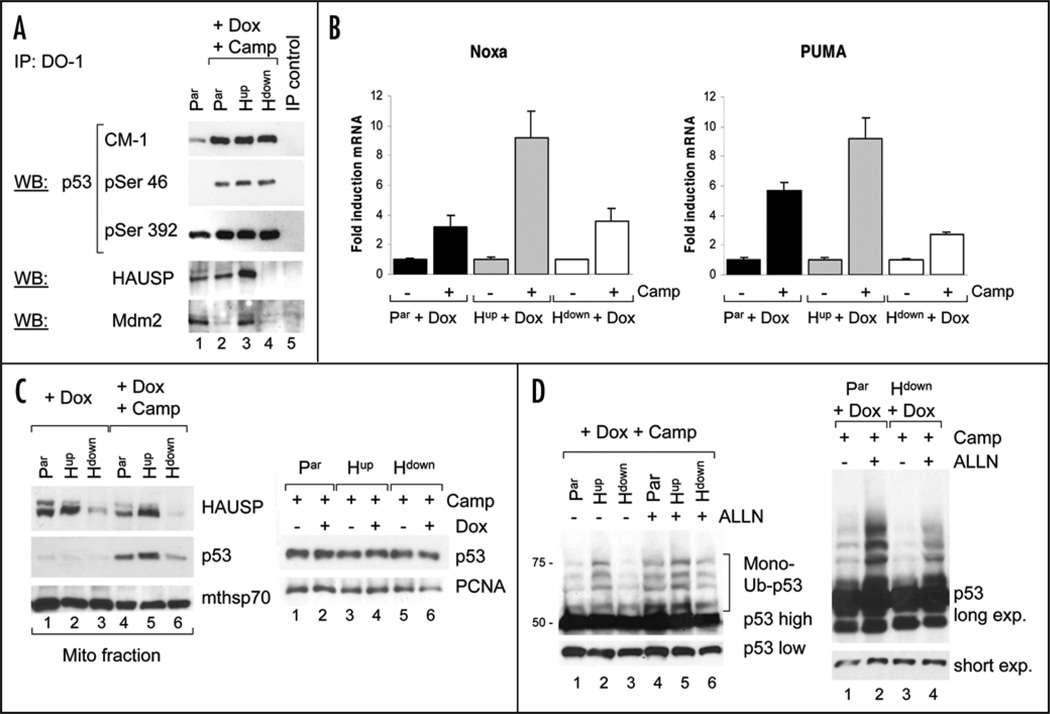
Figure 4.
HAUSP downregulation inhibits stress-induced translocation of p53 to mitochondria. (A) Apoptosis-associated p53 phosphorylation is intact upon HAUSP downregulation. Cells pretreated with Dox were stressed with Camptothecin for 5 hrs and immunoprecipitated with DO1. Input into immunoblots was adjusted for equal amounts of immunoprecipitated p53 in stressed cells (see CM-1 lanes). (B) p53 transactivation function upon HAUSP downregula-tion. Real time qRT-PCR of cells induced with Dox for 48 hrs. Average fold induction of Noxa and Puma after treatment with Camptothecin for 4 hrs (Noxa) and 9 hrs (Puma). Whiskers indicate standard error of triplicates. (C) Left, HAUSP downregulation inhibits stress-induced p53 translocation to mitochondria. Immunoblots of purified mitochondria from cells pretreated with Dox, followed by Camptothecin stress for 4 hrs. Mitochondrial mthsp70 as loading control. Right, immunoblot from total cell lysates of stressed cells +/− Dox. PCNA as loading control. (D) HAUSP downregulation decreases p53 monoubiquitination in response to stress. Cells induced with Dox were treated with Camptothecin for 1 hr, followed by the proteosome inhibitor ALLN for 6 hrs. Input into immu-noblot was normalized for equal amounts of p53 (short exposure). Long exposure of the same blot reveals the difference in monoubiquitination.
These results were confirmed in xenograft assays in vivo, in which half the mice were irradiated with a single dose of 7.5 Gy on day 13 and sacrificed 24 hrs later to evaluate their acute irradiation response. As shown by TUNEL staining (Fig. 3D), tumors with HAUSP downregulation were completely resistant to apoptosis in response to DNA damage. On the other hand, parental and tumors with HAUSP upregulation responded with the same very modest degree of apoptosis. Taken together with the fact that irradiation further inhibits the already suppressed growth of tumors with HAUSP downregulation despite their lack of apoptosis (Fig. 2B and C), these results strongly suggest that the marked decrease in tumor size after irradiation in both Hup and Hdown tumors is due to growth arrest rather than to apoptosis.
As validation, protein analysis of pooled tumor lysates from each animal demonstrated that Dox-control of HAUSP up and down-regulation properly functioned in these xenografts (Fig. 3E). In agreement with the observed growth suppression, basal p53 levels in tumors with HAUSP downregulation and to a greater extent HAUSP upregulation were elevated compared to parental tumors (Fig. 3E, compare lanes 1, 3 and 5). Again, this is consistent with the behavior of these cells in tissue culture (Fig. 1A) and indicates that the growth inhibition by HAUSP upregulation is most likely largely p53-mediated, while in case of HAUSP downregulation p53-independent factors might play a greater role. Similarly, in response to irradiation HAUSP up and downregulation both stabilized wild type p53 levels in the tumors (Fig. 3E). Of note, HAUSP upregulation correlated with the highest irradiation-induced p53 stabilization among the three tumor types. As expected, basal Mdm2 levels were markedly increased in HAUSP-upregulated tumors compared to parental tumors, while they were decreased in HAUSP-downregulated tumors. Also, as a p53 target gene, Mdm2 was induced upon irradiation (Fig. 3E). In sum, these in vivo tumor data recapitulate our results from tissue culture studies (Fig. 1A).
The apoptosis resistance associated with HAUSP downregulation in vitro and in vivo was rather unexpected, given that downregulation of HAUSP markedly suppressed cell proliferation and tumor growth, indicating an effective growth arrest at least in part via p53-mediated p21 induction (Figs. 1B–D and 2A). Thus, HAUSP depletion would be expected to also induce p53’s transcriptional pro-apoptotic action. Moreover, HAUSP-depleted cells displayed the same extent of stress-induced p53 stabilization as parental cells in vitro (Fig. 2A) and in vivo (Fig. 3E), suggesting that the lack of apoptosis is not due to a failure to stabilize p53 upon Camptothecin treatment or irradiation. Yet, at least in theory HAUSP might be required upstream of a signaling cascade that induces pro-apoptotic p53 modifications. To rule out this possibility, we analyzed hallmark p53 residues broadly linked to an apoptotic response that undergo stress-induced phosphorylation. As the most prominent, p53 Ser46 phosphorylation has been linked to DNA damage-induced apoptosis and is mediated by HIPK2 as well as p38MAPK and PKCδ.20 In growth arrest conditions, HIPK2 is degraded by Mdm2. In contrast, after apoptotic stimuli Mdm2 degradation prevents HIPK2 degradation, which then induces p53Ser46 phosphorylation.21 However, as seen in Figure 4A and data not shown, p53 phosphorylation patterns on Ser46, Ser392, Ser15 and Ser9 were intact in Camp-treated Hdown cells and identical to the pattern in parental and Hup cells. Reflecting the difference in basal HAUSP levels, p53 in Hup cells co-precipitated more HAUSP than in parental or Hdown cells, in which the p53-HAUSP complex was undetectable (Fig. 4A). Moreover, in accordance with the literature, the Mdm2-p53 complex in Hdown and parental cells was disrupted upon Camp treatment (Fig. 4A bottom, compare lanes 1, 2 and 4).22 Interestingly, in Hup cells, Mdm2 is stabilized so much that despite Camp treatment the Mdm2-p53 complex was still detectable (lane 3).
Since resistance to apoptosis in Hdown cells was not due to differences in apoptotic p53 phosphorylations, we next performed quantitative realtime RT-PCR to detect a potential inhibitory effect of low HAUSP levels on p53’s transactivation function of relevant apoptotic targets. To this end, we analyzed Dox-treated cells stressed with Camptothecin or mock-treated. Dox treatment alone did not change induction of Noxa or PUMA in parental cells (Suppl. Fig.). As shown in Figure 4B, Dox-induced HAUSP upregulation increased transcription of Noxa and PUMA after Camp treatment compared to their respective untreated cells (>9 fold induction in Hup cells) as well as to the treated parental cells. This likely contributes to the slight increase in apoptosis in Hup compared to parental cells in culture (Fig. 3A). On the other hand, Hdown cells showed a similar induction pattern to parental cells, albeit the PUMA response was decreased (Fig. 4B). Overall, we cannot exclude that the decrease in the induction of PUMA and maybe other pro-apoptotic genes not examined in this study might contribute to the lack of apoptosis after HAUSP downregulation, although it does not appear to play a major role.
HAUSP downregulation decreases Camptothecin-induced mitochondrial translocation of p53
In addition to its transcriptional pathway, p53 also promotes apoptosis through transcription-independent mechanisms, predominantly via its action at the mitochondria.23–26 We recently showed that Mdm2-mediated monoubiquitination of stress-stabilized cytoplasmic p53 promotes its mitochondrial translocation as a transcription-independent mechanism to induce apoptosis in response to stress.19,27 However, the active form of p53 at the mitochondria appears to be nonubiquitinated. Thus, p53 needs to be deubiquitinated by mitochondrial HAUSP upon its arrival via formation of a stress-induced mitochondrial p53-HAUSP complex which we showed exists naturally in stressed cells.19 Hence, we analyzed the effect of altered HAUSP levels on Camptothecin-induced mitochondrial translocation of p53, because (1) HAUSP levels affect the ubiquitination status of p53, which in turn is important for its mitochondrial translocation; (2) HAUSP’s deubiquitinase activity converts monoubiquitinated p53 into its active nonubiquitinated form at the mitochondria; (3) mitochondrial translocation of p53 occurs during apoptosis but not during growth arrest.24 Thus, a decrease in p53’s mitochondrial translocation in HAUSP-depleted cells would only affect p53’s apoptotic function but leave its growth arrest function intact, which could explain the behavior we observed in Hdown cells.
As we previously showed in other cancer lines, HAUSP was also constitutively present at purified mitochondria in this colon carcinoma series (Fig. 4C left, lanes 1–3).19 Proportional to their total cellular levels, Dox-induced HAUSP upregulation increased mitochondrial HAUSP levels compared to parental cells, whereas HAUSP downregulation decreased them. Camp treatment did not have any significant effect on mitochondrial HAUSP levels (Fig. 4C left). Of note though, HAUSP downregulation significantly decreased Camptothecin-induced mitochondrial translocation of p53 compared to parental cells (compare lanes 4 and 6), whereas HAUSP upregulation increased mitochondrial p53 translocation (compare lanes 4 and 5). These results confirm our previous notion that HAUSP has an important pro-apoptotic function in the mitochondrial p53 pathway.19 Moreover, the fact that total cellular p53 levels in Camp-treated cells were similar and independent of HAUSP levels (Fig. 4C right) shows that the observed difference in mitochondrial translocation is not a consequence of differences in total p53 levels, but due to the specific role of HAUSP in promoting mitochondrial p53 translocation. Together, these results suggest that decreased mitochondrial translocation of p53 contributes to the observed apoptosis resistance after HAUSP downregulation. To address the effect of altered HAUSP levels on p53 monoubiquitination that promotes its mitochondrial translocation, we evaluated the status of p53 ubiquitination by stressing Dox-pretreated cells with Camptothecin. For enhanced visualization of ubiquitinated p53 species, experiments were also performed in the presence of proteosome inhibitor ALLN. The Input into p53 immunoblotting was carefully normalized for equal amounts of non-ubiquitinated p53 (Fig. 4D). Indeed, the characteristic ladder of p53 monoubiquitination with species between 50 and 75 kDa was markedly reduced in HAUSP-downregulated cells compared to parental cells, visible even without proteosome inhibitor, but especially well with ALLN (Fig. 4D left and right, lanes 1 and 3). In contrast, HAUSP upregulation resulted in higher monoubiquitination of p53 compared to parental cells (Fig. 4D left, lanes 1 and 2), consistent with increased mitochondrial translocation of p53 (Fig. 4C, lanes 4 and 5). Interestingly, proteosome inhibition in Hup cells only led to a slight increase in monoubiquitinated p53 species (Fig. 4D left, lanes 2 and 5), indicating low p53 turnover due to preferentially mono- rather than poly-ubiquitinated p53 species in these cells. In contrast, Hdown cells displayed a significant increase in monoubiquitinated p53 after proteosome inhibition, suggesting that here p53 is less stable due to increased degradation (Fig. 4D left, lanes 3, 6 and right lanes 3, 4). In sum, our data support the notion that HAUSP downregulation induces resistance to apoptosis, at least in part due to a decrease in p53 monoubiquitination and therefore reduced mitochondrial p53 translocation upon Camp treatment.
Discussion
This is the first study to provide some validation for the basic notion of manipulating HAUSP protein levels as a rational target in wtp53 tumors to activate p53 pathways. To the best of our knowledge, it is also the first study on HAUSP manipulation in vivo. Interestingly, while HAUSP downregulation had been proposed as a new therapeutic strategy in cancer treatment based on biochemical studies,12,15,16 our results indicate that the therapeutic window, at least in theory, expands into both directions of HAUSP down as well as HAUSP upregulation. We show that HAUSP up and downregulation both stabilize basal p53 levels in unstressed cancer cells. However, the effect of HAUSP upregulation is much more robust, reproducibly resulting in significant p53 stabilization, induction of growth arrest and apoptosis (our data and refs. 4, 17 and 18). In contrast, the effect of HAUSP downregulation is dependent on the relative stoichiometry of p53, Mdm2 and HAUSP and can even lead to opposite results, depending on the extent of HAUSP downregulation. While partial reduction of HAUSP indeed destabilizes p53, near complete and complete ablation of HAUSP, on the contrary, stabilizes p53 due to destabilization of Mdm2 (our data and ref. 6).
Importantly, HAUSP up and downregulation both inhibit cell proliferation in vitro and tumor growth in vivo (Fig. 1B and C). The anti-proliferative effect of HAUSP upregulation was mainly due to p53 stabilization and activation with induction of the cell cycle inhibitor p21 (Figs. 1A and 2A). This confirms earlier, mainly non-isogenic studies showing that HAUSP upregulation causes growth arrest in wt p53 cells but not in p53 null cells.4 However, HAUSP downregulation inhibited cell growth in vitro more strongly than HAUSP upregulation (Fig. 1B), although p53 stabilization in Hdown cells was slightly lower than in Hup cells (Figs. 1A and 2A) and p21 levels in Hdown cells were not elevated compared to parental cells (Fig. 2A). Thus, p53-independent factors appear to contribute to the growth suppression by HAUSP downregulation. HAUSP negatively regulates the transcriptional activity of FOXO4, a transcriptional factor that induces cell cycle arrest via induction of the cell cycle inhibitor p27(Kip1) and downregulation of cyclin D.13,28 Hence, downregulation of HAUSP likely enhances the anti-proliferative effect of FOXO4. Moreover, several proteins involved in cell cycle control are downregulated after HAUSP knock-down, like Calgranulin A (S100A8) and Calgranulin B (S100A9).14 Of note, S100A8 and S100A9 proteins can activate the MAP kinase and NFkB signaling pathways,29 thereby promoting cell proliferation and are overexpressed in a variety of human cancers including colon carcinoma.30 In sum, while the growth-inhibiting effect of HAUSP upregulation seems to be mainly p53-dependent,4 the growth-inhibiting effect of HAUSP downregulation appears to be p53-dependent and -independent, likely including a decrease in pro-proliferative proteins and increase in anti-proliferative proteins.
Interestingly, tumor growth suppression upon altered HAUSP levels seems to be predominantly due to growth arrest rather than apoptosis. Unfortunately, tumor staining with the proliferation marker Ki67 did not show any significant difference between the tumor types due to high variability between individual tumors of the same cellular origin depending on its location and thus blood supply (anterior tumors were always bigger than posterior hip), as well as variability of different fields within the same tumor.
Our FACS data do not show any significant change in cell cycle distribution after HAUSP manipulation in both directions (data not shown), suggesting that cells might leave the active cell cycle and enter the quiescent G0 state.
Yet, apoptosis can at best play a minor role in mediating the observed growth suppression since HAUSP upregulation does not provide any significant gain of basal or irradiation-induced apoptosis compared to parental tumors, whereas HAUSP downregulation, on the contrary, even correlates with resistance to irradiation-induced apoptosis (Fig. 3D). Thus, the additional tumor shrinkage after irradiation is largely, if not exclusively, also due to growth suppression.
Nevertheless, the novel finding that HAUSP downregulation is associated with resistance to Camptothecin- and irradiation-induced apoptosis is surprising and raises questions about the mechanistic role of HAUSP in the apoptotic stress response, which has not been studied. We recently proposed p53 monoubiquitination as a mechanism to promote mitochondrial p53 translocation and thus mitochondrial apoptosis in response to stress.19,27 Here we show that HAUSP upregulation enhances monoubiquitination of p53 and thus mitochondrial translocation, whereas HAUSP downregulation decreases p53 monoubiquitination and reduces mitochondrial translocation (Fig. 4C and D). The reason for this seeming paradox lies in the effect of HAUSP on Mdm2. In unstressed cells, HAUSP upregulation stabilizes Mdm2 because it prevents the degradation of ubiquitinated Mdm2.6 Mdm2 monoubiquitinates p53, which then gets polyubiquitinated and rapidly degraded (Fig. 5 left). Of note, monoubiquitinated proteins in general are stable since efficient proteasomal degradation minimally requires a multiubiquityl chain at least 4 subunits per single lysine residue.31 Moreover, HAUSP might preferentially deubiquitinate polyubiquitinated p53 to mono-ubiquitinated p53, thereby preventing p53 degradation while leading to an accumulation of stable monoubiquitinated p53 that translocates to mitochondria upon Camp treatment (Fig. 5 right). In case of HAUSP downregulation, there is less monoubiquitinated p53 before the onset of stress due to destabilization of Mdm2. However, there are still other ubiquitin ligases (like COP1, Pirh2, ARF-BP1) and the E4 ligase p300 that could mediate polyubiquitination and subsequent degradation of p53. Under stress conditions, the residual HAUSP activity might not be sufficient to deubiquitinate polyubiquitinated to monoubiquitinated p53 in order to prevent its degradation. Hence, in Hdown cells p53 is less stable and there is less monoubiquitinated p53 available for mitochondrial translocation. Apart from its effect on ubiquitination, HAUSP might also function as a receptor for p53 at the mitochondria. Our previous data support a model in which mitochondrial HAUSP deubiquitinates and thus activates p53 upon its arrival at the mitochondria via a stress-induced mitochondrial p53-HAUSP complex.19 Consequently, HAUSP upregulation increases the amount of this mitochondrial receptor (Fig. 4C, upper) and thus mitochondrial p53 translocation, whereas HAUSP downregulation decreases it. In conclusion, our results indicate an important role for HAUSP in the mitochondrial apoptotic pathway. Yet, further studies are necessary to elucidate the exact mechanism by which HAUSP promotes mitochondrial translocation of p53.
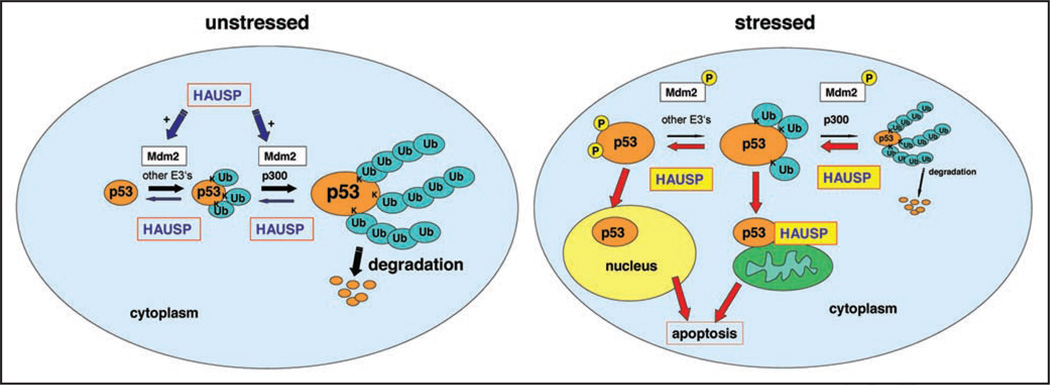
Figure 5.
Model of HAUSP regulation of p53 ubiquitination in unstressed and stressed cells. Left, p53 ubiquitination in unstressed cells. p53 is (multi)monoubiquitinated by Mdm2 and other E3 ligases mainly in the cytoplasm. The E4 ligase p300 together with Mdm2 mediate p53 polyubiquitina-tion.35,36 Polyubiquitinated p53 is unstable and rapidly degraded via 26S proteosomes, which ensures low p53 levels in unstressed conditions. In the absence of stress, HAUSP mainly deubiquitinates und thus stabilizes Mdm2, while to a minor extent HAUSP also deubuitinates p53, either via a direct p53-HAUSP complex or a triple HAUSP-Mdm2-p53 complex.12 Right, Under stress conditions, p53 and Mdm2 are phosphorylated, disrupting both the p53-Mdm2 complex and the Mdm2-HAUSP complex.11,22 Consequently, Mdm2 undergoes accelerated ubiquitination and degradation, while p53 is stabilized due to protection from Mdm2 ubiquitination. Moreover, HAUSP deubquitinates p53, thereby further stabilizing p53. We propose that HAUSP preferentially deubiquitinates unstable, polyubiquitinated p53 and converts it to monoubiquitinated p53, which is stable. Monoubiquitination promotes mitochondrial translocation of p53, thereby contributing to the apoptotic stress response. Upon arrival at the mitochondria, mitochondrial HAUSP deubiquitinates p53 via a stress-induced complex and thus might serve as a p53 receptor.19 Non-ubiquitinated p53 is imported into the nucleus where it transcriptionally activates apoptotic target genes.33
Another factor contributing to apoptotic resistance after HAUSP downregulation might be a decrease in PUMA induction in Camp-treated Hdown cells compared to Camp-treated parental cells. Our RT-PCR results suggest that p53 in stressed Hdown cells, while being transactivation-competent for some p53 target genes like Noxa, might be somewhat transactivation-deficient for other pro-apoptotic target genes like PUMA, resulting in an impaired apoptotic stress response (Fig. 4B). Moreover, we do not exclude that p53-independent factors might also play a role in apoptosis resistance, since HAUSP has been shown to directly or indirectly affect the levels of a broad spectrum of proteins. For instance, downregulation of HAUSP correlates with reduced expression of pro-apoptotic proteins like Alix/HP95 and ASP2,14 which could contribute to impair the apoptotic stress response in a HAUSP-deficient tumor environment.
In conclusion, the role of HAUSP in tumor suppression in vivo appears to be more complex than previously thought. Not only is the interplay between HAUSP, p53 and Mdm2 non-linear and dynamic, but also p53-independent factors seem to be involved. Therefore, more in vivo studies are necessary to evaluate the effect of HAUSP manipulation on tumor growth and response to chemo-and radiotherapy and as potential target in cancer treatment. So far only a single clinical study looked at HAUSP expression in primary human tumors.32 In this study, 45% of non-small cell lung carcinomas, especially adenocarcinomas, (72 of 131 cases) showed reduced HAUSP expression that was associated with lower p53, p21 and Bax levels. Furthermore, adenocarcinoma patients with either mutant p53 or reduced HAUSP expression had a significantly poorer survival than patients with normal HAUSP levels and wild type p53, leading the authors to speculate that reduced HAUSP levels play an important role in lung carcinogenesis through p53-dependent pathways. Further correlative investigations in clinical tumor samples are clearly warranted. Our data suggest that broad changes in HAUSP levels modulate tumor growth and apoptotic sensitivity in vivo. Since upregulation of HAUSP would be pharmacologically difficult, it supports small molecule HAUSP inhibitors in cancer therapy, if anything.12 However, one has to keep in mind that HAUSP inhibitors, albeit inhibiting tumor growth, might impair the apoptotic response to adjuvant chemo-/radiotherapy. On the one hand, this might limit the effect of radiotherapy to mere growth arrest rather than apoptosis of tumor cells. On the other hand, the resistance to stress-induced apoptosis associated with HAUSP downregulation might protect healthy tissues from acute irradiation damage. Thus, HAUSP inhibitors might reduce the adverse side effects of chemo- and radiotherapy and therefore be of use in combination with other anti-cancer treatments. Further studies in normal cells will need to test this notion.
Material and Methods
Cell culture and reagents
The wild-type p53 human colon carcinoma cell line LS174TR1 (here called parental Par) was used to derive a subline called (LS89) that stably overexpresses Dox-inducible HAUSP (Myc-epitope tagged), here called Hup.7 Another subline (LS126) stably expresses Dox-inducible HAUSP RNAi, called Hdown and was previously described.7 All cells were cultured in DMEM plus 10% FBS and induced with Doxycycline (1 µg/ml) for 48–72 h where indicated. Cells were subjected to 10 µM Camptothecin (Sigma), 25 µM ALLN (Calbiochem) or 50 µg/ml Cycloheximide (Sigma) where indicated. For cell proliferation assays, 104 cells were seeded onto 6 well plates in triplicates and counted at the indicated time points.
Immunoblotting and immunoprecipitation
Immunoblotting and immunoprecipitation were performed as previously described.33 The following antibodies were used: monoclonal DO1 (Santa Cruz), polyclonal rabbit CM1 (Vector Labs), p53 Phospho-Ser 46 and Ser 392 (Cell Signaling), HAUSP (Calbiochem), HDAC1, hsp90, mthsp70 (all ABR), p21 (Santa Cruz), PARP (Cell Signaling) PCNA (Santa Cruz). Mitochondria were purified by sucrose density gradients as described.34
TUNEL assay
Apoptosis was assessed using the TUNEL TMR red and TUNEL POD kit (Roche) according to the manufacture’s protocol.
Realtime qRT-PCR
RNA was isolated with Trizol and 4 µg of RNA was reverse transcribed using random primers and SuperScript II Reverse Transcriptase (Invitrogen). Realtime qRT-PCR was performed in a MJ Research DNA Engine Opticon 2 using Qiagen QuantiTect SyBr Green Mix. Primer sequences are shown in supplemental material. Each sample was run in triplicates. Expression of target genes was normalized for Cyclophilin A expression as internal efficiency control. The basal target gene expression in Dox-induced cells was arbitrarily set to one for each cell line. Fold induction was calculated as the ratio between normalized gene expression in Dox + Camptothecin-treated cells versus cells just primed with Dox.
Nude mice xenograft assay
Cells were induced in vitro with Dox for 72 h and then injected subcutaneously into nude mice (106 cells per tumor; 6 paravertebral tumors per mouse). All mice received Dox with their drinking water (2 mg/ml in 5% sucrose) ad libitum. Tumor sizes were measured every other day. In one experiment, 3 out of 6 mice were irradiated with 7.5 Gy or mock treated on day 13 and sacrificed 24 h later. In a second experiment, 3 out of 6 mice were subjected to fractionated irradiation on day 9, 12 and 14 (4 Gy per fraction) or mock treated and sacrificed on day 20.
Supplementary Material
Footnotes
Supplementary materials can be found at: www.landesbioscience.com/supplement/BeckerCC7-9-Sup.pdf
References
- 1.Nakamura S, Roth JA, Mukhopadhyay T. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol Cell Biol. 2000;20:9391–9398. doi: 10.1128/mcb.20.24.9391-9398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 4.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 5.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:86. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 6.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 7.Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Brazhnik P, Kohn KW. HAUSP-regulated switch from auto- to p53 ubiquitination by Mdm2 (in silico discovery) Math Biosci. 2007;210:60–77. doi: 10.1016/j.mbs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 11.Meulmeester E, Pereg Y, Shiloh Y, Jochemsen AG. ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle. 2005;4:1166–1170. doi: 10.4161/cc.4.9.1981. [DOI] [PubMed] [Google Scholar]
- 12.Brooks CL, Li M, Hu M, Shi Y, Gu W. The p53—Mdm2—HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene. 2007;26:7262–7266. doi: 10.1038/sj.onc.1210531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 14.Kessler BM, Fortunati E, Melis M, Pals CE, Clevers H, Maurice MM. Proteome Changes Induced by Knock-Down of the Deubiquitylating Enzyme HAUSP/USP7. J Proteome Res. 2007 doi: 10.1021/pr0702161. [DOI] [PubMed] [Google Scholar]
- 15.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheon KW, Baek KH. HAUSP as a therapeutic target for hematopoietic tumors (review) Int J Oncol. 2006;28:1209–1215. [PubMed] [Google Scholar]
- 17.Lim SK, Shin JM, Kim YS, Baek KH. Identification and characterization of murine mHAUSP encoding a deubiquitinating enzyme that regulates the status of p53 ubiquitination. Int J Oncol. 2004;24:357–364. [PubMed] [Google Scholar]
- 18.Yoo KJ, Lee HJ, Lee H, Lee KY, Lee SH, Chung HM, Baek KH. Expression and functional analyses of mHAUSP regulating apoptosis of cervical adenocarcinoma cells. Int J Oncol. 2005;27:97–104. [PubMed] [Google Scholar]
- 19.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. Embo J. 2007 doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 21.Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell. 2007;25:739–750. doi: 10.1016/j.molcel.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 23.Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24:6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–67212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 25.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 26.Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488:110–115. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- 27.Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle. 2007;6:1718–1723. doi: 10.4161/cc.6.14.4503. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, Medema RH. Cell cycle inhibition by FoxO forkhead transcription factors involves down-regulation of cyclin D. Mol Cell Biol. 2002;22:7842–7852. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermani A, De Servi B, Medunjanin S, Tessier PA, Mayer D. S100A8 and S100A9 activate MAP kinase and NFkappaB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp Cell Res. 2006;312:184–197. doi: 10.1016/j.yexcr.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Cross SS, Hamdy FC, Deloulme JC, Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46:256–269. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- 31.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masuya D, Huang C, Liu D, Nakashima T, Yokomise H, Ueno M, Nakashima N, Sumitomo S. The HAUSP gene plays an important role in non-small cell lung carcinogenesis through p53-dependent pathways. J Pathol. 2006;208:724–732. doi: 10.1002/path.1931. [DOI] [PubMed] [Google Scholar]
- 33.Becker K, Marchenko ND, Maurice M, Moll UM. Hyperubiquitylation of wild-type p53 contributes to cytoplasmic sequestration in neuroblastoma. Cell Death Differ. 2007;14:1350–1360. doi: 10.1038/sj.cdd.4402126. [DOI] [PubMed] [Google Scholar]
- 34.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria.A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 35.Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 36.Li M, Brooks CL, Wu Baer F, Chen D, Baer R, Gu W. Mono-versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.