Abstract
Gastric cancer is the second most common cause of cancer-related death in the world, representing a major global health issue. The high mortality rate is largely due to the lack of effective medical treatment for advanced stages of this disease. Recently next-generation sequencing (NGS) technology has become a revolutionary tool for cancer research, and several NGS studies in gastric cancer have been published. Here we review the insights gained from these studies regarding how use NGS to elucidate the molecular basis of gastric cancer and identify potential therapeutic targets. We also discuss the challenges and future directions of such efforts.
Keywords: somatic mutation, gastric cancer, gene fusion, therapeutic target, next-generation sequencing, RNA sequencing
1. Background
Gastric cancer refers to a tumor that originates in any part of the stomach. Currently the fourth most common malignancy in the world, gastric cancer is second to lung cancer as the leading cause of cancer mortality, causing about 800,000 deaths per year [1; 2]. The incident rate of gastric cancer appears to vary greatly across geography, ethnicity and gender [3]. The mortality rate attributed to gastric cancer has recently declined in North America and Europe; however, gastric cancer remains the most common cancer type in countries of East Asia [4], representing the highest cancer incident rate among males and females in Korea, the most common cancer among females in China, and the most common cancer among males in Japan [5].
Conventional treatment options for gastric cancer include surgery, chemotherapy, radiation therapy, or combinations thereof known as multimodality therapy. If gastric cancer is diagnosed in an early stage, curative treatment can be achieved through complete surgical removal of the tumor tissue [6]. However, because gastric cancer causes few symptoms in its early development, a diagnosis is usually made after the cancer reaches an advanced stage. Moreover, even after having a gastric tumor surgically removed, many patients will experience disease recurrence and die within a few months to years. The 5-year relative survival rate of gastric cancer has not improved during the past 35 years, and remains stubbornly at 20-30%. Thus, the high mortality rate attributed to gastric cancer is due to the lack of both early detection methods and effective medical treatment for advanced stages of the disease [6].
Gastric cancer is characterized by a high level of biological heterogeneity, with each patient exhibiting a distinct genetic and molecular profile [7]. Histologically, the majority of gastric malignancies are adenocarcinomas that can be further categorized as diffuse (poorly differentiated) or intestinal (well-differentiated) types, each with distinct epidemiological and genetic patterns, and the former subtype carrying a worse prognosis [7; 8; 9]. Etiologically, gastric cancer is associated with the combined effects of environmental factors and susceptible genetic variants, including the accumulation of genetic and epigenetic alterations [7; 8]. Diet and lifestyle factors have been shown to significantly affect gastric cancer risk, with greater risk associated with tobacco smoking and obesity [10]. The infectious agent Helicobacter pylori is closely related to the most common types of gastric adenocarcinoma [11], and is the main risk factor in 65-80% of gastric cancer cases. H. pylori induces generalized mutations and genomic instability in the host DNA [12], and this possibility may increase the diversity of oncogenic mechanisms in gastric cancer.
Given the diverse routes to gastric oncogenesis, it is unlikely that a “magic bullet” treatment exists. Therefore, targeted therapy based on the biology of the individual patient is highly attractive. This involves first identifying the biological molecules that contribute to the development or maintenance of the tumor, then specifically targeting those oncogenic mechanisms with an anti-tumor treatment regimen. Such therapies attempt to inactivate specific oncogenic mechanisms that are critical to the survival of tumor cells, while sparing normal gastric cells, thereby maximizing the benefits and minimizing the side effects. In early 2010, trastuzumab, a humanized monoclonal antibody directed against the extracellular domain of the HER2/neu receptor, was approved for the treatment of gastric cancer. Trastuzumab became the first targeted agent to be used in combination with chemotherapy as first-line treatment of ERBB2-positive advanced and metastatic gastric cancer [13; 14]. A relatively large proportion of gastric tumors (2~27%) harbor ERBB2 amplifications and may respond to this treatment regimen [15; 16]. Several molecular targeted agents associated with a survival advantage in other cancer types are now under clinical investigation for the treatment of gastric cancer, including inhibitors of EGFR, MET, FGFR, VEGF, and PI3K [17; 18]. For example, in a single-arm phase II clinical trial for patients with gastric and gastroesophageal adenocarcinoma, Shah et al. demonstrated that the addition of bevacizumab, a monoclonal antibody against VEGF, to a therapeutic regimen of irinotecan and cisplatin achieved a median time-to-disease progression of 8.3 months, which was an improvement of 75% over that of the historical control regimen [19]. Pinto and colleagues demonstrated a response rate of 44.1% and median time-to-disease progression of 8 months in advanced gastric cancer using an irinotecan-based regimen in combination with EGFR-targeting cetuximab [20]. However, these targeted therapies, whether approved or in clinical development, have focused on a few well-known oncogenic genes, and their efficacy in gastric cancer is not yet known. Developing novel therapeutic strategies to specifically target gastric cancer requires a comprehensive dissection of the molecular mechanisms underlying the initiation and progression of this disease.
2. Brief overview of next-generation sequencing technology
Next-generation sequencing (NGS) is a powerful technology for elucidating the pathogenesis of human cancer and identifying potential therapeutic targets [21]. NGS is also known as second-generation sequencing, which describes its relation to capillary-based Sanger sequencing, the first-generation sequencing technology [22]. NGS includes the various implementations of cyclic-array sequencing [23]. Among the most widely used commercial NGS platforms are the 454 Genome Sequencer (Roche Applied Science), Solexa technology (Illumina) and the SOLiD platform (Life Sciences). Although some technical differences exist, the concept of these NGS platforms is quite similar: the sample DNA is first randomly fragmented, followed by in vitro ligation of common adaptor sequences, thereby forming a library; then an array of millions of spatially fixed PCR colonies are generated, each consisting of many copies of a single library fragment; and finally, enzyme-driven biochemistry and imaging-based data processing is performed in parallel. Thus, given a sample, NGS generates millions of short reads in an extremely efficient fashion. In the downstream analysis, the short read sequences are mapped to the source genome to generate a nucleotide-resolution read distribution, from which various biological features about the molecules in the sample can be inferred. Depending on the type of input materials, the major NGS applications include DNA sequencing (DNA-seq), RNA sequencing (RNA-seq) and chromatin immunoprecipitation sequencing (ChIP-seq).
The initial applications of NGS focused on sequencing the genomes of individual people or transcriptomes of various tissues. The more recent applications of NGS to study cancer biology [24] have revolutionized cancer research in several ways. First, NGS is able to generate unbiased, comprehensive rather than biased, limited catalogs of various aberrations in the cancer genomes. For example, pre-NGS sequencing studies on cancer somatic mutations examined only a small set of well-known cancer genes, such as TP53, EFGR and KRAS; and these focused studies had little power to discover novel cancer genes [24]. Second, unlike “analog” signals in earlier sequencing technology, the NGS signaling is based on the count of the short reads. This digital-like signal is extremely good for quantification [25]. Thus, high sequence coverage provides sufficient information to detect biological signals in heterogeneous samples such as cancer tissues. Finally, with the commercialization of NGS platforms, the cost of DNA sequencing has decreased dramatically, reaching a level of ~$1,000 per personal genome [26]. This low cost has facilitated the clinical applications by incorporating personal genomic information.
A handful of NGS studies in gastric cancer have been published over the last two years (summarized in Table 1). In this article, we review the insights gained from these studies regarding the use of NGS to identify potential therapeutic targets in gastric cancer, and discuss the challenges and future directions of such efforts.
Table 1.
Next-generation sequencing studies in gastric cancer
| Study | Method | Samples | Aberration type | Main Findings |
|---|---|---|---|---|
| Wang et al. [30] | Whole-exome sequencing | 22 tumor and matched normal pairs | Point mutations, small indels | Frequent inactivating mutations in ARIDA1 |
| Zang et al. [31] | Whole-exome sequencing | 15 tumor and matched normal pairs | Point mutations, small indels | Frequent inactivating mutations ARIDA1, FAT4 |
| Zang et al. [32] | Whole-kinome sequencing | 14 GC cell lines 3 tumor and normal pairs | Single nucleotide variations, gene fusions, and copy number variations | Recurrent inactivating mutations in MAP2K4, gene fusions involving CDK12 and ERBB2 |
| Holbrook et al. [33] | Targeted DNA sequencing (384 genes) | 44 tumor samples (36 with matched normal) | Point mutations | Genetic alternation in the WNT, Hedgehog, cell cycle, DNA damage and epithelial-to-mesenchymal transition pathways |
| Kim et al. [40] | Whole transcriptome RNA-sequencing | 24 tumor samples and 6 normal samples | mRNA expression, miRNA expression, recurrent somatic mutations | AMPKα2 as a potential therapeutic target in Asian patients |
| Riberiro-dos-Santos [42] | Small RNA Sequencing | Healthy gastric tissue | / | 15 most highly expressed miRNAs in gastric tissue |
| Li et al. [43] | Small RNA sequencing | One pair of tumor and normal samples | microRNA expression | Biased selection of arm miRNAs from the same pre-miRNAs |
3. Targeted DNA-seq in gastric cancer
Although the first applications of NGS in cancer were almost all whole-genome sequencing studies [24], targeted DNA-seq has become a more popular approach for identifying somatic mutations in cancer genomes. Through a DNA capture technique based on hybrid selection, targeted DNA-seq focuses on a proportion of the genome, such as exons. Although this approach misses some aberrations that could be detected in whole-genome sequencing, such as copy number alternations, interchromosomal rearrangements and inversions, targeted DNA-seq represents a cost- and resource-efficient approach, allowing for the identification of somatic mutations of greatest potential interest. Given the mutations detected in a tumor sample, the key goal is to distinguish “driver” mutated genes from “passenger” mutated genes. The somatic mutations of driver genes are those that play critical roles in cancer pathophysiology and thus represent high-quality targets; whereas those from passenger genes result from the underlying genomic instability associated with cancer and have little phenotypic effects. In general, driver genes are expected to harbor more somatic mutations than would occur randomly. Since the mutation rate in the cancer genome depends on various factors, such as coverage, base composition, gene expression [27], and replication time [28], it is essential to take these factors into consideration when assessing the statistical significance of mutations observed in a gene relative to the “background rate.” Importantly, it would be desirable to experimentally validate the functional impact of mutated genes using functional assays such as cell viability or cell transformation assays [29].
Wang and colleagues published the first exome-sequencing study in gastric cancer [30], sequencing 22 matched pairs of gastric cancer and healthy gastric tissue. Using a driver gene score, they identified 20 genes as top candidate drivers, including previously known drivers such as TP53, PTEN, and CTNNB1. One major finding from the study is the high mutation frequency of ARID1A, a key member of the SWI-SNF complex that has been associated with frequent mutations in several other cancer types. Interestingly, Wang et al. also found chromatin modification and cell junction organization pathways to be the most perturbed pathways, and 59% of the gastric cancer samples under survey to have mutations in histone-modifying proteins. Then, using Sanger sequencing to further evaluate ARID1A in additional samples of gastric cancer, they found that the somatic mutation rate and spectrum of ARID1A significantly varies among different molecular subtypes of gastric cancer; and clinically, alterations in ARID1A are associated with better prognosis in a stage-independent manner. Interestingly, ARID1A was also identified as a driver gene in a later exome-sequencing study in which Zang et al. sequenced 15 matched sample pairs of gastric cancer and performed similar computational analyses [31]. Importantly, they provided direct functional evidence supporting a tumor suppressor role of ARID1A: knockdown mediated by ARID1A shRNA or siRNA increased the cell proliferation in the cell lines with wild-type ARID1A. In contrast, in the ARID1A-deleted lines, no such effects of shRNA-mediated silencing were detected, and re-expression of ARID1A suppressed the cell proliferation [31]. These studies thus highlight the importance of chromatin remodeling genes in gastric tumorigenesis. In addition to ARID1A, Zang et al. identified FAT4, a member of the E-cadherin family, as a strong candidate driver gene. Using the same experimental approach in cell lines as done previously for ARIDA1, they showed that FAT4 functions as a tumor suppressor [31]. Fig.1 shows a comparison of top candidate driver genes identified by these two exome-sequencing studies.
Fig.1. A comparison of most significantly mutated genes identified in the two whole-exome sequencing studies.
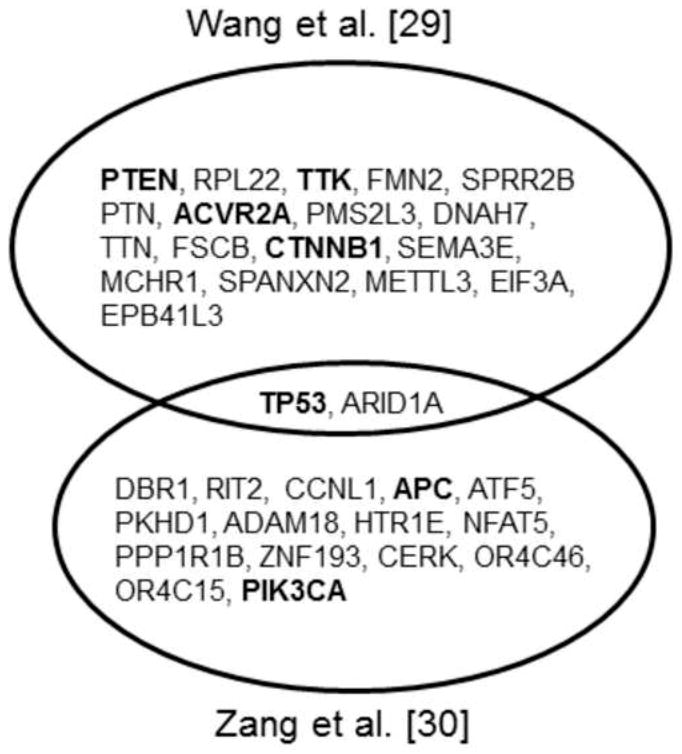
Top significantly mutated genes (FDR < 0.2) were obtained from each study, respectively; and genes previously known to associate with gastric cancer are shown in bold.
Although the exome sequencing studies provided an unbiased way to assess the mutation patterns in all protein-coding genes (~20,000) in the human genome, novel candidate driver genes thereby obtained are often not good candidates for drug discovery. This is because (i) the driver gene itself is not readily targeted by a drug or (ii) no sufficient prior knowledge has been accumulated about the genes, making further functional or clinical investigation very difficult. Therefore, using DNA-seq to target a subset of “druggable” or “important” genes becomes a more effective way to discover therapeutic targets. Two such studies have been recently published. Zang et al. characterized the protein-coding regions of 537 kinases in 14 commonly studied cell lines using NGS, and detected more than 300 novel kinase SNVs [32]. A family-wise analysis further revealed a significant SNV enrichment in MAPK-related genes. In particular, siRNA knockdown and overexpression experiments identified MAP2K4 as a tumor suppressor gene in this disease. Holbrook et al. performed targeted DNA sequencing on 384 genes belonging to various pathways known to be important in cancer [33]. Their study revealed mutations in the RAS/RAF/MEK/ERK pathway, PI3K/AKT pathway, WNT pathway, and the hedgehog pathway, suggesting novel therapeutic opportunities. For example, KRAS G12D mutations imply a treatment option for MEK inhibitors [34]; and inhibitors of the hedgehog pathway, such as GDC-0449, are in clinical development as therapeutic targets [35]. In addition, the mutations identified in the study also suggested novel therapeutic targets such as the thyrotropin receptor (TSHR) and the Rho-associated coiled-coil containing protein kinases (ROCK1 and ROCK2).
4. Transcriptomic RNA-seq in gastric cancer
Before the NGS era, hybridization-based microarrays were the main approach for transcriptomic studies in gastric cancer. Using miRNA expression microarrays, Volinia et al. [36] and Ueda et al. [37] evaluated aberrant miRNA expression signatures in gastric tumor samples from Italian and Japanese patients, respectively; and Cui et al. used exon arrays to identify differentially expressed mRNAs as potential biomarkers in Chinese patients [38]. In recent years, RNA-seq has become a revolutionary approach for transcriptome profiling, supplanting microarrays with rapid speed [25]. Compared with microarrays, NGS-based RNA-seq has several advantages in cancer transcriptome profiling. First, RNA-seq provides an unbiased approach to profiling all the transcribed molecules in a sample, which is not limited to the previously known or annotated transcripts. Second, RNA-seq accommodates a large dynamic range of expression levels, thereby allowing for a much more accurate quantification of genes with very low or high expression levels [25]. Third, in addition to gene-level expression quantification, RNA-seq can detect other types of transcriptional signals, including alternative splicing, transcriptional starts/stops, gene fusion, and expressed alleles [39].
We recently published the first comprehensive RNA-seq study in gastric cancer. We applied a whole-transcriptome sequencing approach to 24 samples of gastric tumors and 6 noncancerous tissue specimens obtained from Asian patients, generating 680 million informative short reads to quantitatively characterize gene expression [40]. We used two sequencing protocols to respectively sequence the transcripts of different lengths. Beyond the conventional analysis, one unique aspect of the study is that we developed a multilayered integrative analysis to identify various types of transcriptional aberrations associated with different stages of gastric cancer, including differentially expressed mRNAs, recurrent somatic mutations, and key differentially expressed microRNAs. Through this computational approach, we identified the central metabolic regulator AMPKα2 (PRKAA2) as a potential functional target in Asian gastric cancer. This gene shows a differential loss of mRNA level in tumor stage I/II relative to that of noncancerous gastric tissue or advanced tumor stages. Through the perturbation experiments in gastric cell lines (metformin-based activation and siRNA-mediated knockdown), we demonstrated the functional relevance of AMPKα2 loss for the AMPK signaling pathway, with downstream consequences that increase both HNF4α and HIF-1α. Consistent with our study, AMPKα2 has been recently shown to suppress embryonic fibroblast transformation and tumorigenesis in animal models [41]. Thus, AMPKα2 may represent a promising therapeutic target for gastric cancer.
Abnormal microRNA (miRNA) expression is the hallmark of many human cancer types, so one important application of RNA-seq in cancer research is to identify miRNAs that play an important role in tumorigenesis. By incorporating the expression data of protein-coding and miRNAs, we identified six key miRNAs in gastric cancer (miR-548d-3p, miR-20b, miR-135b, miR-140-3p, miR-93, and miR-19a) [40]. These key miRNAs not only show significant expression variations across different sample groups, but also have detectable repression effects on the expression of their target genes. In addition to our study, two RNA-seq studies focusing on miRNA profiling have been published. Ribeiro-dos-Santos et al. sequenced a small RNA library of normal stomach tissue and identified 15 highly expressed miRNAs [42]. Using the Solexa platform, Li et al. sequenced small RNAs from one pair of noncancerous/tumor gastric tissues [43]. Interestingly, they found that the 5p arm and 3p arm miRNAs derived from the same pre-miRNAs have different tissue preferences (noncancerous tissue vs. tumor tissue), implying a novel mechanism regulating mature miRNA selection. Although usually miRNAs are not direct therapeutic targets, identification of the key miRNAs would greatly help elucidate the molecular basis of tumor progression and aid in selecting potential therapeutic targets in gastric cancer.
5. Gene fusion detection in gastric cancer
Gene fusion refers to a hybrid gene formed from two genes as a result of translocation, interstitial deletion, or chromosomal inversion. Gene fusion can disrupt the original functions of the partner genes, generating a gene product with a new function, or it can greatly change the expression level of the partner genes (e.g., through fusion to a strong promoter). Since gene fusion can introduce dramatic functional consequences in gene function, a number of fused genes have been identified as therapeutic targets, such as EML4-ALK in non-small cell lung cancer [44; 45]. Both DNA-seq and RNA-seq are able to detect gene fusion, and for that purpose, paired-end sequencing is particularly powerful.
Through whole-kinome sequencing, Zang et al. found two genome rearrangements involving ERBB2, a well-known proto-oncogene and the therapeutic target of tratuzumab, in the MKN7 gastric cell line [32]. One event involves a 169-kb deletion fusing the CDK12 exon 13 to the ERBB2 intron 4; and the other event results from a 106-kb deletion fusing the NEU-ROD2 exon to the ERBB2 exon 8 (Fig. 2A and 2B). To our knowledge, RNA-seq has not been used to detect gene fusions in gastric cancer. However, evaluating prostate cancer using RNA-seq, Palanisamy et al. detected BRAF and RAF1 rearrangements; and then, using break-apart FISH probes, they revealed that exon 8 of the BRAF gene was fused with exon 5 of the AGTRAP gene (angiotensin II, type I receptor-associated protein) in gastric cancer, resulting in the formation of a 597-amino acid fusion protein [46] (Fig. 2C). This result suggests that targeting RAF and MEK inhibitors may be useful in a subset of patients with this type of gene fusion. Moreover, RNA-seq can detect targetable gene fusion across different cancer types.
Fig.2. Gene fusions discovered in gastric cancer by NGS studies.
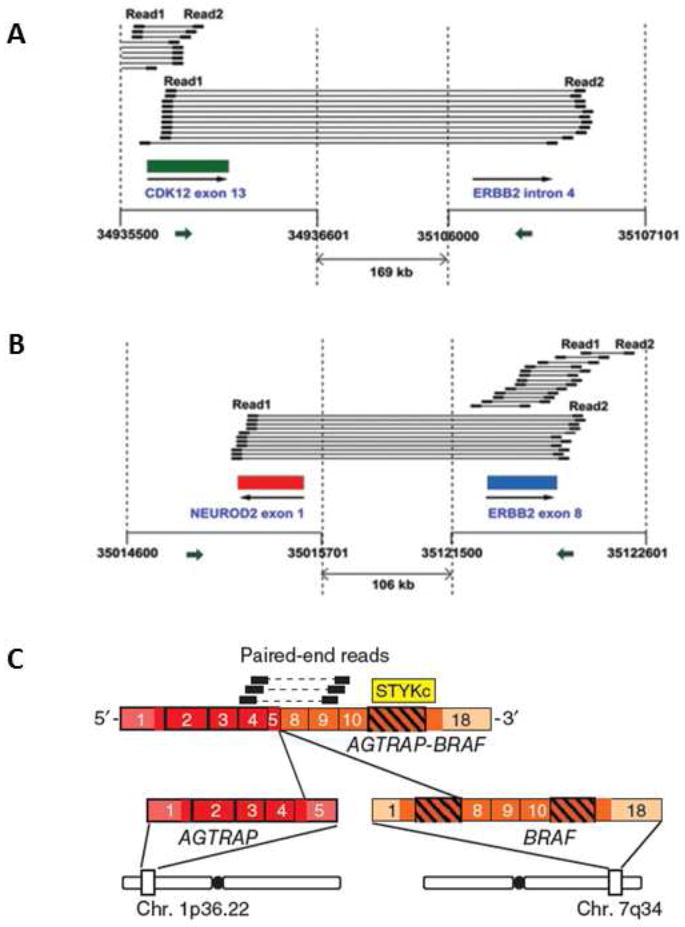
(A) A fusion between CDK12 exon 12 and ERBB2 intron 4 in MKN7 cells; (B) a fusion between NEUROD2 exon 1 and ERBB2 exon 8 in MKN17 cells; and (C) a fusion between AGTRAP exon 5 and BRAF exon 8 in a gastric tumor sample. (A) and (B) are reproduced with permission from Zang et al. [32], and (C) is reproduced from Palanisamy et al. [46]
6. Challenges and future directions
Although the above-mentioned NGS studies provide tremendous insights into the molecular basis of gastric cancer and identify novel potential therapeutic targets, these individual lab-driven studies have limitations. First, these studies are based on small sample sizes, ranging from a handful of samples to 30 at most, which is likely due to cost and resource constraints. Because gastric cancer is a heterogeneous disease in which each cancer patient exhibits a distinct genetic and molecular profile, there is limited statistical power to accurately detect prevalent therapeutic targets based on a relatively small sample cohort. For example, only two candidate driver mutated genes (TP53 and ARID1A) were simultaneously identified by both the exome-sequencing studies [29, 30] (Fig. 1). Second, DNA-seq or RNA-seq is usually employed as the only approach to characterize genomic alternations in these studies. Gastric cancer is a complex disease, involving interactions between multiple layers of aberrations. To understand how an alteration in a driver gene functions in the context of the tumor, and thereby identify and prioritize high-quality therapeutic targets, it is essential to integrate multi-dimensional genomic profiles of gastric cancer. Third, the functional data on driver genes discovered in these NGS studies are still sparse. Due to resource constraints in any individual lab, usually only one or two genes are chosen for further functional investigation. In many cases, the choice is somewhat subjective or is biased by the investigator’s expertise. Ideally, a high-throughput functional assay will be used to systematically examine the functional consequences of genomic aberrations, as demonstrated by Liang et al. [47] Fourth, the huge size of the datasets obtained through NGS make them difficult to access.
Two recently initiated consortium projects have largely overcome these four issues. One project is The Cancer Genome Atlas (TCGA), sponsored by the U.S. National Cancer Institute and the National Human Genome Research Institute. For each selected cancer type, TCGA will systematically characterize ~500 patient samples using different genomic profiling techniques, including exome sequencing, SNP arrays, copy number variation profiling, DNA methylation profiling, miRNA expression profiling, mRNA-seq, and RPPA-based protein expression. Importantly, user-friendly public access is provided for datasets generated by TCGA. The first cancer types evaluated in data released by TCGA are glioblastoma [48] and ovarian cancer [49]. Gastric cancer has been selected for the second phase. A similar effort has been undertaken by the International Cancer Genome Consortium (ICGC), with the aim of systematically studying 25,000 cancer genomes at the genomic, epigenomic, and transcriptomic levels for at least 50 cancer types. Compared with TCGA, one unique feature of the ICGC is the involvement of patients from different countries. As one of the selected cancer types, the samples of gastric cancer come from patients in both China and the United States. Over the next few years, we expect these consortium-based cancer genomic projects to be valuable resources for discovering novel therapeutic targets for gastric cancer.
Four important research areas are of particular interest for the application of NGS to gastric cancer samples. First, the incidence of gastric cancer varies greatly by ethnic group; thus, it is of interest to determine the extent to which the molecular basis of gastric cancer depends on the ethnic background of the individual (e.g., East Asian vs. Caucasian). NGS datasets from the ICGC would be very useful to address this topic. Second, in addition to intertumoral heterogeneity (between patients with gastric cancer), intratumoral heterogeneity (within a single gastric tumor) presents an additional challenge for effective treatment, and may contribute to drug resistance. NGS can address this topic using two powerful approaches: (i) ultra-deep sequencing of the primary tumor to detect rare subclones; and (ii) low-depth sequential characterization of the tumor to identify dominate clones [50]. Third, as RNA-seq allows for the detection of splicing variants in an unprecedented manner, it is of interest to investigate the role of aberrant splicing in gastric tumorigenesis. Some unexpected targets may come from splicing variants. Fourth, using NGS to systematically identify gene fusion in gastric cancer is another approach with tremendous potential for discovering novel therapeutic targets.
Acknowledgments
This work was supported in part by grants from the U.S. National Institutes of Health: a Career Development Award through the Uterine SPORE grant (NIH/NCI P50 CA098258), TCGA GDAC funding (NIH/NCI U24 CA143883), and the Cancer Center Support Grant to MD Anderson Cancer Center (NIH/NCI P30 CA016672). This work was also supported by UTMDACC G.S. Hogan Gastrointestinal Research Fund and the Lorraine Dell Program in Bioinformatics for Personalization of Cancer Medicine, through grants to H. Liang. The funders had no role in data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay Jea. CancerBase. International Agency for Research on Cancer Lyon; 2010. GLOBOCAN 2008, cancer incidence and mortality worldwide. [Google Scholar]
- 3.Volk J, Parsonnet J. In: Epidemiology of Gastic Cancer and Helicobater pylori. Wang T, Fox J, Giraud A, editors. The Biology of Gastric Cancers, Springer; 2009. [Google Scholar]
- 4.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. Ca-Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 5.McDonald M, Hertz RP, Pitman Lowenthal SW Pfizer Facts. The Burden of Cancer in Asia. Pfizer; USA: p. 2008. [Google Scholar]
- 6.Yamashita K, Sakuramoto S, Nemoto M, Shibata T, Mieno H, Katada N, Kikuchi S, Watanabe M. Trend in gastric cancer: 35 years of surgical experience in Japan. World journal of gastroenterology : WJG. 2011;17:3390–3397. doi: 10.3748/wjg.v17.i29.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng L, Wang L, Ajani J, Xie K. Molecular basis of gastric cancer development and progression. Gastric cancer : official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2004;7:61–77. doi: 10.1007/s10120-004-0277-4. [DOI] [PubMed] [Google Scholar]
- 8.Resende C, Thiel A, Machado JC, Ristimaki A. Gastric cancer: basic aspects. Helicobacter. 2011;16(Suppl 1):38–44. doi: 10.1111/j.1523-5378.2011.00879.x. [DOI] [PubMed] [Google Scholar]
- 9.Yasui W, Yokozaki H, Fujimoto J, Naka K, Kuniyasu H, Tahara E. Genetic and epigenetic alterations in multistep carcinogenesis of the stomach. Journal of gastroenterology. 2000;35(Suppl 12):111–115. [PubMed] [Google Scholar]
- 10.Compare D, Rocco A, Nardone G. Risk factors in gastric cancer. Eur Rev Med Pharmaco. 2010;14:302–308. [PubMed] [Google Scholar]
- 11.Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V, Wo WIARCM. A review of human carcinogens-Part B: biological agents. Lancet Oncol. 2009;10:321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 12.Machado AM, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter pylori infection generates genetic instability in gastric cells. Biochimica et biophysica acta. 2010;1806:58–65. doi: 10.1016/j.bbcan.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Ruschoff J, Kang YK, Investigators TT. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 14.Okines AFC, Cunningham D. Trastuzumab in gastric cancer. Eur J Cancer. 2010;46:1949–1959. doi: 10.1016/j.ejca.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Grabsch H, Sivakumar S, Gray S, Gabbert HE, Muller W. HER2 expression in gastric cancer: Rare, heterogeneous and of no prognostic value - conclusions from 924 cases of two independent series. Cellular oncology : the official journal of the International Society for Cellular Oncology. 2010;32:57–65. doi: 10.3233/CLO-2009-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wainberg ZA, Anghel A, Desai AJ, Ayala R, Luo T, Safran B, Fejzo MS, Hecht JR, Slamon DJ, Finn RS. Lapatinib, a dual EGFR and HER2 kinase inhibitor, selectively inhibits HER2-amplified human gastric cancer cells and is synergistic with trastuzumab in vitro and in vivo. Clin Cancer Res. 2010;16:1509–1519. doi: 10.1158/1078-0432.CCR-09-1112. [DOI] [PubMed] [Google Scholar]
- 17.Arkenau HT. Gastric cancer in the era of molecularly targeted agents: current drug development strategies. Journal of cancer research and clinical oncology. 2009;135:855–866. doi: 10.1007/s00432-009-0583-7. [DOI] [PubMed] [Google Scholar]
- 18.Ku GY, Ilson DH. Esophagogastric cancer: targeted agents. Cancer treatment reviews. 2010;36:235–248. doi: 10.1016/j.ctrv.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Shah MA, Ramanathan RK, Ilson DH, Levnor A, D’Adamo D, O’Reilly E, Tse A, Trocola R, Schwartz L, Capanu M, Schwartz GK, Kelsen DP. Multicenter phase II study of irinotecan, cisplatin, and bevacizumab in patients with metastatic gastric or gastroesophageal junction adenocarcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:5201–5206. doi: 10.1200/JCO.2006.08.0887. [DOI] [PubMed] [Google Scholar]
- 20.Pinto C, Di Fabio F, Siena S, Cascinu S, Rojas Llimpe FL, Ceccarelli C, Mutri V, Giannetta L, Giaquinta S, Funaioli C, Berardi R, Longobardi C, Piana E, Martoni AA. Phase II study of cetuximab in combination with FOLFIRI in patients with untreated advanced gastric or gastroesophageal junction adenocarcinoma (FOLCETUX study) Ann Oncol. 2007;18:510–517. doi: 10.1093/annonc/mdl459. [DOI] [PubMed] [Google Scholar]
- 21.Guan YF, Li GR, Wang RJ, Yi YT, Yang L, Jiang D, Zhang XP, Peng Y. Application of next-generation sequencing in clinical oncology to advance personalized treatment of cancer. Chinese journal of cancer. 2012;31:463–470. doi: 10.5732/cjc.012.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 23.Shendure J, Ji H. Next-generation DNA sequencing. Nature biotechnology. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 24.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nature reviews Genetics. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss JA. How to get genomes at one ten-thousandth the cost. Nature biotechnology. 2008;26:1113–1115. doi: 10.1038/nbt1008-1113. [DOI] [PubMed] [Google Scholar]
- 27.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, Anderson KC, Ardlie KG, Auclair D, Baker A, Bergsagel PL, Bernstein BE, Drier Y, Fonseca R, Gabriel SB, Hofmeister CC, Jagannath S, Jakubowiak AJ, Krishnan A, Levy J, Liefeld T, Lonial S, Mahan S, Mfuko B, Monti S, Perkins LM, Onofrio R, Pugh TJ, Rajkumar SV, Ramos AH, Siegel DS, Sivachenko A, Stewart AK, Trudel S, Vij R, Voet D, Winckler W, Zimmerman T, Carpten J, Trent J, Hahn WC, Garraway LA, Meyerson M, Lander ES, Getz G, Golub TR. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. Human mutation rate associated with DNA replication timing. Nature genetics. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn WC, Weinberg RA. Rules for making human tumor cells. The New England journal of medicine. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- 30.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY, Lee SP, Ho SL, Chan AK, Cheng GH, Roberts PC, Rejto PA, Gibson NW, Pocalyko DJ, Mao M, Xu J, Leung SY. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nature genetics. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 31.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nature genetics. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 32.Zang ZJ, Ong CK, Cutcutache I, Yu W, Zhang SL, Huang D, Ler LD, Dykema K, Gan A, Tao J, Lim S, Liu Y, Futreal PA, Grabsch H, Furge KA, Goh LK, Rozen S, Teh BT, Tan P. Genetic and structural variation in the gastric cancer kinome revealed through targeted deep sequencing. Cancer research. 2011;71:29–39. doi: 10.1158/0008-5472.CAN-10-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holbrook JD, Parker JS, Gallagher KT, Halsey WS, Hughes AM, Weigman VJ, Lebowitz PF, Kumar R. Deep sequencing of gastric carcinoma reveals somatic mutations relevant to personalized medicine. Journal of translational medicine. 2011;9:119. doi: 10.1186/1479-5876-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CF, Griffiths S, Rodriguez-Suarez E, Pierce A, Unwin RD, Jaworska E, Evans CA, S JG, Whetton AD. Assessment of downstream effectors of BCR/ABL protein tyrosine kinase using combined proteomic approaches. Proteomics. 2010;10:3321–3342. doi: 10.1002/pmic.201000176. [DOI] [PubMed] [Google Scholar]
- 35.Amin SH, Tibes R, Kim JE, Hybarger CP. Hedgehog antagonist GDC-0449 is effective in the treatment of advanced basal cell carcinoma. The Laryngoscope. 2010;120:2456–2459. doi: 10.1002/lary.21145. [DOI] [PubMed] [Google Scholar]
- 36.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui JA, Chen YB, Chou WC, Sun LK, Chen L, Suo JA, Ni ZH, Zhang M, Kong XX, Hoffman LL, Kang JS, Su YY, Olman V, Johnson D, Tench DW, Amster IJ, Orlando R, Puett D, Li F, Xu Y. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39:1197–1207. doi: 10.1093/nar/gkq960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nature biotechnology. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YH, Liang H, Liu X, Lee JS, Cho JY, Cheong JH, Kim H, Li M, Downey TJ, Dyer MD, Sun Y, Sun J, Beasley EM, Chung HC, Noh SH, Weinstein JN, Liu CG, Powis G. AMPKalpha modulation in cancer progression: multilayer integrative analysis of the whole transcriptome in Asian gastric cancer. Cancer research. 2012;72:2512–2521. doi: 10.1158/0008-5472.CAN-11-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phoenix KN, Devarakonda CV, Fox MM, Stevens LE, Claffey KP. AMPKalpha2 Suppresses Murine Embryonic Fibroblast Transformation and Tumorigenesis. Genes & cancer. 2012;3:51–62. doi: 10.1177/1947601912452883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeiro-dos-Santos A, Khayat AS, Silva A, Alencar DO, Lobato J, Luz L, Pinheiro DG, Varuzza L, Assumpcao M, Assumpcao P, Santos S, Zanette DL, Silva WA, Jr, Burbano R, Darnet S. Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach. PloS one. 2010;5:e13205. doi: 10.1371/journal.pone.0013205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li SC, Liao YL, Ho MR, Tsai KW, Lai CH, Lin WC. miRNA arm selection and isomiR distribution in gastric cancer. BMC genomics. 2012;13(Suppl 1):S13. doi: 10.1186/1471-2164-13-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nature Reviews Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 45.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara SI, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–U563. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 46.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, Han B, Cao Q, Cao X, Suleman K, Kumar-Sinha C, Dhanasekaran SM, Chen YB, Esgueva R, Banerjee S, LaFargue CJ, Siddiqui J, Demichelis F, Moeller P, Bismar TA, Kuefer R, Fullen DR, Johnson TM, Greenson JK, Giordano TJ, Tan P, Tomlins SA, Varambally S, Rubin MA, Maher CA, Chinnaiyan AM. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nature medicine. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, Dogruluk T, Lu Y, Liu X, Gu C, Guo W, Scherer SE, Carter H, Westin SN, Dyer MD, Verhaak RG, Zhang F, Karchin R, Liu CG, Lu KH, Broaddus RR, Scott KL, Hennessy BT, Mills GB. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012 doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Network CGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meric-Bernstam F, Mills GB. Overcoming implementation challenges of personalized cancer therapy. Nature reviews Clinical oncology. 2012;9:542–548. doi: 10.1038/nrclinonc.2012.127. [DOI] [PubMed] [Google Scholar]


