Abstract
The saliva of blood sucking insects contains potent pharmacologically active components that assist them in counteracting the host hemostatic and inflammatory systems during blood feeding. In addition, sand fly salivary proteins affect host immunity and have the potential to be a vaccine against Leishmania infection. In the present study, the salivary gland transcripts of Lutzomyia ayacuchensis, a vector of cutaneous leishmaniasis in Ecuadorian and Peruvian Andes, were analyzed by sequencing randomly selected clones of the salivary gland cDNA library of this sand fly. This resulted in the identification of the most abundant transcripts coding for secreted proteins. These proteins were homologous to the salivary molecules present in other sand flies including the RGD-containing peptide, PpSP15/SL1 family protein, yellow-related protein, putative apyrase, antigen 5-related protein, D7 family protein, and 27 kDa salivary protein. Of note, homologues of maxadilan, an active vasodilator abundantly present in saliva of Lutzomyia longipalpis, were not identified. This analysis is the first description of salivary proteins from a sand fly of the subgenus Helcocyrtomyia and from vector of cutaneous leishmaniasis in the New World. The present analysis will provide further insights into the evolution of salivary components in blood sucking arthropods.
Keywords: Lutzomyia ayacuchensis, Salivary gland, Transcript, Bioinformatics, cDNA library
1. Introduction
Phlebotomine sand flies are hematophagous insects of the family Psychodidae in the order Diptera, and approximately 80 species can transmit Leishmania protozoa, the causative agents of leishmaniasis (Munstermann, 2005; Kato et al., 2010a). During blood-feeding, sand flies inject saliva containing various physiologically active substances including anticoagulants, vasodilators, and inhibitors of platelet aggregation to facilitate successful acquisition of a blood meal (Ribeiro, 1995; Ribeiro and Francischetti, 2003; Valenzuela, 2005). The infection of mammalian hosts with Leishmania protozoa occurs during the blood feeding of an infected female sand fly where the parasites are co-inoculated into the hosts together with sand fly saliva (Rohoušová and Volf, 2006; Oliveira et al., 2009). Sand fly saliva was shown to facilitate the transmission of Leishmania to mammalian hosts and to exacerbate the disease (Titus and Ribeiro, 1988; Theodos et al., 1991; Lima and Titus, 1996). Alternatively, cellular immune response to the salivary components protects the host from Leishmania infection (Belkaid et al., 1998, 2000; Kamhawi et al., 2000; Valenzuela et al., 2001; Gomes et al., 2008; Oliveira et al., 2008; Tavares et al., 2011). Humoral immunity to sand fly salivary components was shown in the case of Leishmania (Viannia) braziliensis to exacerbate infection (Oliveira et al., 2008). In addition, recent studies are showing that sand fly salivary proteins can be used as a specific marker of sand fly exposure and to use them to evaluate potential risks of Leishmania transmission (Teixeira et al., 2010; Vlkova et al., 2011). The profile of salivary components has been defined in 8 Old World Phlebotomus species; Phlebotomus (P.) papatasi [a vector of Leishmania (Leishmania) major] (Valenzuela et al., 2001), P. ariasi [a vector of L. (L.) infantum] (Oliveira et al., 2006), P. perniciosus [a vector of L. (L.) infantum] (Anderson et al., 2006), P. argentipes [a vector of L. (L.) donovani] (Anderson et al., 2006), P. duboscqi [a vector of L. (L.) major] (Kato et al., 2006), P. arabicus [a vector of L. (L.) tropica] (Hostomská et al., 2009), P. tobbi [a vector of L. (L.) infantum] (Rohoušová et al., 2012), and P. sergenti [a vector of L. (L.) tropica] (Rohoušová et al., 2012). On the other hand, the salivary components of the New World Lutzomyia species have been analyzed only in Lutzomyia (Lu.) longipalpis, a vector of L. (L.) infantum [a synonym of L. (L.) chagasi] causing visceral leishmaniasis (Charlab et al., 1999; Valenzuela et al., 2004). To date, no information is available on the repertoire of salivary transcripts from vectors of the New World cutaneous leishmaniasis. In the present study, to obtain further insight into the salivary components of Lutzomyia species, the salivary gland transcriptome analysis was performed on Lu. ayacuchensis, which is a proven vector of L. (L.) mexicana (Takaoka et al., 1990; Kato et al., 2005, 2008) and L. (V.) peruviana (Caceres et al., 2004; Kato et al., 2008), causative agents of cutaneous leishmaniasis in Andean areas of Ecuador and Peru, respectively. This is the first report of salivary proteins from a sand fly of the subgenus Helcocyrtomyia and from vector of cutaneous leishmaniasis in the New World.
2. Materials and methods
2.1. Lutzomyia ayacuchensis salivary glands
Sand flies were captured using protected human bait at Huigra (2°20′S, 78°58′W; 1200 m above sea level) in the Department of Chimborazo, Ecuador, where Andean-type cutaneous leishmaniasis caused by L. (L.) mexicana is endemic. The sand flies were dissected and identified at the species level based mainly on the morphology of their spermathecae (Young and Duncan, 1994). Salivary glands of Lu. ayacuchensis were dissected and transferred to tubes containing RNAlater (Ambion, Austin, TX) and kept at −20 °C.
2.2. Construction of salivary gland cDNA library
Lu. ayacychensis salivary gland mRNA was isolated from 38 pairs of salivary glands using the Micro FastTrack mRNA isolation kit (Invitrogen, San Diego, CA). The PCR-based cDNA library was prepared following the instructions for the SMART cDNA library construction kit (BD-Clontech, Palo Alto, CA) with some modifications (Valenzuela et al., 2004). The quality of the cDNA was checked by agarose gel electrophoresis and the absence of smaller fragments derived from degraded mRNA was confirmed. The obtained cDNA library was fractionated using a Chromaspin 1000 column (BD-Clontech) into small (approximately 400–800 bp), medium (approximately 800–1200 bp) and large (>1200 bp) transcripts based on their electrophoresis profiles on a 1.1% agarose gel. Pooled fractions were ligated into Lambda TriplEx2 vector (BD-Clontech) and packaged into lambda phage (Stratagene, La Jolla, CA).
2.3. Sequence analysis of cDNA library
Single isolated plaques were picked from the plate using sterile wooden sticks and placed into 50 µl of water. Amplification of cDNA was performed in a volume of 15 µl using a pair of primers, PT2F1 (5′-AAG TAC TCT AGC AAT TGT GAG C-3′) and PT2R1 (5′-CTC TTC GCT ATT ACG CCA GCT G-3′), Premix Taq (Takara Bio, Shiga, Japan) and 3 µl of template DNA. After an initial denaturation at 75 °C for 3 min and following 95 °C for 4 min, PCR amplification was performed with 35 cycles of denaturation (95 °C, 1 min), annealing (50 °C, 1 min) and polymerization (72 °C, 2 min). PCR products were cleaned using Multiscreen PCR cleaning plates (Millipore Corporation, Bedford, MA) and used as templates for cycle-sequencing kit (Applied Biosystems, Foster City, CA) with PT2F3 primer (5′-TCT CGG GAA GCG CGC CAT TGT-3′). Cycle-sequencing products were cleaned using sephadex and MultiScreen HV plates (Millipore Corporation), dried and stored at −20 °C. Sequencing was performed on an ABI 3730xl DNA sequencer (Applied Biosystems).
2.4. Bioinformatics
Expressed sequence tags (ESTs) were trimmed of primer and vector sequences and clustered. The ESTs were grouped based on nucleotide homology of 95% identity over 100 residues using the BLASTn algorithm (Altschul et al., 1997). The assembly of the ESTs into transcript contigs was done using the CAP3 algorithm, generating a consensus sequence (Huang, 1992). Contigs and singletons (contig containing only one sequence) were compared using BLASTx or BLASTn (Altschul et al., 1997) of the non-redundant (NR) protein database of the National Center of Biological Information (NCBI), the gene ontology database (GO) (Ashburner et al., 2000), and the Conserved Domains Database (CDD) that includes all Pfam (Bateman and Birney, 2000), SMART (Schultz et al., 1998) and COG protein domains in the NCBI (Marchler-Bauer et al., 2002). Additionally, contigs were compared using BLASTn (Altschul et al., 1997) to custom databases of mitochondrial (mit-pla) and rRNA (rrna) nucleotide sequences. Identification of putative secreted proteins was conducted using SignalP server (Bendtsen et al., 2004).
2.5. T cell epitope prediction
For the T cell epitope prediction, ProPred MHC Class-II Binding Peptide Prediction Server (Singh and Raghava, 2001) was utilized. HLA class II-binding peptides were searched on the 51 different HLA-DR alleles, and the promiscuous epitopes were selected from the Lu. ayacuchensis salivary protein sequences tested that were predicted to bind at least 20 alleles.
2.6. Phylogenetic analysis
The sequences that had homologies with secreted proteins by BLASTx analyses were aligned with CLUSTAL W software (Thompson et al., 1994) and examined using the program MEGA (Molecular Evolutionary Genetics Analysis) version 4.0 (Tamura et al., 2007). Phylogenetic trees by the neighbor-joining method were constructed with the distance algorithms available in the MEGA package. Bootstrap values were determined on 1000 replicates of the data sets.
3. Results and discussion
3.1. Sequencing of Lutzomyia ayacuchensis salivary gland cDNA library
Lu. ayacuchensis salivary gland cDNA library was constructed, and sequencing was performed on randomly selected 1152 clones. As a result, 768 high-quality sequences were obtained. Three categories of expressed genes were derived from the manual annotation of the contigs: secreted, housekeeping and unknown. The putative secreted category comprised 45.7% of the clusters and 74.6% of the total sequences. The high ratio of transcripts encoding secreted proteins were also reported in other phlebotomine sand flies (Valenzuela et al., 2001; Oliveira et al., 2006; Anderson et al., 2006; Kato et al., 2006; Hostomská et al., 2009; Rohoušová et al., 2012). The housekeeping category had 35.0% of the clusters and 16.3% of the total sequences. Finally, the category of ‘‘unknowns”comprised 19.3% of the cluster and 9.1% of the sequences.
3.2. Housekeeping genes
The clusters of sequences attributed to housekeeping genes (94 clusters with 125 sequences in total) were further divided into 18 subgroups, according to their possible function (Table 1). The two largest subgroups were associated with “Energy production and conversion” (24 sequences in 20 clusters) and “Translation, ribosomal structure and biogenesis” (23 sequences in 19 clusters). Six sequences in 6 clusters, which represent conserved proteins with unknown function, were classified as “unknown conserved”. Other sequences were identified with homology to housekeeping genes and were associated with transport, metabolism, signal transduction, and cell structure, among other potential activities.
Table 1.
Functional classification of housekeeping genes expressed in Lutzomyia ayacuchensis salivary glands
| Type of transcripts | Cluster | % | Sequence | % |
|---|---|---|---|---|
| Energy production and conversion | 20 | 21.3 | 24 | 19.2 |
| Translation, ribosomal structure and biogenesis | 19 | 20.2 | 23 | 18.4 |
| General function prediction only | 9 | 9.6 | 10 | 8.0 |
| Signal transduction mechanisms | 8 | 8.5 | 8 | 6.4 |
| Posttranslational modification, protein turnover, chaperones | 7 | 7.4 | 14 | 11.2 |
| Lipid transport and metabolism | 5 | 5.3 | 5 | 4.0 |
| Amino acid transport and metabolism | 4 | 4.3 | 4 | 3.2 |
| Chromatin structure and dynamics | 2 | 2.1 | 2 | 1.6 |
| Inorganic ion transport and metabolism | 2 | 2.1 | 2 | 1.6 |
| Intracellular trafficking, secretion, and vesicular transport | 2 | 2.1 | 3 | 2.4 |
| RNA processing and modification | 2 | 2.1 | 2 | 1.6 |
| Secondary metabolites biosynthesis, transport and catabolism | 2 | 2.1 | 2 | 1.6 |
| Transcription | 2 | 2.1 | 14 | 11.2 |
| Carbohydrate transport and metabolism | 1 | 1.1 | 3 | 2.4 |
| Cell cycle control, cell division, chromosome partitioning | 1 | 1.1 | 1 | 0.8 |
| Cell wall/membrane/envelope biogenesis | 1 | 1.1 | 1 | 0.8 |
| Defense mechanisms | 1 | 1.1 | 1 | 0.8 |
| Unknown conserved | 6 | 6.4 | 6 | 4.8 |
| Total | 94 | 100.0 | 125 | 100.0 |
3.3. Putative secreted proteins
The transcripts coding for secreted proteins were further analyzed using the BLASTx program for comparison to the NCBI non-redundant protein database. Table 2 lists clusters for the most abundant transcripts coding for the putative secreted salivary gland proteins from Lu. ayacuchensis. The table was arranged from the most abundant to the least abundant transcripts found in the cDNA library. The nomenclature for the transcripts on the cDNA library is the following: Lay = Lu. ayacuchensis, S = salivary glands, and the number (i.e. 45) denotes the contig number on the cDNA library where a contig is a cluster of identical transcripts. Many of the isolated transcripts code for proteins previously identified from the saliva of sand flies, Lu. longipalpis and P. perniciosus, including RGD-containing peptide, 14 kDa salivary protein, antigen 5-related protein, yellow-related protein and SL1-like protein (Table 2). Table 3 shows the classification of transcripts coding for putative secreted proteins in Lu. ayacuchensis salivary glands. Of the 573 transcripts associated with putative secreted proteins, the most abundant transcript (18.7%) had homology with that of RGD-containing peptide from salivary glands of Lu. longipalpis. The following abundant transcripts were SL1-like protein (14.3%) and 14 kDa salivary protein (12.4%), both of which belong to a family of PpSP15/SL1 originally identified as 15 kDa protein from P. papatasi saliva. Additional transcripts coding for secreted proteins include homologs to yellow-related protein (11.1%), putative apyrase (7.9%), D7-related salivary protein (7.3%), 27 kDa salivary protein (6.6%), antigen 5-related protein (6.5%), 16.4 kDa salivary protein (5.6%), and others (Table 3).
Table 2.
Most abundant salivary gland transcripts from Lutzomyia ayacuchensis
| Cluster | Sequence name | No. of seq in cluster |
Mature Mw (kDa) |
pI | Best match to NR protein database |
Accession No. | |||
|---|---|---|---|---|---|---|---|---|---|
| Accession No. | Organism | E value | % Identity | ||||||
| LayS45 | RGD-containing peptide | 90 | 5.3 | 3.44 | AAD32196 | Lu. longipalpis | 3E-07 | 47 | AK416785 |
| LayS37 | 14 kDa Salivary protein | 58 | 14.1 | 8.21 | ABA43057 | P. perniciosus | 6E-39 | 50 | AK416778 |
| LayS81 | Antigen 5-related protein | 24 | 28.5 | 9.08 | AAD32191 | Lu. longipalpis | 1E-135 | 80 | AK416820 |
| LayS24 | Yellow-related protein | 23 | 43.5 | 9.37 | AAS05318 | Lu. longipalpis | 0 | 78 | AK416770 |
| LayS59 | SL1-like protein | 19 | 13.8 | 8.22 | AAD32197 | Lu. longipalpis | 1E-51 | 64 | AK416798 |
| LayS91 | 27 kDa Salivary protein | 17 | 27.2 | 10.14 | AAS16906 | Lu. longipalpis | 2E-92 | 59 | AK416828 |
| LayS117* | Yellow-related protein | 16 | AAD32198 | Lu. longipalpis | 4E-77 | 66 | AB744655 | ||
| LayS118 | Yellow-related protein | 14 | 43.8 | 8.05 | AAD32198 | Lu. longipalpis | 1E-164 | 67 | AK416844 |
| LayS127 | 16.4 kDa Salivary protein | 11 | 16.4 | 8.05 | ABB00903 | Lu. longipalpis | 1E-29 | 39 | AK416852 |
| LayS21 | Putative apyrase | 11 | 35.7 | 9.19 | AAD33513 | Lu. longipalpis | 1E-123 | 65 | AK416767 |
| LayS142 | 9.3 kDa Protein | 10 | 9.3 | 4.22 | XP 001238639 | Eimeria tenella | 0.012 | 44 | AK416859 |
| LayS103 | D7-related salivary protein | 9 | 26.6 | 8.81 | ABA43052 | P. perniciosus | 7E-86 | 59 | AK416840 |
| LayS36 | 14 kDa Salivary protein | 9 | 14.1 | 8.21 | ABA43057 | P. perniciosus | 2E-39 | 50 | AK416777 |
| LayS6 | 11.5 kDa Salivary protein | 8 | 11.5 | 6.67 | AAS16919 | Lu. longipalpis | 2E-07 | 33 | AK416753 |
| LayS102 | D7-related salivary protein | 8 | 26.6 | 8.81 | ABA43052 | P. perniciosus | 7E-86 | 59 | AK416839 |
| LayS19 | Putative apyrase | 8 | 35.8 | 9.09 | AAD33513 | Lu. longipalpis | 1E-127 | 65 | AK416765 |
| LayS58 | SL1-like protein | 8 | 13.8 | 8.22 | AAD32197 | Lu. longipalpis | 1E-51 | 64 | AK416797 |
| LayS68 | SL1-like protein | 8 | 13.9 | 8.61 | AAD32197 | Lu. longipalpis | 7E-52 | 64 | AK416807 |
| LayS90 | 27 kDa Salivary protein | 8 | 26.6 | 10.08 | AAS16906 | Lu. longipalpis | 1E-90 | 58 | AK416827 |
| LayS101 | D7-related salivary protein | 7 | 26.6 | 8.81 | ABA43052 | P. perniciosus | 2E-86 | 59 | AK416838 |
| LayS66 | SL1-like protein | 6 | 13.9 | 8.61 | AAD32197 | Lu. longipalpis | 1E-51 | 64 | AK416805 |
| LayS20 | Putative apyrase | 6 | 35.5 | 9.00 | AAD33513 | Lu. longipalpis | 1E-128 | 65 | AK416766 |
| LayS22 | Yellow-related protein | 6 | 43.4 | 9.39 | AAS05318 | Lu. longipalpis | 1E-125 | 75 | AK416768 |
| LayS17 | Putative apyrase | 5 | 35.5 | 9.11 | AAD33513 | Lu. longipalpis | 1E-128 | 65 | AK416763 |
| LayS18 | Putative apyrase | 5 | 36.0 | 9.46 | AAD33513 | Lu. longipalpis | 1E-128 | 65 | AK416764 |
| LayS23 | Yellow-related protein | 5 | 43.5 | 9.37 | AAS05318 | Lu. longipalpis | 0 | 78 | AK416769 |
| LayS26 | 33.6 kDa Salivary protein | 5 | 33.7 | 8.95 | AAX55751 | P. ariasi | 6E-91 | 53 | AK416772 |
| LayS44 | RGD-containing peptide | 5 | 5.3 | 3.47 | AAD32196 | Lu. longipalpis | 8E-07 | 46 | AK416784 |
| LayS71 | SL1-like protein | 5 | 13.8 | 8.22 | AAD32197 | Lu. longipalpis | 2E-50 | 64 | AK416810 |
| LayS72 | SL1-like protein | 5 | 13.8 | 8.43 | AAD32197 | Lu. longipalpis | 3E-51 | 64 | AK416811 |
| LayS100 | D7-related salivary protein | 4 | 26.6 | 8.61 | ABA43052 | P. perniciosus | 4E-86 | 59 | AK416837 |
| LayS131 | 16.4 kDa Salivary protein | 4 | 16.4 | 8.32 | ABB00903 | Lu. longipalpis | 7E-26 | 39 | AK416856 |
| LayS3 | Allergen-related protein | 4 | 26.0 | 5.68 | XP 001865175 | Culex quinquefasciatus | 8E-31 | 35 | AK416750 |
| LayS64 | SL1-like protein | 4 | 13.9 | 8.61 | AAD32197 | Lu. longipalpis | 5E-48 | 59 | AK416803 |
| LayS97 | D7-related salivary protein | 4 | 26.6 | 8.81 | ABA43052 | P. perniciosus | 2E-86 | 59 | AK416834 |
| LayS128 | 16.4 kDa Salivary protein | 3 | 16.4 | 8.05 | ABB00903 | Lu. longipalpis | 1E-29 | 39 | AK416853 |
| LayS129 | 16.4 kDa Salivary protein | 3 | 16.4 | 7.69 | ABB00903 | Lu. longipalpis | 2E-29 | 40 | AK416854 |
| LayS16 | Putative apyrase | 3 | 36.6 | 9.34 | AAD33513 | Lu. longipalpis | 1E-125 | 64 | AK416762 |
| LayS27 | 33.6 kDa Salivary protein | 3 | 33.7 | 8.69 | AAX55751 | P. ariasi | 1E-90 | 52 | AK416773 |
| LayS46 | RGD-containing peptide | 3 | 5.3 | 3.44 | AAD32196 | Lu. longipalpis | 1E-07 | 49 | AK416786 |
| LayS47 | RGD-containing peptide | 3 | 5.3 | 3.44 | AAD32196 | Lu. longipalpis | 2E-07 | 47 | AK416787 |
| LayS70 | SL1-like protein | 3 | 13.9 | 7.98 | AAD32197 | Lu. longipalpis | 5E-51 | 64 | AK416809 |
| LayS77 | Antigen 5-related protein | 3 | 28.6 | 9.21 | AAD32191 | Lu. longipalpis | 1E-135 | 80 | AK416816 |
| LayS89 | 27 kDa Salivary protein | 3 | 26.6 | 10.08 | AAS16906 | Lu. longipalpis | 5E-90 | 58 | AK416826 |
| LayS98 | D7-related salivary protein | 3 | 26.6 | 8.81 | ABA43052 | P. perniciosus | 3E-86 | 59 | AK416835 |
| LayS130 | 16.4 kDa Salivary protein | 2 | 16.4 | 8.05 | ABB00903 | Lu. longipalpis | 2E-29 | 40 | AK416855 |
| LayS132 | 16.4 kDa Salivary protein | 2 | 16.4 | 8.05 | ABB00903 | Lu. longipalpis | 2E-29 | 40 | AK416857 |
| LayS2 | Allergen-related protein | 2 | 26.0 | 5.46 | XP 001865175 | Culex quinquefasciatus | 5E-30 | 34 | AK416749 |
| LayS60 | SL1-like protein | 2 | 13.9 | 7.98 | AAD32197 | Lu. longipalpis | 7E-51 | 64 | AK416799 |
| LayS61 | SL1-like protein | 2 | 13.8 | 8.22 | AAD32197 | Lu. longipalpis | 1E-51 | 65 | AK416800 |
| LayS62 | SL1-like protein | 2 | 13.8 | 8.52 | AAD32197 | Lu. longipalpis | 6E-51 | 64 | AK416801 |
| LayS63 | SL1-like protein | 2 | 13.8 | 8.23 | AAD32197 | Lu. longipalpis | 1E-51 | 64 | AK416802 |
| LayS65 | SL1-like protein | 2 | 13.8 | 7.98 | AAD32197 | Lu. longipalpis | 1E-51 | 64 | AK416804 |
| LayS67 | SL1-like protein | 2 | 13.8 | 8.35 | AAD32197 | Lu. longipalpis | 1E-50 | 64 | AK416806 |
| LayS69 | SL1-like protein | 2 | 13.9 | 8.22 | AAD32197 | Lu. longipalpis | 4E-51 | 64 | AK416808 |
| LayS7 | 11.5 kDa Salivary protein | 2 | 10.8 | 8.36 | AAS16919 | Lu. longipalpis | 2E-07 | 34 | AK416754 |
| LayS78 | Antigen 5-related protein | 2 | 28.4 | 9.16 | AAD32191 | Lu. longipalpis | 1E-133 | 80 | AK416817 |
| LayS79 | Antigen 5-related protein | 2 | 28.4 | 9.08 | AAD32191 | Lu. longipalpis | 1E-135 | 80 | AK416818 |
| LayS80 | Antigen 5-related protein | 2 | 28.5 | 9.01 | AAD32191 | Lu. longipalpis | 1E-135 | 80 | AK416819 |
| LayS92 | 27 kDa Salivary protein | 2 | 27.3 | 10.14 | AAS16906 | Lu. longipalpis | 9E-91 | 58 | AK416829 |
| LayS93 | 27 kDa Salivary protein | 2 | 27.2 | 10.14 | AAS16906 | Lu. longipalpis | 1E-91 | 58 | AK416830 |
| LayS95 | D7-related salivary protein | 2 | 26.6 | 8.81 | ABA43052 | P. perniciosus | 5E-86 | 59 | AK416832 |
| LayS96 | D7-related salivary protein | 2 | 26.6 | 8.81 | ABA43052 | P. perniciosus | 1E-85 | 58 | AK416833 |
| LayS99 | D7-related salivary protein | 2 | 26.6 | 8.81 | ABA43052 | P. perniciosus | 6E-86 | 59 | AK416836 |
Truncated in the 5′ region.
Table 3.
Classification of transcripts coding for putative secreted proteins in Lutzomyia ayacuchensis salivary glands
| Cluster | Sequence | % Sequence | |
|---|---|---|---|
| RGD-containing peptide | 10 | 107 | 18.7 |
| SL1-like protein | 25 | 82 | 14.3 |
| 14 kDa Salivary protein | 6 | 71 | 12.4 |
| Yellow-related protein | 5 | 64 | 11.1 |
| Putative apyrase | 13 | 45 | 7.9 |
| D7-related salivary protein | 9 | 42 | 7.3 |
| 27 kDa Salivary protein | 11 | 38 | 6.6 |
| Antigen 5-related protein | 9 | 37 | 6.5 |
| 16.4 kDa Salivary protein | 13 | 32 | 5.6 |
| 9.3 kDa Protein | 3 | 16 | 2.8 |
| 11.5 kDa Salivary protein | 4 | 12 | 2.1 |
| 33.6 kDa Salivary protein | 3 | 9 | 1.6 |
| Allergen-related protein | 2 | 6 | 1.0 |
| Others | 9 | 12 | 2.1 |
| Total | 122 | 573 | 100.0 |
3.3.1. RGD-containing peptide
The most abundant sequences found in the Lu. ayacuchensis salivary gland transcriptome contain C-terminal RGD (Arg-Gly-Asp) sequence; a motif that has been shown to bind integrins such as αvβ3 and αvβ5 (Hynes, 1992). Ten transcripts of RGD-containing peptide (LayS38–LayS47) were identified in the cDNA library of Lu. ayacuchensis salivary glands. These Lu. ayacuchensis RGD-containing molecules have a predicted molecular mass of 5.3 kDa in the mature form and a calculated isoelectric point of 3.44 (Table 2). The only homologous protein has been identified in the salivary gland transcriptome of Lu. longipalpis (LuloRGD), and speculated to act as an inhibitor of platelet aggregation via its RGD motif; although, the function has not yet been characterized (Charlab et al., 1999). Alignment of the RGD-containing peptide showed a high level of homology among contigs, and an RGD-motif in the C-terminal was strictly conserved in all molecules (Fig. S1).
3.3.2. PpSP15/SL1 family
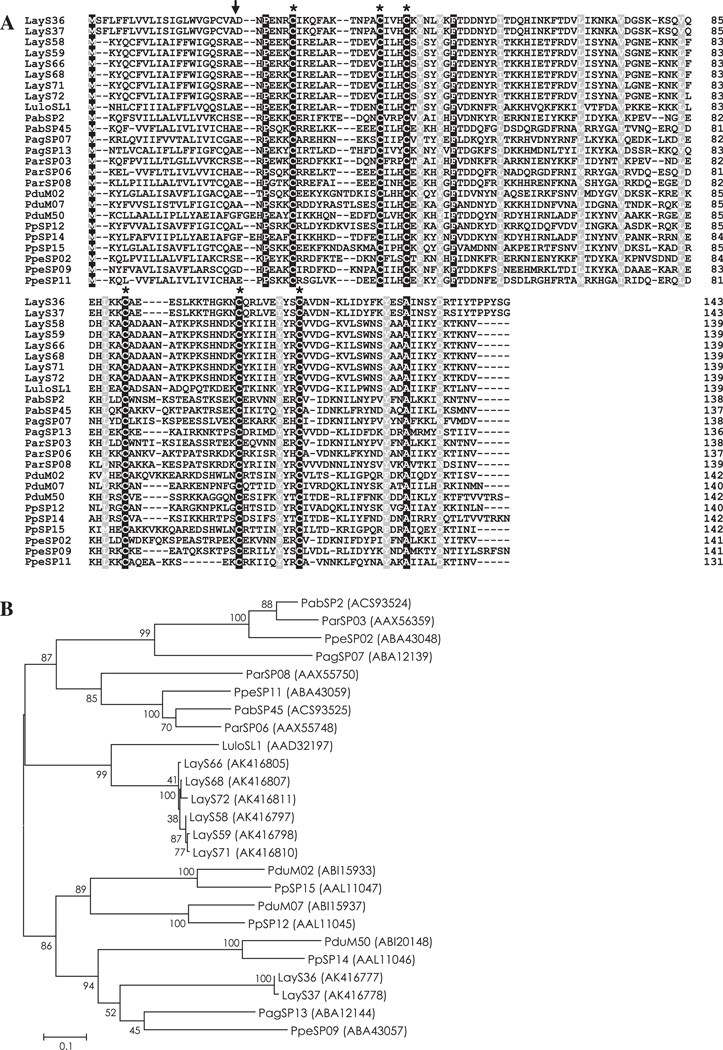
The PpSP15/SL1 family of proteins has been reported only in sand flies and the biological function during blood-feeding remains unknown. Interestingly, immunization of mice with PpSP15 protected against Leishmania major infection co-injected with P. papatasi saliva, suggesting that this family of protein is a vaccine candidate for Leishmania infection (Valenzuela et al., 2001). Two groups of molecules were identified from the Lu. ayacuchensis salivary gland cDNA library, SL-1-like proteins and 14 kDa salivary proteins (PpSP14-like protein), as members of the PpSP15/SL1 family. Twenty-five contigs (LayS48–LayS72) showed the highest homology with SL1 protein from Lu. longipalpis, and six contigs (LayS32–LayS37) coding for a 14 kDa salivary protein were more homologous to the 14.6 kDa salivary protein from P. perniciosus (PpeSP09) (Table 2). Alignment of PpSP15/SL1 family showed a high level of homology among SL-1-like proteins and among 14 kDa salivary proteins (Figs. S2 and S3). When both groups of proteins from Lu. ayacuchensis were aligned with PpSP15/SL1 family proteins from other sand flies, only six cysteine residues and three other amino acids (P, F, and A) were conserved in the amino acid sequence of mature proteins (Fig. 1A), reflecting the divergence among PpSP15/SL1 family proteins in sand flies (Anderson et al., 2006). Since the proteins of this family are immunogenic and potential vaccines for Leishmania infection (Valenzuela et al., 2001), T cell epitopes were analyzed using ProPred MHC Class-II Binding Peptide Prediction Server. We identified two potential T cell epitopes, YRITKKHIE and IIHYYRCVV, in Lu. ayacuchensis SL-1-lile proteins (Fig. S2), and two potential epitopes, IKNKAVDGS and FKYESAINSY, in 14 kDa salivary proteins (Fig. S3). A phylogenetic analysis of sand fly PpSP15/SL1 family proteins revealed that SL-1-like proteins and 14 kDa salivary proteins from Lu. ayacuchensis constructed separate clades (Fig. 1B).
Fig. 1.
(A) Sequence alignment of PpSP15/SL1 family proteins from Lutzomyia (Lu.) ayacuchensis (LayS36, LayS37, LayS58, LayS59, LayS66, LayS68, LayS71, and LayS72) together with those from Phlebotomus (P.) arabicus (Pab), P. argentipes (Pag), P. ariasi (Par), P. duboscqi (Pdu), P. papatasi (Pp), P. perniciosus (Ppe), and Lu. longipalpis (Lulo). Black-shaded amino acids represent identical amino acids and gray-shaded amino acids represent conserved amino acids. Dashes indicate gaps introduced for maximal alignment. Asterisks at the top of the amino acids denote conserved cysteine residues, and an arrowhead indicates the predicted signal peptide cleavage site. (B) Phylogenetic tree analysis of amino acid sequences of PpSP15/SL1 family proteins from sand flies. Accession numbers are in parentheses and node values indicate branch support.
3.3.3. Yellow-related protein
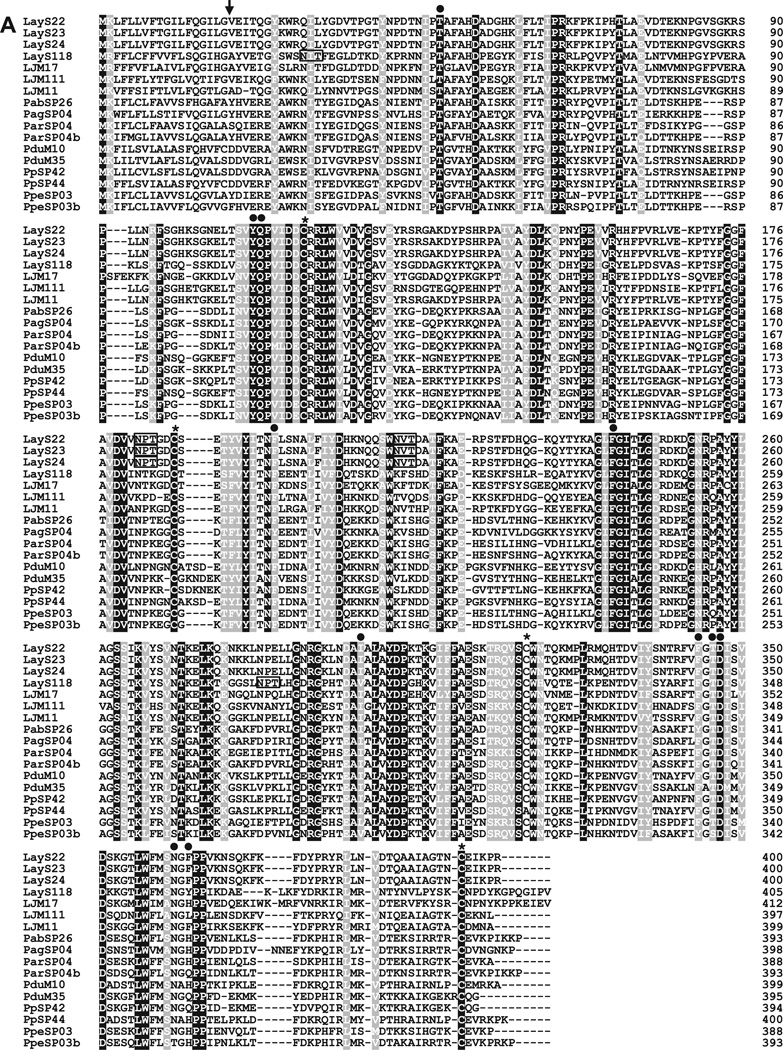
Yellow-related proteins are abundantly expressed in salivary glands of sand flies (Valenzuela et al., 2001; Oliveira et al., 2006; Anderson et al., 2006; Kato et al., 2006; Hostomská et al., 2009; Rohoušová et al., 2012). The proteins of this family are immunogenic and host antibody responses to this protein can be a potential marker for sand fly exposure (Teixeira et al., 2010; Vlkova et al., 2011). Interestingly, prior DNA vaccination of mice with PpSP44 coding for a 44 kDa yellow-related protein from P. papatasi saliva primed strong humoral immunity and exacerbated subsequent L. major infection in the presence of sand fly saliva (Oliveira et al., 2008). In contrast, a yellow-related protein from Lu. longipalpis saliva, LJM11, conferred protective cellular immunity in mice against L. major infection plus sand fly saliva. Other Lu. longipalpis yellow-related proteins, LJM111 and LJM17, were not protective, suggesting that structural features are a determinant for the host immunity against yellow-related proteins (Xu et al., 2011). Regarding the biological function, yellow-related proteins, LJM11, LJM111 and LJM17, were shown to act as high affinity binders of proinflammatory biogenic amines such as serotonin, catecholamines and histamine, suggesting that the proteins play a role for the reduction of inflammation during sand fly blood-feeding (Xu et al., 2011). In the Lu. ayacuchensis cDNA library, five contigs (LayS22–LayS24, LayS117 and LayS118) are putative yellow-related proteins, of which LayS117 was truncated in the 5′ region. When these proteins were aligned with salivary yellow-related proteins from other sand fly species, four cysteine residues were conserved in the amino acid sequence of mature proteins (Fig. 2A and S4). In addition, an amino acid motif, T-x(52,63)-Y-Q-x(85,90)-[FY]-x(44,46)-F-x(54)-[IVL]-x(45,46)-[FY]-x-[TS]-D-x(13)-[NT]-x-[QHFL], recently identified in the ligand binding pocket of yellow-related proteins (Xu et al., 2011) were conserved in those from Lu. ayacuchensis (Fig. 2A), suggesting that these proteins work as anti-inflammatory agents in Lu. ayacuchensis saliva by binding biogenic amines. Two potential N-glycosylation sites were present in yellow-related proteins from Lu. ayacuchensis, but the positions of LayS118 were different from those of others (LayS22–LayS24) (Fig. 2A). T cell epitopes were analyzed on the immunogenic proteins of this family, and we identified seven potential epitopes in LayS22–LayS24 (Fig. S4A) and 5 potential epitopes in LayS118 (Fig. S4B). A phylogenetic analysis of sand fly yellow-related proteins revealed that LayS22, LayS23 and LayS24 were closer related to LJM11 whereas LayS118 were closer related to LJM17 from Lu. longipalpis saliva (Fig. 2B). The result suggested that effect on the host immunity of LayS118 is different from those of LayS22, LayS23 and LayS24.
Fig. 2.
(A) Sequence alignment of yellow-related proteins from Lu. ayacuchensis (LayS22, LayS23, LayS24, and LayS118) together with those from P. arabicus (Pab), P. argentipes (Pag), P. ariasi (Par), P. duboscqi (Pdu), P. papatasi (Pp), P. perniciosus (Ppe), and Lu. longipalpis (LJM). Black-shaded amino acids represent identical amino acids and gray-shaded amino acids represent conserved amino acids. Dashes indicate gaps introduced for maximal alignment. Asterisks at the top of the amino acids denote conserved cysteine residues, and an arrowhead indicates the predicted signal peptide cleavage site. Closed circles show conserved amino acids contained in the ligand binding pocket of yellow-related protein family. Potential N-glycosylation sites of Lu. ayacuchensis yellow-related proteins are boxed. (B) Phylogenetic tree analysis of amino acid sequences of yellow-related proteins from sand flies. Accession numbers are in parentheses and node values indicate branch support.
3.3.4. Apyrase
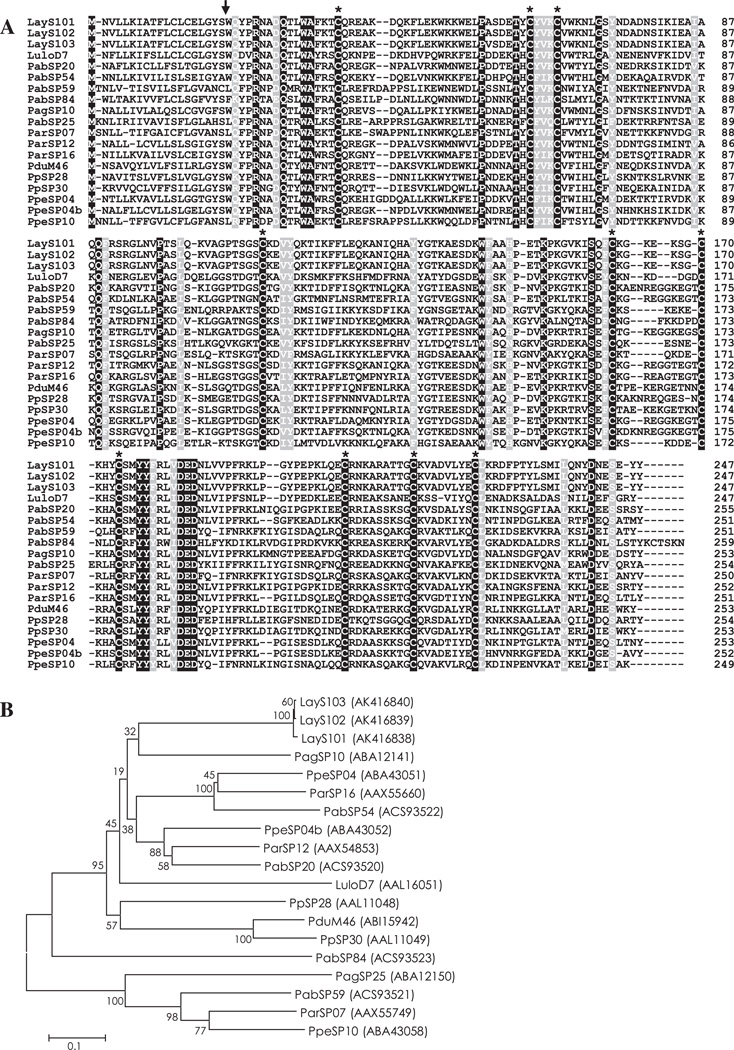
Apyrases are nucleoside triphosphate-diphosphohydrolases present in a variety of organisms. In the saliva of blood sucking arthropods, apyrases function to hydrolyze ADP in a Ca2+-dependent manner and inhibit ADP-induced platelet aggregation to facilitate blood feeding (Ribeiro and Francischetti, 2003; Faudry et al., 2004). Apyrases identified in sand fly saliva belong to the Cimex apyrase family (Valenzuela et al., 2001; Hamasaki et al., 2009). Thirteen contigs (LayS8–LayS14 and LayS16–LayS21) were homologous to putative apyrase from Lu. longipalpis (LuloAPY). A multiple sequence alignment analysis together with LuloAPY confirmed the close relationship among putative apyrases of Lu. ayacuchensis (Fig. S5).
3.3.5. D7 family
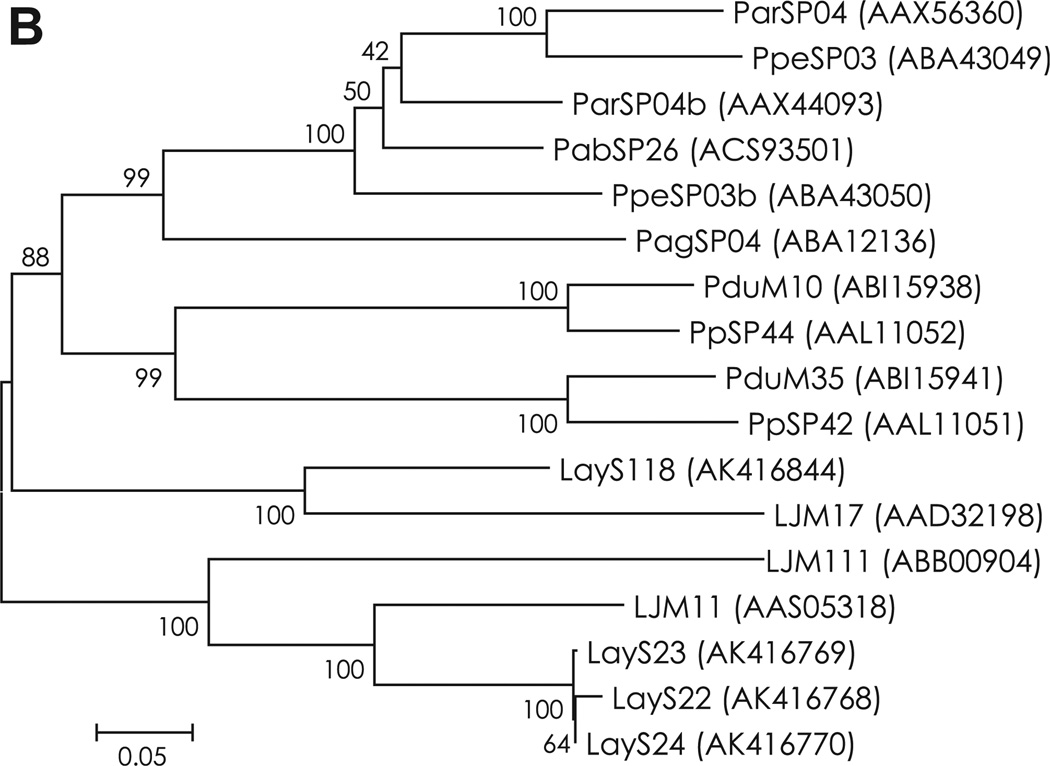
D7 family proteins are abundantly expressed in salivary glands of blood-feeding Diptera such as mosquitoes, sand flies, black flies, and biting midges (James et al., 1991; Valenzuela et al., 2002; Campbell et al., 2005; Andersen et al., 2009). To date, the function of sand fly D7 proteins remains to be elucidated; however, several D7 family proteins have been functionally characterized in mosquitoes. Hamadarin from Anopheles stephensi saliva was identified as a blood coagulation inhibitor affecting the activation of the plasma contact system (Isawa et al., 2002). Salivary D7 proteins from Anopheles gambiae and Aedes aegypti were characterized as binders of biogenic amines such as serotonin, histamine and norepinephrine (Calvo et al., 2006). Nine contigs (LayS95–LayS103) encoding a 26.6 kDa protein were homologous to the 27 kDa D7-related salivary protein from P. argentipes (PagSP10) and P. perniciosus (PpeSP04b). In a multiple sequence alignment analysis of D7 family proteins from sand flies, 10 cysteine residues were strictly conserved in the amino acid sequence of mature proteins (Fig. 3A and Fig. S6). Alignment and phylogenetic analysis together with PpeSP04b showed that D7 family proteins of Lu. ayacuchensis had close relationships (Fig. 3B).
Fig. 3.
(A) Sequence alignment of D7 family proteins from Lu. ayacuchensis (LayS101–LayS103) together with those from P. arabicus (Pab), P. argentipes (Pag), P. ariasi (Par), P. duboscqi (Pdu), P. papatasi (Pp), P. perniciosus (Ppe), and Lu. longipalpis (Lulo). Black-shaded amino acids represent identical amino acids and gray-shaded amino acids represent conserved amino acids. Dashes indicate gaps introduced for maximal alignment. Asterisks at the top of the amino acids denote conserved cysteine residues, and an arrowhead indicates the predicted signal peptide cleavage site. (B) Phylogenetic tree analysis of amino acid sequences of D7 family proteins from sand flies. Accession numbers are in parentheses and node values indicate branch support.
3.3.6. 27 kDa Salivary protein
The protein family is also known as PpSP32-like family. Eleven contigs (LayS83–LayS93) encoding a 27 kDa protein were homologous to the 29.2 kDa salivary protein from Lu. longipalpis (Table 2). These transcripts are also present in the salivary glands from Old World sand flies such as P. ariasi, P. duboscqi, P. arabicus, P. argentipes, P. papatasi, P. perniciosus, P. tobbi, and P. sergenti (Valenzuela et al., 2001; Oliveira et al., 2006; Anderson et al., 2006; Kato et al., 2006; Hostomská et al., 2009; Rohoušová et al., 2012). To date, no homologues of these proteins have been reported in organisms other than sand flies and the biological function remains to be clarified. Although no conserved domains were found in the amino acid sequences, these proteins contain glycine-rich sequences in the middle region (amino acids 66–179 in LayS91) (Fig. S7).
3.3.7. Antigen 5-related protein
This family of proteins belongs to the cysteine-rich secretory proteins (CRISPs) and is related to venom allergens in social wasps and ants (Lu et al., 1993; Hoffman, 1993; King and Spangfort, 2000). Transcripts coding for the members of this protein family have been identified in the salivary glands of blood sucking insects such as mosquitoes (Francischetti et al., 2002; Arcà et al., 2007), sand flies (Valenzuela et al., 2001; Oliveira et al., 2006; Anderson et al., 2006; Kato et al., 2006; Hostomská et al., 2009; Rohoušová et al., 2012) and triatomine bugs (Ribeiro et al., 2004; Santos et al., 2007; Assumpção et al., 2008; Kato et al., 2010b). Although the antigen 5 family of protein has been widely identified in blood-sucking arthropods, the function in their saliva has yet to be determined. Nine contigs (LayS73–LayS81) from Lu. ayacuchensis salivary glands were included in this family. A multiple sequence alignment analysis of antigen 5-related proteins with that of Lu. longipalpis revealed the close relationship among the proteins of Lu. ayacuchensis and strict conservation of 12 cysteine residues in mature forms (Fig. S8).
3.3.8. 16.4 kDa Salivary protein
Thirteen contigs (LayS120–LayS132) coded for 16.4 kDa protein containing a C-type lectin/C-type lectin-like domain (Table 2). This putative domain may function as a Ca2+-dependent carbohydrate-binding pocket involved in extracellular matrix organization, pathogen recognition and cell-to-cell interactions (Weis et al., 1998). Homologous proteins with molecular weight of 16.2–16.5 kDa have been identified from Lu. longipalpis saliva (Charlab et al., 1999; Valenzuela et al., 2004), and they were similar to the previously described anticoagulant from Lu. longipalpis saliva (Charlab et al., 1999). A recombinant protein of mature LayS127 protein was expressed in Escherichia coli, and its activity on coagulation pathway was assessed; however, the protein inhibited neither intrinsic nor extrinsic coagulation pathways (data not shown). At present, the biological function of these proteins from Lu. ayacuchensis remains to be clarified. In a multiple sequence alignment analysis of Lu. ayacuchensis 16.4 kDa proteins together with Lu. longipalpis 16.5 kDa protein, three cysteine residues were strictly conserved in the amino acid sequence of mature proteins (Fig. S9).
3.3.9. 11.5 kDa Salivary protein
Four contigs encoding 10.7–11.9 kDa proteins (LayS4–LayS7) were identified in the cDNA library of Lu. ayacuchensis salivary glands. The different size of these proteins was mainly resulted from the number of ‘‘SSDGSSG”repeat sequences with unknown function (Fig. S10). The only homologous protein has been identified in salivary glands of Lu. longipalpis (9 kDa salivary protein) with unknown function. The amino acid sequences of these proteins are rich in serine from the middle to the C-terminal regions (Fig. S10).
3.3.10. Other putative secreted proteins
Three contigs (LayS25–LayS27) coding for a 33.6 kDa protein had homology with a 34 kDa salivary protein from P. ariasi. Three contigs (LayS141–LayS143) coded for a 9.3 kDa protein. Three singletons (LayS109–LayS111) coding for a 15.9 kDa protein had homology with a 15.5 kDa salivary protein from P. argentipes with unknown function. Two contigs (LayS2 and LayS3) coded for a 25.9 kDa protein homologous to allergen from Culex quinquefasciatus belonging to the sperm-coating protein (SCP) superfamily. Two singletons (LayS167 and LayS168) and one singleton (LayS153) coded for homologous proteins with a 32.4 kDa salivary protein and a 71 kDa salivary protein from Lu. longipalpis, respectively, although the sequence of LayS168 was truncated in the 5′ region. A singleton (LayS147) coded for homologous protein with the 43.7 kDa salivary protein from Lu. longipalpis, a putative endonuclease, but the sequence was truncated in the 5′ region. Singletons, LayS106 and LayS215, coded for homologous protein with the 27 kDa salivary protein from P. ariasi and trypsin inhibitor like cysteine rich domain containing protein from Drosophila, respectively.
4. Conclusions
The present study identified abundant salivary gland transcripts of Lu. ayacuchensis, a proven vector of L. (L.) mexicana (Takaoka et al., 1990; Kato et al., 2005, 2008) and L. (V.) peruviana (Caceres et al., 2004; Kato et al., 2008) causative agents of cutaneous leishmaniasis in Andean areas of Ecuador and Peru, respectively. The relatively divergent transcripts resulting in the generation of a variety of contigs noted for each component probably reflects population of the sand flies used in this study; that is, our cDNA library was constructed from field-captured Lu. ayacuchensis while most studies prepared the cDNA libraries from colonized insects. To date, the salivary components of the New World Lutzomyia species have been analyzed only in Lu. longipalpis (Charlab et al., 1999; Valenzuela et al., 2004), a subgenus Lutzomyia species, transmitting L. (L.) infantum, a causative agent of visceral leishmaniasis (Young and Duncan, 1994). On the other hand, Lu. ayacuchensis is a vector species of cutaneous leishmaniasis and belongs to the subgenus Helcocyrtomyia (Young and Duncan, 1994). Therefore, the present analysis is the first description of salivary proteins of a sand fly in the subgenus Helcocyrtomyia and a vector of cutaneous leishmaniasis in the New World.
In the present analysis, transcripts homologous to maxadilan, an active vasodilator abundantly present in saliva of Lu. longipalpis, were not identified. Maxadilan was originally isolated from saliva of Lu. longipalpis (Lerner et al., 1991), and its homologue was reported from Lu. neivai (Aires et al., 2005). Conversely, maxadilan-related proteins have never been identified in Old World Phlebotomus species (Valenzuela et al., 2001; Oliveira et al., 2006; Anderson et al., 2006; Kato et al., 2006; Hostomská et al., 2009). Phlebotomus sand flies except for P. duboscqi lack adenosine deaminase (ADA) that hydrolyze adenosine and adenosine monophosphate (AMP) (Oliveira et al., 2006; Anderson et al., 2006; Kato et al., 2006; Hostomská et al., 2009), but contain large amounts of adenosine and AMP, very active vasodilators, in their saliva (Ribeiro et al., 1999; Katz et al., 2000; Ribeiro and Modi, 2001). On the other hand, Lu. longipalpis has ADA (Charlab et al., 1999, 2000; Valenzuela et al., 2004) and lacks adenosine and AMP in the saliva (Ribeiro et al., 1989). Therefore, with an exception, salivary vasodilators of sand flies are considered to be adenosine and AMP in Phlebotomus species and maxadilan in Lutzomyia species. Unexpectedly, neither ADA nor maxadilan were identified in Lu. ayacuchensis, which is similar to Phlebotomus species, suggesting that salivary vasodilators of this species may be adenosine and AMP. Further analysis of small molecules in the saliva of Lu. ayacuchensis will elucidate this issue.
In conclusion, the most abundant proteins of Lu. ayacuchensis saliva were identified in this study. These results will provide further insights into the evolution of salivary components in blood sucking arthropods. In addition, the cDNAs and future recombinant proteins prepared from these transcripts will result in the discovery of vaccine candidates and markers of sand fly exposure as well as novel pharmacologically active compounds.
Supplementary Material
Acknowledgements
We are grateful to Dr. José M. C. Ribeiro (Vector Biology Section, Laboratory of Malaria and Vector Research, NIAID, NIH, USA) for the development and training of all custom bioinformatics programs used in this research. This study was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (Grant Nos. 18256004, 10037385 and 23580424).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version, at http://dx.doi.org/10.1016/j.meegid.2012.08.024
References
- Aires JM, Chociay MF, Nascimento MM, Figueiredo JF, Roselino AM. Maxadilan (MAX) - Salivaly protein of Lutzomyia longipalpis: detection of antibodies anti-MAX in American tegmentar leishmaniasis (ATL), and genetic and protein expression of MAX in Lutzomyia neivai. An. Bras. Dermatol. 2005;80:S333–S338. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–4002. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JF, Pham VM, Meng Z, Champagne DE, Ribeiro JMC. Insight into the sialome of the Black Fly, Simulium vittatum. J. Proteome Res. 2009;8:1474–1488. doi: 10.1021/pr8008429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Oliveira F, Kamhawi S, Mans BJ, Reynoso D, Seitz AE, Lawyer P, Garfield M, Pham M, Valenzuela JG. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC Genomics. 2006;7:52. doi: 10.1186/1471-2164-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Francischetti IMB, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JMC. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem. Mol Biol. 2007;37:107–127. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assumpção TC, Francischetti IMB, Andersen JF, Schwarz A, Santana JM, Ribeiro JMC. An insight into the sialome of the blood-sucking bug Triatoma infestans, a vector of Chagas’ disease. Insect Biochem. Mol Biol. 2008;38:213–232. doi: 10.1016/j.ibmb.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E. Searching databases to find protein domain organization. Adv. Protein Chem. 2000;54:137–157. doi: 10.1016/s0065-3233(00)54005-4. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Kamhawi S, Modi G, Valenzuela J, Noben-Trauth N, Rowton E, Ribeiro J, Sacks DL. Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J. Exp. Med. 1998;188:1941–1953. doi: 10.1084/jem.188.10.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Valenzuela JG, Kamhawi S, Rowton E, Sacks DL, Ribeiro JMC. Delayed-type hypersensitivity to Phlebotomus papatasi sand fly bite: an adaptive response induced by the fly? Proc. Natl. Acad. Sci USA. 2000;97:6704–6709. doi: 10.1073/pnas.97.12.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Caceres AG, Villaseca P, Dujardin JC, Bañuls AL, Inga R, Lopez M, Arana M, Le Ray D, Arevalo J. Epidemiology of Andean cutaneous leishmaniasis: incrimination of Lutzomyia ayacuchensis (Diptera: psychodidae) as a vector of Leishmania in geographically isolated, upland valleys of Peru. Am. J. Trop. Med. Hyg. 2004;70:607–612. [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JMC. Function and evolution of a mosquito salivary protein family. J. Biol. Chem. 2006;281:1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Vandyke KA, Letchworth GJ, Drolet BS, Hanekamp T, Wilson WC. Midgut and salivary gland transcriptomes of the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae) Insect Mol. Biol. 2005;14:121–136. doi: 10.1111/j.1365-2583.2004.00537.x. [DOI] [PubMed] [Google Scholar]
- Charlab R, Valenzuela JG, Rowton ED, Ribeiro JMC. Toward an understanding of the biochemical and pharmacological complexity of the saliva of a hematophagous sand fly Lutzomyia longipalpis. Proc. Natl. Acad. Sci. USA. 1999;96:15155–15160. doi: 10.1073/pnas.96.26.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlab R, Rowton ED, Ribeiro JMC. The salivary adenosine deaminase from the sand fly Lutzomyia longipalpis. Exp. Parasitol. 2000;95:45–53. doi: 10.1006/expr.2000.4503. [DOI] [PubMed] [Google Scholar]
- Faudry E, Rocha PS, Vernet T, Lozzi SP, Teixeira AR. Kinetics of expression of the salivary apyrases in Triatoma infestans. Insect Biochem. Mol Biol. 2004;34:1051–1058. doi: 10.1016/j.ibmb.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Francischetti IMB, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JMC. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. J. Exp. Biol. 2002;205:2429–2451. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Gomes R, Teixeira C, Teixeira MJ, Oliveira F, Menezes MJ, Silva C, de Oliveira CI, Miranda JC, Elnaiem DE, Kamhawi S, Valenzuela JG, Brodskyn CI. Immunity to a salivary protein of a sand fly vector protects against the fatal outcome of visceral leishmaniasis in a hamster model. Proc. Natl. Acad. Sci. USA. 2008;105:7845–7850. doi: 10.1073/pnas.0712153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki R, Kato H, Terayama Y, Iwata H, Valenzuela JG. Functional characterization of a salivary apyrase from the sand fly, Phlebotomus duboscqi, a vector of Leishmania major. J Insect Physiol. 2009;55:1044–1049. doi: 10.1016/j.jinsphys.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DR. Allergens in Hymenoptera venom. XXV: The amino acid sequences of antigen 5 molecules and the structural basis of antigenic cross-reactivity. J. Allergy Clin. Immunol. 1993;92:707–716. doi: 10.1016/0091-6749(93)90014-7. [DOI] [PubMed] [Google Scholar]
- Hostomská J, Volfová V, Mu J, Garfield M, Rohousová I, Volf P, Valenzuela JG, Jochim RC. Analysis of salivary transcripts and antigens of the sand fly Phlebotomus arabicus. BMC Genomics. 2009;10:282. doi: 10.1186/1471-2164-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. A contig assembly program based on sensitive detection of fragment overlaps. Genomics. 1992;14:18–25. doi: 10.1016/s0888-7543(05)80277-0. [DOI] [PubMed] [Google Scholar]
- Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Isawa H, Yuda M, Orito Y, Chinzei Y. A mosquito salivary protein inhibits activation of the plasma contact system by binding to factor XII and high molecular weight kininogen. J. Biol. Chem. 2002;277:27651–27658. doi: 10.1074/jbc.M203505200. [DOI] [PubMed] [Google Scholar]
- James AA, Blackmer K, Marinotti O, Ghosn CR, Racioppi JV. Isolation and characterization of the gene expressing the major salivary gland protein of the female mosquito, Aedes aegypti. Mol. Biochem. Parasitol. 1991;44:245–253. doi: 10.1016/0166-6851(91)90010-4. [DOI] [PubMed] [Google Scholar]
- Kamhawi S, Belkaid Y, Modi G, Rowton E, Sacks D. Protection against cutaneous leishmaniasis resulting from bites of uninfected sand flies. Science. 2000;290:1351–1354. doi: 10.1126/science.290.5495.1351. [DOI] [PubMed] [Google Scholar]
- Kato H, Uezato H, Katakura K, Calvopiña M, Marco JD, Barroso PA, Gomez EA, Mimori T, Korenaga M, Iwata H, Nonaka S, Hashiguchi Y. Detection and identification of Leishmania species within naturally infected sand flies in the andean areas of Ecuador by a polymerase chain reaction. Am. J. Trop. Med. Hyg. 2005;72:87–93. [PubMed] [Google Scholar]
- Kato H, Anderson JM, Kamhawi S, Oliveira F, Lawyer PG, Pham VM, Sangare CS, Samake S, Sissoko I, Garfield M, Sigutova L, Volf P, Doumbia S, Valenzuela JG. High degree of conservancy among secreted salivary gland proteins from two geographically distant Phlebotomus duboscqi sandflies populations (Mali and Kenya) BMC Genomics. 2006;7:226. doi: 10.1186/1471-2164-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Cáceres AG, Gomez EA, Mimori T, Uezato H, Marco JD, Barroso PA, Iwata H, Hashiguchi Y. Molecular mass screening to incriminate sand fly vectors of Andean-type cutaneous leishmaniasis in Ecuador and Peru. Am. J. Trop. Med. Hyg. 2008;79:719–721. [PubMed] [Google Scholar]
- Kato H, Gomez EA, Cáceres AG, Uezato H, Mimori T, Hashiguchi Y. Molecular epidemiology for vector research on leishmaniasis. Int. J. Environ. Res Public Health. 2010a;7:814–826. doi: 10.3390/ijerph7030814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jochim RC, Gomez EA, Sakoda R, Iwata H, Valenzuela JG, Hashiguchi Y. A repertoire of the dominant transcripts from the salivary glands of the blood-sucking bug, Triatoma dimidiata, a vector of Chagas disease. Infect. Genet. Evol. 2010b;10:184–191. doi: 10.1016/j.meegid.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz O, Waitumbi JN, Zer R, Warburg A. Adenosine, AMP, and protein phosphatase activity in sandfly saliva. Am. J. Trop. Med. Hyg. 2000;62:145–150. doi: 10.4269/ajtmh.2000.62.145. [DOI] [PubMed] [Google Scholar]
- King TP, Spangfort MD. Structure and biology of stinging insect venom allergens. Int. Arch. Allergy Immunol. 2000;123:99–106. doi: 10.1159/000024440. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Ribeiro JMC, Nelson RJ, Lerner MR. Isolation of maxadilan, a potent vasodilatory peptide from the salivary glands of the sand fly Lutzomyia longipalpis. J. Biol. Chem. 1991;266:11234–11236. [PubMed] [Google Scholar]
- Lima HC, Titus RG. Effects of sand fly vector saliva on development of cutaneous lesions and the immune response to Leishmania braziliensis in BALB/c mice. Infect Immun. 1996;64:5442–5445. doi: 10.1128/iai.64.12.5442-5445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Deadman JJ, Williams JA, Kakkar VV, Rahman S. Synthetic RGD peptides derived from the adhesive domains of snake-venom proteins: evaluation as inhibitors of platelet aggregation. Biochem J. 1993;296:21–24. doi: 10.1042/bj2960021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Ariel N, Bryant SH. Comparison of sequence and structure alignments for protein domains. Proteins. 2002;48:439–446. doi: 10.1002/prot.10163. [DOI] [PubMed] [Google Scholar]
- Munstermann LE. Phlebotomine sand flies, the Psychodidae. In: Marquardt WC, Black WC, Freier JE, Hagedorn HH, Hemingway J, Higgs S, James AA, Kondratieff B, Moore CG, editors. Biology of Disease Vectors. second ed. San Diego, CA: Elsevier; 2005. pp. 41–151. [Google Scholar]
- Oliveira F, Kamhawi S, Seitz AE, Pham VM, Guigal PM, Fischer L, Ward J, Valenzuela JG. From transcriptome to immunome: identification of DTH inducing proteins from a Phlebotomus ariasi salivary gland cDNA library. Vaccine. 2006;24:374–390. doi: 10.1016/j.vaccine.2005.07.085. [DOI] [PubMed] [Google Scholar]
- Oliveira F, Lawyer PG, Kamhawi S, Valenzuela JG. Immunity to distinct sand fly salivary proteins primes the anti-Leishmania immune response towards protection or exacerbation of disease. PLoS Negl. Trop. Dis. 2008;2:e226. doi: 10.1371/journal.pntd.0000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira F, Jochim RC, Valenzuela JG, Kamhawi S. Sand flies, Leishmania, and transcriptome-borne solutions. Parasitol. Int. 2009;58:1–5. doi: 10.1016/j.parint.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect. Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- Ribeiro JMC, Francischetti IMB. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu. Rev. Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Modi G. The salivary adenosine/AMP content of Phlebotomus argentipes Annandale and Brunetti, the main vector of human kala-aza. J Parasitol. 2001;87:915–917. doi: 10.1645/0022-3395(2001)087[0915:TSAACO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Vachereau A, Modi GB, Tesh RB. A novel vasodilatory peptide from the salivary glands of the sand fly Lutzomyia longipalpis. Science. 1989;243:212–214. doi: 10.1126/science.2783496. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Katz O, Pannell LK, Waitumbi J, Warburg A. Salivary glands of the sand fly Phlebotomus papatasi contain pharmacologically active amounts of adenosine and 5′-AMP. J. Exp. Biol. 1999;202:1551–1559. doi: 10.1242/jeb.202.11.1551. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Andersen J, Silva-Neto MA, Pham VM, Garfield MK, Valenzuela JG. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem. Mol. Biol. 2004;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Rohoušová I, Volf P. Sand fly saliva: effects on host immune response and Leishmania transmission. Folia Parasitol. 2006;53:161–171. [PubMed] [Google Scholar]
- Rohoušová I, Subrahmanyam S, Volfová V, Mu J, Volf P, Valenzuela JG, Jochim RC. Salivary gland transcriptomes and proteomes of Phlebotomus tobbi and Phlebotomus sergenti, vectors of leishmaniasis. PLoS Negl. TropDis. 2012;6:e1660. doi: 10.1371/journal.pntd.0001660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A, Ribeiro JMC, Lehane MJ, Gontijo NF, Veloso AB, Sant’Anna MR, Nascimento Araujo R, Grisard EC, Pereira MH. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae) Insect Biochem. Mol. Biol. 2007;37:702–712. doi: 10.1016/j.ibmb.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Raghava GP. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- Takaoka H, Gomez EA, Alexander JB, Hashiguchi Y. Natural infections with Leishmania promastigotes in Lutzomyia ayacuchensis (Diptera: Psychodidae) in an Andean focus of Ecuador. J. Med. Entomol. 1990;27:701–702. doi: 10.1093/jmedent/27.4.701. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tavares NM, Silva RA, Costa DJ, Pitombo MA, Fukutani KF, Miranda JC, Valenzuela JG, Barral A, de Oliveira CI, Barral-Netto M, Brodskyn C. Lutzomyia longipalpis saliva or salivary protein LJM19 protects against Leishmania braziliensis and the saliva of its vector, Lutzomyia intermedia. PLoS Negl. Trop. Dis. 2011;5:e1169. doi: 10.1371/journal.pntd.0001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira C, Gomes R, Collin N, Reynoso D, Jochim R, Oliveira F, Seitz A, Elnaiem DE, Caldas A, de Souza AP, Brodskyn CI, de Oliveira CI, Mendonca I, Costa CH, Volf P, Barral A, Kamhawi S, Valenzuela JG. Discovery of markers of exposure specific to bites of Lutzomyia longipalpis, the vector of Leishmania infantum chagasi in Latin America. PLoS Negl. Trop. Dis. 2010;4:e638. doi: 10.1371/journal.pntd.0000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodos CM, Ribeiro JMC, Titus RG. Analysis of enhancing effect of sand fly saliva on Leishmania infection in mice. Infect. Immun. 1991;59:1592–1598. doi: 10.1128/iai.59.5.1592-1598.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus RG, Ribeiro JMC. Salivary gland lysates from the sand fly Lutzomyia longipalpis enhance Leishmania infectivity. Science. 1988;239:1306–1308. doi: 10.1126/science.3344436. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG. Blood-feeding arthropod salivary glands and saliva. In: Marquardt WC, Black WC, Freier JE, Hagedorn HH, Hemingway J, Higgs S, James AA, Kondratieff B, Moore CG, editors. Biology of Disease Vectors. second ed. San Diego, CA: Elsevier; 2005. pp. 377–386. [Google Scholar]
- Valenzuela JG, Belkaid Y, Garfield MK, Mendez S, Kamhawi S, Rowton ED, Sacks DL, Ribeiro JMC. Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J. Exp. Med. 2001;194:331–342. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG, Charlab R, Gonzalez EC, de Miranda-Santos IK, Marinotti O, Francischetti IMB, Ribeiro JMC. The D7 family of salivary proteins in blood sucking diptera. Insect Mol. Biol. 2002;11:149–155. doi: 10.1046/j.1365-2583.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Garfield M, Rowton ED, Pham VM. Identification of the most abundant secreted proteins from the salivary glands of the sand fly Lutzomyia longipalpis, vector of Leishmania chagasi. J. Exp. Biol. 2004;207:3717–3729. doi: 10.1242/jeb.01185. [DOI] [PubMed] [Google Scholar]
- Vlkova M, Rohousova I, Drahota J, Stanneck D, Kruedewagen EM, Mencke N, Otranto D, Volf P. Canine antibody response to Phlebotomus perniciosus bites negatively correlates with the risk of Leishmania infantum transmission. PLoS Negl. Trop. Dis. 2011;5:e1344. doi: 10.1371/journal.pntd.0001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol. Rev. 1998;163:19–34. doi: 10.1111/j.1600-065x.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- Xu X, Oliveira F, Chang BW, Collin N, Gomes R, Teixeira C, Reynoso D, My Pham V, Elnaiem DE, Kamhawi S, Ribeiro JMC, Valenzuela JG, Andersen JF. Structure and function of a ‘‘yellow”protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection. J. Biol. Chem. 2011;286:32383–32393. doi: 10.1074/jbc.M111.268904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DG, Duncan MA. Memoirs of the American Entomological Institute. Vol. 54. Gainsville, FL: Associated Publishers—American Entomological Institute; 1994. Guide to the Identification and Geographic Distribution of Lutzomyia Sand Flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.