Abstract
While numerous recent advances have contributed to our understanding of excitatory synapse formation, the processes that mediate inhibitory synapse formation remain poorly defined. Previously, we discovered that RNAi-mediated knockdown of a Class 4 Semaphorin, Sema4D, led to a decrease in the density of inhibitory synapses without an apparent effect on excitatory synapse formation. Our current work has led us to new insights about the molecular mechanisms by which Sema4D regulates GABAergic synapse development. Specifically, we report that the extracellular domain of Sema4D is proteolytically cleaved from the surface of neurons. However, despite this cleavage event, Sema4D signals through its extracellular domain as a membrane-bound, synaptically localized protein required in the postsynaptic membrane for proper GABAergic synapse formation. Thus, as Sema4D is one of only a few molecules identified thus far that preferentially regulates GABAergic synapse formation, these findings have important implications for our mechanistic understanding of this process.
Keywords: Sema4D, GABAergic, proteolytic cleavage, inhibitory synapse
Introduction
The formation of synapses, which are sites of cell-cell communication between neurons, is a complex process that is regulated by a number of transmembrane and membrane-associated proteins (Dalva et al. 2007). The majority of studies on mammalian synapse development thus far have focused on the hippocampus, where glutamate is the major excitatory neurotransmitter and γ-aminobutyric acid (GABA) is the major inhibitory neurotransmitter. Historically, greater attention has been paid to the molecules that direct formation of glutamatergic synapses, although recently, we and others have identified proteins important for GABAergic synapse formation (Shin et al. 2003; Paradis et al. 2007; Chih & Engelman 2005; Terauchi et al. 2010; Vicario-Abejón et al. 1998; Pietro Fazzari et al. 2010). The Semaphorins are one such family of proteins that have emerged as important regulators of synapse development throughout the mammalian nervous system. In addition to our studies implicating Semaphorin signaling in GABAergic synapse formation (Paradis et al. 2007), studies have implicated other Semaphorin family members in glutamatergic synapse formation or elimination (Morita et al. 2006; Yamashita et al. 2007; Tran et al. 2009; O’Connor et al. 2009; Paradis et al. 2007)
Semaphorins are a large family of transmembrane and secreted glycoproteins that were originally identified as axon guidance molecules (Kolodkin et al. 1993; Kumanogoh & Kikutani 2004). Of the eight total Semaphorin subclasses, five are expressed in mammals and all share a conserved extracellular Sema domain (Kumanogoh & Kikutani 2004; Love et al. 2003). The Class 4 Semaphorins in particular have been shown to play roles in the immune response as well as neural development (Shi et al. 2000; Zhu et al. 2007; Kumanogoh et al. 2002; Paradis et al. 2007; Oinuma et al. 2010). Our previous work demonstrated that the Class 4 Semaphorin Sema4D mediates GABAergic synaptic development both in vitro and in vivo without an apparent effect on glutamatergic synapse development (Paradis et al. 2007; Kuzirian et al. 2013). In addition, the Class 4 Semaphorin Sema4B mediates both GABAergic and glutamatergic synapse development, suggesting a conserved role for Class 4 Semaphorins in the regulation of mammalian CNS synaptogenesis (Paradis et al. 2007). The molecular mechanisms by which Semaphorins regulate synaptogenesis, and the subcellular localization of these ligands in the nervous system, are not well understood.
The Sema4D protein consists of a short cytoplasmic tail, transmembrane domain, and extracellular Ig and Sema domains (Shi et al. 2000; Furuyama et al. 1996; Hall et al. 1996). Cleavage of Sema4D in non-neuronal cells occurs at the cell surface, putatively between the transmembrane domain and Ig domain (Elhabazi et al. 2001), resulting in an extracellular soluble fragment and an intracellular C-terminal fragment (Zhu et al. 2007; Basile et al. 2007). Although a recent study demonstrated that the extracellular domain of Sema4D is sufficient to drive functional GABAergic synapse formation (Kuzirian et al. 2013), whether cleavage of the Sema4D extracellular domain occurs from the neuronal cell surface, and its implication for signal transduction in the nervous system, has not been addressed until now. In addition, the C-terminal, cytoplasmic domain of Sema4D has no known function and thus it remains an open question as to whether or not signaling through the intracellular domain of Sema4D also influences synapse development.
As a means to gain insight into the molecular mechanisms that instruct GABAergic synaptic development in the rodent hippocampus, we investigated the domains within Sema4D that are required to mediate GABAergic synapse development. We constructed multiple chimeras of Sema4D by replacing different domains of Sema4D with that of the transmembrane protein CD4, a small single pass protein involved in T-cell activation. Using this approach we discovered that Sema4D signaling through its N-terminal extracellular domain is absolutely required to promote GABAergic synapse formation. In addition, we observed that while Sema4D is proteolytically cleaved in the mammalian brain, this event is not required for Sema4D to regulate synaptogenesis, suggesting that it signals as a membrane-bound molecule. Consistent with this model, we demonstrate that Sema4D is localized to the synaptic membrane in the mammalian hippocampus. Taken together, our data establishes that Sema4D is a synaptic protein that can regulate GABAergic synaptogenesis exclusively through its extracellular domain and as a membrane-bound molecule.
Results
The extracellular, N-terminal domain of Sema4D is required for GABAergic synapse formation
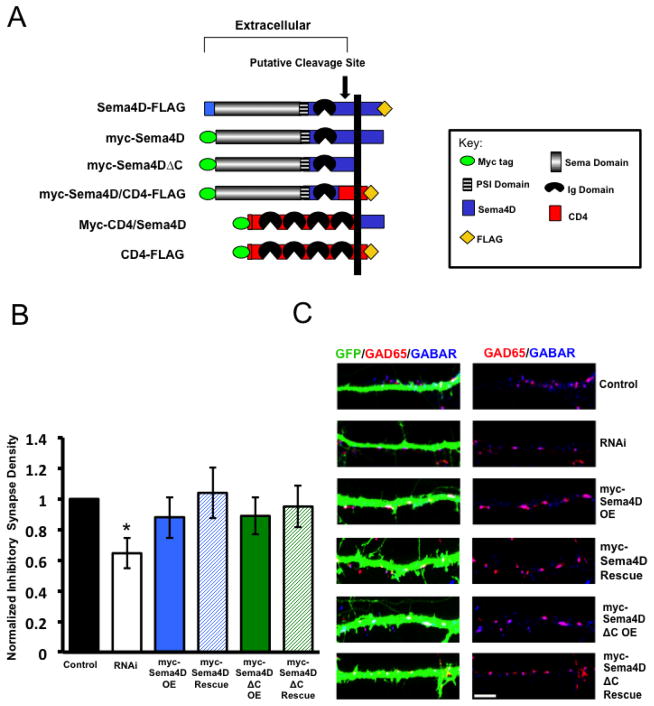
In order to determine the signal transduction mechanism(s) by which Sema4D regulates synapse development, we asked if the intracellular domain of Sema4D was required to mediate GABAergic synapse formation. To address this question, we generated epitope-tagged, RNAi-resistant Sema4D cDNA constructs harboring either a deletion or a swap between regions of Sema4D and the transmembrane immune receptor CD4 (Fig. 1A). To assess the role of the intracellular domain of Sema4D in GABAergic synapse development, we asked if our myc-Sema4DΔC construct could rescue decreases in GABAergic synapse density caused by RNAi-mediated knockdown of endogenous Sema4D. To do this, we co-transfected cultured hippocampal neurons at DIV4 with GFP and either an empty vector (“Control”), our Sema4D specific shRNA (“RNAi”), a Sema4D cDNA that had been rendered resistant to knockdown by RNAi by introduction of silent point mutations (“OE”), or both our shRNA and a RNAi-resistant Sema4D construct, in this case either wild type myc-Sema4D or myc-Sema4DΔC (“Rescue”) (Fig. 1B, C). We then fixed the cultures at DIV14 and stained for the inhibitory pre- and postsynaptic markers GAD65 and the γ2 subunit of the GABAA receptor, respectively. An inhibitory GABAergic synapse was defined as the overlap between GAD65 and GABAAR γ2 puncta onto a GFP-expressing neuron (Paradis et al. 2007).
Figure 1. The intracellular C-terminus is not required for Sema4D to regulate GABAergic synapse formation.
(a) Diagram illustrating the epitope-tagged Sema4D and CD4 deletion and chimeric constructs used throughout this study. The myc epitope was inserted at the N-terminus while the FLAG epitope was inserted at the C-terminus of the peptides. The putative cleavage site between the Ig domain and transmembrane domain of Sema4D (Elhabazi et al 2001) is indicated by arrow.
(b) Quantification of inhibitory GABAergic synapse density at DIV14 as defined by overlapping GAD65/GABARγ2 puncta onto hippocampal neurons transfected with either an empty vector (“Control,” n= 47 neurons), an shRNA targeting Sema4D (“RNAi,” n= 35 neurons), overexpression of a myc tagged Sema4D RNAi resistant cDNA alone (“myc-Sema4D OE,” n=42 neurons), overexpression of a myc-tagged Sema4DΔC RNAi resistant cDNA alone (“myc-Sema4DΔC OE,” n=23 neurons), co-transfection of a the myc-Sema4D RNAi resistant cDNA along with an shRNA targeting Sema4D (“myc-Sema4D Rescue,” n= 43 neurons), or co-transfection of the myc-Sema4DΔC RNAi resistant cDNA along with an shRNA targeting Sema4D (“myc-Sema4DΔC Rescue,” n=31 neurons). Asterisk indicates p<0.05 using a univariate ANOVA compared to rescue conditions and control. Other pertinent p values include: p=0.665 control vs. myc-Sema4D rescue and p=0.244 control vs. myc-Sema4DΔC Rescue. Error bars denote standard error (SEM).
(c) (Left) Immunostaining against GAD65 (red) and GABARγ2 (blue) proteins on a stretch of GFP-positive dendrite from a representative neuron transfected with the constructs indicated on far right; overlapping puncta onto the transfected neuron appear white. (Right) GAD65 and GABARγ2 immunostaining in the absence of the GFP signal; overlapping puncta appear magenta. Scale bar =5um.
We reasoned that if Sema4DΔC was able to rescue the decrease in GABAergic synapse density caused by RNAi-mediated knockdown of endogenous Sema4D, then the intracellular C-terminus of Sema4D is not required for its ability to modulate inhibitory synaptogenesis. As a control, we demonstrated that expression of our RNAi resistant myc-Sema4D, which contains the wildtype Sema4D C-terminus, was able to rescue decreased GABAergic synapse density caused by RNAi of endogenous Sema4D (Fig. 1B and 1C). When we expressed our RNAi resistant myc-Sema4DΔC mutant that lacked the intracellular C-terminus, we observed that it also rescued the decrease in GABAergic synapse density caused by RNAi of endogenous Sema4D (Fig. 1B and 1C). Therefore, we conclude that the intracellular C-terminus of Sema4D is not required for Sema4D to mediate GABAergic synapse development in the mammalian hippocampus.
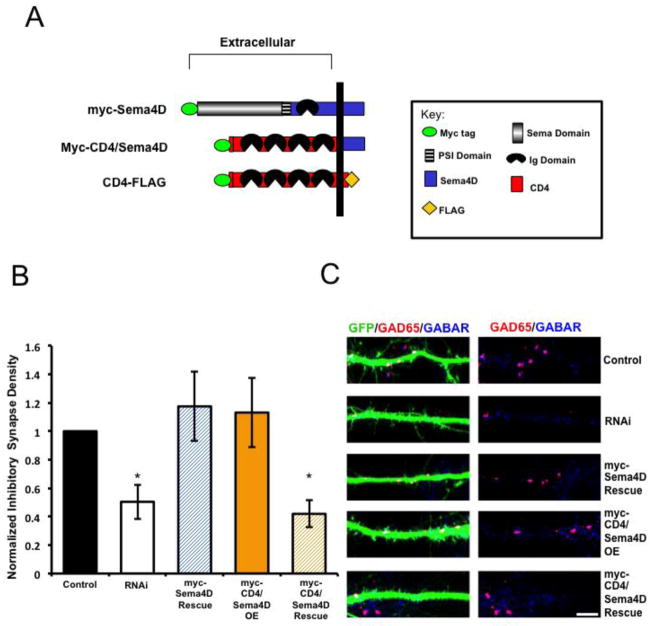
We next asked if the extracellular domain is required for Sema4D to regulate GABAergic synapse formation. To begin, we determined that overexpression of full-length CD4-FLAG on its own does not affect GABAergic synapse density in primary hippocampal cultures (Fig. S1). Further, we determined that expression of CD4-FLAG fails to rescue the Sema4D shRNA-dependent decrease in GABAergic synapse density (Fig. S1). Next, we constructed a chimeric protein consisting of the myc-tagged, extracellular and transmembrane domain of CD4 fused to the C-terminal domain of Sema4D (Fig. 2A). We co-transfected the myc-CD4/Sema4D chimera with our Sema4D shRNA hairpin and observed that myc-CD4/Sema4D failed to rescue the shRNA-dependent decrease in GABAergic synapse density (Figs. 2B and 2C). Therefore, taken together with our findings described in Figure 1, we conclude that the extracellular N-terminal domain of Sema4D is absolutely required to regulate GABAergic synapse formation in the mammalian hippocampus.
Figure 2. The extracellular domain of Sema4D is required for GABAergic synapse formation.
(a) Diagram illustrating the pertinent epitope-tagged Sema4D and CD4 chimeric constructs used in these experiments.
(b) Quantification of inhibitory GABAergic synapse density at DIV14 as defined by overlapping GAD65/GABARγ2 puncta onto hippocampal neurons transfected with either an empty vector (“Control,” n= 21 neurons), an shRNA targeting Sema4D (“RNAi,” n= 19 neurons), co-transfection of a myc-Sema4D RNAi resistant cDNA along with an shRNA targeting Sema4D (“myc-Sema4D Rescue,” n= 19 neurons), overexpression of a myc-tagged CD4/Sema4D RNAi resistant cDNA alone (“myc-CD4/Sema4D OE,” n=16 neurons), or co-transfection of the myc-CD4/Sema4D RNAi resistant cDNA along with an shRNA targeting Sema4D (“myc-CD4/Sema4D Rescue,” n=29 neurons). Asterisk indicates p<0.05 using a univariate ANOVA compared to myc-Sema4D rescue condition and control; Other pertinent p values include: p=0.777 for Control vs. myc-Sema4D Rescue and p=0.700 RNAi vs. myc-CD4/Sema4D Rescue. Error bars denote standard error (SEM).
(c) (Left) Immunostaining against GAD65 (red) and GABARγ2 (blue) proteins on a stretch of GFP-positive dendrite from a representative neuron transfected with the constructs indicated on far right; overlapping puncta onto the transfected neuron appear white. (Right) GAD65 and GABARγ2 immunostaining in the absence of the GFP signal; overlapping puncta appear magenta. Scale bar =5um.
The Class 4 Semaphorin family members (there are 6 mammalian Class 4 homologs) are distinguishable from other classes of Semaphorins both by their sequence homology and protein domain structures. A comparison of primary amino acid sequences of the entire extracellular domain (including both the Sema and Ig domains) of the Class 4 Semaphorins reveals no greater than 44% identity at the amino acid level between any two Class 4 homologs (Sema4C vs. Sema4G). Further, this analysis reveals that Sema4C and Sema4G are similarly related to Sema4D (42.9% and 39% amino acid identities, respectively) and that Sema4D and one of its next closest homologs, Sema4A, are 36.8% identical at the amino acid level throughout the extracellular domain. By contrast, we found that the extracellular domains of Sema4D and a Class 5 Semaphorin family member, Sema5A, are only 28.6% identical at the amino acid level. Given these facts, we sought to determine if other Class 4 homologs, such as Sema4A, could functionally compensate for Sema4D knockdown, and thus rescue the observed decrease in GABAergic synapse density. We found that co-expression of a cDNA encoding the full-length Sema4A protein with the Sema4D shRNA was not able to rescue the decrease in GABAergic synapse density (Fig S2). This suggests that although Class 4 Semaphorins share some amino acid identity throughout the Sema and Ig domains, it is unlikely that other Class 4 Semaphorins are able to compensate for the function of Sema4D in mediating GABAergic synapse formation.
Sema4D is proteolytically cleaved in hippocampal neurons
Our studies indicate that the extracellular domain of Sema4D is both necessary (Fig. 1, 2, and S2) and sufficient (Kuzirian et al. 2013) to mediate GABAergic synapse formation. Therefore, we next asked whether Sema4D was signaling through its extracellular domain as a membrane bound molecule, or via proteolytic cleavage-mediated release of the extracellular domain from the membrane. While the occurrence of Sema4D cleavage as well as its functional relevance has been documented in non-neuronal cell types (Basile et al. 2007; Watanabe et al. 2001) no such observation has been made in the mammalian CNS. Therefore we first sought to determine whether Sema4D is proteolytically cleaved in the rodent hippocampus.
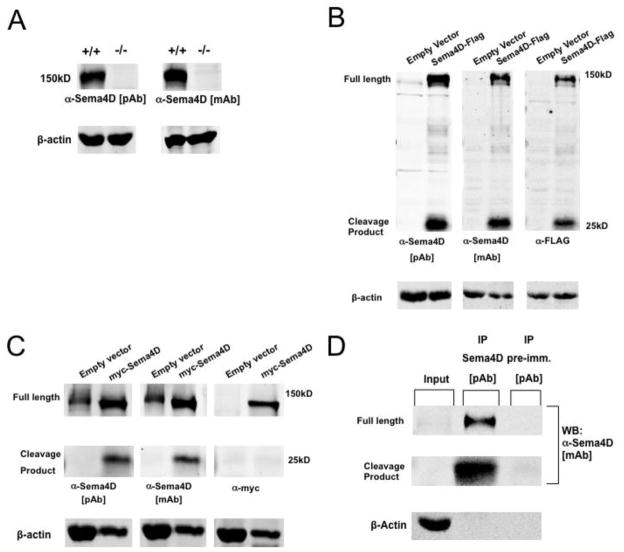
We began by generating a polyclonal antibody that recognizes the C-terminal domain of Sema4D in order to detect endogenous Sema4D protein. We confirmed the specificity of our antibody by Western blot analysis of hippocampal lysates harvested from P8 wild-type mice and Sema4D−/− mice (Shi et al. 2000). As shown in Figure 3A, we detected a band at approximately 150kD in hippocampal lysates from wild-type mice that was absent from hippocampal tissue isolated from Sema4D−/− mice, and is the correct molecular weight to represent full-length Sema4D (Taniguchi et al. 2009; Zhu et al. 2007; Elhabazi et al. 2001; Basile et al. 2007). In addition, we also probed our blots with a commercially available monoclonal Sema4D antibody that also recognizes the intracellular C-terminal domain of Sema4D (Fig. 3A). Consistent with the results using our homemade polyclonal antibody, we detected the presence of a ~150kD band in hippocampal lysates isolated from wild-type mice that was absent in hippocampal lysates isolated from Sema4D−/− mice.
Figure 3. Sema4D is cleaved in hippocampal neurons.
(a) Western Blot analysis of endogenous Sema4D protein expression in hippocampal lysates isolated from either wildtype (+/+) or Sema4D knockout (−/−) mice at postnatal day 8 using both polyclonal (pAb) and monoclonal (mAb) antibodies raised against the C-terminus of Sema4D. Full length Sema4D protein runs at approximately 150kD.
(b) Western blot analysis of lysates from rat hippocampal neurons infected with either empty lentiviral vector or lentivirus containing the Sema4D-FLAG construct, probed with monoclonal and polyclonal antibodies which recognize Sema4D, as well as a monoclonal antibody which recognizes the FLAG epitope. The presence of the Sema4D C-terminal cleavage product was detected at approximately 25kD.
(c) Western blot analysis of lysates from rat hippocampal neurons infected with either empty lentiviral vector or lentivirus containing the myc-Sema4D construct probed with monoclonal and polyclonal antibodies which recognize Sema4D, as well as a monoclonal antibody which recognizes the myc epitope. As expected, the presence of the 25kD Sema4D C-terminal cleavage product was not detected with the monoclonal antibody, which recognizes the myc epitope.
(d) Western blot analysis of hippocampal tissue lysates isolated from postnatal day 1 rat pups and subjected to immunoprecipitation with either a polyclonal antibody which recognizes Sema4D, or the corresponding pre-immune sera. Blots are probed with a monoclonal antibody, which recognizes Sema4D.
To determine if Sema4D is proteolytically cleaved in the rodent hippocampus, we used lentivirus-mediated gene transduction to overexpress a recombinant form of Sema4D containing a FLAG epitope fused to the intracellular, C-terminus (Sema4D-FLAG) or a form of Sema4D containing a myc epitope fused to the extracellular, N-terminus (myc-Sema4D) in primary rat hippocampal cultures (Fig. 1A). We chose this overexpression approach in order to unambiguously label the N- and C-termini of Sema4D as well as to help ensure that the visualization of any cleavage product would not fall below the detection threshold of our antibody. We performed Western blot analysis on cellular lysates of DIV7 cultured hippocampal neurons (6 days post-infection) and probed with our polyclonal Sema4D antibody, the commercially available monoclonal antibody, and an antibody that recognizes the FLAG epitope (Fig. 3B). We detected a ~150kD Sema4D band representing full-length Sema4D and a 25kD band representing a putative cleavage product with all antibodies (Fig. 3B). In addition, we used lentivirus-mediated gene transduction to overexpress myc-Sema4D in primary hippocampal cultures (Fig. 3C). We were also able to detect the 25kD cleavage product using both the monoclonal and polyclonal antibodies that recognize the C-terminus of Sema4D, but as expected we were unable to detect this product using an antibody that recognizes the N-terminal myc epitope. This suggests that the 25kD product represents the intracellular, C-terminal domain of Sema4D after cleavage.
We were unable to detect the endogenous 25kD cleavage product when probing straight lysates of hippocampal cultured neurons (Figs. 3B and 3C, anti-Sema4D antibodies, empty vector lanes at 25kD). Thus, to determine if endogenous Sema4D is proteolytically cleaved in the mammalian hippocampus, we performed immunoprecipitation experiments of Sema4D from hippocampal lysates obtained from postnatal day 1 rat pups (Fig. 3D), a time that corresponds to GABAergic synaptogenesis in the hippocampus (Kuzirian & Paradis 2011; Ben-Ari et al. 2007). When Sema4D was immunoprecipitated using our polyclonal Sema4D antibody, we once again observed a 150kD band representing full-length Sema4D as well as a 25kD C-terminal fragment representing the cleavage product (Fig. 3D). As a negative control, we performed the immunoprecipitation using the pre-immune serum corresponding to our polyclonal antibody and using this serum, we were unable to precipitate either full-length Sema4D or the C-terminal cleavage product. Taken together, these results demonstrate for the first time that Sema4D is cleaved in rat hippocampal neurons both in vitro and in vivo, and suggest the possibility that the extracellular domain of Sema4D signals after cleavage from the cell surface.
Membrane-bound Sema4D can regulate GABAergic synaptogenesis in the mammalian hippocampus
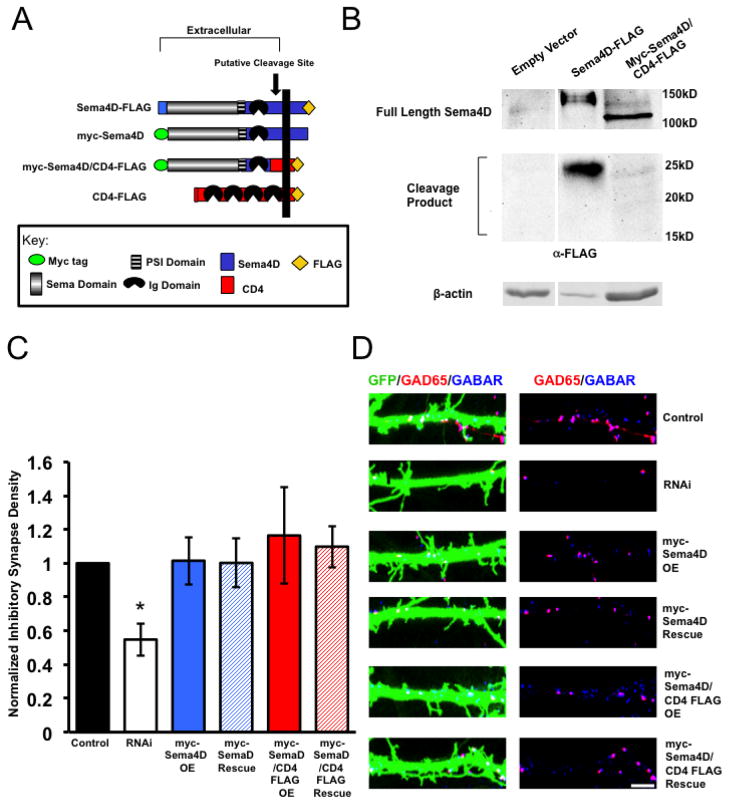
Next, we sought to determine if cleavage of Sema4D was required for its ability to regulate GABAergic synapse formation. To address this issue, we utilized our myc-Sema4D/CD4-FLAG construct, in which the putative cleavage site, transmembrane domain, and C-terminus of Sema4D was replaced with the transmembrane and intracellular domain of CD4-FLAG (Fig. 4A). The cleavage site of Sema4D is reported to lie between the Ig domain and transmembrane domain of Sema4D (Elhabazi et al. 2001); myc-Sema4D/CD4-FLAG lacks this region. To test if the myc-Sema4D/CD4-FLAG protein was proteolytically cleaved, we expressed this construct using lentiviral-mediated gene transduction in primary hippocampal cultures and performed Western blotting assay to monitor the appearance of a C-terminal fragment using the anti-FLAG antibody. As shown in Figure 4B, we observed an approximate 120kD band corresponding to the predicted molecular weight of the full-length myc-Sema4D/CD4-FLAG protein. In addition, we also detected this 120kD band using an anti-myc antibody, which recognizes the N-terminus of myc-Sema4D/CD4-FLAG, leading us to conclude that the 120kD band represents the intact, full length chimeric protein. Furthermore, while we could detect a 25kD C-terminal cleavage product when we overexpressed Sema4D-FLAG, we did not observe a cleavage product of any size between 15kD–25kD when we expressed myc-Sema4D/CD4-FLAG in our primary hippocampal cultures (predicted size is 18kD). From these results, we conclude that myc-Sema4D/CD4-FLAG is not cleaved in cultured hippocampal neurons.
Figure 4. Sema4D promotes GABAergic synaptogenesis as a membrane bound molecule.
(a) Diagram illustrating the pertinent epitope-tagged Sema4D and CD4 chimeric constructs used in these experiments. The myc-Sema4D/CD4-FLAG fusion was constructed such that the CD4 peptide sequence replaces the putative cleavage site between the Ig domain and transmembrane domain of Sema4D (Elhabazi et al 2001).
(b) Western Blot analysis of lysates from rat hippocampal neurons infected with lentivirus containing either an empty vector, Sema4D-Flag, or myc-Sema4D/CD4-FLAG. Western blots were probed with an anti-FLAG antibody, which targets the FLAG epitope on the C-terminus of Sema4D-FLAG and myc-Sema4D/CD4-FLAG. While a 25kD C-terminal cleavage product is detectable in neurons expressing Sema4D-FLAG no cleavage product of any size ranging from 15kD to 25kD was detected in neurons expressing myc-Sema4D/CD4-FLAG.
(c) Quantification of inhibitory GABAergic synapse density at DIV14 as defined by overlapping GAD65/GABARγ2 puncta onto rat hippocampal neurons transfected with either an empty vector (“Control,” n= 18 neurons), an shRNA targeting Sema4D (“RNAi,” n= 29 neurons), overexpression of a myc tagged Sema4D RNAi resistant cDNA alone (“myc-Sema4D OE,” n=23 neurons), overexpression of a myc-tagged Sema4D/CD4-FLAG RNAi resistant cDNA alone (“myc-Sema4D/CD4-FLAG OE,” n=29 neurons), co-transfection of a the myc-Sema4D RNAi resistant cDNA along with an shRNA targeting Sema4D (“myc-Sema4D Rescue,” n= 33 neurons), or co-transfection of the myc-Sema4D/CD4-FLAG RNAi resistant cDNA along with an shRNA targeting Sema4D (“myc-Sema4D/CD4-FLAG Rescue,” n=26 neurons). Asterisk indicates p<0.05 compared to control and Sema4D/CD4-FLAG rescue conditions using a univariate ANOVA; Other pertinent p values include: p=0.875 for Control vs. myc-Sema4D Rescue and p=0.466 for Control vs. myc-Sema4D/CD4-FLAG Rescue. Error bars denote standard error (SEM).
(d) (Left) Immunostaining against GAD65 (red) and GABARγ2 (blue) proteins on a stretch of GFP-positive dendrite from a representative neuron transfected with the constructs indicated on far right; overlapping puncta onto the transfected neuron appear white. (Right) GAD65 and GABARγ2 immunostaining in the absence of the GFP signal; overlapping puncta appear magenta. Scale bar = 5um.
To determine if proteolytic cleavage of Sema4D is required for its ability to regulate GABAergic synapse development, we asked if myc-Sema4D/CD4-FLAG could rescue the decrease in GABAergic synapse density that is observed with Sema4D knockdown by RNAi. We co-transfected myc-Sema4D/CD4-FLAG with our shRNA hairpin and observed that myc-Sema4D/CD4-FLAG rescued the Sema4D shRNA-dependent decrease in GABAergic synapse density (Fig. 4C and 4D). Taken together, this data shows that despite cleavage of Sema4D from the neuronal cell surface, this event is not required for Sema4D to regulate GABAergic synapse development and further, reveals that Sema4D can signal as a membrane-bound molecule.
Sema4D is enriched at the synaptic membrane
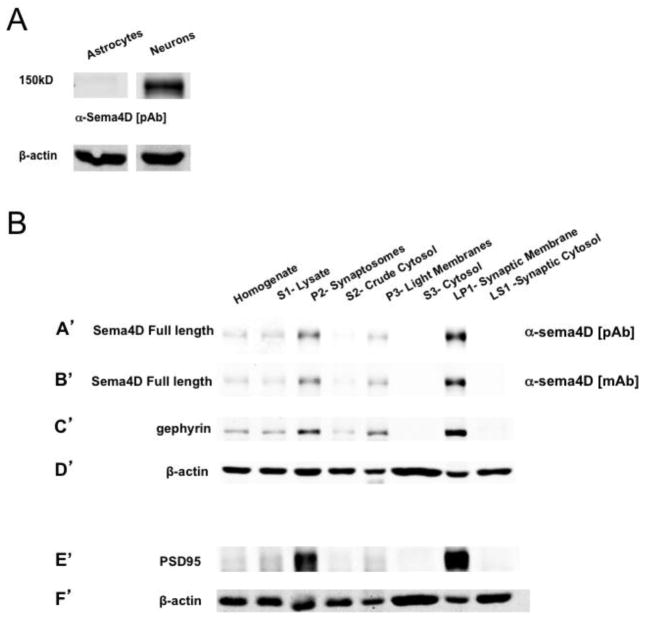
Given that our results point to a mechanism whereby Sema4D signals through its extracellular domain to promote GABAergic synapse formation as a membrane-bound molecule, we sought to determine the subcellular localization of Sema4D. Unfortunately, neither of our anti-Sema4D antibodies were capable of specifically recognizing Sema4D in fixed cells as determined by a comparison between immunostaining of hippocampal neurons isolated from either wild-type or Sema4D−/− mice (S. Paradis, unpublished observations). Therefore, we took a biochemical approach and began by asking if Sema4D was expressed in neurons, astrocytes, or both. To accomplish this, we used Western blotting to compare Sema4D expression in lysates taken from primary cultures of astrocytes to lysates from primary cultures of hippocampal neurons (Fig. 5A). At DIV16, we could not detect any Sema4D protein in astrocyte lysates, suggesting that Sema4D is primarily expressed in neurons in our culture system (Fig. 5A) and consistent with other studies (Furuyama et al. 1996; Masuda et al. 2004).
Figure 5. Sema4D is localized to the synaptic membrane.
(a) Western blot analysis of lysates from DIV16 primary cultures of astrocytes isolated from P1 rat pups or hippocampal neurons isolated from E18 rat pups.
(b) Western blot analysis of fractionated cellular lysates (see Fig S1) taken from hippocampal tissue of postnatal day 12 (P12) rat pups. (A′–D′) Western blot probed with (A′) polyclonal anti-sema4D, (B′) monoclonal anti-sema4D, (C′) anti-gephyrin, (D′) anti-β-actin. (E′–F′) Western blot analysis of same lysates as in A′–D′ probed with (E′) anti-PSD95, (F′) anti-β-actin. The same blot as in A′–D′ could not be reprobed with anti-PSD95 due to similar molecular weights of PSD95 and gephyrin.
Next, we performed a subcellular fractionation assay (see Fig. S3, adapted from (Robbins et al. 2010; Dunah & Standaert 2001)) on hippocampal tissue isolated from P12 rat pups, a time of robust GABAergic synapse formation (Kuzirian & Paradis 2011; Ben-Ari et al. 2007). The synaptic fractions generated in by this protocol contain both pre- and post-synaptic membranes and cytosol from both GABAergic and gluatamatergic synapses (Robbins et al. 2010; Li et al. 2007). Using both the polyclonal and monoclonal Sema4D antibodies characterized earlier, we probed fractionated lysates and analyzed the subcellular distribution of full length, endogenous Sema4D protein by Western blotting (Fig. 5B). We observed a strong enrichment of full length Sema4D in isolated synaptic compartments (P2-synaptosome lanes in Fig. 5BA′ and B′) as well as in isolated synaptic membranes (LP1-synaptosomal membrane lanes in Fig. 5BA′ and B′) compared to non-fractionated, cellular homogenates (Homogenate lanes in Fig. 5BA′ and B′). Correspondingly we observed that Sema4D was nearly absent from cytosolic and synaptic cytosolic fractions, consistent with a membrane localization of Sema4D (S2 crude cytosol and S3 cytosol lanes respectively in (Fig. 5BA′ and B′). As expected, the inhibitory and excitatory postsynaptic scaffolding proteins gephyrin and PSD95 also showed enrichment in both the synaptosome and synaptic membrane fractions but were not found in the cytosolic fractions, confirming the purity of each fractionated subcellular compartment (Fig. 5BC′ and E′). In addition, the cytosolic protein β-actin showed no enrichment in the synaptosome fraction but a strong presence in the cytosolic fraction (Fig. 5BD′ and F′; (Chen et al. 2011)). Taken together, these results provide the first evidence of synaptic localization of a Semaphorin in vivo and demonstrate that endogenous Sema4D is preferentially localized to the synaptic membrane in the rodent hippocampus.
Because we observe that endogenous Sema4D is enriched at the synapse, we examined the subcellular localization of our myc-tagged Sema4D chimeric constructs, which are sufficient to regulate GABAergic synapse development in hippocampal neurons (Fig. 1 and Fig. 4). We performed subcellular fractionation on cultured hippocampal neurons overexpressing a myc-tagged Sema4D protein. Using western blotting, we detected robust expression of full length myc-Sema4D, myc-Sema4DΔC, and myc-Sema4D/CD4-FLAG in both lysates and P2 synaptosome fractions, confirming that these proteins are present at the synapse (Figure S4). However, in contrast to endogenous Sema4D, the myc-tagged Sema4D constructs do not appear to be enriched at the synapse (compare Figs. 5BA′, B′ to Fig. 5C), consistent with our previous observations using immunocytochemistry to detect myc-tagged Sema4D (Paradis et al. 2007). We postulate that the lack of enrichment is due to overexpression from the RSV promoter used to drive the myc-Sema4D constructs. Nonetheless, the fact that all three of the myc-tagged Sema4D constructs are able to rescue the decrease in GABAergic synapse density observed with Sema4D knockdown and do not act as dominant-negative signaling molecules (Figs. 1, 4) indicates that at least a subset of myc-Sema4D, myc-Sema4DΔC, and myc-Sema4D/CD4-FLAG proteins are localized to the appropriate subcellular compartment.
Sema4D is required for proper GABAergic but not glutamatergic synapse development
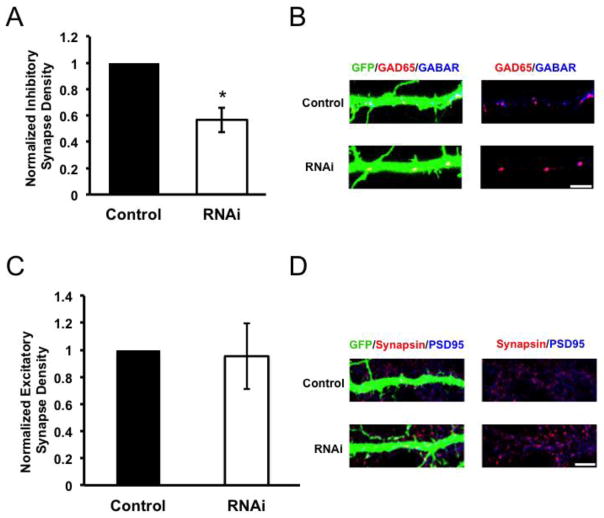
While our biochemical fractionation establishes that Sema4D is localized to synapses (Fig. 5), this technique is unable to distinguish between localization to glutamatergic or GABAergic synapses. Thus, we chose to revisit the possibility that Sema4D may also be required for proper glutamatergic synapse development. To this end, we performed RNAi-mediated knockdown of Sema4D in cultured hippocampal neurons. Neurons transfected with either an empty vector (“Control”) or our Sema4D shRNA hairpin (RNAi) were fixed and stained for either GABAergic synapses or glutamatergic synapses. GABAergic synapses were visualized by immunostaining against the pre-synaptic marker GAD65 and the post-synaptic marker GABAAR γ2, as described earlier. Glutamatergic synapses were visualized by immunostaining against pre-synaptic vesicle-associated protein synapsin I and the excitatory post-synaptic scaffolding protein PSD95. We observed that while RNAi of Sema4D caused a robust decrease in GABAergic synapse density onto hippocampal neurons, in parallel cultures no such effect was observed on glutamatergic synapse density (Figure 6A–D). Therefore, from this data we conclude that Sema4D is selectively required for proper GABAergic synapse development but not glutamatergic synapse development, consistent with previously reported data (Paradis et al. 2007).
Figure 6. Sema4D is required for proper GABAergic but not glutamatergic synapse development.
(a) Quantification of inhibitory GABAergic synapse density at DIV14 as defined by overlapping GAD65/GABARγ2 puncta onto rat hippocampal neurons transfected with either an empty vector (“Control,” n= 47 neurons) or an shRNA targeting Sema4D (“RNAi,” n= 70 neurons). Asterisk indicates p<0.05 compared to control condition using a univariate ANOVA, error bars denote standard error (SEM).
(b) (Left) Immunostaining against GAD65 (red) and GABARγ2 (blue) proteins on a stretch of GFP-positive dendrite from a representative neuron transfected with the constructs indicated on far left; overlapping puncta onto the transfected neuron appear white. (Right) GAD65 and GABARγ2 immunostaining in the absence of the GFP signal; overlapping puncta appear magenta. Scale bar =5μm.
(c) Quantification of excitatory Glutamatergic synapse density at DIV14 as defined by overlapping synapsin/PSD95 puncta onto rat hippocampal neurons transfected with either an empty vector (“Control,” n=37 neurons) or an shRNA targeting Sema4D (“RNAi,” n= 41 neurons). Error bars denote standard error (SEM), p=0.908 between conditions by univariate ANOVA.
(d) (Left) Immunostaining against Synapsin (red) and PSD95 (blue) proteins on a stretch of GFP-positive dendrite; overlapping puncta onto the transfected neuron appear white. (Right) Synapsin and PSD95 immunostaining in the absence of the GFP signal; overlapping puncta appear magenta. Scale bar =5μm.
Discussion
Our study provides novel insight into the molecular mechanism by which Sema4D regulates GABAergic synapse development in the mammalian hippocampus: Sema4D signals as a synaptically localized, membrane-bound molecule through its extracellular N-terminus to promote synapse development. First, we show that a Sema4D protein, which lacks a C-terminus, is sufficient to rescue Sema4D RNAi-mediated decrease in GABAergic synapse development. In addition, a Sema4D protein which lacks its N-terminus fails to rescue the Sema4D RNAi-mediated decrease in GABAergic synapse development. Taken together, these data demonstrate that the extracellular domain of Sema4D is absolutely required to promote GABAergic synapse formation. Interestingly, we also found that although Sema4D is cleaved in neurons, this cleavage event is entirely dispensable for the synaptogenic ability of Sema4D. We also observe that endogenous Sema4D is found at the synapse and preferentially localized to the synaptic membrane in the hippocampus. Although a number of Semaphorins in addition to Sema4D have been implicated in synapse formation, our study is the first to demonstrate that a Semaphorin is enriched at the synapse in vivo. Taken together, these results suggest that Sema4D signals as a membrane-bound, synaptically-localized molecule to promote GABAergic synapse formation.
Our findings also demonstrate that Sema4D is expressed in neurons in the early postnatal hippocampus, a key finding that clarifies potential models of Sema4D action. Other studies have suggested that Sema4D is expressed by mature myelinating oligodendrocytes in the cortex (Moreau-Fauvarque et al. 2003; Taniguchi et al. 2009) and inhibits axon growth in the postnatal CNS (Moreau-Fauvarque et al. 2003). Indeed, a subset of studies have suggested that while Sema4D is expressed in neuronal populations in the embryo (Worzfeld et al. 2004; Masuda et al. 2004; Moreau-Fauvarque et al. 2003; Furuyama et al. 1996), in the postnatal mouse cortex and spinal cord Sema4D mRNA is restricted to mature oligodendrocytes (Worzfeld et al. 2004; Moreau-Fauvarque et al. 2003; Maier et al. 2011). Our analysis of Sema4D expression establishes that at least in the embryonic and early postnatal hippocampus, Sema4D is mostly, if not exclusively, expressed in neurons (Fig. 3 and Fig. 5). In addition, in the hippocampus, mature oligodendrocytes are not detectable until P10 and neuronal myelination does not begin until P17 (Kimoto et al. 2009; Meier et al. 2004). Therefore, because our expression studies encompass ages ranging from neurons cultured from E18 animals (Fig. 3B, C and Fig. 5A), to hippocampal tissue lysates from P1 animals (Fig. 3D) and hippocampal tissue lysates from P12 (Fig. 5B), the possible contribution of Sema4D expression from oligodendrocytes is negligible.
Because our data suggests that proteolytic cleavage of Sema4D is dispensable for its ability to regulate GABAergic synapse formation in the mammalian hippocampus, the question still remains of the functional relevance of Sema4D cleavage in the CNS. In addition to our findings in neurons (Fig. 3), other studies have shown that Sema4D is proteolytically cleaved from the surface of non-neuronal cells by matrix metalloproteinases (Basile et al. 2004; Zhu et al. 2007; Elhabazi et al. 2001). In lymphocytes, proteolytic cleavage of Sema4D is required to induce T-cell activation, suggesting that proteolytic cleavage of Sema4D may be important for its signaling (Wang et al. 2001). Although recent work from our lab demonstrated that addition of soluble, recombinant Sema4D extracellular domain is sufficient to promote GABAergic synapse formation (Kuzirian et al. 2013), it is entirely possible that Sema4D signals as a membrane-bound molecule in vivo.
An intriguing possibility is that proteolytic cleavage of Sema4D and its subsequent release from the membrane serves as an axon guidance mechanism early in development; studies have demonstrated that addition of soluble Sema4D is sufficient to induce growth cone collapse of hippocampal neurons (Swiercz et al. 2002; Oinuma et al. 2010). Later, during the first few postnatal weeks while synapses are forming, membrane-bound Sema4D may instruct synapse formation. The axon guidance protein EphB2 regulates synapse formation in the mammalian hippocampus (Dalva et al. 2000) and also requires proteolytic cleavage by matrix metalloproteinases to mediate axon guidance (K.-T. Lin et al. 2008). Furthermore, netrin family axon-guidance molecules also display a similar developmental pattern by requiring metalloprotease dependent cleavage to regulate axon guidance before modulating synapse formation (J. C. Lin et al. 2003; Galko & Tessier-Lavigne 2000; Kim et al. 2006). Therefore, it is possible that proteolytic cleavage could be acting as a regulatory mechanism to delineate pathfinding signals from synaptogenic signals during nervous system development.
Because Sema4D does not require its intracellular domain to regulate synapse formation, it is possible that Sema4D does not need to interact with intracellular scaffolding proteins nor undergo intracellular post-translational modifications such as phosphorylation to regulate GABAergic synapse development. We postulate that the specificity of Sema4D activity arises via interaction(s) with receptor(s) on the appropriate presynaptic interneuron. Indeed, the high affinity Sema4D receptor PlexinB1 (Tamagnone et al. 1999) is required for the ability of the soluble, extracellular domain of Sema4D to promote GABAergic synapse formation (Kuzirian et al. 2013). Thus, we favor a model whereby Sema4D, localized to the postsynaptic membrane of GABAergic synapses, promotes synapse formation via a trans-synaptic interaction with PlexinB1 expressed on the presynaptic terminal of GABAergic interneurons. In the future, it will be interesting to determine the precise, subcellular localization of the PlexinB1 protein and how Sema4D may be interacting with PlexinB1 to mediate GABAergic synapse formation in the mammalian hippocampus.
Methods and Materials
Sequence alignment
Amino acid sequences comprising the extracellular domain of each mammalian Class 4 Semaphorin was aligned compared, and percent identity determined using the ClustalW alignment tool in MegaAlign software (DNAstar). Percent homology was directly calculated from the alignment. Percent identity between Sema4D and Sema5A was calculated by a pairwise comparison in MegaAlign. Protein Dendogram was generated directly from alignment in MegaAlign software.
Viral Transduction of Neuronal Cultures
Neurons were isolated from Long Evans embryonic day 18 rat pups and cultured at a density of 1M cells/well in 12-well plates that were coated overnight at 37°C with poly-D-lysine (20ug/mL) and laminin (3.4ug/mL). Neurons were plated and grown in Neurobasal media with B27 supplement (Invitrogen) with a complete exchange of media occurring 24 hours after plating. 24–48 hours after plating, neuron cultures were transduced with lentivirus expressing Sema4D constructs (titers ranged between 10^5 and 10^6 plaque forming units (PFU)/ul) using the pFRIG lentiviral expression vector (a gift from Carlos Lois, University of Massachusetts Medical School). Transduced cultures were then lysed in Sample Buffer (6% SDS, bromophenol blue, 1M Tris pH 6.8, glycerol) at day in vitro (DIV) 7 or subjected to membrane fractionation (see below) where indicated.
Immunoprecipitation and Western Blotting
For immunoprecipitation of Sema4D, postanatal day 1 rat pups were sacrificed by decapitation and hippocampal tissue was isolated under a dissection microscope. All samples were prepared on ice and centrifuged at 4°C. Isolated hippocampi were homogenized in 1ml RIPA Buffer (50mM Tris pH 7.5, 150mM NaCl, 1% Triton X-100, 0.1% SDS, 1mM EDTA) and centrifuged at 14,000xg for 20min. Samples were then incubated with either our polyclonal Sema4D antibody or the pre-immune serum (see below) overnight at 4°C with agitation. The next day, protein-G beads (Sigma-Aldrich) were added to samples for 4.5 hours and samples were rotated at 4°C. Samples were then spun down to collect beads and attached protein. Beads were washed 3x with PBS and resuspended in sample buffer to be prepared for Western blotting.
For Western blotting, samples were loaded onto a 12% SDS-PAGE gel and subjected to electrophoresis. The proteins were then transferred to a nitrocellulose membrane, blocked in a 5% milk/TBST solution for 1h, then incubated overnight at 4°C in primary antibody [anti-Sema4D pAb (1:5000), anti-Sema4D mAb (BD Biosciences, 1:500), anti-β-actin (Abcam, 1:5000), anti-gephyrin (Synaptic Systems, 1:250), anti-PSD95 (abcam, 1:500), anti-FLAG (Sigma, 1:100), anti-myc (Sigma, 1:500)]. The following day, the membrane was washed two times for 10 min in TBST then incubated in appropriate secondary antibody [anti-mouse IR800CW (Licor; 1:10000) or anti-rabbit IR680LT (Licor; 1:10000) for 2h at room temperature. After another two 10 minute washes in TBST, the blot was developed using the Odyssey Western Blotting System (Licor). Blots were scanned with laser intensities adjusted to produce signal strength within the dynamic range of detection and avoid saturated pixels.
Antibody Generation
The polyclonal antibody recognizing Sema4D was generated by inoculating rabbits (Covance Research Products) against a GST-Sema4D fusion protein containing amino acids 774–841 (Sema4D GENE ID 20354) from the C-terminus of Sema4D. The anti-serum was verified by screening lysates of hippocampal tissue isolated from either Sema4D+/+ or Sema4D−/− mice (Shi et al. 2000).
Membrane Fractionation Preparations
Biochemical Fractionation was performed as described previously (Robbins et al. 2010; Dunah & Standaert 2001) (also diagrammed in Supplementary Fig. 3). Briefly, postnatal day 12 rats were sacrificed by decapitation and hippocampal tissue was isolated under a dissection microscope. All samples were prepared on ice and centrifuged at 4°C. Hippocampal tissue was homogenized in 2ml Sucrose Buffer (320mM Sucrose with 10mM HEPES pH 7.4) and centrifuged at 800xg. The resulting supernatant (S1) was centrifuged again at 10,000 xg to recover a P2 synaptosomal pellet and a crude cytosolic supernatant (S2). To recover the light membrane fraction (P3) and cytosolic fraction (S3), S2 was centrifuged at 163,000xg for 2h. The resulting P3 pellet and S3 supernatant were collected as the light membrane and cytosolic fraction respectively. To recover the LP1 (synaptic membrane) and LS1 (synaptic cytosol), the washed P2 pellet was hypo-osmotically lysed and centrifuged at 26,000xg. The resulting LP1 pellet and LS1 supernatant were collected as the synaptic membrane and synaptic cytosolic fractions respectively. Samples were diluted in sample buffer and analyzed by Western blotting. Equal loading of samples in each lane was determined before loading by a Bradford Assay (Bio-Rad Protein Assay).
Neuronal Cultures and Transfection for Synapse Density Assays
Astrocyte feeder layers were prepared in the following manner. Astrocytes were isolated from P1 rat cortex by plating disassociated cells at low density in DMEM + 10% FBS on uncoated 10cm tissue culture dishes. The media was completely exchanged one day and five days after plating to remove cells that did not adhere to the bottom of the dish. After 7–10 days when the glia had become confluent, the glia were trypsinized and plated at low density on 12mm glass coverslips coated overnight at 4°C with poly-D-lysine (20ug/mL) and laminin (3.4ug/mL) in 24 well plates. Dissociated hippocampal neurons from E18 rats were plated at a density of ~100,000 cells/well onto the monolayer of confluent glia grown on the 12 mm glass coverslips in Neurobasal media with B27 supplement (Invitrogen). AraC (Sigma) was added to a final concentration of 5uM before, during or 24 hours after plating. At DIV4, neurons were transfected by the calcium phosphate method (Xia et al. 1996) with 500ng GFP plasmid/well. For control conditions, neurons were also tranfected with empty pSuper vector at 15ng/well or empty pCMV vector at 150ng/well. For RNAi conditions, neurons were transfected with pSuper-shRNA plamids containing an shRNA against Sema4D [see (Paradis et al. 2007)] at 15ng/well. For overexpression and rescue conditions, neurons were transfected with the RNAi-resistant Sema4D cDNA, the corresponding Sema4D/CD4 chimera construct, full-length CD4, or full-length Sema4A at 150ng/well alone or in combination with Sema4D shRNA.
Quantification of Imaging
Image acquisition and quantification were performed in a blinded manner. Twelve-bit images of neurons were acquired on an Olympus Fluoview 300 confocal microscope using a 60x objective. Within each experiment, images were acquired with identical settings for laser power, detector gain, and amplifier offset. Images were acquired as a z-stack (5–20 optical sections and 0.5 um step size). Maximum intensity projections were created from each stack.
For GAD65/GABAAR γ2 experiments, synapse density was quantified as the overlap of GFP, α-GAD65 (Millipore, 1:1000) and α-GABAAR γ2 (Millipore, 1:100) staining using MetaMorph image analysis software. For each experiment, the threshold for the GAD65 and GABARγ2 was determined visually using an image of a control neuron. The threshold was chosen such that all punctate staining would be included in the analysis. This threshold was then applied across all images within the experiment. The threshold for GFP was determined independently for each image. A binary mask including all pixels above the threshold was created for all channels for each image and the “logical and” function was used to determine regions of triple co-localization at least one pixel in size. To calculate synapse density, this number was divided by the area of the neuron as measured using the GFP mask minus the cell body. Approximately 5–20 images from 2–3 separate coverslips were acquired and analyzed for each condition within an experiment for a total of at least three experiments. For synapsin/PSD95 experiments, synapse density was quantified as the overlap of GFP, α-synapsin (Millipore, 1:500) and α-PSD95 (Neuromab, 1:500) staining and analysis was performed using the same method as done for GAD65/GABAAR γ2 experiments.
Synapse density values within each experiment were normalized to account for the variation in antibody staining and neuronal density from experiment to experiment. Within an experiment, the average synapse density value was obtained for the control and for experimental conditions. The normalized value of each experiment is the average experimental value divided by the average control value. See (Paradis et al. 2007) for details of this conversion. To assess statistical significance, a univariate ANOVA was performed on the compiled data. In rescue experiments where data samples had unequal variance, planned contrast was performed following a univariate ANOVA to assess statistical significance. All statistical analysis was performed using SPSS. Error bars denote standard error.
Supplementary Material
Acknowledgments
We thank Dr. Michael Marr 2nd at Brandeis University for technical assistance, assistance with data interpretation, and critical reading of the manuscript. We also thank Dr. Atsushi Kumanogoh from Osaka University for generously providing us with Sema4D and Sema4A cDNA constructs. This work was supported by National Institutes of Health Grant (to S. Paradis) R01NS065856, Research Grant No. 5-FY09-125 from the March of Dimes Foundation (S.P), P30NS45713 for Core Facilities for Neurobiology at Brandeis University, as well as funding provided by Richard and Susan Smith Family Foundation, Chestnut Hill, MA (S.P).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Basile JR, et al. Class IV semaphorins promote angiogenesis by stimulating Rho-initiated pathways through plexin-B. Cancer research. 2004;64(15):5212–5224. doi: 10.1158/0008-5472.CAN-04-0126. [DOI] [PubMed] [Google Scholar]
- Basile JR, et al. MT1-MMP controls tumor-induced angiogenesis through the release of semaphorin 4D. The Journal of biological chemistry. 2007;282(9):6899–6905. doi: 10.1074/jbc.M609570200. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y, et al. GABA: A Pioneer Transmitter That Excites Immature Neurons and Generates Primitive Oscillations. Physiological Reviews. 2007;87(4):1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- Chen CY, et al. Sarm1, a negative regulator of innate immunity, interacts with syndecan-2 and regulates neuronal morphology. The Journal of Cell Biology. 2011;193(4):769–784. doi: 10.1083/jcb.201008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Engelman H. Control of Excitatory and Inhibitory Synapse Formation by Neuroligins. Science (New York, NY) 2005 doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- Dalva MB, et al. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103(6):945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nature reviews Neuroscience. 2007;8(3):206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunah AW, Standaert DG. Dopamine D1 receptor-dependent trafficking of striatal NMDA glutamate receptors to the postsynaptic membrane. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2001;21(15):5546–5558. doi: 10.1523/JNEUROSCI.21-15-05546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhabazi A, et al. Biological activity of soluble CD100. I. The extracellular region of CD100 is released from the surface of T lymphocytes by regulated proteolysis. Journal of immunology (Baltimore, Md: 1950) 2001;166(7):4341–4347. doi: 10.4049/jimmunol.166.7.4341. [DOI] [PubMed] [Google Scholar]
- Furuyama T, et al. Identification of a novel transmembrane semaphorin expressed on lymphocytes. The Journal of biological chemistry. 1996;271(52):33376–33381. doi: 10.1074/jbc.271.52.33376. [DOI] [PubMed] [Google Scholar]
- Galko MJ, Tessier-Lavigne M. Function of an axonal chemoattractant modulated by metalloprotease activity. Science (New York, NY) 2000;289(5483):1365–1367. doi: 10.1126/science.289.5483.1365. [DOI] [PubMed] [Google Scholar]
- Hall KT, et al. Human CD100, a novel leukocyte semaphorin that promotes B-cell aggregation and differentiation. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(21):11780–11785. doi: 10.1073/pnas.93.21.11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, et al. NGL family PSD-95–interacting adhesion molecules regulate excitatory synapse formation. Nature neuroscience. 2006;9(10):1294–1301. doi: 10.1038/nn1763. [DOI] [PubMed] [Google Scholar]
- Kimoto H, et al. Alterations of Glial Cells in the Mouse Hippocampus During Postnatal Development. Cellular and Molecular Neurobiology. 2009;29(8):1181–1189. doi: 10.1007/s10571-009-9412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75(7):1389–1399. doi: 10.1016/0092-8674(93)90625-z. [DOI] [PubMed] [Google Scholar]
- Kumanogoh A, Kikutani H. Biological functions and signaling of a transmembrane semaphorin, CD100/Sema4D. Cellular and Molecular Life Sciences (CMLS) 2004;61(3):292–300. doi: 10.1007/s00018-003-3257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumanogoh A, et al. Class IV semaphorin Sema4A enhances T-cell activation and interacts with Tim-2. Nature. 2002;419(6907):629–633. doi: 10.1038/nature01037. [DOI] [PubMed] [Google Scholar]
- Kuzirian MS, Paradis S. Emerging themes in GABAergic synapse development. Progress in neurobiology. 2011;95(1):68–87. doi: 10.1016/j.pneurobio.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzirian MS, et al. The Class 4 Semaphorin Sema4D Promotes the Rapid Assembly of GABAergic Synapses in Rodent Hippocampus. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33(21):8961–8973. doi: 10.1523/JNEUROSCI.0989-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. Two pools of Triton X-100-insoluble GABAA receptors are present in the brain, one associated to lipid rafts and another one to the post-synaptic GABAergic complex. Journal of neurochemistry. 2007;102(4):1329–1345. doi: 10.1111/j.1471-4159.2007.04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, et al. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nature neuroscience. 2003;6(12):1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- Lin K-T, et al. Ephrin-B2-induced cleavage of EphB2 receptor is mediated by matrix metalloproteinases to trigger cell repulsion. The Journal of biological chemistry. 2008;283(43):28969–28979. doi: 10.1074/jbc.M804401200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love CA, et al. The ligand-binding face of the semaphorins revealed by the high-resolution crystal structure of SEMA4D. Nature structural biology. 2003;10(10):843–848. doi: 10.1038/nsb977. [DOI] [PubMed] [Google Scholar]
- Maier V, et al. Semaphorin 4C and 4G are ligands of Plexin-B2 required in cerebellar development. Molecular and Cellular Neuroscience. 2011;46(2):419–431. doi: 10.1016/j.mcn.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, et al. Sema4D stimulates axonal outgrowth of embryonic DRG sensory neurones. Genes to Cells. 2004;9(9):821–829. doi: 10.1111/j.1365-2443.2004.00766.x. [DOI] [PubMed] [Google Scholar]
- Meier S, et al. Myelination in the hippocampus during development and following lesion. Cellular and Molecular Life Sciences (CMLS) 2004;61(9):1082–1094. doi: 10.1007/s00018-004-3469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau-Fauvarque C, et al. The transmembrane semaphorin Sema4D/CD100, an inhibitor of axonal growth, is expressed on oligodendrocytes and upregulated after CNS lesion. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23(27):9229–9239. doi: 10.1523/JNEUROSCI.23-27-09229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita A, et al. Regulation of dendritic branching and spine maturation by semaphorin3A-Fyn signaling. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2006;26(11):2971–2980. doi: 10.1523/JNEUROSCI.5453-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TP, et al. Semaphorin 5B mediates synapse elimination in hippocampal neurons. Neural Development. 2009;4(1):18. doi: 10.1186/1749-8104-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oinuma I, et al. Semaphorin 4D/Plexin-B1 stimulates PTEN activity through R-Ras GTPase-activating protein activity, inducing growth cone collapse in hippocampal neurons. The Journal of biological chemistry. 2010;285(36):28200–28209. doi: 10.1074/jbc.M110.147546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, et al. An RNAi-Based Approach Identifies Molecules Required for Glutamatergic and GABAergic Synapse Development. Neuron. 2007;53(2):217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzari Pietro, et al. Control of cortical GABA circuitry development by Nrg1 and ErbB4 signalling. Nature. 2010;464(7293):1376–1380. doi: 10.1038/nature08928. Available at: http://www.nature.com/nature/journal/v464/n7293/abs/nature08928.html. [DOI] [PubMed] [Google Scholar]
- Robbins EM, et al. SynCAM 1 Adhesion Dynamically Regulates Synapse Number and Impacts Plasticity and Learning. Neuron. 2010;68(5):894–906. doi: 10.1016/j.neuron.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, et al. The class IV semaphorin CD100 plays nonredundant roles in the immune system: defective B and T cell activation in CD100-deficient mice. Immunity. 2000;13(5):633–642. doi: 10.1016/s1074-7613(00)00063-7. [DOI] [PubMed] [Google Scholar]
- Shin H, et al. Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. The Journal of biological chemistry. 2003;278(13):11393–11401. doi: 10.1074/jbc.M211874200. [DOI] [PubMed] [Google Scholar]
- Swiercz JM, et al. Plexin-B1 Directly Interacts with PDZ-RhoGEF/LARG to Regulate RhoA and Growth Cone Morphology. Neuron. 2002;35(1):51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- Tamagnone L, et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99(1):71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, et al. Sema4D deficiency results in an increase in the number of oligodendrocytes in healthy and injured mouse brains. Journal of neuroscience research. 2009;87(13):2833–2841. doi: 10.1002/jnr.22124. [DOI] [PubMed] [Google Scholar]
- Terauchi A, et al. Distinct FGFs promote differentiation of excitatory and inhibitory synapses. Nature. 2010;465(7299):783–787. doi: 10.1038/nature09041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TS, et al. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 2009;462(7276):1065–1069. doi: 10.1038/nature08628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario-Abejón C, et al. Neurotrophins induce formation of functional excitatory and inhibitory synapses between cultured hippocampal neurons. The Journal of Neuroscience. 1998;18(18):7256–7271. doi: 10.1523/JNEUROSCI.18-18-07256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Functional soluble CD100/Sema4D released from activated lymphocytes: possible role in normal and pathologic immune responses. Blood. 2001;97(11):3498–3504. doi: 10.1182/blood.v97.11.3498. [DOI] [PubMed] [Google Scholar]
- Watanabe C, et al. Enhanced immune responses in transgenic mice expressing a truncated form of the lymphocyte semaphorin CD100. Journal of immunology (Baltimore, Md: 1950) 2001;167(8):4321–4328. doi: 10.4049/jimmunol.167.8.4321. [DOI] [PubMed] [Google Scholar]
- Worzfeld T, et al. Plexin-B family members demonstrate non-redundant expression patterns in the developing mouse nervous system: an anatomical basis for morphogenetic effects of Sema4D during development. The European journal of neuroscience. 2004;19(10):2622–2632. doi: 10.1111/j.0953-816X.2004.03401.x. [DOI] [PubMed] [Google Scholar]
- Xia Z, et al. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1996;16(17):5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, et al. Regulation of spine development by semaphorin3A through cyclin-dependent kinase 5 phosphorylation of collapsin response mediator protein 1. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27(46):12546–12554. doi: 10.1523/JNEUROSCI.3463-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, et al. Regulated surface expression and shedding support a dual role for semaphorin 4D in platelet responses to vascular injury. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(5):1621–1626. doi: 10.1073/pnas.0606344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.