Abstract
Though xenografts are used extensively for drug development in breast cancer, how well xenografts reflect the breadth of primary breast tumor subtypes has not been well characterized. Moreover, few studies have compared the gene expression of xenograft tumors to the primary tumors from which they were derived. Here we investigate whether the ability of human breast tumors (n = 20) to create xenografts in immune-deficient mice is associated with breast cancer immunohistochemical (IHC) and intrinsic subtype. We also characterize how precisely the gene expression of xenografts reprises that of parent breast tumors, using hierarchical clustering and other correlation-based techniques applied to Agilent 44K gene expression data from 16 samples including four matched primary tumor-xenograft pairs. Of the breast tumors studied, 25 % (5/20) generated xenografts. Receptor and intrinsic subtype were significant predictors of xenograft success, with all (4/4) triple-negative (TN) tumors and no (0/12) HR+Her2– tumors forming xenografts (P = 0.0005). Tumor cell expression of ALDH1, a stem cell marker, trended toward successful engraftment (P = 0.14), though CDK5/6, a basal marker, did not. Though hierarchical clustering across the 500 most variable genes segregated human breast tumors from xenograft tumors, when clustering was performed over the PAM50 gene set the primary tumor-xenograft pairs clustered together, with all IHC subtypes clustered in distinct groups. Greater similarity between primary tumor-xenograft pairs relative to random pairings was confirmed by calculation of the within-pair between-pair scatter ratio (WPBPSR) distribution (P = 0.0269), though there was a shift in the xenografts toward more aggressive features including higher proliferation scores relative to the primary. Triple-negative breast tumors demonstrate superior ability to create xenografts compared to HR+ tumors, which may reflect higher proliferation or relatively stroma-independent growth of this subtype. Xenograft tumors’ gene expression faithfully resembles that of their parent tumors, yet also demonstrates a shift toward more aggressive molecular features.
Keywords: Mouse model, Breast cancer, Xenograft, Receptor subtype, Intrinsic subtype, ALDH1, CDK5/6, PAM50
Introduction
Xenografts are experimental models created by transplanting tissue from one organism into an organism of another species that have been used to study cancer pathogenesis and drug development for several decades [1]. The ability of a tumor to successfully engraft as a xenograft in a host organism reflects the ability of its cells to survive in a new microenvironment, as engrafted tumors must establish a relationship with the host stroma, recruit a blood supply, and locally invade host tissue.
Human breast tumors transplanted into immune-suppressed mice have historically low rates of engraftment relative to other tumor types, on the order of 10–50 % [2, 3]. These rates have been modestly improved by the addition of matrigel [4] and estrogen supplementation, and the development of alternative mouse host models [5].
Xenografts are used extensively for drug development and validation in breast cancer [6]. Though a growing number of studies have examined how faithfully xenografts recapitulate the tumors from which they were derived [7–11], the fidelity of the xenograft models relative to primary tumors remains controversial [12]. Moreover, until recently xenografts were rarely developed from primary tumors; instead, cell lines or samples from metastatic sites such as pleural effusions are preferentially used due to ease of access to such samples and increased engraftment rate, respectively.
Breast cancer is a heterogeneous disease with well-characterized receptor subtypes, defined by estrogen receptor (ER), progesterone receptor (PR), and Her-2/neu (Her-2) immunohistochemical (IHC) expression, which have both predictive and prognostic implications. In particular, patients with tumors of the TN subtype (ER, PR, and Her-2 negative) do not benefit from targeted therapies directed at ER and Her-2, and are at greater risk of early recurrence and death than are patients with tumors that are ER or PR positive (hormone receptor positive, HR+). HR+ patients tend to recur later if at all, with 50 % of their recurrences occurring more than 5 years from diagnosis [13].
Over the last decade, gene expression analysis using DNA microarrays has defined a set of molecular subtypes that is overlapping but distinct from the IHC receptor subtypes [14]. These subtypes were initially categorized by their expression of genes typical of cells of the lumen or basal layer of normal breast tissue and include Luminal A, Luminal B, Basal, Her2, and Normal-like subtypes. In addition to ER, PR, and Her-2, expression of other IHC tumor markers also defines overlapping but distinct stratifications of breast tumors from the molecular sub-types. Tumor cell expression of aldehyde dehydrogenase 1 (ALDH1), a putative marker of cancer stem cells, is significantly correlated with basal and Her-2 molecular sub-types [15] and a poor prognostic sign [16].
The goal of this study was to create xenografts from human breast primary tumors in immune-deficient mice and to characterize how faithfully the gene expression of xenografts recapitulates that of the breast tumors from which they were created by gene expression analysis. In addition, we were interested in assessing the relationship between IHC and intrinsic subtype, as well as stem cell and basal IHC markers such as ALDH1 positivity and CK5/6 staining, with xenograft formation.
Materials and methods
Tumor collection and xenograft formation
Breast tumor specimens were collected from 20 breast cancer patients who provided consent under a protocol approved by the University of California, San Francisco, CA, institutional review board between 2005 and 2007. Breast tumor fragments were implanted subcutaneously in 6–8-week-old NOD/SCID mice (Harlan Laboratories, Madison, WI): tumor specimens were minced with scissors into small (2 mm3) fragments and implanted s.c. using a 10-gauge Trochar needle through a small incision on the animal's right dorsal flank. Recipient NOD/SCID mice were anesthetized by i.p. injection of a ketamine (75 mg/kg)–xylazine (5 mg/kg) mixture. 17β-estradiol 0.36 mg pellets were implanted subcutaneously in recipient mice. Of note, 2 of the 20 tumor fragments were frozen prior to implantation. Tumors were assessed for growth by visual inspection over 1–3 months. Once engrafted, successful solid tumor xenografts were serially passaged using the same technique [17]. Average doubling time of the tumors in vivo was ~ 11 days. Passage two or three was used for the gene expression analysis.
Clinical characteristics
Patient history, physical examination, and clinical pathologic data (including tumor grade, size, lymph node status, IHC estrogen, progesterone, and Her-2/neu expression data) were retrieved from patient medical records and maintained, de-identified, in password-protected databases.
Gene expression arrays
RNA was isolated from frozen tumor samples with the RNAdvance Tissue kit following manufacturer's instructions (Agencourt Bioscience Corporation, Beverly, MA). Sample preparation, labeling, and array hybridizations were performed according to standard protocols from the UCSF Shared Microarray Core Facilities and Agilent Technologies (http://www.arrays.ucsf.edu and http://www.agilent.com). Total RNA quality was assessed using a Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA was amplified and labeled with Cy3-CTP using the Agilent low-RNA input fluorescent linear amplification kits following the manufacturer's protocol. Amplifications were repeated using the Sigma whole transcriptome amplification kits following the manufacturer's protocol (Sigma-Aldrich, St Louis, MO), and subsequent Cy3-CTP labeling was performed using NimbleGen one-color labeling kits (Roche-NimbleGen Inc, Madison, WI). The size distribution and quantity of the amplified product were assessed using the Agilent 2100 Bioanalyzer and the Nanodrop ND-8000 (Nanodrop Technologies, Inc., Wilmington, DE); the labeled DNA was assessed using the Nandrop 8000, and equal amounts of Cy3 labeled target were hybridized to Agilent human whole genome 4 × 44K Ink-jet arrays. Hybridizations were performed for 14 h, according to the manufacturer's protocol. Arrays were scanned using the Agilent microarray scanner, and raw signal intensities were extracted with Feature Extraction v10.1 software.
Statistical analysis
Raw expression data were normalized by quantile normalizing [18] background adjusted data (normal-exponential convolution model [19]) and then log2 transforming. All tumor samples in the study were compared by unsupervised hierarchical clustering over the 500 most variable genes. Unsupervised hierarchical clustering was also performed over the PAM50 gene set, and tumors were classified into the five intrinsic subtypes (Luminal A, Luminal B, Basal, Her2, or Normal-like) and risk of relapse (ROR) and proliferation scores computed as described [20]. Relationships between tumor profiles were also visualized using multidimensional scaling [21] implemented in MATLAB© (http://www.mathworks.com/). Xenograft-primary pairs were compared to all other possible pairs by the within-pair between-pair scatter ratio (WPBPSR), the ratio of the dissimilarities between matched pairs to the dissimilarities between randomly matched tumors [22], and by correlation analysis. The statistical significance of the WPBPSR was determined by a permutation test in which a WPBPSR was computed for each possible random pairing between xenografts and primaries, followed by a t test to test for significance of the true WPBSPSR relative to the random pairs distribution. Similarly, a t test was applied to the Pearson correlation coefficients of xenograft-primary pairs compared to the correlation coefficients of all random pairs of xenografts and xenograft-generating primary tumors. Association between receptor or intrinsic subtype and xenograft-forming capability was performed using Fisher's exact method.
Immunohistochemistry
Primary breast tumors were stored in paraffin and cut in 5-μm sections for IHC studies. For ALDH1 and CK5/6, the sections were deparaffinized in xylene, then rehydrated in graded alcohol. The sections were incubated in citrate buffer pH 6.0 (Dakocytomation) for antigen enhancement. For ALDH1, ALDH1 antibody (clone 44, BD biosciences) was used at a 1:40 dilution and incubated for 90 min. PE-conjugated secondary antibody (red color in the staining) was used at a 1:250 dilution and incubated for 20 min and cover-slipped. For CK5/6, CK 5/6 antibody (clone D5/16 D4, Chemicon) was used at a 1:50 dilution and incubated overnight. Biotinylated secondary antibody was used for 30 min. Sections were examined with a fluorescent microscope (Olympus FV-500 Confocal). Only those cases showing >5 % tumor cell positivity were regarded as CK5/6 positive. Sections were considered positive for ALDH1 staining if a few tumor cells were positive in a given area of tumor section, per published method [16]. Association analysis between IHC or other markers and xenograft formation capability were performed using Fisher's exact method.
Results
Engraftment of breast tumors
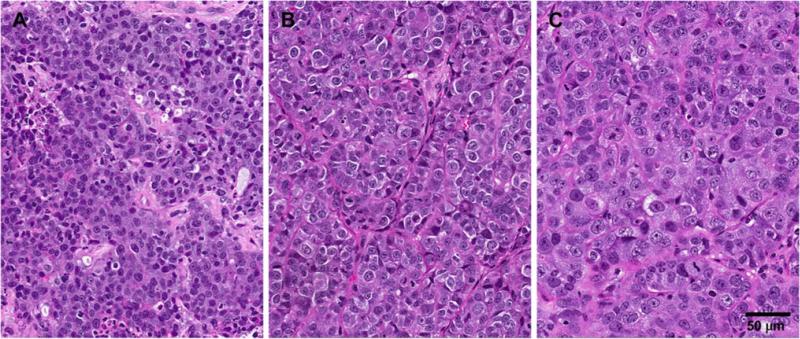
Twenty tumors of breast origin were included in this study, including 18 primary invasive ductal carcincomas, 1 axillary metastasis, and 1 ductal carcinoma in situ, comedo type presenting as an 8-cm mass. Of the invasive ductal carcinomas, two contained a DCIS component and one was mixed with invasive lobular carcinoma. Tumors were all larger than 3 cm, were of mixed receptor type and grade (see Table 1), and were all surgically excised at a single institution, UCSF, where they were sectioned and sent to OncoMed for xenograft development. 25 % (5/20) of the human breast tumors in this study successfully established stable xenografts in NOD/SCID mice. This observed engraftment rate is consistent with other publications reporting successful stable engraftment of 10–50 % of human breast tumors [2, 3]. Tumors were passaged in vivo from one generation of mice to the next without any intervening cell culture. The morphological characteristics were maintained between each passage (Fig. 1).
Table 1.
Primary breast tumor characteristics and their association with xenograft formation
| Primary tumor characteristic | Prevalence n/total (%) | Xenograft + n/total (%) | Xenograft – n/total (%) | Xenograft formation P value (Fisher) |
|---|---|---|---|---|
| HR+/Her2– | 12/20 (60) | 0/12 (0) | 12/12 (100) | 0.0036** |
| HR–/Her2+ | 4/20 (20) | 1/4 (25) | 3/4 (75) | 1 |
| TN (HR–/Her2–) | 4/20 (20) | 4/4 (100) | 0/4 (0) | 0.001** |
| Luminal A | 5/10 (50) | 1/5 (20) | 4/5 (80) | 0.524 |
| Basal | 3/10 (30) | 3/3 (100) | 0/3 (0) | 0.033** |
| ALDH1+ | 3/20 (15) | 2/3 (66.6) | 1/3 (33.3) | 0.14* |
| CK5/6+ | 7/19 (6.3) | 2/7 (28.6) | 5/7 (71.4) | 1 |
| Node positive | 11/20 (55) | 2/11 (18.2) | 9/11 (81.8) | 0.617 |
| Higher grade (II/III) | 14/17 (82.4) | 5/14 (35.7) | 9/14 (64.3) | 0.515 |
| Distant recurrence | 8/20 (40) | 3/8 (37.5) | 5/8 (62.5) | 0.347 |
Primary breast tumor characteristics and their association with xenograft formation. RNA of sufficient quality was isolated and hybridized to expression arrays with success from 10/20 tumors included in this study.
indicates statistically significant associations between tumor characteristics and xenograft formation
indicates a non-significant trend
Fig. 1.
Hematoxylin and eosin stain of B34 primary tumor (a), xenograft passage 1 (b), and xenograft passage 2 (c) tumors. The morphological characteristics of this tumor were maintained between each passage. Primary tumor consisted of moderately poorly differentiated tumor cells with small regions of human stroma (a). Passaged tumors consisted of highly proliferative moderately poorly differentiated tumor cells with well-vascularized murine stroma (b, c)
IHC and intrinsic receptor subtype predicts xenograft engraftment
All human breast tumors were evaluated for receptor subtype, defined by hormone receptor (HR) status (ER or PR IHC expression) and Her-2 status by IHC expression. In this study, 60 % (12/20) of the tumors were HR+/Her2–, 20 % (4/20) were HR–/Her2+, and 20 % (4/20) were TN, roughly reflecting the incidence of these subtypes in the general breast cancer population. Associations between subtype and xenograft formation were assessed using Fisher's exact method. This analysis identified receptor subtype of the original tumor to be a significant predictor of ability to make a xenograft (P = 0.00053). No HR+/Her2– tumors created xenografts (0/12; P = 0.0036), whereas 100 % of TN tumors created xenografts (4/4; P = 0.001). RNA of sufficient quality was isolated and hybridized to expression arrays with success from 10/20 tumors included in this study. On the basis of the expression data, each tumor was also classified as belonging to one of the five intrinsic subtypes (Luminal A, Luminal B, Basal, Her2, or Normal-like) by gene expression analysis as described in Materials and Methods section and [20]. The majority of tumors (5/10) were Luminal A subtype, and no tumor in this study was classified as Luminal B subtype. Interestingly, though intrinsic subtype also correlates with engraftment, the association was weaker than that for receptor subtype (P = 0.071). Basal subtype significantly associated with xenograft formation (3/3; P = 0.033), but Luminal A subtype assignment less clearly precluded engraftment (1/5; P = 0.524). Thus, among the small set of tumors in this study, TN receptors and basal classification predicted engraftment, HR+/Her2– receptors precluded engraftment despite estrogen supplementation, and an “outlier” HR–/Her2+ tumor classified as Luminal A generated a xenograft.
Association between grade, nodal status, and outcome with successful engraftment
Among the tumors in this study, 18 % (3/17) were low grade (grade I), 35 % (6/17) were intermediate grade (grade II), and 47 % (8/17) were high grade. Three tumors, including the two DCIS cases, did not have grade data. Tumors that were intermediate or high grade (grade II or III) were more likely to create xenografts than were low-grade (I) tumors, though this trend was not statistically significant (35.7 % high grade (5/14) vs. no low grade (0/3); P = 0.515). Other variables related to aggressive clinical behavior that one might expect to associate with the capacity of tumors to engraft, such as positive node status or eventual distant recurrence, did not significantly associate with engraftment (P = 0.617 and P = 0.347, respectively), though we observe that 3/5 of patients whose tumors formed xenografts eventually developed breast cancer recurrence and died of their disease (Table 1).
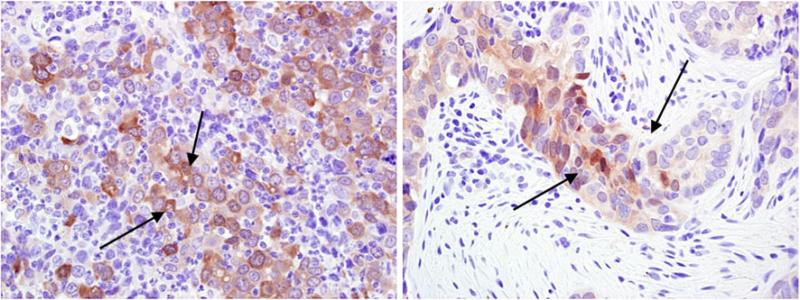
Stem cell marker ALDH1 trends toward association with xenograft establishment
Tumor cell ALDH1 staining, a marker of cancer “stem cell-ness” expected to predict engraftment success, was seen in 15 % (3/20) of tumors in this study (Fig. 2). ALDH1 positive tumors trended toward successful xenotransplantation, as 66 % (2/3) of tumors that were ALDH1 + formed xenografts, whereas only 17.6 % (3/17) of ALDH1 tumors formed xenografts, though this association was not statistically significant (P = 0.14) (Table 1). CK5/6 staining, reportedly associated with basal-like breast cancer, was seen in 36.8 % (7/19) of primary tumors. This marker did not associate with xenograft formation (P = 1), inconsistent with the observation that basal subtype as determined by gene expression analysis yielded a statistically significant association (P = 0.033; Table 1).
Fig. 2.
Aldehyde dehydrogenase 1 staining. DAB staining (arrow) is ALDH1 staining in tumor cells, a marker of cancer stem cells. Two cases positive for ALDH1 are shown above
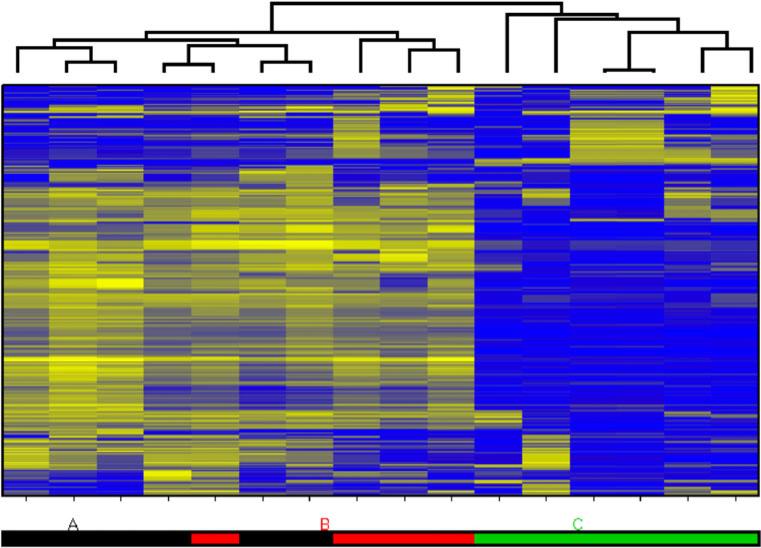
Highest variance genes segregate xenograft tumors from primary tumors irrespective of receptor subtype
Hierarchical clustering of gene expression from 16 tumor samples, including four matched primary tumor-xenograft pairs, across the 500 overall most variable genes distinctly separated human breast tumors from xenograft tumors (Fig. 3). The difference in gene expression between human and mouse was greater than the difference between breast tumors from distinct individuals, as human breast tumors clustered more closely with breast tumors of other individuals than with their related xenografts. Clustering on the basis of tumor-xenograft pairings, or even on the basis of receptor subtype, was not evident.
Fig. 3.
Heatmap plot of hierarchical clustering result using top 500 most variable genes determined by the standard deviation of log-intensities across all arrays in the experiment. Group A is human breast primary tumors that created xenografts, group B is primary tumors that did not create xenografts, and group C is xenografts
To explore the nature of the differences in gene expression between xenografts and primary tumors that might be driving the segregated clustering, we performed a (paired) one-way ANOVA analysis and found that the expression differences between the xenografts and their parent primary tumors are dominated by genes involved in immune response and cell adhesion (296 probes with FDR < 0.05; 647 probes with B > 0, where B is the log posterior odds ratio of differential expression [23]; DAVID [24] pathway enrichment results in Table S2). These results suggest that the “species difference” between xenografts and the primaries that gave rise to them—a result of different host species environments potentially compounded by array hybridization of mouse mRNA from mouse stromal cells within the xenograft—is manifested mostly in the immune and extracellular matrix (ECM) components of the tumor microenvironment.
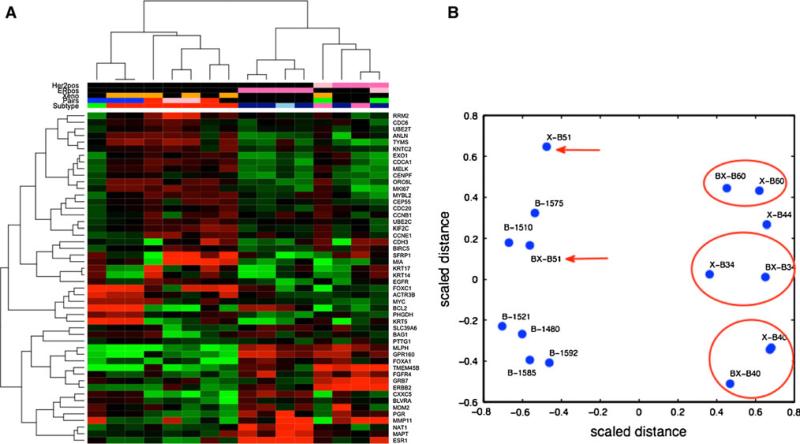
Breast cancer-specific PAM50 genes cluster xenograft tumors with primary tumors according to receptor subtype
The PAM50 gene set, a collection of genes with known prognostic and predictive significance in breast cancer, contains genes involved in estrogen signaling, proliferation, angiogenesis, apoptosis, Her2 signaling, immune reactivity, metabolism, cell motility, ECM, and others [20]. This gene set has been widely used to classify breast cancers into the intrinsic subtypes [20]. When hierarchical clustering was performed over the PAM50 gene set, the primary tumors and xenograft tumors no longer segregated as seen in Fig. 3, but rather blended together in mixed clusters. In addition, primary and xenograft tumors clustered in groups defined by receptor expression, with all ER+, Her-2+, and TN tumors clustered in distinct groups that included the xenografts derived from tumors with those features (Fig. 4a). Therefore, the expression of breast cancer-specific genes in xenografts sufficiently maintains similarities to the tumors from which they were derived to override the strong effect of the immune and ECM species difference seen in the 500 most variable genes.
Fig. 4.
a Unsupervised clustering of primary tumors and xenografts over PAM-50 genes. Heatmap rows are PAM-50 genes, columns are individual tumors. Top dendrogram depicts relationships among tumors by gene expression. b Two-dimensional representation of distance between all samples by multidimensional scaling of gene expression for PAM-50 genes. Three xenograft-primary pairs are circled in red; the fourth pair (BX-B51 and X-B51) is indicated by red arrows. Color key for the sample annotation strips in (a): Her2pos and ERpos: dark pink = positive, black = negative, light pink = missing; Xeno: orange = xenograft; black = primary tumor; Pairs: xenograft-primary pairs are same color; subtype: red = Basal; dark blue = LumA; light blue = LumB; pink = Her2; green = normal
Xenograft tumors have breast cancer-specific gene expression that is more similar to the primary tumors that gave rise to them than to other tumors
Ideally, a xenograft tumor model is more similar to the primary tumor it was derived from than to any other tumor, xenograft or primary. Hierarchical clustering performed over the PAM50 breast cancer-specific gene set cancer shows that most tumor-xenograft pairs cluster together (Fig. 4a). To further visualize the relationships between tumors, we applied multidimensional scaling to represent the relationships between the all tumors in the study with regard to expression of all PAM50 genes, projected on a two-dimensional plane. Inter-tumor distances between matched xenograft and primary pairs appear smaller than distances between all other combinations for three of four xenograft-primary pairs (Fig. 4b). For a fourth pair, the inter-tumor distance between the xenograft and the primary tumor was not the shortest distance between the xenograft and other primary tumors, though the primary tumors that were near the pair clustered together on the dendrogram.
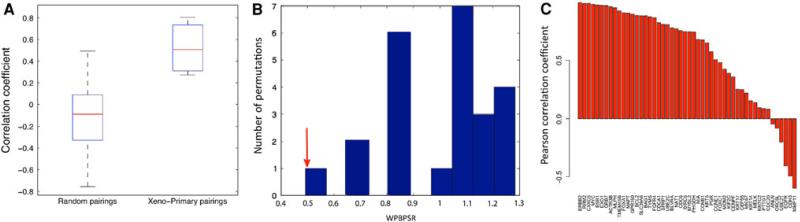
To further investigate the similarity of xenografts to the primary tumors from which they were derived, we compared matched xenograft-primary pairs to all other possible xenograft-primary combinations (Fig. 5a). The correlation coefficients of the expression profiles of the matched pairs were significantly closer to 1 than random pairings (P = 0.0056). We also analyzed the dissimilarities between matched pairs relative to the dissimilarities between randomly matched tumors by the WPBPSR (described above), and determined that the similarity of the matched xenograft-primary pairs was significantly greater (WPBPSR = 0.45) than all random pairs generated (P = 0.027, Fig. 5b).
Fig. 5.
a Correlation coefficients of PAM50 gene expression of xenograft-primary pairs compared to all possible pairs of xenografts and xenograft-generating primary tumors. (P = 0.0056). b Within-pair between-pair scatter ratio. Xenograft-primary pairs indicated with red arrow. c Correlation barplot of individual PAM50 genes. The height of each bar represents the Pearson correlation coefficient of xenograft-primary pairs for a gene
Since not all tumor-xenograft pairs are nearest neighbors in the clustering, we were interested in determining which PAM50 genes were most and least correlated across pairs. To investigate, we calculated the Pearson correlation coefficients between xenografts and parent primaries for each PAM50 gene, and found that 64 % (32/50) of the probes have a correlation coefficient greater than 0.5 and 32 % (21/50) have a correlation coefficient greater than 0.8. Among the least correlated genes were MMP11, CDH3, EGFR, and ANLN, all genes involved in cell–cell adhesion and cellular interactions with the ECM (Fig. 5c).
Xenograft tumors shift toward more aggressive features
Despite the overall similarity between primary-xenograft pairs, there are some additional observable shifts in PAM50 gene expression in xenograft tumor expression suggestive of an epithelial to mesenchymal transition (EMT) toward greater aggressiveness or at least increased cell–stroma adhesion compared to the parent primary tumor. For example, the ROR scores are increased in the xenografts compared to the primary tumors, both as a whole and relative to the primaries that generated them (ROR and ROR-PC are HIGH for all five xenograft tumors but a mix of LOW (2), MEDIUM (1), and HIGH (1) in their parent tumors; see Supplementary material). Also, the non-TN tumor that generated a xenograft, B51, had the original subtype HR–/Her2+ but demonstrated positive gene expression of estrogen receptor. In engrafted form, this tumor retains ESR1 expression but at a lower level (from a positive value of 1.747 to weakly positive 0.313), and the transcriptional subtype switches from LumA to Her2 (both subtypes associated with ERBB2 amplifications, but the latter with a poorer prognosis). More generally, ER trended toward decreased expression in the xenografts relative to the primaries, even in the TN tumors, with 3/4 showing decreased ER expression though this trend was not statistically significant (P = 0.103). In addition, proliferation signature scores increased in 75 % of xenografts relative to their parent primaries, though this trend did not reach statistical significance in this small patient subset (3/4; P = 0.071). One apparent switch of subtype is the TN tumor B40 which transitions from “normal” to “basal,” possibly reflecting increased aggressiveness of the tumor after xenograft passaging (see Supplementary material).
Discussion
This study describes the establishment of five xenograft models of breast cancer created from human breast primary tumors implanted in immune-deficient mice and contrasts primary tumors that gave rise to xenografts with those that did not. Of note, the xenografts in this study were generated predominantly from primary breast tumors from patients with early stage breast cancer, as opposed to tissue from distant recurrence sites in patients with metastatic disease. The rate of engraftment (25 %) was consistent with other studies and reflects the optimization of technique, including careful selection of mouse model and use of subcutaneous estrogen supplementation to increase xenograft yield.
Triple-negative (TN) and basal breast primary tumors demonstrated superior ability to adapt and thrive as xenografts in a novel environment relative to HR+ and luminal breast tumors, consistent with other studies [25]. This difference may reflect inherently more proliferative activity in TN breast cancer, consistent with its association with higher grade histology and more aggressive early clinical behavior than HR+ cancer. Alternatively, the xenograft model may lack factors necessary for local invasion and tumor viability in HR + disease, depending on interactions with stroma, the host immune system, the hormonal milieu, or other host factors. The observation that HR+ tumors did not engraft as xenografts despite estrogen supplementation suggests that estrogen-mediated signaling is not sufficient to increase the metastatic potential of these tumors, though it is also possible that the effect of exogenous estrogen differs from that of endogenous hormones.
Expression of ALDH1 also increased the likelihood of creating a xenograft, an observation that is consistent with the association of ALDH1 and cancer stem cells [16], since the requisite tumorigenicity in the xenograft model resembles the cancer stem cell characteristic of enhanced cell–cell adhesive interactions and junctions, and other mesenchymal invasive and angiogenic capabilities. However, it is possible that the association between ALDH1 and engraftment is an artifact of preferential expression of this marker in TN tumors. CK5/6 positivity in the primary tumors, a cytokeratin stain associated with basal phenotype, did not associate with engraftment potential despite the “basality” of the primaries, a surprising finding though in line with a prior study showing lack of expression of CK5/6 in a basal xenograft made from a CK5/6 positive basal primary [26]. Discrepancy between CK5/6 IHC staining and gene expression by PCR has previously been demonstrated. In this study, the basal gene expression phenotype, which is a composite of many genes including CK5, is a more sensitive marker for ability to form a xenograft than CK5/6 positivity.
This study demonstrates that the breast cancer-specific gene expression of xenografts recapitulates that of the tumors from which they are derived, a finding in concordance with prior work [11]. In particular, the similarity in gene expression over the PAM50 genes between xenografts and their parent tumors overwhelmed the strong immune-and cell adhesion-driven species association initially observed in unsupervised clustering over the 500 most variable genes, in that tumor-xenograft pairs were much more similar to each other than xenografts were to other xenografts. Xenografts clustered closely with their parent tumors in unsupervised hierarchical clustering, and strong associations were again seen in small intertumor distances between primary tumors and corresponding xenografts in multidimensional scaling and significantly stronger correlation among xenograft pairs relative to random pairs in the WPBPS ratio. Notably, PAM50 genes that were the least correlated between xenograft–tumor pairs mediate interactions with the tumor microenvironment.
However, despite the overall similarity between xenograft tumors and their parent primaries with respect to breast cancer-specific PAM50 gene expression, the xenografts, as a group, have a shift toward more aggressive features relative to their primary tumors of origin. This shift can be observed in the form of higher proliferation scores in the xenograft tumors relative to the parent primaries, along with higher risk of relapse (ROR) classification and lower estrogen receptor expression. This observation suggests that the tumors that successfully engrafted as xenografts may have done so as a result of increased adaptability and metastatic potential compared to the tumors that did not engraft. Yet, the actual patient outcomes data in this study do not support this idea. Though most xenograft-producing primaries did in fact metastasize to generate distant recurrences, supporting the hypothesis that xenograft potential might be connected to metastatic potential, data showing that fully half of the recurrences in our patient group occurred in HR+/Her2–tumors that did not engraft belies this supposition and suggests that successful engraftment of even highly aggressive HR+ tumors likely requires preconditions in the host stroma not required for TN tumor engraftment.
A recent study compared the molecular features of a primary inflammatory TN breast tumor, a brain metastasis, and a xenograft created from the primary and found that the xenograft retained all primary tumor mutations but displayed a mutation enrichment pattern that resembled the metastasis [12]. This study proposes that secondary tumors including xenografts and metastases may arise from a minority of cells within the primary tumor [12]. Though we did not analyze for mutation or copy number aberration, the changes in gene expression we observed are consistent with this study and others showing that xenografts tend toward enrichment in basal and mesenchymal phenotypes and depleted in luminal phenotypes, implying that xenografts might be clonal expansions of the more basal-like mesenchymal subpopulations in the tumors [7]. Thus, despite xenograft and primary tumor co-clustering, the xenografts in this study might fail to recapitulate intratumor heterogeneity of individual tumors, though more analysis is required to test this hypothesis. Further, if xenografts selectively accentuate the features essential to metastasis, this may provide clues for more effective and targeted therapy. In addition, the aggressiveness of HR+/Her2– tumors may be under-represented by the xenograft model because these tumors are dominated by luminal phenotypes and lack mesenchymal phenotypes that spur engraftment, yet still clinically demonstrate high metastatic potential.
In conclusion, this study demonstrates that breast cancer-specific gene expression of xenografts recapitulates that of the tumors from which they are derived and supports the use of xenografts as a molecularly representative model system for cancer research. This is particularly true for applications such as testing targeted drug therapy for TN breast cancer, with the above caveats concerning mesenchymal phenotype enrichment and what that might imply about the difficulty in recapitulating intratumor heterogeneity. HR+/Her2– tumors are not well represented in this study, but their inability to grow as xenografts may provide some clue as to what drives their growth and how best to treat them. The models created in this study are in current use by OncoMed Pharmaceuticals Inc. to develop and test monoclonal antibody therapies directed at pathways deranged in the breast tumors. These specific xenograft models add to the growing compendium of primary breast tumor xenografts, collectively representing an advance over previous models that were created from cell lines or metastatic tumor samples in their faithful reflection of clinically and histologically aggressive early stage disease.
Supplementary Material
Acknowledgments
The authors would like to thank the patients who participated in the study. We would like to thank Angie Park for her xenograft development work at OncoMed. This work was supported by funding from OncoMed Pharmaceuticals, Inc. the National Cancer Institute Specialized Program of Research Excellence in Breast Cancer, the Doris Duke Charitable Foundation and the NIH/NCRR/OD UCSF-CTSI Grant Number TL1 RR024129. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Conflicts of interest Ann Kapoun and John Lewicki are employees and stock holders of OncoMed Pharmaceuticals, Inc.
Contributor Information
Laura A. Petrillo, Department of Medicine, University of California, San Francisco, CA 94115, USA
Denise M. Wolf, Department Laboratory Medicine, University of California, San Francisco, CA 94115, USA
Ann M. Kapoun, OncoMed Pharmaceuticals, Inc., Redwood City, CA, USA
Nicholas J. Wang, Oregon Health & Science University, Portland, OR, USA
Andrea Barczak, Functional Genomics Core, University of California, San Francisco, CA, USA.
Yuanyuan Xiao, Functional Genomics Core, University of California, San Francisco, CA, USA.
Hasan Korkaya, Department of Internal Medicine, University of Michigan, Ann Arbor, MI, USA.
Frederick Baehner, Department of Pathology, University of California, San Francisco, CA, USA.
John Lewicki, OncoMed Pharmaceuticals, Inc., Redwood City, CA, USA.
Max Wicha, Department of Internal Medicine, University of Michigan, Ann Arbor, MI, USA.
John W. Park, Department Laboratory Medicine, University of California, San Francisco, CA 94115, USA
Paul T. Spellman, Oregon Health & Science University, Portland, OR, USA
Joe W. Gray, Oregon Health & Science University, Portland, OR, USA
Laura van't Veer, Department Laboratory Medicine, University of California, San Francisco, CA 94115, USA.
Laura J. Esserman, Department of Surgery, University of California, San Francisco, CA, USA
References
- 1.Giovanella B, Stehlin J, Williams L, Shih-Shun L, Shepard R. Heterotransplantation of human cancers into nude mice. Cancer. 1978;42:2269–2281. doi: 10.1002/1097-0142(197811)42:5<2269::aid-cncr2820420527>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 2.Steel GG, Courtenay VD, Peckham MJ. The response to chemotherapy of a variety of human tumour xenografts. Br J Cancer. 1983;47:001–013. doi: 10.1038/bjc.1983.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rae-Venter B, Reid LM. Growth of human breast carcinomas in nude mice and subsequent establishment in tissue culture. Cancer Res. 1980;40:95–100. [PubMed] [Google Scholar]
- 4.Mehta R, Graves J, Hart G, Shilkaitis A, Das Gupta T. Growth and metastasis of human breast carcinomas with Matrigel in athymic mice. Breast Cancer Res Treat. 1993;25:65–71. doi: 10.1007/BF00662402. [DOI] [PubMed] [Google Scholar]
- 5.Clarke R. Human breast cancer cell line xenografts as models of breast cancer&mdash;the immunobiologies of recipient mice and the characteristics of several tumorigenic cell lines. Breast Cancer Res Treat. 1996;39:69–86. doi: 10.1007/BF01806079. [DOI] [PubMed] [Google Scholar]
- 6.Pegram M, Ngo D. Application and potential limitations of animal models utilized in the development of trastuzumab (Herceptin®): a case study. Adv Drug Deliv Rev. 2006;58(5-6):723–734. doi: 10.1016/j.addr.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Keller PJ, et al. Mapping the cellular and molecular heterogeneity of normal and malignant breast tissues and cultured cell lines. Breast Cancer Res. 2010;12(5):R87. doi: 10.1186/bcr2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reyal F, et al. Molecular profiling of patient-derived breast cancer xenografts. Breast Cancer Res. 2012;14(1):R11. doi: 10.1186/bcr3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valdez KE, et al. Human primary ductal carcinoma in situ (DCIS) subtype-specific pathology is preserved in a mouse intraductal (MIND) xenograft model. J Pathol. 2011;225(4):565–573. doi: 10.1002/path.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moestue SA, et al. Distinct choline metabolic profiles are associated with differences in gene expression for basal-like and luminal-like breast cancer xenograft models. BMC Cancer. 2010;10:433. doi: 10.1186/1471-2407-10-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergamaschi A, et al. Molecular profiling and characterization of luminal-like and basal-like in vivo breast cancer xenograft models. Mol Oncol. 2009;3(5-6):469–482. doi: 10.1016/j.molonc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foulkes WD, Smith IE, Reis-Filho JS. Triple negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 14.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 15.Resetkova E, Reis-Filho J, Jain RK, Mehta R, Thorat MA, Nakshatri H, Badve S. Prognostic impact of ALDH1 in breast cancer: a story of stem cells and tumor microenvironment. Breast Cancer Res Treat. 2010;123(1):97–108. doi: 10.1007/s10549-009-0619-3. [DOI] [PubMed] [Google Scholar]
- 16.Ginestier C, Hur M, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalerba P, Dylla SJ, Park I, Liu R, Wang X, Cho R, Hoey T, Gurney A, Huang E, Simeone D, Shelton A, Parmiani G, Castelli C, Clarke M. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolstad BM, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 19.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, et al., editors. Bioinformatics and computational biology solutions using R and bioconductor. Springer; New York: 2005. [Google Scholar]
- 20.Parker JS, Michael M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seber GAF. Multivariate observations. John Wiley & Sons; Hoboken, NJ: 1984. [Google Scholar]
- 22.Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, Glas AM, Perou CM, Van't Veer LJ. Molecular portraits and 70-gene prognosis signature are preserved throughout the meta-static process of breast cancer. Cancer Res. 2005;65(20):9155–9158. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 23.Lonnstedt I, Speed T. Replicated microarray data. Stat Sin. 2002;12(1):31–46. [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocol. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Marangoni E, et al. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin Cancer Res. 2007;13(13):3989–3998. doi: 10.1158/1078-0432.CCR-07-0078. [DOI] [PubMed] [Google Scholar]
- 26.de Plater L, et al. Establishment and characterisation of a new breast cancer xenograft obtained from a woman carrying a germline BRCA2 mutation. Br J Cancer. 2010;103(8):1192–1200. doi: 10.1038/sj.bjc.6605900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.