Abstract
Physiological and pathological roles for small non-encoding miRNAs (microRNAs) in the cardiovascular system have recently emerged and are now widely studied. The discovery of widespread functions of miRNAs has increased the complexity of gene-regulatory processes and networks in both the cardiovascular system and cardiovascular diseases. Indeed, it has recently been shown that miRNAs are implicated in the regulation of many of the steps leading to the development of cardiovascular disease. These findings represent novel aspects in miRNA biology and, therefore, our understanding of the role of these miRNAs during the pathogenesis of cardiovascular disease is critical for the development of novel therapies and diagnostic interventions. The present review will focus on understanding how miRNAs are involved in the onset and development of cardiovascular diseases.
Keywords: cardiovascular disease, diabetes, dyslipidaemia, hypertension, microRNA (miRNA)
INTRODUCTION
miRNAs (microRNAs) are short (~19–25 nucleotides in length) single-stranded non-encoding RNAs that are well conserved in eukaryotic organisms [1,2]. By base pairing with complementary sites within target mRNA, they act as negative regulators of gene expression by inhibiting translation and/or inducing specific mRNA degradation [1,3,4]. The first miRNAs were discovered in the early 1990s; however, they were not recognized as a distinct class of post-transcriptional biological regulators until the early 2000s [5–7]. miRNAs have been identified as being encoded by the human genome, with newly identified miRNAs being reported on a regular basis. miRNA genes can be transcribed from their own promoters (intergenic) as independent transcription units or within the host protein-encoding genes (intronic) [1,8,9].
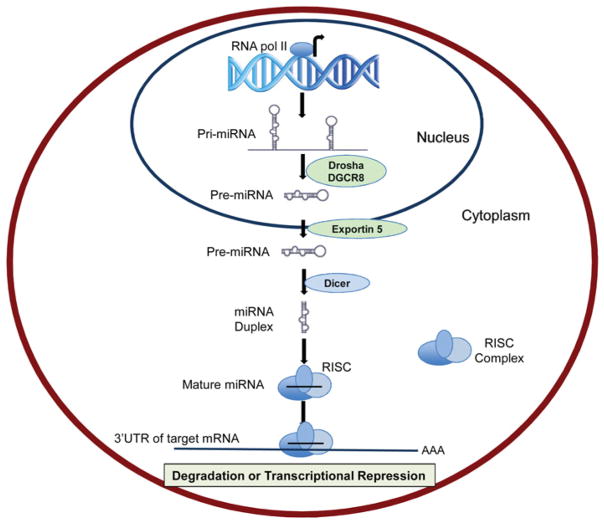
The biogenesis of miRNAs starts with RNA polymerase II-mediated transcription from the genome, leading to the formation of a pri-miRNA (primary miRNA transcript) of variable length depending on the locus [9,10] (Figure 1). The double-stranded pri-miRNA is cleaved by a large nuclear ribonuclease complex, DROSHA/DGCR8, generating a pre-miRNA (precursor miRNA) that is ~70 nucleotides in length and has a hairpin-like secondary structure [10,11]. Following nuclear processing by DROSHA/DGCR8, the pre-miRNA is then exported to the cytoplasm via the nuclear transport receptor, exportin-5, dependent pathway [12,13]. Once in the cytoplasm, the pre-miRNA is subject to further enzymatic processing by Dicer (another ribonuclease III enzyme) and its co-factors, generating the final short double-stranded miRNA strand (~22 nucleotides in length) [14]. The double-stranded miRNA (miRNA duplex) is composed of a mature miRNA guide strand (functional) and a passenger strand, called miRNA*. After unwinding of the duplex, the guide strand is loaded into the RISC (RNA-induced silencing complex), a multiprotein complex containing AGO2 (Argonaute 2) and other associated proteins, whereas the passenger strand is normally degraded (Figure 1) [15]. Once in the RISC, the mature miRNA seed region (nucleotides at position 2–8) participates in gene silencing by interacting with the 3′-UTR of the target mRNA [16,17]. This interaction leads predominantly to the down-regulation of the targeted mRNA (Figure 1).
Figure 1. Schematic representation of miRNA biogenesis.
miRNA biogenesis is initiated with the processing of primary miRNA transcripts in the nucleus by DROSHA/DGCR8, to generate a pre-miRNA. Pre-miRNA is exported to the cytoplasm by Exportin 5 and further processed into a miRNA duplex by Dicer. One of the single stands (mature miRNA) is incorporated into RISC and binds the 3′-UTR of the target miRNA leading to mRNA degradation or translational repression. RNA pol II, RNA polymerase II.
Pioneering studies have also uncovered the presence of circulating extracellular miRNAs, which are stable in the plasma and not associated with cells [18]. The levels of these miRNAs can differ with disease states [18–20]. Previous findings showed that extracellular miRNAs can associate with lipid-based carriers including HDLs (high-density lipoproteins) [21,22]. HDLs were found to contain distinct miRNA signatures and HDL miRNA profiles were altered with cardiovascular disease in humans and mice [22]. Furthermore, the miRNAs present on HDLs can be delivered to cells, including human hepatocellular carcinoma cells (Huh-7) and baby hamster kidney cells overexpressing SR-BI (scavenger receptor class B, type I) [22]. As plasma HDL-C (HDL-cholesterol) levels inversely correlate with cardiovascular risk [23], the discovery of HDL-associated miRNAs and the role of HDLs in miRNA transport represents an important finding linking lipoproteins and miRNAs to cardiovascular diseases.
The aim of the present review is to (i) summarize what is known about the involvement of miRNAs in the onset of cardiovascular disease, and (ii) illustrate how miRNAs can regulate the development of cardiovascular diseases. We will discuss the involvement of miRNAs in dyslipidaemia, hypertension, insulin resistance and diabetes, CAD (coronary artery disease), vascular inflammation, MI (myocardial infarction), and heart failure.
miRNAs AND THE ONSET OF CARDIOVASCULAR DISEASE
miRNAs and dyslipidaemia
Elevated levels of HDLs protect against coronary heart disease [23], whereas elevated levels of serum cholesterol (hypercholesterolaemia), lipids (hyperlipidaemia) and TAG [triacylglycerols (triglycerides)] (hypertriglyceridaemia) promote CAD [24,25]. Cholesterol biosynthesis in the liver is fundamentally important for lipid and lipoprotein synthesis and metabolism [26,27] and miRNAs have recently been found to modulate these processes [28–30].
We have recently identified miR-27b as a candidate post-transcriptional hub of lipid metabolism genes [30]. miR-27b was experimentally determined to target 27 of 151 lipid-associated genes in human hepatoma cell lines, including NDST1 (heparan sulfate N-deacetylase/N-sulfotransferase 1), ANGPTL3 (angiopoietin-like 3), PPARG (peroxisome proliferator-activated receptor γ) and GPAM (glycerol-3-phosphate acyltransferase 1) [30]. Using small RNA sequencing and real-time PCR, we found miR-27b and its target genes to be significantly modulated in mouse livers in response to diet-induced hyperlipidaemia [30]. miR-27b was found to be significantly increased in response to elevated plasma TAGs and hepatosteatosis, whereas ANGPTL3 and GPAM, both targets of miR-27b, were significantly repressed in the livers of mice fed on a high-fat diet [30]. ANGPTL3 is secreted from the liver and inhibits the hydrolysis of TAGs by lipoprotein lipase [31]. GPAM is the first committed enzymatic step in de novo TAG biosynthesis in the liver [32,33]. Therefore, during hypertriglyceridaemia and hepatosteatosis, hepatic miR-27b increases, which leads to inhibition of de novo TAG biosynthesis. These results support a role for miRNAs in the hepatic response to diet-induced hypertriglyceridaemia.
It has been demonstrated that miR-21 and miR-27b negatively regulate PPARα, which is mainly expressed in the liver and regulates the expression of multiple genes involved in fatty acid transport, catabolism and energy homoeostasis [31]. PPARα protein levels in human liver-derived cell lines were found to be decreased by the overexpression of miR-21 and miR-27b, but not by that of miR-22, miR-24, miR-181a and let-7a [31]. In addition to miR-21 and miR-27b, it has been reported in another study that PPARα is also regulated by miR-10b in the human hepatocyte cell line L02 [34]. This later study was performed using L02 cells cultured with high concentrations of non-esterified (free) fatty acids (as a non-alcoholic fatty acid liver disease model). Contrary to what has been reported for miR-21 levels, which are highly expressed in the human liver, miR-10b levels are up-regulated in a human hepatocyte cell line by treatment with high concentrations of non-esterified fatty acids [34]. PPARA (PPARα) is the direct target of miRNA-10b and PPARα protein levels were significantly decreased after overexpressing miR-10b in L02 cells incubated with high concentrations of non-esterified fatty acids [34]. It has therefore been speculated that the role of miR-10b in PPARα regulation might be specific to certain pathological conditions such as non-alcoholic fatty acid liver disease.
miRNAs and cholesterol metabolism
Cellular cholesterol homoeostasis is tightly regulated and achieved through a delicate balance of cholesterol biosynthesis, cholesterol efflux from cells to acceptors in the extracellular space and cellular cholesterol uptake through scavenger receptors, such as the LDLR [LDL (low-density lipoprotein) receptor] and LRPs (LDLR-related proteins). Cellular cholesterol homoeostasis is essential for functional signal transduction, membrane integrity, cell proliferation, lipid metabolism and many other key processes. SREBP2 (sterol-regulatory-element-binding protein 2) transcriptionally regulates most of the enzymes in the cholesterol biosynthetic pathway, as well as the LDLR, the major route of cholesterol entry into cells [35]. Although multiple ATP-binding cassette transporters and other transmembrane proteins mediate cholesterol efflux from cells into the extracellular space, ABCA1 (ATP-binding cassette transporter A1) is the major route of hepatic cholesterol and phospholipid efflux to apoA-I (apolipoprotein A-I), the main HDL apolipoprotein. This results in the formation of discoidal HDLs that are the precursors of the mature spherical HDLs that predominate in the plasma [36]. Likewise, ABCA1-mediated cholesterol efflux from cholesterol-loaded macrophages is a key mechanism for the prevention of foam cell formation and ultimately the development of atherosclerosis [37].
The most widely studied miRNA feedback network in the area of cholesterol metabolism is miR-33a/b [38–43]. miR-33a is harboured within intron 16 of SREBP2, and is thus co-transcribed with this master lipid transcription factor [40–42]. Previous evidence supports miR-33a as a key mediator in the cellular response to depleted cholesterol stores. In low cholesterol states, an intricate sterol-sensing network is activated that cleaves SREBP2 from the endoplasmic reticulum, allowing it to enter the nucleus where it transcriptionally activates the cholesterol biosynthetic pathway and up-regulates LDLR expression, thus increasing cellular cholesterol synthesis and LDL-cholesterol uptake [44]. These events are accompanied by the co-transcriptional activation of SREBP2 and miR-33a, the latter of which directly targets and decreases ABCA1 mRNA levels, which represses cholesterol efflux from the cells. Interestingly, miR-33a and miR-33b are localized within the introns of SREBP genes, therefore they collaborate with their protein encoding host transcripts to control cholesterol and lipid metabolism. In addition to ABCA1, miR-33a/b targets multiple other genes involved in cholesterol metabolism, including ABCG1 (ATP-binding cassette transporter G1; cholesterol efflux), NPC1 (Niemann–Pick C1; cholesterol storage), ABCB11 (ATP-binding cassette transporter B11; bile secretion) and ATP8B1 (phospholipid-transporting ATPase IC; bile acid secretion) [40–42,45]. miR-33a/b was also found to play a key role in fatty acid β-oxidation by directly targeting CROT (carnitine O-octanyl transferase), CPT1A (carnitine palmitoyltransferase 1A) and HADHB (hydroxyacyl-CoA dehydrogenase-3-ketoacyl-CoA thiolase-enoyl-CoA hydratase β-subunit) [38,46]. Similar to the regulation of cellular cholesterol levels by SREBP2/miR-33b, SREBP1/miR-33a promotes fatty acid synthesis and antagonizes fatty acid oxidation. In non-human primates, inhibition of miR-33a/b resulted in reduced VLDL (very-low-density lipoprotein) secretion and a reduction in plasma TAG levels [47]. Likewise, inhibition of miR-33 in mice was found to increase ABCA1 expression and plasma HDL-C levels. Plasma HDL-C levels are also increased in non-human primates in which miR-33a/b is inhibited [43,47].
miR-122 was the first miRNA to be identified as having an important role in the regulation of lipid metabolism [48,49]. Anti-miR-122 therapy in mice and non-human primates resulted in a significant reduction in cholesterol levels [29,48]. These results have recently been confirmed in miR-122-deficient mouse models [50,51]. The mechanisms by which miR-122 induces its effects on lipid metabolism are still poorly understood, but previous studies have demonstrated that genes involved in cholesterol biosynthesis, such as HMGCR (3-hydroxy-3-methylglutaryl-CoA reductase) and HMGCS1 (3-hydroxy-3-methylglutaryl-CoA synthase 1) are down-regulated by this miRNA [50–52]. In addition to its effects on cholesterol biosynthesis, miR-122 seems to be involved in TAG metabolism, with the TAG secretion rate being significantly reduced in miR-122-deficient mice [50].
In the liver, miR-122 accounts for 70–80% of the miRNA, depending on the technique used [53]. As such, miR-122 has been widely studied in the liver and has been found to play a role in hepatitis C, inflammation and hepatocellular carcinoma as well as iron, glucose, lipid and cholesterol metabolism [29,50,51,54–58]. Many cholesterol-regulating genes, including genes in the HMGCR, SQLE (squalene expoxidase) and lipoprotein synthesis/microsomal transfer protein pathways are inversely associated with miR-122 [29,50,52]. Although, miR-122 indirectly regulates cholesterol and lipid homoeostasis, its direct intermediary targets are currently unknown. Interestingly, miR-370 was found to regulate lipid metabolism both though direct targeting of lipid genes and modulation of miR-122 [59].
Recently, another miRNA, miR-144, has been reported to regulate the expression of ABCA1 in macrophages and hepatocytes [60]. Indeed, miR-144 expression was up-regulated in LXR (liver X receptor) agonist (T090)-treated mouse peritoneal macrophages. Human monocytes (THP-1) and human hepatic (Huh-7) cells treated with T090 also showed increased expression of miR-144 [60]. miR-144 is an intergenic miRNA located in the same locus as miR-451, which is also induced by stimulating mouse primary macrophages, THP-1 and Huh-7 cells, with T090 [60]. As expected, overexpression of miR-144 in mouse peritoneal macrophages inhibited ABCA1 expression. Although ABCG1 is not a direct target of miR-144, ABCG1 mRNA levels were also down-regulated in macrophages transfected with miR-144. This may have been an indirect effect involving the retinoid X receptor β, which is a predicted target for miR-144 [60]. Further evidence of the involvement of miR-144 in the regulation of ABCA1 expression was obtained by transfecting J774 murine macrophages with miR-144, which inhibited the efflux of cholesterol to apoA-I. On the other hand, antagonism of endogenous miR-144 in J774 murine macrophages increased ABCA1 expression as well as the efflux of cellular cholesterol to apoA-I [60]. miR-144 is widely expressed in mouse tissues and is particularly abundant in the liver, spleen and aorta. A recent study has reported a role for miR-144 in regulating lipoprotein metabolism and showed that the overexpression or inhibition of miR-144 reduces and increases circulating HDL levels respectively [60]. The injection of miR-144 mimic nanoparticles into mice significantly reduced ABCA1 and ABCG1 mRNA and protein levels in the liver. The inhibition of ABCA1 expression after 6 days of treatment with miR-144 particles also decreased total cholesterol and HDL-C levels without changing the TAG or cholesterol distribution in other lipoproteins. Finally, in vivo inhibition of miR-144 with miR-144 inhibitor-conjugated nanoparticles showed increased liver ABCA1 protein expression and plasma HDL-C levels [60].
Other miRNAs that have been reported to play key roles in lipid metabolism include miR-106, miR-26 and miR-758, which target and repress ABCA1 in multiple cell types including macrophages and hepatocytes [61–63]. Similar to miR-33a/b, miR-758 was also found to be sensitive to cellular sterol levels and was down-regulated in response to cholesterol loading [61]. miR-1, miR-206 and miR-613 have all recently been reported to directly target LXRα and, thus, repress lipogenesis [64,65]. miR-613 suppresses lipogenesis by inhibiting expression of LXRα and its target genes, including SREBP1c, FAS (fatty acid synthase), ChREBP (carbohydrate responsive element-binding protein) and ACC (acetyl-CoA carboxylase) [65,66]. Although NR1H3 (encoding LXRα) is a target of miRNAs, recent evidence suggests that LXRα also controls the expression of specific miRNAs, with miR-26 being repressed by LXRα in macrophages [63]. Most interestingly, miR-26 was also found to directly target the LXRα genes, ABCA1 and ARL7 (ADP-ribosylation factor-like 7). ARL7 has been shown to be induced by cholesterol loading and participates in apoA-I-dependent cholesterol export [63].
Multiple miRNAs have been reported to target receptors associated with lipoprotein or cholesterol uptake. miR-146a represses TLR4 (Toll-like receptor 4) signalling, and inhibits oxLDL (oxidized LDL) cholesterol uptake in macrophages [67]. Most interestingly, oxLDL stimulation was found to decrease miR-146a expression [67]. miR-155, a miRNA associated with inflammation, was also found to repress lipid uptake in oxLDL-stimulated dendritic cells through targeting and down-regulation of the scavenger receptor, CD36, and the lectin-type LOX-1 (oxidized LDL receptor 1). miR-155 also regulates LOX-1 and SR-A (scavenger receptor A) pathways in macrophages [68]. Most interestingly, oxLDL stimulation was previously found to up-regulate miR-155, along with miR-9, miR-146a, miR-125a-5p and miR-146b-5p, in monocytes [69]. In that study, miR-125-5p repressed lipid uptake through targeting of ORP6 (oxysterol-binding protein-like 9) in monocytes [69]. In a separate study, inhibition of miR-155 was found to increase both lipid uptake and inflammation in oxLDL-stimulated THP-1 macrophages [70]. miR-125, miR-455-5p, miR-185, miR-96 and miR-223 have all been reported to target SR-BI and reduce HDL-C uptake [22,71,72]. Recently, miR-217 was found to mediate ethanol-induced SIRT1 (sirtuin 1) repression and contribute to ethanol-induced lipid accumulation and fatty acid synthesis [73]. Chronic ethanol feeding in mice also caused a significant increase in hepatic miR-217 levels [73]. Similar to miR-33, miR-217 is a key modulator of fatty acid oxidation and fatty acid synthesis [73].
Recently, there has been significant interest in identifying single nucleotide polymorphisms and functional variants within miRNAs themselves, and in mRNA target sites [74–78]. A recent study has found a gain-of-function variant in the LPL (lipoprotein lipase) gene that appears to abrogate a miR-410 target site [79]. The LPL variant rs13702 minor allele was found through meta-analyses to be significantly associated with increased HDL-C and decreased TAG levels [79]. miR-467b has also been reported to directly target LPL and inhibit lipid accumulation in macrophages [80]. Although over 1400 miRNAs have been identified in humans, each cell type typically contains 100–300 unique miRNAs. Most are transcribed in the cell; however, some are likely transferred to the cells from the extracellular compartment via lipoproteins or microvesicles [81].
miRNAs in insulin resistance and diabetes
Insulin resistance and T2D (Type 2 diabetes mellitus) are major risk factors of cardiovascular disease, together with associated endothelial dysfunction and micro- and macro-vascular complications [82]. Insulin resistance, a condition where tissues such as muscle and adipose tissue fail to adequately respond to the physiological actions of insulin, often progresses to T2D and coronary heart disease [83]. This is usually seen when pancreatic β-cells are unable to produce sufficient insulin to maintain normal blood sugar levels when insulin resistance is present, leading to hyperglycaemia. The inability of the β-cells to produce sufficient insulin under hyperglycaemic conditions is what characterizes the transition from insulin resistance to T2D [83]. It has been demonstrated that plasma miRNA levels are changed in patients with T2D. Previous studies have shown that plasma levels of miR-24, miR-21, miR-20b, miR-15a, miR-126, miR-191, miR-197, miR-223, miR-320, miR-486, miR-150 and miR-29b are lower in patients with T2D, whereas miR-28-3p and miR-375 tend to be elevated [84,85].
miR-126 is an miRNA that is consistently associated with T2D. In a population study, reduced levels of miR-126 in plasma were systematically associated with T2D [84]. One of the mechanisms by which miR-126 is involved in the development of insulin resistance is through the inhibition of IRS1 (insulin receptor substrate 1) [86]. This miRNA plays an important role in maintaining vascular integrity, angiogenesis and wound repair [87,88]. It also facilitates VEGF (vascular endothelial growth factor) signalling by repressing two negative regulators of the VEGF pathway: SPRED1 (Sprouty-related protein) and PIK3R2 (phosphoinositol-3 kinase regulatory subunit 2)/p85-β [87]. miR-126 is the most important miRNA in endothelial apoptotic bodies, where it is significantly reduced by a high glucose concentration [84,89]. miR-126 in apoptotic bodies is responsible for their cardioprotective properties. Indeed, miR-126-carrying, but not miR-126-deficient, apoptotic bodies confer a protection against diet-induced atherosclerosis in the carotid arteries of apoE−/− mice, which is associated with a reduced infiltration of macrophages into the artery wall and an increase in the number of smooth muscle cells [89].
miRNAs play key roles in the regulation of insulin secretion through both pancreatic development and insulin exocytosis. Enrichment of pancreatic β-cells with miR-375, was found to negatively regulate insulin exocytosis and secretion [90]. The mechanisms by which miR-375 modulates insulin secretion are independent of changes in glucose metabolism and intracellular Ca2+ signalling, but are related to a direct effect on insulin exocytosis through the repression of Mtpn (myotrophin), a gene implicated in actin depolymerization [90]. Mtpn is a predicted and validated target of miR-375 and the inhibition of miR-375 induces the repression of Mtpn which could contribute to the defect in exocytosis [90]. On the other hand, inhibition of miR-375 expression enhances insulin secretion [90]. Furthermore, homozygous deletion of miR-375 in mice is associated with hyperglycaemia due to decreased total pancreatic β-cell mass and plasma insulin levels [90]. miR-375 levels are elevated in people with T2D [85].
miR-9 is an islet-specific miRNA that is an important regulator of insulin secretion [91]. miR-9 may increase insulin secretion from β-cells by regulating expression of the nuclear protein Sirt1 in vivo during glucose-dependent insulin secretion [92]. Sirt1 is a nuclear NAD-dependent protein deacetylase, the expression of which is known to fluctuate in tissues such as the liver, white adipose tissue, brown adipose tissue and muscle under different metabolic conditions such as calorie restriction and starvation [93]. miR-9 targets and regulates Sirt1 expression in insulin-secreting β-cells. Interestingly, Sirt1 levels are down-regulated during glucose-stimulated insulin secretion in vivo in pancreatic β-islets, consistent with miR-9 levels being up-regulated. This targeting is relevant in diabetes as it highlights the functional interplay between insulin secretion, miRNAs and Sirt1 expression [92].
Increased miR-29a and miR-29b levels in the muscle, fat and liver of diabetic Goto–Kakizaki rats are associated with insulin resistance [94]. miR-29a and miR-29b are also highly expressed in the pancreatic islets of diabetic mice and are involved in insulin release through the modulation of Mct1 (monocarboxylate transporter 1) in the plasma membrane [95]. In pancreatic β-cells, elevated glucose concentrations stimulate mitochondrial oxidative metabolism to raise intracellular ATP/ADP levels, prompting insulin secretion. Mct1 enables circulating pyruvate/lactate to enter β-cells, where it acts as a substrate for mitochondrial oxidation, leading to an increase in the cytosolic ATP/ADP ratio. The inhibition of miR-29a in primary mouse islets with LNAs (locked nucleic acids) increases Mct1 mRNA levels, demonstrating that miR-29a contributes to the β-cell-specific silencing of the Mct1 transporter and may thus affect insulin release [95].
miR-223 is up-regulated in insulin-resistant human hearts and is involved in glucose uptake in cardiomyocytes [96]. miR-223 also increases cellular glucose uptake through the up-regulation of Glut4 (glucose transporter 4) protein expression. This effect is independent of PI3K (phosphoinositide 3-kinase) signalling and AMP kinase activity [96]. Overexpression of miR-223 in vitro also inhibits the insulin-stimulated phosphorylation of Akt and GSK3β (glycogen synthase kinase 3 β-subunit) in cardiomyocytes [96].
miR-124a is also abundant in pancreatic β-cells and is implicated in islet development, through the regulation of FOXA2 (forkhead box A2), a transcription factor important for pancreatic development and β-cell differentiation, and RAB27A, a GTPase involved in insulin secretion [97,98]. FOXA2 regulates genes involved in glucose metabolism and insulin secretion, including the ATP-sensitive K+ (KATP) channel subunits Kir6.2 and Sur-1. Correspondingly, miR-124a overexpression decreases, and anti-miR-124a increases, Kir6.2 and Sur-1 mRNA levels [98]. Furthermore, miR-124a modified basal and glucose- or KCl-stimulated changes in intracellular free Ca2+ concentrations in MIN6 and INS-1 β-cells without affecting the secretion of insulin, indicating an altered sensitivity of the β-cell exocytotic machinery to Ca2+ [98].
miRNAs also regulate insulin resistance by modulating insulin-signalling pathways in target tissues. As the predominant action of insulin in the liver is to prevent gluconeogenesis through a cascade of phosphorylation events that terminate with diminished PEPCK (phosphoenolpyruvate carboxykinase) promoter activation [99], it has been demonstrated that miR-29a inhibits insulin actions in the liver by preventing the insulin-mediated inhibition of PEPCK gene expression. This effect is mediated by a direct targeting of the regulatory p85α subunit of PI3K [99]. Furthermore, miR-29 was shown to subsequently inhibit insulin-mediated glucose import by 3T3-L1 adipocytes through mechanisms that involve indirect down-regulation of Akt activation [94].
miRNAs AND CARDIOVASCULAR DISEASES
CAD (coronary artery disease)
A recent study has shown that circulating levels of vascular- and inflammation-associated miRNAs are significantly down-regulated in patients with CAD [100]. Interestingly, most of the highly expressed miRNAs and significantly down-regulated miRNAs in the circulation of patients with CAD are expressed in ECs (endothelial cells). These include miR-126, members of the miR-17~92 cluster (miR-17, miR-20a and miR-92a), miR-130a, miR-221, members of the let-7 family (let-7d), miR-21 and miR-145. miR-155 is also significantly decreased in the plasma of patients with CAD [100]. On the other hand, cardiac muscle-enriched miRNAs, such as miR-133a and miR-208a, are increased in these patients [100]. This indicates that these miRNAs could potentially be used as biomarkers in patients with CAD.
Proliferation of VSMCs (vascular smooth muscle cells) makes a significant contribution to vascular neointimal formation. The role of miRNAs in VSMC proliferation has been extensively studied and miRNAs are key determinants of VSMC differentiation and phenotypic switching [101]. miR-143/145 have been shown to up-regulate the VSMC proliferative response to balloon injury in rat carotid arteries through alterations in cytoskeletal dynamics and organization [102]. Therefore restoration of miR-145 in balloon-injured arteries via an adenovirus expressing miR-145 inhibits neointimal growth [101]. It has also been shown that miR-143/145-enriched vesicles from endothelial cells decrease atherosclerotic fatty lesion formation in the aortas of apoE−/− mice [103].
miR-21 is also associated with VSMC proliferation and vascular neointimal lesion formation and is significantly up-regulated in atherosclerotic plaques [104,105]. It has been shown that anti-sense oligonucleotide inhibition of miR-21 reduces neointima formation following vascular balloon injury through a mechanism that involves increased levels of the pro-apoptotic proteins Bcl-2 and Pten (phosphatase and tensin homologue) [104].
miR-221/222 are highly expressed, but have opposing effects in VSMCs and ECs. Although miR-221/222 are involved in VSMC proliferation and migration, they have anti-migratory effects in ECs [106,107]. CDKN1B (encoding p27Kip1; cyclin-dependent kinase inhibitor 1B), CDKN1C (encoding p57Kip2; cyclin-dependent kinase inhibitor 1C) and the proto encogene c-Kit are target genes of miR-221/222 in both VSMCs and ECs; however, p27Kip1 and p57Kip2 are highly expressed in VSMCs, but not in ECs. In contrast, c-Kit is highly expressed in ECs, but not in VSMCs. The cellular effects of these target genes are also distinct. p27Kip1 and p57Kip2 have been shown to induce anti-proliferative effects on VSMCs and ECs. In contrast, c-Kit has pro-proliferative properties in these vascular cells. Therefore in VSMCs miR-221/222 were found to have pro-proliferative and pro-migratory effects by targeting CDKN1B and CDKN1C [108]. In ECs, in contrast, miR-221/222 were found to have anti-migratory effects by targeting c-Kit and transcription 5A, as well as endothelial and nitric oxide synthase [107]. These opposing effects of miR-221/222 have also been observed in vivo, where they increase neointimal growth, but decrease re-endothelialization in a balloon injury rat carotid artery model [106].
miR-126 is another miRNA that may also exert cardioprotective effects. miR-126, which is enriched in apoptotic bodies, has been shown to mediate the atheroprotective effects of endothelial apoptotic bodies. miR-126 in endothelial apoptotic bodies mediates these effects by targeting RGS16 (regulator of G-protein 16) to induce the expression of the CXC chemokine CXCL12 through its CXCR4 (CXC receptor 4) [89]. In the context of arterial injury, CXCL12, through CXCR4, is implicated in the recruitment of progenitor cells [identified by the presence of Sca-1 (stem cell antigen-1) and the absence of lineage markers on their surface] from the bone marrow to the damaged tissue in order to compensate for apoptosis processes [109,110]. Finally, results from this study showed that miR-126-carrying, but not miR-126-deficient, apoptotic bodies conferred a protection against diet-induced atherosclerosis in the carotid artery [89].
miR-195 also plays an important role in VSMC proliferation and migration, as well as in neointimal formation [111]. miR-195, introduced by adenovirus, reduces neointimal formation in balloon-injured rat carotid arteries [111].
miRNAs in hypertension
The persistent elevation of systemic blood pressure, or systemic hypertension, is classified as ‘essential’ in 90–95% of cases, when there are no obvious medical causes. The remaining causes of hypertension involve various identifiable medical conditions affecting the kidneys, the heart or the endocrine system. Patients with systemic hypertension have an elevated incidence of cardiovascular disease and heart failure [112]. Several mechanisms are involved in the pathogenesis of hypertension, including increased vascular tone, the hyperactivation of the RAAS (renin–angiotensin–aldosterone system), vascular endothelial dysfunction, VSMC and cardiac hypertrophy, and increased activity of the sympathetic nervous system; all of which are risk factors for cardiovascular diseases [113–115]. miRNAs have been shown to be involved in all these processes [116].
Expression of miR-143 and miR-145 is decreased in acute and chronic vascular stress [117]. Mice lacking both miR-143 and miR-145 have decreased blood pressure due to reduced vasoconstriction [102], impaired vasodilation and decreased medial thickness [118]. Neointima formation in response to vascular injury is profoundly impaired in mice lacking these miRNAs due to disarray of actin stress fibres and diminished migratory activity of VSMCs [102]. Indeed, the miR-143 and miR-145 cluster plays an important role in vascular differentiation and vascular function. Furthermore, it has been demonstrated that the miR-143/145 cluster is required for the acquisition of the contractile phenotype for VSMCs in mice [119]. Because the miR-143/miR-145 cluster is expressed mostly in the VSMC compartment, both during development and postnatally, the loss of miR-143 and miR-145 expression induces incomplete differentiation of VSMCs, leading to structural modifications of the aorta [117]. Overexpression of miR-143 and miR-145 on the other hand decreased neointimal formation in a rat model of acute vascular injury [117].
Although miRNAs are highly expressed in the heart, miR-21 was shown to be the most significantly up-regulated miRNA in mouse hypertrophic hearts and is aberrantly increased in acute MI and vascular neointimal lesions [104,117,120–122]. miR-21 is involved in cardiac hypertrophy, remodelling and fibrosis in response to pressure overload [123]. In a mouse model of pressure overload of the left ventricle, silencing of miR-21 using a specific miR-21 antagomir reduced cardiac ERK (extracellular-signal-regulated kinase)/MAPK (mitogen-activated protein kinase) activity, and attenuated cardiac hypertrophy, fibrosis and cardiac dysfunction [123]. However, the involvement of miR-21 in cardiac fibrosis and hypertrophy seems to be more complex, as other studies have shown that targeted deletion of miR-21 in mice, or inhibition of miR-21 with 8-mer LNAs, were not sufficient to improve fibrotic lesions or hypertrophic responses to cardiac stress stimuli [124].
miR-208a is a cardiac-specific miRNA that is involved in stress-dependent cardiac growth [125]. It has been shown that the transgenic overexpression of miR-208a induces hypertrophic growth in mice [126]. On the other hand, therapeutic inhibition of miR-208a using LNA-based therapy in the Dahl hypertensive rat model improves cardiac survival and function and prevents cardiac remodelling in hypertension-induced heart failure [127].
MI and heart failure
MI induced by coronary artery occlusion is accompanied by cardiac remodelling at the site of infarction injury. This remodelling process involves fibrous tissue formation and extracellular matrix deposition. These processes are mediated by cardiac fibroblasts [128]. miRNAs were identified to play potential roles in post-MI-induced cardiac remodelling [129]. Among these miRNAs, the miR-29 family was found to be dramatically down-regulated in the region of fibrous tissue formation in mice post-MI induced by the occlusion of the left coronary artery. The down-regulation of miR-29 correlated with the up-regulation of miR-29 targets which include various collagen and extracellular matrix protein genes, such as fibrillin 1, collagen type I, α1 and α2, and collagen type III α1 [129].
miR-199b is up-regulated in the hearts of animal models of cardiac hypertrophy and has been shown to play a role in heart failure and cardiac hypertrophy [130]. One major intracellular signalling pathway involved in heart failure is the activation of the pro-hypertrophic phosphatase calcineurin and its downstream transcriptional effector NFAT (nuclear factor of activated T-cells) [131,132]. miR-199b promotes calcineurin/NFAT-mediated cardiac hypertrophy by active down-regulation of its direct target gene DyrK1a [dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 1a] [130]. The in vivo injection of the miR-199b antagomir (chemically modified antisense oligonucleotide specific for miR-199b) prevented the development of cardiac disease in mice subjected to transverse aortic constriction pressure overload [130].
miR-499 is a cardiac abundant miRNA and is down-regulated in response to hypoxic and ischaemic stress in cardiomyocytes [133]. It has been shown that under ischaemia/reperfusion, the hearts of miR-499 transgenic mice showed less apoptosis and reduced infarct size when compared with control mice. The cardioprotective properties of miR-499 result from the fact that this miRNA prevents cardiomyocyte apoptosis by targeting the α and β isoforms of calcineurin [133]. Calcineurin is a serine and threonine protein phosphatase that is known to dephosphorylate pro- and anti-apoptotic factors, leading to their activation and inactivation respectively [134]. On the other hand, the knockdown of miR-499 using a cholesterol-modified antagomir (an antisense oligonucleotide with a cholesterol moiety modification) induces myocardial apoptosis and increases the infarct size [133].
Vascular inflammation
Endothelial activation and vascular inflammation are considered to be the first steps in atherosclerotic lesion development and cardiovascular disease. It has been shown that pro-inflammatory cytokines, such as TNF-α (tumour necrosis factor-α), increase the expression of adhesion molecules in ECs, which recruit inflammatory cells such as monocytes to the site of inflammation [135,136]. Adhesion molecule expression is mainly mediated by NF-κB (nuclear factor κB) signalling pathways [137].
It has recently been shown that activation of ECs with TNF-α decreases miR-181b expression [138]. This study shows that the overexpression, both in vitro and in vivo, of miR-181b blocks the expression of adhesion molecules such as VCAM-1 (vascular adhesion molecule 1). Furthermore, the systemic administration of miR-181b mimetics decreases EC activation and leucocyte recruitment in LPS (lipopolysaccharide)-induced lung injury. miR-181b represses the nuclear translocation of NF-κB by targeting importin-α3. These results suggest that the inhibitory effects of miR-181b on TNF-α-induced expression of adhesion molecules are mediated by the inhibition of NF-κB nuclear translocation [138].
Previous studies have demonstrated that miR-126 and miR-195 are also involved in vascular inflammation. Indeed, miR-126, which is highly expressed in ECs, supresses VCAM-1 expression in ECs and decreases leucocyte binding to TNF-α-activated ECs [139]. miR-195 significantly reduces the synthesis of IL (interleukin)-1β, IL-6 and IL-8 in rat VSMCs [111].
Inflammation-induced atherogenesis also involves components of the innate immune system (macrophages and dendritic cells) and of the adaptive immune system (T-lymphocytes) [140]. It has been demonstrated that miRNAs are expressed in activated B-cells, T-cells, macrophages and dendritic cells [141] and miRNA-dependent regulators of immune cells are involved in the control of vascular inflammation and atherosclerosis [142,143]. For example, miR-155, miR-146a and miR-29a are up-regulated in patients with CAD and miR-125a decreases the secretion of inflammatory cytokines such as IL-2, IL-6 and TNF-α from oxLDL-stimulated monocyte-derived macrophages [62,142].
Among the above miRNAs, previous studies have indicated that miR-155, a typical multi-functional miRNA, plays a crucial role in immunity, inflammation and cardiovascular disease [143]. miR-155 is involved in the prevention of atherosclerotic lesion development and progression, and is significantly up-regulated in both arteries and mononuclear cells in a mouse model of atherosclerosis [142]. One of the major mechanisms underlying the anti-atherogenic effects of miR-155 is likely to be the inhibition of inflammation, which involves MAP3K10 (mitogen-activated protein kinase kinase kinase 10) [142]. miR-155 was also shown to be up-regulated in the aorta of apoE−/− mice fed on a high-fat diet for 3–10 months [143,144]. In situ hybrizidation studies established that miR-155 in atherosclerotic plaques was derived from macrophages and smooth muscle cells. This study also showed that the polarization of murine bone marrow derived macrophages into pro-inflammatory M1-type macrophages by stimulation with LPS and IFN-γ (interferon γ) induced miR-155 expression [144].
miR-223 regulates progenitor proliferation as well as granulocyte differentiation and activation during inflammation [145]. A recent role for miR-223 relates to its ability to regulate macrophage polarization. Indeed miR-223 levels are dramatically elevated in bone marrow-derived macrophages after treatment with IL-4 to induce anti-inflammatory M2 macrophages, whereas treatment with LPS to induce M1 pro-inflammatory macrophages leads to the slight decrease in miR-223 [146]. Although miR-223 was initially thought to be restricted to myeloid cells regulating multiple inflammatory genes in monocytes and macrophages [145,147,148], several groups have now reported functional miR-223 expression in non-myeloid cell types, including cardiomyocytes, hepatocytes and ECs [96,149]. Hepatic miR-223 levels in mice are also significantly increased in post ischaemia/reperfusion injury [150]. miR-223 was also shown to protect against diet-induced adipose tissue inflammatory response [146].
CONCLUSIONS AND FUTURE DIRECTIONS
miRNAs play a central role in the onset and development of cardiovascular disease and their discovery has ushered in an entirely new set of drug targets that can be used to identify potential novel therapeutic strategies to treat dyslipidaemia, diabetes and cardiovascular disease. Although the field is developing very rapidly, we are most likely only at the beginning of our ability to understand the complexity and full repertoire of the post-transcriptional regulation of gene expression by miRNAs. The novel finding that miRNAs are present in plasma highlights their potential use as disease biomarkers.
One of the challenges for the future use of miRNA-based therapies arises from the fact that many miRNAs modulate multiple target genes (100 or more) involved in multiple cellular processes, and the manipulation of a single miRNA can lead to therapeutic benefits, but also to pathological effects. Using miRNA-targeted therapies for the prevention and treatment of cardiovascular disease clearly warrants further genetic and pharmacological investigation. The outcomes of such studies are awaited with interest.
Abbreviations
- ABC
ATP-binding cassette transporter
- ANGPTL3
angiopoietin-like 3
- apo
apolipoprotein
- ARL7
ADP-ribosylation factor-like 7
- CAD
coronary artery disease
- CXCR4
CXC receptor 4
- EC
endothelial cell
- FOXA2
forkhead box A2
- GPAM
glycerol-3-phosphate acyltransferase 1
- HDL
high-density lipoprotein
- HDL-C
HDL-cholesterol
- HMGCR
3-hydroxy-3-methylglutaryl-CoA reductase
- IL
interleukin
- LDL
low-density lipoprotein
- LDLR
LDL receptor
- LNA
locked nucleic acid
- LOX-1
oxidized LDL receptor 1
- LPL
lipoprotein lipase
- LPS
lipopolysaccharide
- LXR
liver X receptor
- Mct1
monocarboxylate transporter 1
- MI
myocardial infarction
- miRNA
microRNA
- Mtpn
myotrophin
- NFAT
nuclear factor of activated T-cells
- NF-κB
nuclear factor κB
- p27Kip1
cyclin-dependent kinase inhibitor 1B
- p57Kip2
cyclin-dependent kinase inhibitor 1C
- oxLDL
oxidized LDL
- PEPCK
phosphoenolpyruvate carboxykinase
- PI3K
phosphoinositide 3-kinase
- PPAR
peroxisome proliferator-activated receptor
- pre-miRNA
precursor miRNA
- pri-miRNA
primary miRNA transcript
- RISC
RNA-induced silencing complex
- SIRT1
sirtuin 1
- SR-BI
scavenger receptor class B, type I
- SREBP
sterol-regulatory-element-binding protein
- T2D
Type 2 diabetes mellitus
- TAG
triacylglycerol
- TNF-α
tumour necrosis factor α
- VCAM-1
vascular adhesion molecule 1
- VEGF
vascular endothelial growth factor
- VSMC
vascular smooth muscle cell
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18:131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 6.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 7.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 8.Ladewig E, Okamura K, Flynt AS, Westholm JO, Lai EC. Discovery of hundreds of mirtrons in mouse and human small RNA data. Genome Res. 2012;22:1634–1645. doi: 10.1101/gr.133553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 13.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 15.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 16.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler. Thromb Vasc Biol. 2011;31:2383–2390. doi: 10.1161/ATVBAHA.111.226696. [DOI] [PubMed] [Google Scholar]
- 20.Sun X, Zhang M, Sanagawa A, Mori C, Ito S, Iwaki S, Satoh H, Fujii S. Circulating microRNA-126 in patients with coronary artery disease: correlation with LDL cholesterol. Thromb J. 2012;10:16. doi: 10.1186/1477-9560-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vickers KC, Remaley AT. Lipid-based carriers of microRNAs and intercellular communication. Curr Opin Lipidol. 2012;23:91–97. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barter P, Gotto AM, LaRosa JC, Maroni J, Szarek M, Grundy SM, Kastelein JJ, Bittner V, Fruchart JC. HDL cholesterol, very low levels of LDL cholesterol, and cardiovascular events. N Engl J Med. 2007;357:1301–1310. doi: 10.1056/NEJMoa064278. [DOI] [PubMed] [Google Scholar]
- 24.Shoulders CC, Jones EL, Naoumova RP. Genetics of familial combined hyperlipidemia and risk of coronary heart disease. Hum Mol Genet. 2004;13:R149–R160. doi: 10.1093/hmg/ddh069. [DOI] [PubMed] [Google Scholar]
- 25.Kincer JF, Uittenbogaard A, Dressman J, Guerin TM, Febbraio M, Guo L, Smart EJ. Hypercholesterolemia promotes a CD36-dependent and endothelial nitric-oxide synthase-mediated vascular dysfunction. J Biol Chem. 2002;277:23525–23533. doi: 10.1074/jbc.M202465200. [DOI] [PubMed] [Google Scholar]
- 26.Min HK, Kapoor A, Fuchs M, Mirshahi F, Zhou H, Maher J, Kellum J, Warnick R, Contos MJ, Sanyal AJ. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab. 2012;15:665–674. doi: 10.1016/j.cmet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao C, Dahlman-Wright K. Liver X receptor in cholesterol metabolism. J Endocrinol. 2010;204:233–240. doi: 10.1677/JOE-09-0271. [DOI] [PubMed] [Google Scholar]
- 28.Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metab. 2010;21:699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Vickers KC, Shoucri BM, Levin MG, Wu H, Pearson DS, Osei-Hwedieh D, Collins FS, Remaley AT, Sethupathy P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kida K, Nakajima M, Mohri T, Oda Y, Takagi S, Fukami T, Yokoi T. PPARα is regulated by miR-21 and miR-27b in human liver. Pharm Res. 2011;28:2467–2476. doi: 10.1007/s11095-011-0473-y. [DOI] [PubMed] [Google Scholar]
- 32.Shimizugawa T, Ono M, Shimamura M, Yoshida K, Ando Y, Koishi R, Ueda K, Inaba T, Minekura H, Kohama T, Furukawa H. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J Biol Chem. 2002;277:33742–33748. doi: 10.1074/jbc.M203215200. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Afroza H, Rader DJ, Jin W. Angiopoietin-like protein 3 inhibits lipoprotein lipase activity through enhancing its cleavage by proprotein convertases. J Biol Chem. 2010;285:27561–27570. doi: 10.1074/jbc.M110.144279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng L, Lv GC, Sheng J, Yang YD. Effect of miRNA-10b in regulating cellular steatosis level by targeting PPAR-α expression, a novel mechanism for the pathogenesis of NAFLD. J Gastroenterol Hepatol. 2010;25:156–163. doi: 10.1111/j.1440-1746.2009.05949.x. [DOI] [PubMed] [Google Scholar]
- 35.Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oram JF. ATP-binding cassette transporter A1 and cholesterol trafficking. Curr Opin Lipidol. 2002;13:373–381. doi: 10.1097/00041433-200208000-00004. [DOI] [PubMed] [Google Scholar]
- 37.Tall AR. Role of ABCA1 in cellular cholesterol efflux and reverse cholesterol transport. Arterioscler Thromb Vasc Biol. 2003;23:710–711. doi: 10.1161/01.ATV.0000068683.51375.59. [DOI] [PubMed] [Google Scholar]
- 38.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, Leclercq IA, MacDougald OA, Bommer GT. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, et al. microRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci USA. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci USA. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 45.Allen RM, Marquart TJ, Albert CJ, Suchy FJ, Wang DQ, Ananthanarayanan M, Ford DA, Baldan A. miR-33 controls the expression of biliary transporters, and mediates statin- and diet-induced hepatotoxicity. EMBO Mol Med. 2012;4:882–895. doi: 10.1002/emmm.201201228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 50.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 53.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 54.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castoldi M, Vujic Spasic M, Altamura S, Elmen J, Lindow M, Kiss J, Stolte J, Sparla R, D’Alessandro LA, Klingmuller U, et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J Clin Invest. 2011;121:1386–1396. doi: 10.1172/JCI44883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S, Zhu J, Fu H, Wan J, Hu Z, Liu S, Li J, Tie Y, Xing R, Zhu J, et al. Hepato-specific microRNA-122 facilitates accumulation of newly synthesized miRNA through regulating PRKRA. Nucleic Acids Res. 2012;40:884–891. doi: 10.1093/nar/gkr715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen J, Friedman JR. miR-122 regulates hepatic lipid metabolism and tumor suppression. J Clin Invest. 2012;122:2773–2776. doi: 10.1172/JCI63966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu J, Xu Y, Hao J, Wang S, Li C, Meng S. MiR-122 in hepatic function and liver diseases. Protein Cell. 2012;3:364–371. doi: 10.1007/s13238-012-2036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1α and affects lipid metabolism. J Lipid Res. 2010;51:1513–1523. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramirez CM, Rotllan N, Vlassov AV, Davalos A, Li M, Goedeke L, Aranda JF, Cirera-Salinas D, Araldi E, Salerno A, et al. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ Res. 2013;112:1592–1601. doi: 10.1161/CIRCRESAHA.112.300626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez CM, Davalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, Suarez Y, Fernandez-Hernando C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez-Hernando C, Kim J. MiR-106b impairs cholesterol efflux and increases Aβ levels by repressing ABCA1 expression. Exp Neurol. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun D, Zhang J, Xie J, Wei W, Chen M, Zhao X. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 2012;586:1472–1479. doi: 10.1016/j.febslet.2012.03.068. [DOI] [PubMed] [Google Scholar]
- 64.Zhong D, Huang G, Zhang Y, Zeng Y, Xu Z, Zhao Y, He X, He F. MicroRNA-1 and microRNA-206 suppress LXRα-induced lipogenesis in hepatocytes. Cell. Signaling. 2013;25:1429–1437. doi: 10.1016/j.cellsig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 65.Zhong D, Zhang Y, Zeng YJ, Gao M, Wu GZ, Hu CJ, Huang G, He FT. MicroRNA-613 represses lipogenesis in HepG2 cells by downregulating LXRα. Lipids Health Dis. 2013;12:32. doi: 10.1186/1476-511X-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sacco J, Adeli K. MicroRNAs: emerging roles in lipid and lipoprotein metabolism. Curr Opin Lipidol. 2012;23:220–225. doi: 10.1097/MOL.0b013e3283534c9f. [DOI] [PubMed] [Google Scholar]
- 67.Yang K, He YS, Wang XQ, Lu L, Chen QJ, Liu J, Sun Z, Shen WF. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting Toll-like receptor 4. FEBS Lett. 2011;585:854–860. doi: 10.1016/j.febslet.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 68.Chen T, Yan H, Li Z, Jing T, Zhu W, Ge J, Zheng X, Pan X, Yan H, Zhu J. MicroRNA-155 regulates lipid uptake, adhesion/chemokine marker secretion and SCG2 expression in oxLDL-stimulated dendritic cells/macrophages. Int J Cardiol. 2011;147:446–447. doi: 10.1016/j.ijcard.2010.10.133. [DOI] [PubMed] [Google Scholar]
- 69.Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, Wang C. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83:131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 70.Huang RS, Hu GQ, Lin B, Lin ZY, Sun CC. MicroRNA-155 silencing enhances inflammatory response and lipid uptake in oxidized low-density lipoprotein-stimulated human THP-1 macrophages. J Investig Med. 2010;58:961–967. doi: 10.231/JIM.0b013e3181ff46d7. [DOI] [PubMed] [Google Scholar]
- 71.Hu Z, Shen WJ, Kraemer FB, Azhar S. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol Cell Biol. 2012;32:5035–5045. doi: 10.1128/MCB.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang L, Jia XJ, Jiang HJ, Du Y, Yang F, Si SY, Hong B. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol Cell Biol. 2013;33:1956–1964. doi: 10.1128/MCB.01580-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yin H, Hu M, Zhang R, Shen Z, Flatow L, You M. MicroRNA-217 promotes ethanol-induced fat accumulation in hepatocytes by down-regulating SIRT1. J Biol Chem. 2012;287:9817–9826. doi: 10.1074/jbc.M111.333534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arnold M, Ellwanger DC, Hartsperger ML, Pfeufer A, Stumpflen V. Cis-acting polymorphisms affect complex traits through modifications of microRNA regulation pathways. PLoS ONE. 2012;7:e36694. doi: 10.1371/journal.pone.0036694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park YS, Jeon YJ, Lee BE, Kim TG, Choi JU, Kim DS, Kim NK. Association of the miR-146aC>G, miR-196a2C>T, and miR-499A>G polymorphisms with moyamoya disease in the Korean population. Neurosci Lett. 2012;521:71–75. doi: 10.1016/j.neulet.2012.05.062. [DOI] [PubMed] [Google Scholar]
- 76.Fan C, Chen C, Wu D. The association between common genetic variant of microRNA-499 and cancer susceptibility: a meta-analysis. Mol Biol Rep. 2013;40:3389–3394. doi: 10.1007/s11033-012-2416-z. [DOI] [PubMed] [Google Scholar]
- 77.Wang F, Ma YL, Zhang P, Yang JJ, Chen HQ, Liu ZH, Peng JY, Zhou YK, Qin HL. A genetic variant in microRNA-196a2 is associated with increased cancer risk: a meta-analysis. Mol Biol Rep. 2012;39:269–275. doi: 10.1007/s11033-011-0735-0. [DOI] [PubMed] [Google Scholar]
- 78.Kapeller J, Houghton LA, Monnikes H, Walstab J, Moller D, Bonisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F, et al. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967–2977. doi: 10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]
- 79.Richardson K, Nettleton JA, Rotllan N, Tanaka T, Smith CE, Lai CQ, Parnell LD, Lee YC, Lahti J, Lemaitre RN, et al. Gain-of-function lipoprotein lipase variant rs13702 modulates lipid traits through disruption of a microRNA-410 seed site. Am J Hum Genet. 2013;92:5–14. doi: 10.1016/j.ajhg.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tian GP, Chen WJ, He PP, Tang SL, Zhao GJ, Lv YC, Ouyang XP, Yin K, Wang PP, Cheng H, et al. MicroRNA-467b targets LPL gene in RAW 264.7 macrophages and attenuates lipid accumulation and proinflammatory cytokine secretion. Biochimie. 2012;94:2749–2755. doi: 10.1016/j.biochi.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 81.Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol. 2013;33:186–192. doi: 10.1161/ATVBAHA.112.300139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nathan DM. Long-term complications of diabetes mellitus. N Engl J Med. 1993;328:1676–1685. doi: 10.1056/NEJM199306103282306. [DOI] [PubMed] [Google Scholar]
- 83.Paternostro G, Camici PG, Lammerstma AA, Marinho N, Baliga RR, Kooner JS, Radda GK, Ferrannini E. Cardiac and skeletal muscle insulin resistance in patients with coronary heart disease. A study with positron emission tomography. J Clin Invest. 1996;98:2094–2099. doi: 10.1172/JCI119015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 85.Zhao H, Guan J, Lee HM, Sui Y, He L, Siu JJ, Tse PP, Tong PC, Lai FM, Chan JC. Up-regulated pancreatic tissue microRNA-375 associates with human type 2 diabetes through β-cell deficit and islet amyloid deposition. Pancreas. 2010;39:843–846. doi: 10.1097/MPA.0b013e3181d12613. [DOI] [PubMed] [Google Scholar]
- 86.Ryu HS, Park SY, Ma D, Zhang J, Lee W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS ONE. 2011;6:e17343. doi: 10.1371/journal.pone.0017343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signaling. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 90.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 91.Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 92.Ramachandran D, Roy U, Garg S, Ghosh S, Pathak S, Kolthur-Seetharam U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic β-islets. FEBS J. 2011;278:1167–1174. doi: 10.1111/j.1742-4658.2011.08042.x. [DOI] [PubMed] [Google Scholar]
- 93.Kanfi Y, Peshti V, Gozlan YM, Rathaus M, Gil R, Cohen HY. Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 2008;582:2417–2423. doi: 10.1016/j.febslet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 94.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol. 2007;21:2785–2794. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 95.Pullen TJ, da Silva Xavier G, Kelsey G, Rutter GA. miR-29a and miR-29b contribute to pancreatic β-cell-specific silencing of monocarboxylate transporter 1 (Mct1) Mol Cell Biol. 2011;31:3182–3194. doi: 10.1128/MCB.01433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res. 2010;86:410–420. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 97.Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 2008;389:305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- 98.Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R, Rutter GA, Van Obberghen E. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic β-cell lines. J Biol Chem. 2007;282:19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 99.Pandey AK, Verma G, Vig S, Srivastava S, Srivastava AK, Datta M. miR-29a levels are elevated in the db/db mice liver and its overexpression leads to attenuation of insulin action on PEPCK gene expression in HepG2 cells. Mol Cell Endocrinol. 2011;332:125–133. doi: 10.1016/j.mce.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 100.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 101.Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 104.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 105.Raitoharju E, Lyytikainen LP, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kahonen M, Karhunen PJ, et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 2011;219:211–217. doi: 10.1016/j.atherosclerosis.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 106.Liu X, Cheng Y, Yang J, Xu L, Zhang C. Cell-specific effects of miR-221/222 in vessels: molecular mechanism and therapeutic application. J Mol Cell Cardiol. 2012;52:245–255. doi: 10.1016/j.yjmcc.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 108.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104:476–487. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ponomaryov T, Peled A, Petit I, Taichman RS, Habler L, Sandbank J, Arenzana-Seisdedos F, Magerus A, Caruz A, Fujii N, et al. Induction of the chemokine stromal-derived factor-1 following DNA damage improves human stem cell function. J Clin Invest. 2000;106:1331–1339. doi: 10.1172/JCI10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zernecke A, Schober A, Bot I, von Hundelshausen P, Liehn EA, Mopps B, Mericskay M, Gierschik P, Biessen EA, Weber C. SDF-1α/CXCR4 axis is instrumental in neointimal hyperplasia and recruitment of smooth muscle progenitor cells. Circ Res. 2005;96:784–791. doi: 10.1161/01.RES.0000162100.52009.38. [DOI] [PubMed] [Google Scholar]
- 111.Wang YS, Wang HY, Liao YC, Tsai PC, Chen KC, Cheng HY, Lin RT, Juo SH. MicroRNA-195 regulates vascular smooth muscle cell phenotype and prevents neointimal formation. Cardiovasc Res. 2012;95:517–526. doi: 10.1093/cvr/cvs223. [DOI] [PubMed] [Google Scholar]
- 112.Kannel WB. Blood pressure as a cardiovascular risk factor: prevention and treatment. JAMA, J Am Med Assoc. 1996;275:1571–1576. [PubMed] [Google Scholar]
- 113.Touyz RM, Tabet F, Schiffrin EL. Redox-dependent signalling by angiotensin II and vascular remodelling in hypertension. Clin Exp Pharmacol Physiol. 2003;30:860–866. doi: 10.1046/j.1440-1681.2003.03930.x. [DOI] [PubMed] [Google Scholar]
- 114.Gullapalli N, Bloch MJ, Basile J. Renin-angiotensin-aldosterone system blockade in high-risk hypertensive patients: current approaches and future trends. Ther Adv Cardiovasc Dis. 2010;4:359–373. doi: 10.1177/1753944710384430. [DOI] [PubMed] [Google Scholar]
- 115.Giles TD, Berk BC, Black HR, Cohn JN, Kostis JB, Izzo JL, Jr, Weber MA. Expanding the definition and classification of hypertension. J Clin Hypertens. 2005;7:505–512. doi: 10.1111/j.1524-6175.2005.04769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Batkai S, Thum T. MicroRNAs in hypertension: mechanisms and therapeutic targets. Curr Hypertens Rep. 2012;14:79–87. doi: 10.1007/s11906-011-0235-6. [DOI] [PubMed] [Google Scholar]
- 117.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Norata GD, Pinna C, Zappella F, Elia L, Sala A, Condorelli G, Catapano AL. MicroRNA 143–145 deficiency impairs vascular function. Int J Immunopathol Pharmacol. 2012;25:467–474. doi: 10.1177/039463201202500216. [DOI] [PubMed] [Google Scholar]
- 119.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dong S, Cheng Y, Yang J, Li J, Liu X, Wang X, Wang D, Krall TJ, Delphin ES, Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am. J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 124.Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 126.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, Stack C, Latimer PA, Olson EN, van Rooij E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Swynghedauw B. Molecular mechanisms of myocardial remodeling. Physiol Rev. 1999;79:215–262. doi: 10.1152/physrev.1999.79.1.215. [DOI] [PubMed] [Google Scholar]
- 129.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.da Costa Martins PA, Salic K, Gladka MM, Armand AS, Leptidis S, el Azzouzi H, Hansen A, Coenen-de Roo CJ, Bierhuizen MF, van der Nagel R, et al. MicroRNA-199b targets the nuclear kinase Dyrk1a in an auto-amplification loop promoting calcineurin/NFAT signalling. Nat Cell Biol. 2010;12:1220–1227. doi: 10.1038/ncb2126. [DOI] [PubMed] [Google Scholar]
- 131.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 132.Bourajjaj M, Armand AS, da Costa Martins PA, Weijts B, van der Nagel R, Heeneman S, Wehrens XH, De Windt LJ. NFATc2 is a necessary mediator of calcineurin-dependent cardiac hypertrophy and heart failure. J Biol Chem. 2008;283:22295–22303. doi: 10.1074/jbc.M801296200. [DOI] [PubMed] [Google Scholar]
- 133.Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, Li YR, Li PF. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- 134.Olson EN, Williams RS. Calcineurin signaling and muscle remodeling. Cell. 2000;101:689–692. doi: 10.1016/s0092-8674(00)80880-6. [DOI] [PubMed] [Google Scholar]
- 135.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 136.Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol. 2007;7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 137.Gareus R, Kotsaki E, Xanthoulea S, van der Made I, Gijbels MJ, Kardakaris R, Polykratis A, Kollias G, de Winther MP, Pasparakis M. Endothelial cell-specific NF-κB inhibition protects mice from atherosclerosis. Cell Metab. 2008;8:372–383. doi: 10.1016/j.cmet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 138.Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L, Hunninghake GM, Vera MP, Blackwell TS, Baron RM, Feinberg MW. MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J Clin Invest. 2012;122:1973–1990. doi: 10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hansson GK, Libby P. The immune response in atherosclerosis: a double-edged sword. Nat Rev Immunol. 2006;6:508–519. doi: 10.1038/nri1882. [DOI] [PubMed] [Google Scholar]
- 141.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 142.Zhu J, Chen T, Yang L, Li Z, Wong MM, Zheng X, Pan X, Zhang L, Yan H. Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10. PLoS ONE. 2012;7:e46551. doi: 10.1371/journal.pone.0046551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nazari-Jahantigh M, Wei Y, Noels H, Akhtar S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest. 2012;122:4190–4202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 146.Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125:2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 147.Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, Kawamura A, Nakamura K, Takeuchi T, Tanabe M. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- 148.Hussein K, Busche G, Muth M, Gohring G, Kreipe H, Bock O. Expression of myelopoiesis-associated microRNA in bone marrow cells of atypical chronic myeloid leukaemia and chronic myelomonocytic leukaemia. Ann Hematol. 2011;90:307–313. doi: 10.1007/s00277-010-1072-4. [DOI] [PubMed] [Google Scholar]
- 149.Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M, Zhu Y, Zhao Q, Dong YW, Shao K, et al. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS Lett. 2012;586:1038–1043. doi: 10.1016/j.febslet.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 150.Yu CH, Xu CF, Li YM. Association of MicroRNA-223 expression with hepatic ischemia/reperfusion injury in mice. Dig Dis Sci. 2009;54:2362–2366. doi: 10.1007/s10620-008-0629-8. [DOI] [PubMed] [Google Scholar]