Use of photoinactivation, cisternae-specific fluorescence recovery, and high-resolution microscopy shows that the membrane tethers GRASP65 and GRASP55 on early and late Golgi membranes, respectively, are critical to the specific, homotypic fusion of the membranes on which they reside.
Abstract
Homotypic membrane tethering by the Golgi reassembly and stacking proteins (GRASPs) is required for the lateral linkage of mammalian Golgi ministacks into a ribbon-like membrane network. Although GRASP65 and GRASP55 are specifically localized to cis and medial/trans cisternae, respectively, it is unknown whether each GRASP mediates cisternae-specific tethering and whether such specificity is necessary for Golgi compartmentalization. Here each GRASP was tagged with KillerRed (KR), expressed in HeLa cells, and inhibited by 1-min exposure to light. Significantly, inactivation of either GRASP unlinked the Golgi ribbon, and the immediate effect of GRASP65-KR inactivation was a loss of cis- rather than trans-Golgi integrity, whereas inactivation of GRASP55-KR first affected the trans- and not the cis-Golgi. Thus each GRASP appears to play a direct and cisternae-specific role in linking ministacks into a continuous membrane network. To test the consequence of loss of cisternae-specific tethering, we generated Golgi membranes with a single GRASP on all cisternae. Remarkably, the membranes exhibited the full connectivity of wild-type Golgi ribbons but were decompartmentalized and defective in glycan processing. Thus the GRASP isoforms specifically link analogous cisternae to ensure Golgi compartmentalization and proper processing.
INTRODUCTION
Intracellular organelles connected by membrane-trafficking pathways allow cargo molecules to sequentially move between way stations optimized for processing. The abundant membrane-trafficking connecting compartments must be tightly controlled to prevent rapid breakdown of compartment integrity. Of critical importance is specificity during membrane fusion. Without this specificity, mistakes in the fusion of vesicles with compartments and in compartments with other compartments would quickly intermix membranous organelles. Membrane tethers—proteins that form a transient molecular bridge between membranes before fusion—are believed to contribute to the specificity of fusion because tethering complexes contact the two membranes with specificity (Whyte and Munro, 2002). However, there is very little direct evidence showing that tethers affect fusion specificity and compartment integrity.
The assembly of the mammalian Golgi apparatus into a ribbon-like membrane network provides an important example of organelle tethering and fusion. Whereas other cell types have many isolated Golgi stacks distributed throughout the cytoplasm, mammalian Golgi stacks are collected near the minus ends of microtubules and laterally linked by dynamic fusion events to form the Golgi ribbon (Ladinsky et al., 1999). Golgi stacks consist of flattened cisternae, each with high membrane curvature at their perimeters, called Golgi rims. Membrane tubules extend from the rims forming the lateral linkages between adjacent stacks. Based on their appearance in electron micrographs, the contact areas are termed noncompact zones (Rambourg et al., 1987). In addition to the tubular connections, the noncompact zones also contain vesicles. The lateral linkage is considered a product of homotypic fusion because analogous cisternae connect to one another (Thorne-Tjomsland et al., 1998). That is, membrane continuity is observed among most cis cisternae in a Golgi ribbon, but cis cisternae rarely exhibit connections to medial or trans cisternae. Whether and how the contacts are restricted to homotypic fusion and whether this preserves compartment integrity are not known.
The linkages forming the Golgi ribbon are physiologically important, in part, because they promote uniform distribution of Golgi enzymes throughout the entire network of their respective cisternae (Puthenveedu et al., 2006). For example, in a Golgi assembly assay, the individual Golgi stacks that arise in the cell periphery are differentially enriched in Golgi markers, and then, as the stacks move inward and link to form the ribbon, these markers become evenly distributed. If the linkages are blocked from forming, there is a disproportioning of the markers. As a consequence there are defects in glycan processing presumably due to underprocessing in isolated Golgi stacks containing diminished levels of processing enzyme (Puthenveedu et al., 2006). From an evolutionary perspective, the need for homotypic lateral fusion to distribute Golgi enzymes may have arisen when the Golgi acquired the characteristic of moving inward on microtubule tracks. Inward Golgi movement physically separates mammalian Golgi membranes from endoplasmic reticulum (ER) exit sites. In contrast, the distributed ministacks of other cell types are positioned immediately adjacent to sites of ER exit and may use recycling through the ER to ensure proper enzyme distribution. Rapid recycling due to proximity to ER exit sites can also explain why Golgi ministacks in mammalian cells generated by microtubule depolymerization show full glycosylation competence (Xiang et al., 2013). Thus the specificity of homotypic, cisterna–cisterna fusion is hypothetically required for both Golgi compartmentalization and proper processing.
The Golgi reassembly and stacking proteins (GRASPs) are believed to be the membrane tethers involved in fusion of analogous cisternae to form the Golgi ribbon (Wilson and Ragnini-Wilson, 2010; Klumperman, 2011; Vinke et al., 2011). GRASP65 is localized to cis cisternae, and GRASP55 is localized to medial and trans cisternae (Shorter et al., 1999). GRASP65 localization depends on its N-terminal myristic acid and its ability to bind to GM130 on the cis-Golgi (Barr et al., 1998; Shorter and Warren, 1999; Puthenveedu et al., 2006). GRASP55 localization involves its N-terminal myristic acid and its binding to med/trans-Golgi–localized proteins, including golgin-45 (Short et al., 2001). Knockdown of either GRASP unlinks the ribbon and causes defective glycosylation (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008; Xiang and Wang, 2010; Xiang et al., 2013). Each protein specifically self-interacts using its N-terminal GRASP domain (Wang et al., 2005). The GRASP domain is a tandem set of atypical postsynaptic density–Drosophila disk large tumor suppressor–zonula occludens-1 (PDZ) domains (Truschel et al., 2011). The binding groove of the first PDZ domain mediates the self-interaction by binding an internal PDZ ligand present on the surface of the second PDZ domain (Sengupta et al., 2009; Truschel et al., 2011). At least for GRASP65, the binding groove of the second PDZ domain binds a C-terminal PDZ ligand in its Golgi-localized receptor, GM130 (Bachert and Linstedt, 2010). Thus the model for cisternae-specific membrane tethering during ribbon formation invokes isoform-specific localization and self-interaction of the GRASP proteins, thereby providing an opportunity to test the role of membrane tethering in the specificity of membrane fusion and compartment maintenance.
However, given the small distances and dynamic aspects involved, testing the cisternae-specific roles of the GRASP proteins is challenging. We sought to overcome these limitations by using a combination of chromophore-assisted light inactivation (CALI), fluorescence recovery after photobleaching (FRAP), and high-resolution structured illumination microscopy (SIM). KillerRed (KR)-mediated CALI of either GRASP protein coupled with FRAP of cisternae-specific markers reported Golgi ribbon integrity in the immediate moments after GRASP inactivation and showed that GRASP65 and GRASP55 mediate Golgi ribbon formation in the early and late Golgi cisternae, respectively. Further, SIM analysis of Golgi markers in cells expressing a single GRASP throughout the Golgi indicated that compartmentalization of the Golgi ribbon depends on the separation of duty by the GRASP isoforms.
RESULTS
Rapid GRASP inactivation blocks Golgi ribbon formation
Although the Golgi ribbon is unlinked by depletion of either GRASP using siRNA (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008; Xiang and Wang, 2010), these experiments involve a long time course of inhibition, leaving open the question of whether Golgi ribbon integrity acutely depends on GRASP function. Recently we demonstrated a rapid and specific block of ER exit using a 30-s illumination the KR-tagged COPII component Sec13 (Jarvela and Linstedt, 2012b). Irradiation of KR with green light (561 ± 20 nm) results in the generation of reactive oxygen species (ROS) that can break bonds and denature proteins (Jay, 1988; Surrey et al., 1998). ROS have a half maximal distance of 4 nm, meaning that this damage is spatially limited to the tagged protein and its interacting factors (Bulina et al., 2006). Thus our strategy here was to express either GRASP65 or GRASP55 tagged with KR, inactivate the tagged protein using 561-nm light, and then immediately assess integrity of the Golgi ribbon. Because the GRASP proteins form homo-oligomers, our hope was that irradiation would inactivate not only the expressed chimeric construct, but also its endogenous counterpart. An important aspect of this technique is confirming the proper localization and expression level of the KR constructs, and this was carried out on a cell-by-cell basis before each trial (Supplemental Figure S1). Only cells that exhibited at least 50% of their total GRASP-KR fluorescence on the Golgi, which was demarcated using the Golgi enzyme N-acetylgalactosaminyltransferase T2 tagged with the green fluorescent protein (GalNAcT2-GFP; Storrie et al., 1998), were chosen and illuminated to specifically bleach the KR fluorescence (Supplemental Figure S2). KillerRed-induced oxidative damage may cause any of a number of changes to the proteins that it affects (Jay, 1988; Liao et al., 1994; Surrey et al., 1998). Sometimes this includes cleavage, but not always. Because of this uncertainty and because we were limited to microscope-based inactivation, we were unable to directly assay for damage, and we could not estimate the percentage of GRASP molecules and binding partners damaged by irradiation under conditions of the experiment.
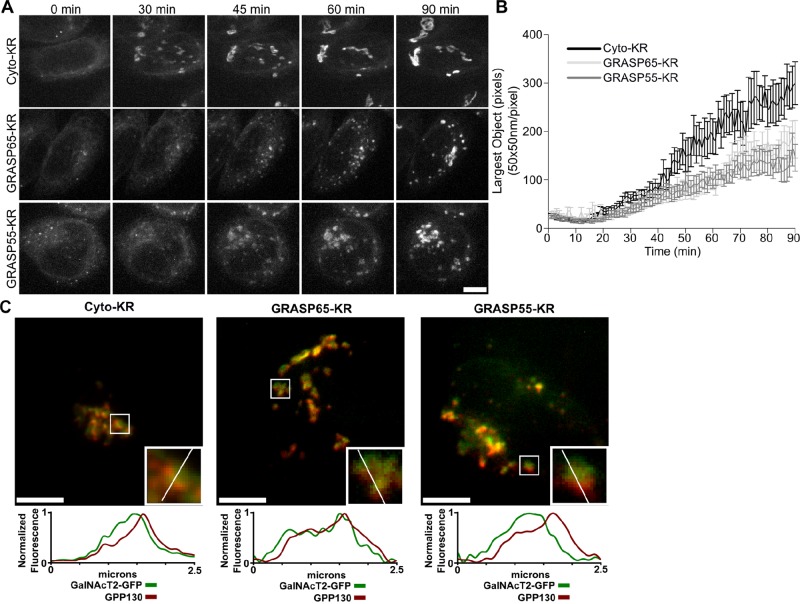
Several permutations of the experiment were used. In the first, we monitored Golgi assembly upon brefeldin A washout in cells after inactivation of either of the GRASPs. Golgi reassembly involves many factors and several distinct steps. Thus whether reassembly proceeds normally up to, but not including, a point at which GRASP proteins link the Golgi is a test of whether inactivation is specific and effective and supports a direct role for GRASPs in linking. The C-terminus of either GRASP was tagged with KR and expressed in cells expressing GalNAcT2-GFP to mark Golgi membranes. GalNAcT2-GFP is present in all cisternae of the stack (Storrie et al., 1998). A cytoplasmic version of KR (Cyto-KR) was used as a control. The constructs were irradiated immediately after drug washout, and GalNAcT2-GFP was imaged at 1-min intervals to follow Golgi assembly. In irradiated cells expressing Cyto-KR, GalNAcT2-GFP emerged from the ER and accumulated in punctate structures that moved inward, eventually coalescing into a Golgi ribbon (Figure 1A and Supplemental Movie S1). Similar to nonirradiated cells or cells not expressing KR constructs, the entire assembly occurred in ∼90 min. In contrast, irradiated cells expressing either GRASP-KR protein showed GalNAcT2-GFP exiting from the ER and moving inward in puncta, but these objects failed to coalesce into an intact Golgi ribbon (Figure 1A and Supplemental Movie S1). The experiment was quantified by determining the size of the largest GalNAcT2-GFP object at each time point for each cell analyzed (Figure 1B). As can be seen, the GRASP-KR–inactivated cells failed to reestablish a Golgi of normal size; instead they exhibited a collection of mostly perinuclear Golgi fragments. These Golgi fragments had the characteristic fluorescence pattern of stacked Golgi membranes, with the fluorescence of cis-Golgi–localized GPP130 being slightly offset from that of GalNAcT2-GFP (Figure 1C). The resulting pattern was essentially identical to that observed after depletion of either GRASP using small interfering RNA (siRNA), in which the membranes were shown by electron microscopy to be stacked but unlinked (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008). In comparison to the siRNA experiments, the relatively short time course of inactivation by KR supports a direct role for the GRASPs in establishing the Golgi ribbon.
FIGURE 1:
Inactivation of GRASP proteins blocks Golgi ribbon formation. (A) Cells stably expressing GalNAcT2-GFP were transfected with KR constructs. After 2 d, GalNAcT2-GFP was redistributed to the ER with 20 μg/ml brefeldin A for 20 min. Cells were then quickly washed and placed in imaging medium and irradiated with 535- to 580-nm light at an intensity of 2 W/cm2 for 30 s. Average value z-projections of GalNAcT2-GFP fluorescence are shown for the indicated times after inactivation. Scale bar, 5 μm. (B) Golgi assembly was quantified by determining the size of the largest GalNAcT2-GFP object at each time point, with the threshold set above the level of GalNAcT2-GFP fluorescence in the ER (n = 10, mean ± SEM). (C) Cells were fixed 90 min after BFA washout and stained for GPP130. Fluorescence intensity across the width of the Golgi objects (white line, inset) was measured and compared for the Golgi markers GPP130 (red) and GalNAcT2-GFP (green). Scale bar, 5 μm.
In the second permutation, we asked whether inactivation of either GRASP would affect an intact Golgi and, if so, with what time course. As a control we used a previously described construct (Jarvela and Linstedt, 2012b) in which KR contains at its C-terminus the membrane anchor of the Golgi protein giantin (KR-Gtn), yielding a Golgi-localized and cytoplasmically disposed KR. The control or GRASP-KR constructs were expressed in GalNAcT2-GFP–expressing cells and irradiated, followed by live-cell imaging of GalNAcT2-GFP at 1-min intervals. The results clearly indicated that Golgi fragmentation consistent with unlinking took place after irradiation of either GRASP but not after irradiation of the control (Figure 2A and Supplemental Movie S2). To quantify the experiment, we determined the number of GalNAcT2-GFP objects at each time point in each cell analyzed. The number of objects was also determined for every frame of a premovie acquired before the inactivation. The resulting plot shows that the GalNAcT2-GFP pattern yields a small number of objects in untreated or control inactivated cells, whereas there was a large, statistically significant increase in object number in GRASP-KR–inactivated cells (Figure 2B). The fragmentation became detectable at around 10 min after inactivation and continued to increase for the next 20 min. As addressed in the next section, this analysis may underestimate the rate because Golgi unlinking is only detectable once individual Golgi objects move far enough away from one another to be resolved. Nevertheless, the fact that fragmentation became detectable within 10–30 min of GRASP-KR inactivation provides strong evidence that ongoing function of these proteins is required to maintain integrity of the Golgi ribbon.
FIGURE 2:
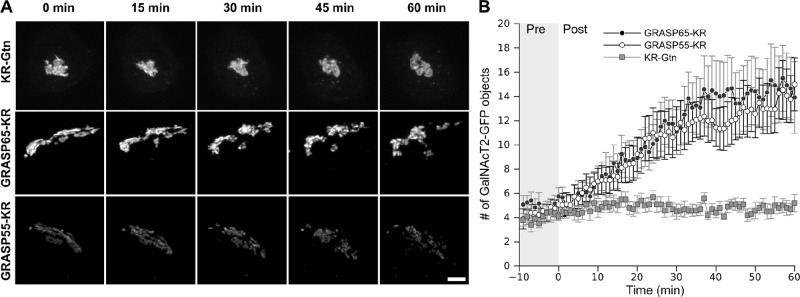
GRASP inactivation blocks Golgi ribbon maintenance. (A) Cells expressing the indicated KR constructs were irradiated with 535- to 580-nm light at an intensity of 2 W/cm2 for 30 s. We acquired z-stacks at 1-min intervals for 10 min before and 60 min after inactivation. Maximum value Z-projections are shown for the indicated time points. Scale bar, 5 μm. (B) The total number of discrete Golgi objects at each time point was quantified using the ImageJ Analyze particles function (n = 10, mean ± SEM).
GRASP inactivation unlinks specific Golgi cisternae
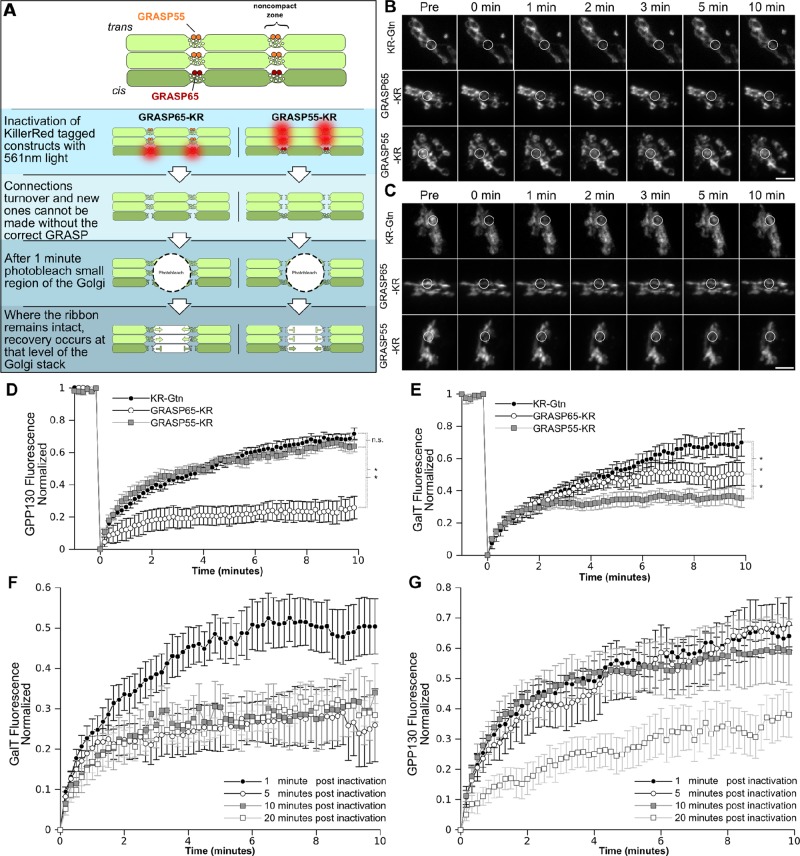
In the third and final permutation of the KR experiment, we addressed the question of whether the two GRASP proteins play compartment-specific roles in linking the Golgi ribbon. For this purpose we analyzed compartment-specific integral proteins. GFP-tagged GPP130 (GPP130-GFP) was used to monitor the cis-Golgi (Mukhopadhyay et al., 2010), and a yellow fluorescent protein–tagged galactosyltransferase transmembrane domain (GalT-YFP) was used to monitor the trans-Golgi (Jarvela and Linstedt, 2012b). To increase temporal sensitivity, we assessed Golgi ribbon connectivity using FRAP. We previously demonstrated that intact Golgi ribbons show rapid recovery of GalNAcT2-GFP fluorescence, whereas unlinked Golgi ribbons do not (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008). Thus our plan was to follow KR inactivation with photobleaching of the GFP and YFP signals in a small region of the Golgi and then record recovery with rapid 10-s acquisition intervals. To test for an immediate effect on Golgi integrity, we carried out the photobleaching 1 min after the KR inactivation. Compartment-specific linking by GRASP65 predicts that its inactivation would produce loss of FRAP for cis-localized GPP130-GFP but not for trans-localized GalT-YFP (Figure 3A). The opposite outcome is predicted for inactivation of GRASP55-KR.
FIGURE 3:
GRASPs are required for integrity of distinct levels of the Golgi stack. (A) A schematic of the experiment in which KR constructs were inactivated, and 1 min after inactivation a small segment of the Golgi was photobleached. If the Golgi ribbon remained intact at the cis-Golgi, GPP130-GFP fluorescence was expected to recover. If the trans-Golgi ribbon remained interconnected, GalT-YFP fluorescence was expected to recover. (B, C) Cells expressing the indicated KR constructs were irradiated with 15.8 mW of a 561-nm laser for 45 μs/pixel repeated six times. GPP130-GFP fluorescence to mark the cis-Golgi (B) or GalT-YFP fluorescence to mark the trans-Golgi (C) was recorded at 10-s intervals for 1 min, and then the outlined segment was bleached with 27.3-mW, 488-nm laser light for 20 μs repeated 60 times. Then fluorescence recovery was recorded for a further 10 min at 10-s intervals. Average value z- projections are shown for the indicated time points. Scale bar, 5 μm. (D, E) GPP130-GFP (D) and GalT-YFP (E) fluorescence was measured in the bleached zone and plotted vs. time (n = 10 cells, mean ± SEM). Results were compared for statistical significance using a two-tailed Students t test (*p < 0.05; n.s., not significant). (F) Trans marker GalT-YFP FRAP after being carried out at the indicated times postinactivation of GRASP65-KR (n > 5, mean ± SEM). (G) Cis marker GPP130-GFP FRAP after being carried out at the indicated times postinactivation of GRASP55-KR (n > 5, mean ± SEM).
The results for the cis-Golgi marked by GPP130-GFP are shown in Figure 3B. As expected, rapid recovery was observed after inactivation of the control construct KR-Gtn. Inactivation of GRASP55-KR was also followed by rapid recovery of GPP130-GFP fluorescence, indicating that the cis-Golgi remained intact. In contrast, inactivation of GRASP65-KR led to a pronounced block in recovery. The intimate role of GRASP65 in linking cis cisternae is strongly supported by these data, given that the FRAP component of the assay was initiated only 1 min after inactivation and that recovery was significantly delayed even at the earliest time points. The results for the trans-Golgi marker GalT-YFP also supported cisternae-specific roles for the GRASPs but were less pronounced (Figure 3C). GRASP65-KR inactivation yielded recovery that was similar to the control, yet the final extent of recovery was somewhat reduced. GRASP55-KR inactivation clearly had a more profound effect on GalT-GFP recovery than did either the KR-Gtn or GRASP65, and this block was also significant compared with its lack of effect on GPP130-GFP. There are several possible explanations for the residual recovery in the inactivated cells, including imprecise targeting of the constructs and recycling of the markers by vesicular trafficking.
Because acute inactivation of either GRASP protein yielded cisternae-specific unlinking, whereas longer-term depletion of either GRASP unlinks the entire Golgi ribbon, we tested whether acute inactivation would lead to unlinking of all cisternae if given sufficient time. For example, if GRASP65 initiates linking as Golgi cisternae elongate on the cis face of the ribbon and GRASP55 maintains these linkages as cisternae mature, then loss of GRASP65 function would ultimately destabilize the entire ribbon. Indeed, whereas GRASP65-KR inactivation had no effect on the trans marker GalT-YFP recovery at 1 min postinactivation, by 5 min there was already a marked reduction, and this was also observed at 10 and 20 min postinactivation (Figure 3D). The level of loss in recovery was similar to that reached by GRASP55-KR inactivation, suggesting that disruption at the cis face propagates to the late Golgi. Of interest, inactivation of GRASP55-KR also propagated, but this occurred with a slower time course. GRASP55-KR inactivation had no effect on cis marker GPP130-GFP recovery at 1, 5, and 10 min postinactivation, but recovery became impaired by 20 min postinactivation (Figure 3E). This time course matched that shown earlier (Figure 2, A and B) for Golgi unlinking as assessed by fragmentation of GalNAcT2-GFP fluorescence into resolvable Golgi objects. One possibility is that once late Golgi cisternae become physically separated from one another, GRASP65 can no longer effectively initiate linking of cisternae. In any case, these results reveal a rapid time course leading from the acute disruption phenotype to the longer-term knockdown phenotype, and taken together, the photoinactivation experiments strongly support a direct role for the GRASP proteins in cisternae-specific linking.
“Single-GRASP” Golgi yields intact ribbon with loss of compartmentalization
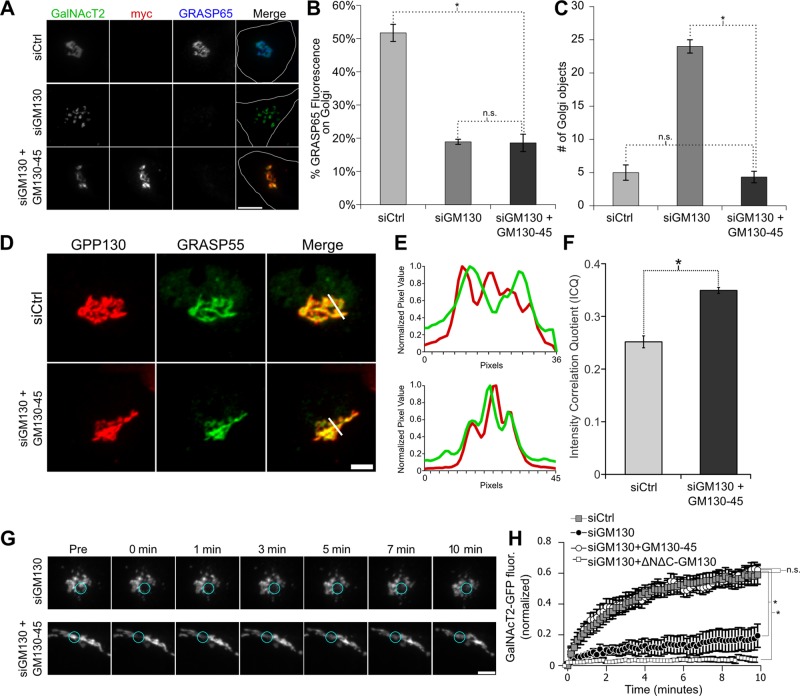
Unlike mammalian cells, which express two GRASP proteins and generate Golgi ribbon networks, nonvertebrate cells express a single GRASP and typically contain isolated ministacks (Wei and Seemann, 2010). To gain insight into why two GRASP proteins might be required in HeLa cells, we attempted to generate Golgi membranes containing a single GRASP localized to all cisternae. First, we took advantage of the fact that GRASP65 can be removed from Golgi membranes by depletion of its membrane receptor GM130 (Puthenveedu et al., 2006). As expected, transfection with GM130 siRNA caused loss of GRASP65 and unlinked the Golgi (Figure 4, A–C). Second, we expressed in these cells a myc-tagged GM130 replacement construct in which we substituted the GRASP55-binding site from golgin-45 in place of the GM130 C-terminal GRASP65-binding site. As expected, the GM130-45 chimera was Golgi localized and failed to recruit GRASP65 (Figure 4, A and B). Of significance, the continuity of the Golgi ribbon was rescued in cells expressing the GM130-45 chimera as assayed by determining the number of resolvable Golgi objects (Figure 4C). The golgin-45 segment was taken from the last 13 residues of the golgin-45 C-terminus. When targeted to the mitochondrial outer membrane, these residues were sufficient to recruit GRASP55 (Supplemental Figure S3). In addition, cells expressing the GM130-45 chimera showed a significant increase in GRASP55 on cis-Golgi membranes as indicated by colocalization with endogenous GPP130 (Figure 4, D–F). Thus rescue of linking was presumably accomplished by recruiting GRASP55 instead of GRASP65 to the cis-Golgi. Indeed, the reformed ribbon in these cells showed a dramatic recovery of GalNAcT2-GFP fluorescence after bleaching, whereas little recovery occurred in GM130-depleted cells not expressing the rescue construct (Figure 4, G and H). If full-length golgin-45 was expressed instead of the chimera, there was no rescue of Golgi linking (Supplemental Figure S4). In addition, if GRASP65 was depleted instead of GM130, the chimeric GM130-45 construct was still able to rescue linking (Supplemental Figure S5). In sum, a single GRASP protein localized to all Golgi cisternae is sufficient to stably link Golgi cisternae into a ribbon structure.
FIGURE 4:
The GM130-45 chimera rescues Golgi ribbon integrity after GM130 knockdown. (A) Immunofluorescence staining for cells stably expressing GalNAcT2-GFP that were transfected with control or GM130-specific siRNA 96 h before fixation. Where indicated, the cells were also transfected with the myc-tagged GM130-45 chimera 12 h before fixation. Scale bar, 5 μm. (B) The amount of GRASP65 fluorescence present on the Golgi was determined using GalNAcT2-GFP as a mask. The percentage of the total cellular pool that is present on the Golgi is shown for each condition (n = 10, mean ± SEM; *p < 0.05; n.s. = p > 0.05). (C) The number of Golgi objects present for each condition determined by counting discrete GalNAcT2-GFP–positive objects (n = 10, mean ± SEM; *p < 0.05; n.s., not significant). (D) Average value z-projections of GRASP55 and GPP130 immunofluorescence images in control and mycGM130g45tail-rescued–expressing cells. Expression of GM130-45 increases colocalization of GRASP55 with cis-Golgi marker GPP130, as shown by profile plots (E) and the intensity correlation quotient (F). (G) Time course of Golgi fluorescence recovery after photobleaching in siGM130 and GM130-45–rescued cells. Blue circles encompass the photobleached region. Scale bar, 5 μm. (H) Fluorescence levels in bleached region measured and plotted vs. time in siCtrl-treated cells and siGM130 cells only and siGM130 cells transfected with GM130-45 or ∆N∆C-GM130 (n = 10 cells, mean ± SEM; *p < 0.05; n.s., not significant).
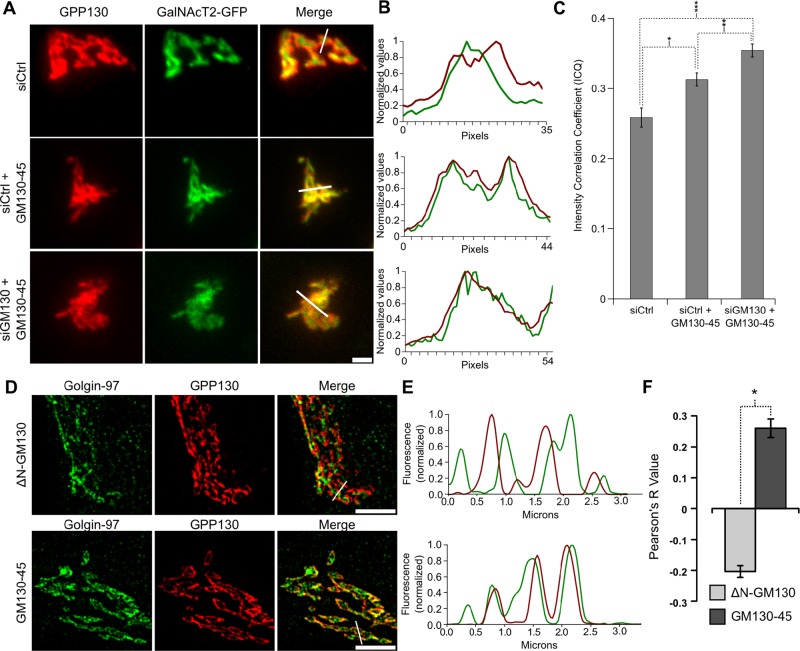
On the basis of this conclusion, we tested whether there might be a functional deficit in HeLa cell Golgi ribbons linked by a single GRASP protein. To test for defective compartmentalization in the “single-GRASP” Golgi, we first analyzed GPP130 and GalNAcT2-GFP by standard confocal microscopy. As expected, in control cells the Golgi markers showed significant side-by-side, rather than colocalized, staining that was evident both in the raw images (Figure 5A) and in profile plots taken across the Golgi ribbon (Figure 5B). In contrast, the markers coalesced in the Golgi ribbons of cells in which the GM130-45 chimera was expressed to replace the depleted GM130. Significantly, this was also the case for cells simply expressing the chimera. That is, expression of the chimera to recruit GRASP55 to the cis-Golgi was dominant negative in the sense that it caused loss of compartmentalization without depletion of GM130 or loss of GRASP65. Quantification of marker colocalization across many cells using the intensity correlation quotient (Li et al., 2004) supported the conclusion that the chimeric construct caused loss of compartmentalization whether expressed on its own or after depletion of GM130, with the latter condition yielding the greatest effect (Figure 5C).
FIGURE 5:
Expression of the GM130-45 chimera increases marker colocalization. (A) Average value z-projections of immunofluorescence images of endogenous GPP130 and stably expressed GalNAcT2-GFP in control knockdown cells, control knockdown cells expressing GM130-45, and GM130 knockdown cells rescued with GM130-45. Scale bar, 5 μm. (B) Profile plots of normalized pixel intensity of GPP130 (red) and GalNAcT2-GFP (green) corresponding to the region marked with white lines in A. (C) Colocalization of endogenous GPP130 and GalNAcT2-GFP across experimental conditions as measured by ICQ (n ≥ 15 cells, mean ± SEM; *p = 0.002, **p = 0.016, ***p = 8.2 × 10−7). (D) HeLa cells were transfected with ∆NGM130 or GM130-45. Shown is a representative single z-slice of a three-dimensional structured illumination image showing fluorescence patterns of endogenous GPP130 (red) and golgin-97 (green). Scale bar, 5 μm. (E) Normalized values of pixel intensity for GPP130 (red) and golgin-97 (green) along the region indicated by the white lines in D. (F) Colocalization of endogenous GPP130 and golgin-97 is described by the Pearson's correlation coefficient for cells transfected with ∆N GM130-WT or GM130-45 (n ≥ 15, mean ± SEM; *p = 2.9 × 10−11).
To substantiate these results, we also analyzed cells expressing the GM130-45 chimera by SIM. Here, to accentuate the degree of compartmentalization in control cells and restrict the analysis to endogenous proteins, we compared GPP130 to the trans-Golgi network marker golgin-97. In untransfected cells or cells expressing wild-type GM130 as a control, the separation of the early and late Golgi markers was striking in both the raw images (Figure 5D) and profile plots taken across the Golgi ribbon (Figure 5E). In cells expressing the GM130-45 chimera the side-by-side staining was greatly diminished, and there was a much higher incidence of colocalization. This was supported by calculation of Pearson's coefficient (Figure 5F). Both the mean and the range of the data show the significant increase in colocalization in cells expressing the GM130-45 chimera. Of importance, there was no apparent loss of cisternal stacking under these conditions, as Golgi stacks were evident in electron micrographs of cells expressing the chimeric golgin GM130-45 (Supplemental Figure S6).
Perturbed compartmentalization is associated with glycan-processing defects
To test whether the apparent loss of Golgi compartmentalization in these cells might cause a glycosylation defect, we used a simple assay for efficient terminal glycosylation (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008). GS-II lectin recognizes terminal N-acetyl-d-glucosamine, which is a processing intermediate that becomes masked in the late Golgi by the further addition of galactose and sialic acid (Kornfeld and Kornfeld, 1985). Thus a decrease in efficiency of these later steps leads to increased GS-II labeling at the cell surface. Control cells showed little surface GS-II staining, whereas cells expressing the GM130-45 chimera yielded a clear signal, indicating a defect in processing efficiency upon chimera expression (Figure 6A). As observed previously (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008), the staining was partly punctate, but the reason for this is unknown. Quantification indicated that surface levels of GS-II staining increased by greater than twofold (Figure 6B). In conclusion, a single GRASP localized throughout the Golgi in HeLa cells is sufficient to maintain Golgi ribbon architecture, but the resulting membranes are decompartmentalized, and this likely causes inefficient glycosylation of cargo en route to the cell surface.
FIGURE 6:
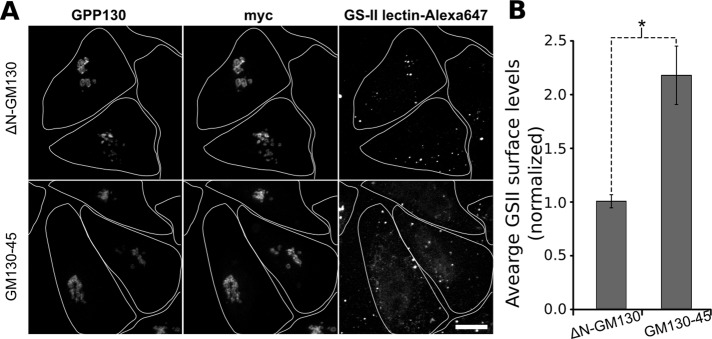
Decreased glycan processing in cells expressing the GM130-45 chimera. (A) Cell surface staining for terminal N-acetyl-d-glucosamine levels was performed using Alexa 647–conjugated GSII-lectin. Cells were transfected with the GM130-45 construct were grown for 84 h and then treated with trypsin and passed onto new coverslips for 12 h before fixation. The Golgi marked by GPP130 and myc-tagged ∆N-GM130 or GM130-45 construct levels are also shown. Maximum value z-projections are shown for each channel. Cell boundaries are outlined to the extent that they were evident from a low level of background staining detected upon overexposure in the anti-myc channel. A small degree of fluorescent signal was present beyond these boundaries and most likely represents lectin binding to cells whose boundaries escaped detection or lectin binding to cell debris. Scale bar, 10 μm. (B) Average staining of GSII-lectin normalized to the mean value of control cells (n > 15 cells, mean ± SEM). Only fluorescence within identified cell boundaries was quantitated. Statistical significance was determined using a single-tailed paired Student's t test (*p = 0.00001).
DISCUSSION
The Golgi ribbon is dynamically maintained by the tubular membrane contacts linking analogous cisternae. These contacts continuously break and reform, and they are also reversibly or irreversibly broken during processes such as cell division (Hidalgo Carcedo et al., 2004; Colanzi and Corda, 2007; Feinstein and Linstedt, 2007; Sengupta and Linstedt, 2010; Rabouille and Kondylis, 2007), directional changes in cell migration (Bisel et al., 2008), apoptosis (Lane et al., 2002), and intracellular bacterial infection (Heuer et al., 2009). The precarious nature of the linkages is further highlighted by the many experimental manipulations (Mironov et al., 2004; Marra et al., 2007) that perturb them, including microtubule depolymerization (Kreis, 1990; Cole et al., 1996). Our findings indicate that the GRASP proteins play direct and cisternae-specific roles in sustaining these linkages. Further, whereas a single GRASP, if localized throughout the Golgi, is capable of linking the membranes into a ribbon, the two GRASPs are needed to compartmentalize the membranes. Thus Golgi ribbon formation involves lateral membrane contacts that require specificity conferred by the GRASP proteins to maintain compartmentalization.
Although the GRASP proteins are involved in other processes (reviewed in Vinke et al., 2011; Jarvela and Linstedt, 2012a), the evidence that they play a direct role in cisternae-specific tethering in cultured cells is now extensive. The localization of GRASP65 is biased to early cisternae and GRASP55 to late cisternae (Shorter and Warren, 1999). Inhibition of expression of either GRASP specifically disrupts Golgi ribbon formation (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008; Xiang and Wang, 2010). That is, under these conditions the Golgi membranes remain trafficking competent, stacked, compartmentalized, and mostly juxtanuclear but fail to establish the ribbon-like membrane network. Perhaps most important, acute inactivation of GRASP65 has the immediate effect of unlinking cis cisternae, whereas rapid inhibition of GRASP55 first unlinks trans cisternae (Figure 3). The GRASP proteins exhibit tethering activity in assays using beads (Wang et al., 2003, 2005), mitochondria (Sengupta et al., 2009; Sengupta and Linstedt, 2010), and liposomes (our unpublished observations). The mechanism is homotypic oligomerization via a unique PDZ interaction (Truschel et al., 2011) such that either GRASP exclusively binds itself, and trans interactions across membranes are favored over cis interactions in the same membrane (Bachert and Linstedt, 2010). Phosphorylation or phosphomimetic mutation of the GRASP proteins blocks tethering activity and prevents Golgi ribbon formation (Jesch et al., 2001; Wang et al., 2003; Preisinger et al., 2005; Feinstein and Linstedt, 2007, 2008; Sengupta and Linstedt, 2010; Bisel et al., 2008; Duran et al., 2008; Tang et al., 2010; Truschel et al., 2012).
A key motivation of the present work using acute inactivation by KillerRed is to counter the adaptability and compensation of cells during longer-term inhibition. The potential for such adaptation is even greater in developing organisms. Thus acute inactivation is more likely to reveal a reaction's true and direct dependences than inhibition experiments such as knockout or knockdown. Evidence that inactivation was specific in our experiments includes the following. Control KillerRed constructs did not perturb Golgi organization. KillerRed constructs targeting factors acting upstream of ribbon formation yielded expected and distinct upstream phenotypes in the Golgi assembly assay (Jarvela and Linstedt, 2012b). GRASP inactivation also yielded the expected phenotype in the Golgi assembly assay (Figure 1). The GRASP isoforms yielded compartment-specific affects upon inactivation that are consistent with their localizations in the Golgi (Figure 3). Finally, the compartment-specific unlinking caused by inactivation led, over time, to a full unlinking of the Golgi ribbon, which is consistent with that observed after knockdown.
These experiments do not distinguish among the varied concepts of Golgi membrane dynamics. Nevertheless, the observation that ablation of either GRASP first yields a cisternae-specific loss of membrane continuity, followed by a loss of lateral continuity among all cisternae, is consistent with cisternal maturation in which GRASP65 establishes ribbon continuity and GRASP55 maintains it. Precisternal membranes coalescing out of a multitude of ER–Golgi intermediate compartment sites (Mironov et al., 2003) would use tethering by GRASP65 as part of their transformation into the interconnected cis cisternae of the Golgi ribbon. As these membranes mature, recycling of GRASP65 (Marra et al., 2001) would ensure further rounds of pre–cisternal membrane maturation. In addition, as the cis membranes mature to medial they would acquire GRASP55 via recycling from later compartments, so that the lateral connections established by GRASP65 are maintained in the medial and trans-Golgi. Recycling of GRASP55 from medial membranes as they mature into trans-cisternae may cause a reduced continuity among cisternae at the most trans position of the Golgi ribbon (Rambourg et al., 1997; Jackson, 2009). As mentioned, the ultimate loss of all ribbon connectivity by acute inhibition of either GRASP agrees with RNA interference experiments showing that each GRASP is required. These findings imply interdependence in the function of the two GRASPs. It may be that, over time, net adhesivity within the noncompact zones is critical to the existence of stable connections. Thus loss of either the initiating or the maintaining GRASP would ultimately cause full unlinking of the Golgi ribbon.
Why are there two GRASP proteins in mammalian cells? Our experiments show that in HeLa cells the GRASPs mediate cisternae-specific linking within the Golgi ribbon and that establishment of the ribbon with a single GRASP causes defects in Golgi compartmentalization. Our interpretation is that the “single-GRASP” Golgi condition yielded net adhesivity that was sufficient to form the Golgi ribbon, but that the presence of the single GRASP on all cisternae promoted an increase in contact between nonanalogous cisternae. As a consequence, the rims of cis, medial, and trans cisternae fused with one another at a sufficient level to cause mixing of what are normally compartmentalized Golgi proteins. In light of this view, it is noteworthy that the number of cisternae per stack can vary greatly among differing cell types (Rambourg et al., 1997), and yet, to our knowledge, all cells with Golgi membranes express, at most, two GRASP proteins (Shorter et al., 1999; Kondylis et al., 2005; Behnia et al., 2007; Kinseth et al., 2007; Struck et al., 2008; Yelinek et al., 2009). It follows that whether there are two or 10 cisternae making up the stacks that are linked into a Golgi ribbon, there are, at most, two GRASPs available to maintain its compartmentalization. This can be understood, in part, due to the fact that Golgi proteins are distributed in gradients spanning multiple cisternae that probably define no more than three functional compartments (Dunphy and Rothman, 1985; Mellman and Simons, 1992; Rabouille et al., 1995). As stated in the preceding paragraph, we hypothesize that GRASP65 initiates lateral contacts as cis cisternae form and these contacts are maintained in later cisternae by GRASP55. Possibly, for Golgi ribbons with many cisternae, a gradient distribution of the GRASPs is sufficient to ensure separation of Golgi membranes into two or three functionally distinct domains. That is, multiple early cisternae may be in continuity at their rims via GRASP65 homotypic connections and multiple late cisternae via GRASP55 homotypic connections.
What about cell types that express only a single GRASP and yet maintain a compartmentalized Golgi? These cell types have multiple, distributed Golgi stacks rather than a single ribbon-like membrane network. It may be that GRASPs are not needed to maintain compartmentalization if Golgi membranes are not brought together to form the Golgi ribbon. The Golgi rims of isolated Golgi stacks have little opportunity for tubule-mediated fusion, among analogous cisternae or not, because there are no adjacent stacks. In contrast, the potential for such fusion contacts is plentiful within a Golgi ribbon, necessitating a mechanism ensuring homotypic fusion so that compartmentalization is not degraded.
In conclusion, rapid inactivation coupled with dynamic imaging and high-resolution microscopy reveal cisternal-specific functions of GRASP65 and GRASP55 in continuity, compartmentalization, and function of the Golgi ribbon.
MATERIALS AND METHODS
Cloning
GRASP55-KR was created by PCR amplification of KR from pKillerRed-N (Evrogen, Moscow, Russia) and replacement of the myc tag at the ClaI and XbaI sites of pCS2-GRASP55myc (Jesch et al., 2001). GRASP65-KR was created by substituting its full-length cDNA generated by PCR at the BamHI and ClaI sites of GRASP55-KR. KR-Gtn, Cyto-KR, and GalT–YFP were as described (Jarvela and Linstedt, 2012b). GPP130-GFP (Mukhopadhyay et al., 2010) and T20-GFP (Sengupta et al., 2009) were as described. T20-GFP-G45 was created by inserting sequence of the last 13 amino acids of golgin-45 into T20-GFP. The myc-tagged GM130 rescue constructs ∆N-GM130 and ∆N∆C-GM130, lacking the N-terminal p115 and C-terminal GRASP65 binding sites, respectively, were as described (Puthenveedu et al., 2006). The GM130-45 chimera was created by using a loop-in modification of the QuikChange protocol (Stratagene, La Jolla, CA) to insert sequence encoding the last 13 amino acids of golgin-45 and a stop codon in place of that encoding the last 13 residues of ∆N-GM130.
Cell culture and transfection
HeLa cells and HeLa cells stably expressing GalNAcT2–GFP were cultured in MEM (Thermo Scientific, Waltham, MA) with 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA). Before experiments, cells were grown until 30–50% confluent in a 60-mm dish and transfected with JetPEI (PolyPlus, Illkirch, France) according to the manufacturer's protocol. After 24 h, cells were suspended using trypsin and dispersed onto 35-mm coverslips, and 36–72 h posttransfection the cells were imaged. siRNA experiments were performed using Interferin (PolyPlus) according to the manufacturer's protocol. Replacement constructs were transfected 24 h postknockdown using JetPrime (PolyPlus), and 72 h later cells were fixed or prepared for live imaging as needed. For Golgi assembly experiments, cells were treated with brefeldin A (Sigma-Aldrich, St. Louis, MO) at 20 μg/ml for 20 min. The cells were washed with 10 volumes of cold phosphate-buffered saline (PBS), and coverslips were then transferred to imaging chambers and imaging medium was added. KR constructs were inactivated within 90 s of drug washout before normal imaging.
Cell imaging
Brefeldin A washout experiments and steady-state Golgi inactivation were performed with a Zeiss Axiovert 200 at 37°C with a 100× Plan-Apo numerical aperture (NA) 1.4 oil objective (Zeiss, Thornwood, NY) attached to an UltraView spinning-disk confocal system (Perkin Elmer-Cetus, Shelton, CT). Photoinactivation was achieved with epifluorescent light at an intensity of 2 W/cm2 for 30 s. We acquired z-slices at 300-nm intervals encompassing the full range of the cell. FRAP experiments and immunofluorescence confocal images were acquired with Andor iQ2 spinning-disk confocal system at the Molecular Biosensors and Imaging Center at Carnegie Mellon University. CALI of KR constructs was achieved by drawing an ROI encompassing the perimeter of the cell. Each pixel was irradiated with 15.8 mW of a 561-nm laser for 45 μs/pixel repeated six times. Photobleaching of GFP or YFP constructs was achieved using 27.3 mW of 488-nm laser power for 20 μs/pixel repeated 60 times, and fluorescence recovery was recorded for a further 10 min at 10-s intervals. Stacks of fluorescence were acquired for the full range of the cell with z-slices at 200-nm intervals. SIM images were acquired with a Nikon A1 Structured Illumination Microscope using a CFI Apo TIRF 100× oil (NA 1.49) objective with Nikon Elements Software at the Center for Biologic Imaging at the University of Pittsburgh (Pittsburgh, PA). Three-dimensional z-stacks were acquired of fixed cells with slices every 100 nm, encompassing the Golgi fluorescence.
Immunofluorescence
Cells adhered to glass coverslips were fixed (3% paraformaldehyde in PBS at pH 7.4) at room temperature for 20 min. They were then rinsed five times in PBS and permeabilized and blocked in 0.5% Triton X-100, 50 mM glycine, and 5% fetal bovine serum in PBS for 30 min. Cells were incubated with one or two of the primary antibodies diluted in block for 30 min and washed again five times in PBS. Secondary antibodies diluted in block were then added to the cells for 30 min. Cells were again rinsed five times and mounted onto slides using either 80% glycerol in Tris pH 7.4 or Gelvatol mounting medium (10.5% polyvinyl alcohol and 21% glycerol in 100 mM Tris, pH 8.5). Antibodies were rabbit anti-GRASP55 (Feinstein and Linstedt, 2007), rabbit anti-GRASP65 (Bachert and Linstedt, 2010), rabbit anti-GPP130 (Puri and Linstedt, 2003), mouse anti-GPP130 (Linstedt et al., 1997), mouse anti–golgin-97 (Life Technologies, Carlsbad, CA), and mouse anti-myc (Evan et al., 1985). Secondary antibodies labeled with Alexa 488, Alexa 568, and Alexa 647, as well as Alexa 647–labeled GSII-lectin, were obtained from Life technologies. GSII lectin surface staining was performed after washing cells 5× in ice-cold PBS. Cells were incubated with 5 μg of GSII lectin/ml in ice-cold PBS containing 0.5 mM MgCl2 and 1 mM CaCl2 and 1% bovine serum albumin for 20 min at 4ºC. Cells were then washed 5× with the same buffer and fixed and stained as just described above.
Image analysis
ImageJ (National Institutes of Health, Bethesda, MD) was used to carry out most of the analysis. All images were background subtracted using the Subtract BG from the ROI plug-in and selecting a region outside the cells. FRAP was measured using FRAP profiler on average projections of confocal stacks, using the freehand selection tool to select the bleached area postbleach and normalized to total cellular fluorescence. Largest objects and total number of objects were determined by using the Analyze particles function in ImageJ. Colocalization of confocal images was performed using the Intensity correlation analysis plug-in (Li et al., 2004). Colocalization analysis for SIM images was performed using the Coloc 2 plug-in incorporated into Fiji (Schindelin et al., 2012) after background subtraction. GRASP65 on the Golgi was the summed fluorescence in the Golgi region, demarcated by GalNAcT2-GFP, as a percentage of the summed fluorescence in the entire cell.
Electron microscopy
HeLa cells in 35-mm dishes were transfected with ∆N-GM130 or GM130-45 and left to grow for 48 h. Expression of myc-tagged ∆N-GM130 or GM130-45 was confirmed using a fluorescent cotransfection marker and visualized through a 20× objective. Regions were then circled by scratching the plastic dish, and the dishes were fixed in 2% gluteraldehyde/PBS for 30 min at room temperature. The glutaraldehyde was removed and replaced with three changes of PBS, followed by 45-min fixation with 2% potassium permanganate in distilled water (dH2O). The potassium permanganate was removed, and the dishes were washed with three changes of dH2O, followed by dehydration in an ascending series of ethanol (50, 70, 80, 90%, and three changes of 100%). The samples were infiltrated in a 1:1 mixture of Epon-Araldite and 100% ethanol. After 30 min, the mixture was exchanged with 100% Epon-Araldite and was held overnight in a desiccator. The next day, the Epon-Araldite was replaced, and the dishes were held in the desiccator at room temperature for 24 h. The dishes were placed in an oven at 30°C, and the temperature was increased 10°C every 24 h until the dishes were at 60°C. The culture dishes were removed, and the encircled groups of cells were cut out of the disk of epoxy with a jeweler's saw. The epoxy was glued to a BEEM capsule and trimmed to isolate the group of encircled cells. Thin (100 nm) sections were cut using a diamond knife (Delaware Diamond Knives, Wilmington, DE) on a Reichert-Jung Ultracut E ultramicrotome. The sections were stained with lead citrate for 1 min. The grids were viewed on an Hitachi H-7100 transmission electron microscope (Hitachi High Technologies America, Pleasanton, CA) operating at 75 kV. Digital images were obtained using an AMT Advantage 10 CCD Camera System (Advanced Microscopy Techniques, Danvers, MA) and ImageJ software.
Supplementary Material
Acknowledgments
We thank Collin Bachert for help with the constructs, Joseph Suhan for expert technical aid with electron microscopy, the Molecular Biosensors and Imaging Center at Carnegie Mellon University, and the Center for Biologic Imaging at the University of Pittsburgh. Funding was provided by National Institutes of Health Grant GM095549 to A.D.L.
Abbreviations used:
- CALI
chromophore-assisted light inactivation
- ER
endoplasmic reticulum
- FRAP
fluorescence recovery after photobleaching
- GalNAcT2-GFP
N-acetylgalactosaminyltransferase T2–green fluorescent protein
- GalT-YFP
galactosyltransferase–yellow fluorescent protein
- GRASP
Golgi reassembly and stacking protein
- KR
KillerRed
- PDZ
postsynaptic density–Drosophila disk large tumor suppressor–zonula occludens-1
- SIM
structured illumination microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0395) on November 13, 2013.
REFERENCES
- Bachert C, Linstedt AD. Dual anchoring of the GRASP membrane tether promotes trans pairing. J Biol Chem. 2010;285:16294–16301. doi: 10.1074/jbc.M110.116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 1998;17:3258–3268. doi: 10.1093/emboj/17.12.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Barr FA, Flanagan JJ, Barlowe C, Munro S. The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J Cell Biol. 2007;176:255–261. doi: 10.1083/jcb.200607151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisel B, Wang Y, Wei JH, Xiang Y, Tang D, Miron-Mendoza M, Yoshimura S, Nakamura N, Seemann J. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol. 2008;182:837–843. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- Colanzi A, Corda D. Mitosis controls the Golgi and the Golgi controls mitosis. Curr Opin Cell Biol. 2007;19:386–393. doi: 10.1016/j.ceb.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy WG, Rothman JE. Compartmental organization of the Golgi stack. Cell. 1985;42:13–21. doi: 10.1016/s0092-8674(85)80097-0. [DOI] [PubMed] [Google Scholar]
- Duran JM, Kinseth M, Bossard C, Rose DW, Polishchuk R, Wu CC, Yates J, Zimmerman T, Malhotra V. The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol Biol Cell. 2008;19:2579–2587. doi: 10.1091/mbc.E07-10-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol Biol Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein TN, Linstedt AD. GRASP55 regulates Golgi ribbon formation. Mol Biol Cell. 2008;19:2696–2707. doi: 10.1091/mbc.E07-11-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer D, Rejman Lipinski A, Machuy N, Karlas A, Wehrens A, Siedler F, Brinkmann V, Meyer TF. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- Hidalgo Carcedo C, Bonazzi M, Spano S, Turacchio G, Colanzi A, Luini A, Corda D. Mitotic Golgi partitioning is driven by the membrane-fissioning protein CtBP3/BARS. Science. 2004;305:93–96. doi: 10.1126/science.1097775. [DOI] [PubMed] [Google Scholar]
- Jackson CL. Mechanisms of transport through the Golgi complex. J Cell Sci. 2009;122:443–452. doi: 10.1242/jcs.032581. [DOI] [PubMed] [Google Scholar]
- Jarvela T, Linstedt AD. Golgi GRASPs: moonlighting membrane tethers. Cell Health Cytoskeleton. 2012a;4:37–47. [Google Scholar]
- Jarvela T, Linstedt AD. Irradiation-induced protein inactivation reveals Golgi enzyme cycling to cell periphery. J Cell Sci. 2012b;125:973–980. doi: 10.1242/jcs.094441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay DG. Selective destruction of protein function by chromophore-assisted laser inactivation. Proc Natl Acad Sci USA. 1988;85:5454–5458. doi: 10.1073/pnas.85.15.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch SA, Lewis TS, Ahn NG, Linstedt AD. Mitotic phosphorylation of Golgi reassembly stacking protein 55 by mitogen-activated protein kinase ERK2. Mol Biol Cell. 2001;12:1811–1817. doi: 10.1091/mbc.12.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinseth MA, Anjard C, Fuller D, Guizzunti G, Loomis WF, Malhotra V. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- Klumperman J. Architecture of the mammalian Golgi. Cold Spring Harb Perspect Biol. 2011;3:a005181. doi: 10.1101/cshperspect.a005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, Spoorendonk KM, Rabouille C. dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol Biol Cell. 2005;16:4061–4072. doi: 10.1091/mbc.E04-10-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kreis TE. Role of microtubules in the organisation of the Golgi apparatus. Cell Motil Cytoskeleton. 1990;15:67–70. doi: 10.1002/cm.970150202. [DOI] [PubMed] [Google Scholar]
- Ladinsky MS, Mastronarde DN, McIntosh JR, Howell KE, Staehelin LA. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J Cell Biol. 1999;144:1135–1149. doi: 10.1083/jcb.144.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane JD, Lucocq J, Pryde J, Barr FA, Woodman PG, Allan VJ, Lowe M. Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J Cell Biol. 2002;156:495–509. doi: 10.1083/jcb.200110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC, Roider J, Jay DG. Chromophore-assisted laser inactivation of proteins is mediated by the photogeneration of free radicals. Proc Natl Acad Sci USA. 1994;91:2659–2663. doi: 10.1073/pnas.91.7.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Mehta A, Suhan J, Reggio H, Hauri HP. Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol Biol Cell. 1997;8:1073–1087. doi: 10.1091/mbc.8.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra P, Maffucci T, Daniele T, Tullio GD, Ikehara Y, Chan EK, Luini A, Beznoussenko G, Mironov A, De Matteis MA. The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat Cell Biol. 2001;3:1101–1113. doi: 10.1038/ncb1201-1101. [DOI] [PubMed] [Google Scholar]
- Marra P, Salvatore L, Mironov A, Jr, Di Campli A, Di Tullio G, Trucco A, Beznoussenko G, Mironov A, De Matteis MA. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130. Mol Biol Cell. 2007;18:1595–1608. doi: 10.1091/mbc.E06-10-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Simons K. The Golgi complex: in vitro veritas. Cell. 1992;68:829–840. doi: 10.1016/0092-8674(92)90027-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, et al. ER-to-Golgi carriers arise through direct en bloc protrusion and multistage maturation of specialized ER exit domains. Dev Cell. 2003;5:583–594. doi: 10.1016/s1534-5807(03)00294-6. [DOI] [PubMed] [Google Scholar]
- Mironov AA, et al. Dicumarol, an inhibitor of ADP-ribosylation of CtBP3/BARS, fragments Golgi non-compact tubular zones and inhibits intra-Golgi transport. Eur J Cell Biol. 2004;83:263–279. doi: 10.1078/0171-9335-00377. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Bachert C, Smith DR, Linstedt AD. Manganese-induced trafficking and turnover of the cis-Golgi glycoprotein GPP130. Mol Biol Cell. 2010;21:1282–1292. doi: 10.1091/mbc.E09-11-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C, Korner R, Wind M, Lehmann WD, Kopajtich R, Barr FA. Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signalling. EMBO J. 2005;24:753–765. doi: 10.1038/sj.emboj.7600569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S, Linstedt AD. Capacity of the Golgi apparatus for biogenesis from the endoplasmic reticulum. Mol Biol Cell. 2003;14:5011–5018. doi: 10.1091/mbc.E03-06-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Hui N, Hunte F, Kieckbusch R, Berger EG, Warren G, Nilsson T. Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J Cell Sci. 1995;108:1617–1627. doi: 10.1242/jcs.108.4.1617. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondylis V. Golgi ribbon unlinking: an organelle-based G2/M checkpoint. Cell Cycle. 2007;6:2723–2729. doi: 10.4161/cc.6.22.4896. [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Hermo L, Segretain D. Tridimensional structure of the Golgi apparatus of nonciliated epithelial cells of the ductuli efferentes in rat: an electron microscope stereoscopic study. Biol Cell. 1987;60:103–115. doi: 10.1111/j.1768-322x.1987.tb00550.x. [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Hermo L, Segretain D. Three-dimensional structure of the Golgi apparatus in mammalian cells. In: Berger EG, Roth J, editors. In: The Golgi Apparatus, Molecular and Cell Biology Updates. Basel: Birkhauser; 1997. pp. 37–61. [Google Scholar]
- Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta D, Linstedt AD. Mitotic inhibition of GRASP65 organelle tethering involves Polo-like kinase 1 (PLK1) phosphorylation proximate to an internal PDZ ligand. J Biol Chem. 2010;285:39994–40003. doi: 10.1074/jbc.M110.189449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta D, Truschel S, Bachert C, Linstedt AD. Organelle tethering by a homotypic PDZ interaction underlies formation of the Golgi membrane network. J Cell Biol. 2009;186:41–55. doi: 10.1083/jcb.200902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short B, Preisinger C, Korner R, Kopajtich R, Byron O, Barr FA. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001;155:877–883. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Warren G. A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J Cell Biol. 1999;146:57–70. doi: 10.1083/jcb.146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 1999;18:4949–4960. doi: 10.1093/emboj/18.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B, White J, Rottger S, Stelzer EH, Suganuma T, Nilsson T. Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck NS, et al. Plasmodium falciparum possesses two GRASP proteins that are differentially targeted to the Golgi complex via a higher- and lower-eukaryote-like mechanism. J Cell Sci. 2008;121:2123–2129. doi: 10.1242/jcs.021154. [DOI] [PubMed] [Google Scholar]
- Surrey T, Elowitz MB, Wolf PE, Yang F, Nedelec F, Shokat K, Leibler S. Chromophore-assisted light inactivation and self-organization of microtubules and motors. Proc Natl Acad Sci USA. 1998;95:4293–4298. doi: 10.1073/pnas.95.8.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Wang Y. The role of GRASP65 in Golgi cisternal stacking and cell cycle progression. Traffic. 2010;11:827–842. doi: 10.1111/j.1600-0854.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne-Tjomsland G, Dumontier M, Jamieson JC. 3D topography of noncompact zone Golgi tubules in rat spermatids: a computer-assisted serial section reconstruction study. Anat Rec. 1998;250:381–396. doi: 10.1002/(SICI)1097-0185(199804)250:4<381::AID-AR1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Truschel ST, Sengupta D, Foote A, Heroux A, Macbeth MR, Linstedt AD. Structure of the membrane-tethering GRASP domain reveals a unique PDZ ligand interaction that mediates Golgi biogenesis. J Biol Chem. 2011;286:20125–20129. doi: 10.1074/jbc.C111.245324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truschel ST, Zhang M, Bachert C, Macbeth MR, Linstedt AD. Allosteric regulation of GRASP protein-dependent Golgi membrane tethering by mitotic phosphorylation. J Biol Chem. 2012;287:19870–19875. doi: 10.1074/jbc.M111.326256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinke FP, Grieve AG, Rabouille C. The multiple facets of the Golgi reassembly stacking proteins. Biochem J. 2011;433:423–433. doi: 10.1042/BJ20101540. [DOI] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G. Mapping the functional domains of the Golgi stacking factor GRASP65. J Biol Chem. 2005;280:4921–4928. doi: 10.1074/jbc.M412407200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JH, Seemann J. Unraveling the Golgi ribbon. Traffic. 2010;11:1391–1400. doi: 10.1111/j.1600-0854.2010.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- Wilson C, Ragnini-Wilson A. Conserved molecular mechanisms underlying homeostasis of the Golgi complex. Int J Cell Biol. 2010;2010:758230. doi: 10.1155/2010/758230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang X, Nix DB, Katoh T, Aoki K, Tiemeyer M, Wang Y. Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nat Commun. 2013;4:1659. doi: 10.1038/ncomms2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelinek JT, He CY, Warren G. Ultrastructural study of Golgi duplication in Trypanosoma brucei. Traffic. 2009;10:300–306. doi: 10.1111/j.1600-0854.2008.00873.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.