A gap remains in the understanding of how nucleoporins are coordinately produced and assembled into macromolecular pore complexes. Here two vertebrate SUMO proteases are found to be important for proper assembly of nuclear pores and maintenance of homeostatic levels of certain nucleoporins.
Abstract
Nuclear pore complexes are composed of ∼30 different proteins, each present at the pore in multiple copies. Together these proteins create specialized channels that convey cargo between the cytoplasm and the nuclear interior. With the building blocks of nuclear pores identified, one challenge is to decipher how these proteins are coordinately produced and assembled into macromolecular pore structures with each cell division. Specific individual pore proteins and protein cofactors have been probed for their role in the assembly process, as well as certain kinases that add a layer of regulation via the phosphorylation status of nucleoporins. Other posttranslational modifications are candidates for coordinating events of pore assembly as well. In this study of two pore-associated small ubiquitin-like modifier (SUMO) proteases, sentrin/SUMO-specific protease 1 (SENP1) and SENP2, we observe that many nucleoporins are mislocalized and, in some cases, reduced in level when SENP1 and SENP2 are codepleted. The pore complexes present under these conditions are still capable of transport, although the kinetics of specific cargo is altered. These results reveal a new role for the pore-associated SENPs in nucleoporin homeostasis and in achieving proper configuration of the nuclear pore complex.
INTRODUCTION
The small ubiquitin-like modifier (SUMO) peptide moiety is posttranslationally conjugated to target proteins and can influence many aspects important to protein function, including the potential repertoire of protein–protein interactions, subcellular localization, and stability (Wilkinson and Henley, 2010). Like many other posttranslational modifications, an important facet of sumoylation is its dynamic nature, with SUMO proteases (or sentrin-specific proteases [SENPs]) being enzymes that catalyze removal of SUMO, in addition to their role in the maturation of SUMO precursors (Yeh, 2009). In mammalian cells, a family of six related SENPs was originally identified (Hickey et al., 2012), but other proteins are newly appreciated to have SUMO protease activity as well (Schulz et al., 2012; Shin et al., 2012). SENP1 and SENP2 are unique in their localization at the nuclear envelope, although this is not their sole intracellular localization (Hang and Dasso, 2002; Zhang et al., 2002; Bailey and O'Hare, 2004). Nuclear pore complexes (NPCs) are prominent features of the nuclear envelope, with thousands present in proliferating mammalian cells. These macromolecular complexes serve as conduits between the cytoplasm and nucleus, allowing for both diffusion of small molecules and active, bidirectional transport of cargo bearing specific signals. NPCs are also sites of association for SENP1 and SENP2 (Hang and Dasso, 2002; Zhang et al., 2002; Goeres et al., 2011; Chow et al., 2012).
NPC architecture broadly comprises a central channel, which spans the inner and outer nuclear membranes, as well a basket-like structure projecting off the nuclear side and filaments emanating from the cytoplasmic side (Raices and D'Angelo, 2012). Both these peripheral features and the central core are eightfold symmetric. NPCs are extremely large (∼60 MDa), yet they have only ∼30 unique components. The large structure is achieved by copy number of the Nups within it, with each constituent present at least eight times, but often in 16 or 32 copies (Rout et al., 2000). Many of these individual components are part of protein subcomplexes that together form the building blocks of the pore. The Nup107-160 complex, consisting of Nup160, Nup107, Nup96, Nup75, Nup47, Nup37, Sec13, and Seh1, is an important scaffold in the central channel. ELYS associates with this subcomplex, playing a pivotal role in recruiting it, as well as other Nups, to newly forming pores during postmitotic nuclear formation (Rasala et al., 2008). POM121 is an integral membrane protein of the vertebrate pore, providing an anchor to the membrane. Nup98 is also positioned centrally, with contact points facing both nucleus and cytoplasm (Chatel et al., 2012). Nup358/RanBP2 is a constituent of the cytoplasmic filaments, and the regulatory protein RanGAP is targeted to the pore via association with Nup358—of interest, in a SUMO-dependent manner (Mahajan et al., 1997). Nup153 is a key constituent of the nuclear basket and is responsible for the localization of other basket residents, specifically Tpr and Nup50 (Hase and Cordes, 2003).
SENP1 and SENP2 have both been found to interact with the nucleoporin Nup153 via a dual interface that includes a bridging interaction with the soluble transport receptor importin α and a SUMO-mediated mechanism (Hang and Dasso, 2002; Zhang et al., 2002; Goeres et al., 2011; Chow et al., 2012). Interactions between SENP2 and the nuclear pore have also been more broadly examined in an affinity purification–mass spectrometry approach (Goeres et al., 2011). Intriguingly, this revealed that SENP2 associates not just with Nup153 (and its partner protein Nup50), but also with Nup358 and members of the Nup107-160 complex. Moreover, interactions with Nup153 and the Nup107 complex were mapped to distinct regions of SENP2 that can independently direct SENP2 to the nuclear rim. Interactions with the NPC are believed to contribute to SENP1/SENP2 localization to the nuclear envelope and restrict/sequester their enzymatic activity to this site (Hang and Dasso, 2002). Interaction with the Nup107-160 complex is also important to kinetochore targeting at mitosis (Cubenas-Potts et al., 2013). Ulp1p, the yeast homologue of SENP1/SENP2, similarly localizes to the nuclear envelope, in part mediated through interaction with nucleoporins (Takahashi et al., 2000; Panse et al., 2003; Zhao et al., 2004; Lewis et al., 2007). Ulp1p and its connection to the NPC are implicated in aspects of mRNA surveillance and DNA repair (Lewis et al., 2007; Palancade et al., 2007). In Drosophila, pore-associated Ulp1 regulates nucleocytoplasmic localization of sumoylated cargo (Smith et al., 2004). Specific roles for SENP1 and SENP2 at the nuclear pore complex are also possible in mammalian cells but have been elusive. Here we seek to better understand the role of SENP1 and SENP2, particularly with respect to the nuclear pore complex.
RESULTS
SENP1 and SENP2 regulate the proper localization of nucleoporins
To first survey broadly for a functional connection between SENP1/SENP2 and the NPC, we used the monoclonal antibody 414, which recognizes the FG-rich nucleoporins Nup358, Nup214, Nup153, and Nup62. Consistent with the idea that SENP1 and SENP2 have a regulatory role that affects the NPC, codepletion of SENP1 and SENP2 resulted in severe mislocalization of the mAb414-reactive nucleoporins (Figure 1A, b and c). Rather than being largely restricted to the nuclear rim and nucleoplasm, the mAb414 signal was additionally present both in a cytoplasmic haze and at distinct cytoplasmic foci. This result was obtained using two independent small interfering RNA (siRNA) oligo sets to deplete SENP1 and SENP2. Depletion of SENP2 alone had a similar but much less robust phenotype, whereas the mAb414 staining pattern looked unaltered after depletion of SENP1 (Figure 1A, e and f). The synergistic effect of depleting SENP1 and SENP2 simultaneously indicates that the presence of one pore-associated SENP may compensate for the other. To test the specificity of this synergism, we codepleted SENP1 and SENP3, which in contrast led to little to no mislocalization of mAb414-reactive nucleoporins (Figure 1A, d). Tracking the SENPs by immunoblot confirmed that their levels were altered by the siRNA treatments (Figure 1C). Although SENP3 levels appear somewhat depressed in this particular SENP1+2–knockdown experiment, this does not contribute to the phenotype, as it was not always observed (see later discussion of Figure 6B) and has little effect when directly targeted for depletion (Figure 1A, d).
FIGURE 1:

Codepletion of SENP1 and SENP2 leads to mislocalization of certain nuclear pore proteins. (A) HeLa cells were transfected with siRNA oligos and, after 48 h, fixed for indirect immunofluorescence with mAb414 antibody to detect FG-rich nucleoporins. The specific oligo treatments were: (a) control, (b) SENP1 and SENP2, (c) an independent set of oligos directed against SENP1 and SENP2, (d) SENP1 and SENP3, (e) SENP1 only, and (f) SENP2 only. (B) Samples similar to those in A were subjected to immunofluorescence to detect Nup153. (C) Lysates from some of these same conditions (control, SENP1 and SENP2, and SENP1 and SENP3) were subjected to Western analysis using antibodies against SENP1, SENP2, SENP3, or importin β. SENP2 has different isoforms (Kadoya et al., 2000; Nishida et al., 2001), and two protein species detected by the SENP2 antibody are depleted by two independent SENP2 siRNA oligos. A third protein species (*) detected by the SENP2 antibody does not decrease with either SENP2 siRNA oligo set and thus is likely to be nonspecific reactivity. Bar, 10 μm.
FIGURE 6:
SENP1 and SENP2 are required for robust nuclear import. (A) Cells treated with independent siRNA oligo sets against SENP1+2 (siSP1+2), siRNA oligos against SENP1+3 (siSP1+3), or control (siControl) were transfected with a plasmid encoding a chimeric RGG protein. Cells were then treated with dexamethasone (DEX) at indicated time points and fixed for immunofluorescence analysis using GFP antibody. (B) Cells subjected to a subset of the foregoing conditions were processed for Western analysis using antibodies against indicated proteins. (C) Data from three experiments were quantified and the mean with SD graphed.
Given the intimate connections characterized between Nup153 and SENP1/SENP2 (Hang and Dasso, 2002; Zhang et al., 2002; Chow et al., 2012), we next probed the localization of this nucleoporin in cells depleted of SENP1 and SENP2 alone and in combination. In each case, however, the localization of Nup153 was unaltered (Figure 1B). This underscores the specificity of the mAb414-localization phenotype and points toward other FG-nucleoporins as ones that are mistargeted in the absence of SENP1/2. Indeed, probing for Nup358 and Nup62 individually revealed that these nucleoporins display phenotypes similar to that seen with the mAb414 antibody (Figure 2). Extending this analysis to members of the Nup107-Nup160 complex (Nup133, Nup96), ELYS, and Nup98 demonstrated widespread disruption of nucleoporin localization, with only ELYS retaining a near-normal pattern (Figure 2j).
FIGURE 2:

Differential localization of nucleoporins upon codepletion of SENP1 and SENP2. HeLa cells treated with control oligo (siControl) or oligos targeted to SENP1 and SENP2 (siSP1+2) were subjected to immunofluorescence analysis using antibodies against Nup358 (a, b), Nup62 (c, d), Nup133 (e, f), Nup96 (g, h), ELYS (i, j), and Nup98 (k, l). Bar, 10 μm.
In the course of this analysis, we used different conditions for fixation. One major difference was whether a light detergent extraction was included early in the procedure. Incorporating this step in the protocol gave rise to a different appearance in the SENP1/2-depletion phenotype that may offer clues to the defects in nucleoporin fate. First, when 0.5% Triton-X was present before methanol fixation, the cytoplasmic foci became more apparent, suggesting that these are not easily solubilized (shown for Nup62 and Nup133 in Figure 3, d and h). This also suggests that a portion of the cytoplasmically mislocalized nucleoporins are in an extractable state, seen as a cytoplasmic haze. Second, much of the nuclear signal was restored when detergent was present, suggesting that epitopes may be masked in aberrant ways when SENP1 and SENP2 are depleted (Figure 3, d and h). This could be due to intramolecular epitope masking, perhaps driven by sumoylation. It is also possible that epitopes are masked by protein–protein interactions promoted by sumoylation of nucleoporins or partner proteins. Although the molecular basis for this observation is not clear, it points to an interesting aspect to pursue further. Overall, decreased levels of pore-associated SENPs leads to significant alteration in nucleoporin distribution and potentially to differences in their association with other factors.
FIGURE 3:

Different fixation conditions reveal changes in where and how nucleoporins are detected after codepletion of SENP1 and SENP2. Control (siControl) or SENP1+2 (siSP1+2)–depleted HeLa cells were treated with PHEM buffer (a, c, e, g) or PHEM buffer containing 0.5% Triton X-100 (b, d, f, h) before fixation. Cells were then subjected to immunofluorescence analysis using antibody against Nup62 (a–d) or Nup133 (e–h). Bar, 10 μm.
SENP1 and SENP2 are not required for localization of the transmembrane proteins POM121 and Sun1 or expansion of the interphase nuclear envelope
Both the transmembrane nucleoporin POM121 and the inner nuclear envelope protein Sun1 have been shown to participate in NPC assembly (Antonin et al., 2005; Liu et al., 2007; Funakoshi et al., 2011; Shaulov et al., 2011; Talamas and Hetzer, 2011). To determine whether their localization depends on the presence of SENP1/2, we next probed for Sun1 and POM121 after their depletion. The distribution of both Sun1 and POM121 appeared relatively unperturbed under these conditions compared with cells transfected with control oligo (Figure 4, A and B), although the localization of proteins probed simultaneously, RanGAP and Nup62, respectively, were clearly disrupted when SENP1 and SENP2 were depleted. We noted that Sun1 levels were sometimes particularly reduced in individual cells, although this was independent of the alterations in nucleoporin localization (Supplemental Figure S1). When SENP1 and SENP2 were depleted, POM121 levels were in the range seen in control conditions, but its levels were noted to be reproducibly in the low end of this range.
FIGURE 4:

Localization of the transmembrane proteins POM121 and Sun1 is not dependent on SENP1/2 levels, nor is interphase expansion of the nuclear envelope. (A) Control (siControl) or SENP1+2 (siSP1+2)–depleted HeLa cells were subjected to immunofluorescence analysis using antibodies against RanGAP1 and SUN1. (B) A set of similarly treated samples was probed with antibodies specific for Nup62 and Pom121. (C) Cells transfected with siRNA oligo(s) were incubated with 2 mM thymidine for 48 h before immunofluorescence analysis of Nup62 and Pom121. Bar, 10 μm.
To focus on events at the nuclear envelope at interphase, namely membrane expansion and interphase pore assembly itself (Doucet et al., 2010; Dultz and Ellenberg, 2010; Maeshima et al., 2010), we cultured depleted cells with thymidine in order to arrest DNA replication. This prevents cell cycle progression; instead, nuclei continue to grow in size. Under these conditions, siControl and siSENP1+2–treated cells appeared very similar with respect to nuclear expansion and POM121 localization pattern (Figure 4C), suggesting that membrane growth and certain steps in pore addition are independent of SENP1 and SENP2.
Codepletion of SENP1 and SENP2 reduces the expression level of multiple nucleoporins
Reduced expression of particular NPC components can lead to defective NPC assembly and mislocalization of nucleoporins similar to the phenotype observed upon the codepletion of SENP1 and SENP2 (Wu et al., 2001; Boehmer et al., 2003; Mitchell et al., 2010). SUMO proteases, through interplay with the ubiquitin pathway, can affect protein stability (Praefcke et al., 2012). We therefore probed for alterations in the level of nucleoporins after depletion of SENP1 and SENP2. We did not observe significant changes in expression level for Nup62 or the pore-associated protein RanGAP. However, Sun1, POM121, Nup133, Nup98, and Nup96 display lower levels (Figure 5, lanes 2 and 3). Although Nup96 was noted to be targeted by ubiquitin-mediated proteolysis during mitosis (Chakraborty et al., 2008), this is not likely to account for the observations here because the decrease is seen visually in interphase cells (Figure 2h). Alterations in level did not always correspond with whether there was a change in subcellular distribution. For instance, Nup62 levels are unaffected by SENP1/SENP2 depletion (Figure 5), although this nucleoporin has a striking change in distribution (Figures 2 and 4). Conversely, Nup153 and POM121 levels decrease to different degrees after depletion of SENP1/2 (Figure 5), although these nucleoporins maintain localization to the nuclear rim (Figure 1). Whether there is a causal relationship in those cases in which a nucleoporin is both mislocalized and reduced in level remains to be determined. The mechanisms that underlie the reduction in nucleoporin levels also remain to be elucidated. Of interest, inhibition of proteasome activity did not have a noticeable effect on nucleoporins in this context (Supplemental Figure S2). What is clear, however, is that SENP1 and SENP2 are required for nucleoporin homeostasis (maintenance of relative levels of nucleoporins typical for the cell and growth conditions).
FIGURE 5:
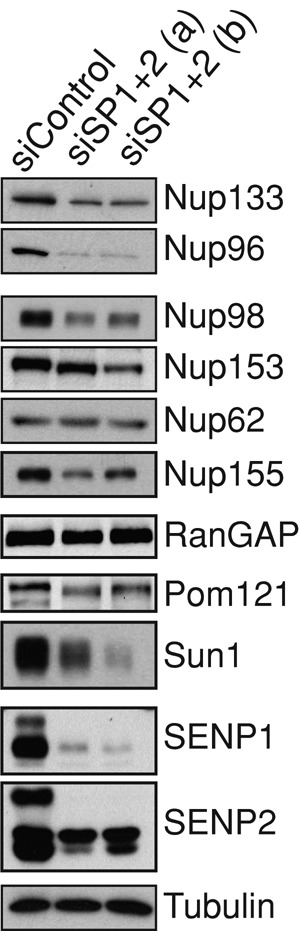
Reduced expression of SENP1 and SENP2 results in lower levels of specific nucleoporins. Cells treated with independent siRNA oligo sets against SENP1+2 or control oligo were subjected to Western analysis using antibodies that recognize the protein indicated.
NPCs remain functional in the absence of SENP1 and SENP2, but specific pathways have altered capacity
Despite significant disruption of nucleoporin biology when SENP1 and SENP2 are reduced, in all cases there was persistence of nucleoporins at the nuclear rim, indicating that nuclear pore complex assembly was still taking place at some level. This raises the question of whether these NPCs have normal function. To address this issue, we used a chimeric Rev–green fluorescent protein (GFP)–glucocorticoid receptor protein (RGG; Love et al., 1998) to monitor nucleocytoplasmic trafficking. This reporter is cytoplasmically sequestered in the absence of cognate steroid and, after addition of dexamethasone, gains access to soluble transport factors. Strong import signals then drive its localization equilibrium to the nucleus. We assessed these initial stages of import and found that the kinetics of RGG nuclear accumulation was slower after SENP1+2 depletion. This is illustrated in Figure 6A and quantified from multiple experiments in Figure 6C. Of note, there was no alteration in import kinetics when either of these pore-associated SENPs was depleted individually (unpublished data) or when SENP1 and SENP3 were codepleted (Figure 6, A and C). RGG has import signals recognized by both importin α and importin 7 (Freedman and Yamamoto, 2004). We did not detect changes in the levels of these factors (Figure 6B), suggesting that changes at the pore itself may underlie the difference in import kinetics.
To next evaluate export capacity, we first treated cells for 3 h with dexamethasone. We then removed dexamethasone from the media. Under these conditions the nuclear export signal in Rev is used to guide export, but once in the cytoplasm, RGG is again sequestered. Here we found that the kinetics of export was very similar between the conditions tested (Figure 7). Thus nuclear pore complexes retain a normal capacity to mediate trafficking by exportin 1. Together the results of this analysis suggest that NPCs are assembled into functional units after depletion of SENP1 and SENP2, despite significant perturbations in the levels and localization of several nucleoporins. This is in line with the robust nature of NPC assembly previously observed (Stavru et al., 2006a, b; Mitchell et al., 2010). It also suggests that the defect in import kinetics is due to an alteration in pore configuration that affects import but not export (see Discussion).
FIGURE 7:

Codepletion of SENP1 and SENP2 does not affect nuclear export. (A) Cells treated with siRNA against SENP1+2 (siSP1+2(a)) or control siRNA (siControl) were transfected with a plasmid encoding a chimeric RGG protein. After treating cells with dexamethasone (DEX) for 3 h to induce RGG nuclear import, the cells were washed to remove DEX. Cells were then fixed at the indicated time points to assess the distribution of the RGG reporter with GFP antibody. (B) Nuclear export was quantified from three experiments and the mean with SD graphed.
DISCUSSION
Genetic and proteomic approaches have yielded a comprehensive understanding of nuclear pore complex composition, and structural and functional studies have added significantly to our understanding of pore architecture and function. Yet little is known about how the level of individual nucleoporins is controlled, how stoichiometric balance of Nups is achieved, and how the process of pore formation itself is regulated. Depleting a particular pore protein experimentally sometimes leads to decreased levels of other nucleoporins (Boehmer et al., 2003), but this is not always the case (Mitchell et al., 2010), even when the protein targeted for depletion is key to NPC formation. Here we found that two of the SUMO-specific proteases, SENP1 and SENP2, play a role in NPC assembly and influence nucleoporin levels, revealing a new regulatory node in nucleoporin biology.
Phosphorylation is the only posttranslational mark directly demonstrated to influence NPC assembly. Initially, nuclear pore proteins were observed to be targets of mitotic phosphorylation, a modification hypothesized to contribute to dispersal of the nuclear pore at mitosis (Macaulay et al., 1995; Favreau et al., 1996). More recently, this has been dissected at a precise level, with the characterization of specific residues in Nup98 targeted by the kinases NEK6 and NEK7 to trigger the release of Nup98 from the NPC (Laurell et al., 2011). This particular alteration at the pore has major ramifications, as it allows greater permeability of the NPC, a step known to occur at mitosis onset (Lenart and Ellenberg, 2006). Phosphorylation was also more recently characterized to be an important aspect of pore assembly and function during interphase. In particular, cyclin-dependent kinases were identified to control the assembly of NPCs during interphase nuclear expansion (Maeshima et al., 2010), and ERK was found to target nuclear pore proteins to modulate transport (Kosako et al., 2009). These and other findings are likely just the beginning of a complex network of specific phosphorylation events that modulate many aspects of pore function. Of importance, regulation of nucleoporin function and/or assembly into the macromolecular nuclear pore by posttranslational modification is almost certainly not restricted to phosphorylation. Indeed, other types of posttranslational modification are present on nuclear pore proteins, including glycosylation (Davis and Blobel, 1987), acetylation (Choudhary et al., 2009), and sumoylation (Golebiowski et al., 2009; Chow et al., 2012). The functional consequences of these modifications are not fully characterized, although more recent molecular characterization points to a critical role for glycosylation in modulating the permeability barrier function of Nup98 (Labokha et al., 2013).
Often a key aspect of posttranslational modification is its reversibility, exemplified by the vast roles that phosphatases play in the spatiotemporal control of biological processes, including nuclear pore assembly (Onischenko et al., 2005). Here we focused in on the sumoylation pathway by assessing enzymes responsible for removing SUMO modification. In particular, the mammalian SUMO proteases SENP1 and SENP2 have been noted for some time to be localized to the nuclear rim and more specifically to the nuclear pore complex itself (Hang and Dasso, 2002; Zhang et al., 2002; Bailey and O'Hare, 2004). Interactions between SENP2, and in some cases SENP1, with Nup153, the Nup107 complex, and Nup358 have been reported (Hang and Dasso, 2002; Zhang et al., 2002; Goeres et al., 2011; Chow et al., 2012). The SUMO modification pathway has been implicated in regulating nucleocytoplasmic transport of certain cargo. This role has been explained by modulation of the SUMO status of the cargo itself and in some cases the soluble receptors involved in transport (e.g., Kindsmuller et al., 2007; Shitashige et al., 2008; Rothenbusch et al., 2012). The results reported here expand the role of these pore-associated SENPs to the assembly of fully formed and functioning NPCs, although, of note, this role is most robustly revealed by depleting SENP1 and SENP2 simultaneously (Figure 1). Other macromolecular complexes, such as kinetochores and ribosomes, similarly rely on a balance of sumoylation/desumoylation, with distinct SUMO proteases playing pivotal roles (Panse et al., 2006; Yun et al., 2008; Mukhopadhyay et al., 2010; Finkbeiner et al., 2011).
One implication of the data presented here is that the cycle of sumoylation on specific proteins contributes to pore protein homeostasis and nuclear pore assembly. When the levels of SENP1 and SENP2 are reduced experimentally, this creates conditions in which particular substrates of these enzymes can be more persistently sumoylated. This, in fact, has been shown to be the case for the pore protein Nup153 (Chow et al., 2012) but is likely to be true for multiple targets relevant to this pathway. Consistent with the idea that sumoylation of the pore proteins could underlie the alterations observed here, several mammalian nucleoporins (Nup358, Nup205, Nup155, Nup153, Nup133, Nup107, Nup96, Nup93, RanGAP, Tpr) have been identified as targets of sumoylation in proteomics approaches (Li et al., 2004; Vertegaal et al., 2006; Blomster et al., 2009; Golebiowski et al., 2009; Matafora et al., 2009). In addition, Nup205, Nup155, Nup93, and Nup210 were found to be polysumoylated (Bruderer, 2011). Yet, although we monitored many nucleoporins by Western analysis in this study, we did not see shifts in migration indicative of sumoylation. Further efforts, including enriching sumoylated proteins by immunoprecipitation and blocking proteasomal degradation with MG132 treatment after SENP1+2 knockdown, still did not yield a pattern of nucleoporin sumoylation (unpublished data). This negative result does not rule out a role for nucleoporin sumoylation as a regulatory step in NPC biogenesis, as it should be noted that even with desumoylation repressed, this modification can be difficult to detect, and it is often the case that modification of a minor protein population at any one time has a functional effect (Hay, 2005). As this avenue is pursued in the future, however, it will be important to look comprehensively at both nucleoporins and soluble factors that affect NPC assembly. Perhaps the interaction of SENP1 and SENP2 with nucleoporins is critical for bringing these enzymes into proximity with such assembly factors where sumoylation status may guide coordination of events. It is also formally possible that SENP1 and SENP2 themselves serve as assembly factors independent of their enzymatic activity.
After depletion of SENP1 and SENP2, we observed a decrease in the kinetics of nuclear import (Figure 6) but not a complete loss of pore function. This is underscored by the lack of change in nuclear export kinetics (Figure 7). Further, the observation that nuclei continued to expand when cells depleted of SENP1 and SENP2 were arrested in interphase (Figure 4C) is consistent with the presence of nuclear pores that exert transport and barrier functions. Using different conditions of fixation revealed that certain nucleoporins were present at their usual location but less accessible to detection, implying that the lack of SENP1 and SENP2 leads to alterations that reconfigure or mask antigen exposure rather than completely relocate nucleoporins (Figure 3). The changes in pore organization and function observed after depletion of SENP1 and SENP2 indicate that these proteases contribute to nuclear transport, perhaps by controlling the configuration of nuclear pore complexes or modulating the interactions that can take place with pore components to promote particular types of traffic. It is also possible that particular components of the soluble transport machinery (via their SUMO status) are functionally modulated by these enzymes, affecting the kinetics of transport for particular cargo. Overall the pore-associated SUMO proteases contribute to the fidelity of nuclear pore assembly and the robustness of nuclear import, suggesting an important means of contributing to cellular fitness.
It is interesting that heat shock was previously reported to modulate the sumoylation status of several nucleoporins (Golebiowski et al., 2009). The results presented here suggest that such stress-induced changes in nucleoporin sumoylation status may trigger a reconfiguration of NPCs. With the physiological significance of these SUMO proteases found in contexts ranging from cancer (Wang et al., 2013) to cardiac function (Kang et al., 2010), it will be important to consider how the roles of SENP1 and SENP2 in nuclear pore function and nucleoporin homeostasis influence a broad range of situations.
MATERIALS AND METHODS
siRNA transfection
siRNAs were introduced into HeLa cells using Lipofectamine RNAi Max (Invitrogen, Carlsbad, CA) as previously described (Chow et al., 2012). In the case in which a single gene was targeted, 10 nM siRNA oligo was used; when two genes were targeted, this was performed with 5 nM respective oligos. Oligos directed against SENP1 and SENP2, as well as the control siRNA oligo, were synthesized in the University of Utah DNA/Peptide Shared Resource and are as described in Chow et al. (2012). SENP3 oligo (Yun et al., 2008) was purchased from Qiagen (Germantown, MD).
Plasmid constructs and antibodies
Nup153 antibody (hZ) was generated by the Ullman lab; antibodies against Nup133 and Elys were a kind gift from Douglass Forbes (University of California, San Diego). Nup96 and Nup98 were provided by the Powers lab. Antibodies against the SENPs and RanBP2/Nup358 were made in the Dasso lab. Other antibodies were obtained from commercial sources as follows: GFP (Ab290; Abcam, Cambridge, MA), importin α (610485; BD Transduction, San Jose, CA), Importin β (610559; BD Transduction), Importin 7 (SC-365231; Santa Cruz Biotechnology, Dallas, TX), 414 (MMS-120P; Covance, Princeton, NJ), Nup62 (610497; BD Transduction, Irvine, CA), POM121 (GTX 102128; GeneTex), RanGAP-1 (33-0800; Zymed), Nup155 (GTX120945; GeneTex), and Sun1 (Ab124770; Abcam).
Indirect immunofluorescence analysis
HeLa cells transfected with siRNA oligos were seeded on coverslips coated with fibronectin. After 48 h of siRNA treatment, cells were fixed with either −20°C methanol for 4 min or 4% paraformaldyhyde (in 1× phosphate-buffered saline [PBS]) for 10 min followed by 10 min of permeabilization with 0.2% Triton X-100 (in 1× PBS). For prepermeabilization experiments, before fixation, cells were incubated with either PHEM buffer (60 mM 1,4-piperazinediethanesulfonic acid, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mM ethylene glycol tetraacetic acid, 4 mM MgSO4) or PHEM buffer containing 0.5% Triton X-100 for 5 min. Cells were then gently washed twice with cold 1× PBS and fixed with −20°C MeOH. After fixation, coverslips were incubated with blocking buffer (3% fetal bovine serum, 0.02% Triton X-100, 1× PBS) for 30 min. Primary antibodies, diluted in blocking buffer, were then incubated with samples for 1 h at room temperature. Cells were washed three times at 5-min intervals with washing buffer (1.5% bovine serum albumin, 0.02% Triton X-100, 1× PBS) before incubation with secondary antibodies for 30 min at room temperature. Cells were washed as described and mounted with Prolong antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen).
Western analysis
HeLa cells treated with siRNA were harvested after 48 h with lysis buffer (0.25% Triton X-100, 60 mM β-glycerophosphate, 10 mM sodium orthovanadate, 2× Roche Complete Protease Inhibitor, 40 mM N-ethylmaleimide). Equal protein levels of lysate in SDS loading buffer were subjected to standard Western analysis.
Import and export assays
HeLa cells, 3 × 104, seeded on fibronectin-coated coverslips were transfected with either control or SENP1/2 siRNA oligos (10 nM). At 24 h post–siRNA transfection, cells were replenished with fresh media and cotransfected with 50 ng of a plasmid encoding a chimeric Rev-GFP-glucocorticoid receptor protein and 100 ng of pBluescript. At 24 h post–DNA transfection, cells were either mock treated or treated with 250 nM of dexamethasone for 10 or 60 min and fixed with −20°C MeOH and processed for import assay analysis. For export assay 24 h posttransfection, cells were treated with 250 nM dexamethasone for 3 h. Cells were then washed three times with 1× PBS, replenished with fresh media, and subsequently fixed with −20°C MeOH at the 0-, 10-, 20-, 30-, and 60-min time points. Fixed samples were processed for indirect immunofluorescence using α-GFP antibody. The 40× images were acquired with a Zeiss Axioskop 2 microscope. Image analysis and intensity measurements were performed on raw images (non–background subtracted) of specified time points in ImageJ (National Institutes of Health, Bethesda, MD). Freehand selections were used to measure the raw integrated intensity of nuclei and total fluorescence in GFP images.
Supplementary Material
Acknowledgments
We thank Douglass Forbes for providing antibodies. This work was supported by National Institutes of Health Grants R01 GM61275 (K.U.) and RO1 GM059975 (M.P.) and National Institute of Child Health and Human Development intramural funds (Projects Z01 HD001902 and Z01 HD008816 to M.D.). Shared resources used in this project are supported in part by P30 CA042014 awarded to the Huntsman Cancer Institute.
Abbreviations used:
- NPC
nuclear pore complex
- SENP
sentrin/SUMO-specific protease
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0256) on November 6, 2013.
*Present address: Jackson Laboratory, Bar Harbor, ME 04609.
REFERENCES
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell. 2005;17:83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Bailey D, O'Hare P. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J Biol Chem. 2004;279:692–703. doi: 10.1074/jbc.M306195200. [DOI] [PubMed] [Google Scholar]
- Blomster HA, Hietakangas V, Wu J, Kouvonen P, Hautaniemi S, Sistonen L. Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol Cell Proteomics. 2009;8:1382–1390. doi: 10.1074/mcp.M800551-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci USA. 2003;100:981–985. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 2011;12:142–148. doi: 10.1038/embor.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, et al. Nucleoporin levels regulate cell cycle progression and phase-specific gene expression. Dev Cell. 2008;15:657–667. doi: 10.1016/j.devcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel G, Desai SH, Mattheyses AL, Powers MA, Fahrenkrog B. Domain topology of nucleoporin Nup98 within the nuclear pore complex. J Struct Biol. 2012;177:81–89. doi: 10.1016/j.jsb.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chow KH, Elgort S, Dasso M, Ullman KS. Two distinct sites in Nup153 mediate interaction with the SUMO proteases SENP1 and SENP2. Nucleus. 2012;3:349–358. doi: 10.4161/nucl.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubenas-Potts C, Goeres JD, Matunis MJ. SENP1 and SENP2 affect spatial and temporal control of sumoylation in mitosis. Mol Biol Cell. 2013;24:3483–3495. doi: 10.1091/mbc.E13-05-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci USA. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in Metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol. 2010;191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau C, Worman HJ, Wozniak RW, Frappier T, Courvalin JC. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 1996;35:8035–8044. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011;30:1067–1078. doi: 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ND, Yamamoto KR. Importin 7 and importin alpha/importin beta are nuclear import receptors for the glucocorticoid receptor. Mol Biol Cell. 2004;15:2276–2286. doi: 10.1091/mbc.E03-11-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T, Clever M, Watanabe A, Imamoto N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol Biol Cell. 2011;22:1058–1069. doi: 10.1091/mbc.E10-07-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres J, Chan PK, Mukhopadhyay D, Zhang H, Raught B, Matunis MJ. The SUMO-specific isopeptidase SENP2 associates dynamically with nuclear pore complexes through interactions with karyopherins and the Nup107-160 nucleoporin subcomplex. Mol Biol Cell. 2011;22:4868–4882. doi: 10.1091/mbc.E10-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- Hang J, Dasso M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem. 2002;277:19961–19966. doi: 10.1074/jbc.M201799200. [DOI] [PubMed] [Google Scholar]
- Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell. 2003;14:1923–1940. doi: 10.1091/mbc.E02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoya T, Kishida S, Fukui A, Hinoi T, Michiue T, Asashima M, Kikuchi A. Inhibition of Wnt signaling pathway by a novel axin-binding protein. J Biol Chem. 2000;275:37030–37037. doi: 10.1074/jbc.M005984200. [DOI] [PubMed] [Google Scholar]
- Kang X, Qi Y, Zuo Y, Wang Q, Zou Y, Schwartz RJ, Cheng J, Yeh ET. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindsmuller K, Groitl P, Hartl B, Blanchette P, Hauber J, Dobner T. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc Natl Acad Sci USA. 2007;104:6684–6689. doi: 10.1073/pnas.0702158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosako H, Yamaguchi N, Aranami C, Ushiyama M, Kose S, Imamoto N, Taniguchi H, Nishida E, Hattori S. Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat Struct Mol Biol. 2009;16:1026–1035. doi: 10.1038/nsmb.1656. [DOI] [PubMed] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hulsmann BB, Urlaub H, Baldus M, Gorlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Lenart P, Ellenberg J. Monitoring the permeability of the nuclear envelope during the cell cycle. Methods. 2006;38:17–24. doi: 10.1016/j.ymeth.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Lewis A, Felberbaum R, Hochstrasser M. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J Cell Biol. 2007;178:813–827. doi: 10.1083/jcb.200702154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, Stadtman ER, Yang DC, Chock PB. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc Natl Acad Sci USA. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Pante N, Misteli T, Elsagga M, Crisp M, Hodzic D, Burke B, Roux KJ. Functional association of Sun1 with nuclear pore complexes. J Cell Biol. 2007;178:785–798. doi: 10.1083/jcb.200704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Sweitzer TD, Hanover JA. Reconstitution of HIV-1 rev nuclear export: independent requirements for nuclear import and export. Proc Natl Acad Sci USA. 1998;95:10608–10613. doi: 10.1073/pnas.95.18.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Maeshima K, et al. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat Struct Mol Biol. 2010;17:1065–1071. doi: 10.1038/nsmb.1878. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Matafora V, D'Amato A, Mori S, Blasi F, Bachi A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol Cell Proteomics. 2009;8:2243–2255. doi: 10.1074/mcp.M900079-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Arnaoutov A, Dasso M. The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol. 2010;188:681–692. doi: 10.1083/jcb.200909008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida T, Kaneko F, Kitagawa M, Yasuda H. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting beta-catenin degradation. J Biol Chem. 2001;276:39060–39066. doi: 10.1074/jbc.M103955200. [DOI] [PubMed] [Google Scholar]
- Onischenko EA, Gubanova NV, Kiseleva EV, Hallberg E. Cdk1 and okadaic acid-sensitive phosphatases control assembly of nuclear pore complexes in Drosophila embryos. Mol Biol Cell. 2005;16:5152–5162. doi: 10.1091/mbc.E05-07-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Liu X, Garcia-Rubio M, Aguilera A, Zhao X, Doye V. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol Biol Cell. 2007;18:2912–2923. doi: 10.1091/mbc.E07-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Kressler D, Pauli A, Petfalski E, Gnadig M, Tollervey D, Hurt E. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 2006;7:1311–1321. doi: 10.1111/j.1600-0854.2006.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse VG, Kuster B, Gerstberger T, Hurt E. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat Cell Biol. 2003;5:21–27. doi: 10.1038/ncb893. [DOI] [PubMed] [Google Scholar]
- Praefcke GJ, Hofmann K, Dohmen RJ. SUMO playing tag with ubiquitin. Trends Biochem Sci. 2012;37:23–31. doi: 10.1016/j.tibs.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Raices M, D'Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell. 2008;19:3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenbusch U, Sawatzki M, Chang Y, Caesar S, Schlenstedt G. Sumoylation regulates Kap114-mediated nuclear transport. EMBO J. 2012;31:2461–2472. doi: 10.1038/emboj.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S, et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012;13:930–938. doi: 10.1038/embor.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulov L, Gruber R, Cohen I, Harel A. A dominant-negative form of POM121 binds chromatin and disrupts the two separate modes of nuclear pore assembly. J Cell Sci. 2011;124:3822–3834. doi: 10.1242/jcs.086660. [DOI] [PubMed] [Google Scholar]
- Shin EJ, Shin HM, Nam E, Kim WS, Kim JH, Oh BH, Yun Y. DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep. 2012;13:339–346. doi: 10.1038/embor.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitashige M, Satow R, Honda K, Ono M, Hirohashi S, Yamada T. Regulation of Wnt signaling by the nuclear pore complex. Gastroenterology. 2008;134:1961–1971, 1971.e1–4. doi: 10.1053/j.gastro.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Smith M, Bhaskar V, Fernandez J, Courey AJ. Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugates. J Biol Chem. 2004;279:43805–43814. doi: 10.1074/jbc.M404942200. [DOI] [PubMed] [Google Scholar]
- Stavru F, Hulsmann BB, Spang A, Hartmann E, Cordes VC, Gorlich D. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol. 2006a;173:509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F, Nautrup-Pedersen G, Cordes VC, Gorlich D. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. J Cell Biol. 2006b;173:477–483. doi: 10.1083/jcb.200601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Mizoi J, Toh EA, Kikuchi Y. Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J Biochem. 2000;128:723–725. doi: 10.1093/oxfordjournals.jbchem.a022807. [DOI] [PubMed] [Google Scholar]
- Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol. 2011;194:27–37. doi: 10.1083/jcb.201012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- Wang Q, Xia N, Li T, Xu Y, Zou Y, Zuo Y, Fan Q, Bawa-Khalfe T, Yeh ET, Cheng J. SUMO-specific protease 1 promotes prostate cancer progression and metastasis. Oncogene. 2013;32:2493–2498. doi: 10.1038/onc.2012.250. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, van Deursen JM. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc Natl Acad Sci USA. 2001;98:3191–3196. doi: 10.1073/pnas.051631598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET. SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol. 2008;183:589–595. doi: 10.1083/jcb.200807185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wu CY, Blobel G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J Cell Biol. 2004;167:605–611. doi: 10.1083/jcb.200405168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



