Telomeres use distinct mechanisms to mediate cohesion between sister chromatids. However, the motivation for a specialized mechanism is not well understood. Fluorescence in situ hybridization and live-cell imaging show that persistent sister chromatid cohesion at telomeres triggers a prolonged anaphase in normal human cells and cancer cells.
Abstract
Telomeres use distinct mechanisms (not used by arms or centromeres) to mediate cohesion between sister chromatids. However, the motivation for a specialized mechanism at telomeres is not well understood. Here we show, using fluorescence in situ hybridization and live-cell imaging, that persistent sister chromatid cohesion at telomeres triggers a prolonged anaphase in normal human cells and cancer cells. Excess cohesion at telomeres can be induced by inhibition of tankyrase 1, a poly(ADP-ribose) polymerase that is required for resolution of telomere cohesion, or by overexpression of proteins required to establish telomere cohesion, the shelterin subunit TIN2 and the cohesin subunit SA1. Regardless of the method of induction, excess cohesion at telomeres in mitosis prevents a robust and efficient anaphase. SA1- or TIN2-induced excess cohesion and anaphase delay can be rescued by overexpression of tankyrase 1. Moreover, we show that primary fibroblasts, which accumulate excess telomere cohesion at mitosis naturally during replicative aging, undergo a similar delay in anaphase progression that can also be rescued by overexpression of tankyrase 1. Our study demonstrates that there are opposing forces that regulate telomere cohesion. The observation that cells respond to unresolved telomere cohesion by delaying (but not completely disrupting) anaphase progression suggests a mechanism for tolerating excess cohesion and maintaining telomere integrity. This attempt to deal with telomere damage may be ultimately futile for aging fibroblasts but useful for cancer cells.
INTRODUCTION
Sister chromatids are held together from the time of their replication in S phase until their separation in anaphase by cohesin, a ring complex comprising Smc1, Smc3, and Scc1 (Anderson et al., 2002; Haering et al., 2002), and a peripheral subunit Scc3, which in vertebrates exists as two related isoforms, SA1 and SA2 (Losada et al., 2000; Sumara et al., 2000) with distinct functions; SA1 is required for telomere and SA2 for centromere cohesion (Canudas and Smith, 2009; Remeseiro et al., 2012; Bisht et al., 2013). Cohesion between sister chromatids is crucial at centromeres, where it serves to resist the pulling forces of microtubules and allow bipolar attachment of sister kinetochores to the mitotic spindle poles (Tanaka et al., 2000). Once this attachment is achieved for all chromosomes, the cohesin ring is released by proteolytic cleavage of the Scc1 subunit; sister chromatids then separate and move to the poles (Waizenegger et al., 2000; Hauf et al., 2001). This irreversible regulatory step governs the metaphase-to-anaphase transition. Cohesin is also distributed along chromosome arms, but unlike centromere cohesin, arm cohesin is removed in prophase by phosphorylation of the SA2 subunit (Hauf et al., 2005). Resolution of sister chromatid cohesion between telomeres also occurs in early mitosis (like arm cohesion) but has an additional requirement for the poly(ADP-ribose) polymerase (PARP) tankyrase 1 (Dynek and Smith, 2004; Hsiao and Smith, 2008).
Mammalian telomeres comprise tandem arrays of TTAGGG repeats and the telomere-specific complex shelterin (Palm and de Lange, 2008). SA1 is enriched at human telomeres (Bisht et al., 2013), where it associates with the shelterin subunit TRF1 (the double-strand TTAGGG repeat–binding protein) and its partner TIN2 (Canudas et al., 2007). In cells depleted of SA1 or TIN2, telomere cohesion cannot be established or maintained (Canudas and Smith, 2009; Bisht et al., 2013). TRF1 is a target of PARsylation by tankyrase 1 (Smith et al., 1998). Tankyrase 1 localizes to telomeres by binding to TRF1 early in mitosis concomitant with resolution of telomere cohesion and release of SA1 from telomeres (Bisht et al., 2012, 2013). In tankyrase 1–depleted mitotic cells, SA1 remains on telomeres and sister telomeres remain cohered, despite normal resolution of sister chromatid arms and centromeres (Dynek and Smith, 2004; Bisht et al., 2013). Sister telomere cohesion can be rescued by wild-type but not PARP-dead tankyrase 1, indicating that PARsylation-dependent remodeling of telomeres is required for resolution of sister telomere cohesion (Bisht et al., 2013).
Here we report that excess telomere cohesion at mitosis, induced by several different methods or occurring naturally in aging cells, leads to a delay in anaphase progression. We speculate that cells use anaphase delay as a means to survey and maintain telomere integrity before exiting mitosis.
RESULTS
Tankyrase 1 inhibition by the small-molecule inhibitor XAV939 leads to prolonged anaphase in tumor cells and normal cells
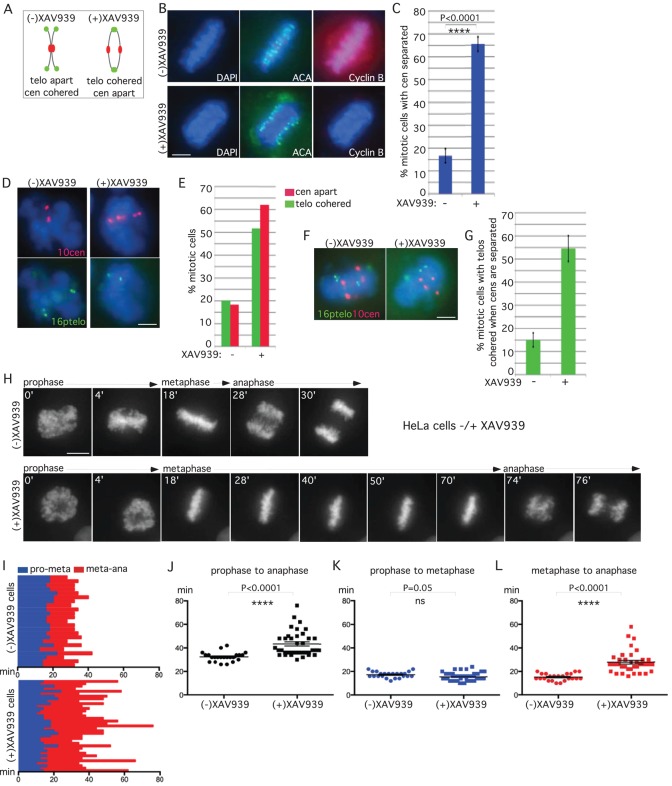
RNA interference (RNAi)–mediated depletion of tankyrase 1 in HeLa cells led to persistent telomere cohesion in mitosis (Dynek and Smith, 2004; Canudas et al., 2007; Bisht et al., 2013) and robust mitotic arrest (Dynek and Smith, 2004; Chang et al., 2005). A time course analysis of TNKS1 small interfering RNA (siRNA)–treated HeLa cells showed that anaphase-like figures (with sister telomeres cohered and centromeres separated) accumulated at early time points (24 h; Dynek and Smith, 2004). However, at later times (36–48 h), cells exhibited a complex phenotype with misaligned chromosomes and aberrant spindle structures, making it difficult to interpret the mitotic arrest. Confounding the issue, RNAi-mediated depletion of tankyrase 1 in most other human cell lines tested led to persistent telomere cohesion but not mitotic arrest (Hsiao and Smith, 2009).
We decided to use more refined approaches to analyze mitotic defects in tankyrase 1–depleted cells. We reasoned that the anaphase-like figures observed at early time points in RNAi-treated HeLa cells reflected the initial response of cells entering mitosis in the absence of tankyrase 1. To focus on this window, we took advantage of the tankyrase 1–specific small-molecule inhibitor XAV939 (Huang et al., 2009). HeLa cells were synchronized by a double-thymidine block, released into S phase in the presence or absence of XAV939 for 10 h, and subjected to immunofluorescence analysis (Figure 1B). The mitotic index (determined by 4′,6-diamidino-2-phenylindole [DAPI] staining) indicated that XAV939 treatment did not induce mitotic arrest. However, staining with anti-centromere antibody (ACA) revealed a 3.5-fold increase in XAV939-treated mitotic cells with separated centromeres compared with control (Figure 1, B and C). Moreover, 100% of cells with separated centromeres showed a loss in cyclin B, indicating progression to anaphase. To analyze the status of the centromere and telomeres together, we isolated cells at the 10-h time point by mitotic shake-off and analyzed them by fluorescence in situ hybridization (FISH) using centromere (10cen)- and telomere (16ptelo)- specific probes. As shown in Figure 1, D and E, XAV939 induced a loss in centromere cohesion, concomitant with persistent telomere cohesion, indicating that the majority of mitotic cells were in anaphase. To further support the conclusion that cells with cohered sister telomeres have initiated anaphase, we scored only those cells displaying centromere separation for persistent telomere cohesion. As shown in Figure 1, F and G, XAV939-treated cells (displaying separated centromeres) show a 3.6-fold increase in cohered telomeres compared with control.
FIGURE 1:
Tankyrase 1 inhibition by the small-molecule inhibitor XAV939 leads to prolonged anaphase. (A) Schematic diagram depicting persistent telomere cohesion and loss of centromere cohesion induced by XAV939 treatment. (B, C) Anaphase figures are increased in XAV939-treated cells. HeLaI.2.11 cells were synchronized with a double-thymidine block, released into S phase in the presence or absence of XAV939 for 10 h, fixed on coverslips in methanol, and (B) analyzed by immunofluorescence with anti-ACA (green) and cyclin B (red) antibodies. DNA was stained with DAPI (blue). Scale bar, 5 μm. (C) Graphical representation of the percentage of mitotic cells with centromeres separated. Values are means ± SD, derived from three independent experiments (n = 19–30 mitotic cells each of 200–212 total cells each). Student's t test was used to calculate the p value (****p ≤ 0.0001). (D, E). XAV939 induces loss of centromere cohesion with persistent telomere cohesion. HeLaI.2.11 cells were synchronized with a double-thymidine block, released into S phase in the presence or absence of XAV939 for 10 h, isolated by mitotic shake-off, and analyzed by (D) centromere (red) and telomere (green) FISH. DNA was stained with DAPI (blue). Scale bar, 5 μm. (E) Graphical representation of the frequency of mitotic cells with centromeres apart and telomeres cohered (n = 50–60 cells each). (F, G) Telomere separation is delayed in cells that have separated centromeres. (F) Cells were treated and processed as in D, but telomere cohesion was scored only in cells that had separated centromeres. (G) Graphical representation of the frequency of mitotic cells with centromeres separated that show cohered telomeres. Values are means ± SEM, derived from two independent experiments (n = 100 cells each). (H–L) Live-cell imaging indicates that XAV939 induces anaphase delay. (H) Time-lapse video live-cell imaging of HeLa-H2B-GFP cells synchronized by a double-thymidine block, released in the presence or absence of XAV939 for 7 h, and imaged for 6 h. Progression from prophase to anaphase for individual cells. Scale bar, 5 μm. (I– L) Graphical summaries of individual mitotic cells (n = 23–37 cells each) shown as (I) a time line and (J–L) scatterplots with calculated mean value ± SEM. Student's t test was used to calculate p values (ns, p ≥ 0.05; ****p ≤ 0.0001).
We next used live-cell imaging to measure the time cells spent in anaphase. HeLa-H2B-green fluorescent protein (GFP) cells were synchronized by a double-thymidine block, released into S phase in the presence or absence of XAV939 for 7 h, and analyzed by live-cell imaging. A representative example is shown in Figure 1H and Supplemental Movie S1. In both control and XAV939-treated cells chromosomes aligned on the metaphase plate at the 18-min time point. However, whereas in control cells chromosomes separated at the 28-min time point, in XAV939-treated cells chromosomes struggled and did not separate until the 74-min time point. The time of progression through mitosis for each individual cell analyzed by live imaging is shown in Figure 1I. Scatterplot analysis shows that XAV939-treated cells spent significantly more time in mitosis (prophase to anaphase) than control cells (Figure 1J). Progression from prophase to metaphase was similar (Figure 1K), whereas progression from metaphase to anaphase was significantly increased in XAV939-treated cells compared with control (Figure 1L), indicating a delay in anaphase.
To analyze the response of normal human cells, IMR90-H2B-GFP cells at early population doubling (PD) (24) were synchronized by a double-thymidine block, released in the presence or absence of XAV939 for 7 h, and analyzed by live-cell imaging. A representative example is shown in Supplemental Figure S1A and Supplemental Movie S2. The time of progression through mitosis for each individual cell analyzed by live imaging is shown in Supplemental Figure S1B. Scatterplot analysis shows that XAV939-treated cells spent significantly more time in mitosis due to a delay in anaphase (Supplemental Figure S1, C–E).
Tankyrase 1 inhibition by siRNA leads to prolonged anaphase
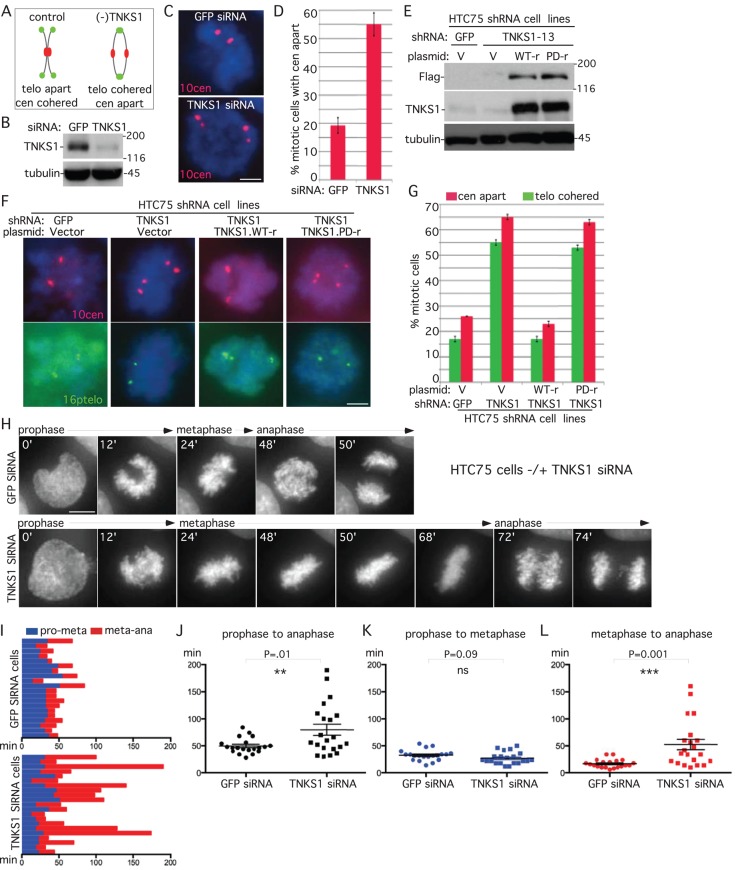
Our studies indicated that despite the lack of obvious mitotic arrest, XAV939-treated HeLa cells delayed in anaphase. To determine whether this was the case for another tumor cell line, we asked whether tankyrase 1–depleted HTC75 cells (shown previously to have persistent telomere cohesion but no mitotic arrest; Hsiao and Smith, 2009) exhibited centromere separation. HTC75 cells were treated with GFP or TNKS1 siRNA for 48 h and analyzed by immunoblot (Figure 2B) and FISH after mitotic shake-off with a centromere-specific probe. As shown in Figure 2, C and D, centromeres were separated in tankyrase 1–depleted cells, indicative of anaphase. To determine whether tankyrase 1 overexpression could rescue the anaphase-like cells, we used HTC75 cells lines stably expressing short hairpin RNA (shRNA) against GFP or TNKS1 (Hsiao and Smith, 2009) and transfected them with shRNA-resistant plasmids (Figure 2E). TNKS1 shRNA cells exhibit a loss in centromere cohesion, concomitant with persistent telomere cohesion that was rescued by wild-type (but not PARP-dead) tankyrase 1 (Figure 2, F and G).
FIGURE 2:
Tankyrase 1 depletion by siRNA leads to anaphase delay in HTC75 cells. (A–D) Centromeres separate in tankyrase 1–depleted cells. (A) Schematic diagram depicting persistent telomere cohesion and loss of centromere cohesion induced by TNKS1 depletion. (B–D) HTC75 cells treated with GFP or TNKS1 siRNA for 48 h were analyzed by (B) immunoblot and (C) centromere FISH (red) after mitotic shake-off. (D) Graphical representation of the frequency of mitotic cells with centromeres apart. Values are means ± SEM derived from two independent experiments (n = 30–100 cells each). (E–G) Wild-type (WT) but not PARP-dead tankyrase 1 rescues centromere separation. Stable HTC75 cell lines expressing GFP or TNKS1 shRNA were transfected with a vector control or siRNA resistant (r) TNKS1 WT or PARP-dead plasmids and analyzed by (E) immunoblot and (F) centromere (red) and telomere (green) FISH after mitotic shake-off. (G) Graphical representation of the frequency of mitotic cells with telomeres cohered and centromeres apart. Values are means ± SEM, derived from two independent experiments (n = 50–146 cells each). (C, F) DNA was stained with DAPI (blue). Scale bars, 5 μm. (H–L) Time-lapse video live-cell imaging of a HTC75-H2B-GFP cell line 36 h after transfection with GFP or TNKS1 siRNA. (H) Progression from prophase to anaphase for individual cells. Scale bar, 5 μm. (I–L) Graphical summaries of individual mitotic cells (n = 20–21 cells from two independent experiments) shown as (I) a time line and (J–L) scatterplots with calculated mean value ± SEM. Student's t test was used to calculate p values (ns, p ≥ 0.05; **p ≤ 0.01; ***p ≤ 0.001).
To determine whether the loss in centromere cohesion reflected anaphase delay, we analyzed HTC75-H2B-GFP cells treated for 48 h with GFP or TNKS1 siRNA by live-cell imaging. A representative example is shown in Figure 2H and Supplemental Movie S3. In both control and TNKS1 siRNA–treated cells chromosomes align on the metaphase plate at the 24-min time point. However, whereas in control cells chromosomes separate at the 48-min time point, in TNKS1 siRNA cells chromosomes struggle and do not separate until the 72-min time point. The time of progression through mitosis for each individual cell analyzed by live imaging is shown in Figure 2I. Scatterplot analysis shows that TNKS1-depleted cells spent significantly more time in mitosis (prophase to anaphase) than control cells (Figure 2J). Progression from prophase to metaphase was similar in GFP- and TNKS1-depleted cells (Figure 2K), whereas progression from metaphase to anaphase was significantly increased in TNKS1-depleted cells compared with control, indicating a delay in anaphase (Figure 2L).
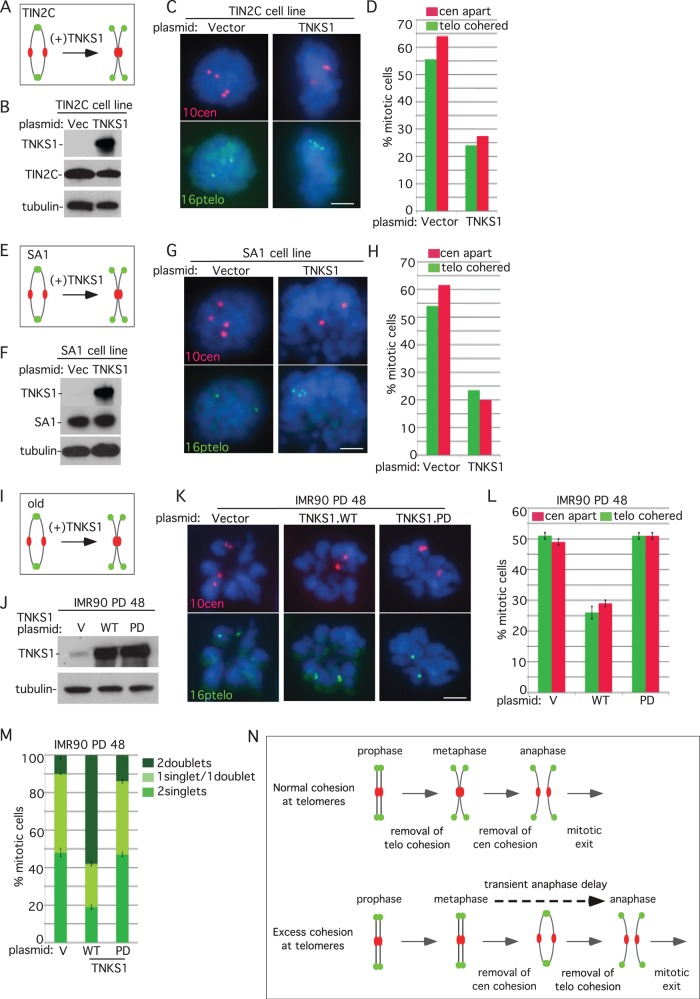
Persistent cohesion induced by overexpression of SA1 or TIN2 induces a prolonged anaphase
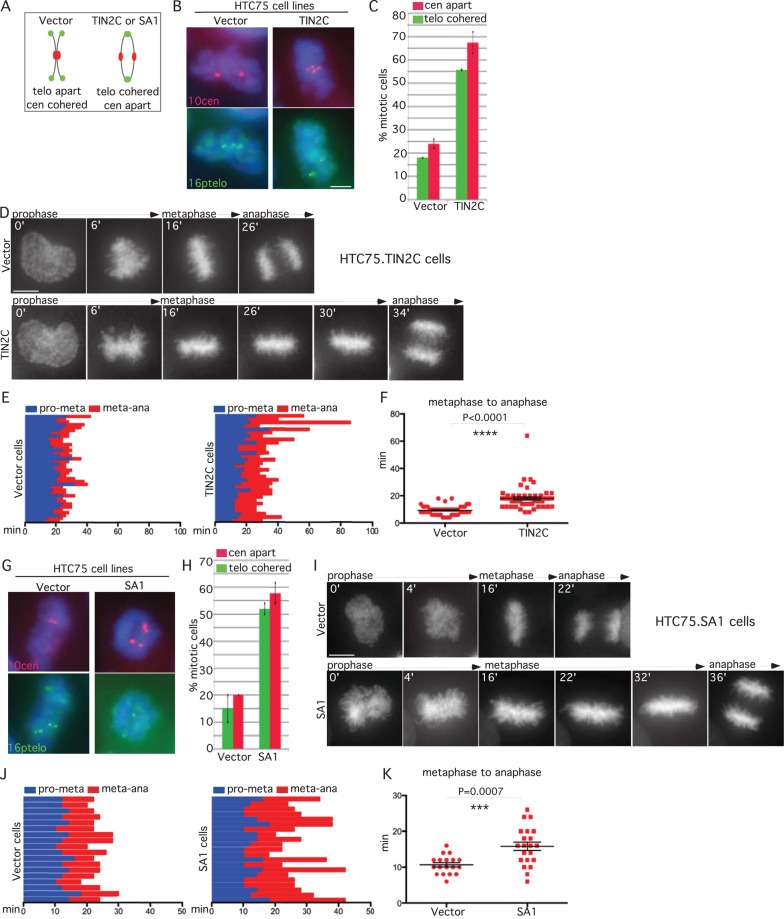
Our studies thus far indicate that inhibition or depletion of tankyrase 1 in multiple cell lines leads to delay in anaphase, consistent with the notion that persistent telomere cohesion triggers anaphase delay. However, because tankyrase 1 has multiple localizations and functions (Hsiao and Smith, 2008), it was possible that tankyrase 1 depletion induced anaphase delay by some other (non–telomere based) mechanism. To address this question, we used different methods to induce persistent sister telomere cohesin at mitosis. We showed previously that sister telomeres cohered in cells overexpressing an N-terminally truncated allele of the shelterin subunit TIN2 (TIN2C; Canudas et al., 2011). To determine whether TIN2C mitotic cells were delayed in anaphase, we analyzed the centromere status. HTC75 cells stably expressing a vector control or TIN2C were isolated by mitotic shake-off and analyzed by FISH. As shown in Figure 3, A–C, the induced persistent telomere cohesion was accompanied by a loss in centromere cohesion. Live-cell imaging of HTC75.V-H2B-GFP versus HTC75.TIN2C-H2B-GFP cell lines showed that TIN2C-overexpressing cells delay in anaphase (Figure 3D and Supplemental Movie S4). The time of progression through mitosis for each individual cell analyzed by live imaging is shown in Figure 3E. Scatterplot analysis showed that progression from metaphase to anaphase was significantly increased in TIN2C cells (Figure 3F).
FIGURE 3:
TIN2C or SA1 overexpression leads to anaphase delay. (A) Schematic diagram depicting persistent telomere cohesion and loss of centromere cohesion induced by TIN2C or SA1 overexpression. (B, C, G, H) TIN2C and SA1 overexpression leads to a loss in centromere cohesion. HTC75 cells stably expressing (B, C) V or TIN2C or (G, H) V or SA1 were analyzed by (B, G) centromere (red) and telomere (green) FISH after mitotic shake-off. DNA was stained with DAPI (blue). Scale bars, 5 μm. (C, H) Graphical representation of the frequency of mitotic cells with centromeres apart and telomeres cohered. Values are means ± SEM, derived from two independent experiments (C, n = 50–56 cells each; H, n = 50–60 cells each). (D–F, I–K) Live-cell imaging indicates that TIN2C or SA1 overexpression induces anaphase delay. Time-lapse video live-cell imaging of (D–F) HTC75.Vector-H2B-GFP and HTC75.TIN2C-H2B-GFP or (I–K) HTC75.Vector-H2B-GFP and HTC75.SA1-H2B-GFP cell lines. (D, I) Progression from prophase to anaphase for individual cells. Scale bars, 5 μm. (E, F, J, K) Graphical summaries of individual mitotic cells (E, F; n = 49–56 cells each from two independent experiments) and (I–K; n = 18–21 cells each) shown as (E, J) a time line and (F, K) a scatterplot with calculated mean value ± SEM. Student's t test was used to calculate p values (***p ≤ 0.001; ****p ≤ 0.0001).
In a second approach we used a cell line overexpressing the cohesin subunit SA1, which we showed previously displayed persistent telomere cohesion in mitosis (Bisht et al., 2013). Cells from HTC75 cell lines stably expressing vector or SA1 were isolated by mitotic shake-off and analyzed by FISH. As shown in Figure 3, G and H, the SA1-induced persistent telomere cohesion was accompanied by a loss in centromere cohesion, indicating anaphase delay. Live-cell imaging of HTC75.V-H2B-GFP versus HTC75.SA1-H2B-GFP cell lines showed that SA1-overexpressing cells delay in anaphase (Figure 3I and Supplemental Movie S5). The time of progression through mitosis for each individual cell analyzed by live imaging is shown in Figure 3J. Scatterplot analysis showed that SA1 cells were delayed in anaphase (Figure 3K).
Normal human cells approaching senescence exhibit anaphase delay
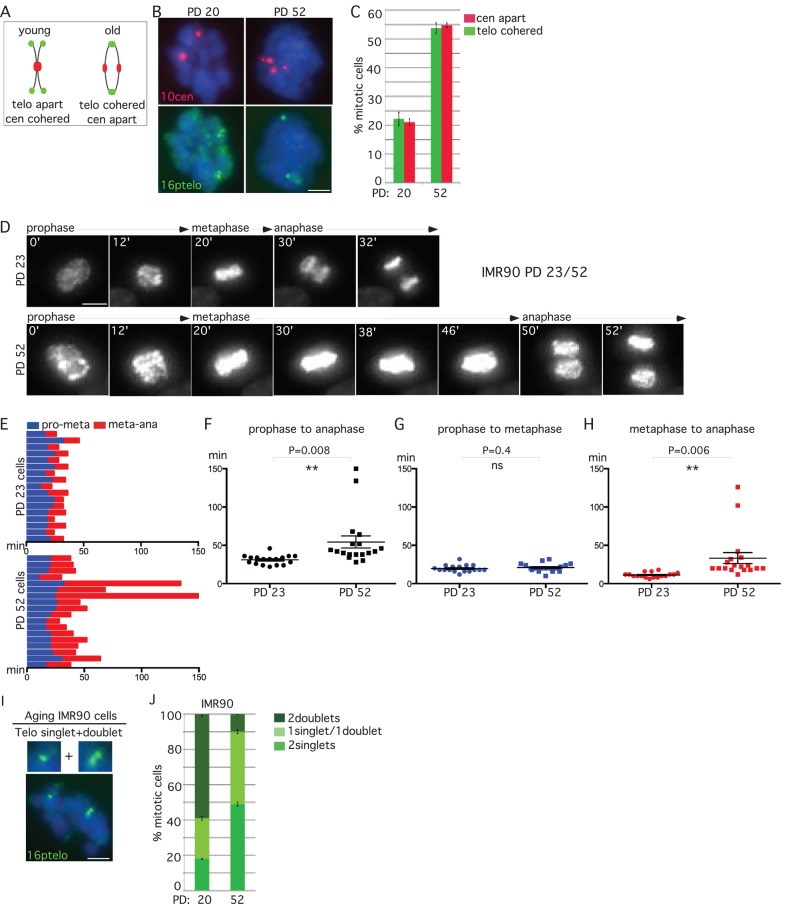
Normal human fibroblasts accumulate persistent telomere cohesion in mitosis at late population doublings before senescence (Ofir et al., 2002; Yalon et al., 2004). We thus wondered whether aging fibroblasts also underwent anaphase delay. IMR90 cells from early (20) and late (52) PDs were isolated by mitotic shake-off and analyzed by FISH. As shown in Figure 4, A–C, aging fibroblasts show loss of centromere cohesion concomitant with an increase in persistent telomere cohesion.
FIGURE 4:
Replicative aging leads to anaphase delay. (A–C) Centromeres separate concomitant with persistent telomere cohesion in aging IMR90 cells. (A) Schematic diagram depicting persistent telomere cohesion and loss of centromere cohesion young vs. old cells. (B, C). IMR90 cells from PD 20 and 52 analyzed by (B) centromere (red) and telomere (green) FISH after mitotic shake-off. DNA was stained with DAPI (blue). Scale bar, 5 μm. (C) Graphical representation of the frequency of mitotic cells with centromeres apart and telomeres cohered. Values are means ± SEM, derived from two independent experiments (n = 45–50 cells each). (D–H) Time-lapse video live-cell imaging of IMR90-H2B-GFP cell line at PD 23 and 52. (D) Progression from prophase to anaphase for individual cells. Scale bar, 5 μm. (E–H) Graphical summaries of individual mitotic cells (n = 17–18 cells) shown as (E) a time line and (F–H) scatter plots with calculated mean value ± SEM. Student's t test was used to calculate p values (ns, p ≥ 0.05; **p ≤ 0.01). (I, J) Aging cells show loss of synchrony in sister telomere separation. (I) Aging IMR90 cells analyzed by telomere (green) FISH after mitotic shake-off exhibit one singlet/one doublet. DNA was stained with DAPI (blue). Scale bars, 5 μm. (J) Graphical representation of the frequency of mitotic cells with one singlet/one doublet, two singlets, or two doublets in IMR90 cells at PD 20 or 52. Values are means ± SEM, derived from two independent experiments (n = 50 cells each).
To determine whether the loss in centromere cohesion manifested as anaphase delay in aging cells, we analyzed IMR90-H2B-GFP cells from early (23) and late (52) PDs by live-cell imaging. A representative example (shown in Figure 4D and Supplemental Movie S6) indicates anaphase delay in aging cells. The time of progression through mitosis for each individual cell analyzed by live imaging is shown in Figure 4E. Scatterplot analysis showed that aging fibroblasts spent significantly more time in mitosis than control cells due to a delay in anaphase (Figure 4, F–H).
Our telomere FISH analysis of aging cells revealed an increase in mitotic cells containing one singlet/one doublet (Figure 4I) rather than the more typical two singlets, suggesting a loss in synchrony in the resolution of sister telomeres. We thus scored cells for three phenotypes: two singlets, one singlet/one doublet, and two doublets. Comparison between PD 20 and 52 shows a twofold increase in the one singlet/one doublet phenotype in aging cells, similar to the two-singlet phenotype (Figure 4J). Indeed, only 10% of aging cells exhibit synchronous loss of telomere cohesion, that is, two sets of telomere doublets.
Tankyrase 1 rescues persistent cohesion and anaphase delay in aging fibroblasts
Together our studies suggest a balance between factors that promote and resolve telomere cohesion. We thus asked whether tankyrase 1 overexpression could rescue the excess cohesion induced by overexpression of TIN2C. The HTC75.TIN2C cell line was transfected with a vector control or tankyrase 1 (Figure 5B) and analyzed by FISH. As shown in Figure 5, C and D, overexpression of tankyrase 1 rescued persistent telomere cohesion and restored normal centromere cohesion, indicating a rescue of the anaphase delay. Similarly, overexpression of tankyrase 1 in the HTC75.SA1 cell line rescued persistent telomere cohesion and restored normal centromere cohesion (Figure 5, E–H).
FIGURE 5:
Tankyrase 1 overexpression rescues persistent telomere cohesion, regardless of the mechanism of induction. (A–H) Tankyrase 1 overexpression rescues persistent telomere cohesion and loss of centromere cohesion in TIN2C- or SA1-overexpressing cells. (A, E) Schematic diagrams. (B–D) HTC75.V or TIN2C and (F–H) HTC75.V or SA1 cells were transfected for 24 h with a vector or TNKS1 plasmid and analyzed by (B, F) immunoblot and (C, G) centromere (red) and telomere (green) FISH after mitotic shake-off. DNA was stained with DAPI (blue). Scale bars, 5 μm. (D, H) Graphical representation of the frequency of mitotic cells with centromeres apart and telomeres cohered (D, n = 50–56 cells each; H, n = 50–60 cells each). (I–L) Wild-type but not PARP-dead tankyrase 1 rescues telomere cohesion and centromere separation in aging IMR90 cells. (I) Schematic diagram. (J–L) IMR90 cells stably expressing tankyrase 1 WT or PARP dead or a vector control were generated by lentiviral infection at an early PD (20). Cell lines were grown for multiple PDs and then, before senescence (PD 48), analyzed by (J) immunoblot and (K) centromere (red) and telomere (green) FISH after mitotic shake-off. DNA was stained with DAPI (blue). Scale bar, 5 μm. (L) Graphical representation of the frequency of mitotic cells with telomeres cohered and centromeres apart. Values are means ± SEM, derived from two independent experiments (n = 50 cells each). (M) Loss of synchrony in sister telomere separation in aging IMR90 cells can be rescued by wild-type but not PARP-dead tankyrase 1. Graphical representation of the frequency of mitotic cells with one singlet/one doublet, two singlets, or two doublets in IMR90 cells expressing vector, TNKS1.WT, or TNKS1.PARP dead at PD 48. Values are means ± SEM, derived from two independent experiments (n = 50 cells each). (N) Model showing that excess telomere cohesion leads to anaphase delay.
We next asked whether tankyrase 1 could rescue the persistent cohesion phenotype in aging fibroblasts. IMR90 cells stably expressing tankyrase 1 (wild type or PARP dead) or a vector control were generated by lentiviral infection at early PD (20). Cell lines were grown for multiple PDs and then analyzed at late PD (48) by immunoblot (Figure 5J) and isolated by mitotic shake-off and analyzed by FISH. As shown in Figure 5, K and L, wild-type (but not PARP-dead) tankyrase 1 rescued persistent telomere cohesion and restored normal centromere cohesion. Moreover, overexpression of wild-type (but not PARP-dead) tankyrase 1 rescued the one singlet/one doublet phenotype in aging fibroblasts (Figure 4M). Together these data indicate that overexpression of tankyrase 1 can offset the anaphase delay and concomitant loss in synchrony of sister telomere separation that occurs in aging fibroblasts.
DISCUSSION
We showed that excess telomere cohesion at mitosis (induced by multiple distinct mechanisms) triggers anaphase delay. Normally, telomere cohesion is removed during prophase at the same time as arm cohesion; subsequent removal of centromere cohesion at metaphase leads to anaphase progression and mitotic exit (Figure 5N, top). However, under conditions that lead to excess cohesion at telomeres, arm and centromere cohesion is removed normally, but telomeres remain cohered and cells delay in anaphase. Ultimately, telomere cohesion is resolved, and cells exit mitosis (Figure 5N, bottom).
Our studies suggest that there are opposing forces that regulate cohesion at telomeres. Tankyrase 1 localizes to telomeres in prophase, leading to loss of SA1 at telomeres and resolution of telomere cohesion (Bisht et al., 2012, 2013). We showed previously that when tankyrase 1 is depleted, SA1 remains on telomeres in mitosis (Bisht et al., 2013). Tankyrase 1 may be limiting for telomere cohesion resolution since overexpression of SA1 alone is sufficient to induce excess cohesion in mitosis even in the presence of tankyrase 1 (Bisht et al., 2013). Indeed, our observation that overexpression of tankyrase 1 can rescue the persistent cohesion induced by SA1 (or TIN2C) overexpression indicates a dynamic balance of the process. We showed here that we can affect this balance by manipulating expression of various proteins. In addition, there appears to be a natural mechanism that promotes excess cohesion. Normal human cells display persistent telomere cohesion and anaphase delay before senescence. The observation that we can rescue this excess cohesion by overexpression of tankyrase 1 indicates that it likely results from an imbalance in the pathway. Whether the excess telomere cohesion at mitosis in aging cells is due to changes in tankyrase 1 PARP activity or localization, increased recruitment or stability of proteins that promote telomere cohesion, or yet to be identified factors remains to be determined.
What is the purpose of the anaphase delay? For insights we can draw parallels between budding yeast and human cells. The highly repetitive sequences (telomeres and rDNA) in budding yeast rely on distinct mechanisms for their resolution at mitosis. In budding yeast, telomeres and rDNA remain connected in a cohesin-independent manner until mid anaphase, when they are resolved, dependent on the Cdc14 phosphatase and condensin (D'Amours et al., 2004; Sullivan et al., 2004). In human cells telomere resolution normally occurs in prophase. However, in the absence of tankyrase 1, telomeres remain connected in a cohesin ring–independent manner (Bisht et al., 2013), and resolution does not occur until mid anaphase. Anaphase delay in human cells may be a response to defective or damaged telomeres. Excess cohesion accumulates at telomeres as human cells age, and we showed that this is accompanied by anaphase delay. Studies in budding yeast show that excess cohesion prolongs anaphase, measured as a loss in the synchrony in resolution of sister chromatids (Lyons and Morgan, 2011). We observe a similar loss in synchrony in separation of telomeres in aging cells.
As human cells age, telomeres shorten and accumulate DNA damage (d'Adda di Fagagna et al., 2003; Fumagalli et al., 2012). The accompanying persistent telomere cohesion could be a response to this damage, and the ensuing anaphase delay may provide an opportunity for repair. Aging fibroblasts (which undergo telomere shortening due to the absence of telomerase) ultimately succumb to the DNA damage and senesce. However, tumor cells (which can maintain their telomeres due to the presence of telomerase) may benefit from the opportunity to assess and repair dysfunctional telomeres.
MATERIALS AND METHODS
Plasmids
Tankyrase1 cDNA under the cytomegalovirus (CMV) promoter TT20 (Smith et al., 1998) was used for transfection of HTC75 TIN2C or SA1 cell lines. For the rescue experiments tankyrase1, wild type or PARP dead, was inserted into a modified pLKO.1ps vector containing a CMV promoter (pLSJH). pLSJH.TNKS1WT and PARP dead were rendered resistant to TNKS1-13 shRNA (Hsiao and Smith, 2009); the T at nucleotide position 1716 in the tankyrase 1 coding sequence was replaced with C by site-directed mutagenesis using the oligonucleotide 5′-GAAAGAGCCCATAATGACGTCATGGAAGTTCTGCAT-3′ and the Stratagene (Santa Clara, CA) QuikChange Site-Directed Mutagenesis Kit.
Cell lines
For live-cell imaging, stable cell lines expressing histone H2B-GFP were generated by retroviral infection. To create stable cell lines, amphotropic retroviruses were generated by transfecting pBABE-H2B-GFP (plasmid 26790; Addgene, Cambridge, MA) into phoenix amphotropic cells (ATCC) using calcium phosphate precipitation. HTC75 cells (an HT1080-derived clonal cell line; van Steensel and de Lange, 1997), IMR90 PD 20 (ATCC), TIN2C (an HTC75 cell line overexpressing TIN2C; Canudas et al., 2011), and SA1 (an HTC75 cell line overexpressing SA1; Bisht et al., 2013) were infected and selected in 2 μg/ml puromycin as described (Houghtaling et al., 2004). HeLa.H2B-GFP cells were described previously (Kanda et al., 1998).
Additional cell lines used for FISH and immunofluorescence analysis were stable HTC75 cell lines expressing GFP or TNKS1-13 shRNA (Hsiao and Smith, 2009) and HeLaI.2.11 (van Steensel et al., 1998).
Chromosome-specific FISH
Cells were fixed and processed as described previously (Dynek and Smith, 2004). Briefly, cells were fixed twice in methanol:acetic acid (3:1) for 15 min, cytospun (Shandon Cytospin) at 2000 rpm for 2 min onto slides, rehydrated in 2× saline–sodium citrate (SSC) at 37°C for 2 min, and dehydrated in an ethanol series of 70, 80, and 95% for 2 min each. Cells were denatured at 75°C for 2 min and hybridized overnight at 37°C with a subtelomeric fluorescein isothiocyanate (FITC)–conjugated probe (16ptelo) and/or a tetramethylrhodamine isothiocyanate (TRITC)–conjugated chromosome 10 centromere probe (10cen) from Cytocell (Cambridge, UK). Cells were washed in 0.4× SSC at 72°C for 2 min and in 2× SSC with 0.05% Tween 20 at room temperature for 30 s. DNA was stained with 0.2 μg/ml DAPI. The distance between FISH signals was measured using OpenLab software (Perkin Elmer, Waltham, MA).
Cell synchronization and XAV939 treatment
HeLaI.2.11, Hela-H2B-GFP, and IMR90-H2BGFP cells were grown in the presence of 2 mM thymidine for 16 h, washed three times with PBS, and released into fresh medium. After 10 h the medium was replaced with medium containing 2 mM thymidine, and the cells were incubated for 16 h, washed three times with PBS, and released into fresh medium containing reduced serum (0.5%) with or without 1 µM XAV939 for 10 h.
Cell extracts
Cells were resuspended in four volumes of TNE buffer (10 mM Tris, pH 7.8, 1% Nonidet P-40, 0.15 M NaCl, 1 mM EDTA, and 2.5% protease inhibitor cocktail [Sigma, St. Louis, MO]) and incubated for 1 h on ice. Suspensions were pelleted at 8000 × g for 15 min. Twenty-five micrograms (determined by Bio-Rad [Hercules, CA] protein assay) of supernatant proteins was fractionated by SDS–PAGE and analyzed by immunoblotting.
Immunoblot analysis
Immunoblots were incubated separately with the primary antibodies rabbit anti–tankyrase 1 762 (1 μg/ml; Scherthan et al., 2000), rabbit anti-Flag (0.4 μg/ml; Sigma), mouse anti–α-tubulin ascites (1:10,000; Sigma), mouse anti-SA1 (0.5 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA), or rabbit anti-TIN2C 701 (0.5 μg/ml; Houghtaling et al., 2004), followed by horseradish peroxidase–conjugated donkey anti-rabbit or anti-mouse immunoglobulin G (1:2500; Amersham, Pittsburgh, PA). Bound antibody was detected by Super Signal West Pico (Thermo Scientific, Waltham, MA).
siRNA and plasmid transfection
siRNA transfections were performed with Oligofectamine (Invitrogen) according to the manufacturer's protocol for 36–48 h. The final concentration of siRNA was 100 nM. The following siRNAs (synthesized by Dharmacon Research, Pittsburgh, PA) were used: TNKS1 (5′- AACAAUUCACCGUCGUCCUCU-3′) described previously (Dynek and Smith, 2004) and GFP Duplex I. For plasmids, cells were transfected with Lipofectamine 2000 (Invitrogen, Grand Island, NY) according to the manufacturer's protocol for 24 h.
Indirect immunofluorescence
Cells were fixed in methanol at −20°C for 10 min, blocked in 1% bovine serum albumin in PBS, and incubated with the following antibodies: human ACA serum (1:4000) or rabbit cyclin B (0.2 μg/ml; Santa Cruz Biotechnology). Primary antibodies were detected with FITC- or TRITC-conjugated donkey anti-human or anti-rabbit antibodies (1:100; Jackson Laboratories, Bar Harbor, ME). DNA was stained with DAPI (0.2 μg/ml).
Image acquisition
Images were acquired using a microscope (Axioplan 2; Carl Zeiss, Thornwood, NY) with a Plan Apochrome 63×/numerical aperture (NA) 1.4 oil immersion lens (Carl Zeiss) and a digital camera (C4742-95; Hamamatsu Photonics, Bridgewater, NJ). Images were acquired and processed using Openlab software (Perkin Elmer).
Live-cell imaging
Cell lines stably expressing H2B-GFP (HeLa, HTC75, HTC75.V, HTC75.TIN2C, HTC75.SA1, IMR90 PD 23 or 52) were imaged in CO2-independent Liebovitz's L-15 medium (Life Technologies, Grand Island, NY) for 4–6 h with an Applied Precision (Issaquah, WA) PersonalDV live-cell imaging system mounted on an Olympus (Center Valley, PA) IX-71 inverted microscope fitted with a Zeiss Plan Neofluor 40×/1.30 NA oil objective lens for HeLa and HTC75 cell lines and with a Olympus UPlanApo 20×/0.75 NA objective lens for IMR90 cell lines. Temperature was maintained at 37°C with an imaging chamber. Images were acquired at 2-min intervals with a CoolSnap HQ2 charge-coupled device camera using Softworx software. For time-lapse movies (Supplemental Movies S1–S6) images were compiled into movies using ImageJ software (National Institutes of Health, Bethesda, MD). All movies were played back at 7 frames/s.
Statistical analysis
Statistical analysis was performed using Prism 6 software (GraphPad, La Jolla, CA). Student's t test was applied. Data are shown as mean ± SEM or ± SD; p < 0.05 was considered significant, and actual p values are depicted in the figures.
Supplementary Material
Acknowledgments
We thank members of the Smith lab and Tom Meier for comments on the manuscript and helpful discussion. Live-cell images were collected in the NYU Langone Medical Center Microscopy Core. Research was supported by the National Cancer Institute of the National Institutes of Health under award number R01CA116352.
Abbreviations used:
- FISH
fluorescence in situ hybridization
- GFP
green fluorescent protein
- PARP
poly(ADP-ribose) polymerase
- PD
population doubling
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0479) on October 30, 2013.
REFERENCES
- Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht KK, Daniloski Z, Smith S. SA1 binds directly to DNA via its unique AT-hook to promote sister chromatid cohesion at telomeres. J Cell Sci. 2013;126:3493–503. doi: 10.1242/jcs.130872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht KK, Dudognon C, Chang WG, Sokol ES, Ramirez A, Smith S. GDP-mannose-4,6-dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-ribose) polymerase activity. Mol Cell Biol. 2012;32:3044–3053. doi: 10.1128/MCB.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Houghtaling BR, Bhanot M, Sasa G, Savage SA, Bertuch AA, Smith S. A role for heterochromatin protein 1{gamma} at human telomeres. Genes Dev. 2011;25:1807–1819. doi: 10.1101/gad.17325211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26:4867–4878. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Smith S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol. 2009;187:165–173. doi: 10.1083/jcb.200903096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol. 2012;14:355–365. doi: 10.1038/ncb2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90:83–92. doi: 10.1016/j.biochi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Hsiao SJ, Smith S. Sister telomeres rendered dysfunctional by persistent cohesion are fused by NHEJ. J Cell Biol. 2009;184:515–526. doi: 10.1083/jcb.200810132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SM, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons NA, Morgan DO. Cdk1-dependent destruction of Eco1 prevents cohesion establishment after S phase. Mol Cell. 2011;42:378–389. doi: 10.1016/j.molcel.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir R, Yalon-Hacohen M, Segev Y, Schultz A, Skorecki KL, Selig S. Replication and/or separation of some human telomeres is delayed beyond S-phase in pre-senescent cells. Chromosoma. 2002;111:147–155. doi: 10.1007/s00412-002-0199-z. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Carretero M, Martinez P, Drosopoulos WC, Canamero M, Schildkraut CL, Blasco MA, Losada A. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Jerratsch M, Li B, Smith S, Hulten M, Lock T, de Lange T. Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores. Mol Biol Cell. 2000;11:4189–4203. doi: 10.1091/mbc.11.12.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Higuchi T, Katis VL, Uhlmann F. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 2004;117:471–482. doi: 10.1016/s0092-8674(04)00415-5. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Yalon M, Gal S, Segev Y, Selig S, Skorecki KL. Sister chromatid separation at human telomeric regions. J Cell Sci. 2004;117:1961–1970. doi: 10.1242/jcs.01032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.