Clustered N-WASP binds directly to actin-filament barbed ends and can either slow individual filament growth or processively accelerate the assembly of bundled actin filaments. This novel Arp2/3-independent mechanism of N-WASP likely plays a role in invadopodia and podosome formation, in which both N-WASP and actin filaments are tightly clustered.
Abstract
Neuronal Wiskott–Aldrich syndrome protein (N-WASP)–activated actin polymerization drives extension of invadopodia and podosomes into the basement layer. In addition to activating Arp2/3, N-WASP binds actin-filament barbed ends, and both N-WASP and barbed ends are tightly clustered in these invasive structures. We use nanofibers coated with N-WASP WWCA domains as model cell surfaces and single-actin-filament imaging to determine how clustered N-WASP affects Arp2/3-independent barbed-end assembly. Individual barbed ends captured by WWCA domains grow at or below their diffusion-limited assembly rate. At high filament densities, however, overlapping filaments form buckles between their nanofiber tethers and myosin attachment points. These buckles grew ∼3.4-fold faster than the diffusion-limited rate of unattached barbed ends. N-WASP constructs with and without the native polyproline (PP) region show similar rate enhancements in the absence of profilin, but profilin slows barbed-end acceleration from constructs containing the PP region. Increasing Mg2+ to enhance filament bundling increases the frequency of filament buckle formation, consistent with a requirement of accelerated assembly on barbed-end bundling. We propose that this novel N-WASP assembly activity provides an Arp2/3-independent force that drives nascent filament bundles into the basement layer during cell invasion.
INTRODUCTION
Invadopodia (Mueller and Chen, 1991) and related podosomes (Tarone et al., 1985) are specialized ventral cellular projections that can degrade the extracellular matrix via localized activity of matrix metalloproteases. Both structures contain tightly-packed actin filaments that depend on neuronal Wiskott–Aldrich syndrome protein (N-WASP) for their generation (Linder et al., 1999; Mizutani et al., 2002; Lorenz et al., 2004; Co et al., 2007). N-WASP is believed to organize invadopodia primarily through its Arp2/3-activating activity (Linder et al., 2000; Lorenz et al., 2004; Li et al., 2010), which generates a branched actin network (Baldassarre et al., 2006) reminiscent of dendritic nucleation in a moving cell's lamellipodium (Pollard et al., 2000; Pollard and Borisy, 2003; Pollard and Cooper, 2009). Despite the presence of Arp2/3, both invadopodia and podosomes contain a mix of bundled and branched actin (Gavazzi et al., 1989; Schoumacher et al., 2010). Invadopodia are rich in actin cross-linking proteins (Schoumacher et al., 2010), and the actin-bundling protein fascin stabilizes invadopodia and enhances substrate degradation (Li et al., 2010). Ultrastructure studies of podosomes show tight clusters of actin filaments (Luxenburg et al., 2007). Thus actin clusters or bundles may play an important role in the extension of both structures into the substrate.
In addition to activating Arp2/3, the C-terminal WWCA region of N-WASP binds to fast-growing actin-filament barbed ends (Egile et al., 1999; Laurent et al., 1999; Co et al., 2007; Hu and Kuhn, 2012). For Arp2/3 activation, the central (C) and acidic (A) regions stabilize the short-pitch helix conformation of arp2 and arp3 subunits (Padrick et al., 2011; Xu et al., 2011), while the two verprolin/WASP homology 2 domains (V, W, or WH2) bind to actin monomers (Marchand et al., 2001; Chereau et al., 2005) and actin barbed ends (Rebowski et al., 2008; Gaucher et al., 2012). Thus N-WASP may also organize actin barbed ends in podosomes and invadopodia independent of its role in Arp2/3 activation.
We recently showed that in the presence of Arp2/3 complex, nanofibers coated with glutathione S-transferase (GST)–WWCA domains from N-WASP generated both branched and bundled actin networks (Hu and Kuhn, 2012). This one-dimensional analogue of a two-dimensional cell membrane formed actin networks reminiscent of those produced at the leading edge of an invasive cell. In this reconstruction, filament bundles arose spontaneously from the dendritic network and appeared to attach processively to WWCA tethers. This attachment generated enough force to buckle filaments within the bundle. Because N-WASP generated a mix of branched and bundled protruding actin, N-WASP's barbed-end binding activity may play an important role in the extension of invadopodia or podosomes into the basement layer. However, it was unclear whether N-WASP attached to barbed ends directly or indirectly through Arp2/3.
Given our observation of bundle tethering to N-WASP–coated nanofibers, we sought to determine whether actin filaments could bind processively to tethered WWCA domains in the absence of Arp2/3 and how WWCA binding affects barbed-end polymerization rates. Here we show that N-WASP coated nanofibers bind to de novo–nucleated actin-filament barbed ends in two regimes. Individual barbed ends bound to N-WASP-coated nanofibers grew at or below the diffusion-limited polymerization rate, depending on whether the nanofiber acted as a barrier to filament growth. However, clustered barbed ends bound to nanofibers could grow substantially faster than the diffusion-limited polymerization rate. Thus, like Ena/vasodilator-stimulated phosphoprotein (VASP) and formin proteins, N-WASP WWCA domains can processively attach to growing barbed-end bundles and increase their diffusion-limited elongation rate. This new activity for N-WASP provides an important new mechanism for cell invasion into the substrate.
RESULTS
Individual barbed-end capture is nonprocessive
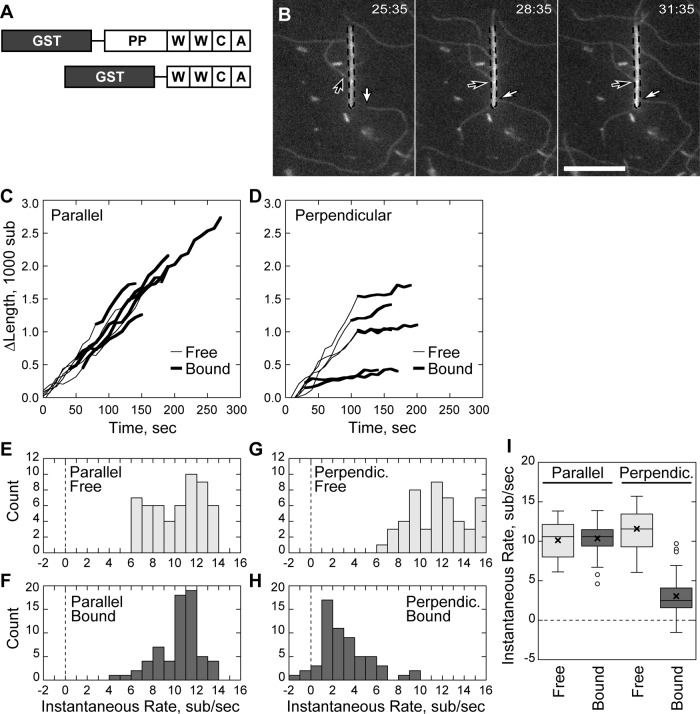
We used total internal reflection fluorescence (TIRF) microscopy of single actin filaments (Amann and Pollard, 2001; Kuhn and Pollard, 2005, 2007) to dissect actin barbed-end capture by the C-terminus of N-WASP in the absence of Arp2/3 complex. Glass nanofibers were coated with GST-tagged constructs of the WWCA domain of N-WASP having both native WH2 domains. The first construct tested contained the native polyproline region (PP) of bovine N-WASP followed by the native WWCA C-terminal Arp2/3 activator domain (Figure 1A). Addition of fluorescently labeled bovine serum albumin (BSA) to coated nanofibers blocked nonspecific binding and highlighted nanofibers for microscopy. Coated nanofibers and fluorescently labeled actin monomers were added to chambers coated with a low concentration of rigor myosin to tether filaments along their length. Myosin attachments appeared as inflection points that served as fiduciary marks to separate barbed- and pointed-end measurements (Kuhn and Pollard, 2005).
FIGURE 1:
Nonbundled-filament barbed-end capture is not processive. Conditions as follows: 1.5 μM (33–40% labeled) Mg-ATP actin, nanofibers coated with 10–20 μM GST-PP-WWCA, 1 mM MgCl2, 10 mM imidazole, pH 7, 50 mM KCl, 1 mM EGTA, 100 mM DTT, 0.2 mM ATP, 0.25% methyl cellulose, 15 mM glucose, 20 μg/ml catalase, and 100 μg/ml glucose oxidase. Slides coated with NEM-inactivated myosin II. (A) The NWASP constructs used in the study. (B) Elongating barbed ends (arrows) attached to a GST-PP-WWCA–coated, fluorescein isothiocyanate–labeled BSA blocked nanofiber (dashed outline), polymerizing either perpendicularly (white arrow) or in parallel (black arrow). (C) Growth of barbed ends parallel to the nanofiber before and after capture. (D) Growth of barbed ends attached perpendicular to the nanofiber before and after capture. (E–H) Histogram of smoothed growth rates before (E) and after (F) parallel capture of barbed ends or before (G) and after (H) perpendicular capture. (I) Box-and-whisker plot of growth rates of parallel and perpendicular captured filaments. Scale bar, 10 μm.
GST-PP-WWCA–coated nanofibers captured growing barbed ends of individual, de novo–nucleated actin filaments. At low filament densities, we identified two types of barbed-end capture with unique behaviors: end capture parallel to the long axis of the nanofiber and end capture perpendicular to the long axis. Filament barbed ends that encountered the nanofiber at a shallow angle (Figure 1B; Supplemental Movie 1) continued to grow along the nanofiber long axis after attachment and were designated as parallel captures. A small fraction of barbed ends (13 ± 24%, N = 24 filaments, mean ± SD) encountered the nanofiber at right angles and remained attached to the same location (Figure 1B). These interactions were designated as perpendicular captures.
We measured filament length over time to assay barbed-end growth before and after capture. Parallel captured barbed ends grew along the nanofiber at the same rate before and after binding (Figure 1C), whereas perpendicular captured barbed ends grew at a substantially reduced rate while attached to the same location on the nanofiber (Figure 1D). To quantify the slow, saltatory growth of perpendicular-bound barbed ends over time, we determined the smoothed, instantaneous growth rates at each time point. Parallel captured barbed ends in 1.5 μM actin monomers grew at 10.1 ± 2.3 s−1 (N = 6 filaments) before capture and at 10.4 ± 1.8 s−1 (N = 6) after capture (Figure 1, E, F, and I), consistent with theoretical rates for labeled actin (Pollard, 1986; Kuhn and Pollard, 2005). In contrast, filaments captured perpendicular to the nanofiber slowed from 11.5 ± 2.5 s−1 (N = 6) to 3.1 ± 2.3 s−1 (N = 6) after capture (Figure 1, G–I). In control experiments, filament barbed ends did not interact with BSA-coated nanofibers (Supplementary Figure S1; Supplemental Movie 6). Thus the parallel growth along N-WASP–coated nanofibers and the substantial reduction in growth of perpendicular capture filaments was due to N-WASP binding rather than nonspecific interaction of barbed ends with nanofibers.
Rapid processive elongation of N-WASP–bound barbed ends
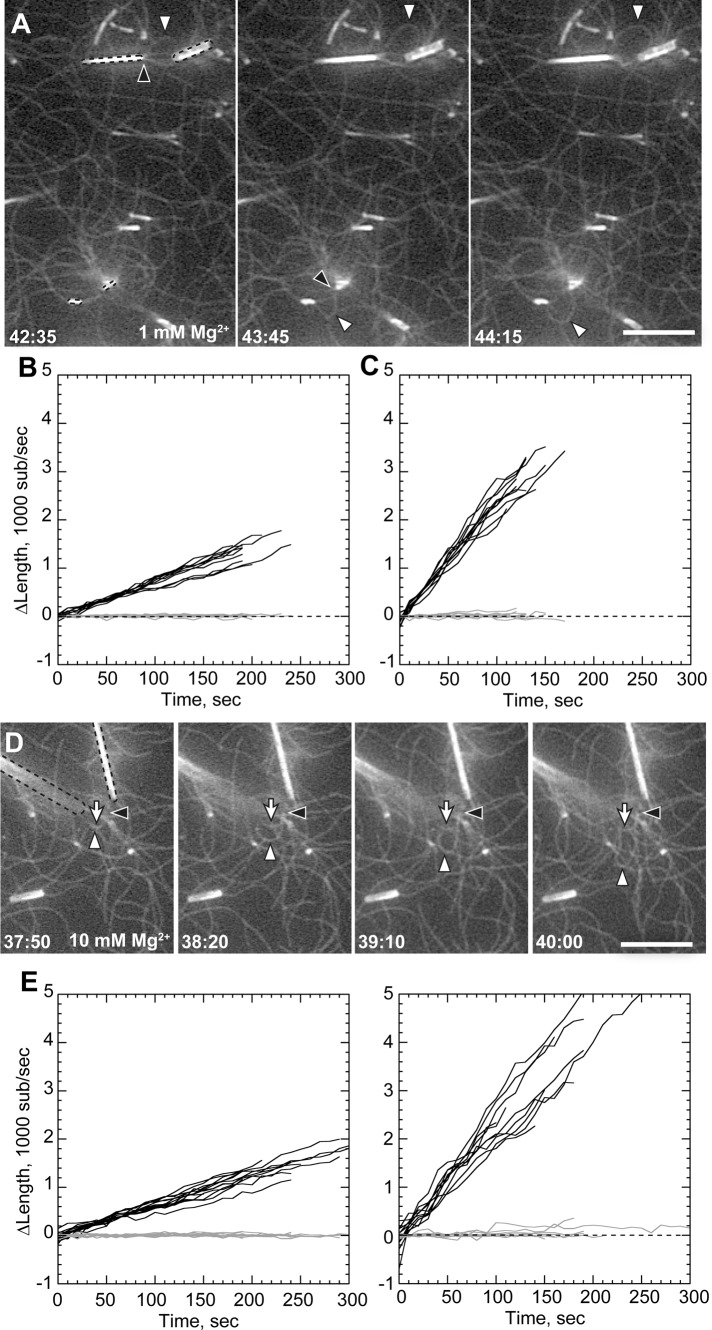
At high filament densities, some nanofiber-associated filaments grew faster than their neighbors to form prominent buckles and loops (Figure 2A; Supplemental Movie 2). We measured the elongation rates of both nanofiber-associated buckling filaments and unattached background filaments in the same experiment (Table 1). Strikingly, buckling barbed ends grew 3.3-fold faster than background filaments. Background barbed ends grew at the theoretical rate (Pollard, 1986; Kuhn and Pollard, 2005) of 6.89 ± 0.13 s−1 (Figure 2B), whereas buckling barbed ends grew at an average rate of 22.42 ± 0.39 s−1 (Figure 2C).
FIGURE 2:
Filament bundling enhances rate of processive elongation. Conditions as in Figure 1, except 1 or 1.2 μM actin as indicated and total MgCl2 concentration of 1 mM (A–C) or 10 mM (D–F). (A) Processive association of actin filaments to GST-PP-WWCA–coated nanofibers (dotted outlines) in 1.2 μM Mg-ATP-actin and 1 mM Mg2+, showing buckling of rapidly elongating filaments (white arrowheads) from compression between barbed ends (black arrowheads) and NEM-myosin attachment points. (B, C) Elongation of barbed (black) and pointed (gray) ends of untethered filaments (B) or tethered buckles (C) in 1 mM Mg2+. (D) Buckling filament (white arrowheads) with barbed end (black arrowheads) overlapping a nonbuckling filament (white arrow) attached to the end of a nanofiber (dotted outlines) in 10 mM Mg2+. (E, F) Elongation of untethered filaments (E) or tethered filaments (F) in 10 mM Mg2+. Barbed-end growth rates shown in Table 1. Scale bars, 10 μm.
TABLE 1:
Average filament elongation rate.
| Barbed ends | Tether | Actin (μM) | Mg2+ (mM) | Seeds (nM) | Profilin (μM) | Barbed-end rate (subunits/s) | Number of filaments | Rate of enhancement |
|---|---|---|---|---|---|---|---|---|
| Tethered | PP-WWCA | 1.2 | 1 | — | — | 22.42 ± 0.39 | 10 | 3.25 |
| Free | PP-WWCA | 1.2 | 1 | — | — | 6.89 ± 0.13 | 10 | |
| Tethered | PP-WWCA | 1 | 10 | — | — | 21.05 ± 0.39 | 10 | 3.44 |
| Free | PP-WWCA | 1 | 10 | — | — | 6.12 ± 0.08 | 10 | |
| Tethered | PP-WWCA | 1 | 10 | 75 | 1 | 14.28 ± 0.18 | 7 | 1.93 |
| Free | PP-WWCA | 1 | 10 | 75 | 1 | 7.40 ± 0.11 | 13 | |
| Tethered | WWCA | 1 | 1 | — | — | 21.45 ± 0.52 | 10 | 3.14 |
| Free | WWCA | 1 | 1 | — | — | 6.83 ± 0.09 | 10 | |
| Tethered | WWCA | 1 | 10 | — | — | 21.74 ± 0.70 | 10 | 3.45 |
| Free | WWCA | 1 | 10 | — | — | 6.29 ± 0.11 | 10 | |
| Tethered | WWCA | 1 | 10 | 75 | 1 | 25.45 ± 1.09 | 10 | 3.45 |
| Free | WWCA | 1 | 10 | 75 | 1 | 7.37 ± 0.28 | 10 |
Filament buckling was rare in 1 mM Mg2+ but more frequent when Mg2+ was raised to induce filament side-to-side association (bundling). Based on previous evidence that filament bundling by divalent cations may mediate processive barbed-end attachments to N-WASP (Hu and Kuhn, 2012), we increased buffer Mg2+ concentration to 10 mM to generate actin bundles (Tang and Janmey, 1996; Hu and Kuhn, 2012). In 10 mM Mg2+, nanofibers mediated more frequent rapid filament growth and buckling (Figure 2D; Supplemental Movie 3). However, high Mg2+ did not change speed of accelerated barbed-end growth once it began. As with lower Mg2+, nanofiber-associated barbed ends grew 3.4-fold faster than unattached barbed ends in 10 mM Mg2+.
Accelerated filaments did not remain bundled along their entire length as they grew. Instead, their barbed ends were frequently oriented parallel to the long nanofiber axis and its parallel captured filaments. In some cases, we found accelerated barbed ends that were directly bundled to other filaments (Figure 2, A and D, black arrowheads; Supplemental Movie 4). Thus parallel association of barbed ends (end bundling) likely plays a role in accelerated barbed-end growth.
Profilin bound to the proline-rich region slows barbed-end processivity
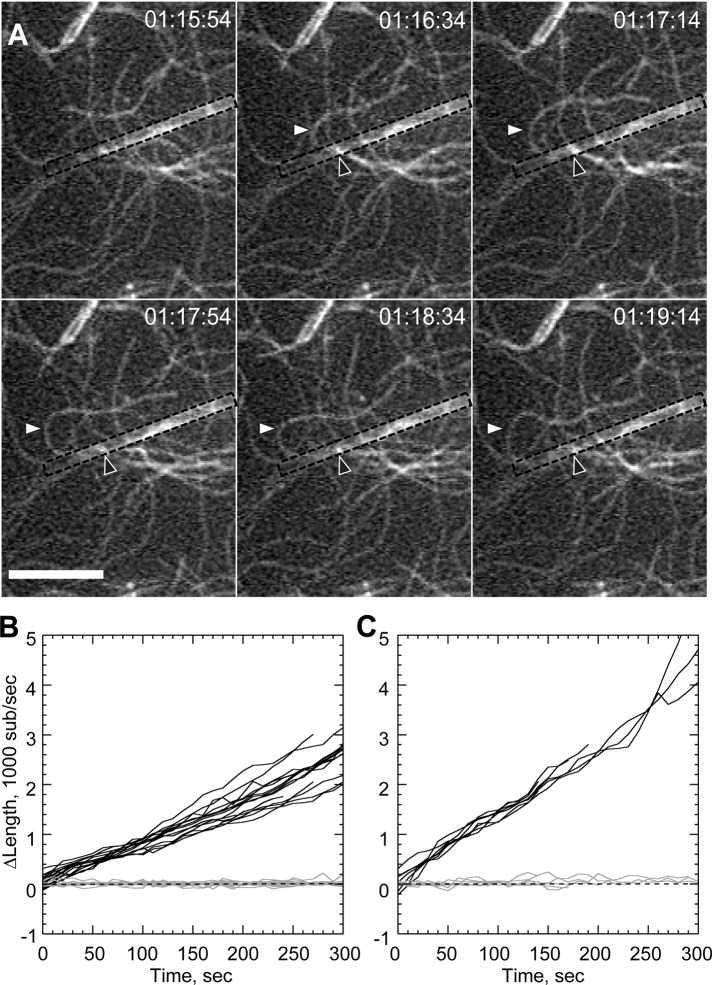
The proline-rich region of PP-WWCA might recruit profilin–actin heterodimers (Mullins et al., 1998; Suetsugu et al., 1998) for insertion at the barbed end, similar to formin proteins (Romero et al., 2004). To test whether profilin modulated processivity, we repeated the barbed-end capture experiments with profilin–actin, high Mg2+, and short actin seed filaments to overcome the suppression of de novo filament nucleation by profilin. Nanofibers generated filament buckles (Figure 3A; Supplemental Movie 5), but the acceleration was reduced by profilin. Buckling filament barbed ends grew at an average rate of 14.28 ± 0.18 s−1 (Figure 3C), compared with 7.4 ± 0.11 s−1 for unattached barbed ends (Figure 3B). This 1.9-fold acceleration was significantly lower (p < 0.001) than the 3.4-fold acceleration seen in the absence of profilin (Table 1).
FIGURE 3:
Profilin reduces rate of bundle-mediated processive elongation. Conditions as in Figure 2 with 1 μM actin, 10 mM total Mg2+, 1 μM profilin, and 75 nM actin seeds. (A) Time-lapse image of sustained processive association of actin filaments on GST-PP-WWCA–coated glass nanofibers in presence of profilin. White arrowheads indicate buckling of a rapidly growing filament. Black arrowheads indicate approximate position of barbed-end attachment point. (B, C) Elongation of untethered (B) or tethered filaments (C) in 10 mM Mg2+. Barbed-end growth rates shown in Table 1. Scale bar, 10 μm.
The proline-rich domain is not required for processivity
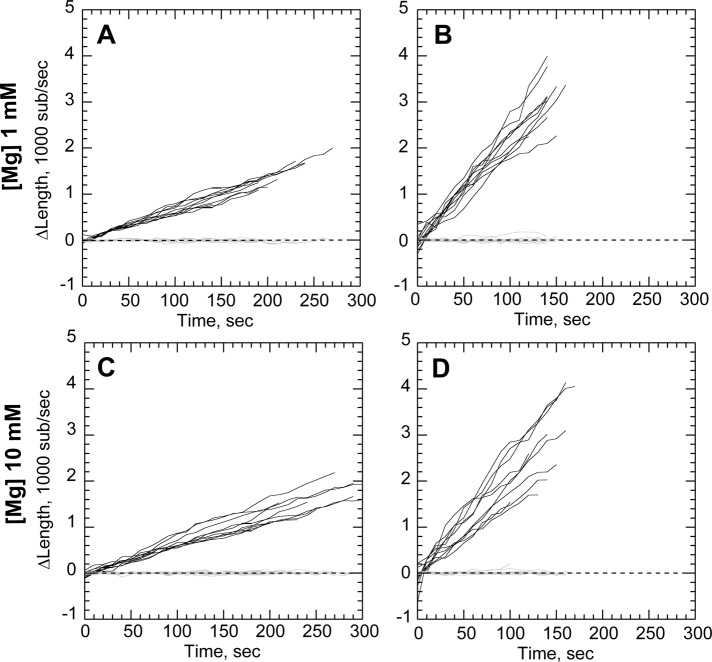
The decrease in profilin–actin assembly rates could be due either to slower incorporation of profilin–actin dimers at the barbed end or through direct interaction between profilin and the proline-rich region of N-WASP. We therefore designed a GST N-WASP construct (Figure 1A) lacking the polyproline domains to determine its effect on accelerated processivity. We found that GST-WWCA–coated nanofibers generated filament buckles in both high (10 mM) and low (1 mM) Mg2+ (Figure 4). GST-WWCA–tethered ends grew 3.14-fold faster than untethered ends in low Mg2+ and 3.45-fold faster in high Mg2+. These acceleration rates were not significantly different (p = 0.1083) in the two Mg2+ concentrations tested. The polyproline region thus does not have a major role in processive barbed-end acceleration of actin alone.
FIGURE 4:
N-WASP proline-rich domains are not required for processive elongation. (A–D) Conditions as in Figure 2, except nanofibers coated with GST-WWCA. (A, B) Elongation of untethered (A) or tethered filaments (B) in 1 μM actin and 1 mM Mg2+. (C, D) Elongation of untethered (C) or tethered filaments (D) in 1 μM actin 10 mM Mg2+. Growth rates shown in Table 1.
To determine whether profilin affected barbed-end processivity of GST-WWCA constructs, we repeated the foregoing experiments in the presence of profilin–actin and actin seeds. As with GST-PP-WWCA constructs, GST-WWCA constructs accelerated barbed-end assembly in the presence of profilin–actin (Table 1). However, the 3.45-fold rate enhancement was equal to the rate enhancement seen in the absence of profilin. Thus the slower barbed-end acceleration rate seen with construct containing the polyproline region was due to specific interactions between the proline-rich region of N-WASP and profilin–actin.
Rare processive elongation events were not evident in bulk assays
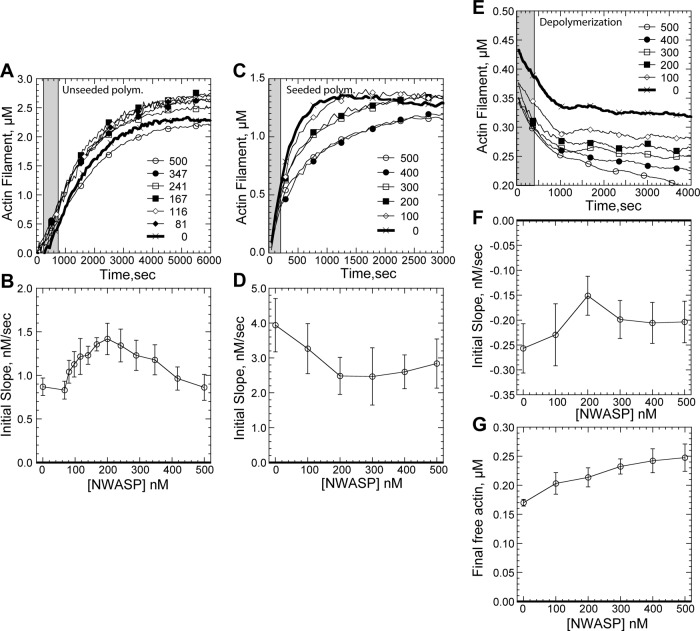
Given the rarity of these processive elongation events, we sought to determine whether processive elongation would be evident or hidden in bulk actin polymerization assays under typical conditions (1 mM Mg2+). Moderate concentrations of GST-WWCA (200 nM) enhanced the initial polymerization rate of 2.5 μM pyrenyl–actin (Figure 5, A and B). However, this enhancement was likely due to GST-WWCA–mediated nucleation rather than barbed-end acceleration. Although isolated N-WASP WWCA peptides do not nucleate filaments (Gaucher et al., 2012), dimerization contributed by the GST tag (Lim et al., 1994) effectively doubles the number of WH2 domains available for nucleation. Like the actin nucleators Spire with four WH2 domains (Quinlan et al., 2007) or Cobl's three WH2 domains (Ahuja et al., 2007), GST dimerization could contribute to the weak nucleation activity we found at moderate GST-WWCA concentrations.
FIGURE 5:
Bulk assays of N-WASP barbed-end binding. Conditions are as follows: 1.5–5 μM (30–97% labeled) Mg-ATP actin, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0. (A) Representative de novo polymerization of 2.5 μM pyrenyl–actin in the indicated nanomolar concentration of GST-WWCA. Gray band indicates period for initial slope estimation. (B) Average (N = 6) de novo initial polymerization rates in a range of GST-WWCA concentrations. (C) Polymerization of 1.5 μM pyrenyl–actin from 0.4 μM seeds. Gray band indicates period for initial slope estimation. (D) Average (N = 8) initial seeded polymerization rates over a range of nanomolar GST-WWCA concentrations. (E) Pyrenyl–actin depolymerization in the presence of indicated nanomolar GST-WWCA. Pyrenyl–actin, 5 μM, was polymerized and then diluted 20-fold in the indicated final nanomolar concentration of GST-WWCA. Gray band indicates period for initial slope estimation. (F) Average (N = 6) initial seeded polymerization rates over a range of GST-WWCA concentrations. (G) Average (N = 6) final free actin concentrations over a range of GST-WWCA concentrations.
In support of a nucleation-based increase in initial rate seen at moderate GST-WWCA concentrations, high concentrations of GST-WWCA (500 nM) abolished the initial rate enhancement, consistent with WWCA sequestration of actin monomers (Gaucher et al., 2012). Furthermore, when GST-WWCA nucleation was overwhelmed with prepolymerized actin seeds, we found no concentration-dependent rate enhancement (Figure 5, C and D). Instead, high concentration of GST-WWCA decreased the actin assembly rate.
This decrease in actin polymerization at high GST-WWCA concentrations could come from monomer sequestration or N-WASP–mediated changes to actin barbed-end kinetics. We therefore tested the effects of GST-WWCA on actin depolymerization. A 20-fold dilution of actin filaments prepolymerized from 5 μM ATP-pyrenyl-actin reduced overall filament concentration to 0.24 μM, and free monomer concentration to <10 nM was used. Under these conditions, monomer binding by GST-WWCA would have little effect on the initial depolymerization rate. Nevertheless, GST-WWCA decreased the actin depolymerization rate (Figure 5, E and F), consistent with previous studies of uncomplexed WWCA peptides (Gaucher et al., 2012). The decreased depolymerization was thus likely mediated by GST-WWCA interaction with filament ends rather than by further depleting actin monomers. In support, GST-WWCA also produced a concentration-dependent increase in the final monomer critical concentration (Figure 5G), consistent with its binding to actin barbed ends.
Taken together, bulk assays support binding of N-WASP to barbed ends but cannot unequivocally show barbed-end acceleration. Bulk pyrene–actin assays are often difficult to interpret, as they only measure the average total filamentous actin content over time. Rare barbed-end acceleration events would only contribute a small fraction to total pyrene–actin filament mass. Thus the crucial but rare acceleration activity of N-WASP would be discernible only through TIRF microscopy and could have easily been missed in previous studies.
Filament bundling increases frequency of N-WASP–mediated barbed-end acceleration
Although Mg2+ did not significantly alter barbed-end acceleration rates, nanofiber-mediated buckles appeared earlier in reactions with high (10 mM) Mg2+. We therefore scored the buckle initiation time and normalized against the total nanofiber length in each experiment (Figure 6). Buckles appeared at significantly higher frequencies in 10 mM Mg2+ than they did in 1 mM Mg2+ for both N-WASP constructs (Table 2). Furthermore, the N-WASP construct containing the polyproline region (PP-WWCA) was less efficient than the shorter construct (WWCA) at initiation barbed-end acceleration in all cases. Once initiated, both constructs accelerated polymerization to the same extent. This difference in initiation efficiency may lie in the difference between tether lengths. Thus the range of motion of nearby clustered WWCA domains may play a key role in finding and maintaining contact with bundled filament barbed ends. Because both PP-WWCA and WWCA constructs were nonspecifically adhered to glass nanofibers, we cannot rule out differences in the way each construct adhered.
FIGURE 6:
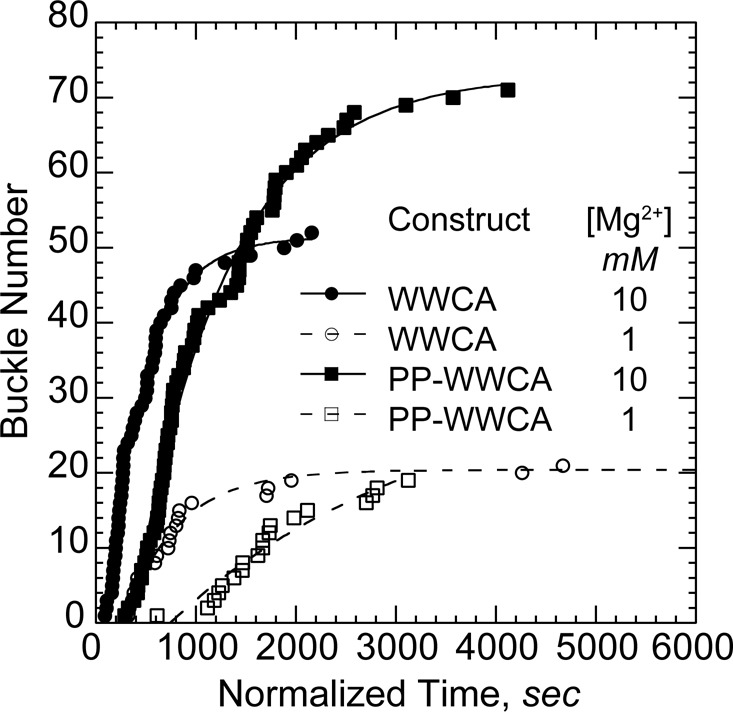
Bundling by Mg2+ increases frequency of buckle initiation. Buckle initiation times were normalized against total nanofiber length in each experiment. Cumulative initiation times over several experiments with the indicated N-WASP construct and Mg2+ levels were fit to a delayed exponential to yield an initiation rate (Table 2). Buckle formation rates increased with polycation concentration for both constructs. WWCA initiation buckles faster than PP-WWCA.
TABLE 2:
Rate of buckle occurrence.
| Construct | Actin (μM) | Mg2+ (mM) | Rate (s−1 μm−1) |
|---|---|---|---|
| PP-WWCA | 1.2 | 1 | (3.9 ± 1.8) × 10−6 |
| PP-WWCA | 1 | 10 | (13.8 ± 0.6) × 10−6 |
| WWCA | 1 | 1 | (73.6 ± 6.7) × 10−6 |
| WWCA | 1 | 10 | (134.4 ± 5.6) × 10−6 |
The primary effect of Mg2+ on N-WASP processivity was likely due to barbed-end bundling. We previously showed that actin filaments form bundles at cellular (1 mM) Mg2+ levels under the same buffer conditions used in the present study (Hu and Kuhn, 2012). However, the fraction of bundled filaments and the speed of bundle initiation increase significantly in 10 mM Mg2+. Furthermore, GST-WWCA–coated particles capture significantly more bundled barbed ends than unbundled barbed ends in higher Mg2+ or when bundling factors are added.
In the present study, N-WASP–generated accelerated filament buckles only appeared when filament density increased and filaments were highly clustered (Figures 2, A and D, and 3A). Adding Mg2+ increased the frequency of initiation of processive WWCA-mediated barbed-end assembly twofold to threefold without significantly modifying the overall rate of this accelerated assembly. In contrast, increased Mg2+ does not increase de novo filament nucleation (Hu and Kuhn, 2012). Barbed ends were generated at the same rate in both high and low Mg2+, and the increased rate of buckle initiation in high Mg2+ was due to increased availability of bundled barbed ends rather than increase in total barbed ends. Taken together, the results indicate that polycations influenced the acceleration of barbed ends by bundling negatively charged filaments and limiting the diffusion of barbed ends away from N-WASP rather than by increasing the number of available barbed ends or enhancing the binding of N-WASP to barbed ends. Bundle-mediated cooperativity of barbed ends thus appears to be a key factor in the transition from diffusion-limited elongation to fast processive assembly.
DISCUSSION
Here we showed that N-WASP–coated nanofibers captured and assembled actin filament barbed ends in two distinct regimes. 1) Individual barbed ends bound to N-WASP assembled at their normal rate when growing along the nanofiber or at a slower rate when growing against the nanofiber. 2) At high filament densities, N-WASP binding promoted rapid, processive barbed-end growth. Processive barbed-end assembly was rare at low filament densities and could not be discerned with traditional bulk pyrene–actin assembly assays. Processive assembly was relatively rare in the 1 mM Mg2+ concentrations used in most actin assays, even at high filament densities. Thus this novel behavior of N-WASP likely escaped notice, as the actin field focused primarily on the Arp2/3-binding activity of N-WASP rather than its interaction with actin filaments.
N-WASP–mediated growth of individual barbed ends at or below the diffusion-limited polymerization rate fits well with current models of WH2-domain association at the barbed end. WASP-family WH2 domains bind to the hydrophobic cleft of actin monomers (Chereau et al., 2005). This binding site is exposed at the barbed end of a filament, allowing WH2-bound actin monomers to add to the barbed end. Once assembled, WH2 domains must dissociate from the hydrophobic cleft to allow further longitudinal actin assembly. Like profilin (Pollard and Cooper, 1984; Kang et al., 1999), soluble N-WASP WWCA (VCA) activator domains do not prevent barbed-end assembly (Egile et al., 1999), implying that WH2 dissociates somewhere in the assembly process.
In contrast to profilin–actin, it is unclear whether WH2 domains dissociate from the terminal barbed-end subunit immediately after WH2-actin addition or after some delay. Our data demonstrated that N-WASP–associated actin assembly was much slower than the diffusion-limited rate if the nanofiber acted as a barrier to monomer addition (perpendicular attachments). Continuous attachment of filaments to N-WASP during periods of slow barbed-end growth implies that WH2 dissociation from the barbed end may be linked to subsequent monomer addition. When further monomer addition was restricted, N-WASP remained attached to the barbed end.
Slow growth of barbed ends against a barrier (perpendicular capture) implies a thermal ratchet model of nonprocessive barbed-end assembly by N-WASP. Here some of the free energy from polymerization goes toward buckling the filament (Dogterom and Yurke, 1997), leading to a decrease in the diffusion-limited assembly rate. In the “tethered ratchet” model of motility (Mogilner and Oster, 2003a, b) and the related “cooperative thermal breakage” model (Alberts and Odell, 2004; Soo and Theriot, 2005), nonpolymerizing actin barbed ends are transiently attached to the leading edge, while polymerizing barbed ends push against the leading edge. Transient filament-to-membrane attachments are broken as the compressive force of polymerization against the membrane is translated through the cross-linked network to the membrane-bound filaments. As we showed, N-WASP can provide this transient attachment to filament barbed ends. Cycles of attachment and detachment were rapid when monomer addition was unrestricted (growth along the nanofiber), whereas N-WASP detachment was slower when monomer addition was restricted (growth against the nanofiber).
In the second assembly regime, buckling filaments remained processively attached to the same location on the nanofiber. Rapid and processive barbed-end assembly fits closely with the “actoclampin” model of Dickinson and Purich (Dickinson and Purich, 2002; Dickinson et al., 2004; Dickinson, 2008), which posits that a hypothetical membrane-bound “actoclampin” molecule maintains constant, processive attachment to the growing barbed end.
The barbed-end assembly rate of buckling filaments can be used to distinguish between processive and nonprocessive polymerization. In the fast-elongation regime, acceleration of barbed ends beyond their natural diffusion-limited rate unequivocally shows processive barbed-end assembly by WWCA domains. Like Ena/VASP (Kovar and Pollard, 2004; Breitsprecher et al., 2008; Hansen and Mullins, 2010) and formin-family proteins (Kovar and Pollard, 2004; Romero et al., 2004; Otomo et al., 2005; Paul and Pollard, 2009), N-WASP WWCA domains can processively attach to growing barbed ends and increase their diffusion-limited elongation rate, as originally postulated by Chereau et al. (2005).
Polyproline regions of WASP-family proteins recruit profilin–actin (Suetsugu et al., 1998). We found that N-WASP construct containing this region accelerated profilin–actin polymerization to a lesser extent (1.9-fold) than it did actin alone (3.4-fold). This reduction was likely due to profilin binding to the polyproline region, as N-WASP constructs lacking this region accelerated barbed-end assembly to the same extent with or without profilin. Reduced acceleration could stem from misaligned insertional assembly of profilin–actin at the barbed end. However, profilin–actin binds to the loading site containing a GPPPP consensus sequence (Mahoney et al., 1997), and the first WH2 domain is a major factor in stabilizing Arp2/3 daughter branches (Suetsugu et al., 1998; Mullins, 2000; Yang et al., 2000). More likely, GPPPP binding to profilin reduces dissociation of profilin from the barbed end required for subsequent longitudinal actin monomer addition. Further experiments with N-WASP constructs lacking one or more GPPPP domains or with different lengths of linker between the N-terminal GPPPP binding site and the first WH2 domain of N-WASP may shed light on the mechanism of N-WASP–mediated profilin–actin insertion at the barbed end.
How is this transition from diffusion-limited elongation to fast processive assembly driven? The active barbed-end–binding domains of VASP and WASP proteins are highly homologous (Ferron et al., 2007; Dominguez, 2009), and VASP requires its tetramerization domain to accelerate barbed ends (Bachmann et al., 1999; Breitsprecher et al., 2008; Hansen and Mullins, 2010). Similarly, formin requires dimerization to processively walk along growing barbed ends (Moseley et al., 2004), and clustered WH2 domains from VopL can assembly barbed ends (Namgoong et al., 2011). Membrane-clustered WWCA domains could similarly walk along bundled barbed ends and insert actin subunits, as we previously proposed (Figure 8 in Hu and Kuhn, 2012). Although our GST-tagged N-WASP constructs likely formed dimers at the nanofiber surface (Padrick et al., 2008), lending a pseudomultimerization to our N-WASP, we did not see fast processive elongation until barbed ends clustered as well. In contrast to VASP and formin, which rely on multimerization for processive actin assembly, N-WASP relies on both clustering and bundling provided by extrinsic factors such as polycations. Although N-WASP is less likely to initiate processive assembly than VASP, it is efficient once started. We propose that filament bundling limits diffusion of a barbed end away from WWCA after WWCA dissociation. WWCA is then free to bind additional actin monomers and efficiently refind the same barbed end for subsequent monomer addition.
Alternatively, the two tandem WH2 domains of N-WASP could provide the two actin-binding sites required for processive stepping. While the C-terminal WH2 domain binds to a filament barbed end, the upstream WH2 domain would bind and add a new monomer. Both WH2 domains bind actin monomers (Rebowski et al., 2008; Gaucher et al., 2012), but structures of tandem WH2 domains from N-WASP show that orientation of the tandem actin monomers is incompatible with longitudinal dimer formation (Rebowski et al., 2008). We do not preclude an intermediate, low-affinity binding state in which longitudinal dimer formation ejects the second WH2 from the hydrophobic cleft at the longitudinal dimer interface. However, it is unclear how the short linker between WH2 domains in N-WASP could provide enough flexibility for stepping toward a new binding site during elongation (Dominguez, 2010). Further studies with extended WH2 linkers, N-WASP constructs lacking one WH2 domain, or WASP's single WH2 domain should elucidate the mechanism of processive elongation.
We demonstrated a novel Arp2/3-independent barbed-end assembly mechanism by the C-terminal of N-WASP. This new activity provides new insight into the formation of cellular structures by N-WASP. Although actin microspikes are enriched with N-WASP (Nakagawa et al., 2001) and N-WASP overexpression can generate filopodia (Miki et al., 1998), the involvement of N-WASP in filopodia generation remains controversial (Snapper et al., 2001; Steffen et al., 2006; Sarmiento et al., 2008). However, N-WASP appears to be required for invadopodia and podosome formation (Linder et al., 1999; Mizutani et al., 2002; Lorenz et al., 2004; Co et al., 2007). We propose that N-WASP promotes transient actin bundles to invasive structures by accelerating their assembly. The simultaneous requirement of both a high density of free barbed ends and their parallel association means that the transition from passive to processive assembly is rare throughout the actin cortex, where free barbed end densities are relatively low. The probability of buckle initiation increases substantially in regions of active barbed-end nucleation where barbed ends are also densely packed, such as within podosomes or nascent invadopodia. Once initiated, the force from this accelerated assembly provides an initial push or “spear tip” for invadopodia to wedge into the basement layer. N-WASP–accelerated bundles that do not rapidly dissipate into the dendritic network are then stabilized by cross-linking proteins such as α-actinin and fascin. VASP and formin are then recruited to these sites to extend invadopodia into the basement layer.
MATERIALS AND METHODS
Protein expression and purification
Actin was purified from rabbit skeletal muscle actin acetone powder through one round of polymerization and depolymerization followed by gel filtration (Spudich and Watt, 1971). Actin was labeled with Oregon Green 488 iodoacetamide (Invitrogen, Grand Island, NY) as described (Kuhn and Pollard, 2005). Actin and labeled actin were stored for 1 mo at 4ºC. Both labeled and unlabeled actins were dialyzed overnight against fresh buffer G (2 mM Tris-Cl, pH 8, 0.2 mM ATP, 1 mM NaN3, 0.1 mM CaCl2, 0.5 mM dithiothreitol [DTT]) and centrifuged at 38,000 × g for 2 h at 4ºC. Actin concentrations were estimated from extinction coefficients as follows: actin, E290 = 26,600 M−1 cm−1 (Kuhn and Pollard, 2005); Oregon Green actin, E290 = 26,600 M−1 cm−1 using the correction A*290 = A290 − 0.16991A491; Oregon Green, E491 = 77,800 M−1 cm−1.
Bovine N-WASP WWCA (A403-D505) was purified as a GST fusion protein as described (Hu and Kuhn, 2012). Bovine N-WASP PP-WWCA (A244-D505) starting at the end of the CRIB domain was cloned into the vector pET-41a (Novagen, Madison, WI) containing N-terminal GST, histidine, and S tags, and the sequence was verified. GST-PP-WWCA was expressed in Rosetta DE3pLysS (Novagen) bacteria grown at 37°C to an A600 of 0.8 and induced with 0.5 mM isopropyl β-d-thiogalactopyranoside overnight at 16°C. Bacteria were pelleted, resuspended in TBSE (20 mM Tris, 200 mM NaCl, 1 mM EDTA, 10 mM 2-mercaptoethanol) supplemented with complete EDTA-free protease inhibitors (Roche, South San Francisco, CA), and pulse sonicated on ice for a total of 150 s. Bacteria were pelleted for 30 min at 46,000 × g, the supernatant was added to glutathione resin (Thermo Scientific, Waltham, MA), and the resin was washed with five volumes of wash buffer (TBSE with 0.1% Thesit). Protein was eluted using 100 mM reduced glutathione, pH 8.0, and gel filtered on a Superdex 200 column (GE Healthcare, Pittsburgh, PA). GST-PP-WWCA concentration was determined using an extinction coefficient E280 = 53080 M−1 cm−1. Both N-WASP constructs were flash frozen in liquid nitrogen and stored at −80ºC.
Nanofiber preparation and coating
Glass nanofibers (200-nm nominal diameter; Johns Mansville, Denver, CO) were broken into smaller fragments in chloroform in a Dounce homogenizer as previously described (Hu and Kuhn, 2012). Nanofibers were centrifuged at 3750 rpm for 10 min, excess chloroform drained, and the remaining chloroform evaporated. Nanofibers were washed with deionized water by low-speed centrifugation and sonicated for 1 h in 1 M KOH in a bath sonicator to remove contaminants. Nanofibers were washed briefly in deionized water, resuspended in 1 M HCl, sonicated for 1 h, and incubated overnight in HCl. Cleaned nanofibers were subsequently pelleted by centrifugation and sonicated for 30 min each in deionized water, 1 mM EDTA, 70% ethanol, and absolute ethanol to dry, with pelleting between each step. Cleaned nanofibers were stored in glass containers in absolute ethanol for up to 6 mo. For coating, ethanol was removed after centrifugation and the remaining ethanol evaporated. Nanofibers were incubated with 20 μM N-WASP in coating buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.3, 0.1 M KCl, 1 mM MgCl2, 1 mM ATP, 0.1 mM CaCl2, 1 mM NaN3, final pH 7.56) overnight. Fluorescein isothiocyanate–labeled BSA (Invitrogen) was added to a final concentration of 0.02 mg/ml to aid visibility, and nanofibers were incubated an additional 5 min. Nanofibers were washed three times with coating buffer and resuspended in coating buffer supplemented with 1 mg/ml low-grade BSA (Sigma-Aldrich, St. Louis, MO) to block subsequent protein addition. Nanofibers were stored in BSA at 4ºC for up to 5 d before use.
TIRF microscopy
Clean glass slides, coverslips, and flow cells were constructed as previously described (Kuhn and Pollard, 2005). For filament tethering, flow cells were coated with 100 nM N-ethylmaleimide (NEM)–inactivated myosin II for 2 min. To prevent nonspecific binding, flow cells were blocked with 1% (wt/vol) BSA for 2 min as described (Kuhn and Pollard, 2005). Unlabeled Mg-ATP-actin and Mg-ATP-actin labeled with Oregon Green 488 were mixed with nanofibers and 2× TIRF buffer (2×: 20 mM imidazole, pH 7, 100 mM KCl, 2 mM MgCl2, 2 mM ethylene glycol tetraacetic acid [EGTA], 200 mM DTT, 0.4 mM ATP, 30 mM glucose, 0.5% methyl cellulose 1500 cP, 40 μg/ml catalase, 0.2 mg/ml glucose oxidase) or 2× high-Mg TIRF buffer (2×: TIRF buffer with 20 mM total MgCl2) to start spontaneous actin assembly. For each experiment 16 μl of reaction mixture was added to the chamber, and the entry and exit ports of flow cell were sealed with warm VALAP (1:1:1 Vaseline/lanolin/paraffin).
Actin seeds
Short actin seeds were created as described (Kuhn and Pollard, 2007) by polymerizing 6 μM unlabeled Mg-ATP-actin at room temperature for 10 min in buffer F (buffer G with 10 mM imidazole, pH 7, 50 mM KCl, 0.105 mM MgCl2, 1 mM EGTA). Seeds were diluted 1:20 in buffer F and vortexed immediately at high speed for 60 s to shear filaments. Seeds were added to a final concentration of 75 nM.
Image acquisition and analysis
TIRF images were collected on an Olympus (Center Valley, PA) upright microscope (BX51WI) using prism-based excitation from a 488-nm solid-state laser (Sapphire; Coherent, Santa Clara, CA) and custom optics. Images were captured by a Rolera-MGI EMCCD camera (QImaging, Surrey, Canada) at 10-s intervals using Micro-Manager open source acquisition software (Edelstein et al., 2010). Images were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD; Schneider and Rasband, 2012). Actin filament barbed- and pointed-end lengths were measured against fiduciary marks provided by NEM-myosin II attachment points as previously described (Kuhn and Pollard, 2005). We measured the lengths of an average of 10 filaments per experiment and at least three experiments per condition unless otherwise indicated.
Pyrenyl–actin assembly assays
Pyrene–actin fluorescence was measured using a fluorescence plate reader (Gemini XPS; Molecular Devices, Sunnyvale, CA). Polymerization was monitored by continuous pyrene fluorescence measurements (λexc = 364 nm, λem = 407 nm) at 22–24°C. All reactions were carried out in a Corning 96-well, half-area, flat-bottom plate. Unseeded actin polymerization assays were monitored over a range of nanomolar concentrations of GST-WWCA. The reaction well was prepared by addition of with 1.5 μl of 100× Antifoam (100×: 0.005% Antifoam-204; Sigma-Aldrich), 2× initial concentration of Mg KMEI (10×: 500 mM KCl, 10 mM MgCl2, 10 mM EGTA, 100 mM imidazole, pH 7.0), and buffer G and GST-WWCA. In the preparatory well, 5 μM Ca-ATP actin (30% pyrene labeled) was mixed 9:1 with ME exchange buffer (10× ME: 10 mM EGTA, 1 mM MgCl2) and incubated on ice for 2 min to form 2× final concentrations of Mg-ATP actin. The reaction was initiated by transferring 75 μl of actin monomer mixture from the preparatory well to the corresponding row in reaction well for a final concentration of 2.5 μM monomeric actin in a 150-μl reaction. The delay from the reaction start until the first data read was recorded manually and added before analysis of kinetic data. Seeded polymerization reactions were similarly produced with 1.5 μM actin (30% pyrene labeled), varying GST-WWCA, and prepolymerized, unlabeled actin seeds. To create actin seeds, 6 μM Mg-ATP actin was polymerized in F buffer (buffer G with 10 mM imidazole, pH 7, 50 mM KCl, 1.1 mM MgCl2, 2 mM EGTA) for 1 h at room temperature. Seeds were vortexed (Vortex Genie 2; VWR Scientific Products, West Chester, PA) at maximum speed for 60 s to break longer filaments. Short seeds were added to the reaction well to give a final concentration of 0.4 μM actin seeds in a 150-μl reaction. For depolymerization assays, actin (5 μM, 88–97% pyrene labeled) was polymerized overnight. The reaction well was prepared by addition of 7.5 μl incubated with varying concentrations of GST-WWCA for 10 min. A 1.5-μl drop of 100× Antifoam was carefully added along the side of each well. The reaction was diluted 20-fold in F-buffer with 2× slow mixing strokes and wide-bore pipette tips to minimize filament breakage. After dilution, fluorescence of pyrene–actin was monitored every 15 s. Initial rates of actin assembly were determined from linear fits of the initial 400 s of depolymerization data.
Buckle occurrence timing
The nanofiber density varied between experiments and therefore influenced the number of nucleation-competent sites in each captured movie. To quantify the frequency of buckle occurrence, we recorded the time after the start of the reaction at which each buckle became visible (buckle initiation). To correct for variations in the number of nucleation sites between experiments, we multiplied the initiation time by the total length of nanofibers in each experiment and divided by the weighted mean of total nanofiber length across all experiments. Normalized initiation times for each condition were combined and sorted to give a cumulative count of the number of buckles. Buckle counts, C, were fit to an exponential growth curve, C{1 – exp[–(t – t0)/τ]}, to find the initiation rate, τ, and delay, t0, for de novo filament nucleation.
Supplementary Material
Acknowledgments
We thank David Kovar for critical reading of the manuscript. This work was supported by an Interfaces in Science Career Award from the Burroughs Wellcome Fund (Award 1003964) and a Research Grant (Award J-991) from the Thomas F. Jeffress and Kate Miller Jeffress Memorial Trust.
Abbreviations used:
- BSA
bovine serum albumin
- GST
glutathione S-transferase
- N-WASP
neuronal Wiskott–Aldrich syndrome protein
- TIRF
total internal reflection fluorescence
- VASP
vasodilator-stimulated phosphoprotein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0781) on November 13, 2013.
REFERENCES
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts JB, Odell GM. In silico reconstitution of Listeria propulsion exhibits nano-saltation. PLoS Biol. 2004;2:e412. doi: 10.1371/journal.pbio.0020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci USA. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Baldassarre M, Ayala I, Beznoussenko G, Giacchetti G, Machesky LM, Luini A, Buccione R. Actin dynamics at sites of extracellular matrix degradation. Eur J Cell Biol. 2006;85:1217–1231. doi: 10.1016/j.ejcb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 2008;27:2943–2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RB. A multi-scale mechanistic model for actin-propelled bacteria. Cell Mol Bioeng. 2008;1:110–121. [Google Scholar]
- Dickinson RB, Caro L, Purich D. Force generation by cytoskeletal filament end-tracking proteins. Biophys J. 2004;87:2838–2854. doi: 10.1529/biophysj.104.045211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RB, Purich DL. Clamped-filament elongation model for actin-based motors. Biophys J. 2002;82:605–617. doi: 10.1016/S0006-3495(02)75425-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M, Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- Dominguez R. Actin filament nucleation and elongation factors—structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. The WASP-homology 2 domain and cytoskeleton assembly. In: Carlier M-F, editor. In: Actin-Based Motility: Cellular, Molecular and Physical Aspects. New York: Springer; 2010. pp. 255–277. [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Chapter 14, Unit 14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier M-F. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin–actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher J-F, Mauge C, Didry D, Guichard B, Renault L, Carlier M-F. Interactions of isolated C-terminal fragments of Neural Wiskott-Aldrich syndrome protein (N-WASP) with actin and Arp2/3 complex. J Biol Chem. 2012;287:34646–34659. doi: 10.1074/jbc.M112.394361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavazzi I, Nermut MV, Marchisio PC. Ultrastructure and gold-immunolabelling of cell-substratum adhesions (podosomes) in RSV-transformed BHK cells. J Cell Sci. 1989;94:85–99. doi: 10.1242/jcs.94.1.85. [DOI] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191:571–584. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Kuhn JR. Actin filament attachments for sustained motility in vitro are maintained by filament bundling. PLoS One. 2012;7:e31385. doi: 10.1371/journal.pone.0031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F, Purich DL, Southwick FS. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J Biol Chem. 1999;274:36963–36972. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J Biol Chem. 2007;282:28014–28024. doi: 10.1074/jbc.M705287200. [DOI] [PubMed] [Google Scholar]
- Laurent V, Loisel TP, Harbeck B, Wehman A, Gröbe L, Jockusch BM, Wehland J, Gertler FB, Carlier M-F. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J Cell Biol. 1999;144:1245–1258. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Dawson JC, Forero-Vargas M, Spence HJ, Yu X, König I, Anderson K, Machesky LM. The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr Biol. 2010;20:339–345. doi: 10.1016/j.cub.2009.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K, Ho JX, Keeling K, Gilliland GL, Ji X, Rüker F, Carter DC. Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV. Protein Sci. 1994;3:2233–2244. doi: 10.1002/pro.5560031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S, Higgs H, Hüfner K, Schwarz K, Pannicke U, Aepfelbacher M. The polarization defect of Wiskott-Aldrich syndrome macrophages is linked to dislocalization of the Arp2/3 complex. J Immunol. 2000;165:221–225. doi: 10.4049/jimmunol.165.1.221. [DOI] [PubMed] [Google Scholar]
- Linder S, Nelson D, Weiss M, Aepfelbacher M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc Natl Acad Sci USA. 1999;96:9648–9653. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M, Yamaguchi H, Wang Y, Singer RH, Condeelis J. Imaging sites of N-WASP activity in lamellipodia and invadopodia of carcinoma cells. Curr Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Luxenburg C, Geblinger D, Klein E, Anderson K, Hanein D, Geiger B, Addadi L. The architecture of the adhesive apparatus of cultured osteoclasts: from podosome formation to sealing zone assembly. PLoS One. 2007;2:e179. doi: 10.1371/journal.pone.0000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney NM, Janmey PA, Almo SC. Structure of the profilin-poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol. 1997;4:953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Miki H, He H, Maruta H, Takenawa T. Essential role of neural Wiskott-Aldrich syndrome protein in podosome formation and degradation of extracellular matrix in src-transformed fibroblasts. Cancer Res. 2002;62:669–674. [PubMed] [Google Scholar]
- Mogilner A, Oster G. Force generation by actin polymerization II: the elastic ratchet and tethered filaments. Biophys J. 2003a;84:1591–1605. doi: 10.1016/S0006-3495(03)74969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A, Oster G. Polymer motors: pushing out the front and pulling up the back. Curr Biol. 2003b;13:R721–R733. doi: 10.1016/j.cub.2003.08.050. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SC, Chen WT. Cellular invasion into matrix beads: localization of beta 1 integrins and fibronectin to the invadopodia. J Cell Sci. 1991;99:213–225. doi: 10.1242/jcs.99.2.213. [DOI] [PubMed] [Google Scholar]
- Mullins RD. How WASP-family proteins and the Arp2/3 complex convert intracellular signals into cytoskeletal structures. Curr Opin Cell Biol. 2000;12:91–96. doi: 10.1016/s0955-0674(99)00061-7. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Miki H, Ito M, Ohashi K, Takenawa T, Miyamoto S. N-WASP, WAVE and Mena play different roles in the organization of actin cytoskeleton in lamellipodia. J Cell Sci. 2001;114:1555–1565. doi: 10.1242/jcs.114.8.1555. [DOI] [PubMed] [Google Scholar]
- Namgoong S, Boczkowska M, Glista MJ, Winkelman JD, Rebowski G, Kovar DR, Dominguez R. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat Struct Mol Biol. 2011;18:1060–1067. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Padrick SB, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108:E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry (Mosc) 1984;23:6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebowski G, Boczkowska M, Hayes DB, Guo L, Irving TC, Dominguez R. X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc Natl Acad Sci USA. 2008;105:10785–10790. doi: 10.1073/pnas.0801650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Clainche CL, Didry D, Egile C, Pantaloni D, Carlier M-F. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Sarmiento C, et al. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J Cell Biol. 2008;180:1245–1260. doi: 10.1083/jcb.200708123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol. 2010;189:541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper SB, et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- Soo FS, Theriot JA. Adhesion controls bacterial actin polymerization-based movement. Proc Natl Acad Sci USA. 2005;102:16233–16238. doi: 10.1073/pnas.0507022102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TE. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T. The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 1998;17:6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JX, Janmey PA. The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. J Biol Chem. 1996;271:8556–8563. doi: 10.1074/jbc.271.15.8556. [DOI] [PubMed] [Google Scholar]
- Tarone G, Cirillo D, Giancotti FG, Comoglio PM, Marchisio PC. Rous sarcoma virus-transformed fibroblasts adhere primarily at discrete protrusions of the ventral membrane called podosomes. Exp Cell Res. 1985;159:141–157. doi: 10.1016/s0014-4827(85)80044-6. [DOI] [PubMed] [Google Scholar]
- Xu X-P, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 2011;31:236–247. doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Huang M, DeBiasio J, Pring M, Joyce M, Miki H, Takenawa T, Zigmond SH. Profilin enhances Cdc42-induced nucleation of actin polymerization. J Cell Biol. 2000;150:1001–1012. doi: 10.1083/jcb.150.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.