Single-molecule speckle microscopy and computational modeling reveal the relation between cortical dynein complex dynamics and microtubule behavior in mammalian cells. Experimentally observed rapid turnover of dynein complexes at the cell cortex facilitates their search to efficiently capture and push microtubules with leading plus ends.
Abstract
Microtubules are under the influence of forces mediated by cytoplasmic dynein motors associated with the cell cortex. If such microtubules are free to move, they are rapidly transported inside cells. Here we directly observe fluorescent protein–labeled cortical dynein speckles and motile microtubules. We find that several dynein complex subunits, including the heavy chain, the intermediate chain, and the associated dynactin subunit Dctn1 (also known as p150glued) form spatially resolved, dynamic speckles at the cell cortex, which are preferentially associated with microtubules. Measurements of bleaching and dissociation kinetics at the cell cortex reveal that these speckles often contain multiple labeled dynein heavy-chain molecules and turn over rapidly within seconds. The dynamic behavior of microtubules, such as directional movement, bending, or rotation, is influenced by association with dynein speckles, suggesting a direct physical and functional interaction. Our results support a model in which rapid turnover of cell cortex–associated dynein complexes facilitates their search to efficiently capture and push microtubules directionally with leading plus ends.
INTRODUCTION
Cytoplasmic dynein, a minus end–directed microtubule motor, is primarily known for its role in driving the majority of retrograde vesicle transport in cells. It is composed of several subunits, including the motor-containing heavy chains, the regulatory dynactin complex, several additional regulatory subunits, and the intermediate chains (ICs), which link the major complex components. There is increasing evidence that dynein complexes play a more general role in the spatial organization of microtubule-dependent cell structures. In particular, a subpopulation called cortical dynein, which is associated with the cell cortex (Dujardin and Vallee, 2002), can produce forces that power directional microtubule movements along the plasma membrane. Because dynein is a minus end–directed motor, microtubules are pushed with leading plus ends via this mechanism. Combined with the stochastic nature of microtubule dynamic instability, such forces can influence the spatial organization of microtubules via self-organization (Dehmelt and Bastiaens, 2011). Mathematical modeling provides the basis for a deeper understanding of such dynamic processes. For example, several models were developed to explain how microtubule-length-dependent forces generated by cortical dynein can instruct the positioning of the microtubule-organizing center (MTOC) relative to cell borders (Vogel et al., 2009; Zhu et al., 2010; Laan et al., 2012).
The role of cortical dynein in directed cell motility (Dujardin et al., 2003) and cellular morphogenesis (Ahmad et al., 2000; Dehmelt et al., 2006; Grabham et al., 2007) is less well understood. These processes are based on cell-shape changes that are believed to be mainly driven by actin-dependent forces in the cell periphery. Microtubules are believed to play a more indirect role by regulating those actin-dependent mechanisms. However, in a previous study we found that dynein can play a direct role in cellular morphogenesis. In that study, we showed that a microtubule-based, dynein-dependent mechanism can induce the formation of new, neurite-like cell protrusions (Dehmelt et al., 2006). This morphogenetic process was powered by dynein-dependent forces pushing MTOC-independent microtubules and was independent of the actin cytoskeleton.
Although this previous study suggested a functional role for cortical dynein in cellular morphogenesis, direct observation of interactions between microtubules and cortical dynein complexes in living mammalian cells was lacking. Furthermore, the association dynamics of dynein complexes with the cell cortex was unknown. To address these open questions, we directly observed cortical dynein and motile microtubules in living cells via total internal reflection fluorescence microscopy (TIRFM). We then built a mathematical model for this dynamic cellular process on the basis of our experimental measurements and known physical properties of the system components and performed stochastic simulations based on this model.
RESULTS
Dynein associates transiently with the cell cortex
To study how cortical dynein affects microtubule motility in cells, we generated constructs to directly observe complex subunits in fixed and living COS7 cells. We particularly focused on observing the dynein heavy chain (Dync1h1), as this subunit contains the motor domain. The N-terminus of Dync1h1 is located in a region that is involved in multiple critical interactions with other dynein complex components and dynein cargoes. In contrast, the extreme C-terminus is located in a region distant from the N-terminal tail, the stalk base, and the primary ATPase site of the motor domain (Roberts et al., 2009). Therefore, to minimally perturb dynein localization and function, we fused enhanced green fluorescent protein (EGFP) to the dynein heavy-chain C-terminus. We used TIRF microscopy to selectively illuminate the cell cortex. To further avoid cytosolic background due to overexpression, we used a truncated cytomegalovirus (CMV) promoter (Watanabe and Mitchison, 2002). Via Western blotting, we estimated that EGFP-Dync1h1 is expressed at a level that was only 2.5-fold higher than that of the endogenous protein (Supplemental Figure S1). This low expression level allowed us to visualize individual, spatially resolved speckles of dynein complexes at the cell cortex. Bleaching in fixed cells (Figure 1A) showed that individual speckles contain, on average, 2.49 ± 0.66 (mean ± SEM; nexperiments = 3; nspeckles = 1982) EGFP-labeled dynein heavy-chain molecules, suggesting that multiple dynein molecules form clusters at the cell cortex. Overnight treatment with nocodazole lead to a further increase in that number (3.69 ± 0.90; nexperiments = 4; nspeckles = 4896), suggesting that microtubules can promote the extraction of dynein from cortical clusters. Given the low EGFP-Dync1h1 expression level estimated according to Supplemental Figure S1, ∼70% of Dync1h1 is labeled with EGFP. However, because speckles usually contained multiple Dync1h1 molecules, the majority (>90%) of speckles are expected to contain at least one EGFP molecule.
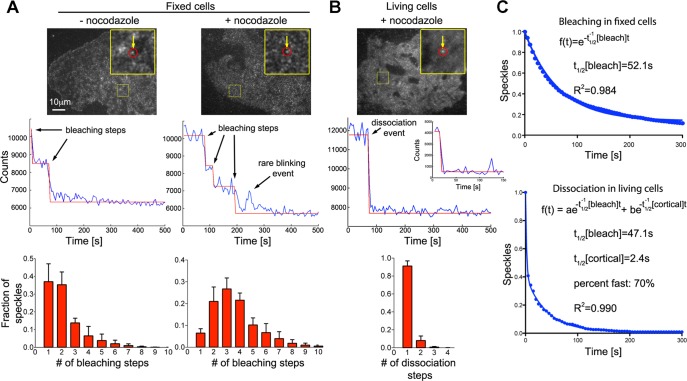
FIGURE 1:
Rapid turnover of dynein complexes at the cell cortex. (A) Bleaching of individual EGFP-labeled dynein heavy-chain (Dync1h1) speckles in fixed COS7 cells (top) shows stepwise decrease in fluorescence intensity (middle). Fitting of a step function to the bleaching kinetics allows estimation of the number of EGFP molecules per speckle, which is displayed as histograms (bottom). After overnight depolymerization of microtubules with 10 μM nocodazole (n = 4896 speckles in four cells), the distribution of bleaching steps per speckle is shifted toward larger numbers compared with control fixed cells (n = 1982 speckles in three cells). (B) Rapid dissociation of EGFP-labeled dynein heavy-chain (Dync1h1) speckles from the cell cortex in living COS7 cells (top). Middle, to best illustrate the steplike dissociation, an unusually stable speckle that dissociates from the cortex within a single video frame after a prolonged delay is shown. The corresponding inset shows the more frequent, rapid dissociation within the first acquired video frames. The distribution of dissociation steps shows that speckles usually dissociate in a single step (bottom left; n = 1599 speckles in four cells). (C) Number of remaining EGFP-Dync1h1 molecules plotted against time. In fixed cells, the bleaching kinetics of initially detected individual EGFP molecules fits well to a single-exponential decay function (the average value was t1/2[bleach] = 46.2 ± 2.0 s, n = 4896 speckles in four cells). In living cells, the kinetics of EGFP-Dync1h1 dissociation does not fit a single-exponential decay (R = 0.94 ± 0.02). Assuming similar bleaching kinetics in fixed and living cells, a fast component, which is due to dynamic interaction of dynein speckles with the cortex, is detected using a double-exponential fit (t1/2[cortical] = 2.6 ± 0.9 s, fraction fast: 59 ± 13%, n = 1599 speckles in four cells; the t1/2 of the slow component was set to the average t1/2[bleach] obtained in fixed cells from the same experimental day). The overall density of speckles was 0.28 ± 0.05 (live cells) or 0.46 ± 0.13 (fixed cells) speckles/μm2 (n = 6 or 4 cells).
Our TIRF measurements in living cells revealed highly dynamic behavior of cortical dynein speckles (Figure 1, B and C) on an increased but homogeneous cytosolic background due to averaging of rapidly diffusing, free EGFP-Dync1h1 molecules. In cells treated with nocodazole the association of these speckles with the cell cortex had a half-life of t1/2[cortical] = 2.6 ± 0.9 s (mean ± SEM; nexperiments = 4; nspeckles = 1599), which is >15-fold faster than the apparent half-life due to bleaching (t1/2[bleach] = 46.2 ± 2.0 s; nexperiments = 4; nspeckles = 4896, Figure 1C). Similar results were obtained in the absence of nocodazole (Supplemental Figure S2).
Dynein-dependent movements of MAP2c-induced short microtubules along the cell cortex
To evaluate the function of these dynamic dynein clusters with respect to microtubule movements, we induced the formation of many short microtubules via washout of the microtubule-depolymerizing drug nocodazole in the presence of the neuronal microtubule-stabilizing protein MAP2c (Dehmelt et al., 2006). In these experimental conditions, nascent short microtubules are formed due to a nucleating activity of MAP2c. Short microtubules then move rapidly and directionally in cells until they encounter an obstacle such as the cell border. Over time, this directional transport converts randomly oriented short microtubules into peripheral microtubule foci via a self-organizing mechanism (Dehmelt et al., 2006; Dehmelt and Bastiaens, 2010, 2011). Our previous work showed that the directional motility of such short microtubules is not due to treadmilling (i.e., combined polymerization and depolymerization at microtubule ends), which is very slow (0.6 μm/min) in the presence of the microtubule-stabilizing protein MAP2c due to acute tubulin depletion. Instead, injection of function-blocking antibodies showed that microtubule movements depended on cytoplasmic dynein. We therefore hypothesized that cortical dynein complexes might push short microtubules to power their sliding (Dehmelt et al., 2006). However, direct evidence for association of motile short microtubules with dynein complexes was lacking. In addition to moving along the cell cortex, short microtubules can transiently move along various additional cellular structures, including mitochondria and intermediate filaments, all of which can also anchor cytoplasmic dynein (Habermann et al., 2001; Helfand et al., 2002). However, short microtubules were able to reach the most peripheral regions of cells, which are largely devoid of those cellular components (Dehmelt et al., 2006).
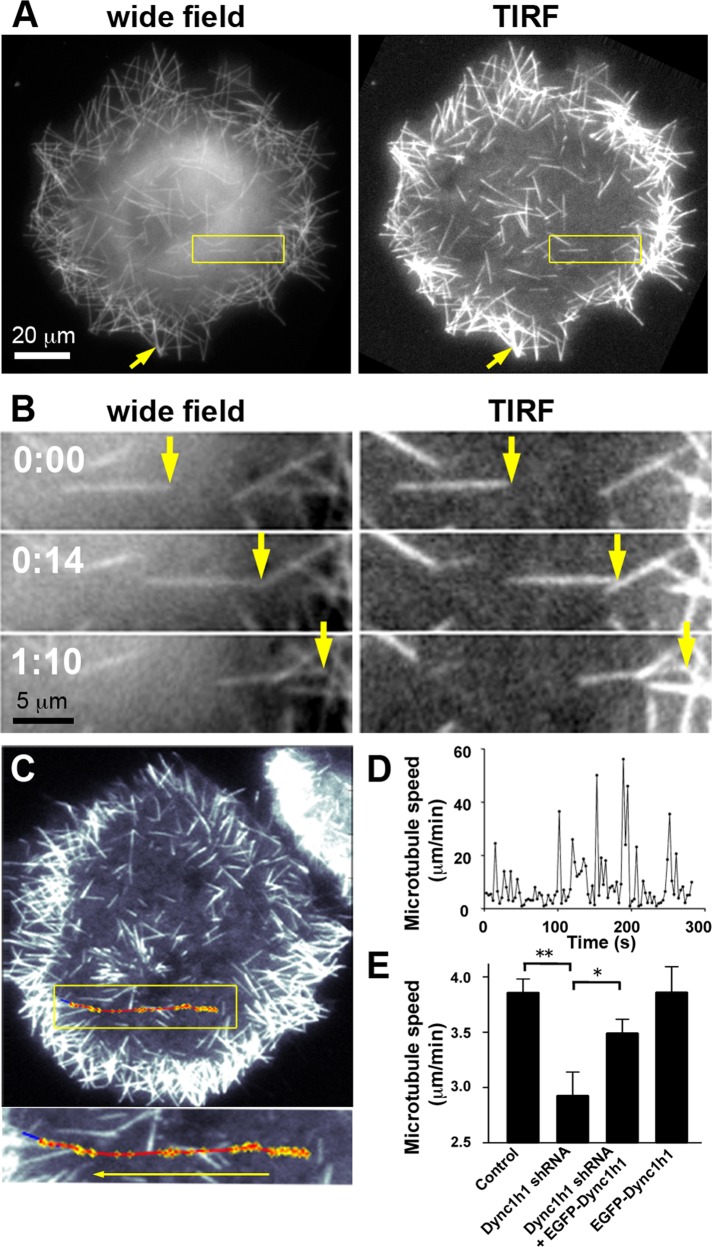
Here we find, by comparison between TIRF and wide-field imaging, that the majority of microtubule movements occurs near the cell cortex (Figure 2, A and B, and Supplemental Movie S1). The observation that microtubules near the cell cortex within the evanescent field of the TIRF microscope (with a depth of ∼300 nm from the glass surface) are highly motile suggests that an activity anchored at the cell cortex, such as cortical dynein, drives short-microtubule motility. As noted previously (Dehmelt et al., 2006), both individual short microtubules and bundles of few or many short microtubules can form after nocodazole washout in the presence of MAP2c. Here we find that nascent microtubules have a very uniform intensity distribution, suggesting that they are mostly composed of single microtubules or bundles that contain only few microtubules. As microtubules accumulate at the cell periphery, those bundles can align to form thicker bundles (Figure 2, A and B). Automated tracking of short microtubules (Figure 2C and Supplemental Figure S3) also reveals intermittent, saltatory bursts of rapid movements that can reach 30–60 μm/min interrupted by much slower, apparently random Brownian motion over variable durations (Figure 2D). This intermittent microtubule motility was reported previously (Dehmelt et al., 2006), but the underlying mechanism was unclear. To provide further evidence for a role of dynein in short-microtubule motility we used RNA interference. We found that short hairpin RNA (shRNA)–mediated knockdown of endogenous Dync1h1 resulted in a significant decrease in average short-microtubule speed that was partially rescued by simultaneous expression of the shRNA-insensitive EGFP-Dync1h1 construct (Figure 2E).
FIGURE 2:
Directional movement of MAP2c-induced short microtubules along the cell cortex after nocodazole washout. (A) COS7 cells were imaged soon after nocodazole washout via wide-field or TIRF microscopy. The yellow arrow points to a more intensely labeled microtubule structure in a peripheral region that was formed during the accumulation of multiple weakly labeled short microtubules in the cell periphery (see also Supplemental Movie S1). (B) Example of a short microtubule that moves directionally near the cell cortex within the evanescent wave of the TIRF microscope (see also Supplemental Movie S1). (C) Automated tracking of short microtubules via TIRF microscopy (see Supplemental Figure S3 and Materials and Methods for details). The trajectory of microtubule movement (red) was overlaid onto the last video frame used for tracking. Blue, final position of tracked short microtubule. Yellow, tracked short microtubule endpoint. (D) Microtubule speed plotted against time reveals saltatory, rapid movements with intermittent pauses characterized by slow directional movements and Brownian motion. (E) Average speed of short microtubules in nocodazole-washout experiments in control Neuro2A cells and Neuro2A cells treated with shRNA targeting Dync1h1 and/or with EGFP-Dync1h1 (mean ± SEM; nexperiments = 8 for control, Dync1h1 shRNA, and Dync1h1 shRNA + EGFP-Dync1h1; nexperiments = 4 for EGFP-Dync1h1 alone). Dynein heavy-chain silencing resulted in a significant decrease in average short microtubule speeds, which was largely recovered by simultaneous expression of shRNA-resistant EGFP-Dync1h1 construct. *p < 0.05; **p < 0.01; one-way analysis of variance.
Microtubules pushed by cortical dynein should move with leading plus ends. Indeed, the microtubule plus-tip marker EB3 always localized to the leading end of motile short microtubules (Supplemental Figure S4 and Supplemental Movie S2). We also tested whether this microtubule motility is unique for the neuronal microtubule regulator MAP2c. Similar sliding movements were also observed if microtubules were decorated with the related neuronal or nonneuronal MAP tau or the microtubule-binding domain (MTBD) of EMAP115 (Supplemental Figure S5 and Supplemental Movie S3). However, the instantaneous directional movements of MAP2c-decorated short microtubules were often larger than those observed with the other MAPs. This could be due to a change in dynein binding to microtubules due to the presence of MAP2c. Alternatively, MAP2c-decorated microtubules might be transported more efficiently due to increased rigidity (Felgner et al., 1997). Bundle formation might further increase rigidity and play an additional role. Indeed, MAP2c-decorated short microtubules were very straight, and therefore cortical dynein mediated forces will be transduced very efficiently into straight directional sliding movements. In addition, due to the stabilizing effect of MAP2c, cells contain many short microtubules, which are less obstructed by collisions and are less likely shifted along organelles or other microtubules outside the narrow TIRF field.
In contrast to MAP2c, the EMAP115-MTBD was shown to have minimal influence on microtubule stability at low or moderate expression levels (Faire et al., 1999), and it does not induce the formation of straight microtubule bundles in cells, suggesting that it does not strongly influence microtubule rigidity. Indeed, EMAP115-MTBD–decorated microtubules appeared less rigid, were frequently bent, and were much longer than MAP2c-decorated short microtubules. Interactions of such flexible, long microtubules with cortical dynein motors will result not only in microtubule translocations, but also in microtubule bending, which is difficult to quantify. In addition, such long microtubules are frequently obstructed by collisions and potentially shifted along other dynein-containing organelles or other microtubules along their extended length. If microtubules were directly labeled with fluorescent tubulin and observed in unperturbed cells (i.e., without nocodazole washout), those effects appeared to be more pronounced due to higher microtubule densities and longer microtubule lengths, and it was even more difficult to interpret dynamic microtubule behaviors in this condition (Supplemental Figure S6A and Supplemental Movie S4). Thus, to analyze the function of cortical dynein complexes more directly, we chose MAP2c-induced motile short microtubules as an assay system, as they should respond to forces transduced from cortical motors predominantly with quantifiable microtubule translocations.
Short microtubules associate with dynein speckles
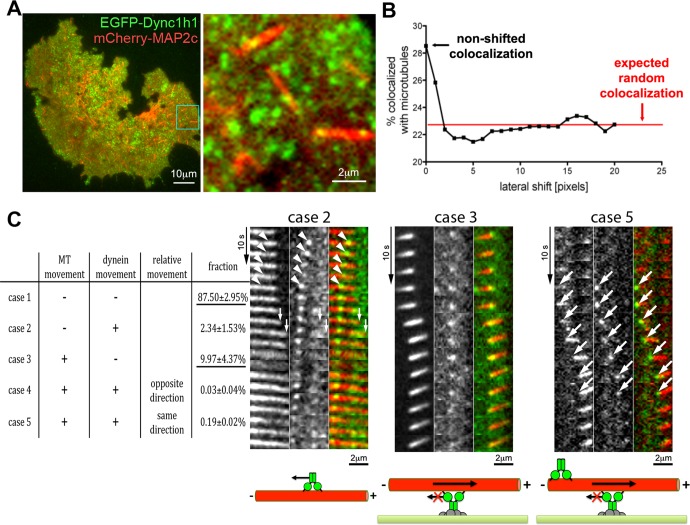
To analyze the relation between dynein speckles and microtubule motility, we used two-color TIRF microscopy. We found that EGFP-labeled dynein heavy-chain speckles preferentially localized to mCherry-MAP2c–labeled short microtubules (Figure 3, A and B). Similar results were obtained using the dynein IC or the dynactin subunit Dctn1 (Supplemental Figure S7A). This suggests that dynein speckles in which those subunits are labeled are able to interact with short microtubules at the cell cortex. The dynamic behavior of dynein speckles, which are colocalized with short microtubules, can be classified into five groups based on their mobility relative to the associated microtubule (Figure 3C). In case 1, neither the dynein speckle nor the short microtubule moves. Over the length of the whole recording, this occurs most frequently (87.50 ± 2.95%, mean ± SEM; nexperiments = 3) and most likely represents colocalization of dynein and microtubules within the diffraction limit (∼320 nm) but not direct, functional interaction within the estimated dynein complex reach distance of ∼82.5 nm (Gennerich et al., 2007). In case 2, the dynein speckle moves along a stationary microtubule, presumably toward the minus end (Supplemental Movie S5). Such observations are compatible with the classic role of cytoplasmic dynein as a directional microtubule motor. However, this case is observed rarely (2.34 ± 1.53%) under the experimental conditions, presumably because observation using TIRF microscopy selectively focuses on relatively immobile, cortex-associated dynein speckles. In two cells, we observed a microtubule aster, which entered the TIRF field near the cell cortex (Supplemental Figure S7B). In agreement with the minus end–directed motility of dynein or its interaction with centrosome components, such asters concentrated dynein speckles in their center. In case 3, stationary dynein speckles colocalize with motile short microtubules. Based on the known minus-end directionality of dynein motors, this case represents short microtubules being pushed by anchored dynein with leading plus ends and trailing minus ends (see drawing in Figure 3C). Of all cases in which movement of microtubules and/or dynein takes place, this case occurs most frequently (9.97 ± 4.37%). In the example shown in Figure 3C, a single stationary dynein speckle is observed, which is relatively stable over time, whereas in other examples stationary speckles transiently appear and disappear at motile microtubules (see Supplemental Movies S6 and S7). We made a similar observation in unperturbed cells that did not express MAP2c (Supplemental Figure S6, B and C); however, the apparent movement of microtubules in that condition is difficult to interpret due to microtubule dynamic instability and potential interactions of long microtubules with other cell components. In the two remaining cases, both dynein speckles and the short microtubules move, either in the opposite (case 4) or the same (case 5) direction. The occurrence of case 4 is negligible (0.03 ± 0.04%), and we rarely observed case 5 (0.19 ± 0.02%). However, when case 5 was detected, dynein speckles were always associated with the trailing microtubule end (Figure 3C and Supplemental Movie S8). This observation can be explained by accumulation of dynein at the trailing minus ends of motile microtubules via their minus end–directed motor activity, where they then piggyback on moving microtubules. Although case 5 is observed very rarely for the dynein heavy chain, it is observed more frequently for the dynactin component Dctn1, especially at higher expression levels (Supplemental Figure S7C). This accumulation thus seems to be more effective for the dynactin subunit Dctn1, presumably due to its ability to bind microtubules independently of the dynein heavy-chain motor domain. Of interest, Dctn1 can accumulate both at plus ends and minus ends (at centrosomes) of microtubules (Vaughan et al., 1999). However, here we observed their accumulation only on the trailing end and thus the microtubule minus end. This is expected, as Dctn1 localization to plus ends depends on microtubule polymerization, which is strongly reduced due to the slow turnover of MAP2c-stabilized microtubules (Umeyama et al., 1993).
FIGURE 3:
Colocalization of dynein complexes with short microtubules. (A) Two-color TIRF microscopic analysis of the EGFP-labeled dynein heavy chain (EGFP-Dync1h1) and mCherry-labeled MAP2c–decorated microtubules in COS7 cells. A subpopulation of dynein speckles (green) overlaps with MAP2c-induced microtubules (red) at the cell cortex. (B) Determination of the percentage of speckles that overlap with MAP2c-decorated short microtubules reveals their preferential association: About 28% of speckles are colocalized with short microtubules. By shifting the dynein channel by >1 pixel relative to the microtubule channel, this apparent colocalization is reduced to the expected value for random overlap, which was calculated to be ∼23%. The observed colocalization was consistently higher than the expected random overlap (4.3 ± 0.6%, p < 0.0001; Student's paired t test; n = 9 cells). See Supplemental Figure S7A for analysis of additional dynein components. (C) Left, five different dynamic behaviors can be distinguished by automated speckle and short-microtubule tracking. The relative fractions for observation of these cases are shown (based on 10,478 three-frame subtrajectories from four cells). Measurements are shown as percentages with SEM. The two most common cases are underlined. Right, examples of observations belonging to these cases are shown above the schematics of their interpretation. The first example illustrates case 2, in which a dynein speckle moves toward the trailing end of a moving short microtubule (white arrowheads). The example for the more frequently observed case 3 shows a short microtubule, which slides along a stationary dynein speckle. In the rarely observed case 5, dynein speckles usually accumulate at the trailing end of motile microtubules and translocate together with them in the same direction (large white arrows; see also Supplemental Figure S7C). For the example of case 2, first the dynein speckle moves leftward (white arrowheads) while the microtubule is stationary, corresponding to case 2, followed by a short rightward movement (white small arrows) of the microtubule and a relatively stationary dynein speckle characteristic of case 3, suggesting that microtubules and dynein speckles can rapidly switch their motile behavior. See also Supplemental Movies S5–S8. MT: microtubule.
Cortical dynein affects short-microtubule behavior
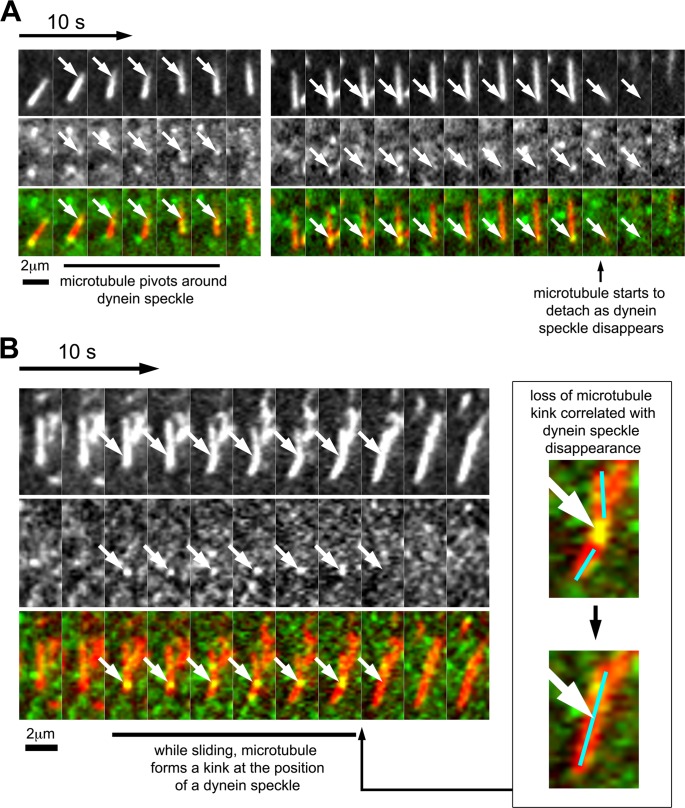
The relatively frequent case of stationary dynein speckles colocalizing with motile short microtubules shows that such microtubules can indeed be pushed by cortically linked dynein complexes. However, short-microtubule movements did not exclusively correlate with detectable speckle association. This could be due to the presence of endogenous dynein or photobleached speckles. Alternatively, dynein complexes could be missed due to their transient cortex association. Nevertheless, our analysis revealed several functional consequences of colocalization between short microtubules with dynein speckles. First, motile short microtubules were observed to pivot around dynein speckles (i.e., they rotated, with the dynein speckle being the apparent anchor, or pivot point of the rotation; Figure 4A, Supplemental Figure S8, and Supplemental Movie S9). This suggests that dynein speckles serve as physical links for short microtubules. This is also observed in the example for case 3 in Figure 3C (Supplemental Movie S7), where the fluorescence intensity of the short microtubule in the TIRF field varies between being maximal at the speckle position and minimal at more distant positions, suggesting a swiveling movement of nonattached microtubule regions away from the cell cortex. Second, occasionally, short microtubules form transient, sharp kinks during their movement (Figure 4B and Supplemental Movie S10). Dynein speckles were often associated with these kinks, suggesting that they might form an anchor point on a microtubule stretched by additional associated motor complexes. Intriguingly, in the case shown in Figure 4B, the microtubule kink is suddenly lost from one frame to another, simultaneous with the disappearance of the dynein speckle at the kink position. This shows that the observed dynein speckles can indeed support a physical link between motile short microtubules and the cell cortex. Furthermore, the strength of this physical link is sufficient to withstand elastic forces of relatively rigid (Felgner et al., 1997), MAP2c-stabilized microtubules. Third, short microtubules that associated with one or more dynein speckles moved slightly faster (4.25 ± 0.09 μm/min; mean ± SEM; n = 1202 subtrajectories) than short microtubules in which no dynein speckles were detected (3.72 ± 0.05 μm/min; mean ± SEM; n = 4665 subtrajectories; see Supplemental Figure S9 for histogram). These observations show that the association with cortical dynein complexes indeed promotes microtubule motility.
FIGURE 4:
Correlation of dynein speckle dynamics with short-microtubule behavior. (A) Sequence showing a short microtubule, which pivots around a dynein speckle. At the first encounter with a dynein speckle, the short microtubule moves only marginally. After this brief delay, the short microtubule swivels around the dynein speckle (white arrows) while moving forward. At a later time point, a visible speckle (white arrows) accumulates at the trailing end of the same short microtubule. The disappearance of this speckle correlates with microtubule detachment from the cell cortex. (B) Sequence of a short microtubule that forms a transient kink during sliding. The short microtubule slides directionally and forms a kink at the position of a bright and persistent speckle (white arrows). This kink is lost from one frame to the other, simultaneous with the loss of this bright speckle (see also enlarged inset). See also Supplemental Figure S8 and Supplemental Movies S9 and S10.
Stochastic model for dynein-mediated microtubule reorganization
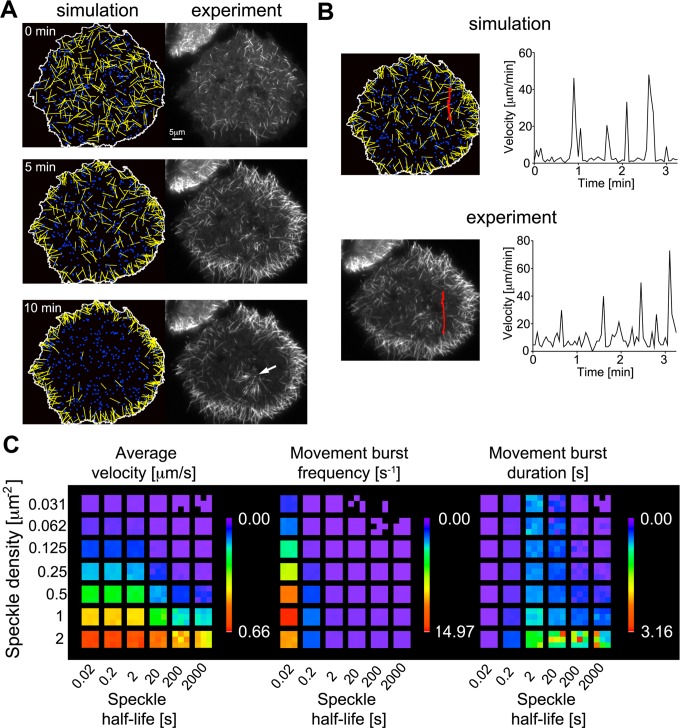
To determine whether the measured characteristics of dynamic, cortically associated dynein speckles can account for the observed dynamic microtubule behavior, we built a computational model. Here the motility of initially randomly oriented short microtubules and the dynamics of cortical dynein complexes are modeled based on our experimental observations and known physical properties of system components. Short microtubules undergo Brownian motion unless they are captured and transiently pushed directionally by cortical dynein. Stochastic simulations show that this model closely mimics experimental observations in two critical aspects: 1) The reorganization of short microtubules to the cell periphery over a time course of several minutes, and 2) the saltatory, intermittent movement bursts of individual short microtubules (Figure 5 and Supplemental Movie S11). Variation of simulation parameters reveals that the frequency of directional movement bursts and the resulting average velocity depend on both speckle density and speckle half-life (Figure 5C). The capture area of short microtubules is ∼0.85 μm2 (5 μm length × 2 × 82 nm dynein reach distance), which is further increased by microtubule movements via Brownian motion to ∼1 μm2. At dynein densities >1 μm−2, dynein turnover does not strongly affect microtubule velocity, as one dynein motor per capture area suffices to drive maximal movements. However, at lower densities, the effect of speckle half-life is more complex. If dynein half-life is less than ∼5 s, dynein complexes frequently dissociate before movements of captured microtubules (at 1 μm/s) complete along their entire length (5 μm), leading to reduced movement burst duration. On the other hand, short half-lives lead to an increase in movement burst frequency due to increased space exploration in the initial capture process and increased movement burst duration due to recapture of moving microtubules. In the range of our experimentally observed speckle densities (0.28 ± 0.05 μm−2 in live cells and 0.46 ± 0.13 μm−2 in fixed cells), our simulations show that the largest gain in average microtubule velocity is observed in the range of experimentally observed speckle half-lives (2.6 ± 0.9 s). In that range, a 10-fold decrease in speckle half-life from 20 to 2 s induces a similar increase in the average velocity as a doubling of the speckle density from 0.25 to 0.5 μm−2 (Table 1). This suggests that the dynamic sampling of space at the cell cortex by stochastic dynein association promotes efficient microtubule transport.
FIGURE 5:
Stochastic simulations of short-microtubule movements. Microtubules (yellow), dynein speckles (blue), and their interactions were modeled based on experimental observations and known physical properties of system components (for details, see Materials and Methods and experimental parameters in Table 2). (A) Stochastic simulations closely mimic the experimentally observed distribution of microtubules over time as they accumulate at the cell periphery (see also Supplemental Movie S11). The initial time point (0 min) is defined as the first observation of nucleated microtubules after nocodazole washout. Immobile microtubules anchored to the microtubule-organizing center observed in the experiment (white arrow) are not included in the simulation. (B) Tracking of individual short microtubules shows that dynamic microtubule behavior, with intermittent bursts and pauses of rapid motility, is observed in both simulations and experiments. (C) Systematic variation of speckle density and speckle half-life in simulations lacking a cell border. Three heat maps visualize the average short-microtubule velocity, as well as the frequency and duration of movement bursts. Each colored subsquare corresponds to one simulation with random initial conditions.
TABLE 1:
Variation of dynein speckle density and turnover in stochastic simulations of microtubule movements.
| Simulation setting | Low density, slow turnover | High density, slow turnover | Low density, fast turnover | High density, fast turnover |
|---|---|---|---|---|
| Speckle density (μm−2) | 0.25 | 0.5 | 0.25 | 0.5 |
| Speckle half-life (s) (turnover rate [s−1]) | 20 (0.0347) | 20 (0.0347) | 2 (0.347) | 2 (0.347) |
| Average microtubule velocity (μm/min) | 3.50 ± 0.14 | 7.67 ± 0.19 | 8.12 ± 0.18 | 15.36 ± 0.15 |
DISCUSSION
Here we directly observed cortical dynein complexes and their effect on short microtubule behavior in living cells. Cortical dynein forms distinct, spatially separable accumulations, or speckles, that often contained multiple fluorescent protein–tagged heavy chains. These dynein speckles associated only transiently with the cell cortex and thus present a high level of turnover at this cellular compartment. Direct, simultaneous observation of dynein speckles and microtubules suggests that cortical dynein can indeed push microtubules if they come in close proximity at the cell cortex.
The rapid kinetics of dynein complex association suggests a potential mechanism by which relatively few cortical motor proteins can perform efficient directional sliding of microtubules. Variation of computer simulation parameters shows that an increase in the turnover rate leads to more efficient microtubule transport over a wide range of speckle densities. This effect is due to more efficient sampling of the cell cortex area to increase the probability of dynein complexes interacting with short microtubules. Thus the overall efficiency of microtubule transport is limited not only by the speed of dynein dependent bursts of movements, but also by the relatively rare events of encounter between sparsely distributed dynein speckles and short microtubules. The sparse distribution of cortical dynein motors also provides an explanation for the experimentally observed intermittent microtubule motility.
Apart from increasing microtubule transport efficiency, the high level of dynein speckle turnover also enables a high level of plasticity in the spatiotemporal regulation of cortical dynein association via upstream signaling pathways. For asymmetric cell division, accumulating evidence supports a mechanism by which dynein is anchored to the cell cortex via interaction with nuclear and mitotic apparatus protein (NuMA; Merdes et al., 1996), which in turn is linked to the G protein α subunit Gαi via the mammalian Pins homologue leucine-glycine-asparagine repeat–enriched protein (LGN; Du and Macara, 2004). Recent studies show a tight spatiotemporal regulation of dynein localization by regulating those interactions by the cell cycle (Collins et al., 2012) and spindle pole–localized polo-like kinase, as well as by chromosome-derived RanGTP gradients (Kiyomitsu and Cheeseman, 2012). Cortical dynein was also proposed to play a role in processes related to cellular morphogenesis, such as directed cell motility (Dujardin et al., 2003), axonal outgrowth (Ahmad et al., 2006), growth cone turning (Grabham et al., 2007), and neurite formation (Dehmelt et al., 2006). It is unclear whether the regulatory mechanisms that control asymmetric cell division also apply to those systems. Of interest, a recent study suggests that two modes of microtubule/dynein interaction exist at the cell cortex, which are controlled by different sets of regulators (Gusnowski and Srayko, 2011). In NuMA/LGN/Gαi-dependent asymmetric cell division, cells are round, and the interaction between microtubules and the cell cortex likely occurs at a very steep angle. In contrast, the morphogenetic processes described here and our observations in this study involve microtubule cortex interactions at a shallow angle, which favors a microtubule sliding motion that is independent of LGN/Gαi but dependent on dynactin (Gusnowski and Srayko, 2011).
In conclusion, we showed how cortical dynein complexes influence motile short microtubules. By combining detailed experimental observations with computational modeling, we identified and characterized a minimal set of components that is sufficient to describe this dynamic cellular system. Our analyses show that rapid turnover of cortical dynein is a central feature of this system and suggest that this feature could facilitate spatiotemporal regulation of dynein activity in cells.
MATERIALS AND METHODS
Plasmid constructs
The vectors pEGFP-N1 and pEGFP-C1 were modified to generate pEGFP-N1-delCMV and pEGFP-C1-delCMV by exchanging the original strong CMV promoter into the much weaker delCMV promoter (Watanabe and Mitchison, 2002) via the restriction sites AseI and NheI.
The vector pEGFP-MAP2c (Ozer and Halpain, 2000) was modified to generate pmCherry-MAP2c-N1 by exchanging the EGFP fluorophore to mCherry (Shaner et al., 2004) via BamHI/HindIII.
The murine dynein heavy-chain gene Dync1h1 was cloned via RT-PCR from P19 cells differentiated into neurons with retinoic acid. RNA was isolated using the RNeasy Kit (Qiagen, Hilden, Germany). Reverse transcription (RT)-PCR was performed using the Phusion RT-PCR Kit (Finnzymes, Espoo, Finland) at 40°C for 30 min with oligo dT primers. First, the partial sequences DHC 1, 2a-c, and 3 were amplified using the Phusion Hot Start DNA Polymerase (Finnzymes) using the following primer sequences: DHC-1 5′-primer, CGC GAG ATC TGG ATA TCG CGA A; DHC-1 3′-primer, ATC AAT CAC AGG AGC TCG CT; DHC-2a 5′-primer, AGC GAG CTC CTG TGA TTG AT; DHC-2a 3′-primer, ACT CGT AAT GCA CTC CCA CC; DHC-2b 5′-primer, GGT GGG AGT GCA TTA CGA GT; DHC-2b 3′-primer, CCT CAC TGA ACC ACA CCA TG; DHC-2c 5′-primer, CAT GGT GTG GTT CAG TGA GG; DHC-2c 3′-primer, CTT GTA GAG CTC CTC GTG GG; DHC-3 5′-primer, CCC ACG AGG AGC TCT ACA AG; and DHC-3 3′-primer, GAT ATC GCG ATC TCA GTG CAC AGA ACG GCA. The partial sequences DHC-2a and DHC-2b were fused to produce DHC-2a+b via overlap extension PCR using the DHC-2a 5′-primer and DHC-2b 3′-primer. The partial sequences DHC-2b and DHC-2c were fused to produce DHC-2b+c via overlap extension PCR using the DHC-2b 5′-primer and DHC-2c 3′-primer. These parts were sequentially cloned into pEGFP-N1-delCMV: First, DHC-1 was inserted via BglII and SacI, followed by insertion of DHC-3 via SacI/SmaI and the compatible combination SacI/NruI. Finally, DHC-2a+b and DHC-2b+c were cloned into the resulting vector via a nondirectional three-fragment ligation using SacI and EcoRI to generate pEGFP-N1-delCMV-Dync1h1. In this construct, EGFP is fused in-frame to the C-terminus of the dynein heavy chain to ensure that only full-length proteins are visualized. The C-terminal primer (DHC-3 3′-primer) only skips the naturally occurring stop codon at the very end of the Dync1h1 cDNA. Sequencing of the full-length construct and comparison to the reference sequence revealed one known single-nucleotide polymorphism (SNP) leading to a variation in the protein sequence (I3856V), 17 known SNPs that do not alter the protein sequence, and six unknown differences in individual nucleotides, which also did not alter the protein sequence.
The dynein IC Dync1i2 was cloned via RT-PCR from undifferentiated P19 cells using the primer sequences (Dync1i2 5’-primer: GAG AAT TCT AAC ATG TCG GAC AAA AGT GA; Dync1i2 3’-primer: GCT GTC GAC CTA GGC AGG GAT TCG GGT AG). The resulting full-length fragment was cloned into pEGFP-C1-delCMV via EcoRI/SalI to generate pEGFP-C1-delCMV-Dync1i2. The cloned product corresponds to isoform C of Dync1i2.
The dynactin component Dctn1 (also known as p150glued) was subcloned from the MGC full-length clone MGC86152 using the following primer sequences: Dctn1 5′-primer, GAG AAT TCG ATG AGT ACG GAG GCA AGC; and Dctn1 3′-primer, GCT GTC GAC TTA GGA GAT GAG GCG ACT G. The resulting full-length fragment was cloned into pEGFP-C1-delCMV via EcoRI/SalI to generate pEGFP-C1-delCMV-Dctn1. The cloned product corresponds to the original full-length MGC clone, which differs from the reference sequence by alternative splicing of exon 1, leading to a 17–amino acid truncation in the very N-terminus. The CAP-Gly domain, which follows shortly after the extreme N-terminus, is identical to the reference sequence.
Dync1h1 and scrambled nontargeting shRNA vectors were obtained from Thermo Scientific (Huntsville, AL; clone TCRN0000091638 targeting the 3′-untranslated region of murine Dync1h1, which is missing in the EGFP-Dync1h1 construct: GAAGGGTTGATGGACTGGAAA) and Addgene (Cambridge, MA), respectively.
Exogenous expression and pharmacological treatment of eukaryotic cells
COS7 and Neuro2A cells were obtained from the American Type Culture Collection (Manassas, VA) and DSMZ (Braunschweig, Germany) and cultured using standard conditions. For microscopy, 50,000 cells were plated on individual glass-bottom dishes (MatTek, Ashland, OR) and transfected using FuGENE 6 or X-tremeGENE 9 (Roche, Mannheim, Germany) on the following day according to the manufacturer's protocol. In some experiments, nocodazole was added 30 min after transfection. The following day, cells were imaged via TIRF microscopy in CO2-independent growth medium containing serum and 4-(2-hydroxyethyl)-1-piperazine-ethanesulfonic acid (HEPES-buffered imaging medium; Pan Biotech, Aidenbach, Germany) at 35°C in a home-built incubation chamber. Extensive washout of nocodazole was performed on the microscope stage.
Western blot analysis
Lysates of transfected or untransfected COS7 or Neuro2a cells were prepared by solubilization in boiling-hot 10% SDS/50 mM Tris-HCl and resolved in ready-to-use 4–15% gradient gels (BioRad, Munich, Germany). Subsequently, proteins were transferred onto polyvinylidine fluoride membranes (Millipore, Schwalbach, Germany) at 100 V for 1 h. After blocking (Odyssey blocking buffer; LI-COR, Lincoln, NE), membranes were incubated with primary antibodies against Dync1h1 (dynein heavy chain rabbit polyclonal antibody R-325 at 1:200; Santa Cruz Biotechnology, Santa Cruz, CA) and GFP (goat antibody 15246 at 1:1000; Abcam). Odyssey IRDye680 donkey anti-rabbit or IRDye800 donkey anti-goat (LI-COR) were used as secondary antibodies. Immunoreactivity was recorded using the LI-COR infrared scanner. α-Tubulin (rabbit antibody 6658 at 1:2500; Abcam) was poststained on the same membrane and used to normalize for gel loading. Band intensities were quantified using ImageJ (National Institutes of Health, Bethesda, MD).
TIRF single-molecule speckle microscopy
For TIRF and wide-field microscopy, an Olympus IX-81 microscope (Olympus, Hamburg, Germany) equipped with a PlanAPO60xOil TIRFM objective (numerical aperture, 1.45) and 1.6 Optovar was used. A triple bandpass dichroic mirror (U-M3TIR405/488/561; Olympus) was combined with Semrock Brightline emission filters (HC 520/35 and HC 629/53; AHF Analysentechnik, Tübingen, Germany) and CellR diode lasers at 488 nm (20 mW) and 561 nm (25 mW). For detection, an electron-multiplying charge-coupled device camera (C9100-13; Hamamatsu, Herrsching am Ammersee, Germany) was used at maximal gain. The laser intensity at the objective aperture was ∼1.3 mW at 488 nm and 0.7 mW at 561 nm. All microscope components were controlled by the CellR software (Olympus). Because the very weak delCMV promoter was used, cells typically expressed very low amounts of the dynein heavy chain. For speckle microscopy, such typical, low-expressing cells were selected. Exposure times of at least 800 ms were required to visualize single-molecule speckles. In general, 2000- to 3000-ms exposures were optimal to balance signal-to-noise against phototoxicity, allowing imaging of several hundred frames without noticeable changes in dynein dynamics or cell morphology. For single-molecule bleach experiments and measurements of dissociation kinetics, exposure times of up to 5000 ms were used to optimize signal-to-noise ratios and facilitate subsequent analysis. All comparisons between matched live and fixed cells in the presence or absence of nocodazole were performed in parallel in imaging medium (Pan Biotech; see earlier description) using identical settings of TIRF angle, exposure time, and laser intensity. Image processing was performed using MATLAB (MathWorks, Ismaning, Germany) or ImageJ, and figures were prepared in Photoshop (Adobe, Munich, Germany). Image manipulations were limited to cropping, scaling, rotating, adjusting levels, and adding clearly identifiable labels or symbols. To correct for lateral drift in the stage position, an image stabilizer plug-in for ImageJ was used (www.cs.cmu.edu/∼kangli/code/Image_Stabilizer.html). The levels in all image panels in time series are adjusted identically. Analysis of the density of endogenous Dync1h1 accumulations was performed via TIRF microscopy in fixed cells after immunocytochemistry using anti-Dync1h1 primary antibodies (Dynein heavy-chain rabbit polyclonal antibody R-325 at 1:50; Santa Cruz Biotechnology) and Alexa 488 goat-anti-rabbit secondary antibodies (Invitrogen, Karlsruhe, Germany) as described earlier (Dehmelt et al., 2006).
Analysis of speckle dynamics
Dynein speckles were detected and tracked using an ImageJ plug-in developed at ETH Zürich (Sbalzarini and Koumoutsakos, 2005). Individual tracks were analyzed to provide frame-to-frame displacement (or displacement over several frames) and speckle detachment from the plasma membrane. The analysis of photobleaching and speckle dissociation kinetics was performed using custom MATLAB scripts. In brief, the background-subtracted maximal and average speckle fluorescence intensity was measured in a 3 × 3 pixel area around the identified speckle over time. Individual bleaching/dissociation steps were recorded if the intensity decreased more than eight times the average noise level. In fixed cells, blinking of EGFP molecules was rarely observed. To be consistent with measurements in living cells, in which repeated binding of a speckle to the same structure cannot be distinguished from blinking, the first loss of fluorescence was considered the bleaching/dissociation event.
Tracking of short microtubules
Image stacks of fluorescently labeled microtubules were first processed using a fast Fourier transform bandpass filter and binarized using ImageJ. Custom MATLAB scripts were used to detect microtubules in individual images and track their movement over time. In brief, binarized microtubule images were skeletonized, and the skeleton branching and end points were detected and segmented via pixel-by-pixel erosion. Detected microtubule segments were connected if they shared a branching point and their orientation was similar. The positions of individual microtubule objects were then tracked over time. For this purpose, microtubule objects in consecutive frames were dilated and linked over the time lapse based on maximal overlap and similarity in orientation and length. Optimal object links were identified by comparing these linking parameters between all microtubule objects within close proximity in both the forward and backward time-lapse sequences. The procedure is summarized in Supplemental Figure S3.
Correlative analysis of speckles and short microtubules
The relationship between dynein heavy-chain speckles and microtubules was assessed using ImageJ and MATLAB. Dynein speckles were considered to be colocalized with microtubules if they were within a distance of ∼0.320 μm. The percentage of colocalized speckles in the original TIRF images was compared with the degree of colocalization in images shifted by a defined number of pixels (Supplemental Figure S7). Only the central region of individual cells was included in this analysis to exclude artifacts based on loss of random overlap beyond cell borders. The value for the expected random colocalization of speckles with microtubules was determined by calculating the microtubule-covered fraction of the cell area.
For microtubules that associate with dynein heavy-chain speckles, five distinct cases were distinguished based on the speed and direction of the identified objects. Dynein speckles were considered to be motile if their velocity based on three-frame displacement exceeded 4.9 μm/min. Because microtubules also moved randomly via Brownian motion, the threshold for motile microtubules was set slightly higher at 8.5 μm/min. Dynein speckles and microtubules were considered to move in the same direction if the orientation of their displacement vectors was within 0 ± 38°. They were considered to move in opposite directions if the orientation of their displacement vectors was within 180 ± 38°.
Five classes of behavior were distinguished:
Both the microtubule and the associated dynein heavy chain speckle were stationary.
The dynein speckle was moving, and the microtubule was stationary.
The microtubule moved, and the dynein speckle was stationary.
The dynein speckle and the microtubule moved in opposite directions.
Both the microtubule and the speckle moved in the same direction.
Additional analysis was performed on these data using custom MATLAB scripts: for case 4, microtubule displacements over three frames were determined and compared with microtubule displacements in which no associated speckle was detected. For case 5, the distance from the leading and trailing microtubule ends (determined based on the movement direction) was measured and plotted as a histogram to determine speckle association with these microtubule ends.
Stochastic simulation of short-microtubule motility
Microtubule behavior (growth and movement) under the conditions of a washout experiment were simulated in silico via custom MATLAB scripts using double-precision, subpixel spatial accuracy and a simulation time step of 10 ms. Simulations included the following objects: 1) the cell border, 2) dynein heavy chain speckles, and 3) short microtubules. Two types of simulations were performed: In one case, the shape of the cell border, average number, and dynamic properties of dynein heavy chain speckles, as well as the total number of short microtubules, were matched to observations from an individual experiment. Additional simplified simulations were performed in which the cell border was omitted to allow free microtubule movements. Dynein speckle plasma membrane association/dissociation dynamics were based on experimental data. The speed of dynein (Toba et al., 2006) and the estimated reach distance (Gennerich et al., 2007) were based on published values. All experimental parameters are summarized in Table 2.
TABLE 2:
Parameters for stochastic simulation of short microtubules.
| Parameter | Simulation shown in Figure 5, A and Ba | Simulations shown in Figure 5Ca | Units | Reference |
|---|---|---|---|---|
| Number of nucleated short microtubules | 262 | 50 | This work | |
| Number of dynein complexes | 434 | Varied with density | This work | |
| Dynein complex density | 0.16 | 0.03125–2 | μm−2 | This work |
| Microtubule length (mean ± SD) | 5.0 ± 0.7 | 5.0 ± 0.7 | μm | This work |
| Dynein speckle turnover rate | 0.5 | 0.005–50 | s−1 | This work |
| Directional microtubule velocity (based on maximal dynein speed) | 48 | 48 | μm/min | Toba et al. (2006) |
| Dynein reach distance | 82.5 | 82.5 | nm | Gennerich et al. (2007) |
| Effective diffusion coefficient for short-microtubule fluctuations at the cell cortex (corresponding to temperature of 35°C and dynamic viscosity of 0.05 Pa⋅s) | 0.016 | 0.016 | μm2/s | Modified from Nedelec (2002) |
| Simulation time step | 0.01 | 0.01 | s |
aValues from this work derived from the cell shown in Figure 5.
The initial positions of nucleated microtubules and dynein speckles were stochastic and evolved based on the following rules:
Microtubules were approximated as stiff rods due to the increase in microtubule rigidity by MAP2c binding (Felgner et al., 1997). Microtubules undergo fluctuations in their position and orientation based on Brownian motion. In brief, otherwise independent fluctuations of the endpoints of short microtubules were constrained by the preset microtubule length. The extent of fluctuations was set by an effective diffusion coefficient (Nedelec, 2002). Because microtubule fluctuations are constrained by the cell cortex, the effective diffusion coefficient of microtubules near the plasma membrane was estimated to be reduced by one order of magnitude compared with free microtubules in solution.
Microtubules move directionally only when associated with dynein speckles. Dynein speckles were considered associated if they were within the reach distance from the microtubule. Thus the interaction between dynein and short microtubules was considered “diffusion” limited. This is a reasonable assumption, given the fixed position of the dynein speckle and the very slow effective diffusion coefficient of short microtubules.
If microtubules were associated with one dynein speckle, Brownian motion was restricted to rotational movements that occur around the pivot point of speckle association.
If microtubules were associated with multiple speckles, Brownian motion was suppressed. The directional movement speed of microtubules was independent on the number of associated speckles.
If a microtubule would move across the cell border, that movement is suppressed.
The dynein motor activity is considered to be processive during the lifetime of speckle/microtubule interactions. This assumption is reasonable because a) the average lifetime of dynein speckle/microtubule interactions is relatively short because of the short length of the MAP2c-decorated microtubules and the high turnover rate of dynein at the cortex, and b) we find that dynein speckles are often composed of multiple dynein molecules, which is expected to increase the overall processivity of individual dynein speckles.
For simplicity, interactions between microtubules are ignored. This simplification is reasonable in most cells, as colliding microtubules can usually quickly overcome their blockage. However, in some cells, a high density of short microtubules can lead to local accumulations that can significantly slow overall microtubule translocation.
Supplementary Material
Acknowledgments
We thank Philippe Bastiaens for helpful suggestions, fruitful discussions, and departmental support. We also thank Perihan Nalbant, Silke Gandor, and Julia Arens for discussions and critical reading of the manuscript; Naoki Watanabe (Kyoto University, Kyoto, Japan) for providing the delCMV promoter; Anna Straube (University of Warwick, Warwick, United Kingdom) for providing the mCherry-EB3; and Shelley Halpain (University of California, San Diego, La Jolla, CA) for providing EGFP-MAP2c constructs. This work was funded by the FORSYS partner initiative of the German Federal Ministry of Education and Research (BMBF, Grant 0315258) to L.D.
Abbreviations used:
- CMV
cytomegalovirus
- EGFP
enhanced green fluorescent protein
- EMAP115
epithelial microtubule-associated protein of 115 kDa (official symbol Map7)
- IC
intermediate chain
- LGN
leucine–glycine–asparagine repeat–enriched protein (official symbol Gpsm2)
- MAP2c
microtubule-associated protein 2c (official symbol Map2)
- MTBD
microtubule-binding domain
- SEM
standard error of the mean
- TIRF
total internal reflection fluorescence
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0376) on October 30, 2013.
*These authors contributed equally to this work.
REFERENCES
- Ahmad FJ, He Y, Myers KA, Hasaka TP, Francis F, Black MM, Baas PW. Effects of dynactin disruption and dynein depletion on axonal microtubules. Traffic. 2006;7:524–537. doi: 10.1111/j.1600-0854.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- Ahmad FJ, Hughey J, Wittmann T, Hyman A, Greaser M, Baas PW. Motor proteins regulate force interactions between microtubules and microfilaments in the axon. Nat Cell Biol. 2000;2:276–280. doi: 10.1038/35010544. [DOI] [PubMed] [Google Scholar]
- Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol Biol Cell. 2012;23:3380–3390. doi: 10.1091/mbc.E12-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehmelt L, Bastiaens PI. Spatial organization of intracellular communication: insights from imaging. Nat Rev Mol Cell Biol. 2010;11:440–452. doi: 10.1038/nrm2903. [DOI] [PubMed] [Google Scholar]
- Dehmelt L, Bastiaens P. Self-organization in cells. In: Meyer-Ortmanns H, Thurner S, editors. In: Principles of Evolution. Heidelberg, Germany: Springer; 2011. pp. 219–238. [Google Scholar]
- Dehmelt L, Nalbant P, Steffen W, Halpain S. A microtubule-based, dynein-dependent force induces local cell protrusions: implications for neurite initiation. Brain Cell Biol. 2006;35:39–56. doi: 10.1007/s11068-006-9001-0. [DOI] [PubMed] [Google Scholar]
- Dujardin DL, Barnhart LE, Stehman SA, Gomes ER, Gundersen GG, Vallee RB. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163:1205–1211. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin DL, Vallee RB. Dynein at the cortex. Curr Opin Cell Biol. 2002;14:44–49. doi: 10.1016/s0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Faire K, Waterman-Storer CM, Gruber D, Masson D, Salmon ED, Bulinski JC. E-MAP-115 (ensconsin) associates dynamically with microtubules in vivo and is not a physiological modulator of microtubule dynamics. J Cell Sci. 1999;112:4243–4255. doi: 10.1242/jcs.112.23.4243. [DOI] [PubMed] [Google Scholar]
- Felgner H, Frank R, Biernat J, Mandelkow EM, Mandelkow E, Ludin B, Matus A, Schliwa M. Domains of neuronal microtubule-associated proteins and flexural rigidity of microtubules. J Cell Biol. 1997;138:1067–1075. doi: 10.1083/jcb.138.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabham PW, Seale GE, Bennecib M, Goldberg DJ, Vallee RB. Cytoplasmic dynein and LIS1 are required for microtubule advance during growth cone remodeling and fast axonal outgrowth. J Neurosci. 2007;27:5823–5834. doi: 10.1523/JNEUROSCI.1135-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusnowski EM, Srayko M. Visualization of dynein-dependent microtubule gliding at the cell cortex: implications for spindle positioning. J Cell Biol. 2011;194:377–386. doi: 10.1083/jcb.201103128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann A, Schroer TA, Griffiths G, Burkhardt JK. Immunolocalization of cytoplasmic dynein and dynactin subunits in cultured macrophages: enrichment on early endocytic organelles. J Cell Sci. 2001;114:229–240. doi: 10.1242/jcs.114.1.229. [DOI] [PubMed] [Google Scholar]
- Helfand BT, Mikami A, Vallee RB, Goldman RD. A requirement for cytoplasmic dynein and dynactin in intermediate filament network assembly and organization. J Cell Biol. 2002;157:795–806. doi: 10.1083/jcb.200202027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Nedelec F. Computer simulations reveal motor properties generating stable antiparallel microtubule interactions. J Cell Biol. 2002;158:1005–1015. doi: 10.1083/jcb.200202051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer RS, Halpain S. Phosphorylation-dependent localization of microtubule-associated protein MAP2c to the actin cytoskeleton. Mol Biol Cell. 2000;11:3573–3587. doi: 10.1091/mbc.11.10.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY, Higuchi H. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc Natl Acad Sci USA. 2006;103:5741–5745. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeyama T, Okabe S, Kanai Y, Hirokawa N. Dynamics of microtubules bundled by microtubule associated protein 2C (MAP2C) J Cell Biol. 1993;120:451–465. doi: 10.1083/jcb.120.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci. 1999;112:1437–1447. doi: 10.1242/jcs.112.10.1437. [DOI] [PubMed] [Google Scholar]
- Vogel SK, Pavin N, Maghelli N, Julicher F, Tolic-Norrelykke IM. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009;7:e1000087. doi: 10.1371/journal.pbio.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- Zhu J, Burakov A, Rodionov V, Mogilner A. Finding the cell center by a balance of dynein and myosin pulling and microtubule pushing: a computational study. Mol Biol Cell. 2010;21:4418–4427. doi: 10.1091/mbc.E10-07-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.