Abstract
Agrobacterium tumefaciens transfers DNA to plant cells as a single-stranded DNA molecule (the T-strand) covalently linked to VirD2 protein. VirD2 contains nuclear localization signal sequences that presumably help direct the T-strand to the plant nucleus. We identified a tomato cDNA clone, DIG3, that encodes a protein that interacts with the C-terminal region of VirD2. DIG3 encodes an enzymatically active type 2C serine/threonine protein phosphatase. Overexpression of DIG3 in tobacco BY-2 protoplasts inhibited nuclear import of a β-glucuronidase-VirD2 nuclear localization signal fusion protein. Thus, DIG3 may be involved in nuclear import of the VirD2 protein and, consequently, the VirD2/transferred DNA complex.
Agrobacterium tumefaciens is a unique soil bacterium that genetically transforms most dicotyledonous and some monocotyledonous plants through an interkingdom transfer of genetic information, the transferred DNA (T-DNA) (1). The bacteria often cause crown gall tumors on infected plants as a result of overproduction of the phytohormones auxin and cytokinin whose synthesis is directed by T-DNA genes expressed in the plant cell. Despite the significance of this plant-microbe interaction for both plant pathology and plant genetic engineering, we currently understand little about plant genes and proteins involved in the transformation process.
In Agrobacterium, the processing and transfer of T-DNA is mediated by bacterial virulence proteins encoded by vir genes present on the tumor-inducing (Ti) plasmid. On vir gene induction by phenolics, T-DNA is processed from the Ti plasmid by the VirD1/VirD2 endonuclease (2-6). VirD2 protein then becomes covalently attached to the 5′ end of the single-stranded T-DNA molecule, the T-strand (4, 5, 7-10). The T-strand may subsequently become coated with another vir gene product, the VirE2 single-stranded DNA-binding protein, to form the T-complex (11). The T-strand and/or T-complex is thought to be the form of T-DNA transferred from Agrobacterium to the plant cell. T-strands have been detected in the cytoplasm of infected plant cells (12), and VirE2 protein has been suggested to protect T-DNA from nuclease attack in plant cells (12, 13).
The Agrobacterium transformation process includes plant cell wall recognition; binding of the bacteria; and the transfer, nuclear translocation, and integration of T-DNA into chromosomal DNA. Details of these processes remain largely unknown. Presumably, they are mediated by several Agrobacterium vir gene products and plant factors. Nam et al. (14) described differences among Arabidopsis thaliana ecotypes in their response to Agrobacterium infection, and these differences were heritable. A genetic basis for susceptibility to Agrobacterium has also been described for other plant species, and the identification of Arabidopsis rat (resistant to Agrobacterium transformation) mutants (15) further confirms the role of plant genes in the transformation process.
The nuclear envelope is one of the plant barriers to T-DNA transfer. Active nuclear import of macromolecules is mediated by signals in the import substrate, i.e., the nuclear localization signal (NLS) in karyophilic proteins, and other factors such as NLS-binding proteins and regulatory factors (16, 17). That any DNA, irrespective of size or sequence, placed between the T-DNA borders can be transferred to the plant nucleus indicates that the signal for nuclear import does not reside in the T-DNA sequence. Rather, the covalent association of VirD2 with the T-strand suggests that signals for nuclear import of T-DNA may reside in the VirD2 protein. Indeed, VirD2 contains two distinct NLSs (18-21). Either NLS can direct import of a reporter fusion protein to the plant cell nucleus (19-23). The C-terminal bipartite NLS is thought to be the only one that functions in T-strand nuclear import (19, 20, 24). When infected by an Agrobacterium strain containing mutations in the C-terminal NLS of VirD2, plant cells showed slightly reduced T-DNA expression and tumorigenicity (23-25), indicating that the VirD2 NLS is partially responsible for nuclear import of T-DNA. Another possible signal for nuclear import of T-DNA may be the NLSs of the VirE2 protein (26). These VirE2 NLSs are also capable of directing β-glucuronidase (GUS) protein into the plant cell nucleus (22, 26). When microinjected into Tradescantia virginiana cells, fluorescently labeled DNA complexed with the VirE2 protein localized to the nucleus (27). Ziemienowicz et al. (28, 29) showed the importance of both VirD2 and VirE2 in the nuclear localization of in vitro synthesized T-complexes in animal and plant cells. However, the overlap of the VirE2 NLS region with the single-stranded DNA-binding region makes it difficult to demonstrate the in vivo function of this NLS region by mutational analysis, although the involvement of VirE2 NLSs in nuclear import of T-DNA has been shown (28-30).
Because the signal for nuclear import of T-DNA is likely carried in both VirD2 and VirE2 proteins, these proteins may serve as targets for modifications that regulate T-DNA nuclear import. There is little information concerning the plant factors that mediate nuclear import of T-DNA. Here, we report the identification of a cDNA clone, DIG3, that encodes a plant type 2C protein phosphatase that interacts specifically with the Agrobacterium VirD2 protein.
Materials and Methods
Bacterial, Yeast, and Plant Cell Growth Conditions. Yeast strains were cultured in the appropriate media containing yeast nitrogen-base and all but the selective amino acids [complete media (CM)] media at 30°C. Escherichia coli strains were cultured in LB medium containing the appropriate antibiotics. A. tumefaciens strains were grown in yeast extract/peptone medium containing the appropriate antibiotics. Antibiotics used were: ampicillin, 100 μg/ml; rifampicin, 10 μg/ml; kanamycin, 50 μg/ml; spectinomycin, 50 μg/ml; and carbenicillin, 100 μg/ml. Tobacco BY-2 cell cultures were grown at room temperature with shaking at 140 rpm in Murashige and Skoog medium (Life Technologies, Grand Island, NY; GIBCO/BRL) containing 88 mM sucrose, 370 mg/liter KH2PO4 (pH 5.7), 1 mg/liter thiamine, and 0.2 mg/liter 2,4-dichlorophenoxyacetic acid.
Interaction Trap. To identify plant proteins that interact with VirD2 protein, we used an interaction trap (31). We constructed the VirD2 bait containing a 668-bp BamHI-SalI fragment of wild-type VirD2 (from pTiA6) sequences in pEG202 (32) digested with BamHI/SalI. The VirD2 sequences in the bait represent the C-terminal half of VirD2 and include the C-terminal bipartite NLS residues. We transformed EGY48 by electroporation (32) with the bait plasmid and screened a tomato cDNA library in the vector pJG4-5. Library transformation of the bait strain was carried out by using a lithium acetate method (32).
The bait strain was determined to be functional for the interaction trap by activation and repression assays, as described (31). Screening and grouping of candidate clones were performed as described (31). Yeast colonies containing candidate interacting cDNA clones were picked for direct PCR to amplify the cDNAs, followed by AluI restriction analysis to classify cDNA groups. The PCR was performed in a 50-μl reaction volume by using Taq polymerase with the primers 5′-TAACGATACCAGCCTCTTG-3′ (forward primer) and 5′-GACAACCTTGATTGGAGAC-3′ (reverse primer) for 30 cycles with the following program: 94°C for 1 min (1 cycle), 92°C for 40 sec, 60°C for 40 sec, 75°C for 1.5 min (29 cycles), and 75°C for 5 min (one cycle).
Generation of the VirD2 (Ser-394→ Ala) mutant. To replace Ser-394 with Ala, the PCR primers 5′-AGCAAGATCTATCGGTACCGAGCAACCGGAAGCTGCTCCAAAGCGTCCGCGT-3′ (forward) and 5′-GCTCTAGAGCTTTCCGAAGAATCACGCA-3′ (reverse) were synthesized. PCR was performed by using plasmid DNA from E1255 (containing virD2) and Pwo polymerase (Boehringer Mannheim). The amplified product containing the mutated KpnI-SalI fragment of the octopine VirD2 sequence was digested with BglII and XbaI and cloned into BglII-XbaI-digested pRTL-2-GUS-NIa (33) to create GUS-VirD2 NLS (Ser-394→Ala).
Assay to Determine Interaction Strength. Three independent yeast colonies for each bait/prey combination were used to determine the interaction strength. Yeast strains harboring interaction plasmids were cultured in liquid CM-Ura-His-Trp medium supplemented with 2% glucose at 30°C for 2 days. Fifty microliters of saturated culture was inoculated into 5 ml of the appropriate selection medium supplemented with either glucose (2%) or galactose/raffinose (2%/1%) and cultured at 30°C until the A600 reached 0.5-0.7. Cells were assayed for β-galactosidase activity as described (34). β-Galactosidase activity was reported as Miller units per milligram of total protein. Protein concentrations were determined by a Bradford assay using BSA as a standard.
In Vitro Interaction Between DIG3 and VirD2. E. coli BL21 (DE3) cultures containing pGEX4T-1, the BamHI-SalI C-terminal fragment of octopine VirD2 (cVirD2), or the full-length octopine VirD2 (fl-VirD2) fused in frame with the GST tag of pGEX4T-1 were induced with 1 mM isopropyl β-D-thiogalactoside at 37°C for 3 h to obtain GST, GST-cVirD2, or GST-flVirD2, respectively. GST, GST-cVirD2, or GST-flVirD2 was separately incubated with glutathione-Sepharose (Sigma). The columns were washed with three bed volumes of PBS. DIG3 protein was purified as a T7-tag fusion protein after cloning the DIG3 cDNA into pET23a by using a T7-tag antibody column purification kit as specified (Novagen). Purified T7-tagged DIG3 fusion protein was passed over each glutathione-Sepharose column three times. The columns were washed with three bed volumes of PBS, and bound proteins were eluted by using cold 5 mM reduced glutathione in 50 mM Tris, pH 8.0. Eluted fractions corresponding to equal amounts of GST, GST-cVirD2, or GST-flVirD2 were used for Western blot analysis with anti-T7 monoclonal antibodies (Novagen).
Screening cDNA Library for Full ORF cDNA Clones. We screened a tomato cDNA library constructed in the λ vector Uni-ZAP XR (Stratagene) according to the manufacturer's manual (Stratagene). PCR-amplified DIG3 DNA was digested by EcoRI and SacI to release the 5′-end fragment (≈500 bp) as a probe that was labeled with 32P-dCTP by using a Multiprime Labeling kit (Amersham Pharmacia).
Protoplast Preparation, Transfection, and Plasmid Construction. We made GUS fusion constructions under the control of a cauliflower mosaic virus 35S promoter with a tobacco etch virus translational leader (33). The plasmids pRTL2-GUS, pRTL2-GUS/NIa, and pRTL2-GUS/cVirD2 have been described (23). We digested pRTL2-GUS/NIa with NcoI, eluted the vector fragment from the gel, and self-ligated the vector to form pYT-6. We amplified DIG3-3 DNA by PCR with Pwo polymerase (Boehringer Mannheim) by using the following primers: DT-3, 5′-ATAGCCATGGTCGATTATGCCTCTCCCGAATTC-3′ (forward) and DT-4, 5′-ACTGCCATGGCATACCAAAGCTTCTCGAG (reverse). Amplified DIG3 with the in-frame start codon ATG was digested with NcoI and cloned into the NcoI site of pYT-6 in the correct orientation to form pYT-7. Protoplasts were prepared, transfected, and stained for GUS activity according to Mysore et al. (23).
Type 2C Serine/Threonine Protein Phosphatase (PP2C) Activity Assay. We performed phosphatase activity assays according to the instructions provided by the manufacturer by using a nonradioactive serine/threonine phosphatase assay system (Promega, catalog no. V2460). The color was allowed to develop for 15 min, and the absorbance was measured at 630 nm with a plate reader. The composition of the buffers used in the assay was: PPTase-2A 5× buffer (250 mM imidazole, pH 7.2/1 mM EGTA/0.1% 2-mercaptoethanol/0.5 mg/ml BSA), PPTase-2B 5× buffer (250 mM imidazole, pH 7.2/1 mM EGTA/50 mM MgCl2/2 mM CaCl2/250 μg/ml calmodulin/0.1% 2-mercaptoethanol), PPTase-2C 5× buffer (250 mM imidazole, pH 7.2/1 mM EGTA/25 mM MgCl2/0.1% 2-mercaptoethanol/0.5 mg/ml BSA).
Results
Identification of DIG3, a Protein That Interacts with the VirD2 NLS. We used an interaction trap approach (31) to search a tomato cDNA library (35) for cDNAs that encode proteins binding specifically to the VirD2 protein. The bait protein consisted of the C-terminal half of VirD2 protein (designated as cVirD2) from the A. tumefaciens octopine-type Ti-plasmid pTiA6 (Fig. 1A). This region of protein contains the NLS region thought to direct the T-strand to the plant nucleus (24). From among 3 × 106 primary yeast transformants, we obtained 500 candidate interacting clones. cDNAs were amplified from these clones by PCR and digested by AluI to classify clones representing cDNAs of the same gene into a family. A total of 28 cDNA families representing different tomato genes that encode proteins that interact with the C-terminal half of VirD2 protein were obtained.
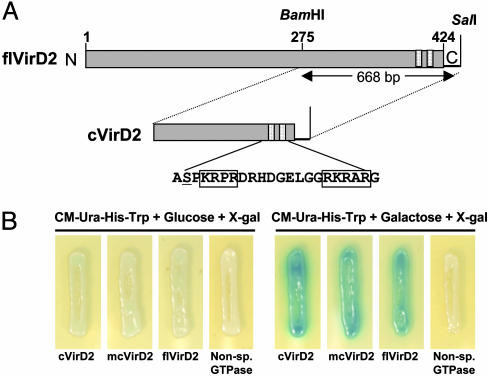
Fig. 1.
Interaction trap to identify cDNAs encoding VirD2-interacting proteins. (A) Schematic diagram of full-length (flVirD2) and the C-terminal half of pTiA6 VirD2 (cVirD2) used as baits. Numbers above the bar indicate amino acid sequence numbers. The amino acid sequence surrounding the C-terminal bipartite NLS (boxed residues) is shown below the bar. Ser-394 is underlined. (B) VirD2-DIG3-3 interaction in yeast. Yeast strains harboring the DIG3-3 prey were transformed with cVirD2, mcVirD2, flVirD2, or an A. nidulans GTPase as a nonspecific bait. The resulting strains were grown for 2 days on CM-Ura-His-Trp + 5-bromo-4-chloro-3-indolyl β-d-galactoside plates containing either glucose or galactose.
We recovered prey plasmids and used them to transform yeast strains that express LexA fused either to full-length VirD2 (designated as flVirD2; Fig. 1A) or the original bait cVirD2 (as a positive control). Transformants were selected on CM-Ura-His-Trp medium and assayed for both galactose-dependent leu2 and lacZ gene activities. We pursued characterization of one positive clone (designated as DIG3) because of its interaction with both cVirD2 and flVirD2 baits. Fig. 1B shows lacZ activities resulting from interaction of flVirD2 or cVirD2 protein with the DIG3 protein in yeast. An unrelated protein (Aspergillus nidulans GTPase) when used as bait did not show interaction with DIG3 in the yeast two-hybrid system (Fig. 1B). To determine the strength of the interaction between DIG3 and the VirD2 baits (cVirD2 and flVirD2), the bait strains were individually transformed with a prey plasmid containing the DIG3 cDNA. The resulting strains were cultured in liquid medium containing CM-Ura-His-Trp supplemented with either glucose or galactose.
Fig. 2A shows that interaction between DIG3 protein and the cVirD2 bait protein resulted in 5,200 ± 400 β-galactosidase units/mg total protein in the yeast strain harboring these plasmids. The flVirD2 bait shows lower (≈800 ± 50 β-galactosidase units/mg total protein) strength of interaction with DIG3. Western blot analysis indicated that similar levels of LexA-cVirD2 and LexA-flVirD2 were produced in the corresponding yeast strains (data not shown). We speculate that the lower number of β-galactosidase units resulting from interaction of flVirD2 with DIG3 may result from conformational differences between flVirD2 and cVirD2. We detected no significant β-galactosidase activity using the nonspecific A. nidulans GTPase bait with the DIG3 prey. The β-galactosidase activity resulting from the interaction of DIG3 with VirD2 depended on galactose induction of the gal promoter in the prey plasmid (Fig. 2A). Consistent with this observation, the Leu+ prototrophic phenotype resulting from the specific interaction of DIG3 and VirD2 baits (either cVirD2 or flVirD2) depended on galactose induction (data not shown). Thus, DIG3 encodes a protein that interacts in yeast specifically with the VirD2 bait.
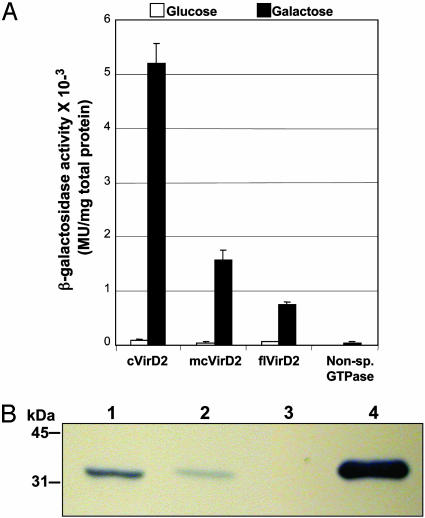
Fig. 2.
Strength and specificity of DIG3-3 interaction with VirD2. (A) Yeast strains harboring the DIG3-3 prey were transformed with cVirD2, mcVirD2, flVirD2, or the A. nidulans GTPase baits and assayed for β-galactosidase activity as described in Materials and Methods. (B) In vitro interaction of DIG3-3 with VirD2. Purified T7-epitope tagged DIG3-3 was passed over glutathione-Sepharose columns that had bound GST, GST-cVirD2, or GST-flVirD2. Bound fractions were eluted and analyzed on Western blots with anti-T7 monoclonal antibodies. Lane 1, GST-cVirD2; lane 2, GST-flVirD2; lane 3, GST; lane 4, purified T7-tagged DIG3-3. Molecular mass markers are indicated in kDa on the left.
To confirm further the interaction between VirD2 and DIG3, an in vitro GST pull-down experiment was performed (Fig. 2B). Equimolar amounts of purified T7 epitope-tagged DIG3 fusion protein were passed over separate glutathione-Sepharose columns to which were bound GST, GST-cVirD2, or flVirD2 proteins. Bound proteins were eluted with reduced glutathione, and Western blot analyses performed with anti-T7 monoclonal antibodies. DIG3 proteins were detected in the bound fractions from GST-cVirD2 (lane 1) and GST-flVirD2 (lane 2) columns but not from the GST column (lane 3), indicating that DIG3 does indeed interact with VirD2 protein. Although bound fractions corresponding to equimolar amounts of GST-cVirD2 and -flVirD2 were used for the Western blot analysis (data not shown), we detected a significantly lower level of DIG3 in the GST-flVirD2 bound fraction in comparison to the GST-cVirD2 bound fraction (Fig. 2B). This result corresponds to our earlier observation that in yeast, flVirD2 interacts with DIG3 to a lesser extent than does cVirD2 (Fig. 2A).
Sequencing the DIG3 cDNA revealed an ≈700-bp ORF followed by a putative polyA addition signal (AATAAA). The fact that we did not find a start codon in the DIG3 ORF suggested that we obtained a partial cDNA clone. We therefore used a 500-bp EcoRI-SacI fragment at the 5′ end of DIG3 as a probe to identify a full-length DIG3 ORF from a tomato λ phage cDNA library. From 1 × 106 phage, four cDNA clones were identified. The longest clone, DIG3-3, contains an in-frame stop codon (TAA) at the 5′ end. There are four putative in-frame start codons for DIG3-3 (Fig. 3, underlined). The last one matches best with the consensus start codon context in eukaryotes: ACCATGG. The cDNA of the original DIG3 isolated from the interaction trap starts two bases after the fourth start codon (xAA; x comes from the EcoRI adapter for library construction). Consequently, if the first ATG were used in vivo as the start codon, the first 47 amino acids are not essential for interaction with VirD2. RNA gel blot analysis revealed an ≈1.0-kb transcript that hybridized to a DIG3 cDNA probe (Fig. 6, which is published as supporting information on the PNAS web site). Genomic DNA blot analysis revealed that DIG3 represents either a single gene or a member of a small gene family in tomato (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 3.
Nucleotide sequence of the DIG3-3-cDNA. The deduced amino acid sequence is indicated below the respective triplet codons. Underlined sequences encode putative in-frame start codons (5′ end) and polyA addition signals (3′ end). *, predicted stop codon.
DIG3 Encodes a Functional PP2C. A computer search of various databases suggested that DIG3 likely encodes a PP2C. The deduced amino acid sequence of DIG3 shows 30-36% identity to PP2Cs from other organisms (Fig. 8, which is published as supporting information on the PNAS web site), including the Arabidopsis ABI1 gene product (36, 37).
To determine whether DIG3 encodes a functional type 2C protein phosphatase, we expressed the original partial DIG3 cDNA in E. coli as an in-frame C-terminal fusion protein with GST. We assayed the purified protein for phosphatase activity by measuring the release of phosphate from a phosphorylated synthetic peptide. Fig. 4A shows that the GST-DIG3 fusion protein, but not GST alone, displayed protein phosphatase activity. This activity could be inhibited by sodium fluoride, indicating that DIG3 encodes a serine/threonine phosphatase. Incubation of the GST-DIG3 fusion protein with the substrate in various reaction buffers (Fig. 4B; see Materials and Methods for assay conditions) indicated that, as suggested by DNA sequence analysis, DIG3 encodes a type 2C protein phosphatase.
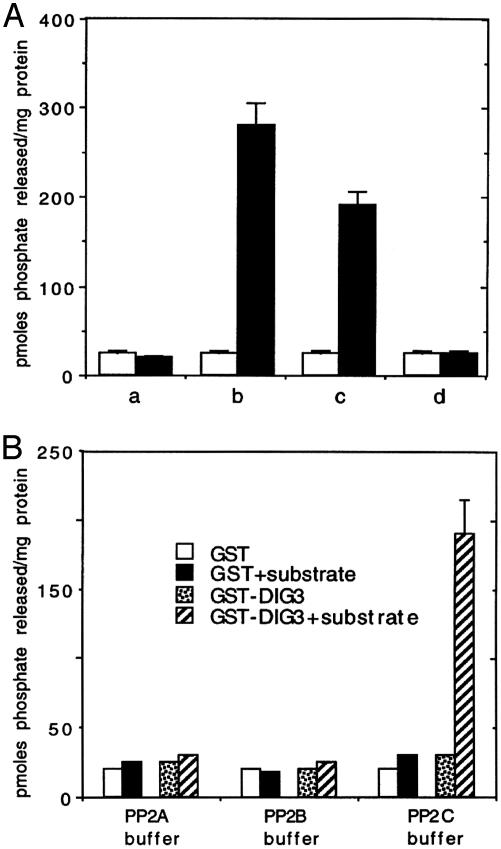
Fig. 4.
Characterization of protein phosphatase activity encoded by a GST-DIG3 fusion protein. (A) The DIG3 protein is a serine/threonine phosphatase. One microgram of GST or GST-DIG3 was incubated with: a, No substrate; b, 100 μM substrate; c, 100 μM substrate plus 1 mM sodium vanadate; d, 100 μM substrate plus 50 mM sodium fluoride. □, GST; ▪, GST-DIG3. (B) The DIG3 protein is a PP2C. One microgram of GST or GST-DIG3 was incubated with or without 100 μM substrate in each of the three buffers specific for PP2A, PP2B, or PP2C activity. The data are average values of three experiments. Error bars represent standard deviations.
DIG3 Can Affect Nuclear Import of VirD2 in Plant Protoplasts. Because DIG3 specifically interacted with the C-terminal half of VirD2 that includes the bipartite NLS residues, we speculated that the encoded PP2C may affect the process of VirD2 nuclear import. To test this hypothesis, we electroporated a gene encoding a GUS-cVirD2 fusion protein into tobacco BY2 protoplasts. The localization of GUS activity exclusively in the nucleus occurred in >80% of the transfected cells (Fig. 5A). We next constructed a plasmid containing DIG3 under the control of the cauliflower mosaic virus 35S promoter and coelectroporated it into tobacco BY2 protoplasts with the gene encoding the GUS-cVirD2 fusion protein. We used a high molar ratio of DIG3 to GUS-cVirD2 constructions (10:1) in these coelectroporation experiments to maximize the probability that GUS-cVirD2 transfected protoplasts also took up the DIG3 construction. Overexpression of DIG3 in tobacco BY2 protoplasts partially inhibited nuclear import of the GUS-cVirD2 fusion protein in the cotransfected protoplasts (Fig. 5B). As opposed to 80-90% of transfected protoplasts that showed exclusive nuclear localization of GUS activity using the GUS-cVirD2 construction alone, only 43% of the coelectroporated protoplasts showed exclusive nuclear localization of GUS activity (Fig. 5B). This result suggests an inhibitory effect of DIG3 on nuclear import of the GUS-cVirD2 fusion protein. We obtained similar results using a higher molar ratio (20:1), suggesting that the expression of DIG3 was saturated in these coelectroporation experiments. When we coelectroporated the vector lacking DIG3 with the GUS-cVirD2 construction, the percentage of transfected protoplasts showing localization of GUS activity exclusively in nuclei remained high. Taken together, these data suggest that DIG3, a PP2C, is involved in the nuclear import of the VirD2/T-DNA complex.
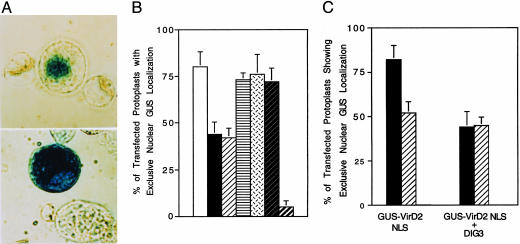
Fig. 5.
(A) Expression of the DIG3 cDNA in tobacco BY-2 protoplasts inhibits nuclear import of a GUS VirD2 NLS fusion protein. Fourteen hours after electroporation, protoplasts were stained with 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-gluc) and visualized by using a phase-contrast microscope. (Upper) Electroporation of a GUS-VirD2 NLS fusion gene. (Lower) Coelectroporation of DIG3 and a GUS-VirD2 NLS fusion gene. (B) Percentage of transfected protoplasts with GUS activity localized exclusively in the nucleus. Approximately 100 X-glucstained protoplasts were scored for each sample, and each experiment was performed three times in molar ratios of 10:1 and 20:1. Coelectroporation controls, vector, and GUSVirD2 NLS or DIG3 and GUS-NIa were performed at a ratio of 20:1. □, GUS-VirD2 NLS; ▪, DIG3 + GUS-VirD2 NLS (10:1);  , DIG3 + GUS-VirD2 NLS (20:1);
, DIG3 + GUS-VirD2 NLS (20:1);  , vector + GUS-VirD2 NLS (20:1);
, vector + GUS-VirD2 NLS (20:1);  , GUS-NIa;
, GUS-NIa;  , DIG3 + GUS-NIa;
, DIG3 + GUS-NIa;  , GUS. (C) A serine residue near the VirD2 C-terminal NLS is involved in nuclear import. Tobacco BY-2 protoplasts were electroporated with either the GUS-VirD2 NLS constructions alone or with the DIG3 gene. X-gluc-stained protoplasts were scored for the percentage of protoplasts that showed exclusive nuclear GUS localization. At least 300 transfected protoplasts were examined for each sample. ▪, GUS-VirD2 NLS;
, GUS. (C) A serine residue near the VirD2 C-terminal NLS is involved in nuclear import. Tobacco BY-2 protoplasts were electroporated with either the GUS-VirD2 NLS constructions alone or with the DIG3 gene. X-gluc-stained protoplasts were scored for the percentage of protoplasts that showed exclusive nuclear GUS localization. At least 300 transfected protoplasts were examined for each sample. ▪, GUS-VirD2 NLS;  , GUS-VirD2 NLS (Ser-394→Ala).
, GUS-VirD2 NLS (Ser-394→Ala).
To examine the specificity of the effect of DIG3 on the VirD2 nuclear import, we coelectroporated the DIG3 construction and a GUS-NIa (a tobacco etch virus protein that is known to contain a plant functional NLS) fusion construction into tobacco BY2 protoplasts. When electroporated with GUS-NIa fusion construction alone, 78% of transfected protoplasts showed exclusive nuclear accumulation of GUS activity. Overexpression of DIG3 did not have an inhibitory effect on the nuclear import of a GUS-NIa fusion protein (Fig. 5B), indicating that the inhibitory effect of DIG3 on VirD2 nuclear targeting is relatively specific.
Ser-394 of VirD2 Is Important for Nuclear Import. The phosphorylation status of serine residues near the NLS of karyophilic proteins may affect nuclear import. Ser-394 precedes the first domain of the VirD2 bipartite NLS and is the only potential phosphorylation target of a PP2C near the NLS. We investigated whether Ser-394 of VirD2 is important for interaction with DIG3 using the yeast two-hybrid assay system. We generated a modified cVirD2 bait (designated hereafter as mcVirD2) in which Ser-394 was changed to alanine. We chose cVirD2, rather than flVirD2, as a bait because it shows a 6-fold greater interaction strength with DIG3 than does the flVirD2 bait, and changes in the strength of interaction with DIG3 resulting from the amino acid substitution in mcVirD2 would be more apparent. mcVirD2 interacts with DIG3 with a 3-fold lower affinity than does cVirD2 (Fig. 2A). Western blot analysis indicated that yeast strains express similar levels of LexA-cVirD2 or LexA-mcVirD2 fusion proteins (data not shown). Thus, Ser-394 is important for modulating the strength of interaction with DIG3.
We further investigated the effects of the Ser-394 to alanine substitution on the nuclear import of the resulting mutant GUS-mcVirD2 protein in tobacco BY-2 protoplasts. Fig. 5C shows that ≈50% of the cells transfected with GUS-mcVirD2 showed exclusive nuclear localization of GUS activity compared to ≈80% of the cells when transfected with the GUS-cVirD2 construction. Coelectroporation of the DIG3 construction with this mutant GUS-mcVirD2 construction had little effect on exclusive nuclear localization of GUS activity. Thus Ser-394, a potential target for phosphorylation/dephosphorylation, is important for the functioning of the VirD2 bipartite NLS.
Discussion
We used an interaction trap to identify a cDNA, DIG3, that encodes a protein that interacts with the A. tumefaciens VirD2 protein. Both amino acid sequence comparisons and enzymatic assays indicated that DIG3 encodes a PP2C. Our results suggest that DIG3 is involved in nuclear import of the Agrobacterium VirD2/T-DNA complex through specific interactions with the C-terminal region of VirD2, which includes the bipartite NLS thought to mediate the nuclear import of the T strand (19, 20, 24). This model is based on our observations that overexpression of DIG3 protein in transfected tobacco BY-2 protoplasts inhibits nuclear transport of a GUS-cVirD2 fusion protein. The data also imply that this effect is relatively specific to the VirD2 NLS because DIG3 does not inhibit the nuclear localization of a GUS-NIa fusion protein in tobacco protoplasts. Our model is strengthened by the observation that an Arabidopsis abi1 mutant (that lacks a PP2C activity) is more susceptible to Agrobacterium-mediated transformation than is the wild-type plant (38). Taken together, our results suggest that the PP2C encoded by DIG3 is a negative regulator of nuclear import of the VirD2/T-DNA complex.
How might DIG3 play a role in nuclear import of the VirD2/T-DNA complex? We suggest two possible mechanisms: (i) Association and dissociation of DIG3 with VirD2 masks and unmasks the NLS. Such a mechanism is used by NF-κB/IkB family proteins (39). (ii) Phosphorylation in the VirD2 NLS region potentiates nuclear import of the VirD2/T-DNA complex. Such a mechanism has been reported for nuclear import of karyophilic proteins in animal and yeast cells (40). There are seven potential phosphorylation sites for PKC, one for casein kinase II (19) and one for protein kinase A in the VirD2 NLS region (cVirD2) used in our two-hybrid screen. There is accumulating evidence for the existence of PKC activity in plant cells (41-43), and among the seven potential PKC sites is one that involves the first lysine residue of the NLS (SPK). This site is well conserved in VirD2 proteins of different Agrobacterium strains. Our data indicate that Ser-394 in the proximity of the VirD2 NLS may be a potential target of DIG3, because mutation of this residue to alanine affected both the strength of interaction with DIG3 and the nuclear import of VirD2. The participation of a protein phosphatase has been directly demonstrated in the regulation of protein nuclear import (44, 45). Our results open the possibility that phosphorylation and dephosphorylation of the VirD2 NLS region may play an important role in tumorigenesis by potentiating nuclear import of the VirD2/T-DNA complex.
A link between VirD2 phosphorylation and the integration of T-DNA was recently suggested by the observation that VirD2 is phosphorylated by the plant nuclear cyclin-dependent activating kinases (CAK2Ms) and interacts with the TATA box-binding protein (TBP) (46). CAK2Ms maybe involved in transcription-coupled DNA repair by recruiting TBP through phosphorylation of the C-terminal regulatory domain of RNA polymerase II. It would be interesting to determine whether the roles of VirD2 in nuclear import of the T complex and T-DNA integration are regulated by the phosphorylation of VirD2.
Very little is known about the physiological functions of PP2C (47). Physiological substrates for PP2C remain to be identified in animals and plants. In fission yeast, three genes (PTC1, 2, 3) encoding PP2Cs have been cloned, and their functions have been implicated in osmoregulation (48, 49). In plants, PP2C activity has been detected in carrot, cauliflower inflorescence, and leaves of pea and wheat (50), and they are mainly cytosolic. Three novel PP2Cs, PP2C-At, ABI1, and kinase-associated protein phosphatase, have been cloned from Arabidopsis (51). DIG3 from tomato is previously undescribed and encodes the smallest PP2C identified so far. Among all PP2Cs identified, the two plant PP2Cs [the ABI1 gene product and the putative PP2C in Arabidopsis (PP2C-At, here as PP2Carath)] are the most closely related to DIG3, with 60% similarity and 36% identity in amino acid sequences (Fig. 8). Whereas the role of PP2C-At is not known, the role of ABI1 and another less closely related plant PP2C, kinase-associated protein phosphatase (KAPP), has been implicated in signal transduction pathways. The ABI1 gene encodes a unique type of PP2C containing a putative Ca2+-binding site in the N-terminal region (36, 37) and may be involved in a signal transduction pathway. KAPP was isolated via an in vitro interaction screen for proteins that interact with RLK5 (52), a membrane-bound receptor kinase, implying a role in a signal transduction pathway.
In a model proposed by Howard and Citovsky (11), a T-DNA molecule with VirD2 protein attached to the 5′ end is protected by the single-stranded DNA-binding protein VirE2. Both VirD2 and VirE2 proteins have plant functional NLSs (19, 26). Whereas the in vivo function of the VirD2 NLS has been detected (24, 25), the function of the VirE2 NLS in T-DNA nuclear import still awaits clarification. However, that a mutation in the octopine-type VirD2 NLS reduced virulence only to 60% of the wild-type (24) suggests that other factors may contribute to the nuclear import of T-DNA. Genetic evidence (30) and the presence of two NLSs in the VirE2 protein make it the best candidate. Assuming that the T complex model is correct, the T-DNA molecule would be coated by many VirE2 proteins and, hypothetically, the many NLSs provided by VirE2 would be more than sufficient for nuclear import of T-DNA. That the NLS on the single VirD2 protein of the T complex is partially responsible for the nuclear import of T-DNA suggests that the VirD2 NLS and VirE2 NLS may function differently. The NLSs of VirD2 and VirE2 may be recognized by different import machineries. In Xenopus oocytes and Drosophila embryos, microinjected VirD2 and VirE2 proteins were localized differently. Whereas fluorescently labeled VirD2 protein localized to the nucleus, VirE2 protein remained in the cytoplasm (53). Alternatively, nuclear import mediated by VirD2 NLS and VirE2 NLS may be subjected to different regulatory mechanisms. Nuclear import of some karyophilic proteins is known to be regulated cell-specifically or developmentally. Some of this regulation takes place by modification of the NLS region of these proteins (40, 44, 45). VirD2 and VirE2 proteins also may adapt existing plant mechanisms to regulate nuclear import. Some experimental observations suggest that nuclear import of VirD2 and VirE2 may be regulated developmentally (22).
The interaction between Agrobacterium and plant cells is the only known natural system involving the interkingdom exchange of genetic information. For Agrobacterium to transform a plant, the T-DNA must be imported into the plant cell nucleus. The nuclear import of proteins is a highly regulated process in eukaryotic cells (17). We speculate that nuclear import of protein/nucleic acid complexes may also be regulated. Our findings suggest that nuclear import of the VirD2/T-DNA complex is regulated, and that this regulation is achieved by interaction with DIG3, probably via phosphorylation and dephosphorylation.
Supplementary Material
Acknowledgments
We thank Dr. Roger Brent (Molecular Sciences Institute, Berkeley, CA) for providing plasmids and strains used for the yeast interaction trap, Drs. Jianmin Zhou and Greg Martin (Purdue University) for providing the tomato cDNA bait library, Dr. Raghothama (Purdue University) for providing the tomato cDNA library, and Dr. Nune Darbinian (Purdue University) for advice on conducting the protoplast electroporation assay. This work was funded by U.S. Department of Agriculture Grants 95-37301-2040 and 98-35304-6675 (to S.B.G.) and a Purdue Research Foundation Fellowship (to Y.T.).
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY534757).
Abbreviations: NLS, nuclear localization signal; T-DNA, transferred DNA; Ti, tumor-inducing; PP2C, type 2C serine/threonine protein phosphatase; GUS, β-glucuronidase; CM, conditioned media.
References
- 1.DeCleene, M. & DeLey, J. (1976) Bot. Rev. 42, 389-466. [Google Scholar]
- 2.Yanofsky, M. F., Porter, S. G., Young, C., Albright, L. M., Gordon, M. P. & Nester, E. W. (1986) Cell 47, 471-477. [DOI] [PubMed] [Google Scholar]
- 3.Jayaswal, R. K., Veluthambi, K., Gelvin, S. B. & Slightom, J. L. (1987) J. Bacteriol. 169, 5035-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stachel, S. E., Timmerman, B. & Zambryski, P. (1987) EMBO J. 6, 857-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albright, L. M., Yanofsky, M. F., Leroux, B., Ma, D. & Nester, E. W. (1987) J. Bacteriol. 169, 1046-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filichkin, S. A. & Gelvin, S. B. (1993) Mol. Microbiol. 8, 915-926. [DOI] [PubMed] [Google Scholar]
- 7.Veluthambi, K., Ream, W. & Gelvin, S. B. (1988) J. Bacteriol. 170, 1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward, E. R. & Barnes, W. M. (1988) Science 242, 927-930. [Google Scholar]
- 9.Young, C. & Nester, E. W. (1988) J. Bacteriol. 170, 3367-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard, E. A., Winsor, B. A., De Vos, G. & Zambryski, P. (1989) Proc. Natl. Acad. Sci. USA 86, 4017-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard, E. & Citovsky, V. (1990) BioEssays 12, 103-108. [Google Scholar]
- 12.Yusibov, V. M., Steck, T. R., Gupta, V. & Gelvin, S. B. (1994) Proc. Natl. Acad. Sci. USA 91, 2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi, L., Hohn, B. & Tinland, B. (1996) Proc. Natl. Acad. Sci. USA 93, 126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nam, J., Matthysse, A. G. & Gelvin, S. B. (1997) Plant Cell 9, 317-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam. J., Mysore, K. S., Zheng, C., Knue, M. K., Matthysse, A. G. & Gelvin, S. B. (1999) Mol. Gen. Genet. 261, 429-438. [DOI] [PubMed] [Google Scholar]
- 16.Raikhel, N. (1992) Plant Physiol. 100, 1627-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macara, I. G. (2001) Microbiol. Mol. Biol. Rev. 65, 57-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrera-Estrella, A., van Montagu, M. & Wang, K. (1990) Proc. Natl. Acad. Sci. USA 87, 9534-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard, E. A., Zupan, J. R., Citovsky, V. & Zambryski, P. (1992) Cell 68, 109-118. [DOI] [PubMed] [Google Scholar]
- 20.Tinland, B., Koukolikova-Nicola, Z., Hall, M. N. & Hohn, B. (1992) Proc. Natl. Acad. Sci. USA 89, 7442-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossi, L., Hohn, B. & Tinland, B. (1993) Mol. Gen. Genet. 239, 345-353. [DOI] [PubMed] [Google Scholar]
- 22.Citovsky, V., Warnick, D. & Zambryski, P. (1994) Proc. Natl. Acad. Sci. USA 91, 3210-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mysore, K. S., Bassuner, B., Deng, X.-B., Darbinian, N. S., Motchoulski, A., Ream, W. & Gelvin, S. B. (1998) Mol. Plant-Microbe Interact. 11, 668-683. [DOI] [PubMed] [Google Scholar]
- 24.Shurvinton, C. E., Hodges, L. & Ream, W. (1992) Proc. Natl. Acad. Sci. USA 89, 11837-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narasimhulu, S. B., Deng, X.-B., Sarria, R. & Gelvin, S. B. (1996) Plant Cell 8, 873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Citovsky, V., Zupan, J., Warnick, D. & Zambryski, P. (1992) Science 256, 1802-1805. [DOI] [PubMed] [Google Scholar]
- 27.Zupan, J. R., Citovsky, V. & Zambryski, P. (1996) Proc. Natl. Acad. Sci. USA 93, 2392-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziemienowicz, A., Gorlich, D., Lanka, E., Hohn, B. & Rossi, L. (1999) Proc. Natl. Acad. Sci. USA 96, 3729-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ziemienowicz, A., Merkle, T., Schoumacher, F., Hohn, B. & Rossi L. (2001) Plant Cell 13, 369-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gelvin, S. B. (1998) J. Bacteriol. 180, 4300-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golemis, E. A., Gyuris, J. & Brent, R. (1994) in Current Protocol in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), Vol. 2, pp. 13.14.1-13.14.17. [Google Scholar]
- 32.Becker, D. M. & Lundblad, V. (1994) in Current Protocol in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), Vol. 2, pp 13.7.1-13.7.2. [Google Scholar]
- 33.Carrington, J. C., Freed, D. D. & Leinicke, A. J. (1991) Plant Cell 3, 953-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. M. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 35.Zhou, J., Loh, Y.- T., Bressan, R. A. & Martin, G. B. (1995) Cell 83, 925-935. [DOI] [PubMed] [Google Scholar]
- 36.Leung, J., Bouvier-Durand, M., Morris, P.-C., Guerrier, D., Chefdor, F. & Giraudat, J. (1994) Science 264, 1448-1452. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, K., Leube, M. P. & Grill, E. (1994) Science 264, 1452-1455. [DOI] [PubMed] [Google Scholar]
- 38.Tao, Y. (1999) Ph.D. thesis (Purdue University, West Lafayette, IN).
- 39.Liou, H. C. & Baltimore, D. (1993) Curr. Opin. Cell Biol. 5, 477-487. [DOI] [PubMed] [Google Scholar]
- 40.Vandromme, M., Gauthier-Rouviere, C., Lamb, N. & Fernandez, A. (1996) Trends Biochem. Sci. 21, 59-64. [PubMed] [Google Scholar]
- 41.Nanmori, T., Taguchi, W., Kinugasa, M., Oji, Y., Sahara, S., Fukami, Y. & Kikkawa, U. (1994) Biochem. Biophys. Res. Comm. 203, 311-318. [DOI] [PubMed] [Google Scholar]
- 42.Karibe, H., Komatsu, S. & Hinano, H. (1995) Plant. Physiol. 95, 127-133. [Google Scholar]
- 43.Subramaniam, R., Despres, C. & Brisson, N. (1997) Plant Cell 9, 653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibasaki, F., Price, E. R., Milan, D. & McKeon, F. (1996) Nature 382, 370-373. [DOI] [PubMed] [Google Scholar]
- 45.Shibasaki, F., Kondo, E., Akagi, T. & McKeon, F. (1997) Nature 386, 728-731. [DOI] [PubMed] [Google Scholar]
- 46.Bako, L., Umeda, M., Tiburcio, A. F., Schell, J. & Koncz, C. (2003) Proc. Natl. Acad. Sci. USA 100, 10108-10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wera, S. & Hemmings, B. A. (1995) Biochem. J. 311, 17-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiozaki, K., Akhavan-Niaki, H., McGowan, C. H. & Russell, P. (1994) Mol. Cell. Biol. 14, 3742-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiozaki, K. & Russell, P. (1995) EMBO J. 14, 492-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mackintosh, C., Coggins, J. & Cohen, P. (1991) Biochem. J. 273, 733-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, R. D. & Walker, J. C. (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 101-125. [DOI] [PubMed] [Google Scholar]
- 52.Walker, J. C. (1994) Plant Mol. Biol. 26, 1599-1609. [DOI] [PubMed] [Google Scholar]
- 53.Guralnick, B., Thomsen, G. & Citovsky, V. (1996) Plant Cell 8, 363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.