SUMMARY
Virus infection-induced global protein synthesis suppression is linked to assembly of stress granules (SGs), cytosolic aggregates of stalled translation preinitiation complexes. To study long-term stress responses, we developed an imaging approach for extended observation and analysis of SG dynamics during persistent hepatitis C virus (HCV) infection. In combination with type 1 interferon, HCV infection induces highly dynamic assembly/disassembly of cytoplasmic SGs, concomitant with phases of active and stalled translation, delayed cell division, and prolonged cell survival. Double-stranded RNA (dsRNA), independent of viral replication, is sufficient to trigger these oscillations. Translation initiation factor eIF2α phosphorylation by protein kinase R mediates SG formation and translation arrest. This is antagonized by the upregulation of GADD34, the regulatory subunit of protein phosphatase 1 dephosphorylating eIF2α. Stress response oscillation is a general mechanism to prevent long-lasting translation repression and a conserved host cell reaction to multiple RNA viruses, which HCV may exploit to establish persistence.
INTRODUCTION
Cells respond to various forms of stress by a global reduction of protein synthesis, which promotes cellular survival by limiting the consumption of energy and nutrients, and reallocating resources to the repair of cellular damage. Translation suppression is induced by the phosphorylation of the α subunit of eukaryotic initiation factor-2 (eIF2α), which delivers initiator tRNAiMet to the small 40S ribosomal subunit (Holcik and Sonenberg, 2005). eIF2α phosphorylation interferes with formation of the eIF2-GTP-tRNAiMet ternary complex and thereby causes stalling of translation initiation. Among the four mammalian eIF2α kinases, protein kinase R (PKR) responds to double-stranded (ds) RNA in the cytoplasm and mediates translation inhibition upon replication of many RNA viruses (García et al., 2007). Translation is reactivated by dephosphorylation of eIF2α through protein phosphatase 1 (PP1) and its regulatory subunit, growth arrest, and DNA-damage-inducible 34 (GADD34), which couples the catalytic subunit of PP1 to eIF2α (Connor et al., 2001). Moreover, the editing enzyme adenosine deaminase acting on RNA 1 (ADAR1) is part of the interferon (IFN)-mediated antiviral responses, and as a direct inhibitor of PKR participates in the restoration of translation (Gélinas et al., 2011).
Global reduction of protein synthesis is intimately linked to assembly of stress granules (SGs), cytosolic aggregates of stalled translation preinitiation complexes (Anderson and Kedersha, 2008). Hence, SGs contain polyadenylated mRNAs bound to eIF4E, eIF4G, eIF3, and 40S, but not 60S ribosomal subunits. Assembly of SGs is driven by aggregation-prone RNA-binding proteins such as T cell internal antigen-1 (TIA-1), TIA-1-related protein (TIAR), and Ras GTPase-activating protein-binding protein 1 (G3BP1) (Anderson and Kedersha, 2008). SGs promote cellular survival by sequestering components of apoptotic signal transduction pathways (Arimoto et al., 2008; Kim et al., 2005). By keeping preinitiation complexes assembled, SGs are also thought to facilitate reactivation of translation when cells recover from stressful conditions.
SGs are dynamically linked to processing-bodies (PBs), cytoplasmic sites at which most enzymes of the general RNA decay machinery are concentrated (Eulalio et al., 2007; Kulkarni et al., 2010). PBs can also function as storage sites of stalled mRNAs that resume translation upon exit from PBs. Given that viruses interfere with the cellular gene and protein expression machinery, it is not surprising that many viruses were found to interact in different ways with both SGs and PBs (Beckham and Parker, 2008). For instance, infection with reoviruses causes SG formation, which is linked to eIF2α phosphorylation as requirement for viral replication (Smith et al., 2006). Early in infection, Semliki Forest virus causes a transient induction of SGs, whereas SGs are suppressed at later stages of infection (McInerney et al., 2005). The same is true for poliovirus, where the viral protease 3C was found to cleave G3BP1, a protein essential for SG formation (White et al., 2007). While these viruses cause acute lytic infections that eventually lead to cell death, it is unclear how chronic infections such as those caused by hepatitis C virus (HCV), a major causative agent of chronic liver diseases, modulate cellular stress responses to allow long-term viral replication and cell survival.
By using chronic HCV infection of Huh7 liver cells as a model system to study long-term virus-induced stress response, we discovered that HCV-infected cells oscillate between active and repressed phases of RNA translation. These phases are marked by the presence of SGs, triggered by dsRNA and controlled by the antagonistic actions of PKR and GADD34.
RESULTS
HCV- and IFN-α-Dependent Induction of SGs and PB Association
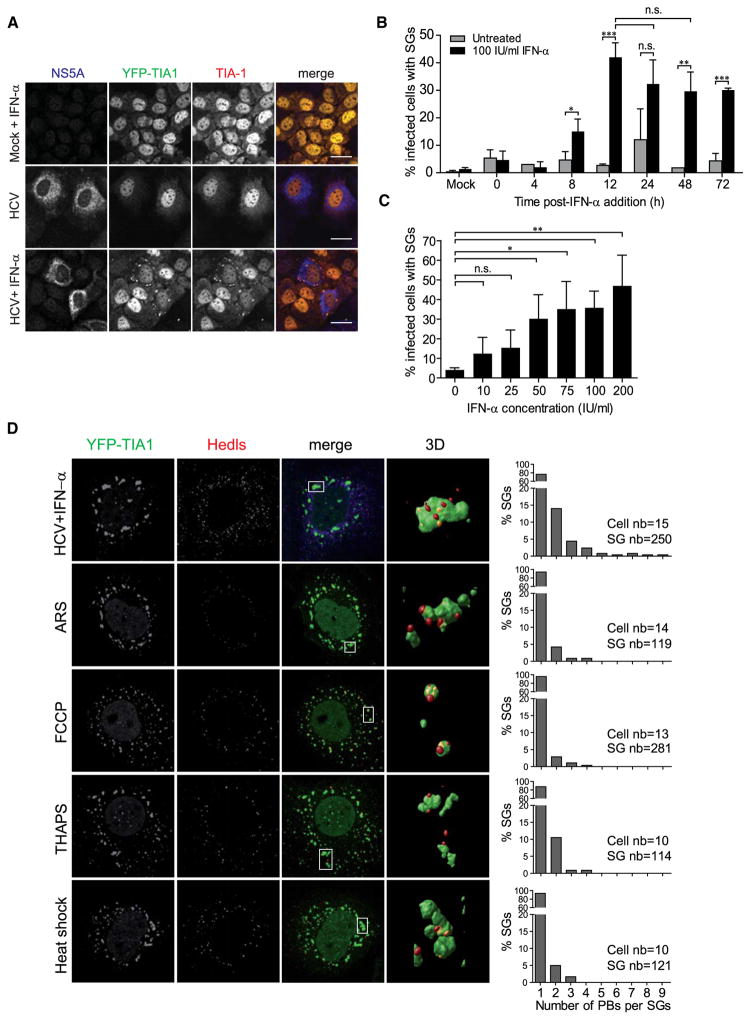
To visualize the cellular stress response induced by HCV, we established a reporter cell line derived from Huh7 human hepatoma cells that stably express the SG marker TIA-1 fused to yellow fluorescent protein (YFP). YFP-TIA1 was validated as bona fide SG marker by colocalization with endogenous SG markers upon exposure to various stress stimuli (Figures S1A and S1B). Importantly, HCV infection and replication in YFP-TIA1 expressing Huh7 cells was indistinguishable from that observed in parental Huh7 cells although production of infectious progeny was reduced at late time points (Figure S1C).
Huh7 cells have an impaired innate antiviral response (Keskinen et al., 1999), which might contribute to SG formation. Since they fail to produce IFN-α in response to virus infection, we decided to add 100 IU/ml IFN-α exogenously, which is a range comparable to the one found in serum of IFN-α treated patients (Vrolijk et al., 2003). Formation of SGs was not observed in mock-infected cells treated for 18 hr with IFN-α (Figure 1A). HCV infection gave rise to a low percentage of SGs in infected cells (Figure 1B). However, the combination of HCV infection and IFN-α treatment triggered a potent stress response indicated by the pronounced formation of SGs. They emerged 8 hr after addition of IFN-α and were detectable 4 hr later in 30%–40% of HCV-infected cells (Figure 1B). Induction of SGs upon HCV infection depended on the dose of IFN-α and was observed in nearly half of HCV-infected cells that were treated with 200 IU/ml IFN-α (Figure 1C). HCV-induced SGs were characterized by the presence of several endogenous SG markers—TIA-1, eIF3B, eIF4G, G3BP1, Hu protein R (HuR), poly(rC)-binding protein 2 (PCBP2)—and also contained cellular mRNAs (Figure 1A and data not shown). Interestingly, SGs also contained the IFN-induced human ds RNA-specific adenosine deaminase (ADAR1), a primarily nuclear protein eventually involved in HCV RNA editing (Taylor et al., 2005) (data not shown). However, SGs did not contain HCV proteins to a detectable level (Figure S1D).
Figure 1. HCV Infection Induces SG Formation in IFN-α-Treated Human Hepatoma Cells.
(A) Huh7 YFP-TIA1 cells (green) were infected with HCV for 24 hr and subsequently cultured with or without IFN-α. Characterization of HCV-induced SGs 18 hr after addition of 100 IU/ml IFN-α. Cells were fixed and stained for NS5A (blue) and TIA-1 (red). Shown are representative confocal micrographs. Scale bars, 25μm. See also Figure S1.
(B) Quantification of the kinetics of HCV-induced SGs. Cells were treated with 100 IU/ml IFN-α and fixed at given time points. NS5A was detected by fluorescence microscopy. Shown are mean values ± SD from three independent experiments with at least 100 cells analyzed each. Statistical significance is given on the top. n.s., no statistical significance.
(C) Quantification of HCV-induced SGs upon treatment with increasing concentrations of IFN-α. Cells were analyzed as described in (B).
(D) Quantification of PB docking to HCV-induced SGs in IFN-α treated cells and comparison to SGs induced by the indicated stresses. The PB marker Hedls (red) was detected by confocal microscopy. Shown are maximum-intensity Z-projections of representative cells. Deconvolved optical sections were analyzed using 3D surface rendering (column “3D”). Red and yellow dots indicate PB docking to the front or the back side of SGs, respectively. A quantification of PB docking to SGs is shown s in the right of each panel. Number of cells and SGs analyzed are given in the top. See also Figure S2.
SGs display dynamic interactions with PBs including the use of common protein components such as Xrn1 (Kedersha et al., 2005). To quantify potential association of PBs with SGs we developed an image-based analysis tool. As illustrated by the specific PB marker Hedls, an enhancer of mRNA decapping (Fenger-Grøn et al., 2005), we observed frequent colocalization of PBs and SGs after HCV infection and IFN-α treatment. Quantification showed that in comparison to other stress conditions, HCV infection combined with IFN-α resulted in enlarged SGs to which significantly more PBs were “docked” (Figures 1D and S2A). Extending the duration of ER stress or heat-shock treatment also resulted in SG enlargement concomitant with a higher number of associated PBs, but SG size never reached that observed upon HCV infection (Figures S2B and S2C). These results revealed that in combination with IFN-α treatment, cells mount a potent stress response to HCV infection, characterized by the formation of enlarged SGs and correlating with an increased number of associated PBs.
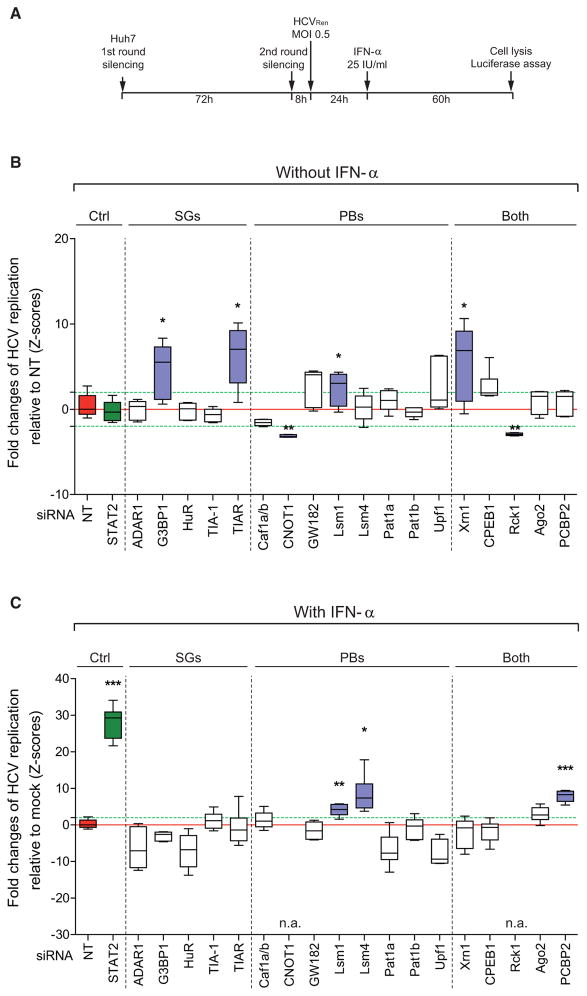
Role of SG Components in Controlling HCV Replication
We next determined whether individual SG components are responsible for the antiviral effect exerted by IFN-α. For this we used an RNAi-based screening assay in which we measured the extent of restoration of HCV replication in IFN-α treated cells upon knockdown of a given SG- or PB-related gene (Figure 2A). In addition, the analogous experimental set-up, but without IFN-α treatment of infected cells, was used to identify SG- and PB-related genes promoting or restricting HCV replication. Selected target genes were earlier reported to be involved in SG formation such as G3BP1, TIA-1, and TIAR, as well as HuR, a protein that promotes mRNA stability. We also included PB-resident proteins such as Caf1a, Caf1b, CNOT1, GW182, Lsm1, Lsm4, Pat1b, and Upf1. Finally, we included factors residing both in SGs and PBs such as Xrn1, CPEB1, Rck, Ago2, and PCBP2. The latter facilitates IRES-mediated RNA translation and enhances the antiviral effect of IFN-α against HCV (Xin et al., 2011). For each of these genes, cell viability and silencing efficiency were determined (Figures S3A and S3B). Knockdown of each individual gene had no effect on SG number and morphology (Figures S3C and S3D), arguing that aggregation properties of unaffected SG components might still be sufficient to allow SG formation. Nevertheless, in the absence of IFN-α G3BP1, TIAR, Lsm1, and Xrn1 were found to limit HCV replication, whereas CNOT1 and Rck1 promoted viral replication (hit selection criteria: Z score > 2; p value < 0.05) (Figure 2B). When SG formation was induced by treatment of HCV-infected cells with IFN-α, we identified the PB components Lsm1 and 4 as repressors of HCV replication (Figure 2C). Likewise, PCBP2 was also found to suppress HCV replication, supporting the notion that PCBP2 facilitates the antiviral activity of IFN-α against HCV. Although we cannot exclude that we missed PB and SG components due to limited knockdown efficiency, our results reveal that SG components G3BP1 and TIAR restrict HCV replication. The fact that with the exception of PCBP2 no SG component was identified with IFN-α treated cells suggests that SG components do not directly contribute to the antiviral activity of IFN-α.
Figure 2. SG Components Promote HCV Replication but Are Dispensable for IFN-α-Mediated Suppression of HCV.
(A) Schematic of the RNAi-based screen set-up to measure rescue of HCV replication in IFN-α treated cells upon knockdown of a given SG- or PB-related gene.
(B and C) Results of RNAi screen. A STAT2-specific siRNA (green box) and a nontargeting siRNA (NT; red box) were included as positive and negative control, respectively. Red and green dashed lines correspond to Z-score = 0 as determined by NT siRNA and to Z-score ± 2, respectively. (B) Boxplots of quantile raw values of HCV replication in absence of IFN-α normalized to cells transfected with NT siRNA. siRNAs leading to an efficient knockdown (Z-score > 2) and a significant change of HCV replication (p value < 0.05) are highlighted with blue boxes. These genes were excluded from the calculation shown in panel (C). (C) Boxplots of quantile ratios between HCV replication in IFN-α treated and untreated cells (mock). Only knockdowns giving rise to a p value < 0.05 and a Z-score > 2 in both calculations (raw values and ratios) were considered as hit candidates. In case of CNOT1 and Rck1, two dependency factors (see B), knockdown and IFN-α treatment reduced replication to nonmeasurable levels and therefore, values could not be determined (n.a., not applicable). See also Figure S3.
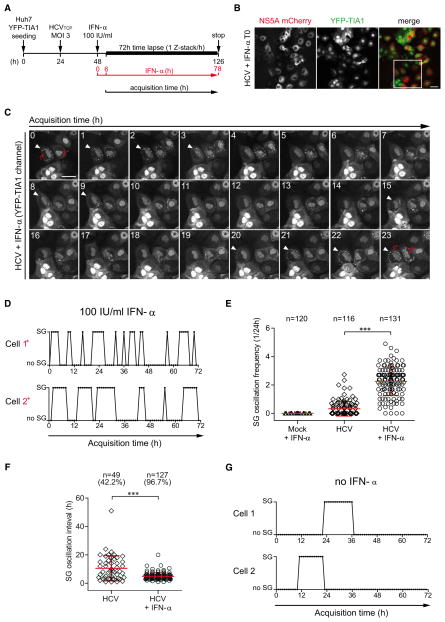
HCV-Induced SGs Oscillate
Having identified several SG proteins as modulators of viral replication on one hand, but SG formation in only ~40% of cells infected with HCV and treated with IFN-α on the other hand, we wondered whether SGs are formed in only a fraction of cells in general or whether this reflects steady-state. Therefore we established a long-term live-cell imaging analysis in which one Z-stack was acquired every hour over a period of 72 hr after HCV infection and subsequent treatment with IFN-α (Figure 3A). Huh7 YFP-TIA1 cells were infected with HCVTCP (Steinmann et al., 2008), a trans-complemented virus encoding a fully functional NS5A-mCherry fusion protein (Figures S4A and S4B), which allowed for the simultaneous visualization of HCV-infected cells and SG formation (Figure 3B). To our surprise, time-lapse analysis revealed a pronounced oscillation of SGs induced by HCV upon IFN-α treatment. As exemplified in the 23 hr time course shown in Figure 3C, SGs frequently disappeared and reappeared in individual cells. These oscillations continued throughout the 72 hr observation period and displayed a cell-specific rhythm rather than a synchronized pattern (Figure 3D). Importantly, ~97% of HCV-infected and IFN-α treated cells displayed SGs at least once during the 72 hr observation period (Figures 3E and 3F), demonstrating that steady-state quantification strongly underestimates cellular stress response (Movie S1). In the absence of IFN-α, SG oscillation was also observed but only in ~40% of HCV-infected cells and with a much lower frequency (Figures 3E, 3F, and 3G; Movie S2). In fact, IFN-α increased oscillation frequency ~7-fold as compared to nontreated cells (2.2 ± 0.09 versus 0.3 ± 0.05, respectively; Figure 3E), concomitant with a reduced mean duration of stress peaks (i.e., oscillation interval; Figure 3F). Neither oscillation frequency of SGs nor total stress duration correlated with the expression level of NS5A-mCherry and, thus, with the degree of viral RNA replication (Figures S4C and S4D). Therefore, the extent of virus replication did not determine the frequency at which SGs are assembled and disassembled. Since SG oscillations were not observed for other stress conditions such as ER and metabolic stress (data not shown) we concluded that SG oscillation might be a specific feature inherent to viral infections.
Figure 3. SGs Induced in HCV-Infected Cells upon IFN-α Treatment Are Highly Dynamic.
(A) Representation of the imaging procedure to visualize the dynamics of HCV-induced SGs. Movies were created from 3D time series by using maximum intensity Z-projections (MIP) and acquired for 72 hr at 1 hr intervals. See also Figure S4.
(B) Representative MIP of the first acquisition time point (T0). White square represents a cropped section shown as example in (C). Scale bar, 50μm.
(C) Cropped section of a MIP of the YFP-TIA1 channel representing the first 23 hr of time-lapse acquisition with 1 hr intervals. Two representative cells (1* and 2*) are labeled. The white arrow indicates phases of detectable SGs in cell 1*.
(D) Dynamics of SGs in HCV-infected cells treated with 100 IU/ml IFN-α. SG oscillation plots indicate presence or absence of SGs in individual infected cells during the 72 hr observation period. Cell 1* and 2* correspond to the ones highlighted in (C). See also Movie S1.
(E and F) Scatter plots of SG oscillation frequency and interval from at least 100 randomly selected HCV-infected cells in presence or absence of IFN-α. Shown in red are mean values ± SD. Statistical significance and the number of analyzed cells (n) are given in the top. (E) SG oscillation frequency is given as the number of stress peaks per 24 hr observation time. (F) SG oscillation interval corresponds to the mean stress peak duration in infected cells showing SGs. Percentage of the total population showing SGs is given in the top (see also Figure S5).
(G) SG oscillation plots of two representative HCV-infected cells in absence of IFN-α. See also Movie S2.
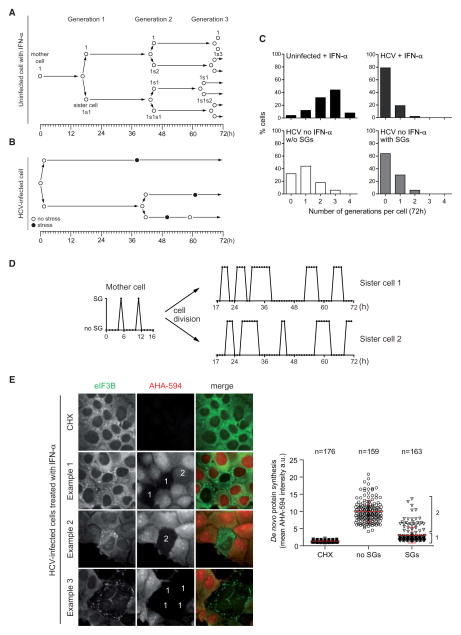
HCV-Induced SG Formation Coincides with Reduced Cell Division and Translation Arrest
To gain further insight into the cell-specific regulation of SG oscillation, we analyzed SG formation in the two sister cells arising after division of a mother cell. In noninfected cells and irrespective of IFN-α treatment, SGs were observed in less than 1% of cells, and cell divisions were found to be synchronized in all sister cells (Figure 4A) with a majority of cells undergoing 2 to 3 divisions during the 72 hr observation period (Figure 4C, upper left panel). Cell division synchrony was disturbed upon HCV infection as the emergence of SGs in individual cells coincided with a lack of cell division in ~80% of infected cells or a significant delay in ~20% of cells (Figures 4B and 4C, upper right panel). In the absence of IFN-α, ~65% of HCV-infected cells showing SGs did not divide (Figure 4C, lower right panel). In contrast, HCV-infected cells that did not display SGs during the 72 hr observation period divided more rapidly with the majority of cells giving rise to at least one generation (Figure 4C, lower left panel). In any case, cell division occurred earliest 10 hr (average 19 hr ± 9) after disassembly of SGs arguing for a “recovery phase” from stress that is required prior to cell division. Moreover, sister cells adopted an individual SG oscillation pattern immediately after division of the mother cell (Figure 4D).
Figure 4. SG Induction by HCV Coincides with Delayed Cell Division and Translational Arrest.
(A and B) Cell division properties of naive or HCV-infected Huh7 YFP-TIA1 cells during the 72 hr observation period. “Mother cell” corresponds to the cell chosen for analysis before its division. Sister cells result from a mother cell division and correspond to one generation. A randomly selected uninfected (A) and HCV-infected (B) cell treated with IFN-α is shown. Black dots represent stress phases.
(C) Quantification of the generation number of Huh7 YFP-TIA1 cells. Shown is a comparison between uninfected (upper left panel) and HCV-infected (upper right panel) cells treated with IFN-α and untreated HCV-infected cells (lower left, cells without SGs, and lower right, cells with SGs).
(D) SG oscillation plot of a representative IFN-α-treated HCV-infected mother cell that divides after a 16 hr observation period.
(E) Reduction of RNA translation in HCV-infected cells during SG periods. Left panel: Huh7 cells infected for 24 hr with HCV and treated 18 hr with IFN-α were incubated 1 hr in methionine-free medium in presence or absence of 200 μM cycloheximide (CHX). De novo synthesized proteins were labeled by incubating cells with Click-iT metabolic labeling reagent coupled to Alexa Fluor 594 (AHA-594, red). After fixation of cells, SGs were immunostained for eIF3B (green). Three exemplary view fields are shown. “1”: cell with SGs and a de novo protein synthesis level reduced to that of cells treated with CHX. “2”: cells with SGs and residual de novo protein synthesis. Right panel: scatter plot of de novo protein synthesis measured by mean fluorescence of AHA-594 intensity and shown as arbitrary unit (a.u.). “1” and “2” refer to cell populations specified above. Shown in red are mean fluorescence intensities ± SD. The number of analyzed cells (n) is given in the top.
In search for a potential mechanism coupling cellular stress (i.e., SG formation) to cell division, we compared translation activity in cells with and without SGs. De novo protein synthesis was visualized in individual cells by metabolic incorporation of a modified amino acid (L-azidohomoalanine, AHA) for 2 hr. After cell fixation, incorporated AHA was coupled to Alexa Fluor 594 (AHA-594). As a control, cells were treated with the translation inhibitor cycloheximide, which completely blocked AHA-594 staining (Figure 4E, left panel). In HCV-infected cells treated with IFN-α, robust translational activity was observed in cells lacking SGs. In sharp contrast, de novo protein synthesis was absent or massively reduced in cells that displayed SGs (Figure 4E, cells 1 and 2; quantification in the right panel). HCV infection in combination with IFN-α thus led to cycles of translation arrest, concomitant with SG formation, which likely delayed progression through the cell cycle.
PKR Is Necessary and Sufficient to Induce SG Oscillation
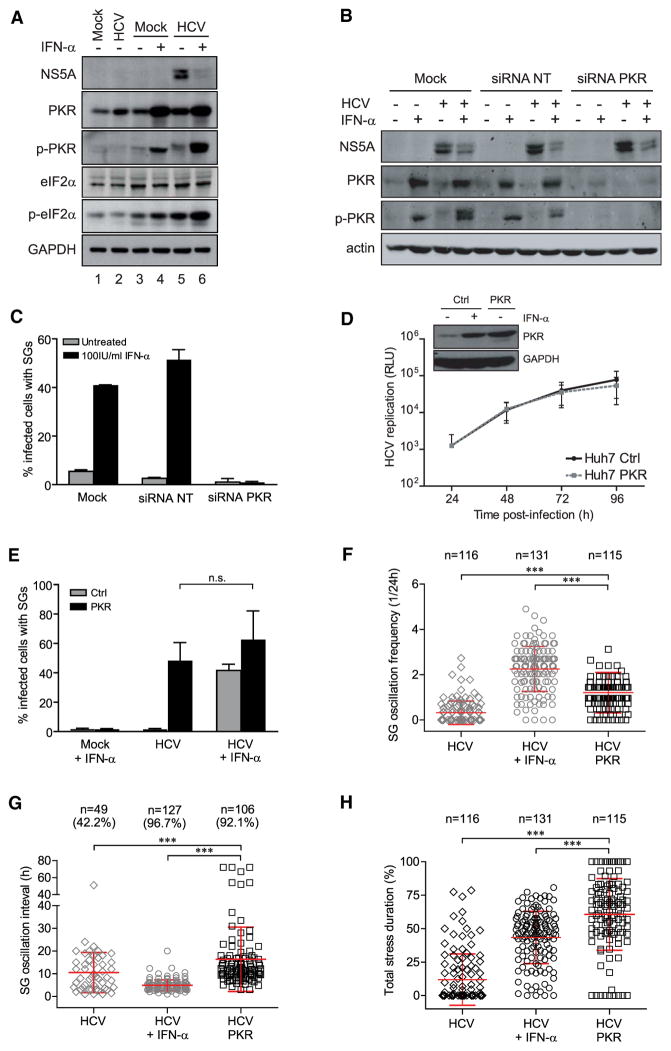
Phosphorylation of eIF2α causes inhibition of translation initiation and is an established hallmark of stress-induced arrest of protein synthesis (Hershey, 1991). Since the eIF2α kinase PKR is activated by dsRNA (Wu and Kaufman, 1997) and induced by IFN-α (Meurs et al., 1990; Thomis et al., 1992), we next tested the role of PKR in SG oscillation triggered by HCV and IFN-α. While PKR protein expression was induced to similar levels upon treatment with IFN-α in the absence and presence of HCV, phospho-PKR and phospho-eIF2α levels showed that PKR activation was much stronger in HCV-infected cells (Figure 5A).
Figure 5. The eIF2α Kinase PKR Is Necessary and Sufficient to Induce SG Formation in HCV-Infected Huh7 Cells.
(A) Western blot analysis of cells infected with HCV for 24 hr (lane 2) or cells infected for 24 hr and cultured for 18 hr in the presence (lane 4, 6) or absence of IFN-α (lane 3, 5). Naive cells cultured for 24 hr in parallel were used as reference (lane 1). (B and C) Transient silencing of PKR in Huh7 YFP-TIA1 cells. Cells were transfected without siRNA (Mock), with nontargeting siRNA (siRNA NT) or with PKR-specific siRNA (siRNA PKR). Twenty four hours later cells were infected with HCV for 24 hr and treated or not with IFN-α for 18 hr. (B) Western blot analysis. (C) Cells were fixed, stained for NS5A, and analyzed. Shown are the mean values ± SD from three independent experiments with at least 100 cells analyzed each.
(D and E) Overexpression of PKR in Huh7 YFP-TIA1 cells. (D) Upper panel: Western blot analysis of Huh7 YFP-TIA1 control cells (Ctrl) treated or not with 100 IU/ml IFN-α for 18 hr or Huh7 YFP-TIA1-PKR cells. Lower panel: characterization of HCV permissiveness of Huh7 YFP-TIA1-PKR cells. Cells were infected with a HCV Renilla luciferase reporter virus, lysed at different time points post-infection and luciferase activity was measured. HCV replication was normalized to the 4 hr values to correct for transfection efficiency (RLU, relative light units). (E) Quantification of HCV-induced SGs in Huh7 YFP-TIA1-PKR cells. Cells were infected with HCV for 24 hr and subsequently treated or not with IFN-α for 18 hr. Cells were fixed, stained for NS5A and analyzed. Shown are the mean values ± SD from three independent experiments with at least 100 cells analyzed each.
(F–H) Scatter plots of SG oscillation frequency, interval, and total stress duration from randomly selected HCV-infected Huh7 YFP-TIA1-PKR cells. Shown in red are mean ± SD. Statistical significance and the number of analyzed cells (n) are given in the top. HCV results (gray symbols) are shown as reference. (F) SG oscillation frequency in HCV infected cells. (G) SG oscillation interval in HCV-infected cells. Number of analyzed cells (n) and corresponding percentage of the total population showing oscillating SGs are given in the top. (H) Total stress duration (i.e., the ratio between total stress time and the observation period for an individual cell) in HCV-infected cells is given in percent. See also Movie S3.
To test the role of PKR in HCV-induced SG formation we employed siRNA-mediated gene silencing (Figure 5B). Of note, suppression of PKR expression fully abrogated the formation of SGs in HCV-infected cells treated with IFN-α (Figure 5C) without affecting viral replication (see NS5A levels in Figure 5B). For the reciprocal experiment, a Huh7 YFP-TIA1 cell pool was generated that stably expresses PKR at levels similar to those observed for endogenous PKR upon induction with 100 IU/ml IFN-α (Figure 5D, upper panel). These PKR overexpressing cells support HCV replication to a level comparable to parental Huh7 YFP-TIA1 cells showing that PKR does not antagonize HCV (Figure 5D, lower panel). While PKR over-expression alone or in combination with IFN-α caused only marginal SG formation in noninfected cells, HCV infection of these cells triggered a massive induction of SGs already in the absence of IFN-α (Figure 5E). In fact, PKR overexpression triggered SG formation to a level comparable to that observed in HCV-infected control cells treated with IFN-α. Combination of IFN-α and PKR overexpression did not enhance SG formation further arguing for saturation. Time-lapse analysis revealed that upon HCV infection ~90% of PKR overexpressing cells displayed SGs. Oscillation frequency was reduced as compared to HCV-infected control cells treated with IFN-α (Figure 5F and Movie S3), concomitant with extended SG oscillation intervals (Figure 5G). These results suggested that the stress response is less dynamic when PKR is expressed ectopically, a conclusion that is reflected by the increased total stress duration of infected cells (Figure 5H). In summary, these results identify PKR as the central IFN-induced gene that is necessary and sufficient for SG formation upon HCV infection. Furthermore, the results confirm that PKR expression alone and SG formation per se do not mediate the antiviral activity of IFN-α.
GADD34 Controls SG Oscillation and Cellular Stress Homeostasis
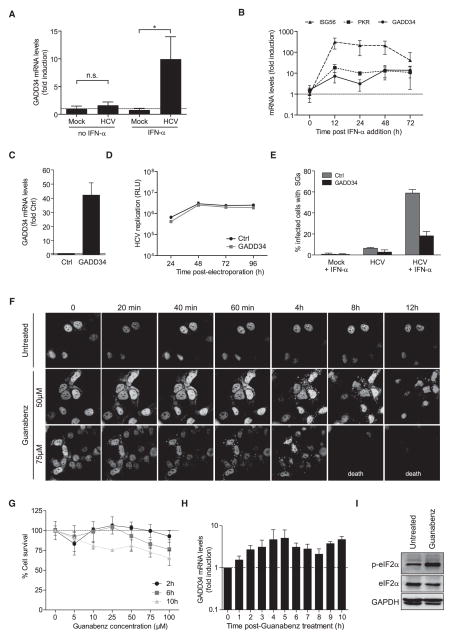
We next asked whether regulation of SG oscillation occurs at the level of eIF2α. In addition to eIF2α kinases, the phosphorylation status of eIF2α is regulated by the antagonistic action of protein phosphatase PP1 and its regulatory subunit GADD34 (Kojima et al., 2003). This phosphatase promotes recovery from translational arrest in a feedback manner as both its transcription and translation is activated in response to eIF2α phosphorylation (Novoa et al., 2003). We therefore speculated that changes in GADD34 expression correlate with the observed SG oscillation pattern. Due to the lack of a specific antibody for detection of GADD34 in our Huh7 cells, we assessed its expression at the mRNA level. Indeed, GADD34 mRNA levels were significantly induced by HCV and IFN-α, while no significant induction was observed by HCV infection alone (Figure 6A). A kinetic analysis of GADD34 and PKR mRNA levels revealed that both were rapidly induced following addition of IFN-α to HCV-infected cells and remained high throughout the 72 hr observation period (Figure 6B). Importantly, upon infection of primary human hepatocytes with HCV, GADD34 was also induced, which in this case was brought about by endogenous IFN-β produced upon viral infection (Figure S5). Thus, IFN-mediated induction of GADD34 is a general feature of liver cells.
Figure 6. GADD34 Controls SG Formation and Cell Viability.
(A) mRNA quantification of GADD34 in Huh7 cells infected with HCV for 24 hr and cultured for 18 hr with or without IFN-α. All values were normalized to GAPDH mRNA levels. Results represent fold induction relative to uninfected cells. Shown are means of triplicate measurements ± SD of a representative experiment. Statistical significance is given in the top. n.s., no statistical significance.
(B) mRNA quantification of PKR and GADD34 in Huh7 cells infected with HCV as given in panel A. Cells were harvested at given time points and mRNA amounts were quantified relative to GAPDH mRNA levels. ISG56 mRNA served as control for IFN-α response of Huh7 YFP-TIA1 cells. Results represent fold induction relative to uninfected cells. Shown are mean values ± SD from three independent experiments. See also Figure S6.
(C–E) Characterization of GADD34 overexpressing cells. (C) Quantification of GADD34 mRNA in parental Huh7 (Ctrl) and Huh7-GADD34 cells, relative to GAPDH mRNA levels. Results represented fold induction relative to Ctrl cells. Shown are means of triplicate measurements ± SD of a representative experiment. (D) Ectopic expression of GADD34 does not affect HCV replication. Cells were transfected with in vitro transcripts of a HCV Renilla reporter virus, lysed 4, 24, 48, 72, and 96 hr later and luciferase activities were determined (Relative Light Units, RLU). (E) Quantification of HCV-induced SGs in Huh7-GADD34 cells (see also Movie S4). Cells were infected with HCV for 24 hr and subsequently treated or not with IFN-α for 18 hr. Cells were fixed, stained for NS5A, and analyzed. Shown are the mean values ± SD from three independent experiments with at least 100 cells analyzed each.
(F) Time-lapse series of Huh7 YFP-TIA1cells that were left untreated or treated with 50 or 75 μM Guanabenz, a GADD34-specific inhibitor. Incubation times with the drug are given in the top.
(G) Cytotoxicity of Guanabenz treatment. Huh7 YFP-TIA1 cells were treated with increasing amounts of Guanabenz for 2, 6, or 10 hr. Cell viability was determined by measuring release of lactate dehydrogenase. Values were normalized to untreated cells (0μM) and are represented as percentage of cell survival.
(H) mRNA quantification of GADD34 in Huh7 YFP-TIA1 cells treated with 50 μM Guanabenz. Cells were collected at given time points and GADD34 mRNA amounts were determined relative to GAPDH mRNA levels. Results represent fold induction relative to untreated cells. Shown are means of triplicate values ± SD. of a representative experiment.
(I) Western blot analysis of Huh7 YFP-TIA1 cells treated for 4 hr with 75 μM Guanabenz (lane 2). Untreated cells were used as reference (lane 1).
To determine the role of GADD34 in SG oscillation, we generated Huh7 cells stably overexpressing GADD34 (Figure 6C). In these cells, SG formation upon HCV infection and IFN-α treatment was significantly reduced despite normal levels of HCV replication (Figures 6D and 6E). Consistently, treatment of noninfected cells with Guanabenz, a recently described specific inhibitor of GADD34 (Tsaytler et al., 2011), induced massive SG formation in 90% of cells without causing cytotoxicity (Figures 6F and 6G, respectively). Of note, SGs formed in noninfected cells in response to Guanabenz did not disassemble and no SG oscillation was observed even though the treatment increased GADD34 transcription (Figure 6H) and eIF2α phosphorylation level (Figure 6I). Higher drug concentrations and longer durations of treatment were cytotoxic, arguing that a strong nonoscillating SG response might lead to cell death. This observation could explain why our attempts to reduce expression of GADD34, a gene essential for cell survival, by using GADD34-specific RNAi strategies failed (data not shown). Moreover, Guanabenz treatment of cells infected with HCV for 24 hr and subsequently treated with IFN-α for 18 hr stopped SG oscillation and led to cell death after 10 hr treatment (Movie S4). These results suggest that cellular stress homeostasis is tightly controlled by a balance between eIF2α kinase and phosphatase activities and that induction of GADD34 is essential to prevent cell death.
SG Oscillation Is Triggered by dsRNA and Is a General Feature of RNA Virus Infection
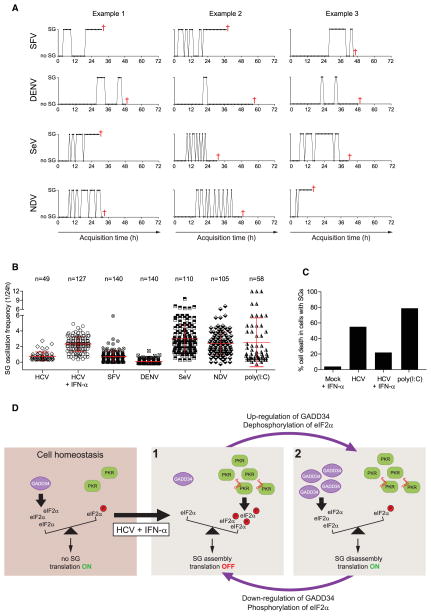
To test if SG oscillation is a general response to infection with RNA viruses, we analyzed whether positive-strand RNA viruses with a 5′ capped genome such as Semliki Forest virus (SFV) and Dengue virus (DENV) as well as negative-strand RNA viruses such as Sendai virus (SeV) and Newcastle Disease virus (NDV) induce SGs similar to those observed with HCV. We note that at variance to HCV, all these RNA viruses are cytolytic. Moreover, since DENV, SeV, and NDV impair the IFN response, for proper comparison all infections were studied in the absence of IFN-α treatment (Randall and Goodbourn, 2008). As expected (McInerney et al., 2005), SFV induced relocalization of YFP-TIA1 from the nucleus to the cytosol and formation of SGs soon after infection (Movie S5). SG oscillation frequency was low and nonoscillating SGs were detected before cell death (Figures 7A and 7B). Likewise, DENV induced very few SGs, which oscillated at low frequency before cell death (Figures 7A and 7B). In contrast, SeV and NDV readily caused formation of SGs with frequencies significantly higher than those detected with HCV-infected cells in the absence of IFN-α (Figures 7A and 7B) and containing canonical SG markers (data not shown). Moreover, we found that fusion of NDV-infected cells, which is mediated by the viral F protein, caused rapid spread of non-oscillating SGs throughout the syncytia, prior to cell death (Movie S6). Except for DENV, which has been reported to dissolve SGs (Emara and Brinton, 2007), the total stress duration in cells infected with all RNA viruses was significantly higher as compared to HCV-infected cells in the absence of IFN-α (Figure S6A).
Figure 7. SG Oscillations Are Triggered by dsRNA.
(A) Oscillation plots of three representative Huh7 YFP-TIA1 cells infected with SFV, DENV, SeV, or NDV. Red cross, cell death.
(B) Scatter plots of SG oscillation frequencies; see also Figure S7 and Movies S5 for SFV; Figure S6 for NDV; and Figure S7 for poly(I:C). Analyses are based on randomly selected Huh7 YFP-TIA1 cells infected with SFV, DENV, SeV, NDV, or transfected with 4 μg poly(I:C). Shown in red are means ± SD. Number of analyzed cells (n) is given in the top. SG oscillation frequencies of HCV-infected cells with SGs are shown as reference.
(C) Quantification of death of Huh7 YFP-TIA1 cells containing SGs. Histograms represent the percentage of death of cells infected with HCV (±IFN-α, respectively), or transfected with poly(I:C). Uninfected cells treated with IFN-α (mock) were used as control.
(D) Model of cell homeostasis and its perturbation by HCV and IFN-α treatment. Left panel: basal levels of phospho-eIF2α are continuously regulated by the PP1 complex and its regulatory subunit GADD34. Under these conditions, sufficient amounts of eIF2α are available for initiation of protein synthesis (translation ON) and SGs are not formed (no SG). This cytoprotective state reflects homeostasis and thus cell survival. 1: Upon HCV infection, dsRNA is formed (red waved lines). Addition of IFN-α limits HCV replication and induces expression of PKR that binds viral dsRNA. Activated PKR phosphorylates eIF2α, thus arresting RNA translation (translation OFF). 2: GADD34 transcription and translation are increased in response to eIF2α hyper-phosphorylation. GADD34 causes rapid dephosphorylation of eIF2α by PP1, restoring RNA translation and promoting SG disassembly and thus stress recovery. Under this condition GADD34 is rapidly downregulated resulting in accumulation of phosphorylated eIF2α. This regulatory circuit sustains SG oscillation that correlate with active and stalled phases of RNA translation.
Having found that SGs are induced upon infection with very diverse RNA viruses, we next determined whether dsRNA per se is sufficient to trigger SG oscillation. To this end we transfected Huh7 YFP-TIA1 cells with high amounts of poly(I:C), a synthetic analog of dsRNA. Poly(I:C) transfection induced massive formation of oscillating SGs clearly detectable 6 hr post transfection (Figure 7B) concomitant with reduced SG oscillation intervals (Figure S6B). Notably, oscillating SGs were still detectable 72 hr after poly(I:C) transfection, arguing for a high intracellular stability of the synthetic RNA. Indeed, purified total RNA extracted from cells at different time points after poly(I:C) transfection was still capable to induce an IFN response as determined with an ISG56 promoter assay (Binder et al., 2007) (Figure S6C). Analogous to what we had observed upon virus infection, the presence of poly(I:C)-induced SGs correlated with delayed cell division (Movie S7) and a rapid and marked increase of GADD34 mRNA levels (Figures S6D and S6E; Figure 6B for HCV).
SG Oscillation and HCV Persistence
Quantification of dying cells that displayed SGs during the 72 hr observation period revealed that in the absence of IFN-α 55% of HCV-infected cells with SGs (Figure 7C), but only 22% of HCV-infected cells that did not form SGs died (data not shown), arguing that SG formation contributes to cell death. This conclusion was supported by results obtained with cells that had been transfected with poly(I:C). In these cases, ~75% of cells with SGs died (Figure 7C). However, IFN-α treatment of HCV-infected cells induced SG formation, concomitant with extended total stress duration (Figure S7B), yet cell death was profoundly reduced and affected only ~18% of cells (Figure 7C). These results suggest that via an IFN-mediated reduction of HCV replication cell survival is prolonged in spite of higher stress induction (see Discussion).
In conclusion, we show that SG formation is a hallmark of infections by RNA viruses. Our data suggest that SG formation is regulated by a delicate balance between dsRNA promoting PKR-mediated phosphorylation of eIF2α and its dephosphorylation by the antagonist GADD34. As a net result cells enter cycles of translational activity and arrest, which critically determine cell survival.
DISCUSSION
HCV infection is known to elicit multiple host cell responses, including oxidative stress (González-Gallego et al., 2011), ER stress (e.g., Li et al., 2009; Joyce et al., 2009; Benali-Furet et al., 2005), and SG formation (Ariumi et al., 2011; Jones et al., 2010). With the aim to study long-term stress responses and their impact on host cell homeostasis, we developed an imaging approach allowing determination of SG dynamics during extended observation periods. This method revealed the highly dynamic and stochastic nature of SG assembly and disassembly and is thus superior to earlier studies relying on static analyses (Souquere et al., 2009) or short-term time-lapse microscopy (Fujimura et al., 2009; Nadezhdina et al., 2010). Importantly, our method avoids illumination-induced cytotoxicity and allowed tracking of motile cells, which are frequent technical limitations of long-term live-cell imaging (Schroeder, 2011). We found that SGs in HCV-infected cells are associated with stalled translation and delayed cell division. In the presence of a stress trigger (such as dsRNA that is sensed by PKR) this process is regulated in a highly dynamic fashion, resulting in rapid switches of eIF2α phopshorylation states and thus, rapid cycles of SG assembly and disassembly (Figure 7D). Phosphorylation of eIF2α by PKR mediates SG formation and translational arrest. This process is antagonized by the upregulation of the regulatory subunit GADD34 of PP1, which allows a rapid dephosphorylation of eIF2α. This negative feedback loop regulation is reminiscent of the mechanism of recovery from ER stress (Ma and Hendershot, 2003; Novoa et al., 2001). Consistent with its role as key regulator of SG oscillation, GADD34 expression is rapidly induced upon stress exposure, but protein amounts are tightly regulated owing to the short half-life of GADD34 (Brush and Shenolikar, 2008; Zhou et al., 2011). Importantly, our analyses reveal that basal levels of GADD34 govern continuous eIF2α dephosphorylation in the absence of exogenous stress and thus determine RNA translation (Figure 7D). In this respect, GADD34 is a central cytoprotective regulator of cellular homeostasis linked to cell survival.
While low SG oscillation frequency was observed in HCV-infected cells, addition of IFN-α dramatically increased SG dynamics. RNA replication of HCV occurs in a membrane-wrapped and nuclease-protected compartment (Quinkert et al., 2005), which probably limits PKR accessibility and sensing of viral dsRNA. This would also explain why HCV replication per se does not interfere with the induction of SGs, e.g., by arsenite treatment (data not shown). However, addition of IFN-α induces PKR expression, thus increasing the likelihood to bind to dsRNA and to trigger a stronger stress response.
Results obtained with poly(I:C) demonstrate that dsRNA alone is sufficient to trigger an oscillating SG response that prevails as long as dsRNA is present. Consistent with its role as IFN-induced eIF2α kinase and dsRNA sensor, PKR was necessary and sufficient for this stress response. Virus-induced PKR activation thus antagonizes GADD34 to potently enhance eIF2α phosphorylation levels, suggesting that SG oscillation results from an HCV-induced shift of the balance of the eIF2α phosphorylation status (Figure 7D). While this model is in full agreement with earlier reports, we cannot exclude that SG formation is induced by a PKR- and eIF2α-independent pathway (White and Lloyd, 2012). However, irrespective of the pathway triggering SG formation in HCV-infected cells treated with IFN-α, SG oscillation avoids extended periods of translational shut off that otherwise leads to cell death (Nakagawa et al., 2000). In contrast, these recovery periods are not sufficient to ensure survival of cells infected by lytic viruses that potently induce cell death by different mechanisms.
Virus-induced formation of SGs has been described as an antiviral host cell defense for viruses requiring cap-dependent translation of their viral RNAs (Emara and Brinton, 2007). In contrast, initiation of translation of the HCV RNA genome is regulated by an IRES supporting protein synthesis under conditions where cap-dependent translation is compromised. Consistent with that our RNAi-based screen revealed no role for PKR or SGs in IFN-mediated suppression of HCV replication. Nevertheless, despite its potent antiviral efficacy, even long-term treatment of HCV-infected cells with IFN-α is not sufficient to clear the virus (Bauhofer et al., 2012). Under conditions of IFN-suppressed HCV replication—that were not reflected in our RNAi screen—we observed that SG oscillation is associated with prolonged cell survival, implying that the balance between host cell stress response and virus replication facilitates HCV persistence. These results suggest that HCV may exploit stress response to establish persistence.
Although SG formation is frequently triggered by viral infection, virus-specific differences in magnitude, dynamics and outcome of the stress response exist (Beckham and Parker, 2008; Schütz and Sarnow, 2007). For instance, unlike Poliovirus, SFV, West Nile virus, and DENV infections cause transient accumulation of SGs early during infection that disappear in the course of the replication cycle owing to virus-mediated blocks in SG formation (McInerney et al., 2005; White et al., 2007; Mazroui et al., 2006; Emara and Brinton, 2007). However, in these studies SGs were monitored only up to 8 hr after infection, at variance to our long-term live-cell imaging in which we observed low-frequency SG oscillations in cells infected with SFV or DENV. Since DENV recruits TIA-1 to the 3′ end of its RNA genome and thus might dissolve SGs (Emara and Brinton, 2007), we also studied DENV-infected Huh7 cells stably expressing GFP-G3BP1 (data not shown). These studies confirmed the oscillation of SGs in DENV-infected cells, thus excluding experimental artifacts due to the use of the TIA-1 marker.
In conclusion our study identified a highly dynamic oscillation of the cellular stress response that is regulated by antagonistic actions of the dsRNA sensor PKR and the phosphatase PP1 controlling eIF2α phosphorylation and, thus, the translation state of the cell. This oscillation prolongs cell survival by avoiding extended phases of translation shut-off. As exemplified here with HCV, viruses may exploit this system to sustain chronicity.
EXPERIMENTAL PROCEDURES
Infection and Induction of SGs
For HCV infection, 1 × 105 Huh7 or Huh7 YFP-TIA1 cells were seeded into a 6-well plate. Cells were infected with HCV at a moi of 5 TCID50 per cell for 24 hr and subsequently treated or not with IFN-α (PBL Laboratories) as described in the main text. For steady-state experiments, cells were treated for 18 hr with 100 IU/ml IFN-α and harvested for further analysis or fixed for immunostaining.
Stress Induction and Drug Treatment
Oxidative stress was induced by treating cells with 0.5 μM Na-Arsenite (Sigma) contained in culture medium for 1 hr at 37°C. Heat shock was induced by incubating cells at 42°C for 20 min or at 44°C for 1 hr. Metabolic stress was induced by incubation of cells with 10 μM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (Sigma) in glucose-free DMEM for 1 hr or 12 hr. ER stress was induced by incubation of cells with DMEM containing 10 μM Thapsigargin (Biotrends) for 1 hr at 37°C. Guanabenz Acetate Salt (Sigma) was used to inhibit GADD34. Live-imaging experiments were performed by using 50 μM or 75 μM Guanabenz for 24 hr as specified in the main text.
RNAi-Based Replication Rescue Screen
Gene silencing in Huh7 cells was achieved by two consecutive transfections. Eight hours post silencing, cells were infected with the HCV Renilla reporter virus for 24 hr and subsequently treated or not with 25 IU/ml IFN-α for 60 hr. Five independent repetitions of the RNAi-based screen were performed. Raw data were subjected to statistical analysis (see Supplemental Experimental Procedures for details).
Long-Term Live-Cell Laser Confocal Imaging
Huh7 YFP-TIA1 cells were infected with HCVTCP for 24 hr. Cells were extensively washed and incubated in microscopy medium containing 25 mM HEPES and 10% FCS. For IFN-α treatment, 100 IU/ml IFN-α were added to the medium. Confocal time series of YFP-TIA1 and NS5A-mCherry acquisition was started 6 hr after medium change and equilibration (see Supplemental Experimental Procedures for details).
Metabolic Labeling of Proteins and Quantification of Fluorescence Intensities
De novo synthesized proteins were quantified by using the Click-iT Protein Kit (Molecular Probes, Life Technologies) and a protocol adapted from Dieterich and colleagues (Dieterich et al., 2010). Cells were further stained for eIF3B (Alexa Fluor 488). Fluorescence intensities were quantified by using the ImageJ software package (see Supplemental Experimental Procedures for details).
Statistical Analyses
Statistical analysis was performed by using the GraphPad Prism software (GraphPad). Statistical significance was calculated by performing a two-tailed Student’s t test (***, p < 0.0001; **, p < 0.001; *, p < 0.01).
Supplementary Material
Acknowledgments
Special thanks to J. Reymann (Screening facility, Bioquant, University of Heidelberg, Germany) and M. Kiesel (Nikon, Germany) for excellent support with microscopy. We thank the Nikon Imaging Center (Heidelberg University) for providing access to their facility. We are grateful to U. Herian for excellent technical assistance, N. Kedersha (Harvard Medical School, Boston, USA) for helpful advice, and O. Fackler (Heidelberg University) for constructive discussions and critical manuscript reading. We thank C. Rice, T. Wakita, M. Harris, G. Sen, R. Zawatski, and G. Kochs for provision of reagents.
This work was supported by the Deutsche Forschungsgemeinschaft (FOR1202 TP1 and TP3 and SFB/TRR77, TP A1, to R.B. and V.L.), the Bundesministerium für Bildung und Forschung (01 KI 0786 to R.B.), the European Union (SysPatho to L.K., R.B., and K.R.), the Cell-Networks Cluster of Excellence and U.S. National Institutes of Health (AI-12520 and -20611to C.E.S.). E.D. was supported by the Viroquant project and P.M. by a Viroquant HBIGS-Stipendium. G.S. was supported by a bridging project grant from the DKFZ-ZMBH Alliance.
Footnotes
Supplemental Information includes six figures, seven movies, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2012.05.013.
References
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol. 2011;85:6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauhofer O, Ruggieri A, Schmid B, Schirmacher P, Bartenschlager R. Persistence of HCV in Quiescent Hepatic Cells Under Conditions of an Interferon-Induced Antiviral Response. Gastroenterology. 2012 doi: 10.1053/j.gastro.2012.04.018. in press. Published online April 20, 2012. http://dx.doi.org/10.1053/j.gastro.2012.04.018. [DOI] [PubMed]
- Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benali-Furet NL, Chami M, Houel L, De Giorgi F, Vernejoul F, Lagorce D, Buscail L, Bartenschlager R, Ichas F, Rizzuto R, Paterlini-Bréchot P. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene. 2005;24:4921–4933. doi: 10.1038/sj.onc.1208673. [DOI] [PubMed] [Google Scholar]
- Binder M, Kochs G, Bartenschlager R, Lohmann V. Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy. Hepatology. 2007;46:1365–1374. doi: 10.1002/hep.21829. [DOI] [PubMed] [Google Scholar]
- Brush MH, Shenolikar S. Control of cellular GADD34 levels by the 26S proteasome. Mol Cell Biol. 2008;28:6989–7000. doi: 10.1128/MCB.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol. 2001;21:6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Brinton MA. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci USA. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Fujimura K, Katahira J, Kano F, Yoneda Y, Murata M. Microscopic dissection of the process of stress granule assembly. Biochim Biophys Acta. 2009;1793:1728–1737. doi: 10.1016/j.bbamcr.2009.08.010. [DOI] [PubMed] [Google Scholar]
- García MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Gélinas JF, Clerzius G, Shaw E, Gatignol A. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J Virol. 2011;85:8460–8466. doi: 10.1128/JVI.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Gallego J, García-Mediavilla MV, Sánchez-Campos S. Hepatitis C virus, oxidative stress and steatosis: current status and perspectives. Curr Mol Med. 2011;11:373–390. doi: 10.2174/156652411795976592. [DOI] [PubMed] [Google Scholar]
- Hershey JW. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce MA, Walters KA, Lamb SE, Yeh MM, Zhu LF, Kneteman N, Doyle JS, Katze MG, Tyrrell DL. HCV induces oxidative and ER stress, and sensitizes infected cells to apoptosis in SCID/Alb-uPA mice. PLoS Pathog. 2009;5:e1000291. doi: 10.1371/journal.ppat.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskinen P, Nyqvist M, Sareneva T, Pirhonen J, Melén K, Julkunen I. Impaired antiviral response in human hepatoma cells. Virology. 1999;263:364–375. doi: 10.1006/viro.1999.9983. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Back SH, Kim V, Ryu I, Jang SK. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol Cell Biol. 2005;25:2450–2462. doi: 10.1128/MCB.25.6.2450-2462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima E, Takeuchi A, Haneda M, Yagi A, Hasegawa T, Yamaki K, Takeda K, Akira S, Shimokata K, Isobe K. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB J. 2003;17:1573–1575. doi: 10.1096/fj.02-1184fje. [DOI] [PubMed] [Google Scholar]
- Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem Soc Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- Li S, Ye L, Yu X, Xu B, Li K, Zhu X, Liu H, Wu X, Kong L. Hepatitis C virus NS4B induces unfolded protein response and endoplasmic reticulum overload response-dependent NF-kappaB activation. Virology. 2009;391:257–264. doi: 10.1016/j.virol.2009.06.039. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljeström P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell. 2005;16:3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs E, Chong K, Galabru J, Thomas NS, Kerr IM, Williams BR, Hovanessian AG. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- Nadezhdina ES, Lomakin AJ, Shpilman AA, Chudinova EM, Ivanov PA. Microtubules govern stress granule mobility and dynamics. Biochim Biophys Acta. 2010;1803:361–371. doi: 10.1016/j.bbamcr.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003;22:1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinkert D, Bartenschlager R, Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Schroeder T. Long-term single-cell imaging of mammalian stem cells. Nat Methods. 2011;8(4, Suppl):S30–S35. doi: 10.1038/nmeth.1577. [DOI] [PubMed] [Google Scholar]
- Schütz S, Sarnow P. How viruses avoid stress. Cell Host Microbe. 2007;2:284–285. doi: 10.1016/j.chom.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Smith JA, Schmechel SC, Raghavan A, Abelson M, Reilly C, Katze MG, Kaufman RJ, Bohjanen PR, Schiff LA. Reovirus induces and benefits from an integrated cellular stress response. J Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci. 2009;122:3619–3626. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- Steinmann E, Brohm C, Kallis S, Bartenschlager R, Pietschmann T. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J Virol. 2008;82:7034–7046. doi: 10.1128/JVI.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Puig M, Darnell ME, Mihalik K, Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol. 2005;79:6291–6298. doi: 10.1128/JVI.79.10.6291-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomis DC, Doohan JP, Samuel CE. Mechanism of interferon action: cDNA structure, expression, and regulation of the interferon-induced, RNA-dependent P1/eIF-2 alpha protein kinase from human cells. Virology. 1992;188:33–46. doi: 10.1016/0042-6822(92)90732-5. [DOI] [PubMed] [Google Scholar]
- Tsaytler P, Harding HP, Ron D, Bertolotti A. Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science. 2011;332:91–94. doi: 10.1126/science.1201396. [DOI] [PubMed] [Google Scholar]
- Vrolijk JM, Kaul A, Hansen BE, Lohmann V, Haagmans BL, Schalm SW, Bartenschlager R. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J Virol Methods. 2003;110:201–209. doi: 10.1016/s0166-0934(03)00134-4. [DOI] [PubMed] [Google Scholar]
- White JP, Lloyd RE. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Cardenas AM, Marissen WE, Lloyd RE. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Wu S, Kaufman RJ. A model for the double-stranded RNA (dsRNA)-dependent dimerization and activation of the dsRNA-activated protein kinase PKR. J Biol Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- Xin Z, Han W, Zhao Z, Xia Q, Yin B, Yuan J, Peng X. PCBP2 enhances the antiviral activity of IFN-α against HCV by stabilizing the mRNA of STAT1 and STAT2. PLoS ONE. 2011;6:e25419. doi: 10.1371/journal.pone.0025419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Brush MH, Choy MS, Shenolikar S. Association with endoplasmic reticulum promotes proteasomal degradation of GADD34 protein. J Biol Chem. 2011;286:21687–21696. doi: 10.1074/jbc.M110.212787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.