Abstract
Increased interest in the pathogenic potential of Yersinia pestis has emerged because of the potential threats from bioterrorism. Pathogenic potential is based on genetic factors present in a population of microbes, yet most studies evaluating the role of specific genes in virulence have used a limited number of strains. For Y. pestis this issue is complicated by the fact that most strains available for study in the Americas are clonally derived and thus genetically restricted, emanating from a strain of Y. pestis introduced into the United States in 1902 via marine shipping and subsequent spread of this strain throughout North and South America. In countries from the former Soviet Union (FSU), Mongolia, and China there are large areas of enzootic foci of Y. pestis infection containing genetically diverse strains that have been intensely studied by scientists in these countries. However, the results of these investigations are not generally known outside of these countries. Here we describe the variety of methods used in the FSU to classify Y. pestis strains based on genetic and phenotypic variation and show that there is a high level of diversity in these strains not reflected by ones obtained from sylvatic areas and patients in the Americas.
INTRODUCTION
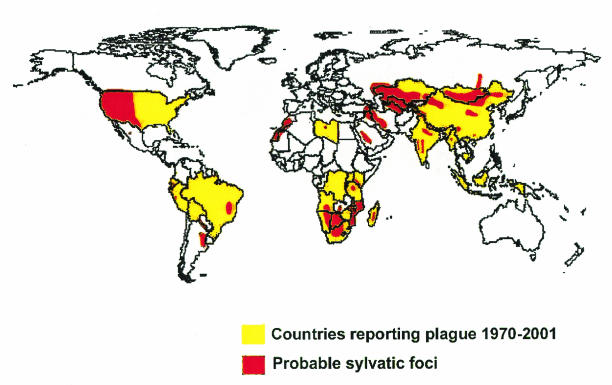
Plague is a zoonotic infection that is spread to humans from natural rodent reservoirs, commonly via the bite of an infected flea. Yersinia pestis, the causative agent of bubonic, septicemic, pneumonic, pharyngeal, cutaneous, and enteric plague as well as plague meningitis, can be found in populations of more than 200 species of wild rodents which inhabit natural plague foci in all the continents save Australia (Fig. 1). Over 80 species of fleas are proven vectors of plague (7-9, 11, 13, 28, 30, 31, 34, 49, 55, 56, 104, 132, 135, 141, 146, 160). Utilizing such a broad host and vector range provides a large opportunity for genetic diversity and natural selective forces to generate considerable variability in the Y. pestis genome. Yet much of what is known about the genetic and phenotypic properties of Y. pestis comes from studies of a limited number of strains commonly found in the Americas, wherein there is very restricted genetic diversity. Thus, much of the pathogenic potential of Y. pestis for humans remains largely unknown, locked away in the multitude of strains circulating in natural foci, many of which are found in isolated regions of Russia and Asia and are not easily accessible to many researchers.
FIG. 1.
Global distribution of plague. Reprinted with permission from K. L. Gage and J. A. Montenieri, Centers for Disease Control and Prevention, Fort Collins, Colo.
Enzootic circulation of Y. pestis in natural plague foci requires active infection of host rodents and growth in the fleas to produce blockage by a large mass of bacilli in the proventriculus, a sphincter-like organ that separates the stomach and esophagus. These factors are essential for continued transmission of the bacilli to new hosts and thus for maintenance of the infectious focus in a natural environment. The organism must be able to resist host defense systems, multiply, and cause bacteremia for further transmission by fleas to a new host. Each of these stages of the Y. pestis life cycle is dependent on elaboration of specific bacterial virulence factors that may act in concert or separately (7-9, 11, 13, 28, 30, 31, 34, 49, 55, 56, 104, 132, 135, 141, 146, 160, 179). Many of the natural plague foci are geographically not connected, resulting in considerable ecological differences needed for Y. pestis to survive and be transmitted in these different environments. This results in considerable diversity in genotype and phenotype among plague isolates from different natural foci (1, 2, 9, 11, 16-18, 23-25, 31, 33, 52, 55, 56, 60, 67-69, 104, 107, 109, 120-122, 127, 130, 132, 135, 140, 141, 169, 172, 177, 179, 181, 184, 198; M. I. Levi, Abstr. Sci. Conf. Natur. Focality Prophyl. Plague Tularemia, p. 72-74, 1962; F. Zhenya, Z. Xiang, L. Yunheng, L. Jun, W. Shenrong, Z. Yaoxing, J. Lingling, and L. Feng, Abstr. 7th Int. Symp. Yersinia, abstr. P-127, Med. Microbiol. [Ned. N. Voor] 6[Suppl. II]: S42, 1998).
As the current concern with use of microbes as agents of bioterrorism grows, it will be essential to gain a fuller understanding of the variety of strains of Y. pestis that can be found in the world and the effectiveness of countermeasures taken to control, prevent, and treat plague infections. Much of what is known is not available in English language publications, since many of the studies are published in Russian language journals. Therefore, we have compiled information from around the world to summarize what is known about diversity in isolates of Y. pestis. Given the natural genetic and phenotypic diversity in an organism that has very high pathogenic potential for humans, it is essential to understand how genetic and phenotypic variation impacts the pathogenesis, diagnosis, treatment, and development of immunologic therapies for plague.
GLOBAL DISTRIBUTION OF YERSINIA PESTIS STRAINS
In spite of being present on all continents save Australia, Y. pestis is not widespread throughout all of the world. Natural plague foci cover 6 to 7% of the dry land of the Earth. While notorious as a cause of disease in Europe for a long period, it is now notably absent from Western and Central Europe as well as Canada and parts of North and South America (Fig. 1). In the most northerly and southerly parts of the world, winter temperatures may preclude maintenance of the plague transmission cycle of flea to rodent. Obviously vigorous quarantine and public health measures in places such as Australia can effectively prevent plague from taking hold in that continent, although the island nation of Madagascar nonetheless now has endemic foci of plague and human plague outbreaks due to introduction of the organism via marine shipping. Quarantine and public health measures probably limit the occurrence of human plague in the more developed countries of the world, but it is not clear how this prevents endemic foci of plague from becoming established in wild rodents. There is obviously much complexity to plague, flea, and rodent biology and the ecology of Y. pestis transmission that would impact the sustainability of the organism in sylvatic foci in different parts of the world.
Geographical Distribution of Plague in the Former Soviet Union
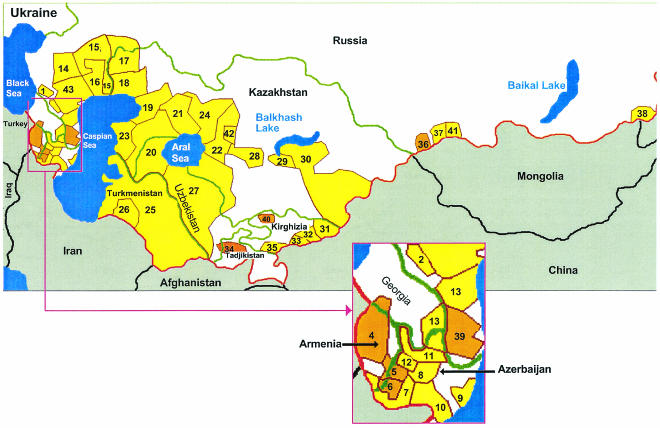
Forty-three natural foci are found in the southern and southeastern regions of the former Soviet Union (FSU) (Fig. 2; Table 1). They cover more than 216 × 106 hectares, or 8.6% of its territory (132). Separation and subsequent classification of these natural plague foci was performed first on the basis of their geographical distribution and then on the basis of the primary infected host found in each focus. Primary hosts from different foci have several ecological characteristics in common with each other, such as a large and steady quantity of the rodent hosts, infectious bacteremia as one of the stages of the disease, flea parasites that serve as active plague vectors, and fleas that can easily survive within rodent's burrows and nests. As a rule, these features are not necessary for infection of secondary hosts. Adaptation for continued transmission of the plague pathogen within different rodent species is assumed to contribute strongly to the emergence of variant Y. pestis subspecies, differentiated by fermentative activity, nutritional requirements, and ability to cause infectious bacteremia and death in different animal species (157).
FIG. 2.
Distribution of natural plague foci in the FSU. Identifications of the plague foci are given in Table 1 and the text. The red line indicates the FSU frontier; green lines indicate the frontiers of the states of the FSU; brown lines indicate boundaries of the plague foci; black lines indicate frontiers of other countries. Plague foci are yellow (for foci containing the main Y. pestis subspecies) or light brown (for foci containing non-main Y. pestis subspecies). This figure is based on references 17, 104, 132, 135, and 169.
TABLE 1.
Relevant characteristics of the natural plague foci in the FSUa
| Focus no. | Focus designation | Epizootic activityb | Main host | Main vector(s) | Country |
|---|---|---|---|---|---|
| Caucasian region | |||||
| 1 | Central Caucasian | Constant | Citellus musicus | Citellophillus tesquorom | Russia |
| 4 | Transcaucasian highland (Leninakan) | Constant | Microtus arvalis | Callopsylla caspia, Nosopsyllus consimilis | Armenia, Georgia |
| 5 | Transcaucasian highland (Pre-Sevan) | Constant | Microtus arvalis | C. caspia, N. consimilis | Armenia, Azerbaijan |
| 6 | Transcaucasian highland (Zanzegur-Karabakh) | Constant | Microtus arvalis | C. caspia, N. consimilis | Armenia, Azerbaijan |
| 7 | Pre-Araks | Recurring | Meriones vinogradov | Xenopsylla conformis | Armenia, Azerbaijan |
| 8 | Bozchel' | Recurring | Meriones libicus | X. conformis, Nosopsyllus laeviceps | Azerbaijan |
| 9 | Kobystan | Recurring | M. libicus | X. conformis, N. laeviceps | Azerbaijan |
| 10 | Mila-Karabakh | Recurring | M. libicus | X. conformis, N. laeviceps | Azerbaijan |
| 11 | Dzheiranchel' | Recurring | M. libicus | X. conformis, N. laeviceps | Azerbaijan |
| 12 | Gyanzha-Kazakh | Recurring | M. libicus | X. conformis, N. laeviceps | Azerbaijan |
| 13 | Iori | Recurring | M. libicus | X. conformis, N. laeviceps | Azerbaijan, Georgia |
| 39 | Dagestan-highland | Recurring | Microtus arvalis | C. caspia | Russia |
| North Pre-Caspian region | |||||
| 2 | Terek-Sunzha | Recurring | Citellus pygmaeus | Neopsylla setosa, C. tesquorum | Russia |
| 3 | Dagestan plain foothill | Recurring | C. pygmaeus | N. setosa, C. tesquorum | Russia |
| 14 | Pre-Caspian northwestern | Recurring | C. pygmaeus | N. setosa, C. tesquorum | Russia |
| 15 | Volga-Ural steppe | Recurring | C. pygmaeus | N. setosa, C. tesquorum | Russia |
| 17 | Trans-Ural | Recurring | C. pygmaeus | N. setosa, C. tesquorum | Russia, Kazakhstan |
| 43 | Pre-Caspian sandy | Recurring | Meriones meridianus | N. laeviceps | Russia |
| 16 | Volga-Ural sandy | Recurring | M. meridianus | X. conformis, N. laeviceps | Russia |
| Central Asian Desert region | |||||
| 18 | Ural-Emba | Constant | Rhombomys opimus | X. skrjabini | Kazakhstan |
| 19 | Pre-Ustyurt | Constant | R. opimus | X. skrjabini | Kazakhstan |
| 20 | Ustyurt | Recurring | R. opimus | X. skrjabini | Kazakhstan, Uzbekistan |
| 21 | North-Pre-Aral | Recurring | R. opimus | X. skrjabini | Kazakhstan, Uzbekistan |
| 22 | Trans-Aral | Recurring | R. opimus | X. skrjabini | Kazakhstan |
| 23 | Mangyshlak | Recurring | R. opimus | X. skrjabini | Kazakhstan |
| 24 | Pre-Aral-Kara-Kum | Constant | R. opimus | X. skrjabini | Kazakhstan |
| 25 | Kara-Kum | Recurring | R. opimus | X. gerbilli | Turkmenistan |
| 26 | Kopet-Dag | Recurring | R. opimus | X. gerbilli | Turkmenistan |
| 27 | Kyzyl-Kum | Recurring | R. opimus | X. gerbilli | Uzbekistan, Kazakhstan |
| 28 | Muyun-Kum | Recurring | R. opimus | X. gerbilli | Kazakhstan |
| 29 | Tau-Kum | Recurring | R. opimus | X. gerbilli | Kazakhstan |
| 30 | Pre-Balkhash | Recurring | R. opimus | X. gerbilli | Kazakhstan |
| 42 | Betpak-Dala | Recurring | R. opimus | X. gerbilli | Kazakhstan |
| Central Asian Mountain region | |||||
| 31 | Sarydzhaz | Recurring | Marmota baibacina | Oropsylla silantiewi | Kirghizia |
| 32 | Upper-Naryn | Recurring | M. baibacina | O. silantiewi | Kirghizia |
| 33 | Aksai | Recurring | M. baibacina | O. silantiewi | Kirghizia |
| 34 | Gissar | Recurring | Microtus carruthersi | C. caspia, Amphipsylla phaiomydis, Frontopsylla elata, N. pleskei | Tadjikistan, Uzbekistan |
| 35 | Alai | Recurring | Marmota caudata | O. silantiewi | Kirghizia |
| 40 | Talas | Recurring | Microtus gregalisc | C. caspia | Kirghizia |
| Siberian region | |||||
| 36 | Mountain-Altai | Recurring | Ochotona pricei | Paradoxopsyllus scorodumovi | Russia |
| 37 | Tuva (Mongun-Taigin) | Recurring | Citellus undulatus | C. tesquorum | Russia |
| 41 | Tuva (Sagly) | Recurring | C. undulatus | C. tesquorum | Russia |
| 38 | Trans-Baikal | Recurring | Citellus dauricus | C. tesquorum | Russia |
Constant activity indicates activity during the entire period of the epizootic observations. Recurring activity indicates that epizootics were interrupted by 2- to 10-year periods between outbreaks.
Plague in the Americas
Y. pestis was first introduced into the United States at the turn of the 20th century through the port of San Francisco during the third pandemic, which is still ongoing. There were hundreds of deaths from plague in the first quarter of the 20th century, with the last major outbreak of pneumonic plague occurring in Los Angeles in 1924 to 1925. Since then, most cases in the United States have been found in individuals living in sylvatic areas close to foci of plague circulation in rodents. The fact that Y. pestis is essentially a recently introduced pathogen into the Americas indicates that the genetic and phenotypic diversity of American isolates is relatively restricted, particularly compared to those from Central and East Asia. Thus, strains of Y. pestis from patients and collections in the Americas that are readily available to investigators are likely to be much less genetically and phenotypically diverse than strains from other parts of the world.
Current Assessment of Plague Diversity
A major study by Achtman et al. (2) proposed that Y. pestis is a recently emerged clone of Yersinia pseudotuberculosis, since the authors found that within five housekeeping and one lipopolysaccharide (LPS) biosynthesis gene there was essentially no genetic diversity among 36 globally diverse Y. pestis strains. The Y. pestis alleles were identical or nearly identical to those in 12 strains of Y. pseudotuberculosis. By taxonomic standards, Y. pestis might be considered to be Y. pseudotuberculosis, but because of large differences in disease manifestations and the role of Y. pestis in human disease and history, this grouping has not been pursued. Plasmid content and perhaps other small genetic differences are thought to account for the different diseases caused by Y. pestis and Y. pseudotuberculosis. Obviously if plasmid content and small genetic differences can account for the differences in the host range and virulence of Y. pestis and Y. pseudotuberculosis, it might be reasonable to assume that similar small differences could arise is the multitude of Y. pestis strains found in natural foci of infection and that this could potentially have important impacts on the pathogenesis of Y. pestis and the manifestations of plague.
Achtman et al. (2) also performed a restriction fragment length polymorphism analysis of 44 strains of Y. pestis, using a probe for the IS100 element, and found by constructing a neighbor-joining phylogenetic tree that three major biovars of Y. pestis, which have been proposed as a basis for further differentiation of Y. pestis strains and have been designated Antiqua, Medievalis, and Oreintalis, were each composed of closely related strains and that all of the strains were derived from a common ancestor. However, a closer analysis of the strains used by Achtman et al. (2) shows that only six of the strains were from European Russia or Central Asia (Kurdistan) and that five of the six were all of the highly related Medievalis biovar. No data were reported for the sixth strain. The remainder of the strains were from non-Central Asian parts of the world. Achtman et al. (2) also noted that Y. pestis, like Mycobacterium tuberculosis, has a relatively uniform genetic structure, but endonuclease restriction analysis suggests more diversity (85, 119). Overall, whether the Y. pestis isolates from diverse natural foci in European Russia and Central Asia share a similar close genetic relatedness has not been addressed, leaving open the possibility that these strains may have virulence properties or pathogenic potential distinct from the more closely related isolates of Y. pestis that have been found to date to have limited genetic diversity.
HISTORICAL METHODS OF DISTINCTION OF YERSINIA PESTIS STRAINS IN EUROPEAN RUSSIA AND CENTRAL ASIA
Intraspecific Taxonomy
Many studies have been carried out over the years by investigators in the FSU to attempt to classify Y. pestis and understand the diversity of phenotypic traits in this species. Early studies (1928) by Bezsonova (23) divided Y. pestis strains into two varieties on the basis of their ability to ferment glycerol (i.e., glycerol positive and glycerol negative). Glycerol-negative strains were reclassified in 1938 by Berlin and Borzenkov (22) into the oceanic variety, since they were usually isolated from rats in seaports, and glycerol-positive strains, termed the continental variety, since such strains were isolated from “wild” rodents, susliks (ground squirrels), gerbils, etc., from natural plague foci. The designation of glycerol-negative plague strains as the oceanic variety reflected their predominant distribution at the time, but it also appears that the source of these strains may have been the Yunnan interior region in China, which is close to the border of present-day Myanmar (Burma), suggesting that glycerol-negative strains were present in southeastern Asia before the outbreak of the third pandemic in the late 19th century. However, a number of investigators have found in the southern part of Vietnam only anthropogenic plague foci in inhabited localities but no natural foci (176). The main flea vector in Vietnam was Xenopsylla cheopis, while the main rodent host was Rattus exulans. The lack of sylvatic plague foci in Vietnam indicates there were not widespread foci of plague throughout Southeast Asia. Nowadays, glycerol-negative strains are found in natural plague foci located in the United States, South Africa, and Southeast Asia as such strains spread to these regions via marine shipping from Hong Kong starting in 1894 and during the following years of the third pandemic (2, 141).
Devignat in 1951 (52) and Tumanskii in 1957 (181) used glycerol fermentation, nitrate reduction, and ammonia oxidation to classify Y. pestis into three intraspecific groups that were named Orientalis, Antiqua, and Medievalis by Devignat (Table 2). The Devignat classification is currently widely used, referring to strains as belonging to biovars, although some Y. pestis strains cannot be classified into any of these three biovars (3, 130, 155, 156, 174). Melibiose fermentation was found by Mollaret and Mollaret (127) to distinguish biovar Orientalis and biovar Antiqua strains, neither of which ferments this sugar, from most biovar Medievalis strains, which do ferment it. However, it should be noted that biovar characteristics are unstable and that one strain can undergo spontaneous phenotypic variation which would cause it to be classified into another biovar (96, 106). Additionally, strains identical in essentially all of their studied characteristics but differing in their biovars may circulate within one rodent population (106; S. V. Balakhonov, personal communication).
TABLE 2.
Subgroups of Y. pestis identified on the basis of glycerol fermentation, nitrate reduction, and ammonia oxidationa
| R. Devignat's varieties | V. M. Tumanskii's varieties | Differences
|
Areas in which strains were isolated | ||||
|---|---|---|---|---|---|---|---|
| Glycerol fermentation | Nitrate reduction | Ammonia oxidationb | Urease activityc | Fermentation of melibiosec | |||
| Antiqua (the suspected cause of Justinian's plague, 541-767 AD) | Marmotaed | + | + | ± | − | − | Central Africa, Central and Northern Asia, China (Manchuria), Mongolia |
| Medievalis (the suspected cause of the Black Death and subsequent epidemics from 1346 to the early 19th century) | Citelli | + | − | − | − | ± | FSU (Caspian Sea region) |
| Orientalis (the suspected cause of the third pandemic that has spread globally via marine shipping from Hong Kong starting in 1894) | Ratti | − | + | ± | ± | − | Burma, Southern China, India, South Africa, South America, California |
This test was used only for V. M. Tumanskii's classification (181).
Varieties were named according to the main host of the corresponding Y. pestis subgroup in a given plague focus. The main (132, 135, 157) or enzootic (141) host is responsible for maintenance of Y. pestis in natural foci, while secondary (132, 135,157) or epizootic (141) hosts are only infected during epizootics, although they may also play a significant epidemiological role in spreading infection. Accidental hosts (132, 135, 157) are animals such as predators and insectivores (polecats, foxes, weasels, shrews, etc.) as well as muskrats, saigas, camels, and domestic cats. The most dangerous accidental hosts in terms of their capacity for transmitting plague to humans are camels and domestic cats (40, 63, 75,132).
Subsequently, new natural plague foci containing Y. pestis variants with some additional characteristics differentiating them from previously described strains were discovered. This led to changes in the intraspecific classification of Y. pestis. The differential biochemical characteristics used in these groupings were nitrate reduction and ammonia oxidation; fermentation of sugars such as rhamnose, arabinose, melibiose, melezitose, maltose, mannose, and trehalose; pesticin-fibrinolytic-coagulase activities; nutritional requirements; susceptibility to pesticin 1; and virulence for mice and guinea pigs (11). All of these classification schemes were published in Russian based on the recovery of plague isolates from around the FSU. While newer, more specific classifications are now available, the diversity in phenotypic characteristics of Y. pestis isolates from the FSU highlights the importance of expanded studies to include such strains in evaluations of factors of plague pathogenesis and host immunity.
A major question concerning these diverse Y. pestis isolates is that of their pathogenic potential for different hosts, including humans. Levi (Abstr. Sci. Conf. Nat. Focality Prophyl. Plague Tularemia, 1962) supplemented Y. pestis intraspecific differentiation with several additional tests that allowed him to designate two additional Y. pestis subgroups, gerbil and vole (Table 3), using such specific characteristics as host-parasite interactions and selective virulence in laboratory animals, including development of bacteremia. However, the induction of bacteremia was not very reproducible, nor could it be routinely applied for diagnosis, and so the use of this test in the taxonomic classification of Y. pestis was discontinued (11).
TABLE 3.
Classification of Yersinia pestis subgroups by M. I. Levia
| Y. pestis variety | Differential characteristics
|
||||||
|---|---|---|---|---|---|---|---|
| Fermentation of:
|
Nitrate reduction | Virulence for guinea pigs | Infectious bacteriemiab
|
||||
| Glycerol | Rhamnose | Pygmy suslik | Left-bank midday gerbilc | Common vole | |||
| Rat's | − | − | + | + | NDe | ND | ND |
| Marmot's | + | − | + | + | + | − | + |
| Suslik's | + | − | − | + | + | − | + |
| Gerbil's | + | − | − | + | + | + | + |
| Vole's | + | + | ±d | − | − | − | + |
This table was compiled from reference 11 and M. I. Levi (Abstr. Sci. Conf. Nat. Focality Prophyl. Plague Tularemia, 1962), modified from reference 11, and used with the permission of the copyright holder (G. P. Aparin and E. P. Golubinskii).
Maintenance of plague in nature is completely dependent on cyclic transmission between fleas and rodents. An infectious bacteremia of at least 104 CFU/ml can ensure ingestion of a number of bacteria sufficient for flea blockage, leading to infection of a new mammalian host (7-9, 11,13, 28, 30, 31, 34, 49, 55, 56, 104, 132, 135, 141, 146,179).
Midday gerbils living on the opposite banks of the Volga river differ in their susceptibilities to lethal plague infection. LD50s for the left-bank midday gerbils, which are living in the natural plague focus, are 2 orders of magnitude more than those of the right-bank population (56).
Presence of a character but not in all strains.
ND, no data.
Numerical Taxonomy and Standardization of Yersinia pestis Classification in the Former Soviet Union
Using numerical taxonomy, I. L. Martinevskii (120) classified Y. pestis into three varieties: mediaasiatica montana (corresponding to biovar Antiqua), mediaasiatica deserta (corresponding to biovar Medievalis), and oceanica (corresponding to biovar Orientalis). He also concluded that strains isolated from common voles in natural foci of infection in the Transcaucasian highlands (Fig. 2, foci 4, 5, and 6) or from Mongolian pikas in the Mountain Altai and Transbaikalian regions (foci 36 and 38) were a different species, Yersinia pestoides, and included three varieties: Yersinia pestoides parvocaucasica, Yersinia pestoides altaica, and Yersinia pestoides transbaicalica, respectively. Surprisingly, after three decades, “Pestoides” reappeared in publications emanating from the United States as a strain designation and as part of the nomenclature used to classify strains imported from the FSU (3, 103, 130, 155, 156; P. L. Worsham and C. Roy, Abstr. 8th Int. Symp. Yersinia, abstr. P-41, 2002), although this classification was practically forgotten in the FSU. While not in use in the FSU, this classification scheme nonetheless further indicates the amount of genetic and phenotypic diversity in plague isolates related to the sylvatic areas in which they are circulating as epizootic pathogens.
Timofeeva (177) proposed a new classification of Y. pestis into subgroups, which was based on numerical taxonomy, and used subspecies as a taxon designator (Table 4). This classification was formulated subject to the International Code of Bacterial Taxonomy and used the nomenclature indicating the main species and subspecies, (i.e., Yersinia pestis subsp. pestis). She further divided the main Yersinia pestis subspecies into two more groups, continental and oceanic. Also, with the help of numerical taxonomy, Peisakhis and Stepanov (140) proposed a classification of Y. pestis strains which were isolated in the FSU into groups based on 25 phenotypic features, some of which are shown in Table 5. Since these numerical taxonomic classifications were constructed to include the Y. pestis isolates that were found in the territory of the FSU and also not generally available outside of the FSU, additional assessments of non-FSU strains were not included.
TABLE 4.
Classification of Yersinia pestis subgroups by L. A. Timofeevaa
| Y. pestis subspecies | Differential characteristics
|
||||||
|---|---|---|---|---|---|---|---|
| Fermentation of:
|
Nitrate reduction | Pesticin, fibrinolytic, and coagulase activities | Susceptibility to pesticin 1 | Virulence for guinea pigs | |||
| Rhamnose | Arabinose | Glycerol | |||||
| pestis (continental group) | − | + | + | ± | + | − | + |
| pestis (oceanic group) | − | + | − | ± | + | − | + |
| altaica | + | − | + | − | + | + | − |
| caucasica | + | + | + | + | − | + | − |
TABLE 5.
Classification of Yersinia pestis subgroups by L. A. Peisakhis and V. M. Stepanova
| Y. pestis subspecies | Differential characteristics
|
Notes | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Fermentation of:
|
Nitrate reduction | Pesticin, fibrinolytic, and coagulase activities | Susceptibility to pesticin 1 | Additional growth factors | Virulence for guinea pigs | ||||
| Rhamnose | Melibiose | Arabinose | |||||||
| pestis (main) | − | ND | ND | + | + | − | ND | + | Initially isolated from rats in Hong Kong in 1894 |
| plana | − | − | + | − | + | − | Leucine, tryptophan | + | Circulates in populations of great gerbils in Central-Asian-Desert foci, midday gerbils in the focus between Volga and Ural rivers, and red-tailed gerbils in the Transcaucasian plain |
| montanatianschanica | − | − | + | + | + | − | Leucine | + | Circulates in populations of gray marmots in Tien Shan mountains and red marmots in Pamirs-Altai mountains |
| montanahissarica | + | + | − | − | + | ± | Leucine | − | Circulates in populations of juniper voles (Microtus carruthersi) and red marmots in Hissarian Ridge (Tadjikistan) |
| montanacaucasica | + | + | + | + | − | + | Arginine, tyrosine, thiamine | − | Circulates in populations of common voles in Transcaucasian highland |
| montanaaltaica | + | + | − | − | + | + | Leucine, arginine | − | Circulates in populations of Mongolian pikas in Mountain Altai |
To bring some standardization to the system of classification of Y. pestis isolates, the conference of experts of the Anti-Plague Establishments of the Soviet Union (Saratov, 1985) recommended classifying all of the variants of the plague pathogen that were isolated from the territory of the FSU and Mongolia (Fig. 2 and 3A) into the “subspecies” Y. pestis subsp. pestis (sometimes referred to as the “main” subspecies), Y. pestis subsp. altaica, Y. pestis subsp. caucasica, Y. pestis subsp. hissarica, and Y. pestis subsp. ulegeica on the basis of the numerical analysis of 60 phenotypic features. In 1998, Sludskii (169) proposed one more intraspecific group, Y. pestis subsp. talassica. The last five subspecies are sometimes referred to as the “nonmain” subspecies (Table 6) and have also been referred to as the “pestoides” group of Y. pestis isolates. The numerical analysis was based on a similarity index (SI), calculated from the formula
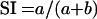 |
where a is the number of coincident signs and b is the number of unmatched classification features. In comparing the properties of strains belonging to the main subspecies, pestis, with strains of other subspecies, the SI was found to be within the range of 0.82 to 0.95. The SI is also sometimes expressed as percent similarity (i.e., SI × 100%). The most significant differences were found between the pestis and caucasica subspecies (SI = 0.82), whereas subspecies altaica and hissarica were found to be closely related (SI = 0.95) (11, 12). It was also found that in general among the five non-pestis subspecies (i.e., the “pestoides” subgroup), subspecies caucasica was classified as biovar Antiqua and subspecies altaica, hissarica, ulegeica, and talassica were biovar Medievalis.
FIG. 3.
Distribution of Y. pestis subspecies (A) and plasmidovars (B) in Mongolia. Symbols for provinces (aymags): 1, Bayanölgie; 2, Uvs; 3, Hovd; 4, Dzavham; 5, Govï-Altay; 6, Hovsgol; 7, Arhangay; 8, Bayanhongor; 9, Bulgan; 10, Övörhangay; 11, Ömnögovï; 12, Selenge; 13, Töv; 14, Dundgovï; 15, Hentiy; 16, Dornogovï; 17, Dornod; 18, Sühbaatar. Panel A reprinted with permission from S. V. Balakhonov (15), Antiplague Research Institute of Siberia and Far East, Irkutsk, Russia. Panel B reprinted with permission from A. Erdenebat (62), Centre for Control and Research of Natural Infectious Diseases, Ulaanbaatar, Mongolia.
TABLE 6.
Taxonomic characters of strains which distinguish different Y. pestis subspecies isolated in the territory of the FSU and Mongolia, and compliance of subspecies with biovarsa
| Y. pestis subspecies | Fermentation of:
|
Nitrate reduction | Urease activity | Pesticin 1 production | Susceptibility to pesticin 1 | Fibrinolytic activity | Coagulase activity | Dependence on nutrition factors
|
Virulence for guinea pigs | Region of circulation | Host(s) | Biovar | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rhamnose | Mellbiose | Arabinose | Glycerol | Melezitose | Leucine | Methionine | Arginine | Thiamine | Cysteine | Pnenylalanine | Threonine | Tyrosine | |||||||||||
| pestis (main) | − | − | + | ± | − | ± | − | + | − | + | + | ± | + | − | − | ± | ± | + | − | + | Central Africa, central and northern Asia, China (Manchuria), Mongolia | Rodents of genera Marmota, Citellus, Meriones, Rattus, etc. | Antiqua |
| altaica | + | + | − | + | ? | − | − | + | + | + | + | + | − | + | − | + | + | ? | ? | − | Mountain Altai | Ochotona pricei | Medievalis |
| caucasicab | + | + | + | + | − | + | − | − | + | − | − | ± | + | + | + | ± | + | + | ? | − | Transcaucasian highland, Mountain Dagestan | Microtus arvalis | Antiqua |
| hissarica | + | + | − | + | ± | − | ± | + | ± | + | + | + | + | − | − | + | + | − | − | − | Hissarian Ridge | Microtus carruthersi | Medievalis |
| ulegeica | + | + | + | + | ? | − | − | + | ±c | + | + | − | − | − | − | + | + | ? | ? | − | Northeast Mongolia, Gobi Desert | Ochotona pricei | Medievalis |
| talassica | + | + | − | + | − | − | + | + | − | + | + | + | ? | + | ? | + | + | ? | + | − | Talassian Ridge | Microtus gregalisd | Medievalis |
This table was compiled from references 11, 52, and 169, modified from reference 11, and used with the permission of the copyright holder (G. P. Aparin and E. P. Golubinskii). Symbols: −, absence of a character; +, presence of a character; ±, presence of a character but not in all strains; ?, no data.
Strains from subspecies caucasica are deficient in plasmid pPst
Strains are susceptible to pesticin of strains from subspecies pestis and altaica and also resistant to pesticin from strains of its own subspecies.
Current Use and Utility of Classification of Yersinia pestis Strains in the Former Soviet Union
The classification of Y. pestis strains into the different subspecies, including the main subspecies pestis and the nonmain subspecies altaica, caucasica, hissarica, ulegeica, and talassica, is currently widely used in the work of the Anti-Plague Establishments of the FSU. Chinese plague experts use their own classification, utilizing the term “ecotype” for different Y. pestis subspecies groups, which differ in some phenotypic properties from the Russian nonmain subspecies (175). However, these classification systems are not currently included in the International Bacterial Nomenclature. Derived from the best studies produced in the Soviet Union over 50 years, the Russian system currently stands up to repeated use in classifying strains isolated in the FSU and Mongolia. New proposals for its improvement appear periodically (67, 78, 109), but none have been felt to be good enough to take the place of the original one. Thus, the phenotypic characteristics listed in Table 6 should be a useful basis for identification and classification of Y. pestis strains until other systems are found to be more useful, practical, or amenable to classification of this genetically diverse group of otherwise closely related bacterial strains.
Limitations of the Classification in Identifying Potentially Virulent Strains of Yersinia pestis
Atypical strains.
Diversity in genotype and phenotype is found even among plague isolates from the same natural focus. Thus, no system of classification is likely to be perfect, having nearly 100% specificity and sensitivity for classifying plague isolates. Some idea of what is already known about atypical strains and how this might impact the classification and virulence of Y. pestis isolates is warranted, particularly with the potential that atypical or engineered strains with a variety of unusual characteristics might reasonably be expected to be a cause of human infection, either accidentally or deliberately.
Most of what is known about the appearance of atypical strains comes from field studies in endemic foci of infection. In the field, the appearance of atypical Y. pestis strains can correlate with different phases of the epizootic cycle (105, 173, 179). So-called atypical strains differ in some of the principal features found in the predominant Y. pestis variant isolated from a given plague focus. Thus, the ecological and other changes that occur during the epizootic cycle can allow nondominant Y. pestis strains to emerge in more consequential numbers in newly infected rodents, potentially providing the opportunity for the generation of new genotypes with altered virulence properties and for the exchange of potential virulence genes among different clones of Y. pestis.
An analysis of the frequency of the appearance of variant forms of Y. pestis isolated from diverse natural plague foci found a low level of variation in the Volga-Ural steppe focus (1.58%) and the Trans-Ural focus (3.3%) (Fig. 2, foci 15 and 17, respectively). In the Gissar (focus 34) and Central Asian Desert (foci 18 to 30 and 42) foci, this index was 6.59 and 6.55%, respectively. In the Volga-Ural sandy focus (focus 16), variant forms were not found. In the Trans-Ural focus (focus 17), variant strains were isolated during all of the phases of the epizootic process with an identical frequency. In the Volga-Ural steppe and Gissar foci (foci 15 and 34, respectively), such strains were recovered only during the height of an acute epizootic spread of Y. pestis. The frequency of strain variation in the early and later stages of an acute epizootic was uniform in the Central Asian Desert foci (foci 18 to 30 and 42). During the phase when the epizootic started to wane, the number of variant isolates increased (173).
In the Ural-Emba focus (focus 18), the greatest number of atypical strains (6.5%) was isolated at the height of the acute epizootic spread. It was found that 24.4% of atypical strains had modified biochemical activities, 17.6% were lysogenized with bacteriophages, 13.5% had modified requirements for growth factors, 14.27% had mutations in the hms (for “hemin storage system”) locus, 16.75% displayed reduced virulence, 9.93% were F1−, 7.14% were Lcr−, 2.36% were pesticin deficient, and 0.4% were resistant to the lytic action of diagnostic phages. A lower percentage of atypical strains was detected during the persistent phase of the epizootic process (195). According to other data (183), different foci were characterized by the presence of high proportions (up to 48%) of strains with reduced virulence in laboratory animals, with 10% of these isolates being essentially avirulent. A total of 0.2 to 1.2% and 0.5 to 2% of the variants from different foci had no autonomous pFra or pPst plasmid; 0.2 to 8.4% of the isolates from different foci did not display Ca2+-deficient growth cessation; and 0.04 to 29.2% were Hms−. These results reinforce the idea that there is a lot of genetic diversity among Y. pestis strains circulating in natural plague foci, with the consequent opportunities for genetic exchange and rapid emergence of new phenotypes.
One problem for making determinations about what represents an atypical strain of Y. pestis is that bacterial systematics has not yet reached a consensus for defining the fundamental unit of biological diversity, the species, let alone the subspecies. Cohan (43) thought that for bacteria, the fundamental unit of biological diversity is not the species but the ecotype, representing the population of organisms occupying the same ecological niche, whose genetic diversity is affected primarily by natural selection and whose diversity can be defined by sequence-based approaches. A typically named bacterial species could contain many ecotypes. One major question of relevance is that of the diversity in the properties of ecotypes that are associated with virulence for humans and animals of economic importance and whether these can be defined such that comprehensive tests for diagnosing plague can be developed, treatments can be validated against the range of strains pathogenic for humans, and active and passive immunotherapies can be deemed to be comprehensive enough to cover the range of pathogenic plague strains.
Phenotypic Variation and Virulence for Laboratory Animals
Unfortunately, many of the phenotypic signs that are listed in Table 6 and used for intraspecific differentiation of Y. pestis strains in the FSU are not absolute for a given subspecies, and thus the variability in the pathogenic potential of these strains can be large, even for strains from the same subspecies. One likely indicator of potential virulence for humans is a high level of virulence in animals. The guinea pig has been the animal of choice for virulence studies in the FSU. However, when tested for virulence in guinea pigs, most Y. pestis subspecies pestis strains are lethal whereas strains of subspecies altaica, caucasica, hissarica, ulegeica, and talassica as a rule exhibit dramatic reductions in virulence or even the complete absence of virulence for these animals (Tables 3 to 6). However, a few isolates belonging to these non-pestis subspecies and circulating in the same geographical region as the poorly virulent strains can kill guinea pigs, with 50% lethal dose (LD50) between 10 and >109 CFU per animal (1, 60, 61, 110, 169, 184). When 40 Y. pestis subspecies caucasica strains isolated within several years from the Transcaucasian highland (Fig. 2, foci 4 to 6) were examined for virulence in guinea pigs, it was shown that Y. pestis isolates recovered from one focus in this region, the Leninakan focus (focus 4), were more virulent for guinea pigs than were strains obtained from another focus in this region, the Zanzegur-Karabakh focus (focus 6) (61). Other investigators also noticed the high variability in guinea pig virulence of individual Y. pestis strains isolated in the Armenian highland focus (foci 4 to 6) (1), the Dagestan-highland focus (focus 39) (60), the Gissar focus (focus 34), the Talas focus (focus 40) (169), and Mongolia (Fig. 3A, provinces no. 1 and 11) (110). Thus, even though there may be considerable homogeneity of different Y. pestis isolates circulating within a natural focus of infection, as determined by membership in a specific taxonomic group, differences in properties, such as lethality for guinea pigs, that may impact their potential virulence for humans is also found.
While many of these strains are not readily classifiable into one of the subspecies and may not be lethal for mammals, they still have the potential to cause considerable morbidity, involving such conditions as pneumonia and bacteremia, that would make them serious pathogens of humans. Intraspecific heterogeneity in virulence, host specificity, and biochemical and physiological traits is also found among isolates outside of the FSU and related Asian areas such as Mongolia. For example, 24 Y. pestis strains from different natural foci in Africa showed significant variability in their phenotypic characteristics and virulence for mice and guinea pigs (Table 7). Given that many studies of Y. pestis pathogenesis performed outside of the FSU and Asian countries have been carried out with a limited number of closely related strains, it cannot be concluded with complete confidence that the virulence factors identified in these strains are essential factors for all strains of Y. pestis or that lethality is necessarily the only outcome in experimentally infected animals that would be predictive of virulence for humans.
TABLE 7.
Relevant characteristics of Y. pestis strains isolated in northern, northwestern, western, and equatorial Africaa
| Region of isolation | No. of strains | Fermentation of:
|
Ammonia oxidation | Additional growth factors | DCLb (CFU) for:
|
|||
|---|---|---|---|---|---|---|---|---|
| Glycerol | Rhamnose | Maltose | Mice | Guinea pigs | ||||
| North Africa (Tunis, Algeria, Morocco) | 5 | − | − | + | − | − | ≤25 | >106 |
| Northwestern Africa (Mauritania) | 2 | − | − | + | − | − | ≤25 | ≤100 |
| Western Africa (Senegal) | 7 | − | − | + | − | − | ≤25 | >106 |
| Equatorial Africa (Kenya, Congo) | 4c | + | − | + | + | Riboflavin | >106 | >106 |
| 3c | + | − | + | + | Not established | >106 | >106 | |
| 2d | + | − | − | − | Lysine | >106 | >106 | |
| 1d | − | − | − | − | Uracil, ornithine, leucine, alanine | >106 | >106 | |
Compiled from reference 121.
DCL, absolutely lethal dose (dosis certa letalis).
Strains isolated in Kenya.
Strains isolated in The Congo Democratic Republic.
Attempts To Predict the Pathogenic Potential of Variant Yersinia pestis Clones
Overall, in spite of trying to standardize classification of Y. pestis by using the large variety of isolates available in the FSU, it is still not feasible to really know which particular set of genetic and phenotypic traits are indicative of virulence for laboratory animals or humans. Kuklev had attempted to use epidemiologic data to determine whether certain strains from specific foci are more likely to cause human illness and/or be transmitted to humans (108). As one might expect, the question of epidemic potential for humans of strains within a given natural focus is more complex than just the issue of the virulence of an individual strain for humans. It obviously also involves such factors as intensity of human contact, population densities of host rodents and fleas, and other factors that are sometimes difficult to quantify. Kuklev (108) proposed the following formula to calculate epidemic potential (EP) of strains of Y. pestis from natural foci:
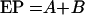 |
where A represents the diffusion and intensity of epizootic manifestations of plague and B represents the intensity of human contact with the environment of the natural focus. The first term, A can be defined as
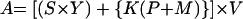 |
where S is the physical area of epizootic manifestation, Y is the intensity of the epizootic in terms of infected animals, K is the proportion of the focus area populated by the main rodent host of the Y. pestis strain, P is the number of rodents within 1 Ha, M is the number of fleas within 1 Ha, and V represents the virulence of Y. pestis strains. Obviously, many of these values must be determined by field and laboratory studies.
The index V is of especial interest since it is an important modifier of the overall equation. It takes into account two findings, laboratory studies measuring virulence for guinea pigs and the ability to ferment rhamnose. The V index runs across a scale of 0.1 to 1.0 and can thus cause up to 10-fold decrease of the total EP index (Table 8).
TABLE 8.
Epidemiologic variants of glycerol-positive Y. pestis from the natural plague foci of the FSUa
| Regional group of foci | Focus no. | Main host | Differentiating findings
|
Vb | |
|---|---|---|---|---|---|
| Virulence for guinea pigs | Rhamnose fermentation | ||||
| Caucasian region | 1 | Citellus musicus | ±c | − | 0.5-1.0 |
| 4-6, 39 | Microtus arvalis | − | + | 0.1 | |
| 7 | Meriones vinogradovi | + | − | 1.0 | |
| 8-13 | Meriones libicus | + | − | 1.0 | |
| North-Pre-Caspian region | 2-3, 14-15, 17 | Citellus pygmaeus | + | − | 1.0 |
| 16 | Meriones meridianus | + | − | 1.0 | |
| 43 | M. meridianus | + | − | 1.0 | |
| Central-Asian-Desert region | 18-30, 42 | Rhombomys opimus | + | − | 1.0 |
| 28 | R. opimus | ±d | − | 0.5 | |
| Central-Asian-Mountain region | 31-33 | Marmota baibacina | + | − | 1.0 |
| 35 | Marmota caudata | + | − | 1.0 | |
| 34 | Microtus carruthersi | − | + | 0.1 | |
| 40 | Microtus gregalis | − | + | 0.1 | |
| Siberian region | 36 | Ochotona pricei | − | + | 0.1 |
| 37 | Citellus undulatus | + | − | 1.0 | |
| 38 | Citellus dauricus | + | − | 1.0 | |
Virulence index.
Y. pestis strains isolated from the right bank of the river Baksan are virulent for guinea pigs, while the majority of those from the left bank of the river have low virulence or are even avirulent for guinea pigs (167).
Y. pestis strains with reduced virulence.
The second term, B, can be defined as
 |
where B1 represents the potential human contact with the fleas of the wild rodents in the field, B2 quantifies the presence of rodents and fleas in human habitations, B3 quantifies the presence of camels and their number (which can transmit plague to humans [40, 63]), B4 quantifies the use of hunting for animals likely to be carrying fleas infected with Y. pestis, B5 quantifies the closeness of the place of residence of rodent hosts to human habitations and contact of human children with rodents, and B6 quantifies the presence of cats and dogs in human habitations. Each of the indexes has its own numerical range and technique for calculation, but in total the EP cannot be more than 100. As with the qualitative estimation of the epidemic potential, more than 50 is high, 25.1 to 50 is intermediate, 5 to 25 is low, and less than 5 is very low.
Kuklev (108) used these formulas to determine the epidemic potential of Y. pestis spread from four different plague foci: Kara-Kum focus (Fig. 2, focus 25), EP = 70.1; Upper-Naryn focus (focus 32), EP = 54.0; Central-Caucasian focus (focus 1), EP = 15.6; and Transcaucasian-highland foci (foci 4 to 6), EP = 2.8. While this type of investigation provides one framework for evaluating the potential virulence of Y. pestis strains found in natural foci, many of these factors would not be relevant to the virulence potential of strains that can be encountered outside of these rural environments. At the moment, there is no one definitive criterion for stating that a given strain of Y. pestis has high or low pathogenic potential for humans, but guinea pig virulence and rhamnose fermentation are the two traits currently known to be the best predictors of likely virulence for humans.
PHENOTYPIC AND GENOTYPIC DIVERSITY
Obviously, with large areas of European Russia and Central Asia sites of endemic foci of zoonotic plague, there is a tremendous opportunity for vast genetic and consequent phenotypic diversity. Some have referred to this as the “metastability of phenotype” (27). Mechanisms giving rise to the metastability of phenotypes include the overall plasmid content, diverse reversible intergenomic realignments that include displacement of IS elements, rearrangements of variable-number tandem repeats (VNTRs) or changes generated during their mismatch repair, integration of plasmids and bacteriophages into the bacterial chromosome, and frameshift mutations in regulatory genes. These intergenomic realignments can be detected by different approaches, and it will be essential in the future to try to determine the effect of genetic variability on the virulence of Y. pestis for humans.
Diversity and Virulence
When virulence is measured in the context of animal infections, the intensity and manifestations of pathogenicity of an individual microbial strain are dependent on many conditions: animal species used for evaluation, immune status of the host, conditions of animal care and feeding, route of infection, and potentially even subtle variations such as the time of day or season when the infectious dose is given (8-10, 100, 141, 144). Since bacteremia in an infected rodent is a necessary condition for Y. pestis transmission to a new host via fleas, strains of Y. pestis maintained in endemic foci must be sufficiently virulent to establish a bacteremic state conducive to transmission (7-9, 30, 31, 34, 89, 104, 141). Long-term studies of isolates from different natural foci indicate that the majority of strains possessed sufficient virulence to cause bacteremia in the main rodent host, although isolates from different foci differ from each other in terms of their virulence for different animal species (1, 11, 12, 15, 22, 33, 55, 56, 101, 104, 106, 107, 118, 123, 151, 166, 167, 169, 177, 179, 184, 198).
Kozlov (106) and Kokushkin (104) discriminated two main variants of glycerol-positive Y. pestis strains obtained from natural plague foci within the FSU which differed in their epidemic potential. (i) The first are “rhamnose fermentation-negative” strains, which were highly virulent for guinea pigs and were isolated from foci with different potentials for epidemic spread into humans (i.e., associated or not with reported cases of human to human plague transmission). These were Y. pestis subsp. pestis. (ii) The second are “rhamnose fermentation-positive” strains, which were of low virulence or avirulent for guinea pigs but caused occasional disease in humans that was not accompanied by outbreaks of human-to-human transmission of infection (Y. pestis subspp. altaica, caucasica, hissarica, ulegeica, and talassica).
Kudinova (107), studying 107 Y. pestis subsp. pestis strains isolated from great gerbils and their fleas in the Ili-Karatal interriver region of the Pre-Balkhash focus (Fig. 2, focus 30), studied the differences in the virulence of strains isolated from two foci separated by 40 km. The strains isolated near the Karoi settlement were virulent not only for great gerbils but also for other laboratory animals (mice and guinea pigs), while the strains isolated from the Bosugen tract (focus 30) could cause death in great gerbils but were avirulent for mice and guinea pigs (LD50 > 108CFU). While the genetic or physiologic basis for this difference was not reported, one might consider differential susceptibility to host defenses, particularly susceptibility to complement-mediated killing, as a potential explanation for the differences in virulence in different rodent species.
Studying 121 Y. pestis subsp. pestis strains isolated from 1971 to 1988 in the Central-Caucasian natural plague focus (Fig. 2, focus 1), Serdyukova (167) found that all of the tested strains were highly virulent (LD50 = 1 to 104 CFU) for their main host, the mountain suslik. Interestingly, Y. pestis strains isolated from the right bank of the river Baksan in this focus (focus 1) that were also found not to require proline for their growth displayed high virulence for both mice and guinea pigs (LD50 = 1 to 104 CFU). On the other hand, only 22% of strains auxotrophic for proline and isolated from the left bank of the river Baksan (focus 1) were virulent for these laboratory animals. Other strains auxotrophic for proline and with high virulence for mountain susliks had low virulence (LD50 = 104 to 108 CFU) or even avirulence (LD50 > 108CFU) for mice or guinea pigs, either singly or in some cases for both of these animal species. Spontaneous proline prototrophs obtained from initially proline-requiring strains maintained the virulence selectivity of their parent strains, indicating that proline requirements were not determinants of Y. pestis virulence for laboratory animals. Nonetheless, these findings point out that endemic strains of Y. pestis maintaining their transmission cycle by infecting comparable animal species can still have marked differences in their virulence for other mammalian hosts. Clearly, then, some of these other hosts could potentially include humans.
Although these studies indicate that there is host specificity for some Y. pestis subsp. pestis strains (33, 107, 167), many of these strains also represent atypical strains even for this subspecies. As with the Y. pestis strains of subspp. altaica, caucasica, hissarica, ulegeica, and talassica which are circulating within diverse populations of voles and Ochotona pricei (a type of pika, which is classified as a lagomorph and thus is related to rabbits and hares), there is more of a tendency to find selective virulence for different animals. Rhamnose fermentation-positive strains tend to be virulent for mice and for some species of wild rodents, but as a rule they have low virulence, or are even avirulent, for guinea pigs, other species of wild rodents (1, 11, 12, 55, 104, 118, 169, 177, 184), and human volunteers in one study (Zhenya et al., Abstr. 7th Int. Symp. Yersinia, 1998).
This large amount of strain heterogeneity in Y. pestis can obviously have a major impact on virulence studies and provides a major challenge for investigators trying to define essential factors involved in Y. pestis virulence. As with any pathogen, choosing a set of strains for virulence studies by using such techniques as directed inactivation of genes relies on choosing properly representative parental strains for the investigations. This standard method for studying microbial pathogenesis can readily determine the contribution of gene products, either singly or in combination, to a virulence phenotype, but the outcome of the experiments is obviously dependent on the overall genetic makeup of the parental strain being studied. Since microbial pathogenesis is complex and multifactorial, with several virulence factors usually acting in concert to produce infection (71), the large array of potential genotypic diversity that strains of Y. pestis can draw upon due to the extensive occurrence of this pathogen in natural foci may explain, in part, the conflicting experimental data on the role in virulence of some Y. pestis pathogenicity factors (Table 9). Use of different parental strains that can possess unidentified allelic variations in genes that are not directly under study but whose products are not necessary for survival within their natural host undoubtedly underlies some of the variability in virulence study outcomes. Elimination or inactivation of one or more genes from such strains could lead to a significant decrease in virulence when tested in laboratory animals, but the parental strain may or may not be particularly representative of strains of Y. pestis with pathogenic potential for humans. Thus, in another strain background, the specific virulence factor may make a relatively minor contribution to pathogenesis. Since many studies of plague pathogenesis have focused on strains isolated from the Americas that possess much less genetic diversity than strains in Russia and Asia, the conclusions from these studies may not be entirely applicable across the board for all strains of Y. pestis with high virulence for humans.
TABLE 9.
LD50 studies with Yersinia pestis harboring mutations in putative virulence genes
| Straina | Virulence characteristicsb
|
Route | Virulence for:
|
Refer- ence | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lcr | Pgm | Pla | F1 | Ymt | pH6 | Mice
|
Guinea pigs
|
|||||
| LD50 (95% confidence intervals) | Avg life duration | LD50 (95% confidence intervals) | Avg life duration | |||||||||
| Y. pestis subsp. pestis | ||||||||||||
| w.t.c 231 (focus 33, FSU) | + | + | + | + | + | + | s.c. | 3 (1-18) | 7.3 (4-8) | 4 (1-22) | 8.1 (5-9) | 59 |
| 2 | 5.7 | 10 | 7.3 | 110 | ||||||||
| 231/830 (psaF::kan) | + | + | + | + | + | − | s.c. | >108 | >1.5 × 1010 | 136 | ||
| 231pPst− (pPst−) | + | + | − | + | + | + | s.c. | 1 (1-4) | 6.9 (4-7) | 4 (1-21) | 8.6 (5-9) | 59 |
| 231Psb− (Hms−Psts) | + | Hms− Psts | + | + | + | + | s.c. | 4 (1-21) | 7.8 (5-8) | 10 (2-24) | 8.9 (6-10) | 59 |
| 231 #3 (Hms− Psts) | + | Hms− Psts | + | + | + | + | s.c. | <316 | No data | No data | No data | 198 |
| 231 #2 (Hms+ Pstr) | + | Hms+ Pstr | + | + | + | + | s.c. | >107 | No data | No data | No data | 198 |
| 231 #4 (Hms− Pstr) | + | Δpgm | + | + | + | s.c. | >107 | No data | No data | No data | 198 | |
| 231Pgm− (Δpgm) | + | Δpgm | + | + | + | + | s.c. | >108 | >1.5 × 1010 | 8 | ||
| M-231 #5 (pPst−) | + | + | − | + | + | + | s.c. | 180 | 8.2 | 140 | 16.5 | 110 |
| M-231 #5-31 (pVK1) | + | + | + | + | + | + | s.c. | 4 | 6.1 | 22 | 8.2 | 110 |
| M-231 #5-31 (pVK2, pst::Tn1) | + | + | + | + | + | + | s.c. | 3 | 5.8 | 18 | 7.8 | 110 |
| w.t. 358 (focus 21, FSU) | + | + | + | + | + | + | s.c. | 7 (1-27) | 4.6 (3-5) | 13 (3-63) | 8.6 (5-9) | 59 |
| + | + | + | + | + | + | Aerosol | No data | No data | 2.1 × 103 (618-10,120) | 8.3 (7-10) | 163 | |
| 358pPst− (pPst−) | + | + | − | + | + | + | s.c. | 1 (1-2) | 5.5 (4-6) | 11 (2-68) | 6.3 (5-9) | 59 |
| + | + | − | + | + | + | Aerosol | No data | No data | 4.2 × 103 (1,916-25,110) | 9.3 (8-11) | 163 | |
| 358Pgm− (Δpgm) | + | Δpgm | + | + | + | + | s.c. | >108 | >1.5 × 1010 | 8 | ||
| parent KM219 (pPst−) (Madagascar) | + | + | − | + | + | + | s.c. | 440 | 9.5 | 9.4 × 105 | 26.6 | 110 |
| KM219-1 (pVK1) | + | + | + | + | + | + | s.c. | 75 | 5.4 | 1.6 × 105 | 16.3 | 110 |
| KM219-2 (pVK2, pst::Tn1) | + | + | + | + | + | + | s.c. | 29 | 5.5 | 1.3 × 105 | 14.8 | 110 |
| parent A-250 (pPst−) (FSU) | + | + | − | + | + | + | s.c. | 2.6 × 103 | 8.7 | 1.5 × 106 | 15.7 | 110 |
| A-250-1 (pVK1) | + | + | + | + | + | + | s.c. | 1.7 × 103 | 6.0 | 6.3 × 103 | 10.6 | 110 |
| A-250-2 (pVK2, pst::Tn1 | + | + | + | + | + | + | s.c. | 3.5 × 103 | 5.3 | 4.7 × 103 | 8.9 | 110 |
| w.t. KIM5+ (Iran/Kurdistan) | + | + | + | + | + | + | s.c. | <10 | No data | No data | No data | 182 |
| KIM5 (Δpgm) | + | Δpgm | + | + | + | + | s.c. | >107 | No data | No data | No data | 182 |
| w.t. KIM1001 (Iran/Kurdistan) | + | + | + | + | + | + | s.c. | 42 | No data | No data | No data | 171 |
| KIM1002 (pPst−) | + | + | − | + | + | + | s.c. | 8.8 × 106 | No data | No data | No data | 171 |
| KIM1008 (Δpla) | + | + | − | + | + | + | s.c. | 6 × 107 | No data | No data | No data | 171 |
| KIM5-3001 (Smr) | + | Δpgm | + | + | + | + | r.o.d | 42 | No data | No data | No data | 117 |
| KIM5-3001.1 (Smr, psaA3::m-Tn3) | + | Δpgm | + | + | + | − | r.o. | 9 × 103 | No data | No data | No data | 117 |
| w.t. CO92 (Colorado, U.S.) | + | + | + | + | + | + | s.c. | 1.9 | No data | No data | No data | 186, 188 |
| + | + | + | + | + | + | i.p. | 14 | No data | No data | No data | 186, 188 | |
| + | + | + | + | + | + | Aerosol | 2.3 × 104 | No data | No data | No data | 186, 188 | |
| CO92 pPst− (pPst−) | + | + | − | + | + | + | s.c. | 1.4 × 106 | No data | No data | No data | 186, 188 |
| + | + | − | + | + | + | i.p. | 7.6 | No data | No data | No data | 186, 188 | |
| + | + | − | + | + | + | Aerosol | 105 | No data | No data | No data | 186, 188 | |
| CO92 Pla-12, #1 | + | + | + | + | + | + | s.c. | 0.81 (0.04-6.86) | No data | No data | No data | 186, 188 |
| CO92 Pla-Δ12, #1 (Δpla) | + | + | − | + | + | + | s.c. | 1.2 × 105 (3.2 × 104-5.1 × 106) | No data | No data | No data | 186, 188 |
| CO92 Pla-Δ12, #2 (Δpla) | + | + | − | + | + | + | s.c. | 3.4 × 104 (6.5 × 103-1.8 × 105) | No data | No data | No data | 186, 188 |
| CO92 Pgm− (Δpgm) | + | Δ-pgm | + | + | + | + | s.c. | ∼107 | No data | No data | No data | 186, 188 |
| + | Δ-pgm | + | + | + | + | Aerosol | ∼106 | No data | No data | No data | 186, 188 | |
| w.t. Alexander (U.S.) | + | + | + | + | + | + | s.c. | 0.61 | No data | No data | No data | 31 |
| + | + | + | + | + | + | i.p. | 0.99 | No data | No data | No data | 31 | |
| + | + | + | + | + | + | i.v. | 0.81 | No data | No data | No data | 31 | |
| Alexander pPst− (pPst−) | + | + | − | + | + | + | s.c. | >5 × 107 | No data | No data | No data | 31 |
| + | + | − | + | + | + | i.p. | 3.8 × 105 | No data | No data | No data | 31 | |
| + | + | − | + | + | + | i.v. | 7.1 | No data | No data | No data | 31 | |
| Alexander Pgm− (Δpgm) | + | Δ-pgm | + | + | + | + | s.c. | >5 × 107 | No data | No data | No data | 31 |
| + | Δpgm | + | + | + | + | i.p. | >5 × 107 | No data | No data | No data | 31 | |
| + | Δ-pgm | + | + | + | + | i.v. | 1.5 | No data | No data | No data | 31 | |
| Y. pestis subsp. caucasia | ||||||||||||
| w.t. 6499 (pPst−) (focus 4-6, FSU) | + | + | − | + | + | + | s.c. | 13 | 5.9 | 1.3 × 108 | 48.1 | 110 |
| 6499-1 (pVK1) | + | + | + | + | + | + | s.c. | 11 | 4.5 | 2.4 × 108 | 26.7 | 110 |
| 6499-2 (pVK2, pst::Tn1) | + | + | + | + | + | + | s.c. | 15 | 6.3 | 1.1 × 108 | 27.1 | 110 |
| 6499 #2 (Hms+ Pstr) | + | Hms+ Pstr | − | + | + | + | s.c. | 1.5 × 103 | No data | No data | No data | 198 |
| 6499 #3 (Hms− Psts) | + | Hms− Psts | s.c. | <316 | No data | No data | No data | 198 | ||||
| 6499 #4 (Hms− Pstr) | + | Δpgm | s.c. | >107 | No data | No data | No data | 198 | ||||
| w.t. Pestoides F (FSU) | + | + | − | + | + | + | s.c. | 3.0 | No data | No data | No data | 186 |
| Y. pestis subsp. altaica | ||||||||||||
| w.t. I-2359 #1 (Hms+ Psts) (focus 36, FSU) | + | + | + | + | + | + | s.c. | <681 | No data | No data | No data | 198 |
| I-2359 #2 (Hms+ Pstr) | + | Hms+ Pstr | + | + | + | + | s.c. | >107 | No data | No data | No data | 198 |
| I-2359 #3 (Hms− Psts) | + | Hms− Psts | + | + | + | + | s.c. | <316 | No data | No data | No data | 198 |
| I-2359 #4 (Hms− Pstr) | + | Δpgm | + | + | + | + | s.c. | >107 | No data | No data | No data | 198 |
| Y. pestis subsp. hissarica | ||||||||||||
| w.t. I-2359 #1 (Hms+ Psts) (focus 34, FSU) | + | + | + | + | + | + | s.c. | <681 | No data | No data | No data | 198 |
| I-2359 #2 (Hms+ Pstr) | + | Hms+ Pstr | + | + | + | + | s.c. | >107 | No data | No data | No data | 198 |
| I-2359 #3 (Hms− Psts) | + | Hms− Psts | + | + | + | + | s.c. | <316 | No data | No data | No data | 198 |
| I-2359 #4 (Hms− Pstr) | + | Δpgm | + | + | + | + | s.c. | >107 | No data | No data | No data | 198 |
| Y. pestis subsp. ulegeica | ||||||||||||
| w.t. I-2359 #1 (Hms+ Psts Mongolia) | + | + | + | + | + | + | s.c. | <681 | No data | No data | No data | 198 |
| I-2359 #2 (Hms+ Pstr) | + | Hms+ Pstr | + | + | + | + | s.c. | >107 | No data | No data | No data | 198 |
| I-2359 #3 (Hms− Psts) | + | Hms− Psts | + | + | + | + | s.c. | <681 | No data | No data | No data | 198 |
| I-2359 #4 (Hms− Pstr) | + | Δpgm | + | + | + | + | s.c. | >107 | No data | No data | No data | 198 |
Where identified, strain origin and/or relevant mutations are noted in parentheses.
For Lcr, a plus sign indicates pLcr plasmid detected and the strain is positive for Ca2+-deficient growth restriction and/or synthesis of LcrV or Yops. For Pgm, a plus sign indicates that the 102-kb pgm locus was present and a functional test for hemin storage was performed; a minus sign for pgm indicates deletion of the entire 102-kb pgm locus; Psts and Pstr indicate sensitive and resistant, respectively, to the bacteriocin pesticin, a presumptive test for the functional pesticin/yersiniabactin receptor, Psn; Hms+ and Hms− indicate positive and negative, respectively, for hemin or Congo red adsorption. For Pla, a plus sign denotes detection of plasmid and Pla activity. For F1, Ymt, and pH6, a plus sign indicates that fraction 1, murine toxin, and pH6 antigens, respectively, were detected.
w.t., wild-type Y. pestis strain.
r.o., retro-orbital injection (equivalent to intravenous injection); s.c., subcutaneous injection; i.p., intraperitoneal injection; i.v., intravenous injection.
Impact of Diversity on the Interrelationship of Host Immune Factors and Virulence
The ability of pathogenic bacteria to survive in the face of host defense systems is intimately linked to virulence (8, 9, 71). The ability of Y. pestis to maintain its transmission cycle in rodents, as well as to infect incidental hosts such as humans, is highly dependent on both rapid growth in the host and effective resistance to host innate immune effectors including bactericidal cationic peptides as well as opsonic and lytic complement proteins. These innate immune factors can rapidly kill bacteria and prevent acute infections (82, 83, 131). Thus, Y. pestis must survive exposure to bactericidal complement conditions within the blood and bactericidal cationic peptides conditions within the phagocytes and must therefore have evolved or acquired complex systems to counteract host defenses (8, 9, 20, 74, 93, 141, 142, 149, 182; A. P. Anisimov and S. V. Dentovskaya, unpublished data). In contrast to other strains of Y. pestis, strains of subsp. caucasica are highly susceptible to the bactericidal activity of 80% human serum (87; Anisimov and Dentovskaya, unpublished), while all Y. pestis strains are able to grow in heat-inactivated human serum or in 80% normal mouse serum (Anisimov and Dentovskaya, unpublished). The lcrV virulence gene leads to immunosuppression by inducing the anti-inflammatory cytokine interleukin-10 via interactions with CD14 and toll-like receptor-2 (169a). It seems likely that similar factors exist in many of the strains of Y. pestis found in natural foci that have comparable effects on the inflammatory responses of their native hosts. Since host innate immune factors such as CD14 and toll-like receptors are fairly conserved across species, there is clear potential for variant LcrV-like proteins to be present in non-American strains of Y. pestis, and such proteins may not be amenable to neutralization by antibodies raised to LcrV vaccines prepared from strains found in the Americas.
Another factor with potential for high variability in chemical and antigenic activity involved in providing resistance to host defenses is LPS, with the oligosaccharides mediating resistance to the bactericidal effect of complement and, along with features of the lipid A, mediating resistance to the effects of antimicrobial peptides. Investigation of the responses to polymyxin B of Y. pestis strains isolated from various foci showed that many are usually highly resistant to polymyxin B (MIC, 200 to 3000 μg/ml) (74, 122). However, Y. pestis strains of subspp. hissarica (122; S. V. Balakhonov, personal communication) and caucasica (74, 122; S. V. Balakhonov, personal communication) and fresh isolates of subsp. altaica are highly sensitive to polymyxin B (MIC, 10 to 25 μg/ml), suggesting differences in the aminoarabinose content in the lipid A (180) or in heptose content of the LPS core in bacteria from subspecies pestis and some non-pestis subspecies (Y. A. Knirel, E. V. Vinogradov, S. N. Senchenkova, N. A. Kocharova, B. Lindner, O. Holst, R. Z. Shaikhutdinova, A. P. Anisimov, and T. A. Gremyakova, Abstr. Carbohydr. Workshop, Güstrow-Rostock, Germany, 2003). The recently published structure of the core oligosaccharide of one strain of Y. pestis (185) has already indicated that variability in structure occurs in relation to growth temperature. Given the high mobility of genetic loci containing genes whose products are enzymes involved in the synthesis of LPS polysaccharie components, there is clearly some potential for variability in this structure in Y. pestis that could impact virulence and host immune capabilities. Overall, it appears that adaptations made by the plague pathogen that allow it to infect specific mammalian species are based, in part, on quantitative and qualitative changes in Y. pestis factors that counteract host immune components, with some of these virulence factors likely to be effective against innate immune resistance in humans.
Plasmid Content
The discovery of Y. pestis plasmids of 9.5 kb (termed pPst, pPla, pPCP1, or pYP), 70 to 75 kb (termed pCD1, pCad, pVW, pYV, or pLcr), and 100 to 110 kb (termed pFra/Tox, pFra, pTox, pMT1, or pYT) (21, 66, 148, 150, 154) made it possible to compare plasmid profiles between strains isolated from different natural plague foci and determine if there was an association of plasmid content with virulence. Plasmids are known to be key factors that determine the virulence of Y. pestis, and the potential for these genetic elements to move among different strains could readily underlie the acquisition and loss of virulence potential. Indeed, characterizing the plasmid content and variations in plasmid sizes has been instrumental in identifying genetic diversity among Y. pestis strains within the FSU and China. There are quite a number of these studies, which demonstrate different types of changes in plasmid content and/or plasmid size, and their results emphasize one of the major mechanisms of generation of genetic diversity in Y. pestis and the potential of this diversity to impact virulence, antigen expression, and susceptibility of strains to diagnostic reagents. Importantly, an understanding of the natural variation in the content and size of plasmids found in Y. pestis will be critical for distinguishing which plasmids are present in given strains from different regions of the world, the contribution made by the different plasmids to the pathogenic potential of strains carrying them, and the question whether newly engineered plasmids with added or rearranged virulence factors have been made and introduced into strains of this organism. Also, knowing something about where different Y. pestis strains carrying different types of plasmids are typically found will be instrumental in epidemiologic identification of sources of outbreaks of disease.
Variation in plasmid content.
One striking finding from research into enzootic strains of Y. pestis in the FSU is that in some populations the plasmid content is quite stable whereas in other populations there is considerable variation in plasmid content among strains isolated from rodents living in close proximity. For example, plasmids in strains isolated from four autonomous foci on the northern border of the Central Asian zone (Fig. 2, foci 36 to 38 and 41) were characterized as stable and independent of the source and time of strain isolation. In contrast, by using agarose gel electrophoresis to determine the plasmid content in strains of Y. pestis subsp. caucasica isolated from common voles and their fleas in Leninakan, Pre-Sevan, Zanzegur-Karabakh, and Dagestan-highland natural plague foci (foci 4, 5, 6, and 39, respectively), it was found that this group was missing the pPst plasmid that carries the genes for pesticin-fibrinolytic-coagulase activities and showed susceptibility to pesticin 1. However, in the nearest plague foci involving primarily Y. pestis subsp. pestis the organisms isolated from susliks (focus 3), gerbils (foci 7, 8, and 11 to 13), or their flea vectors contained, as a rule, the three classical plague plasmids (12, 15, 67-69, 73, 99, 168, 186; L. Bakanidze, D. Tsereteli, M. Kekelidze, I. Velijanashvili, L. Beridze, E. Zangaladze, M. Zakalashvili, and P. Imnadze, Abstr. 8th Int. Symp. Yersinia, abstr. P-63, 2002; Worsham and Roy, Abstr. 8th Int. Symp. Yersinia, 2002; P. L. Worsham and M. Hunter, Abstr. 7th Int. Symp. Yersinia, abstr. P-88, Med. Microbiol. [Ned. N. Voor] 6[Suppl. II]:S34-S35, 1998). Similarly, strains of Y. pestis isolated from mountain susliks and their fleas in the Central-Caucasian natural focus on the left bank of the river Baksan (focus 1) showed that the strains that were auxotrophic for proline carried an additional 3- to 4-MDa plasmid (67, 79, 114, 167, 197) while the majority of strains from the right bank (focus 1) had only three classic plague plasmids and did not require proline for growth. Spontaneous proline prototroph derivatives of initially proline-requiring strains retained the 3- to 4-MDa plasmid (79, 167). Hence, the plasmid was associated with a phenotype but was not responsible for it and was not readily transmissible to nearby Y. pestis strains. The role of the 3- to 4-MDa plasmid in virulence or its potential to be modified is not known.
In Mongolia it was found that the plasmid content of 894 Y. pestis strains isolated from patients, wild mammals, and arthropods were divisible into three distinct populations of strains based on different plasmid contents (Fig. 3B and 4). These were divided into plasmidovars. The first plasmidovar harbors three plasmids with molecular masses of 6 MDa (pPst), 45 to 47 MDa (pCad), and 62 to 65 MDa (pFra). The second plasmidovar contained plasmids with molecular masses of 6 MDa (pPst), 16 MDa (cryptic), 45 to 47 MDa (pCad), and 62 to 65 MDa (pFra). The third plasmidovar harbored plasmids of 8 MDa (pPst), 45 to 47 MDa (pCad) (18, 62), and 62 MDa (pFra) (according to reference 62) or 75 to 80 MDa (pFra) (according to reference 18).
FIG. 4.
Distribution of Y. pestis plasmidovars in some Central Asian natural foci. Reprinted with permission from S. V. Balakhonov (17), Antiplague Research Institute of Siberia and Far East, Irkutsk, Russia.
Plasmidovar classification was associated, to a high but not exclusive degree, with strain source and phenotype, documenting the potential utility of this method for epidemiologic investigations and for contributing to the determination of the pathogenic potential of a given isolate. All of the strains bearing the combination of plasmids designated as the third plasmidovar were isolated from Microtus brandti (a type of vole); 80.9% of these isolates were classified as Y. pestis subsp. altaica. Strains carrying plasmids representative of either the first or second plasmidovar groups were isolated from different rodents (58.6% of the strains, including 2.7% of the strains isolated from M. brandti), fleas (33.1% of the strains), and humans (8.1% of the strains). In this series, 95.2% of the strains with the plasmidovar 2 plasmid profile were Y. pestis subsp. pestis while 4.8% were classified as Y. pestis subsp. ulegeica. Although Y. pestis subsp. pestis can express the plasmidovar 2 profile, expression of the plasmidovar 1 profile appears to be largely, but not exclusively, confined to Y. pestis subsp. pestis (91.6 to 99.3% of strains, depending on the region of isolation). The remainder of the strains expressing the plasmidovar 1 profile were classified as Y. pestis subsp. ulegeica (0.7 to 8.4% of strains) and Y. pestis subsp. altaica (0 to 5.6% of strains).
Plasmid content is sometimes associated with other phenotypic properties of the strains, suggesting some clonality and also providing another tool for diagnostic and epidemiologic investigations. Strains with different plasmid contents can vary in their carbohydrate fermentation activity. The strains of plasmidovar 2 did not metabolize rhamnose and melibiose. The strains of plasmidovar 3 did not ferment arabinose. Amino acid requirements for growth also varied among the strains. Isolates of plasmidovars 1 and 2 required methionine for their growth, while those of plasmidovar 3 required arginine and leucine.
It was also mentioned that during long-term laboratory storage some strains maintained their initial plasmid profiles but nonetheless had changes in other phenotypic characteristics, which caused their reclassification into other subspecies (62). Hence, even storage can lead to genetic and phenotypic changes in Y. pestis.
Many studies based on plague pathogenesis have focused on the role of the three well-characterized Y. pestis plasmids associated with virulence, with molecular massesof about 6 MDa (pPst), 45 to 50 MDa (pCad), and 60 MDa (pFra). Assuming that these are representative of at least one class of virulent plague strains, it would be important to know something about the variation in plasmid content in strains in the FSU, Mongolia, and China harboring these plasmids and how such variation would impact virulence and diagnosis. One study of the plasmid composition of 257 Y. pestis strains from 31 natural foci in the FSU and other countries revealed that 68% of the strains carried the three classic plasmids. In 10% of the strains obtained from different sources, additional cryptic plasmids were detected. A total of 18% of the isolates from the Volga-Ural sandy focus (Fig. 2, focus 16) and 43% of the isolates from the Talas focus (focus 40) also contained cryptic plasmids, usually of 20 MDa. In some strains from Talas, Central-Caucasian, and Pre-Balkhash foci (foci 40, 1, and 30, respectively), as well as from Senegal, Sri Lanka, and Indonesia, cryptic plasmids with molecular masses from 1.6 to 31 MDa were found. In some cases, the absence of one or two typical plasmids was observed.
In some geographic areas in the FSU, Mongolia, and China, plasmids not found in Y. pestis isolates from different parts of the world were identified. For example, 1,020 strains of Y. pestis isolated from 44 counties of Yunnan province in China and the border of China-Myanmar that were analyzed by agarose gel electrophoresis were found to carry plasmids of nine different sizes, having molecular masses of about 3.93 MDa (pYC) 6.05 MDa (pPst), 22.97 MDa (cryptic), 35.65 MDa (cryptic), 45.35 MDa (pCad), 64.82 MDa (pFra), 74.59 MDa, 111.36 MDa, and 129.55 MDa (58). Y. pestis strains carrying the specific combination of plasmids of 3.93, 35.65, and 111.36 MDa were isolated only from Yunnan province. The plasmid content of these 1,020 strains allowed for their division into 10 plasmidovars. These findings emphasize that in the strains isolated from Russia and Asia there is marked variability in plasmid content, which can potentially affect virulence and the effectiveness of host defense mechanisms.
Other variant plasmids have been identified in strains of Y. pestis. Strains isolated in Tuva (Fig. 2, foci 37 and 41) contained an additional 21.5-MDa (cryptic) plasmid (16, 67). Testing of this plasmid using PCR with a set of 10 pairs of primers based on different genes usually found on Y. pestis classical plasmids and in the chromosome indicated that this plasmid contained nucleotide sequences derived from the structural genes for plasminogen activator (pla) and the bacteriocin pesticin (pst) (from pPst) as well as from the structural gene for capsular antigen fraction I (caf1) derived from pFra (S. V. Balakhonov, Abstr. 8th Int. Symp. Yersinia, abstr. P-64, 2002). Overall, Y. pestis clearly has the capacity to harbor a large amount of genetic information in its plasmids, making these structures potential targets of genetic engineering to change virulence properties and diagnostic antigens useful for intervention in an outbreak of plague.
Variation in plasmid size.
Because the classical Y. pestis plasmids can vary in size, this property can be useful for determining the relatedness of strains isolated from different geographic areas. Also, plasmid size has the potential to be easily changed, which could be confusing with regard to whether a plasmid identified in a clinical isolate is a natural variant or potentially an engineered variant with possible additional virulence factors added in. Therefore, understanding the variation in plasmid size found among different Y. pestis isolates is important for differentiating natural variation from engineered variation leading to potentially greater virulence in strains harboring such theoretical constructs. Hence, the pPst plasmid isolated from strains circulating in several natural foci have been found to have additional sequences enlarging it from 6 up to 8 MDa (18, 62) or can even harbor a dimer of the plasmid (42). pCad and especially pFra may also vary in size depending on the geographical origin of the isolate (18, 38, 58, 62, 67-69, 112, 141; Worsham and Roy, Abstr. 8th Int. Symp. Yersinia, 2002; Worsham and Hunter, Abstr. 7th Int. Symp. Yersinia, 1998). The larger variants of plasmid pFra (69 to 190 MDa) were characteristic of the non-pestis subspecies of Y. pestis (15, 67-69, 109; V. Kutyrev, O. Protsenko, G. Smirnov, E. Bulgakova, L. Kukleva, I. Zudina, N. Vidyaeva, and I. Kuzmichenko, Abstr. 8th Int. Symp. Yersinia, abstr. P-9, 2002; Worsham and Roy, Abstr. 8th Int. Symp. Yersinia, 2002; Worsham and Hunter, Abstr. 7th Int. Symp. Yersinia, 1988). Specific plasmid rearrangements in different Y. pestis strains which distinguish Y. pestis biovar Orientalis strains from other biovars have also been demonstrated (147, 152). A more thorough determination of plasmid sizes among a large collection of Y. pestis strains allowed Filippov (67) to distinguish up to 20 plasmidovars, many of which were characteristic of strains circulating in certain natural foci (Table 10). Such natural variation could make it difficult to distinguish highly pathogenic Y. pestis from low-pathogenicity enzootic isolates without a thorough understanding of the implications of size variation.
TABLE 10.
Molecular masses of classical Y. pestis plasmids characteristic of strains from different subspeciesa
| Yersinia pestis subspecies | Biovar | Focus designation/no. | Main host | Molecular mass (MDa) of:
|
||
|---|---|---|---|---|---|---|
| pPst | pCad | pFra | ||||
| Yersinia pestis subsp. pestis | Antiqua | Most of the foci but Tuva and Alai | Rodents of different species | 6.4 | 47 | 67 |
| Medievalis | All of the foci but those of the gerbil's type | Rodents except gerbils | 6.4 | 47 | 67 | |
| Orientalis | Most of the foci | Rodents of different species | 6.4 | 47 | 64 | |
| Antiqua | Tuva/37, 41 | Citellus undulatus | 6.4 | 46 | 67 | |
| Alai/35 | Marmota caudata | 6.4 | 48 | 67 | ||
| Medievalis | All of the gerbil's-type foci | Gerbils of different species | 6.4 | 48 | 67 | |
| Yersinia pestis subsp. altaica | Medievalis | Mountain-Altai/36 | Ochotona pricei | 6.4/-b | 48 | 72 |
| Medievalis | South-Khangaiskii, Mongolia | Microtus brandti | 6.8 | 48 | 72 | |
| Yersinia pestis subsp. ulegeica | Medievalis | Northeast Mongolia, Gobi Desert | Ochotona pricei | 6.4 | 47 | 67 |
| Yersinia pestis subsp. hissarica | Medievalis | Gissar/34 | Microtus carruthersi | 6.4 | 47 | 70 |
| Yersinia pestis subsp. talassica | Medievalis | Talas/40 | Microtus gregalisc | 6.4 | 46 | 70 |
| Yersinia pestis subsp. caucasica | Antiqua | Leninakan/4; Pre-Sevan/5; Zanzegur-Karabakh/6; Dagestan-highland/39 | Microtus arvalis | 50 | 82 | |
Plasmid variation in American strains of Y. pestis.
In contrast, in the Americas there is much less variability observed in the plasmid content of Y. pestis isolates, indicating the much more restricted nature of plague diversity in this part of the world. In Brazil, a profile of four plasmids with molecular masses of 6.4 MDa (pPst), 14.9 MDa (cryptic), 44 MDa (pCad), and 60 MDa (pFra) was found in all of the 26 strains which were isolated from Paraiba State. DNA cleavage with EcoRI further demonstrated the uniform plasmid content of these Y. pestis isolates (112). When 250 Brazilian Y. pestis strains stored in bacterial collections for different periods were analyzed for plasmid content, it was confirmed that the majority of the strains contained a homogeneous pattern composed of the three classic Y. pestis plasmids: pPst, pCad, and pFra. More recent analysis of the plasmid patterns of 53 Y. pestis isolates from three plague foci from the state of Ceara, Brazil, showed that 39 strains had the three classical plasmids while seven isolates had an additional plasmid that was longer than 90 kb. The other seven lacked all or some of the plasmids (38). Fourteen of these strains with different plasmid content were analyzed by Southern blotting with probes for the genes pla (from pPst), lcrV (from pCad), and caf1 (from pFra). The pla probe reacted with pPst and with bands of about 35 kb. The lcrV probe reacted with pCad and also with bands that migrated slower than the pFra plasmid itself, as well as with bands that migrated between pFra and pCad. The caf1 probe hybridized with the pFra plasmid and to bands that migrated slower than pFra (113; N. C. Leal, M. S. B. da Silva, T. C. A. Leal, and A. M. P. de Almeida, Abstr. 7th Int. Symp. Yersinia, abstr. O-14, Med. Microbiol. [Ned. N. Voor] 6[Suppl. II]:S9, 1998). Ultimately, the Brazilian researchers came to the conclusion that “the Brazilian Yersinia pestis strains displaying atypical plasmid profiles could not represent true wild type spontaneous variants” (113; Leal et al., Abstr. 7th Int. Symp. Yersinia, 1998), since all of the “extra DNA bands, as well as the elimination of some plasmids, resulted from DNA rearrangements during storage” (38). Thus, the restricted diversity in plasmid contents of isolates of Y. pestis from Brazil is consistent with the findings that this organism has had sufficient time to spread throughout the Americas since its introduction in the early 20th century but little time to diversify in natural foci of infection.
A similar conclusion can be drawn from the study of Y. pestis isolates from the United States. Strains isolated from Arizona, California, Colorado, New Mexico, and Texas harbor the three classic Y. pestis plasmids but, interestingly, had an additional plasmid, estimated to be approximately 19 kb long. However, this plasmid was found to be a dimer of a 9.5-kb plasmid (42). Hence, only this minor change in plasmid genotype has been found in these U.S. isolates.
The potential for plasmids of Y. pestis to acquire new genetic information that would complicate diagnosis or affect pathogenesis was emphasized by the discovery of plasmid-mediated high-level resistance to multiple antibiotics, including all of the drugs recommended for plague prophylaxis and treatment, in a clinical isolate of Y. pestis from Madagascar (76). More recently, a new Y. pestis strain with a 40-kb conjugative plasmid mediating high-level resistance to streptomycin was also isolated in Madagascar from a human with bubonic plague. The plasmid and the host bacterium were different from those previously associated with multiple-drug resistance (86). Alarmingly, both of these plasmids can be transferred to antibiotic-susceptible recipients at a high frequency.
Implications of plasmid variation for virulence, diagnosis, and epidemiologic investigations.
What is clear from these studies is that among the strains of Y. pestis found in the FSU and Asia that have arisen in natural foci of infection, there is considerably more genetic diversity, and thus potential for variations in virulence, than is found among isolates of Y. pestis commonly obtained in the Americas. The overall implications of this extensive diversity are currently not known. The potential that there are virulence factors on the plasmids or within the chromosome of these non-American isolates that would be particularly harmful for humans or domestic animals is certainly possible. If some of these factors could substitute for some of the established or known virulence factors, the potential of plague-like illnesses due to strains harboring these alternative genes is real. If diagnostic tests or therapeutic reagents target virulence factors found primarily among American isolates for which there are “alternative“ factors that would not be amenable to detection or neutralization by these tests or drugs, then the utility of such tests and drugs could be circumvented. Knowing something about the natural occurrence of variant plasmids may be instrumental in epidemiologic investigations to trace the source of a plague outbreak and institute proper public health and political responses to such an outbreak. Obviously, there is a strong need to understand better the nature of the genetic diversity among Y. pestis strains, particularly in regard to their impact on infection and immunity in humans and economically important animals.
Why Is Plague Diversity in the Americas So Limited?
Epidemic spread of plague in natural foci of infection is associated with an increase in the genetic diversity of the strains. Since the introduction of Y. pestis into the Americas in the early 1900s and the subsequent spread of the organism throughout this region of the world, which occurred within a few decades, why was not a similar diversity generated? This question has no specific answer, but there are some possible reasons.
One simple explanation is that diversity has arisen in the Americas but has not yet been discovered. For several decades, hundreds of plague experts in the FSU and Mongolia were involved in field research studies of natural plague foci. To find the diversity of Y. pestis strains now known to exist there, the investigators had to analyze literally millions of rodents and fleas during different seasons in all types of natural plague foci. These investigators also had opportunities to investigate Y. pestis strains from other regions of Asia and Africa. In contrast, the effort of researchers in North, Central, and South America involved in plague research in the field was probably considerably more limited, and so there was only a very limited ability to evaluate changes in the population diversity of Y. pestis during different epizootic phases. Also, given the lack of plague in the Americas prior to 1900, the appearance of atypical strains in natural foci is assuredly going to be less frequent since the degree of diversity due to introduction of Y. pestis was limited in comparison to diversity in natural foci in Asia.
Another possibility is that diversity of Y. pestis strains from different foci in the Americas does exist and has been detected but has not yet been described in scientific papers. Some diversity was noted in a paper by Burrows and Gillett (33). However, it appears that the knowledge of diversity that has arisen from genotyping of Y. pestis isolates from the FSU and Asia is not generally shared or appreciated among investigators in the Americas, and there may be more diversity in these strains than what has been described in the literature. Since the importance of a better understanding of plague diversity and its relation to virulence and disease has increased dramatically in the past few years, the observations documented here should serve as a basis for further explorations of the type of genetic diversity that exists among American strains of Y. pestis.
OTHER METHODS OF STRAIN CHARACTERIZATION AND THE IMPACT ON GENETIC DIVERSITY OF YERSINIA PESTIS
Numerous studies have used other methods of characterization of Y. pestis strains to look for genetic variability among isolates from different parts of the world. Evaluation of SpeI macrorestriction pattern polymorphism by pulsed-field gel electrophoresis showed that although Y. pestis strains from different natural foci have the same chromosome size, each of the biovars has its own restriction pattern (119). Pulsed-field gel electrophoresis and restriction digests using SpeI and NotI were successfully used in Madagascar for epidemiologic purposes (C. Buchrieser, B. Rasoamanana, L. Rahalison, S. Chanteau, and E. Carniel, Abstr. 7th Int. Symp. Yersinia, abstr. O-12, Med. Microbiol. [Ned. N. Voor] 6[Suppl. II]:S8, 1998).
Using subtractive hybridization, an analysis of 78 strains of the three Y. pestis biovars obtained from diverse geographical origins was carried out, based on finding six difference regions (DFRs) (from 4.6 to 19 kb in size) (155). This study showed that the Orientalis biovar had a specific DFR profile that was different from that of the two other biovars, Antiqua and Medievalis. The Antiqua and Medievalis biovars also had specific DFR profiles, as well as some that were common between the two biovars. Y. pseudotuberculosis strains possessed their own DFR profile (G. L. Andersen, L. Radnedge, P. Agron, and P. L. Worsham, Abstr. 8th Int. Symp. Yersinia, abstr. P-1, 2002). This result supports the conclusion that the division of Yersinia pestis isolates into biovars is associated with genetic differences among these strains.
A 41.7-kb region unique to Y. pestis was identified by suppression subtractive hybridization with Y. pestis as the tester and Y. pseudotuberculosis as the driver. This region exhibits similarity to putative phage genes, many of which are found in Escherichia coli O157:H7. Within 28 Y. pestis and 5 Y. pseudotuberculosis strains under study, primer pairs designed based on nucleotide sequences from within this region were found to be highly specific for Y. pestis. Using both biovar and subspecies classifications of Y. pestis, one primer pair, 3a, amplified PCR products from all biovars and subspecies, while three other primer pairs, 3b, 3c, and 3d, amplified PCR products from 20 strains of the pestis subspecies in all of the biovar variants of this subspecies, and 4 strains of subspecies caucasica, representing 3 of biovar Antiqua and 1 of an undescribed biovar. However, primer pairs 3b, 3c, and 3d did not react with other representatives of the Medievalis biovar strains of the non-pestis subspecies. None of the primers amplified DNA from Y. pseudotuberculosis (156). Interestingly, this genotyping technique placed Y. pestis subsp. caucasica closer than other non-pestis subspecies to the pestis subspecies, while data obtained by other methods suggest that subsp. caucasica is the phylogenetically most ancient Y. pestis strain (24, 25, 67-69, 73, 130; Worsham and Roy, Abstr. 8th Int. Symp. Yersinia, 2002).
Ribotyping has also been used to elucidate genetic diversity in Y. pestis. Hybridization of EcoRI and EcoRV Y. pestis genome restriction fragments with a 16S-23S rRNA probe from E. coli resulted in the elucidation of 16 ribotypes. Ribotypes designated B and O were characteristic of 65.7% of the strains studied. Strains of biovar Orientalis were of ribotypes A to G, those of biovar Antiqua were of ribotypes F to O, and those of biovar Medievalis were of ribotypes O and P. Great heterogeneity in ribotypes was found among strains isolated in Africa, whereas there was no ribotype heterogeneity among the American strains (85), again pointing out the limited genetic diversity of Y. pestis isolates from the Americas.
Ribotyping was useful for showing both the stability of Y. pestis strains in a given environment and the potential of newly introduced strains to replace indigenous ones. Y. pestis was introduced into Madagascar just under a century ago. When 187 Y. pestis strains isolated mainly from human cases between 1939 and 1996 from different regions of the island were studied, all of those isolated before 1982 were of ribotype B, the ribotype ascribed to the Y. pestis clone that spread globally via marine shipping during the third pandemic. In 1982, 1983, and 1994, strains with new ribotypes, designated R, Q, and T, correspondingly, were isolated on the high-plateau region of Madagascar. Analysis of the NotI genomic restriction profiles and the EcoRV plasmid restriction profiles showed that the new variants could also be distinguished by specific genomic and/or plasmid patterns. The strains of ribotypes Q and R are now well established in their ecosystem and have had a tendency to spread to new geographic areas and dislodge the initial ribotype B strains (84). Thus, when introduced into a new environment, genetically diverse strains of Y. pestis can spread rapidly and even become the dominant genotype in certain areas.
A similar pattern of change in Y. pestis strains was observed in Vietnam. The majority of Y. pestis isolates from Vietnam were biovar Orientalis, with a minority of biovar Medievalis isolated from a local area of the mountainous region of the country (T. T. X. Mai, A. Guiyoule, D. X. Vinh, D. T. N. Tuyet, D. Thung, and E. Carniel, Abstr. 7th Int. Symp. Yersinia, abstr. P-70, Med. Microbiol. [Ned. N. Voor] 6[Suppl. II]:S30-S31, 1998). Ribotyping of 108 Y. pestis strains isolated between 1955 and 1996 indicated that strains isolated in the southern part of Vietnam before 1956 were ribotype E while from 1963 on, all studied strains isolated from different regions of Vietnam were of ribotype G (Mai et al., Abstr. 7th Int. Symp. Yersinia, 1998).
There is considerable diversity in Y. pestis global isolates as determined by VNTR analysis. A tetranucleotide repeat sequence, (CAAA)N, was identified in the genome of Y. pestis. This VNTR region has nine alleles (from 3 to 13 repeats). In 35 Y. pestis strains isolated from different natural plague foci, the Antiqua biovar was the most diverse, with four alleles in 5 strains, while the Orientalis and Medievalis biovars exhibited five alleles in 21 strains and three alleles in 8 strains, respectively (3). Only one allele, allele H, was found in both the Antiqua and Orientalis biovars. The diversity among these VNTR alleles can be calculated, and a diversity value, DV, equal to 1 minus the sum of the squared allele frequencies, can be derived. For the 35 isolates initially studied by VNTR, the DV was 0.82, indicative of a high level of diversity among the studied strains.
Another study of the variability in the (CAAA)N locus among 61 strains isolated from natural foci in the FSU (Table 11) and 8 strains of other origins identified 10 VNTR alleles (from 2 to 14 repeats) and also great diversity (DV = 0.86). This index was shown to vary significantly, from 0.24 in a group of seven strains isolated from voles living in the Leninakan and Zanzegur-Karabakh foci (Fig. 2, foci 4 and 6, respectively) to 0.77 in nine isolates from gerbils living in the Muyun-Kum, Kyzyl-Kum, Pre-Aral-Kara-Kum, Kara-Kum, Mangyshlak, Ural-Emba, Bozchel', and Dzheiranchel' foci (foci 28, 27, 24, 25, 23, 18, 8, and 11, respectively) (174).
TABLE 11.
(CAAA)N alleles of Y. pestis isolates from the FSUa
| Main host | Focus designation/no. | No. of strains | No. of repeatsb |
|---|---|---|---|
| Marmot | Alai/35 | 1 | 6* |
| 1 | 8* | ||
| 1 | 9 | ||
| 2 | 11 | ||
| 1 | 14 | ||
| Aksai/33 | 2 | 11 | |
| 3 | 12 | ||
| Suslik | Trans-Baikal/38 | 2 | 7 |
| 1 | 7* | ||
| 2 | 9 | ||
| 1 | 11 | ||
| Pre-Caspian northwestern/14 | 2 | 9 | |
| Volga-Ural steppe/15 | 1 | 9* | |
| 1 | 13 | ||
| Dagestan-plain-foothill/3 | 1 | 9* | |
| 1 | 12 | ||
| Central-Caucasian/1 | 3 | 6 | |
| 7 | 7 | ||
| 2 | 7* | ||
| Vole | Zanzegur-Karabakh/6 | 4 | 4 |
| 1 | 12 | ||
| Leninakan/4 | 2 | 4 | |
| Talas/40c | 4 | 6 | |
| 1 | 7 | ||
| 2 | 11 | ||
| Gerbil | Kobystan/9 | 1 | 9 |
| 1 | 9* | ||
| 1 | 11 | ||
| Bozchel'/8 | 1 | 12 | |
| Dzheiranchel'/11 | 1 | 9 | |
| 1 | 11 | ||
| Ural-Emba/18 | 1 | 12 | |
| Mangyshlak/23 | 1 | 14 | |
| Pre-Aral-Kara-Kum/24 | 1 | 9* | |
| Kara-Kum/25 | 1 | 11 | |
| Kyzyl-Kum/27 | 1 | 9 | |
| Muyun-Kum/28 | 1 | 9 |
VNTR analysis also showed that there is very limited diversity in Y. pestis strains isolated in the Americas compared with those from Russia and Asia. The allele polymorphism of 42 VNTR loci found among 12 Y. pestis isolates (biovar Orientalis) from Siskiyou County in California showed that there was an average of only 1.8 alleles in these strains compared with an average of 4 alleles among 12 isolates representing all three biovars from a broad worldwide distribution (103).
Another 8-bp VNTR, (ATAGAAAG)N, was found in the sequences flanking the pigmentation locus of Y. pestis. Studies of 56 strains representing the 3 biovars, 22 ribotypes, and an additional 20 non-typed Y. pestis strains revealed 11 alleles. Different strains contained 3 to 12 repeats of the octonucleotide sequence, which included 9 entire repeats with four additional internal bases, AAAG. The Orientalis biovar was the most diverse, with 9 of the 11 alleles, while the Antiqua biovar exhibited 3 alleles (from five to seven repeats) and the Medievalis biovar was exclusively represented by a single allele of four repeats. Five Y. pseudotuberculosis strains under study contained only one copy of this VNTR (L. P. Blackman, Abstr. 8th Int. Symp. Yersinia, abstr. P-2, 2002).
Complete Genome Sequencing
Although none of the atypical Y. pestis strains have been sequenced to date, two typical strains have been (48, 137). The two completely sequenced strains are members of two different classic biovars that carry the standard complement of plasmids harbored by Y. pestis. Comparison of these two genome sequences has offered some insight into the genetic fluidity of this pathogen as well as the genes that may differentiate this acute bacterial pathogen from its enteropathogenic ancestor (2). The results of these genome-sequencing projects further demonstrate how Y. pestis strains may be genetically diverse.
The first complete Y. pestis genome to be reported was that of strain CO92 (137). CO92 was isolated in 1992 from a human case of fatal pneumonic plague acquired in Colorado (54). Like all other American isolates, CO92 is of the Orientalis biovar. Given that Y. pestis CO92 is a recent isolate that is typical in plasmid content and biochemical properties for this biovar, the complete genome sequence can be considered representative of similar isolates commonly found in the Americas. The sequence would be expected to be representative of the general gene content but not necessarily the gene order, as has been suggested by restriction digestion of the genomes of various isolates (84, 85, 94, 119). This conclusion is supported by the relatively few differences found between CO92 and the other completely sequenced strain, Yersinia pestis KIM (Kurdistan Iran Man) (48), which is a biovar Medievalis strain obtained from a geographically distinct region (see below).
Chromosome.
The chromosome of Y. pestis contains many regions that appear to have been acquired by horizontal gene transfer. A total of 21 regions were found to be significantly different in guanine plus cytosine (G + C) content from the majority of the CO92 genome (137). Many of these regions are near tRNA genes and are therefore similar to pathogenicity islands (129). Although the G + C content of the entire CO92 chromosome was 47.6%, these variable regions ranged in G + C content from 32.8 to 56.4%. One of the most interesting regions, with a significantly different G + C content, encodes a putative type III secretion system whose function has yet to be identified. One of the genes within the chromosomal type III secretion system appears to be nonfunctional due to a frameshift mutation in the KIM strain (48). Other apparently recently acquired regions were predicted to encode insect-associated viral pathogenicity factors or toxins, iron acquisition proteins, adhesins, phage remnants, and secretion-associated proteins (137). The insecticidal toxin genes found in Y. pestis CO92 were also reported to be carried in the genome of Y. pseudotuberculosis strain IP 32953, demonstrating an early association between this genus and insects.
On a gross level, there are significant differences between the Medievalis KIM genome and the Orientalis CO92 genome in terms of gene content. Although the Orientalis biovar is thought to have emerged more recently (2, 52), the CO92 genome is ∼50 kb larger than the KIM genome due to one 11-kb insertion as well as additional smaller insertions (48). The number of copies of insertion elements (IS) is larger in CO92, accounting for ∼27 kb of the increased size of this chromosome. The low-G + C island that encodes the insect-associated toxins, Tcc and Tca, includes a putative single-stranded prophage in the genome of strain CO92 that was not found in the genome of strain KIM, further demonstrating a genetic basis for differentiating Y. pestis strains and possibly biovars. Conversely, the genome of strain KIM encodes proteins that are not found in the genome of strain CO92. There were 526 putative proteins that did not match CO92, mostly due to the larger number of pseudogenes. Of the 526 open reading frames ORFs, 208 appeared to encode specific hypothetical proteins in the genome of Y. pestis KIM; that is, they did not match any proteins with known function. The KIM-specific genes also tended to use rare codons compared to “backbone” genes that are in common with E. coli K12, further suggesting the horizontal acquisition of numerous genes from outside Y. pestis (48). Additionally, the strain KIM genome contains six rRNA operons whereas the strain CO92 genome contains only five. Taken together, the above observations demonstrate that the Y. pestis chromosome is still in a state of flux, modified by the gain and loss of genetic information. The fact that Y. pestis KIM contains approximately 208 ORFs not found in the CO92 strain could easily be enough to explain potential differences in host specificity or virulence between the two, although none are currently known. Conversely, the fact the strain CO92 has more than 20 kb of non-IS-associated genetic material not found in strain KIM could also be enough to account for potential differences in the pathogenicity of these two strains. Accordingly, it is reasonable to speculate that genetic differences may help explain, in part, the differences in pathogenesis among the variant Y. pestis strains circulating in the world.
Pseudogenes.
One of the striking features of the Y. pestis CO92 genome was the identification of 149 pseudogenes in the sequence. The fact that the number of pseudogenes classified as potential virulence factors outnumbered the apparently functional genes in that group by a ratio of 3:1 underscores the genetic differences that separate the progenitor enteropathogen from what we now know as Y. pestis. A large number of these pseudogenes resulted from single-base-pair frame shifts and thus have the potential to easily be transformed into functional genes. One basis for the occurrence of pseudogenes may be inferred from the early observation that many Y. pestis strains can be induced to synthesize some metabolic precursors if selective pressure is applied (28). This may arise from maintenance of a reservoir of pseudogenes that could potentially become functional genes with only limited DNA changes, providing the needed genetic material for rapid emergence of strains adapted to new or changed environments. Interestingly, the number of pseudogenes is even larger in Y. pestis KIM (48). The KIM strain is of the putatively older biovar Medievalis (52) and therefore might be expected to encode fewer pseudogenes due to loss of nonessential genetic material if this biovar preceded Orientalis. However, strain KIM was isolated from a distinct region of the world in 1961 and has been passed in the laboratory and out of the natural environment for over 40 years (70). Alternatively, different lineages or environmental pressures may have given rise to these two biovars or strains.
Insertion sequences.
The number of IS elements found in the Y. pestis genome is large, with the highest copy number belonging to the IS200-like element IS1541. The second most abundant IS is IS100, followed by IS 285 and IS1661. The continued mobility of these IS elements is highlighted by a comparison of the two sequenced Y. pestis genomes. A larger number of complete or partial copies of IS1541 (eight more), IS100 (nine more), and IS285 (two more) was noted in strain CO92 compared with strain KIM. The presence of the IS elements, and their activity, probably explain many of the observations that have been made during attempts to genotype Y. pestis. Most notably, the macrorestriction pattern of the genome of at least two strains appears to be extremely unstable even after serial passage (85), although others have not observed these rapid genomic rearrangements in other strains (119). The rearrangement of specific regions of the genome could be detected by PCR after growth of cultures of CO92 (137). This characteristic has been a useful tool for genotyping isolates from geographically related regions within the United States (94). Genome sequencing has identified one of the metabolic lesions responsible for one of the phenotypic characteristics used to differentiate biovars of Y. pestis.
The inability to ferment glycerol is a characteristic of the Orientalis group (52). Motin et al. (130) identified two different deletions within genes involved in glycerol fermentation. One mutation was a deletion that spanned glpK-glpX genes and was found only in Y. pestis isolates from the United States. The second mutation was a 93-bp deletion in glpD and was found in all Orientalis strains examined, suggesting that this lesion is responsible for the general inability of this biovar to ferment glycerol. Thus, deletion mutations have been established that explain at least one of the defining characteristics of biovars of Y. pestis. The mobility of IS elements, along with point mutations and larger deletions, could explain the phenotypic changes and possibly host preferences and pathogenicity differences noted above for some strains of Y. pestis, although these ideas still await experimental confirmation.
The fluidity of the Y. pestis genome is also evidenced by the gene order and G + C content of the DNA that makes up the chromosome. There is a high propensity for the leading strand of replication to have a high-G + C bias in E. coli. This is not true in either strain of Y. pestis that has been sequenced. The G + C bias alternates between leading and lagging strands of DNA (48, 137), indicating inversions of large regions of the chromosome. When strain KIM was compared to CO92, three regions of multiple inversions were noted. This was after identifying 27 regions of the genome that either were directly the same between the two strains or were comparable, with changes due merely to the inversion of one of the regions. Thus, on the large scale, there are twenty-seven blocks of genes that can be rearranged within the Yersinia pestis genome that can be assembled in various ways and still result in an organism that can cause plague in infected animals. There are smaller regions within these twenty-seven that can also be rearranged and add to the complexity of the observed restriction patterns.
Plasmids
The fact that Yersinia virulence is strongly linked to the presence of one particular plasmid points to the importance of this type of element in the evolution of pathogens. More specifically, the fact that typical strains of Y. pestis harbor two additional plasmids underscores this point and partially explains the difference in pathogenesis of this member of the genus. Since many of the atypical strains of Y. pestis have different plasmid profiles compared with the more typical isolates of the pestis subspecies (15-18, 57, 62, 67-69, 79, 167, 196), this also demonstrates the importance of plasmids in the virulence of this organism. Acquisition of new plasmids by emerging strains of Y. pestis is an ongoing process (57). Plasmid-specific differences have been identified as part of the genome sequence projects of strains KIM and CO92.
The “LCR” plasmid.
The Low-Calcium-Response (LCR) plasmid (pLcr) encodes a major group of defined virulence proteins termed Yops (for “Yersinia Outer Proteins”) (29, 44). Curing of this plasmid results in total avirulence of the Y. pestis strains that have been studied (29, 111). The core region involved in Yop secretion is a contiguous group of genes of ∼25 kb that is present on all of the sequenced pLcr plasmids (97, 143, 170). The effector Yops, as well as specific accessory proteins, are located at different places within the plasmid. The known differences between the KIM and CO92 pLcr molecules provide some further insights into Y. pestis strain variation.
pLcr was 70,509 bp long in strain KIM (143) and 70,305 bp long in CO92 (152). A 212-bp deletion of a partial IS285 sequence accounted for most of the difference in size between the two plasmid molecules. The major gross difference was in the location of an IS100 insertion (152). This IS100 insertion occurs within two different degenerate elements (IS21 in CO92 and ISD1 in KIM) and in two different places on the plasmid. In KIM the insertion occurred between yscM and yopH, while in CO92 it occurred between sycE and sycH. However, the location of this IS within pLcr was not biovar specific. This strongly suggests the ongoing mobility of IS elements in Y. pestis. Several other minor differences were noted between the sequences, but the overall gene order was conserved between the KIM and CO92 plasmids except for the position of the IS100 element noted above.
The murine toxin plasmid pFra.
The ∼100-kb plasmid resident in Y. pestis encodes the capsular antigen F1 and the Yersinia murine toxin Ymt (116, 152). This plasmid is generically referred to as pFra. The major difference between the Y. pestis KIM and CO92 pFra sequences is due to IS100 activity. The IS activity resulted in two large deletions of ∼2 kb each from the CO92 pFra plasmid compared to the KIM plasmid (152). Apparently further recombination between the two copies of IS100 present on pFra resulted in a 37-kb region being inverted. Thus, the gene order in this area is different when pFra from strains KIM and CO92 are compared. These two large 2-kb deletions occurred in 11 biovar Orientalis strains but not the Antiqua or Medievalis strains examined (152). Other than a few single-base-pair changes, little difference was seen between the sequences common to KIM and CO92. In fact, the variability in sequence between the two chromosomes was about the same as that seen between two pFra plasmids harbored by two different isolates of KIM held in two different laboratories (116, 152). Thus, similar to the architecture of the chromosome, the major difference between pFra harbored by strains KIM and CO92 is in the gene order and not the overall gene content.
The most striking feature of pFra is that over 50% of the plasmid is >95% identical at the DNA level to a cryptic plasmid harbored by Salmonella enterica serovar Typhi (152). The homology is not contiguous but, rather, is dispersed in large blocks around most of pFra. The G + C content of the shared regions is more similar to that of Salmonella than Yersinia, suggesting that the prototypical plasmid was of enteric origin. pFra and the Salmonella plasmid have a common origin of replication but encode different partitioning regions. No obvious transfer regions are present on pFra or the corresponding plasmid, pHCM2, of Salmonella. The discovery of this high level of DNA homology that is shared between a Y. pestis plasmid and an S. enterica serovar Typhi plasmid indicates the potential for the exchange of genetic material between these two pathogens with different lifestyles (152), although it is also possible that the plasmids were acquired from a separate source by both Y. pestis and Salmonella. The transfer of conjugative plasmids between enteric bacteria and Y. pestis has recently been directly demonstrated to occur inside the flea (91). In light of this information and since pFra has been shown to vary in size when obtained from natural isolates (15, 67-69, 109; V. Kutyrev et al., Abstr. 8th Int. Symp. Yersinia, 2002; Worsham and Roy, Abstr. 8th Int. Symp. Yersinia, 2002; Worsham and Hunter, Abstr. 7th Int. Symp. Yersinia, 1998), it seems important to undertake further investigations of the mechanism of this variability.
The pesticin plasmid pPst.
The pesticin plasmid, pPst, is the smallest (∼9.5 kb) of the typical plasmids harbored by Y. pestis. There are very minor differences between the plasmid in strain KIM and that in strain CO92. These differences were found to be in intergenic regions and not in coding regions on the plasmid (152). However, some strains have been shown to be naturally devoid of this plasmid but are nonetheless virulent (12, 15, 67-69, 73, 99, 168, 186; Bakanidze et al., Abstr. 8th Int. Symp. Yersinia, 2002; Worsham and Roy, Abstr. 8th Int. Symp. Yersinia, 2002; Worsham and Hunter, Abstr. 7th Int. Symp. Yersinia, 1998). The only known virulence factor encoded by pPst is the coagulase/fibrinolysin also known as plasminogen activator. This protein is thought to promote the systemic spread of Y. pestis from peripheral sites (171, 186). The bacteriocin, pesticin, and the pesticin immunity protein add selective pressure for maintenance of the plasmid. There is also a complete copy of IS100 on this plasmid, but it appears to be in the same position on pPst from strains KIM and CO92, unlike the position of this IS on pLcr and pFra (see above). Finally, the pPst plasmid encodes a ColE1-like origin of replication, again suggesting common ancestry between plasmids typically found in enteric organisms and pPst. Given that some strains of Y. pestis do not appear to require this plasmid and its activities for virulence yet other strains do, it will be of interest to determine which features differ between these strains and which are in common between them.
The emerging plasmid pYC.
The acquisition of new plasmids by Y. pestis is a well-documented occurrence (57, 67-69, 76, 86), and, given the involvement of these extrachromosomal elements in the pathogenesis of the organism, it is likely that new virulence factors could be readily introduced into Y. pestis strains. Currently, the only fully sequenced new plasmid that has been isolated from Y. pestis is the 5.9-kb cryptic plasmid designated pYC (57). This small plasmid has been increasingly isolated from strains recovered in an ever-widening area around the Yunnan province of China. The complete DNA sequence did not reveal any obvious potential virulence factors. This plasmid contains 12 potential ORFs, of which only 3 had any significant predicted similarity to proteins of known function. One of these putative proteins was highly similar to the replication protein encoded by a Neisseria plasmid such that this protein and surrounding DNA repeat sequences were designated as the likely origin of replication. The other two putative proteins were highly homologous to the E. coli DinJ1 and DinJ2 DNA damage-inducible proteins. These proteins might help Y. pestis survive under stressful conditions. The last notable feature of pYC was a 70-bp region that was almost identical to sequences found on a Shigella plasmid, again underscoring the linkage between enteric bacteria and gene acquisition by Y. pestis. The fact that the geographic region containing strains of Y. pestis harboring pYC is expanding strongly indicates that this molecule offers a selective advantage to the organism, even without any obvious phenotype such as a bacteriocin activity like that found on pPst or genes encoding antibiotic resistance.
DIAGNOSIS, TREATMENT, AND PREVENTION
The key issue of diversity in Y. pestis isolates relevant to humans is how this impacts on pathogenic potential, diagnosis, treatment, and prevention of plague. In view of the current concern about the potential use of the organism as a weapon of bioterrorism, there is considerable interest in knowing whether there are ways to use the naturally occurring genetic diversity in Y. pestis to increase virulence and/or thwart diagnosis, treatment, or prevention. Unfortunately, we do not have an answer for these questions that can be discerned from published or unclassified studies. Nonetheless, while not meant to be a comprehensive discussion, a brief review of some of the current diagnostic, therapeutic, and prophylactic strategies being used for Y. pestis infection will help illuminate some of the areas needing further study and research in relation to the issue of the large-scale genetic diversity in this organism.
Laboratory Diagnosis
Rapid and accurate laboratory diagnosis of Y. pestis infection is a key to monitoring the presence of this organism in natural foci and in cases of human disease. Samples for analysis can include human blood or lymph node aspirate, bubo fluid, sputum or throat smears, autopsy material, specimens from rodents and their ectoparasites, and even air samples (13, 41, 49, 55, 56, 95, 132, 133, 135, 141, 146). At present, the definitive laboratory diagnosis of plague is made by the isolation in pure culture of Y. pestis, along with appropriate microbiological studies (microscopy of stained preparations, characteristics of growth in liquid and solid media, biochemical characterization, and lysis of pure cultures by specific diagnostic bacteriophages). Additional methods include the detection of Y. pestis antigens with specific antibodies in direct fluorescence tests, or the detection of Y. pestis-specific DNA sequences. Accurate laboratory classification of an isolate as Y. pestis is essential for many reasons: identification of strains recovered from enzootic and infectious human cases of plague, the ability to make a provisional diagnosis of infection when rapid decisions regarding treatment are needed, confirmation of clinical diagnosis, and retrospective diagnosis in appropriate situations. Accurate laboratory identification guarantees the safe control and handling of specimens and is essential for the surveillance of the spread of Y. pestis in natural plague foci (11, 13, 31, 41, 49, 55, 56, 95, 132, 135, 141, 146).
There is a particular concern with diagnostic reagents that rely on the production of specific Y. pestis antigens. Even the recently described rapid detector of potential pathogens (158) relies on antibodies to identify specific antigens of pathogens, which, if altered, could neutralize the effect of the rapid diagnostic sensor. The genetic diversity that Y. pestis can call upon makes it particularly worrisome that high-technology systems relying on using the knowledge of existing Y. pestis antigens, DNA sequences, or the like for diagnosis could be easily thwarted by using genetic techniques to change some of the parameters. Thus, while some of the older biochemical and morphological tests used might seem antiquated in a modern diagnostic laboratory, knowledge of these features of the variant strains of Y. pestis may be critical to diagnosis and epidemiologic investigations of future outbreaks.
Serotyping and Phage Typing
It is commonly thought that Y. pestis, in contrast to its nearest relatives, Y. pseudotuberculosis and Y. enterocolitica, belongs to a single phage type as well as to a single serotype (28, 52, 55). This homogeneity is thought to be due to the absence of O-side chains in the LPS, leading to a rough-type LPS, sometimes referred to as a lipooligosaccharide (55, 141). Since phages and serotyping reagents are often specific to O-side chains, the lack of such structures in Y. pestis may underlie the common belief that there is little value in serotyping or phage typing these strains.
However, there are bacteriophages specifically lytic for the overwhelming majority of Y. pestis strains (41, 55, 133, 135, 141), and they can be used for diagnostic purposes. Strains falling into different Y. pestis subspecies were recently shown to differ in their sensitivity to the rough-LPS-specific FP1 phage. Phage-susceptible strains were found mostly among subspecies pestis and altaica isolates. Among the rest of the subspecies, phage-resistant strains were notably more frequent. Electron microscopy evaluation of phage FP1 adsorption onto bacterial cells showed that adsorption is strain and temperature dependent (Balakhonov, personal communication; R. Z. Shaikhutdinova, T. A. Gremyakova, E. L. Zhilenkov, S. V. Dentovskaya, and A. P. Anisimov, Abstr. 8th Int. Symp. Yersinia, abstr. P-53, 2002). Thus, development of a phage-typing system will have to take these findings into consideration.
A number of investigators have reported on the abilities of monoclonal and polyclonal antibodies to Y. pestis LPS to detect a wide range of Y. pestis strains differing in their geographic origin, plasmid content, and culture temperature (50, 153). Interestingly, differences in serologic specificities of antibodies to LPS from Y. pestis subspecies pestis and caucasica may often be primarily due to temperature-dependent variations in the structural, and hence serologic, properties of lipooligosaccharides (T. A. Gremyakova, personal communication). Recent studies of the structure of the Y. pestis LPS, the first of which was reported recently (185), show variations in both oligosaccharide and lipid A composition based on strain and growth temperature (80, 102; Knirel et al., Abstr. Carbohydr. Workshop, 2003).
With regard to laboratory diagnosis, the way in which the phenotypic and genotypic diversity of Y. pestis isolates found among non-American strains will impact the accurate identification of Y. pestis is a matter for concern. One main modern diagnostic test used is based on a capture enzyme-linked immunosorbent assay to detect the F1 capsular antigen or detection by PCR of the structural gene for this protein, caf1. Synthesis of the F1 capsule occurs when Y. pestis is grown at 37°C (13, 41, 55, 56, 95, 132, 133, 135, 141, 146). However, except in some situations (13), it is generally recommended to grow suspected cultures of Y. pestis obtained from the field or clinical samples on solid media at 28°C (41, 55, 56, 95, 132, 133, 135). Under these growth conditions, F1 protein synthesis decreases by approximately 800- to 1,000-fold compared with its production at 37°C (14). Correspondingly, Y. pestis does not synthesize F1 in the flea vector (39). Therefore, immunodiagnosis based on detection of F1 antigen is useful only after cultivation of bacteria at 37°C.
Of greater concern is the observation that elimination of pFra, which carries the caf1 operon, or mutations in the caf1 operon itself can lead to cessation of capsule formation. Correspondingly, this makes Y. pestis cells undetectable by immunologic tests targeting F1 antigen, yet in some cases it does not decrease the virulence of the strain (47, 59, 72, 110, 111, 163, 187, 194). Time and again, F1 antigen-negative Y. pestis strains are isolated from different animal species (9, 19, 72, 98, 145, 190, 191), in at least one case, such a strain has been isolated from human infection (193). According to data derived from many studies performed by the experts of the Anti-Plague Establishment of the FSU with many epizootic outbreaks, the frequency of isolation of all atypical Y. pestis variants was on the order of 7%. F1-negative variants accounted for up to 15.8% of all atypical strains (9, 104, 195).
The virulence for humans of Y. pestis strains not producing the F1 antigen has spurred interest in the development of new diagnostic tests targeting other Y. pestis antigens that are synthesized in comparable amounts at both the mammalian host temperature (≥37°C) and temperatures commonly found in flea vectors (∼28°C). Such tests could focus on the genes or proteins encoded by ymt, pla, or pst or on the production of LPS (31, 141). In addition, it would be useful if the production of a diagnostic antigen was also necessary for the manifestation of human infection without regard to the route of infection. These could include such factors as Yops, V antigen, PsaA (for “pH Six Antigen“), and components of the system needed for siderophore-dependent acquisition of iron, Ybt (for “yersiniabactin”), or Psn (for “pesticin receptor“) (8, 9, 31, 141). Other potential diagnostic targets are factors necessary for effective transmission of infection, such as Ymt, which ensures colonization of the flea midgut (90), and components of the hemin storage system (Hms), which ensures blockage of the flea proventriculus that is required for effective transmission (89, 92). Although some results of the attempts to develop such diagnostic tests have been published (19, 50, 51, 64, 65), none have led to the development of commercially available diagnostic tests.
Antibiotic Treatment
The specific therapy for Y. pestis infection, as suggested by the World Health Organization Expert Committee on Plague (1970), focuses on the use of tetracycline, streptomycin, and chloramphenicol antibiotics. More recently, the U.S. Working Group on Civilian Biodefense added gentamicin, doxycycline, and ciprofloxacin to this list (95). It was accepted until recently that, as a rule, natural isolates of Y. pestis lacked much in the way of antibiotic resistance, in contrast to many other pathogenic bacteria. The infrequent recovery of drug-resistant natural isolates of Y. pestis (141) was explained by the relative rarity of cases of human plague at present and the acute character of the disease, along with the need for transmission of the organism by fleas, which would limit the contact between Y. pestis and natural reservoirs of antibiotic resistance factors in other microbes (55). However, it was recently shown that in fact the flea midgut was a good environment for conjugative transfer of plasmids mediating high-level resistance to multiple antibiotics, including all of those recommended for plague prophylaxis and treatment (91). This finding may account for isolation of the conjugative plasmid-mediated multidrug-resistant (76) and conjugative plasmid-mediated streptomycin-resistant (86) strains in Madagascar. Naturally occurring strains with undefined mechanisms of antibiotic resistance were also rarely isolated in Vietnam (191) and Mongolia (11, 62).
It has also been suggested (A. P. Anisimov and I. A. Dyatlov, Letter, J. Med. Microbiol. 46:887-889, 1997) that the treatment of plague caused by F1-negative Y. pestis strains was less effective when antibiotics such as doxycycline, ampicillin, and cefoperazone were used (138, 162, 164, 165) since they are unable to enter macrophages, and hence phagolysosomes, in an active form (161). However, in vitro, the F1-negative strains were as susceptible to antibiotics as were wild-type, F1-positive bacteria (138, 162, 164, 165). The F1-negative strains were also resistant to tetracycline, β-lactam agents, and quinolones when inside macrophages cultivated in vitro (138, 162, 164, 165). It has been proposed that the capsular F1 antigen might increase the macrophage membrane permeability for a range of antibiotics and, respectively, that phagocytosed F1-negative bacteria are protected from contact with the antibiotic and are able to multiply within the macrophage (Anisimov and Dyatlov, letter). Chemotherapeutic drugs such as aminoglycosides (streptomycin, kanamycin, tobramycin, gentamicin, and amikacin) and cephalosporins (ceftriaxone and ceftazidim) are recommended for the treatment of plague caused by F1-negative Y. pestis strains. Studies with mice suggest that an increase in the daily doses of less efficient drugs such as cefotaxime, cefoperazone, sulbactam-ampicillin, aztreonam, ciprofloxacin, and rifampin, along with prolongation of the treatment course to up to 7 days, made it possible to increase the protective effects up to 80 to 100%. Doxycycline and ampicillin were not efficient even when used for 10 days in the prophylaxis of plague caused by F1-negative Y. pestis strains (162).
VACCINATION FOR PREVENTION OF PLAGUE
The potential impact of the large genetic and phenotypic diversity in Y. pestis isolates outside of the Americas on current research initiatives is exemplified by the possible effect of such diversity on the development of plague vaccines. Immunization against plague is one major approach being pursued to deal with the potential threats from the use of this organism as a bioterrorist agent. As with any vaccine, successful development is dependent on an understanding of the basic immunologic effectors and target antigens that are involved in mediating high-level immunity. To date, however, this type of information has not been incorporated into any of the extant human plague vaccines, which currently are based on either a live attenuated strain or a killed whole-cell preparation. The live attenuated vaccine is produced in the FSU and is based on Y. pestis strain EV, line NIIEG. This strain is attenuated due to deletion of the 102-kb pgm locus, which includes the hms locus responsible for the ability to store hemin and a cluster of genes needed for production of the siderophore-based yersiniabactin biosynthetic/transport systems. The parental wild-type strain was isolated in Madagascar. The second vaccine is a formalin-fixed virulent Y. pestis strain 195/P (originally isolated in India) that was developed in the United States, although presently it is manufactured only in Australia (124, 134).
Given that these types of vaccines generally have unacceptable side effects when used in large civilian populations, there has been a focus on developing subunit vaccines that offer the advantage of being a defined antigen that possesses the ability to raise protective circulating antibodies and is also less reactogenic than the currently used whole-cell vaccine (31). However, given the genetic and phenotypic diversity of Y. pestis strains, it is not clear if the subunit vaccines under development, based primarily on antigens expressed by American isolates of Y. pestis, would be comprehensive enough to provide extensive coverage against the potential strains that could cause plague in humans. Two major antigens of Y. pestis have been the focus of most of the attention in this regard, the F1 capsule antigen and the LcrV (or simply the V) antigen. The former is synthesized by the caf1 operon and is a protein capsule commonly expressed by many isolates. The second is a component of the type III secretion system, whose activity is thought to be critical to virulence for all isolates of Y. pestis. Since F1 capsule-negative Y. pestis strains have been recovered from at least one case of human infection (193) and there is serologic diversity in the V antigen (159; K. F. Griffin, J. Hill, K. Murray, S. E. C. Leary, R. W. Titball, and E. D. Williamson, Abstr. 7th Int. Symp. Yersinia, abstr. P-89, Med. Microbiol. [Ned. N. Voor] 6[Suppl. II]:S35, 1998; Worsham and Hunter, Abstr. 7th Int. Symp. Yersinia, 1998), there may be some limitations in the comprehensiveness of the current subunit vaccines under evaluation. Some of the potential advantages and limitations of these preparations are presented in Table 12.
TABLE 12.
Relevant characteristics of the current and forthcoming plague vaccines and their componentsa
| Vaccine preparation | Protection of:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Humans
|
Mice from:
|
Guinea pigs from:
|
Nonhuman primates from:
|
|||||||||||||||||
| Reduction of morbidity | Reduction of mortality | w.t.b
|
F1−c
|
V-O:3+d
|
w.t.
|
F1−
|
V-O:3+
|
w.t.
|
F1−
|
V-O:3+
|
||||||||||
| Bubonic plague | Pneumonic plague | Bubonic plague | Pneumonic plague | Bubonic plague | Pneumonic plague | Bubonic plague | Pneumonic plague | Bubonic plague | Pneumonic plague | Bubonic plague | Pneumonic plague | Bubonic plague | Pneumonic plague | Bubonic plague | Pneumonic plague | Bubonic plague | Pneumonic plague | |||
| Live (EV) | + | + | + | + | ± | ? | + | ? | + | + | + | + | + | ? | + | + | ± | ± | ? | ? |
| Killed USP | − | + | + | − | − | − | ? | ? | + | ? | ? | ? | ? | ? | + | − | − | − | ? | ? |
| F1 | ? | ? | + | + | − | − | + | ? | − | − | − | − | ? | ? | + | − | − | − | ? | ? |
| Ve | ? | ? | + | + | + | + | − | ? | ? | ? | ? | ? | ? | ? | + | + | + | + | ? | ? |
| V-O:3f | ? | ? | − | − | − | − | + | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
| BaSoAng | ? | ? | − | ? | ? | ? | ? | ? | ± | ? | + | ? | ? | ? | ? | ? | ? | ? | ? | ? |
| Bh | ? | ? | − | ? | ? | ? | ? | ? | + | ? | + | ? | ? | ? | + | + | ? | ? | ? | ? |
| F1 + V | ? | ? | + | + | + | + | + | ? | + | ? | ? | ? | ? | ? | + | + | + | + | ? | ? |
| F1−V | ? | ? | + | + | + | + | + | ? | ? | ? | ? | ? | ? | ? | + | + | + | + | ? | ? |
| F1 + BaSoAn | ? | ? | + | ? | ? | ? | ? | ? | + | ? | ? | ? | ? | ? | + | + | ? | ? | ? | ? |
| F1 + B | ? | ? | + | ? | ? | ? | ? | ? | + | + | ? | ? | ? | ? | + | + | ? | ? | ? | ? |
Compiled from references 4, 5, 6, 35-37, 45, 46, 88, 115, 125, 126, 139, 178, and 192; Adamovicz et al., Abstr. 8th Int. Symp. Yersinia, 2002; and Titball and Williamson, Abstr. 8th Int. Symp. Yersinia, 2002.
Challenge with wild-type Y. pestis strains.
Challenge with F1-negative Y. pestis strains.
Challenge with Y. pestis strain possessing V-O:3 type of V antigen.
V-Yp type of V antigen.
V-O:3 type of V antigen.
Basic somatic antigen (BaSoAn) or antigen obtained from Y. pestis or Y. pseudotuberculosis (81) by trichloroacetic acid extraction (26).
B antigen is a complex antigen including an undefined polysaccharide(s) that is tightly bound to undefined protein(s) and also includes lipid(s) but lacks endotoxin activity.
In regard to the V antigen, DNA sequencing of the most common serotypes of pathogenic Y. enterocolitica and Y. pseudotuberculosis revealed that two evolutionarily distinct types of V antigen exist in Yersinia spp. One type is expressed by Y. enterocolitica serotype O:8 (designated LcrV-YenO8 or V-O:8); the other type is expressed by Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica serotypes O:3, O:9, and O:5,27 (LcrV-Yps or V-O:3). Antisera to these two variant V antigens are protective only if the immunizing V antigen is of the same type as the V antigen produced by the challenge strain (159). More recently, it was shown that Y. pestis strains possess their own V-antigen type, V-Yp (Griffin et al., Abstr 7th Int. Symp. Yersinia, 1998), although some strains classified in the FSU as Yersinia pestis subsp. caucasica produce the V-O:3 type of V antigen (Worsham and Hunter, Abstr. 7th Int. Symp. Yersinia, 1998). In active vaccination studies, no cross-protection was observed between the three different types of V antigen (Griffin et al., Abstr. 7th Int. Symp. Yersinia, 1998). Overall, it appears that there are at least three, potentially interchangeable V antigen variants, and it is possible that in the diverse population of Y. pestis circulating in Asia there are even more variants.
Newer Plague Vaccines
Currently there are two newer plague vaccines under development. In Britain the F1+V vaccine is composed of the recombinant plague capsule protein F1 plus the V-Yp type of V antigen combined with an aluminum hydroxide adjuvant (178, 192; R. W. Titball and E. D. Williamson, Abstr. 8th Int. Symp. Yersinia, abstr. O-35, 2002). In the United States, a recombinant plague vaccine designated F1-V is a fusion protein composed of the entire capsule protein (F1) and V protein (V-Yp type) of Y. pestis combined with an aluminum hydroxide adjuvant (4, 88; J. J. Adamovicz, G. P. Andrews, C. Bolt, C. L. Wilhelmsen, J. W. Raymond, and L. M. Pitt, Abstr. 8th Int. Symp. Yersinia, abstr. O-40, 2002). However, the data in Table 12 suggest that these vaccines could be ineffective in protecting against plague caused by F1-negative strains, which produce non-Yp types of V antigen. This problem was noted at the 8th International Symposium on Yersinia that was held in 2002 in Turku, Finland, and was considered to be a potentially serious problem that should be addressed by including in subunit vaccines the V-O:8 and V-O:3 types of V antigen or two other antigens designated the B antigen and BaSoAn.
B Antigen and BaSoAn Vaccines
B antigen and BaSoAn are thought to be the major components (35, 36, 45) of the water-insoluble “residual” antigen initiating a classic T-cell-modulated state of cellular immunity that has been highly protective for guinea pigs (28, 32). The B antigen is a complex antigen including an undefined polysaccharide(s) that is tightly bound to undefined protein(s) and also includes a lipid(s) but lacks endotoxin activity. B antigen is produced by Y. pestis only in vivo, while Y. pseudotuberculosis is able to produce it both in vivo and in vitro. Therefore, B antigen for vaccine use is derived from Y. pseudotuberculosis, but the isolation technique has not been published (35).
BaSoAn is also a complex antigen that may be obtained from either Y. pestis or Y. pseudotuberculosis grown under a wide range of culture conditions (45, 81). It is extracted and precipitated by trichloroacetic acid, based on the technique originally outlined by Boivin et al. for endotoxin extraction (26). Interestingly, when this technique is used with Y. pestis or Y. pseudotuberculosis, the resulting preparations lack endotoxic activities (45, 81). It is known that extraction of LPS from a variety of smooth and rough bacterial strains by the Boivin procedure produces preparations in which LPS is complexed through its lipid A moiety to a protein-rich component, lipid-associated protein (53). The proteins in Boivin-type LPS are a mixture of porins and other outer membrane proteins (77) that possibly can block endotoxic activities in BaSoAn. It should be mentioned that Y. pestis LPS obtained by phenol-water extraction and lacking lipid-associated protein (189), when injected simultaneously with F1 antigen, produced an immunosuppressive effect (37). However, whether the B antigen will have acceptable toxicity when tested in humans is not known, while a reportedly well-tolerated and potentially efficacious vaccine composed of F1 antigen plus the BaSoAn material has already been evaluated in humans (46).
OVERALL POTENTIAL IMPLICATIONS OF GENETIC DIVERSITY IN YERSINIA PESTIS FOR HUMAN DISEASE
The presence of this large array of diverse Y. pestis strains in the FSU and Asian countries would serve as a natural laboratory for evaluating the potential of the non-pestis subspecies to cause disease in humans or economically important animals. In the FSU, infection with the non-main subspecies altaica, caucasica, hissarica, ulegeica, and talassica is rare or not reported, suggesting that the potential of these strains to cause epidemic outbreaks is low. However, a low EP is not the same as the potential for high-level virulence for humans, since highly virulent strains in an endemic focus with which humans have little to no contact would be unlikely to be a cause of human infection. Therefore, we cannot directly infer that the low or absent occurrence of many of the variant strains of Y. pestis in human infections in the FSU is indicative of the fact that they are not very pathogenic for humans.
There are a few data suggesting that some of the non-pestis subspecies can cause human disease. Three human cases of bubonic plague, with two of them confirmed by isolation of Y. pestis subsp. caucasica in pure culture, were found in the Transcaucasian-highland focus (two cases in Leninakan natural focus [Fig. 2; focus 4] and one in Zanzegur-Karabakh natural focus [focus 6] (O. V. Ovasapyan, M. T. Shekhikyan, Abstr. Regional Sci. Conf. Epidemiol. Microbiol. Immunol. Bacterial Viral Infect., p. 193-195, 1989). Two of the cases were associated with flea bites, while one (probably unique) case was reportedly caused by a peck from an eagle's beak. All of the patients had the clinical manifestations typical of bubonic plague, such as an acute on set of the disease with symptoms of sudden fever (39.5 to 40°C), chills, and headache followed several hours later by nausea and vomiting. Within 3 weeks, the bubos burst open; their healing continued for 3 to 5 months.
There is only one published, abstract report in English (Zhenya et al., Abstr. 7th Int. Symp. Yersinia pestis 1998), showing that the Y. pestis strains isolated from Microtus brandti (a “non-main subspecies” that was isolated in China and classified using the Chinese classification system of ecotypes for which there is not yet a corresponding subspecies designation in the Russian classification system) had no effect in human volunteers. Unfortunately, no description of the details of those experiments was included in this abstract. Kutyrev (110) described a Y. pestis subspecies ulegeica F1-negative strain I-2422 that was highly virulent for mice and guinea pigs, but the virulence for humans was not described. However, we do know that the studies of the large number of Y. pestis isolates that are found in the FSU, Mongolia, and China indicate a tremendous amount of extant genetic diversity in this species, raising the potential that new introductions of and changes in DNA could lead to changes in phenotypes associated with virulence, as well as in antigens used for diagnosis, as targets of therapies or as vaccines. In any case, the non-pestis subspecies of Y. pestis, which are virulent in some selected rodent species, are the first evolutionary step from low- or medium-virulence Y. pseudotuberculosis to classic, high-virulence Y. pestis subspecies pestis. The comparative study of such strains may clarify the mechanisms of evolution of pathogenicity in Y. pestis.
CONCLUDING REMARKS
The resurgent interest in plague as a pathogen in the context of its potential use as an agent of bioterrorism and the associated increased funding opportunities to study such agents underlie the need to acquire as much information as possible about this organism. However, given that the genetic and phenotypic diversity of strains of Y. pestis that cause disease in the Americas is limited because this organism was introduced into this part of the world just over 100 years ago, it will be important to understand the potential impact for human health and welfare of the genetic and phenotypic variation in the diverse strains isolated from natural foci in the FSU and Asia. The much longer periods of enzootic residence in multiple natural foci in this part of the world provides the ideal setting for genetic variation to be acted on by natural selective forces to allow for the emergence of Y. pestis strains with virulence potentials that are not fully appreciated outside of the FSU, Mongolia, and China. Especially worrisome are human isolates lacking classic virulence factors such as F1 antigen, acquisition of plasmids encoding multiple antibiotic resistances, and the presence of a large pool of genes that could confer increased virulence on Y. pestis strains that are generally not available for study outside of the FSU. Many of these factors will of necessity need to be incorporated into the new and expanded studies that are expected to occur in the next few years in preparation for the worldwide production of appropriate diagnostic and therapeutic agents to counter the threat of bioterrorism. To do this, major cooperation among international investigators is needed and, ultimately, access to the strains must be provided for investigators worldwide. A tremendous amount of knowledge and data resides in laboratories in the FSU that have studied plague and Y. pestis for many decades, particularly strains from the multiple endemic foci that exist, and such knowledge will be most helpful to investigators worldwide in order to counter the modern and real threats of bioterrorism.
Acknowledgments
We thank E. P. Golubinskii for permission to use Tables 3 through 6. The computer graphic efforts of I. A. Anisimov for Fig. 2 and the computer retouching of Fig. 3 and 4 are greatly appreciated. Special thanks are extended to K. L. Gage and J. A. Montenieri for providing Fig. 1, to S. V. Balakhonov for providing Fig. 3A and 4; and to A. Erdenebat for providing Fig. 3B.
This work was financially supported by funding from the International Science and Technology Center (ISTC) partner project 1197, supported by the Cooperative Threat Reduction (CTR) Program of the U.S. Department of Defense. A.P.A. was also supported by the contract 43.600.1.4.0031 from the Ministry for Industry, Science and Technologies of the Russian Federation.
REFERENCES
- 1.Abgaryan, G. P. 1966. Characterization of some Yersinia pestis strains, which were isolated on Armenian Highland from common voles. Ph.D. thesis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 2.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. (Erratum, 97:8192, 2000.) [DOI] [PMC free article] [PubMed]
- 3.Adair, D. M., P. L. Worsham, K. K. Hill, A. M. Klevytska, P. J. Jackson, A. M. Friedlander, and P. Keim. 2000. Diversity in a variable-number tandem repeat from Yersinia pestis. J. Clin. Microbiol. 38:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, G. W. Jr., D. G. Heath, C. R. Bolt, S. L. Welkos, A. M. Friedlander. 1998. Short- and long-term efficacy of single-dose subunit vaccines against Yersinia pestis in mice. Am. J. Trop. Med. Hyg. 58:793-799. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, G. W., S. E. C. Leary, E. D. Williamson, R. W. Titball, S. L. Welkos, P. L. Worsham, and A. M. Friedlander. 1996. Recombinant V antigen protects mice against pneumonic and bubonic plague caused by F1-capsule-positive and F1-capsule-negative strains of Yersinia pestis. Infect. Immun. 64:4580-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews, G. P., D. G. Heath, G. W. Anderson, Jr., S. L. Welkos, and A. M. Friedlander. 1996. Fraction 1 capsular antigen (F1) purification from Yersinia pestis CO92 and from an Escherichia coli recombinant strain and efficacy against lethal plague challenge. Infect. Immun. 64:2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anisimov, A. P. 1999. Factors providing the blocking activity of Yersinia pestis. Mol. Gen. Mikrobiol. Virusol. 4:11-15. [PubMed] [Google Scholar]
- 8.Anisimov, A. P. 2002. Factors of Yersinia pestis proving circulation and persistence of plague pathogen in ecosystems of natural foci. Communication 2. Mol. Gen. Mikrobiol. Virusol. 4:3-11. [PubMed] [Google Scholar]
- 9.Anisimov, A. P. 2002. Yersinia pestis factors, assuring circulation and maintenance of the plague pathogen in natural foci ecosystems. Report 1. Mol. Gen. Mikrobiol. Virusol. 3:3-23. [PubMed] [Google Scholar]
- 10.Anisimova, T. I. 1962. Experimental plague in grey rats (Rattus norvegicus). Ph.D. thesis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 11.Aparin, G. P. and E. P. Golubinskii. 1989. Plague microbiology. Manual. Irkutsk State University, Irkutsk, USSR.
- 12.Aparin, G. P., S. V. Balakhonov, L. A. Timofeeva, and A. I. Logachev. 1987. Numerical analysis of the phenotypic properties and total genomic characteristics of strains of Yersinia pestis related to different subspecies. Zh. Mikrobiol. Epidemiol. Immunobiol. 11:16-20. [PubMed] [Google Scholar]
- 13.Bahmanyar, M., and D. C. Cavanaugh. 1976. Plague manual. World Health Organization, Geneva, Switzerland.
- 14.Baker, E. E., H. Sommer, L. E. Foster, E. Meyer, K. Meyer. 1952. Studies on immunization against plague. I. The isolation and characterization of the soluble antigen of the Pasteurella pestis. J. Immunol. 68:131-145. [PubMed] [Google Scholar]
- 15.Balakhonov, S. V. 1987. Molecular-biological criteria of genome similarity in taxonomy of bacteria from the genus Yersinia. Ph.D. thesis. Russian Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 16.Balakhonov, S. V. 1989. Results of screening of plasmids of Yersinia pestis strains from various regions of endemic Central-Asian plague focus. Mol. Gen. Mikrobiol. Virusol. 4:39-42. [PubMed] [Google Scholar]
- 17.Balakhonov, S. V. 2000. Genome markers of the plague, pseudotuberculosis, cholera, and brucellosis pathogens (epidemiological and diagnostic significance). Sc.D. thesis. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 18.Balakhonov, S. V., S. Tsendzhav, and A. Erdenebat. 1991. New plasmidovars of Yersinia pestis strains isolated in Mongolia. Mol. Gen. Mikrobiol. Virusol. 11:22-29. [PubMed] [Google Scholar]
- 19.Beesley, E. D., and M. J. Surgalla. 1970. Pesticinogeny: a characteristic useful for presumptive identification and isolation of Pasteurella pestis. Appl. Microbiol. 19:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bengoechea, J.-A., B. Lindner, U. Seydel, R. Díaz, and I. Moriyón. 1998. Yersinia pseudotuberculosis and Yersinia pestis are more resistant to bactericidal cationic peptides than Yersinia enterocolitica. Microbiology 144:1509-1515. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Gurion, R., and A. Shafferman. 1981. Essential virulence determinants of different Yersinia species are carried on a common plasmid. Plasmid 5:183-187. [DOI] [PubMed] [Google Scholar]
- 22.Berlin, A. L., and A. K. Borzenkov. 1938. Fermentative characterization of Mongolian B. pestis strains. Fermentation of different carbohydrates, alcohols, and glucosides. Attitude of B. pestis strains toward glycerol. Herald Microbiol. Epidemiol. Parasitol. 17:215-227. [Google Scholar]
- 23.Bezsonova, A. A. 1928. About two varieties of B. pestis, which can be distinguished while growing on glycerol-containing media. Herald Microbiol. Epidemiol. Parasitol. 7:250-253. [Google Scholar]
- 24.Bobrov, A. G. 1995. Studying prevalence and location of mobile elements, IS 100 and IS 285, in yersiniae genomes. Ph.D. thesis. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 25.Bobrov, A. G., and A. A. Filippov. 1997. Prevalence of IS285 and IS100 in Yersinia pestis and Yersinia pseudotuberculosis genomes. Mol. Gen. Mikrobiol. Virusol. 2:36-40. [PubMed] [Google Scholar]
- 26.Boivin, A., I. Mesrobeanu, and L. Mesrobeanu. 1933. Technique pour la préparation des polysaccharide microbiens spécifiques. C. R. Soc. Biol. 113:490-492. [Google Scholar]
- 27.Borinskaya, S. A. and N. K. Yankovskii. 1999. Structure of prokaryotic genomes. Mol. Biol. 33:941-952. [PubMed] [Google Scholar]
- 28.Brubaker, R. R. 1972. The genus Yersinia: biochemistry and genetics of virulence. Curr. Top. Microbiol. Immunol. 57:111-158. [DOI] [PubMed] [Google Scholar]
- 29.Brubaker, R. R. 1983. The Vwa+ virulence factor of yersiniae: the molecular basis of the attendant nutritional requirement for Ca++. Rev. Infect. Dis. 5(Suppl 4):S748-S758. [DOI] [PubMed] [Google Scholar]
- 30.Brubaker, R. R. 1991. Factors promoting acute and chronic disease caused by yersiniae. Clin. Microbiol. Rev. 4:309-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brubaker, R. R. 2000. Yersinia pestis and bubonic plague. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackelbrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. [Online.] Springer Verlag, New York, N.Y. http://www.prokaryotes.com.
- 32.Burrows, T. W. 1963. Virulence of Pasteurella pestis and immunity to plague. Ergeb. Mikrobiol. Immun. Exp. Ther. 37:59-113. [DOI] [PubMed] [Google Scholar]
- 33.Burrows, T. W., and W. A. Gillett. 1971. Host specificity of Brazilian strains of Pasteurella pestis. Nature 229:51-52. [DOI] [PubMed] [Google Scholar]
- 34.Butler, T. 1983. Plague and other Yersinia infections. Plenum Press, New York, N.Y.
- 35.Byvalov, A. A., I. V. Darmov, and V. I. Evstigneev. 1997. Antigen protecting guinea pigs from experimental plague, p. 192-193. In Proceedings of the Scientific and Practical Conference dedicated to the centenary of the Russian Anti-Plague Service, vol. 1. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 36.Byvalov, A. A., V. I. Evstigneev, E. V. Pimenov, and N. T. Vasil'ev. 1997. Immunization of experimental animals against plague with complex preparation including F1 and B antigens, p. 194. In Proceedings of the Scientific and Practical Conference dedicated to the centenary of the Russian Anti-Plague Service, vol. 1. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 37.Byvalov, A. A., V. N. Pautov, Iu. V. Chicherin, V. A. Lebedinskii, and V. I. Evtigneev. 1984. Effectiveness of revaccinating hamadryas baboons with NISS live dried plague vaccine and fraction I of the plague microbe. Zh. Mikrobiol. Epidemiol. Immunobiol. 4:74-76. [PubMed] [Google Scholar]
- 38.Cavalcanti, Y. V., Leal, N. C., and A. M. de Almeida. 2002. Typing of Yersinia pestis isolates from the state of Ceara, Brazil. Lett. Appl. Microbiol. 35:543-547. [DOI] [PubMed] [Google Scholar]
- 39.Cavanaugh, D. C., and R. Randall. 1959. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of fleaborne plague. J. Immunol. 83:348-363. [PubMed] [Google Scholar]
- 40.Christie, A. B., T. H. Chen, S. S. Elberg. 1980. Plague in camels and goats: their role in human epidemics. J. Infect. Dis. 141:724-726. [DOI] [PubMed] [Google Scholar]
- 41.Chu, M. C. 2000. Laboratory manual of plague diagnostic tests. World Health Organization, Geneva, Switzerland.
- 42.Chu, M. C., X. Q. Dong, X. Zhou, and C. F. Garon. 1998. A cryptic 19-kilobase plasmid associated with U.S. isolates of Yersinia pestis: a dimer of the 9.5-kilobase plasmid. Am. J. Trop. Med. Hyg. 59:679-686. [DOI] [PubMed] [Google Scholar]
- 43.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 44.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geuijen, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dal'vadyants, S. M. 1990. Plague chemical vaccine: designing, experimental and clinical substantiation of application for human revaccination. Sc.D. thesis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 46.Dal'vadyants, S. M., V. V. Korolev, V. V. Seroglazov, L. G. Belov, G. M. Sergeeva, K. I. Gerasimova, T. M. Drobysheva, N. P. Mironova, M. S. Verenkov, and N. M. Aleksandrova. 1997. Indices of indirect analytical method for evaluation of efficacy of revaccination of volunteers with live and chemical plague vaccines, p. 201-202. In Proceedings of the Scientific and Practical Conference dedicated to the centenary of the Russian Anti-Plague Service, vol. 1. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 47.Davis, K. J., D. L. Fritz, M. L. Pitt, S. L. Welkos, P. L. Worsham, and A. M. Friedlander. 1996. Pathology of experimental pneumonic plague produced by fraction 1-positive and fraction 1-negative Yersinia pestis in African green monkeys (Cercopithecus aethiops). Arch. Pathol. Lab. Med. 120:156-163. [PubMed] [Google Scholar]
- 48.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM1. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dennis, D. T., K. L. Gage, N. Gratz, J. D. Poland, and E. Tikhomirov. 1999. Plague manual: epidemiology, distribution, surveillance and control. World Health Organization, Geneva, Switzerland.
- 50.Devdariani, Z. L., M. S. Verenkov, V. A. Fedorova, N. S. Solodovnicov, and L. G. Belov. 1993. Identification of Yersinia pestis with varied plasmid composition using monoclonal and polyclonal fluorescent immunoglobulins. FEMS Immunol. Med. Microbiol. 6:31-35. [DOI] [PubMed] [Google Scholar]
- 51.Devdariani, Z. L., V. A. Fedorova, O. V. Gromova, and T. M. Taranenko. 1997. Comparative incidence of detecting specific antibodies to Yersinia pestis capsular antigen and lipopolysaccharide in humans immunised with antipestis vaccine. Clin. Lab. Diag. 4:39-41. [PubMed]
- 52.Devignat, R. 1951. Variétés de l'espèce Pasteurella pestis: nouvelle hyphothèse. Bull. W. H. O. 4:247-263. [PMC free article] [PubMed] [Google Scholar]
- 53.Doe, W. F., S. T. Yang, D. C. Morrison, S. J. Betz, P. M. Henson. 1978. Macrophage stimulation by bacterial lipopolysaccharides. II. Evidence for differentiation signals delivered by lipid A and by a protein rich fraction of lipopolysaccharides. J. Exp. Med. 148:557-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doll, J. M., P. S. Zeitz, P. Ettestad, A. L. Bucholtz, T. Davis, and K. Gage. 1994. Cat-transmitted fatal pneumonic plague in a person who traveled from Colorado to Arizona. Am. J. Trop. Med. Hyg. 51:109-114. [DOI] [PubMed] [Google Scholar]
- 55.Domaradskii, I. V. 1993. Plague: contemporary state, assumptions, problems. Saratov Medical Institute Press, Saratov, Russia.
- 56.Domaradskii, I. V. 1998. Plague. Meditsina Press, Moscow, Russia.
- 57.Dong, X. Q., L. E. Lindler, and M. C. Chu. 2000. Complete DNA sequence and analysis of an emerging cryptic plasmid isolated from Yersinia pestis. Plasmid 43:144-148. [DOI] [PubMed] [Google Scholar]
- 58.Dong, X., F. Ye, and H. Peng. 2001. Geographic distribution and feature of Yersinia pestis plasmid isolated from Yunnan province. Zhonghua Liu Xing Bing Xue Za Zhi 22:344-347. [PubMed] [Google Scholar]
- 59.Drozdov, I. G., A. P. Anisimov, S. V. Samoilova, I. N. Yezhov, S. A. Yeremin, A. V. Karlyshev, V. M. Krasilnikova, and V. I. Kravchenko. 1995. Virulent non-capsulate Yersinia pestis variants constructed by insertion mutagenesis. J. Med. Microbiol. 42:264-268. [DOI] [PubMed] [Google Scholar]
- 60.Dyatlov, A. I. 1989. Evolutional aspects in natural plague focality. Stavropol' Bookish Press, Stavropol', USSR.
- 61.Elkin, Yu. M. and P. A. Petrov. 1974. Paleogenesis of Transcaucasian highland plague focus in connection with dissimilarity in virulence of Yersinia pestis vole's strains from different landscape-geographical Caucasus localities, p. 43-45. In Particularly dangerous infections in the Caucasus. Stavrapol' Research Anti-Plague Institute, Stavropol', USSR.
- 62.Erdenebat, A. 2000. Plasmid content of Yersinia pestis isolated in Mongolia. Ph.D. thesis. Centre for Control and Research of Natural Infectious Diseases, Ulaanbaatar, Mongolia.
- 63.Fedorov, V. N. 1960. Plague in camels and its prevention in the USSR. Bull. W. H. O. 23:275-281. [PMC free article] [PubMed] [Google Scholar]
- 64.Fedorova, V. A., and Z. L. Devdariani. 1998. Study of antigenic determinants of Yersinia pestis lipopolysaccharide using monoclonal antibodies. Mol. Gen. Mikrobiol. Virusol. 3:22-26. [PubMed] [Google Scholar]
- 65.Fedorova, V. A., and Z. L. Devdariani. 2000. Development, characterisation and diagnostic application of monoclonal antibodies against Yersinia pestis fibrinolysin and coagulase. J. Med. Microbiol. 49:261-269. [DOI] [PubMed] [Google Scholar]
- 66.Ferber, D. M., and R. R. Brubaker. 1981. Plasmids in Yersinia pestis. Infect. Immun. 31:839-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Filippov, A. A. 2001. Mobile genetic elements of pathogenic yersiniae. Sc.D. thesis. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 68.Filippov, A. A., N. S. Solodovnikov, L. M. Kookleva, and O. A. Protsenko. 1990. Plasmid content in Yersinia pestis strains of different origin. FEMS Microbiol. Lett. 67:45-48. [DOI] [PubMed] [Google Scholar]
- 69.Filippov, A. A., N. S. Solodovnikov, L. M. Kukleva, and O. A. Protsenko. 1992. Plasmid composition of Yersinia pestis strains from different natural foci. Zh. Mikrobiol. Epidemiol. Immunobiol. 3:10-13. [PubMed] [Google Scholar]
- 70.Finegold, M. J., J. J. Petery, R. F. Berendt, and H. R. Adams. 1968. Studies on the pathogenesis of plague. Blood coagulation and tissue responses of Macaca mulatta following exposure to aerosols of Pasteurella pestis. Am. J. Pathol. 53:99-114. [PMC free article] [PubMed] [Google Scholar]
- 71.Finlay, B. B. and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedlander, A. M., S. L. Welkos, P. L. Worsham, G. P. Andrews, D. G. Heath, G. W. Anderson, Jr., M. L. M. Pitt, J. Estep, and K. Davis. 1995. Relationship between virulence and immunity as revealed in recent studies of the F1 capsule of Yersinia pestis. Clin. Infect. Dis. 21(Suppl. 2):S178-S181. [DOI] [PubMed] [Google Scholar]
- 73.Fursov, V. V., and Y. A. Popov. 1983. Studying of Yersinia pestis “vole” strains by agarose gel electrophoresis, p. 326-327. In Y. G. Suchkov (ed.), Prophylaxis of natural focal infections. Region Administration Press, Stavropol', Russia.
- 74.Fursova, N. K., R. Z. Shaikhutdinova, and A. P. Anisimov. 2002. Strain differences in susceptibility of the plague pathogen to polymyxin B, p. 65-66. In Proceedings of the 4th Russian Conference on Current Problems of Anti-Microbe Chemotherapy, Moscow, Russia.
- 75.Gage, K. L., D. T. Dennis, K. A. Orloski, P. Ettestad, T. L. Brown, P. J. Reynolds, W. J. Pape, C. L. Fritz, L. G. Carter, and J. D. Stein. 2000. Cases of cat-associated human plague in the Western US, 1977-1998. Clin. Infect. Dis. 30:893-900. [DOI] [PubMed] [Google Scholar]
- 76.Galimand, M., A. Guiyoule, G. Gerbaud, B. Rasoamanana, S. Chanteau, E. Carniel, and P. Courvalin. 1997. Multiple antibiotic resistance in Yersinia pestis mediated by a self-transferable plasmid. N. Engl. J. Med. 337:677-680. [DOI] [PubMed] [Google Scholar]
- 77.Goldman, R. C., D. White, and L. Leive. 1981. Identification of outer membrane proteins, including known lymphocyte mitogens, as the endotoxin protein of Escherichia coli O111. J. Immunol. 127:1290-1294. [PubMed] [Google Scholar]
- 78.Gorshkov, O. V., E. P. Savostina, Iu. A. Popov, O. P. Plotnikov, N. A. Vinogradova, and N. S Solodovnikov. 2000. Genotyping Yersinia pestis strains from various natural foci. Mol. Gen. Mikrobiol. Virusol. 3:12-17. [PubMed] [Google Scholar]
- 79.Gramotina, L. I. and S. L. Protsenko. 1994. Low-molecular-weight plasmids—a prospective test for the intraspecific differentiation of Yersinia pestis. Zh. Mikrobiol. Epidemiol. Immunobiol. 6:35-36. [PubMed] [Google Scholar]
- 80.Gremyakova, T. A., E. V. Vinogradov, B. Lindner, N. A. Kocharova, S. N. Senchenkova, A. S. Shashkov, Y. A. Knirel, O. Holst, R. Z. Shaikhutdinova, and A. P. Anisimov. 2003. The core structure of the lipopolysaccharide of Yersinia pestis strain KM218. Influence of growth temperature. Adv. Exp. Med. Biol. 529:229-231. [DOI] [PubMed] [Google Scholar]
- 81.Griboedov, A. V., G. P. Barabash. 1985. Basic somatic antigen or antigen obtained from Yersinia pestis by the Boivin-Mesrobeanu method. Zh. Mikrobiol. Epidemiol. Immunobiol. 3:108-115. [PubMed] [Google Scholar]
- 82.Groisman, E. A. 1994. How bacteria resist killing by host-defence peptides. Trends Microbiol. 2:444-449. [DOI] [PubMed] [Google Scholar]
- 83.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guiyoule, A., B. Rasoamanana, C. Buchrieser, P. Michel, S. Chanteau, and E. Carniel. 1997. Recent emergence of new variants of Yersinia pestis in Madagascar. J. Clin. Microbiol. 35:2826-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guiyoule, A., F. Grimont, I. Iteman, P. A. Grimont, M. Lefevre, and E. Carniel. 1994. Plague pandemics investigated by ribotyping of Yersinia pestis strains. J. Clin Microbiol. 32:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guiyoule, A., G. Gerbaud, C. Buchrieser, M. Galimand, L. Rahalison, S. Chanteau, P. Courvalin, and E. Carniel. 2001. Transferable plasmid-mediated resistance to streptomycin in a clinical isolate of Yersinia pestis. Emerg. Infect. Dis. 7:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gvozdenko, N. A., V. E. Valentsev, B. D. Rublev, and V. I. Ryzhkov. 1992. The sensitivity of bacterial strains of Yersinia pestis isolated from voles to normal human serum. Zh. Mikrobiol. Epidemiol. Immunobiol. 11-12:8-10. [PubMed] [Google Scholar]
- 88.Heath, D. G., G. W. Anderson, Jr., J. M. Mauro, S. L. Welkos, G. P. Andrews, J. Adamovicz, and A. M. Friedlander. 1998. Protection against experimental bubonic and pneumonic plague by a recombinant capsular F1-V antigen fusion protein vaccine. Vaccine 16:1131-1137. [DOI] [PubMed] [Google Scholar]
- 89.Hinnebusch, B. J. 1997. Bubonic plague: a molecular genetic case history of the emergence of an infectious disease. J. Mol. Med. 75:645-652. [DOI] [PubMed] [Google Scholar]
- 90.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and Å. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733-735. [DOI] [PubMed] [Google Scholar]
- 91.Hinnebusch, B. J., M. L. Rosso, T. G. Schwan, and E. Carniel. 2002. High-frequency conjugative transfer of antibiotic resistance genes to Yersinia pestis in the flea midgut. Mol. Microbiol. 46:349-354. [DOI] [PubMed] [Google Scholar]
- 92.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 93.Hitchen, P. G., J. L. Prior, P. C. F. Oyston, M. Panico, B. W. Wren, R. W. Titball, H. R. Morris, and A. Dell. 2002. Structural characterization of lipo-oligosaccharide (LOS) from Yersinia pestis: regulation of LOS structure by the PhoPQ system. Mol. Microbiol. 44:1637-1650. [DOI] [PubMed] [Google Scholar]
- 94.Huang, X. Z., M. C. Chu, D. M. Engelthaler, and L. E. Lindler. 2002. Genotyping of a homogeneous group of Yersinia pestis strains isolated in the United States. J. Clin. Microbiol. 40:1164-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Inglesby, T. V., D. T. Dennis, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, J. F. Koerner, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, M. Schoch-Spana, K. Tonat, and the Working Group on Civilian Biodefense. 2000. Plague as a biological weapon: medical and public health management. JAMA 283:2281-2290. [DOI] [PubMed] [Google Scholar]
- 96.Innokent'eva, T. I. 1969. Virulence raising and changing of the other properties of Yersinia pestis during its passaging through the organism of the Ochotona pricei. Ph.D. thesis. Antiplague Research Institute of Siberia and Far East, Irkutsk, USSR.
- 97.Iriarte, M., and G. R. Cornelis. 1999. The 70-kilobase virulence plasmid of yersiniae, p. 91-126. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence elements. ASM Press, Washington, D.C.
- 98.Isaäcson, M., D. Levy, B. J. Te, W. N. Pienaar, H. D. Bubb, J. A. Louw, and D. K. Genis. 1973. Unusual cases of human plague in Southern Africa. S. Afr. Med. J. 47:2109-2113. [PubMed] [Google Scholar]
- 99.Ivanova, V. S., S. A. Lebedeva, N. A. Goncharova, and G. G. Gurleva. 1990. Screening of plasmids in museum strains of Yersinia pestis isolated from various natural foci. Mol. Gen. Mikrobiol. Virusol. 3:16-88. [PubMed] [Google Scholar]
- 100.Kalabukhov, N. I. 1969. Periodical (seasonal and years') changes in rodents' organisms, their reasons and after-effects. Nauka Press, Leningrad, USSR.
- 101.Kalmykova, L. I. 1984. Biological properties and spectrums of the water-soluble proteins of Yersinia pestis strains isolated from Caucasian natural foci. Ph.D. thesis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 102.Kawahara, K., H. Tsukano, H. Watanabe, B. Lindner, and M. Matsuura. 2002. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 70:4092-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Klevytska, A. M., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kokushkin, A. M. 1995. Social and biological aspects of plague epidemiology. Sc.D. thesis. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 105.Kondrashkina, K. I., N. I. Nikolaev, L. M. Gol'dfarb, V. A. Ivanov, I. I. Kuraev, and N. G. Ponomarev. 1971. Role of atypical strains in mechanisms of self-regulation of plague epizootic process. Probl. Particularly Dangerous Infect. (Saratov) 20:5-17.
- 106.Kozlov, M. P. 1979. Plague (natural focality, epizootology, epidemiological manifestations). Meditsina Press, Moscow, USSR.
- 107.Kudinova, T. P. 1968. Attributes of Yersinia pestis strains isolated in autumn 1963 in Ili-Karatal inter-river region. Ph.D. thesis. Research Anti-Plague Institute of Central Asia, Alma-Ata, USSR.
- 108.Kuklev, E. V. 1999. Quantitative assessment of epidemic potential of natural plague foci. Sc.D. thesis. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 109.Kukleva, L. M., O. A. Protsenko, and V. V. Kutyrev. 2002. Modern concepts on the relationship between the agents causing plague and pseudotuberculosis. Mol. Gen. Mikrobiol. Virusol. 1:3-7. [PubMed] [Google Scholar]
- 110.Kutyrev, V. V. 1992. Genetic analysis of Yersinia pestis virulence factors. Sc.D. thesis. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 111.Kutyrev, V. V., A. A. Filippov, N. Y. Shavina, and O. A. Protsenko. 1989. Genetic analysis and modeling of the virulence of Yersinia pestis. Mol. Genet. Mikrobiol. Virusol. 8:42-47. [PubMed] [Google Scholar]
- 112.Leal, N. C., A. M. P. de Almeida, and L. C. S. Ferreira. 1989. Plasmid composition and virulence-associated factors of Yersinia pestis isolates from a plague outbreak at the Paraiba State, Brazil. Rev. Inst. Med. Trop. Sao Paulo 31:295-300. [DOI] [PubMed] [Google Scholar]
- 113.Leal, N. C., M. Sobreira, T. C. A. Leal, and A. M. P. de Almeida. 2000. Homology among extra-cryptic DNA bands and the typical plasmids in Brazilian Yersinia pestis strains. Braz. J. Microbiol. 31:20-24. [Google Scholar]
- 114.Lebedeva, S. A., A. L. Ntukhachev, V. S. Ivanova, and A. S. Chernyavskaya. 2002. On the question of evolutional development of Yersinia pestis species and epidemiological significance of its representatives, p. 147-149. In Proceedings of the Jubilee Scientific and Practical Conference on Epidemiological Safety at the Caucasus devoted to fiftieth anniversary of the Stavrapol' Research Anti-Plague Institute. Stavrapol' Research Anti-Plague Institute, Stavrapol', Russia.
- 115.Lebedinskii, V. A., Iu. V. Chicherin, V. N. Pautov, V. I. Evstigneev, and A. A. Byvalov. 1982. Experience using fraction I of the plague microbe for revaccinating experimental animals. Zh. Mikrobiol. Epidemiol. Immunobiol. 5:60-63. [PubMed] [Google Scholar]
- 116.Lindler, L. E., G. V. Plano, V. Burland, G. F. Mayhew, and F. R. Blattner. 1998. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect. Immun. 66:5731-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lindler, L. E., M. S. Klempner, and S. C. Straley. 1990. Yersinia pestis pH 6 antigen: genetic, biochemical, and virulence characterization of a protein involved in the pathogenesis of bubonic plague. Infect. Immun. 58:2569-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Logachev, A. I. 1979. Biological properties of Yersinia pestis strains isolated in Mountain Altai. Ph.D. thesis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 119.Lucier, T. S., and R. R. Brubaker. 1992. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J. Bacteriol. 174:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martinevskii, I. L. 1969. Biology and genetic features of plague and plague-related microbes. Meditsina Press, Moscow, USSR.
- 121.Martinevskii, I. L. 1975. Studying of peculiarities of strains of the plague pathogen isolated from certain African regions. Probl. Particularly Dangerous Infect. 42:10-13. [Google Scholar]
- 122.Martinevskii, I. L., and I. S. Arakelian. 1981. Response to polymyxin of plague microbe strains isolated from various natural foci and of their mutants with a decreased requirement in nutritional factors. Antibiotiki 26:687-689. [PubMed] [Google Scholar]
- 123.Mel'nikov, I. F. 1971. Characterization of Yersinia pestis strains isolated from Kyzyl-Kum Desert and interrelations of P+ and P− variants of the plague pathogen with the organisms of hosts and vectors. Ph.D. thesis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 124.Meyer, K. F. 1970. Effectiveness of live or killed plague vaccines in man. Bull. W. H. O. 42:653-666. [PMC free article] [PubMed] [Google Scholar]
- 125.Meyer, K. F., D. C. Cavanaugh, P. J. Bartelloni, and J. D. Marshall, Jr. 1974. Plague immunization. I. Past and present trends. J. Infect. Dis. 129(Suppl.):S13-S18. [DOI] [PubMed] [Google Scholar]
- 126.Meyer, K. F., J. A. Hightower, and F. R. McCrumb. 1974. Plague immunization. VI. Vaccination with the fraction I antigen of Yersinia pestis. J. Infect. Dis. 129(Suppl.):S41-S45. [DOI] [PubMed] [Google Scholar]
- 127.Mollaret, H. H., and C. Mollaret. 1965. Melibiose fermentation in the genus Yersinia and its importance in the diagnosis of the varieties of Yersinia pestis. Bull. Soc. Pathol. Exot. Fil. 58:154-156. [PubMed] [Google Scholar]
- 128.Mollaret, H. H., V. B. Nguyen, M. Vandekerkove, Y. Karimi, and M. Eftekhari. 1964. Sur l'uréase du bacille de Yersin. Ann. Inst. Pasteur 107:424-429. [PubMed] [Google Scholar]
- 129.Morschhauser, J., V. Vetter, L. Emody, and J. Hacker. 1994. Adhesin regulatory genes within large, unstable DNA regions of pathogenic Escherichia coli: cross-talk between different adhesin gene clusters. Mol. Microbiol. 11:555-566. [DOI] [PubMed] [Google Scholar]
- 130.Motin, V. L., A. M. Georgescu, J. M. Elliott, P. Hu, P. L. Worsham, L. L. Ott, T. R. Slezak, B. A. Sokhansanj, W. M. Regala, R. R. Brubaker, and E. Garcia. 2002. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J. Bacteriol. 184:1019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Muller-Eberhard, H.J. 1975. Complement. Annu. Rev. Biochem. 44:695-724. [DOI] [PubMed] [Google Scholar]
- 132.Naumov, A. V., and L. V. Samoilova (ed.). 1992. Manual on plague prophylaxis. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 133.Naumov, A. V., and L. V. Samoilova. 1992. Laboratory diagnostics of plague. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 134.Naumov, A. V., M. Yu. Ledvanov, I. G. Drozdov. 1992. Plague immunology. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 135.Nikolaev, N. I. (ed.). 1972. Manual on plague prophylaxis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 136.Panfertsev, E. A., P. A. Cherepanov, and G. A. Karimova. 1991. Construction of Yersinia pestis strains defective in pH6 synthesis, p. 22-24. In R. V. Borovik (ed.), Proceedings of the XIV Scientific and Practical Conference on new technologies and biosystems: Achievements and perspectives. Medbioekonomika Press, Obolensk, USSR.
- 137.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 138.Pasiukov, V. V., I. V. Ryzhko, R. I. Tsuraeva, E. D. Samokhodkina, and N. V. Roshchina. 1994. Cefoperazone in the prevention and treatment of experimental plague caused by typical and fraction 1-free pathogen strains in white mice. Antibiot. Khimioter. 39:37-40. [PubMed] [Google Scholar]
- 139.Pautov, V. N., I. V. Chicherin, V. I. Evstigneev, A. A. Byvalov, and O. A. Kedrov. 1979. Experimental protection of fraction 1 by the plague microbe. Zh. Mikrobiol. Epidemiol. Immunobiol. 10:37-42. [PubMed] [Google Scholar]
- 140.Peisakhis, L. A., and V. M. Stepanov. 1975. Intraspecific classification of the plague pathogen subject to geographical zoning. Probl. Particularly Dangerous Infect. 42:5-9. [Google Scholar]
- 141.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis — etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Perry, R. D., and R. R. Brubaker. 1983. Vwa+ phenotype of Yersinia enterocolitica. Infect. Immun. 40:166-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Perry, R. D., S. C. Straley, J. D. Fetherston, D. J. Rose, J. Gregor, and F. R. Blattner. 1998. DNA sequencing and analysis of the low-Ca2+ response plasmid pCD1 of Yersinia pestis KIM5. Infect. Immun. 66:4611-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Petrovskaya, V. G. 1967. The problem of bacterial virulence. Meditsina Press, Leningrad, USSR.
- 145.Phillips, A. P., B. C. Morris, D. Hall, M. Glenister, and J. E. Williams. 1988. Identification of encapsulated and non-encapsulated Yersinia pestis by immunofluorescence tests using polyclonal and monoclonal antibodies. Epidemiol. Infect. 101:59-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pollitzer, R. 1954. Plague. W. H. O. Monogr. Ser. 22:1-698. [Google Scholar]
- 147.Popov, I. A., I. I. Iashechkin, and I. G. Drozdov. 1998. Molecular genetic analysis of DNA structure of pFra plasmids in plague pathogen of varying biovar. Genetika 34:198-205. [PubMed] [Google Scholar]
- 148.Popov, Y. A., O. A. Protsenko, P. I. Anisimov, A. M. Kokushkin, and O. T. Mozharov. 1980. Detection of pesticinogenic plasmids in Yersinia pestis by agarose gel electrophoresis, p. 20-25. In P. I. Anisimov (ed.), Prophylaxis of particularly dangerous infections. Communist Press, Saratov, USSR.
- 149.Porat, R., W. R. McCabe, and R. R. Brubaker. 1995. Lipopolysaccharide-associated resistance to killing of yersiniae by complement. J. Endotoxin Res. 2:91-97. [Google Scholar]
- 150.Portnoy, D. A. and S. Falkow. 1981. Virulence-associated plasmids from Yersinia enterocolitica and Yersinia pestis. J. Bacteriol. 148:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Poshevina, G. O. 1974. On the biology of the plague pathogen from Muyun Kum Desert. Ph.D. thesis. Institute of Ministry of Public Health of Kazakh SSR for Epidemiology, Microbiology and Infectious Diseases, Alma-Ata, USSR.
- 152.Prentice, M. B., K. D. James, J. Parkhill, S. G. Baker, K. Stevens, M. N. Simmonds, K. L. Mungall, C. Churcher, P. C. Oyston, R. W. Titball, B. W. Wren, J. Wain, D. Pickard, T. T. Hien, J. J. Farrar, and G. Dougan. 2001. Yersinia pestis pFra shows biovar-specific differences and recent common ancestry with a Salmonella enterica serovar Typhi plasmid. J. Bacteriol. 183:2586-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Prior, J. L., and R. W. Titball. 2002. Monoclonal antibodies against Yersinia pestis lipopolysaccharide detect bacteria cultured at 28C or 37C. Mol. Cell. Probes 16:251-256. [DOI] [PubMed] [Google Scholar]
- 154.Protsenko, O. A., P. I. Anisimov, O. T. Mozharov, N. P. Konnov, and I. A. Popov. 1983. Detection and characterization of the plasmids of the plague microbe which determine the synthesis of pesticin 1, fraction 1 antigen and “mouse” toxin exotoxin. Genetika 19:1081-1090. [PubMed] [Google Scholar]
- 155.Radnedge, L., P. G. Agron, P. L. Worsham, and G. L. Andersen. 2002. Genome plasticity in Yersinia pestis. Microbiology 148:1687-1698. [DOI] [PubMed] [Google Scholar]
- 156.Radnedge, L., S. Gamez-Chin, P. M. McCready, P. L. Worsham, and G. L. Andersen. 2001. Identification of nucleotide sequences for the specific and rapid detection of Yersinia pestis. Appl. Environ. Microbiol. 67:3759-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rall, Y. M. 1965. Natural focality and epizootology of plague. Meditsina, Moscow, USSR.
- 158.Rider, T. H., M. S. Petrovick, F. E. Nargi, J. D. Harper, E. D. Schwoebel, R. H. Mathews, D. J. Blanchard, L. T. Bortolin, A. M. Young, J. Chen, and M. A. Hollis. 2003. A B cell-based sensor for rapid identification of pathogens. Science 11:213-215. [DOI] [PubMed] [Google Scholar]
- 159.Roggenkamp, A., A.M. Geiger, L. Leitritz, A. Kessler, and J. Heesemann. 1997. Passive immunity to infection with Yersinia spp. mediated by anti-recombinant V antigen is dependent on polymorphism of V antigen. Infect. Immun. 65:446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rudnev, G. P. 1940. Clinical picture of plague. Medgiz, Moscow, USSR.
- 161.Russell, A. D., J. R. Furr, and J.-Y. Maillard. 1997. Microbial susceptibility and resistance to biocides. ASM News 63:481-487. [Google Scholar]
- 162.Ryzhko, I. V., E. D. Samokhodkina, R. I. Tsuraeva, A. I. Shcherbaniuk, and N. S. Tsetskhladze. 1998. Characteristics of etiotropic therapy of plague infection induced by atypical strains of F1− phenotype plague microbe. Antibiot. Khimioter. 43:24-28. [PubMed] [Google Scholar]
- 163.Samoilova, S. V., L. V. Samoilova, I. N. Yezhov, I. G. Drozdov, and A. P. Anisimov. 1996. Virulence of pPst+ and pPst− strains of Yersinia pestis for guinea-pigs. J. Med. Microbiol. 45:440-444. [DOI] [PubMed] [Google Scholar]
- 164.Samokhodkina, E. D., I. V. Ryzhko, A. I. Shcherbaniuk, I. V. Kasatkina, R. I. Tsuraeva, and T. A. Zhigalova. 1992. Doxycycline in the prevention of experimental plague induced by plague microbe variants. Ant. Khimioter. 37:26-28. [PubMed] [Google Scholar]
- 165.Samokhodkina, E. D., I. V. Ryzhko, R. I. Tsuraeva, and V. V. Pasiukov. 1994. Beta-lactam antibiotics (ampicillin, cefotaxime) in prevention of experimental plague in albino mice, caused by non-fractioned strains of the pathogen. Antibiot. Khimioter. 39:20-23. [PubMed] [Google Scholar]
- 166.Samygin, V. M. 1982. Characterization of Yersinia pestis strains from the Volga-Ural sandy natural focus and right-bank flood-lands of the Ural River. Ph.D. thesis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 167.Serdyukova, T. V. 1991. Biological peculiarities of the plague pathogen from the Central-Caucasian natural focus. Ph.D. thesis. Russian Research Anti-Plague Institute “Microbe,“ Saratov, Russia.
- 168.Shvedun, G. P., O. P. Plotnikov, O. T. Mozharov, T. K. Merkulova, and O. G. Shishkina. 1990. Evaluation of genetic tests as criteria for the choice of type and reference strains for individual natural plague foci, p. 43-47. In Laboratory diagnostics and genetics of virulence of the agents of particularly dangerous infections. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 169.Sludskii, A. A. 1998. Vole type of natural plague foci (structure and function). Sc.D. thesis. Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 169a.Sing, A., D. Rost, N. Tvardovskaia, A. Roggenkamp, A. Wiedemann, C. J. Kirschning, M. Aepfelbacher, and J. Heesemann. 2002. Yersinia V-antigen exploits toll-like receptor 2 and CD14 for interleukin 10-mediated immunosuppression. J. Exp. Med. 196:1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Snellings, N. J., M. Popek, and L. E. Lindler. 2001. Complete DNA sequence of Yersinia enterocolitica serotype O:8 low-calcium-response plasmid reveals a new virulence plasmid-associated replicon. Infect. Immun. 69:4627-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sodeinde, O. A., Y. V. B. K. Subrahmanyam, K. Stark, T. Quan, Y. Bao, and J. D. Goguen. 1992. A surface protease and the invasive character of plague. Science 258:1004-1007. [DOI] [PubMed] [Google Scholar]
- 172.Stepanov, V. M. 1975. Nutrition factors and their roles in manifestation of virulence and immunogenicity of the plague pathogen. Sc.D. thesis. Russian Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 173.Stepanov, V. M., L. N. Klassovskii, and G. Y. Pak. 1981. Studying of the influence of epizootic process phase upon Yersinia pestis changeability, p. 110-112. In Proceedings of the 11th Inter-Republic Scientific and Practical Conference of Anti-Plague Establishments of Central Asia and Kazakhstan on Plague Prophylaxis. Research Anti-Plague Institute of Central Asia, Alma-Ata, USSR.
- 174.Suchkov, I. Yu., B. N. Mishankin, S. O. Vodopyanov, L. M. Smolikova, and M. V. Shishiyanu. 2002. Genotyping of Yersinia pestis: variability of locus (CAAA)N in natural strains isolated in areas of the former Soviet Union. Mol. Gen. Mikrobiol. Virusol. 4:18-21. [PubMed] [Google Scholar]
- 175.Tan, J, Y. Liu, E. Shen, W. Zhu, W. Wang, R. Li, and L. Yang. 2002. Towards the atlas of plague and its environment in the People's Republic of China: idea, principle and methodology of design and research results. Huan Jing Ke Xue 23:1-8. [PubMed] [Google Scholar]
- 176.Tarasov, M. A., L. T. V. Khyong, N. I. Suntsova, and V. V. Suntsov. 1994. Data on population ecology of background flea species (Insecta, Aphaniptera) and plague vectors in the south part of Viet Nam. Probl. Particularly Dangerous Infect. 74:84-89. [Google Scholar]
- 177.Timofeeva, L. A. 1972. On taxonomy of the plague pathogen. Probl. Particularly Dangerous Infect. 23:15-22. [Google Scholar]
- 178.Titball, R. W., and E. D. Williamson. 2001. Vaccination against bubonic and pneumonic plague. Vaccine 19:4175-4184. [DOI] [PubMed] [Google Scholar]
- 179.Toporkov, V. P., A. V. Podsvirov, and K. B. Yashkulov. 1999. Ecological-epidemiological monitoring of predictors of extreme epidemic situations in the natural plague focus situated at the northwest from the Caspian Sea. Center for State Epidemiologic Control of Kalmyk Republic, Elista, Kalmykia.
- 180.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, C. R. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122-43131. [DOI] [PubMed] [Google Scholar]
- 181.Tumanskii, V. M. 1957. On classification of varieties of the plague pathogen. Zh. Mikrobiol. Epidemiol. Immunobiol. 6:3-7. [PubMed] [Google Scholar]
- 182.Une, T., and R. R. Brubaker. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect. Immun. 43:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Velichko, L. N., and A. M. Kokushkin. 1997. Frequency of isolation of atypical Yersinia pestis strains in the natural plague foci, p. 21. In Proceedings of the Scientific and Practical Conference dedicated to the centenary of the Russian Antiplague Service, vol. 1.Russian Research Anti-Plague Institute “Microbe,” Saratov, Russia.
- 184.Vershinina, T. I. 1986. On population heterogeneity of the plague pathogen from the natural foci of Mountain Altai and northwestern Mongolia. Ph.D. thesis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 185.Vinogradov, E. V., B. Lindner, N. A. Kocharova, S. N. Senchenkova, A. S. Shashkov, Y. A. Knirel, O. Holst, T. A. Gremyakova, R. Z. Shaikhutdinova, and A. P. Anisimov. 2002. The core structure of the lipopolysaccharide from the causative agent of plague, Yersinia pestis. Carbohydr. Res. 337:775-777. [DOI] [PubMed] [Google Scholar]
- 186.Welkos, S. L., A. M. Friedlander, and K. J. Davis. 1997. Studies on the role of plasminogen activator in systemic infection by virulent Yersinia pestis strain CO92. Microb. Pathog. 23:211-223. [DOI] [PubMed] [Google Scholar]
- 187.Welkos, S. L., K. M. Davis, L. M. Pitt, P. L. Worsham, and A. M. Friedlander. 1995. Studies on the contribution of the F1 capsule-associated plasmid pFra to the virulence of Yersinia pestis. Contrib. Microbiol. Immunol. 13:299-305. [PubMed] [Google Scholar]
- 188.Welkos, S., M. L. M. Pitt, M. Martinez, A. Friedlander, P. Vogel, and R. Tammariello. 2002. Determination of the virulence of the pigmentation-deficient and pigmentation-/plasminogen activator deficient strains of Yersinia pestis in non-human primate and mouse models of pneumonic plague. Vaccine 20:2206-2214. [DOI] [PubMed] [Google Scholar]
- 189.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further application of the procedure, vol. 5. Academic Press, Inc., New York, N.Y.
- 190.Williams, J. E., and D. C. Cavanaugh. 1984. Potential for rat plague from nonencapsulated variants of the plague bacillus (Yersinia pestis). Experientia 40:739-740. [DOI] [PubMed] [Google Scholar]
- 191.Williams, J. E., D. N. Harrison, T. J. Quan, J. L. Mullins, A. M. Barnes, and D. C. Cavanaugh. 1978. Atypical plague bacilli isolated from rodents, fleas, and man. Am. J. Public Health 68:262-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Williamson, E. D. 2001. Plague vaccine research and development. J. Appl. Microbiol. 91:606-608. [DOI] [PubMed] [Google Scholar]
- 193.Winter, C. C., W. B. Cherry, and M. D. Moody. 1960. An unusual strain of Pasteurella pestis isolated from a fatal human case of plague. Bull. W. H. O. 23:408-409. [PMC free article] [PubMed] [Google Scholar]
- 194.Worsham, P. L., M.-P. Stein, and S. L. Welkos. 1995. Construction of defined F1 negative mutants of virulent Yersinia pestis. Contrib. Microbiol. Immunol. 13:325-328. [PubMed] [Google Scholar]
- 195.Zakharov, A. I. 1989. Studies of the biology of the plague pathogen from Uralo-Embenskii autonomous focus. Ph.D. thesis. All-Union Research Anti-Plague Institute “Microbe,” Saratov, USSR.
- 196.Zao, M., Z. Ceng, C. Zang, Y. He, and G Thang. 1993. The plasmid profile of Yersinia pestis strains in different natural foci of plague in the People's Republic of China. Zh. Mikrobiol. Epidemiol. Immunobiol. 2:27-31. [PubMed] [Google Scholar]
- 197.Zharinova, N. V., A. F. Bryukhanov, V. V. Zarubin, G. D. Bryukhanova, and Ch. B. Ulukhanov. 2002. Attributes of strains isolated from Central-Caucasian natural plague focus in 1999-2001, p. 99-100. In Proceedings of the Jubilee Scientific and Practical Conference on Epidemiological Safety at the Caucasus devoted to the fiftieth anniversary of the Stavrapol' Research Anti-Plague Institute. Stavrapol' Research Anti-Plague Institute, Stavrapol', Russia.
- 198.Zudina, I. V. 2000. Genetic characterization of the chromosomal pigmentation region of five Yersinia pestis subspecies. Ph.D. thesis. Russian Research Anti-Plague Institute “Microbe," Saratov, Russia.