Abstract
Foot-and-mouth disease (FMD) is a highly contagious disease of cloven-hoofed animals. The disease was initially described in the 16th century and was the first animal pathogen identified as a virus. Recent FMD outbreaks in developed countries and their significant economic impact have increased the concern of governments worldwide. This review describes the reemergence of FMD in developed countries that had been disease free for many years and the effect that this has had on disease control strategies. The etiologic agent, FMD virus (FMDV), a member of the Picornaviridae family, is examined in detail at the genetic, structural, and biochemical levels and in terms of its antigenic diversity. The virus replication cycle, including virus-receptor interactions as well as unique aspects of virus translation and shutoff of host macromolecular synthesis, is discussed. This information has been the basis for the development of improved protocols to rapidly identify disease outbreaks, to differentiate vaccinated from infected animals, and to begin to identify and test novel vaccine candidates. Furthermore, this knowledge, coupled with the ability to manipulate FMDV genomes at the molecular level, has provided the framework for examination of disease pathogenesis and the development of a more complete understanding of the virus and host factors involved.
THE DISEASE AND ITS AGENT
The recent outbreaks of foot-and-mouth disease (FMD) in a number of FMD-free countries, in particular Taiwan in 1997 and the United Kingdom in 2001, have significantly increased public awareness of this highly infectious disease of cloven-hoofed livestock. Furthermore, worldwide concern following the terrorist attacks in the United States has raised the possibility that terrorist organizations or rogue states might target the $100 billion/year U.S. livestock industry by employing the etiologic agent of FMD. These events have directed the efforts of the scientific community to reexamine our knowledge of FMD, the viral agent that causes the disease, and current methods of disease control. In this review, we summarize the history of this disease; present, in detail, our current knowledge of the molecular biology, pathogenesis, and virulence factors; and discuss recent disease outbreaks as well as new disease control strategies.
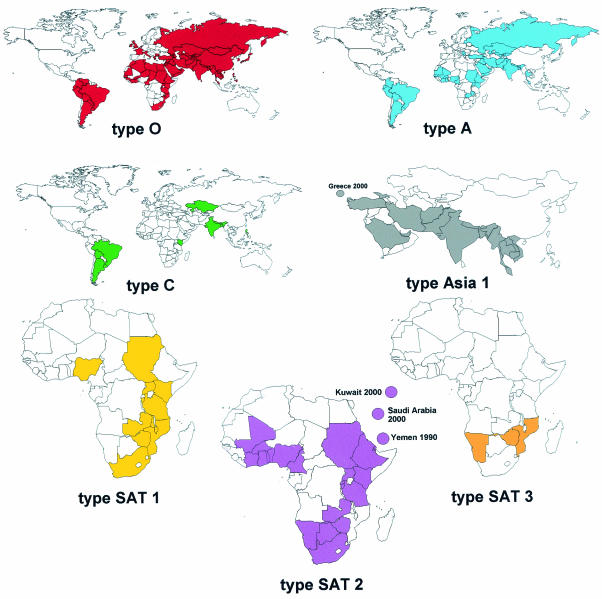
The first written description of FMD probably occurred in 1514, when Fracastorius described a similar disease of cattle in Italy (159). Almost 400 years later, in 1897, Loeffler and Frosch (271) demonstrated that a filterable agent caused FMD. This was the first demonstration that a disease of animals was caused by a filterable agent and ushered in the era of virology. Subsequently it was shown that the agent, FMD virus (FMDV), consists of a single-stranded, plus-sense RNA genome of approximately 8,500 bases surrounded by four structural proteins to form an icosahedral capsid (401). FMDV is the type species of the Aphthovirus genus of the Picornaviridae family. The only other member of this genus is equine rhinitis A virus (240). Seven serotypes (A, O, C, Asia 1, and South African Territories 1, 2, and 3) have been identified serologically, and multiple subtypes occur within each serotype (21).
Outbreaks have occurred in every livestock-containing region of the world with the exception of New Zealand, and the disease is currently enzootic in all continents except Australia and North America (Fig. 1). The disease affects domestic cloven-hoofed animals, including cattle, swine, sheep, and goats, as well as more than 70 species of wild animals, including deer (155), and is characterized by fever, lameness, and vesicular lesions on the tongue, feet, snout, and teats (see “Pathogenesis” below). In sheep and goats the disease is generally mild and can be difficult to distinguish from other common conditions (138, 168). In addition, other vesicular diseases, such as swine vesicular disease (SVD), vesicular stomatitis, and vesicular exanthema of swine, cause signs so similar to those of FMD that differential clinical diagnosis alone can be difficult (21). Although FMD does not result in high mortality in adult animals, the disease has debilitating effects, including weight loss, decrease in milk production, and loss of draught power, resulting in a loss in productivity for a considerable time. Mortality, however, can be high in young animals, where the virus can affect the heart. In addition, cattle, sheep, and goats can become carriers, and cattle can harbor virus for up to 2 to 3 years (70) (see “Carrier state” below).
FIG. 1.
Countries in which FMD was reported to the OIE between 1990 and 2002. The data and maps were compiled by Nick Knowles and can be found at www.iah.bbsrc.ac.uk/virus/picornaviridae/aphthovirus.
FMD is one of the most highly contagious diseases of animals or humans, and FMDV rapidly replicates and spreads within the infected animal, among in-contact susceptible animals, and by aerosol. Disease signs can appear within 2 to 3 days after exposure and can last for 7 to 10 days. FMD is on the A list of infectious diseases of animals of the Office International des Épizooties (OIE) and has been recognized as the most important constraint to international trade in animals and animal products (266). Countries that are free of the disease have introduced a number of measures to retain this status because of the detrimental economic consequences resulting from its presence. The Smoot-Hawley Tariff Act of 1930, which was passed after the last outbreak of FMD in the United States in 1929, contained restrictions on importation of susceptible livestock, fresh meat, and animal products from countries where FMD was present (21). To protect disease-free countries, the OIE has developed control policy recommendations for affected countries to reacquire FMD-free status and therefore participate in international trade. These recommendations favor the more rapid return to FMD-free status if an outbreak is controlled by slaughter and vaccination is not employed. Thus, disease-free countries such as the United States, the United Kingdom, and other Western European countries have adopted control strategies that include inhibition of animal movement and slaughter of infected and in-contact susceptible animals but generally do not include vaccination. However, recent events, specifically the 2001 outbreak in the United Kingdom, have had a profound effect on this control strategy, and new recommendations have evolved and are evolving.
DESCRIPTION OF THE AGENT
Genome Organization
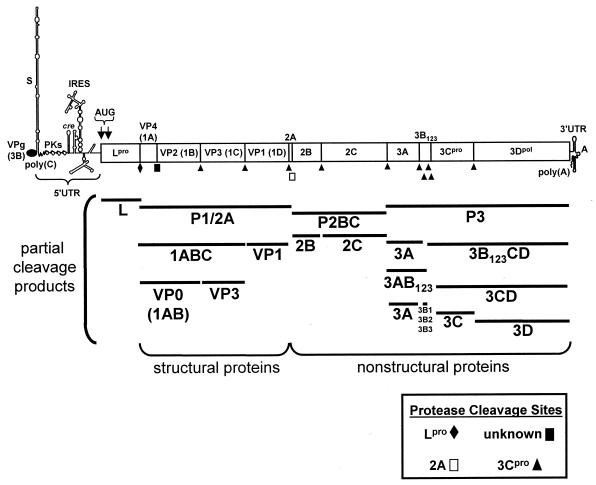
The virion is a 140S particle consisting of a single-stranded RNA genome and 60 copies each of four structural proteins (VP1 [1D], VP2 [1B], VP3 [1C], and VP4 [1A]). The FMDV genome has a basic organization similar to those of other members of the Picornaviridae, and the nomenclature for the viral proteins was established by Rueckert and Wimmer (402). In this review, we refer to the viral structural proteins by their more common designations, VP1 to -4, and to viral nonstructural (NS) proteins mainly by their designations as described by Rueckert and Wimmer (402). Within the virion, there are small amounts of a cleavage precursor of VP2 and VP4, called VP0 (1AB) (401), and one copy of a 23- to 24-amino-acid genome-linked protein, 3B (VPg [we use this designation for the protein]), covalently bound to the 5′ terminus of the RNA (186, 420). The organization of the viral genome is shown in Fig. 2. The RNA is translated as a single long open reading frame (ORF) into a polyprotein, followed by a series of posttranslational proteolytic cleavages to generate both the intermediate and mature structural and NS viral proteins (191, 387, 402).
FIG. 2.
Schematic map of the FMDV genome. The ORF is shown in the boxed area, with the viral proteins named according to the nomenclature of Rueckert and Wimmer (402). Also shown are the functional elements of the genome as described in the text and the partial protein cleavage products. The sites of the primary cleavages and the proteases responsible are indicated. PKs, pseudoknot structures. (Adapted from reference 295.)
Based on the initial cleavage products, the genome ORF is divided into four regions (Fig. 2). The 5′ end, the L region, which encodes the N-terminal component of the polyprotein, contains two in-frame AUG initiation codons that result in the generation of two L proteins, Lab and Lb (387, 419). While both forms of L are synthesized during in vitro translation of viral RNA (48, 418) and in infected cells (102), it has been shown, by using site-directed mutagenesis, that deletion of the second AUG from an FMDV infectious clone abolished viral replication upon transfection of the transcribed RNA into cells, while deletion of the first AUG had no effect on viral replication (84). In addition, Piccone et al. (356) generated synthetic viral genomes lacking the L gene and showed that only the genome that initiated polyprotein synthesis at the second AUG codon produced live virus. These results strongly suggest that Lb may be the biologically functional protein in vivo. The L protein, a papain-like protease (Lpro) (251, 356, 386, 435), is autocatalytically cleaved from the polyprotein at its C terminus (440). A crystallographic structural analysis of Lpro suggested that the self-cleavage reaction might occur intermolecularly, in trans, based on the position of the protein's C terminus within the active site of an adjacent Lpro molecule (194). However, other structural features within the same crystals suggested that an intramolecular cis self-cleavage reaction is also possible (194). In addition, Lpro has unique cation concentration and pH range requirements, due to differences within the molecule's active site, which distinguish it from other papain-like enzymes (193). The Lpro also plays a role in inhibition of host protein synthesis and has been identified as a viral virulence factor (see “Viral translation” and “Virulence factors” below).
Directly downstream of the L region is the P1 region of the genome (Fig. 2), encoding the four viral structural proteins VP4, VP2, VP3, and VP1. Following the P1 region is the P2 region (Fig. 2), encoding three viral NS proteins, 2A, 2B, and 2C, and the P3 region, encoding NS proteins 3A, three copies of VPg, 3Cpro, and 3Dpol. Historically the 2A region was considered part of the P2 region; however, genetic and biochemical evidence has shown that the FMDV 2A peptide is cleaved as a P1-2A precursor (463) (see “Viral translation” below). 3Cpro was identified as a viral protease (252) and is involved in processing the viral polyprotein, while 3Dpol is the viral RNA-dependent RNA polymerase (110, 275, 335, 368-370, 389). The roles that each of these proteins play in viral replication are discussed in Infectious Cycle below.
The FMDV genome also contains untranslated RNA found upstream (5′ untranslated region [5′ UTR]) and downstream (3′ UTR) of the ORF (Fig. 2). The 5′ UTR of FMDV contains about 1,300 bases (157, 191, 387) and can be divided into five functional elements which play roles in virus translation and RNA replication (Fig. 2). The most 5′ segment, the S fragment, encompasses about 360 bases and folds into a long stem-loop (Fig. 2) (77, 203, 336). The function of the S fragment is not known, but analogies with other picornaviral genomes suggest that it may play a role in maintaining genome stability in infected cells (33) and may also be involved in the binding of proteins involved in genome replication (12, 13, 33, 212, 481). There have been some suggestions that the S fragment may affect viral pathogenicity, but currently there is no direct supporting evidence for this (85, 150).
Following the S fragment, there is a poly(C) tract comprising over 90% C residues with a small number of U and A residues (Fig. 2). This segment is over 100 bases in length; however, the length of the poly(C) tract can be extremely variable (108), and, in one case, a tissue culture-adapted virus has been shown to have a poly(C) length of over 400 bases (150). Although an early study suggested that the length of the poly(C) tract was associated with virulence (203), other studies have been unable to correlate poly(C) length with this property of the virus (108). It has been shown that the poly(C) lengths of natural viral isolates are increased by repeated passage in cell culture (150), as is the length of this segment in genetically engineered viruses (381). It is possible to replicate virus with essentially no poly(C), and while this virus is virulent in suckling mice, it has a much higher particle/PFU ratio than viruses containing longer poly(C) tracts (381). The virulence of this virus in susceptible animals has not been determined. While the exact role that the poly(C) tract plays in FMDV replication is unknown, recent studies with poliovirus have shown an association of the host factor, poly(rC) binding protein (PCBP), with the 5′ end of the poliovirus genome (165), which could regulate the switch from translation to genome replication (see “Viral transcription and genome replication” below) (33, 212, 470). Just 3′ of the poly(C) tract there is a series of RNA pseudoknot structures of unknown function (Fig. 2) (149, 381).
Downstream of the pseudoknots there is a short hairpin loop structure, the cis-acting replicative element (cre) (Fig. 2) (294). The cre, which has been identified in the genomes of human rhinoviruses (169, 310, 311), poliovirus (176, 382), and cardioviruses (270), has a stem-loop with a conserved AAACA sequence in the loop region. In contrast to the case for other picornaviruses, where the cre is located within different regions of the ORF, the cre of FMDV is located within the 5′ UTR (294). The cre is essential for RNA genome replication, and its function is discussed in “Viral transcription and genome replication” below.
The region between the cre and ORF contains a series of highly conserved stem-loop structures which together constitute the internal ribosome entry site (IRES) (Fig. 2). All picornavirus mRNAs lack the 7-methyl-G cap structure present at the 5′ ends of most eukaryotic mRNAs. In addition, the 5′ UTR of picornaviral genomes is quite long, and it was demonstrated for poliovirus and encephalomyocarditis virus (EMCV) that ribosomes enter the genome internally at the IRES (231, 351). An IRES element was subsequently identified within the FMDV genome (51, 261). The IRES elements of picornaviruses contain a high degree of secondary and tertiary structure. They have been divided into three groups, based on conserved structure as opposed to primary sequence (359, 360, 385). The IRES element for aphthoviruses, which is similar in structure to the cardiovirus IRES (group 2 IRES), is about 450 nucleotides in length (51, 261) and has been modeled into a five-domain structure (359). IRES elements contain a pyrimidine-rich region at their 3′ ends immediately preceding the AUG translation initiation codon, and FMDV contains pyrimidine-rich regions directly upstream of each of the alternative AUG initiation codons (51, 261, 361).
The 3′ UTR, which follows the ORF termination codon, contains a short stretch of RNA which folds into a specific stem-loop structure (362) followed by a poly(A) tract of variable length carried on the genome (Fig. 2) (141). The 3′ UTR also appears to be important for genome replication (312, 364, 392). This is supported by studies showing that the 3′ UTR can bind a number of picornaviral proteins that are involved in RNA replication (114, 115, 202). Gutierrez and coworkers (195) demonstrated that hybridization of antisense RNA to the 3′ UTR of FMDV did not effect in vitro translation of viral RNA but did inhibit RNA replication in infected cells. In contrast, more recent studies have demonstrated that deletion of the FMDV 3′ UTR reduced the efficiency of in vitro translation (274) and blocked the ability to recover viable virus from transfected cells (413). Replacing the FMDV 3′ UTR with that of the enterovirus, SVD virus, resulted in a nonviable genome (413), suggesting that the 3′ UTR is specific for each picornavirus. The poly(A) tract probably functions in FMDV translation (274) and may also play a role in picornavirus RNA replication (33, 212).
Virus Structure
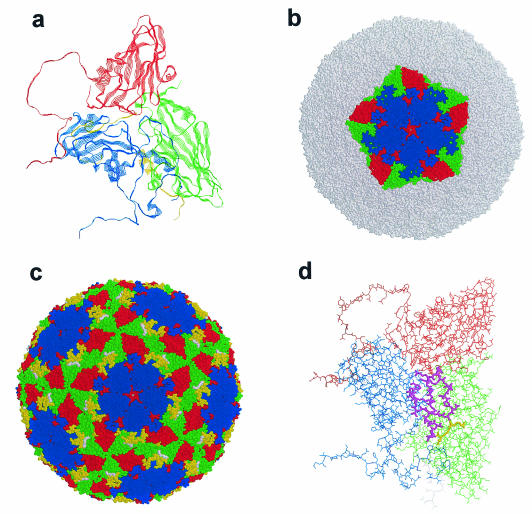
By electron microscopy, the FMD virion appears to be a round particle with a smooth surface and a diameter of about 25 nm (21). The fine structure of the viral capsid has been determined for a number of serotypes by using X-ray crystallographic techniques (2, 117, 227, 263, 264), and the structural features of type O1BFS are shown in Fig. 3. The structural proteins, VP1 to -3, fold into an eight-stranded wedge-shaped β-barrel which fit together to form the majority of the capsid structure (Fig. 3a) (2). The VP4 protein is buried within the capsid and has a myristyl group covalently attached to its N terminus (50, 100). The strands of the β-barrels of VP1 to -3 are connected by loops which form the outer surface of the virion (Fig. 3a) (227). FMDV is distinguished from other picornaviruses by the lack of a surface canyon, or pit, which has been shown to be the receptor binding site for the entero- and cardioviruses (49, 207, 215, 257, 280, 325, 398, 483). Another feature of the virion is the presence of a channel at the fivefold axis which permits the entry of small molecules, such as CsCl, into the capsid, resulting in FMDV having the highest buoyant density of the picornaviruses (Fig. 3c) (2, 227).
FIG. 3.
Structure of the mature type O1BFS FMD virion based on X-ray crystallographic data. The structures shown are based on the data of Acharya et al. (2) and Logan et al. (272). (a) A viral protomer highlighting the β-barrel-and-loop organization of the viral proteins. (b) A pentamer positioned on the virion looking down the fivefold axis. (c) The organization of the entire virion, highlighting the G-H loop (yellow) and the RGD sequence (white). Note the pore located at the top of the fivefold axis (see text). (d) A protomer highlighting the positions of the G-H loop (purple) and RGD sequence (yellow). All structures are representative of the mature virion (cleaved VP0 [see text]), and the viral proteins are colored blue (VP1), green (VP2), and red (VP3). VP4 is buried within the particle and is visible only in panel a, where it is colored yellow. The graphic renderings were done by using RasMole.
Unlike those of other picornaviruses, the FMDV capsid is dissociated at pHs of below 6.5 into 12S pentameric subunits (76). The reason for this instability is thought to be a cluster of His residues at the interface between VP2 and VP3 which become protonated at low pH, weakening the capsid through electrostatic repulsion (116, 147). This low-pH-induced instability of FMDV leads to differences in the mechanism of its uncoating upon infection of cells compared to that for other picornaviruses (see “Early interactions: adsorption, penetration and uncoating” below) and also probably plays a role in the targeting of the virus to specific tissues and organs in susceptible hosts.
Infectious Cycle
FMDV, like other members of the Picornaviridae, has a relatively short infectious cycle in cultured cells. Depending on the multiplicity of infection, newly formed infectious virions begin to appear at between 4 and 6 h after infection. The virus is cytocidal, and infected cells exhibit morphological alterations, commonly called cytopathic effects, which include cell rounding and alteration and redistribution of internal cellular membranes. The virus also causes biochemical alterations, including inhibition of host translation and transcription (401).
Early interactions: adsorption, penetration, and uncoating.
The interactions of FMDV with cells have been extensively studied for many years. It is generally accepted that FMDV receptors, as well as other picornavirus receptors, play a role in tissue and organ tropism which leads to disease pathogenesis (112, 151, 429, 476). FMDV binds rapidly to cells in culture at both 4 and 37°C by using a limited number of receptor sites, which has measured at between 103 and 104 per cell (39). These studies also suggested that while six of the seven serotypes bound to a single class of receptor site, some of the serotypes bound to a second class of receptors which were present at a high copy number (39, 431). Early studies showed that limited trypsin digestion of virus resulted in viral particles which were noninfectious due to the inability to bind to cells in culture (22, 30, 38, 90, 318). Analysis of trypsin-treated virus revealed a single cleavage of the VP1 protein at Arg144 (388), which was later shown to be located within a surface loop connecting the βG and βH strands of the protein (G-H loop) (2, 272) (Fig. 3c and d). These results suggested that this region of the VP1 protein interacted with the cell surface receptor.
In 1984 Pierschbacher and Ruoslahti (357), studying the binding of fibronectin to cells, reported that the tripeptide sequence Arg-Gly-Asp (RGD) was a cellular recognition site on the molecule and that this sequence was also found in the FMDV VP1 protein (358) (Fig. 3d). The fibronectin receptor was subsequently shown to be part of a large family of transmembrane glycoproteins called integrins (449). These molecules are type l membrane glycoproteins, consisting of two subunits (α and β) which are noncovalently bound at the cell surface. They are involved in cell adhesion, cell migration, thrombosis, and lymphocyte interactions (220, 221, 223). In FMDV, while sequences surrounding the RGD sequence within the G-H loop are variable, the RGD sequence itself is highly conserved (353). The first indication that the RGD sequences might be involved in the virus-receptor interaction came from studies showing that small peptides containing RGD could inhibit the binding of virus to cells (40, 158). Direct genetic evidence for this interaction was obtained by mutating or deleting the RGD sequence in infectious cDNA clones, resulting in viral particles which were noninfectious, could not adsorb to susceptible cells, and could not cause disease in susceptible animals (268, 297, 309). Of the 24 known integrin receptors, only 8 use the RGD tripeptide as a recognition sequence, and 5 of those are part of the αV subgroup of integrin receptors (222, 403). The first identification of the integrin receptor for FMDV was made based on comparison of the receptor specificity of the virus with that of a human enterovirus, coxsackievirus A9 (CAV9). This virus contains an extension in its VP1 protein that includes an RGD sequence (92, 93), which was shown to be involved in virus binding to cells in culture (393). The receptor utilized by CAV9 was demonstrated to be the integrin αVβ3 (394), and competition binding studies revealed that CAV9 and antibodies to the αVβ3 integrin were able to inhibit the binding of FMDV to monkey kidney cells (56). These results were confirmed genetically by demonstrating that cells which did not express this integrin and were not susceptible to FMDV infection became permissive for viral infection upon transfection of cDNAs encoding either the human (334) or the bovine (144, 332, 333) αVβ3 integrin. Following these studies, it was further demonstrated that FMDV could also utilize two additional αV integrins, αVβ6 (229) and αVβ1 (228); however, the virus was unable to utilize the integrin αVβ5 as a receptor (144; for reviews, see references 44 and 227). To date, none of the other integrins which utilize the RGD recognition sequence have been analyzed for FMDV receptor activity.
A number of alternative receptors have been shown to mediate FMDV infection in vitro. Antibody-complexed virus can infect cells via Fc receptor-mediated adsorption (42, 293, 297). In addition, an artificial receptor which consists of a single-chain anti-FMDV monoclonal antibody fused to intercellular adhesion molecule 1 (ICAM-1) has been engineered, and this receptor was also able to mediate infection with RGD-deleted virus (380). In 1996, Jackson and coworkers (226) reported that a type O1 virus was able to utilize the glycosaminoglycan heparan sulfate (HS) as a coreceptor. We had previously shown that there appeared to be a second, unidentified receptor present at a high copy number (39, 431), and the report by Jackson et al., (226) was consistent with this finding. It contrasted, however, with our findings that a type A virus could not bind to CHO cells which lack the integrin receptor for FMDV but express HS (293). We reconciled these seemingly opposite findings by showing that tissue culture adaptation of a type O1 virus selects a variant which has a positively charged Arg at residue 56 of VP3 and can grow in CHO cells (409). Interestingly, this variant was relatively avirulent in cattle (409). In contrast, a second variant of this virus containing a His at residue 56 of VP3 could not grow in CHO cells and was relatively virulent in cattle (409). We expanded these studies to show that the virus with the Arg residue required only HS to replicate in CHO cells but that the variant with the His residue required the integrin to replicate in cell culture (334). A similar result was also noted for type C viruses, where multiple passages in tissue culture selected viruses with positive surface charges which can replicate in the absence of the known integrin receptors (26, 27, 290). Structural analysis of HS-complexed type O virus revealed a direct interaction between HS and residue 56 in VP3 (162, 227).
While the penetration and uncoating of FMDV have not been studied in great detail, there have been some observations which suggest possible mechanisms of how they might occur. We and others have shown that after adsorption to the cell surface, the 140S virion breaks down into 12S pentameric subunits, releasing the RNA (37-39, 89). This breakdown does not occur at the cell surface, since particles which are eluted from the cell after adsorption are fully infectious and still sediment at 140S (39). By using a series of lysosomotropic agents, which raise the pH of intracellular endosomes, it has been demonstrated that the virus probably breaks down upon entering an acidic endosome (37, 87, 88). Recently it has been shown that a genetically engineered FMDV, which is unable to perform the maturation cleavage of VP0 to VP2 and VP4 (see “Encapsidation and maturation” below) is noninfectious, can adsorb to cells in culture, and is acid sensitive (253). Thus, the breakdown of 140S virus to pentameric subunits by itself does not lead to productive infection, but there must be other events after the breakdown. These results indicate that the viral receptor is responsible only for docking the virus to the membrane of the susceptible cell and plays no role in viral uncoating, which is consistent with the ability of FMDV to utilize multiple receptors for infection in cell culture.
Viral translation.
Following uncoating, the RNA is released into the cytoplasm by an as-yet-unknown mechanism and begins a round of viral translation. The genome-linked protein VPg is cleaved by a cellular enzyme prior to translation of the incoming RNA (10, 11); however, protein synthesis initiation complexes can be formed with mRNA containing VPg (174). Unlike most host mRNAs, actively translated viral mRNA does not contain a 7-methyl-G cap structure at its 5′ end (187) and initiates protein synthesis internally at the IRES by a cap-independent mechanism (51, 231, 261, 351). Cap-dependent mRNA translation is inhibited in infected cells as the result of the cleavage of the protein synthesis initiation factor eIF4G by Lpro (124, 241). Intact eIF4G acts as a bridge connecting the mRNA cap to the 40S ribosomal subunit. This is accomplished by the binding of the cap binding protein, eIF4E, to the N-terminal domain of eIF4G, while the C-terminal domain binds eIF4A and the 40S ribosomal subunit via eIF3 (262). In contrast, initiation of FMDV RNA translation requires only the Lpro-generated C-terminal eIF4G cleavage product, which binds to the FMDV IRES and interacts with 40S ribosomal subunit-bound eIF4A and eIF3 (273, 414). In addition, eIF4B is bound to the FMDV IRES and has been identified in both 48S preinitiation complexes and 80S ribosomes (273, 314, 343, 405).
The FMDV IRES interacts with a number of cellular proteins, including initiation factors important for normal cellular mRNA translation. A host factor of 57 kDa, subsequently identified as the nuclear polypyrimidine tract binding protein (PTBP) (209, 237, 337, 338), was shown to interact with at least two regions of the IRES (281, 363). Deletion of these two sites inhibited both the binding of the protein and in vitro translation (282). More recently a second host factor, which is required for FMDV IRES-driven translation but not for translation of cardiovirus mRNA (363), has been identified. This 45-kDa protein, IRES-specific trans-acting factor (ITAF45), along with PTBP, is required for the formation of the 48S translation-initiation complex (291, 363). A third host factor, PCBP, which is required for translation of poliovirus RNA (65) has not to date been shown to be involved in FMDV translation. However, the presence of the poly(C) tract upstream of the IRES suggests that it may also play a role in FMDV translation, genome replication, or both. It has also been postulated that PCBP facilitates a circularization of the poliovirus genome to modulate the balance between translation and RNA replication (33, 212).
Interestingly, the 3′ end of the genome may also be required for FMDV translation, since deletion of either the poly(A) tract or the 3′ stem-loop and the poly(A) tract generated noninfectious FMDV RNAs which had a lowered translation efficiency in in vitro translation reactions (274). Furthermore, addition of either the poly(A) tract, the FMDV 3′ stem-loop, or both to a bicistronic construct, driven by the FMDV IRES, stimulated IRES-directed translation in vitro (274).
Following initiation, translation results in the production of a single polypeptide which undergoes a series of cleavages leading to the production of both structural and NS proteins (Fig. 2). The primary cleavage reactions are performed by three different proteases. As discussed above, Lpro autocatalytically cleaves itself from polyprotein. 2A, (an 18-amino-acid peptide), autocatalytically removes itself from the P2 polyprotein, and remains associated with the P1 precursor (192, 408, 463). There has been a suggestion that this cleavage is not a proteolytic event but rather is a modification of the translational machinery by the 2A peptide which allows the release of P1-2A from the ribosome while permitting the synthesis of the downstream proteins to proceed (139, 140). This hypothesis, however, has not been confirmed by other laboratories. Nevertheless, the 2A peptide has been used in nonviral systems to cleave foreign genes from polyproteins (177, 185). All of the other cleavages of the polyprotein, as outlined in Fig. 2 (with the exception of the maturation cleavage [see below]), are performed by 3Cpro (20, 101, 463). This protein is related to the trypsin family of serine proteases (18, 45, 179), and Grubman and coworkers have mapped the active site of FMDV 3Cpro to Cys163, His46, and Asp84 (192). The protease cleaves at a number of different dipeptide sequences, including Gln-Gly, Glu-Gly, Gln-Leu, and Glu-Ser (348, 387).
Viral transcription and genome replication.
Picornavirus RNA replication presents a number of unique challenges. The 5′ end of the genome RNA is covalently linked to VPg, and the 3′ end has a genetically coded poly(A) tail. Thus, the viral RNA-dependent RNA polymerase (3Dpol) must distinguish between viral RNAs and cellular mRNAs, which also contain 3′-terminal poly(A) tracts. In addition, since the mRNA and the genome RNA are the same molecule, with the exception of the genome-linked VPg, there must be a mechanism to distinguish RNAs which are bound for the ribosome and those which will be packaged into virion particles. While there have been very few studies on transcription and replication in the FMDV system, extensive studies on these activities have been performed with enteroviruses.
The first step in picornavirus RNA replication is the synthesis of a minus-strand RNA molecule. This system has not been studied in FMDV; however, the models of RNA replication developed for poliovirus are probably quite similar (350). It is thought that translation of the plus-strand RNA must cease before minus-strand synthesis begins (164). The mechanism of this shutdown of translation of the plus strand is unclear; however, it has been proposed that, in poliovirus-infected cells, when the polymerase precursor (3CD) accumulates in the cell, it binds to the 5′ cloverleaf structure and modifies the affinity of PCBP for the IRES, an interaction which is essential for translation (see “Viral translation” above). Whether 3CD plays a similar role in FMDV RNA replication is not known; however, it has been shown that in FMDV-infected cells, this protein is rapidly cleaved to 3Cpro and 3Dpol (191).
There is still controversy about the initiation of picornavirus minus-strand RNA synthesis. One model proposes that initiation begins after circularization of the genome facilitated by the interactions of poly(A) binding protein (PABP) with the 3′ poly(A) tail and the PCBP-3CD-5′ cloverleaf structure (211). In the FMDV genome, the 5′ interactions probably take place within the S fragment and may also involve the poly(C) tract (see above). Since initiation of minus-strand synthesis occurs in the cytoplasm in the presence of cellular mRNAs, which also contain poly(A) tails, picornaviruses must have developed mechanisms enabling the polymerase to recognize viral RNA. In picornavirus-infected cells, both plus- and minus-strand RNAs are linked to VPg (339), and the presence of a small protein-linked dinucleotide, VPgpUpU, in infected cells (111) suggested that VPg might be a primer for the RNA polymerase. The discovery of the cre provided a rational mechanism for both the uridylylation of VPg and the ability of the polymerase to discriminate viral RNA from cellular mRNAs. The cre is a stem-loop RNA structure, found within the 5′ UTR in the FMDV genome (Fig. 2), with a conserved AAACA motif within the loop (see “Genome organization” above). This conserved motif is required for poliovirus and rhinovirus minus-strand synthesis (176, 311), and the first two A's serve as the template for the synthesis of VPgpU and VPgpUpU and for viral replication initiation in enteroviruses (169, 382). Mutations in this motif within the FMDV cre severely reduced viral replication in cell culture; however, the cre is positionally independent within picornavirus genomes (176, 294, 310, 487). It is not clear at this time whether free VPg is utilized in the initiation step or whether a cleavage precursor (either 3AB, 3ABC, or 3BCD) is needed. The second model, discussed below, postulates that minus-strand synthesis is primed on the poly(A) tail.
Following the initiation reaction, elongation of the minus strand begins, catalyzed by 3Dpol. For this to occur, the initiation complex must translocate to the 3′ end of the plus-strand template. The mechanism by which this occurs is unknown, but one hypothesis suggests that binding of PABP to the poly(A) tract positions this region of the plus strand near the cre (350). The elongation of the nascent strands results in the formation of a double-stranded molecule, the replicative form (RF) (3, 477). Free minus strands are not detectable in vivo.
After formation of the RF, new plus-strand synthesis can begin. In poliovirus-infected cells, the ratio of plus to minus strands is about 50:1 (340), indicating that a single minus strand can be a template for the synthesis of numerous plus strands, resulting in the formation of a partially double-stranded RNA molecule, the replicative intermediate (3). The initiation of plus-strand synthesis from the RF has not been elucidated; however, two possible mechanisms to generate VPgpUp have been suggested. The first proposes either using existing uridylylated VPg, made in abundance during minus-strand synthesis, or uridylylating VPg at the 3′ end of the minus strand (350). The second hypothesis, which also disputes the mechanism for the initiation of minus strands presented above, proposes that VPgpUpU is generated on the 3′ poly(A) tail of the plus strand and utilized for minus-strand synthesis, while cre-generated VPgpUpU is utilized for plus-strand synthesis (33, 321, 330). Since the data which led to the latter hypothesis was generated totally with cell-free systems, it is still uncertain whether these mechanisms are utilized in infected cells. In addition, it has recently been suggested that FMDV cre function can be complemented in trans (456). While more studies are necessary to confirm this result, it should be noted that cre function could not be complemented in trans in either the human rhinovirus (176) or the poliovirus (176, 382) system.
For plus-strand synthesis to proceed, the RF must be unwound. The mechanism for this is also unclear. The picornavirus 2C protein both has ATPase activity (249, 354, 390) and contains helicase motifs (125, 128, 178, 180, 250), but helicase activity has not been demonstrated (355). It has been shown that 2C and a cellular protein (p38) bind to the minus-strand 3′ stem-loop (25, 391, 392), and this may act to destabilize the RF molecule. The possibility of involvement of either a cellular helicase or a nuclear protein has also been suggested, since the RF is infectious when transfected into whole cells (317) but not when transfected into enucleated cells (322). The elongation of the plus strand by 3Dpol also occurs by an unknown mechanism. The complete replication of a picornaviral RNA in a cell-free system, including de novo protein synthesis, genome replication, and encapsidation to produce infectious virus, has been accomplished for poliovirus (32, 211, 316, 461) and EMCV (446).
RNA synthesis occurs within a membranous replication complex, which is derived from membranes of the endoplasmic reticulum and Golgi and contains viral NS proteins encoded by both the P2 (2B, 2BC, and 2C) and P3 (3A and its precursors, 3Cpro, and 3Dpol) regions (58-61, 66, 145, 170, 181, 232, 428, 442, 452, 460, 465). Structures similar to enterovirus replication complexes containing RNA and 3Dpol have been detected in FMDV-infected cells (371, 372). The 2B protein has been shown to enhance membrane permeability and block protein secretion (129, 232, 465, 466), but its role in RNA synthesis is not clear. Protein 2C has been found in membranous aggregates along the periphery of FMDV-infected cells (450) and has been directly implicated in FMDV RNA synthesis by using the picornavirus RNA synthesis inhibitor guanidine hydrochloride (81). FMDV mutants that are resistant to guanidine inhibition had an altered 2C isoelectric point (426), and changes in the viral 2C protein in resistant mutants were directly shown by using recombination and RNase T1 oligonucleotide mapping (427). Infection with picornaviruses results in a rapid inhibition of host cell transcription, which does not appear to be related to the inhibition of host cell translation (235). For FMDV-infected cells it has been shown that histone H3 is cleaved by 3Cpro, and this cleavage does not occur in cells infected by either poliovirus or EMCV (184, 451). In the case of enterovirus infections, transcription catalyzed by RNA polymerases I, II, and III is inhibited, and this inhibition requires the synthesis of 3Cpro, which appears to cleave cellular transcription factors required for the activity of these three enzymes (see reference 119 and references therein). Thus, FMDV may inhibit host cell transcription by a mechanism different from that of the entero- and cardioviruses.
Encapsidation and maturation.
The final steps in the replication cycle are the encapsidation of the plus-strand viral RNA and maturation cleavage of VP0 to VP2 and VP4 to form the mature virion. The mechanisms of encapsidation and maturation are still unresolved and are probably the least studied of all of the steps in the replication cycle. Again, most of the studies have been done with the enteroviruses, and therefore analogies must be drawn with FMDV. In broad terms, the 3Cpro cleavage products of the P1 region are assembled into a protomer structure containing one copy of each of the proteins VP0, VP1, and VP3 (Fig. 3a). Five protomers can assemble into a pentamer (Fig. 3b), and 12 pentamers assemble into the final capsid structure (Fig. 3c). Following encapsidation of the RNA, the maturation cleavage reaction (VP0 to VP2 and VP4) takes place (see below). A number of intermediate particles have been identified in picornavirus-infected cells, including protomers, pentamers, a particle containing RNA with an uncleaved VP0 (provirion) (197, 265), and a particle with an uncleaved VP0 lacking RNA (empty capsid) (190, 485). Two unresolved issues in picornaviral maturation are what signals are necessary for encapsidation of the RNA and what are the roles of the empty capsid and provirion.
Picornaviruses encapsidate only plus-strand RNA, linked to VPg, to the exclusion of all other viral and cellular RNAs (339, 340, 475). In addition, only newly synthesized plus-strand RNAs are encapsidated, indicating that there is a link between active RNA replication and encapsidation (201, 341). Thus, there may be cis-acting packaging signals present within the plus-strand RNA to facilitate encapsidation. The putative packaging signal does not appear to reside within the P1 region of the genome, since defective-interfering particles have been demonstrated in poliovirus-infected cells (198, 236, 279), all of which have deletions within the P1 region. Interestingly, no defective-interfering particles have been detected in FMDV-infected cells (103, 367). In addition, the complete P1 regions of a number of picornaviruses, including FMDV (294), can be deleted and replaced with a reporter gene generating a replicon, which, in the case of poliovirus, can be encapsidated when capsid proteins are provided in trans by a coinfecting virus (15, 28, 308, 373). Further studies, using the poliovirus replicon system, have shown that if the structural proteins of a heterologous picornavirus are provided in trans, packaging of the replicon RNA does not occur (28, 374). These data provide additional evidence for a packaging element, specific for each individual picornavirus, located within the genome outside the P1 region. There has been a single unconfirmed report of heterologous trans encapsidation of the FMDV genome with bovine enterovirus structural proteins (462). Recently, a cis-acting encapsidation element was identified within the stem of a stem-loop beginning four bases from the 5′ end of the genome of a newly discovered picornavirus, Aichi virus (425). This is the first report of such an element within the Picornaviridae.
While naturally occurring provirions have not been demonstrated in FMDV-infected cells, they have been shown to occur during the replication of other picornaviruses (63, 197, 214). There are currently two models of picornavirus assembly. One proposes that pentamers assemble into empty capsids, followed by insertion of the RNA, and the second proposes that pentamers directly interact with the RNA to form the provirion. In either case, it is known that myristylation of the N terminus of VP0 is necessary for capsid formation (16, 286, 323). Studies with FMDV have demonstrated that radioactive label can be chased from structural proteins into protomers, pentamers, empty capsids, and finally virions (485). In addition, poliovirus pentamers can self-assemble into empty capsids in vitro in the absence of viral RNA (287, 347, 395). More recently, however, it has been shown in a cell-free replication system that only poliovirus pentamer structures can interact with newly synthesized RNA to form virions (467), indicating that empty capsids may be either a storage particle for pentamers or a by-product of the assembly reaction.
The final step in virion assembly is the maturation cleavage of VP0 into VP4 and VP2, requiring the presence of viral RNA (19, 215, 230, 398). An aberrant cleavage of VP0 in FMDV empty capsids has been demonstrated, suggesting that viral RNA is required for the proper cleavage event to take place (116, 118). Cleavage is thought to be autocatalytic and results from a conserved His residue in VP2 which activates local water molecules, leading to a nucleophilic attack on the scissile bond and cleavage (34, 118, 213). Maturation cleavage is required for the generation of infectious virus (201, 253, 265). In FMDV, site-directed mutations within VP0 led to the formation of noninfectious provirions that exhibited receptor binding and acid sensitivity, similar to the case for infectious virus (253). Upon acid dissociation, however, the generated pentamers were more hydrophobic than those from mature virions, suggesting that VP0 cleavage may be necessary for release of the RNA into the cytoplasm (253).
Antigenic Variation
The presence of seven serotypes and multiple subtypes and variants has added to the difficulty of laboratory diagnosis and control of FMD. The rise of new variants is inevitably caused by continued circulation of the virus in the field and the quasispecies nature of the RNA genome (134, 206). RNA viruses in general, and FMDV in particular, have very high mutation rates, in the range of 10−3 to 10−5 per nucleotide site per genome replication, due to the lack of error correction mechanisms during RNA replication (135, 142). This high error rate leads to differences of FMDV replicated genomes from the original parental genome of 0.1 to 10 base positions (206), and the quasispecies concept was developed to explain the effects of errors in replication on the evolution of replicating RNA molecules (146). In its simplest terms, the concept envisions that within any population of virus, all genome sequences are not identical, and that selection occurs at the population level rather than at the individual level (134). Thus, there is not a “wild type” as such but rather an observed “average” phenotype which has adapted to and replicates “best” within any given environment. The environment can be in either tissue culture or a particular host species, and in either situation, immunologic pressure or physical conditions such as temperature or pH are influential. Any change in the environment can lead to the emergence of a new “average” phenotypic trait. The quasispecies nature of the FMDV genome was described over 20 years ago (132, 437), and while the concept is studied at the nucleotide level, the variability of FMDV populations is manifested when mutations lead to codon changes resulting in a change in the viral phenotype. Most of this variation occurs within the capsid-coding region of the genome (the P1 region [Fig. 2]) and leads to antigenic variation. While mutations also occur within the NS protein-coding regions of the genome, they are probably less tolerated, since proteins encoded by these regions are necessary for viral replication and changes are more likely to be lethal. In addition to variation caused by mutation, FMDV has been shown to undergo RNA recombination in tissue culture (239, 304, 474). Interestingly, these studies indicated that recombination was more likely to occur within the regions of the genome coding for the NS proteins; however, a more recent study has suggested that RNA recombination within the capsid-coding P1 region of the genome may contribute to genetic diversity in FMDVs isolated from the field (459).
Antigenic variation in the field increases with time and most probably results from immunologic pressure placed on the virus by either the infected or vaccinated host species (134, 205). In addition, antigenic variation in FMDV has also been observed in tissue culture in the absence of immunologic pressure (67, 126, 133, 153, 175, 433), indicating that antigenic sites on the virion may also be involved in other virus functions. Regardless of the mechanism, analysis of both genome sequence and antigenic variation has been invaluable in epidemiological studies of outbreaks and analysis of virus within countries where the disease is enzootic, and, in the case of a possible deliberate introduction of virus, it will also have forensic value in tracking the source (17, 206, 210, 255, 258, 288).
Antigenic sites on the surface of the FMD virion have been identified for five of the seven serotypes of the virus (South African Territories 1 and 3 being the only exceptions) (41, 43, 68, 80, 113, 204, 247, 264, 288, 298, 305, 453, 484). At least four antigenic sites have been identified, involving one or more of the capsid proteins, VP1, VP2, and VP3; however each serotype may not contain all four sites. Interestingly, three of the sites have elements located within the flexible loops which connect the β-sheets of the viral proteins, and at least two sites include the C terminus of VP1 (134). While all of the sites appear to be necessary for a complete immunologic response to either infection or vaccination, the major antigenic site, to which most of the immune response is directed and which is common to all of the serotypes, is located within the G-H loop of VP1 (Fig. 3d) (see “Virus structure” above) (69, 299). This site also contains the RGD receptor-binding recognition sequence (see “Early interactions: adsorption, penetration, and uncoating” above). While this site is clearly the major antigenic site, FMDV antigenic variation is associated with mutations leading to amino acid replacements within all of the known antigenic sites (154, 300). Nevertheless, even though there is extensive antigenic variation within FMDV, the changes are limited to very specific regions of the viral surface. This may be because changes within other regions of the capsid would compromise either viral structural integrity or virus identity (134). The antigenic variation within FMDV makes control extremely difficult, since even the best vaccine may induce immunologic pressure within the population that results in the emergence of a new variant. Furthermore, the observation that antigenic variation can also occur in tissue culture has implications for vaccine production, since a number of tissue culture passages are required to produce vaccine for a new variant, leading to the possibility that the virus eventually utilized as antigen may not provide the antigenic coverage needed.
PATHOGENESIS
While FMD affects a wide variety of cloven-hoofed animals, pathogenesis has been studied mainly in cattle and pigs. Infection of cattle generally occurs via the respiratory route by aerosolized virus (137). Infection can also occur through abrasions on the skin or mucous membranes, but is very inefficient, requiring almost 10,000 times more virus (137). Virus is excreted into the milk of dairy cattle (78, 219, 378) as well as in semen, urine, and feces (137, 243), and calves can become infected by inhaling milk droplets. Infected cattle also aerosolize large amounts of virus, which can infect other cattle in addition to other species (438). A number of studies have suggested that the lung or pharyngeal areas are the sites of initial virus replication (72, 79, 444), with rapid dissemination of the virus to oral and pedal epithelial areas (72, 79, 444), possibly mediated by cells of monocyte/macrophage origin (72). In cattle experimentally infected via aerosol, it was found, by in situ hybridization (ISH), that within the first 24 h, virus was present in respiratory bronchiolar epithelium, subepithelium, and interstitial areas of the lung (74). By 72 h, signal was detected in epithelial cells of the tongue, soft palate, feet, tonsils, and tracheobronchial lymph nodes (74). Other studies, however, have suggested that the pharynx, and not the lungs, may be the initial site of viral replication in infected cattle (8, 79, 489). The conflicting observations about the region of the respiratory tract that is initially infected in cattle exposed to aerosols may be the result of a number of variables, including aerosol particle size, strain of virus, or how the aerosol was generated (8).
Vesicles develop at multiple sites, generally on the feet and tongue, and are usually preceded by fever. Severe lesions often occur in areas subjected to trauma or physical stress, and most animals develop viremia. The incubation period can be between 2 and 14 days, depending on the infecting dose and route of infection (163).
Pigs usually become infected either by eating FMDV-contaminated food, by direct contact with infected animals, or by being placed into areas that had once housed FMDV-infected animals. They are, however, much less susceptible to aerosol infection than cattle (5, 6), yet they excrete far more aerosolized virus than cattle or sheep (6, 7). As in cattle, the incubation period is dependent on the amount of infecting virus and the route of infection, but it is generally 2 days or more. Animals develop fever, viremia, and lesions on the feet and tongue. Foot lesions are the most common finding in pigs, while lesions at other sites occur less frequently (244). Tongue lesions are usually small and less noticeable than those in cattle (244). In young piglets, the infection may be fatal due to myocarditis. Initial replication of the virus occurs at the site through which the virus gains entry, followed by rapid dissemination to most of the epithelial sites within the animal (73, 346). Interestingly, virus can be found at sites where clinical lesions either were not present or do not form (73, 346). While pigs excrete large amounts of aerosolized virus, recent evidence suggests that much more viral replication takes place in the nasal mucosa than in the lungs (346).
Of all of the important livestock species, sheep played the major role in the United Kingdom outbreak of 2001 (see below). Because it is very difficult to make a clinical diagnosis of FMD in sheep (R. De la Rua, G. H. Watkins, and P. J. Watson, Letter, Vet. Rec. 149:30-31, 2001), the disease can be spread to other livestock prior to detection (245). Sheep are highly susceptible to virus infection via aerosol and can excrete airborne virus; however, during outbreaks they are most likely infected by contact with infected animals (245). Clinical disease in sheep is characterized by lesions on the feet and mouth, fever, and viremia. It has been reported, however, that up to 25% of infected sheep may fail to develop lesions, and an additional 20% may form only one lesion (171, 218).
FMDV can also infect a wide variety of wildlife. The risk of spread of the infection by wildlife is controversial and is discussed in two recent review articles (443, 455).
Virulence Factors
In theory, any of the viral structural and NS proteins, elements of the viral RNA, and host proteins and membranes that participate in the viral replication cycle can be considered a virulence factor, since defects in the factor or its absence in the cell may lead to the virus' inability to replicate and cause disease in the host species. Table 1 lists viral and host factors that might be involved in virulence. Some of the factors listed in Table 1 have been shown to play a role in virulence either in the FMDV system or in other picornaviruses (295). It is beyond the scope of this review to examine the roles of all of these factors, so we will discuss only those that have been shown to be directly involved in virulence in susceptible animals.
TABLE 1.
Viral and host factors that may determine virulence
| Function | Viral factors | Host factors |
|---|---|---|
| Adsorption and penetration | Capsid proteins (VP1 to 3) | Integrins, other receptors (?), entry factors (?) |
| Translation | Lpro, IRES elements | eIFs, PTBP, ITAF45 |
| RNA replication | S region, poly(C), cre, 2B, 2C, 3A, 3B, 3Cpro, 3Dpol | Cellular membranes, PCBP, PABP |
| Assembly and maturation | Capsid proteins (VP0, VP1, and VP3), 3Cpro, 2A, viral RNA | Cellular membranes (?) |
It has been recognized for many years that viral receptors play a role in tissue tropism and disease pathogenesis (112, 151, 429, 476). In “Early interactions: adsorption, penetration, and uncoating” above, we discussed the viral integrin receptors that have been identified and the ability of FMDV to utilize alternative receptors. It is known that virus which has had the RGD sequence of the VP1 G-H loop either mutated or deleted cannot replicate in tissue culture or cause disease in animals (268, 297, 309). A type O virus variant adapted to utilize the HS receptor in vitro (see “Early interactions: adsorption, penetration, and uncoating” above) has shown the importance of the virus-integrin interaction in vivo. This virus was shown to be relatively avirulent in cattle, while the wild-type virus, which required only integrin receptors to initiate infection in vitro (334), was virulent in bovines (409). More interestingly, two bovines inoculated with large amounts of the HS binding virus eventually showed signs of FMD. Virus isolated from these animals could only utilize the integrin as a receptor and had lost the ability to interact with HS in vitro (334, 409). It is not clear why FMDV should need three integrin receptors to cause disease or whether it utilizes all or some the integrins in the susceptible hosts. We have recently shown in tissue culture that different serotypes of the virus exhibit altered efficiencies of integrin utilization (144). It is possible that the virus uses different receptors during various stages of the disease. While it appears that the disease process in susceptible animals is mediated by the virus-integrin interaction, a type C virus containing an RGGD sequence has been isolated from a bovine which was not protected from virus challenge following immunization with an experimental peptide vaccine (447, 448). In addition, a tissue culture-adapted type C virus with a genetically engineered RGG sequence, which was unable to bind to heparin, was able to infect cells expressing both HS and integrin receptors and cells which do not express FMDV integrin receptors and HS (26, 27). The ability of these two viruses to cause disease in susceptible animals has not been demonstrated. More interestingly, a tissue culture-adapted derivative of a type O1 virus, isolated in China, was able to replicate in tissue culture in both an integrin- and HS-independent manner and to cause mild disease in pigs (490). Thus, there is a possibility that nonintegrin receptors may be involved in disease pathogenesis. However, with the exception of the virus isolated after challenge from the peptide-vaccinated bovine (447, 448), no natural isolate of FMDV which does not contain the RGD sequence within the VP1 G-H loop has been identified.
Lpro was shown to be a virulence determinant based on experiments with animals with genetically engineered type A12 virus with Lpro deleted (leaderless) (98, 296, 356). It was thought that this virus would be less virulent than the wild-type virus, since the lack of Lpro would lead to the inability of the virus to cleave eIF4G and shut off cellular protein synthesis. While this virus replicated at only a slightly lower rate than wild-type virus in BHK-21 cells (356), it was markedly avirulent when injected into cattle and pigs and was unable to spread to cohoused animals (98, 296). The mechanism of attenuation of leaderless type A12 virus was examined by aerosol exposure of cattle. Wild-type-infected cattle had histologically altered respiratory bronchioles and virus-specific ISH signals in bronchioles by 24 h, and by 72 h they developed clinical disease, including fever and vesicles on the feet and positive ISH signals in epidermal sites corresponding to visible lesion development. In contrast, cattle infected with leaderless virus showed no clinical disease at 72 h and no pulmonary changes at either 24 or 72 h. These animals had only limited positive virus-specific ISH signals in respiratory bronchioles by 24 h and had no evidence of lesions or ISH signals in epithelial tissue by 72 h (74). Thus, the leaderless virus did not appear to replicate well at the site of primary infection and was not able to spread to other sites within the host. It was subsequently shown that infection with wild-type or leaderless virus induced the synthesis of alpha/beta interferon (IFN-α/β) mRNA both in tissue culture (96, 99) and in lung mononuclear cells from aerosol-exposed cattle (71), but in tissue culture, IFN activity was detected only in leaderless-virus-infected cells (96, 99). The latter observation can be attributed to the inability of the leaderless virus to inhibit host translation, including IFN synthesis, and the production of IFN within the infected animal probably inhibited initial amplification and spread of the virus. In contrast, wild-type virus infection blocks capped IFN mRNA translation, allowing the virus to rapidly spread to neighboring cells and systemically prior to the induction of the adaptive immune response.
The role of the 3A protein in viral virulence was demonstrated during studies of the FMDV isolate responsible for an outbreak in Taiwan in 1997 (designated O/Taw/97) (see below). This outbreak was unusual in that only pigs, and not cattle, were affected, and the disease had an unusually high mortality rate in pigs (143, 216). Molecular characterization of the virus revealed that it contained a 10-codon deletion in the C-terminal half of the 3A protein (47). The location of this deletion was similar to that of a 19- to 20-codon deletion found in FMDV passaged in chicken embryos. This virus also exhibited reduced virulence in bovines (172, 410). The role of this deletion in the bovine-attenuated phenotype was confirmed by using reverse genetic analysis (47), and an analysis of viruses circulating in the region for the last 30 years suggested that in addition to the deletion, mutations in the 3A protein in the region surrounding the deletion may also be responsible for the observed phenotype (254). The molecular basis for the porcinophilic phenotype appears to be related to a reduction in viral RNA synthesis, which is manifested to a greater degree in bovine cells than in swine cells (345). The 3A proteins from either the porcinophilic or a bovine-virulent isolate colocalized to RNA replication complexes in either bovine or porcine cells and also caused a disruption of the Golgi apparatus (345). However, as yet, there is no clear picture as to why the 3A deletion should affect FMDV replication in bovine cells more than in swine cells. Nunez and coworkers have also shown that a single amino acid change in the 3A protein was responsible for adaptation of FMDV to guinea pigs; however, the mutation was located in a different region than the deletion associated with the porcinophilic phenotype (342).
It has been suggested that IRES elements, and possibly the host factors that bind to them, affect the pathogenicity and virulence of other picornaviruses (152, 238, 285, 324, 344, 436). In FMDV, a virus rescued from persistently infected BHK-21 cells had two mutations within the IRES, which the authors suggest might have resulted in increased virulence of the virus in tissue culture (292). Some recent preliminary results suggest that the FMDV-specific IRES-binding protein ITAF45 may play a role in virus virulence by controlling virus tropism within tissues of susceptible species (V. O'Donnell, E. Pilipenko, E. Viktorova. R. Roos, and P. Mason, Abstr. 12th Meet. Eur. Study Group Mol. Biol. Picornaviruses, abstr. K11, 2002).
Host Response
The virus elicits a rapid humoral response in either infected or vaccinated animals. Virus-specific antibodies protect animals in a serotype-specific manner against reinfection, or against infection in the case of vaccination, and protection is generally correlated with high levels of neutralizing antibodies (reviewed in references 306 and 415). The response is directed to epitopes on the three external structural proteins (see “Antigenic variation” above), and good protective immunity is apparent between 7 and 14 days after either infection or vaccination. In cattle, the immunoglobulin G1 (IgG1) response predominates over IgG2 (86, 326, 417), and antibody, including IgA, can be detected in upper respiratory secretions early in infection (415, 417). The neutralization of virus within the host may occur by mechanisms similar to those occurring in in vitro neutralization; however, there is a suggestion that macrophages may play a role in clearing the virus from the infected animal by phagocytosis of opsonized virus (306, 307, 383).
The role of cellular immunity in the protection of animals from FMD is still a matter of some controversy. Specific T-cell antiviral responses, involving CD4+ and CD8+ cells, have been observed in cattle and swine following either infection or vaccination (36, 94, 104, 166, 173, 412), and it has been suggested that cell-mediated immunity is involved in clearance of virus from persistently infected animals (see below) (94, 224). The induction of anti-FMDV antibody correlates with a lymphoproliferative response in cattle and swine (104, 412) and is T-cell dependent in mice (105). A recent study of the early acute phase of FMDV infection of swine, prior to the detection of antibody, demonstrated a transient lymphopenia by 2 days after infection involving CD4+, CD8+, and CD4+/CD8+ T cells, which does not appear to be related to either infection of T cells or apoptosis and thus may be caused by alteration of lymphocyte trafficking (36). In addition, T-cell function, as measured by response to mitogens, is either reduced or eliminated (36). Both the number of lymphocytes and the altered T-cell function return to normal levels by 4 days after infection. These results suggest that T cells play a role in virus protection and that the reduction of both T-cell numbers and function enhances viral pathogenesis by allowing the virus to spread within the host, leading to increased viral shedding into the environment. In addition, immunization of both cattle and swine with a replication-competent human adenovirus 5 (Ad5) vector expressing the P1 capsid precursor did not result in the generation of virus-specific neutralizing antibody in the serum but partially protected animals from FMDV challenge (423, 424). The swine developed FMDV-specific T-cell responses (423), but these assays were not performed on the immunized cattle. These results may further indicate a role for cellular immunity in protection from FMDV infection; however, it also possible that innate immune responses may be responsible for the protection seen in these studies.
There has been increasing interest recently concerning the role of the innate immune response of the host to both FMDV infection and vaccination. A number of studies have shown that IFN-α, -β, and -γ may be involved in the host defense against FMDV infection (7, 71, 96, 99, 488) (see “Antiviral approach” below). In addition to the IFNs, other cytokines may also play a role in the host response. In studies of swine which were immunized with a conventional FMD vaccine, it was shown that vaccinated pigs did not appear to exhibit a systemic inflammatory response, but chemotactic activity of plasma on peripheral blood leukocytes increased within the first week after immunization (383). Furthermore, in pigs that either were only vaccinated or were vaccinated and challenged, levels of interleukin-6 (IL-6), IL-8, and IL-12 in plasma increased after vaccination and/or challenge, suggesting monocyte/macrophage activation (29). Although the levels of IL-6 and IL-8 did not appear to be related to protection of pigs upon challenge, IL-12 levels were higher in vaccinated pigs, which were protected from contact challenge, suggesting a role for cytokine-induced monocytic cell activity in protection from acute-phase disease (29).
Carrier State
Following the acute phase of FMDV infection in ruminants, some animals may experience a long asymptomatic persistent infection. In addition, animals which have been successfully vaccinated may also become persistently infected if exposed to infectious virus. These animals are referred to as carrier animals, and the carrier state is a complication which can occur during outbreak situations. In this section we briefly discuss what is known about the carrier state. For more in-depth information, a number of excellent reviews on this topic have been written (7, 415, 416).
Van Bekkum and colleagues first demonstrated that live FMDV could be recovered from esophageal-pharyngeal fluids of cattle during the convalescent phase of FMD (464). Currently, carrier animals are defined as those from which live virus can be isolated at 28 days, or later, after infection (445). In domestic cattle, the carrier state can last as long as 3.5 years, and it has also been identified in sheep and goats but not in pigs (7). African buffalo have been reported to carry live virus for up to 5 years (106), and other cloven-hoofed wildlife may become carriers (see reference 7 and references therein). The number of carrier animals in a population depends on the species, the incidence of infection, and the immune state of the herd (i.e., vaccinated or not vaccinated). In the African buffalo, the carrier rate can be as high as 50 to 70% in the field (106), and rates in cattle and sheep can vary widely, from 15 to 50% (7). In general, the titer of virus in the esophageal-pharyngeal fluids of carrier animals is low, and virus is not consistently recovered from individual animals. Currently, virus isolation from esophageal-pharyngeal fluids is the most sensitive method to detect carrier animals, but reverse transcription-PCR (RT-PCR) assays are being developed to attempt to increase sensitivity. The recovered virus probably originates in the pharynx, which appears to be the target region for persistent infection in cattle (7, 329, 489).
The role of carrier animals in the spread of virus in the field is still controversial. The only direct evidence is that of transmission from African buffalo to cattle during outbreaks in Zimbabwe in the late 1980s and early 1990s (120). In addition, transmission from buffalo to cattle has been obtained experimentally (121), and it has recently been proposed that such transmission might occur through sexual contact (35). There has been no experimental evidence to date indicating that carrier cattle or sheep can transmit virus to uninfected animals. The presence of live virus in esophageal-pharyngeal fluids, however, does make this a real possibility. In addition, the long persistence and replication of the virus in the host animals can lead to genetic variation in the field, possibly being responsible for the generation of new viral variants (167, 411, 457).
The mechanisms for the establishment and maintenance of the carrier state are not well understood, since persistence can occur in animals exposed to virus after either acute disease or vaccination. It does appear that the immune status of the animal probably controls the level of virus replication (7). Alexandersen and colleagues (7) have proposed two mechanisms for the development of persistence in the pharynx. One suggests that FMDV can infect immune system cells, such as macrophages, or other immunologically privileged sites, leading to evasion of the immune response. Baxt and Mason (42) examined viral replication in porcine peripheral blood macrophages and found that virus can infect such cells only when presented as an immune complex, presumably by Fc receptor-mediated adsorption. Furthermore, the infection was abortive and did not lead to the production of new infectious virus. In a more recent study, Rigden and colleagues (384) found that porcine alveolar macrophages also were not able to support viral replication; however, the virus bound to macrophages in the absence of specific antibody. In addition, virus appeared to be internalized by phagocytosis but remained infectious for at least 12 h. The second mechanism proposes that the virus exploits the host response to provide favorable intracellular conditions for long-term persistence, possibly by utilizing cytokine signaling. Studies on the innate immune response (see “Host response” above) should help to define the mechanisms involved in this phenomenon and possibly help in the development of methods to eliminate persistence.
DISEASE OUTBREAKS AND CONTROL MEASURES
Outbreaks
Since the beginning of the 20th century, FMD has been of considerable concern to many countries, and outbreaks or the fear of disease incursions have led to the establishment of institutes to investigate methods to control the disease. In particular, the Insel Reims in Germany in 1909, the Pirbright laboratory in the United Kingdom in 1924, Lindholm Island in Denmark in 1925, the Centro Panamericano de Fiébre Aftósa (PanAftosa) in Brazil in 1951, and the Plum Island Animal Disease Center in the United States in 1953 were opened specifically to study FMD (70).
In the late 19th and early 20th centuries, FMD outbreaks occurred sporadically in Europe, but their occurrence had devastating consequences (31). By the early 1950s however, some countries in Western Europe were experiencing 104 to 105 outbreaks per year (75). At that time, disease control consisted of inhibition of animal movement, slaughter of infected animals, and disinfection. As a result of a concerted effort in the 1930s, especially by Waldmann and colleagues in Germany (469), an inactivated FMD vaccine was developed (70). Vaccine was produced by inactivation of live virus with formalin in the presence of aluminum hydroxide gel. Virus for the Waldmann vaccine was obtained by infecting cattle at the slaughterhouse and collecting epithelium and vesicular fluid (70). Since only limited numbers of animals could be infected, this method was not able to produce the amount of vaccine needed to control the disease in Europe. It was not until Frenkel developed a method to produce virus by infecting bovine tongue epithelium obtained at the time of slaughter of healthy animals (161) that FMD vaccine commercialization became a reality. Systematic vaccination of cattle with this product led to a dramatic reduction in the number of disease outbreaks in Western Europe (75). The observation that FMDV replicates in BHK-21 cells led to more commercially viable large-scale production of virus in suspension cultures. Current vaccines are produced in cell culture, inactivated by treatment with aziridines such as binary ethylenemine (24), and mixed with adjuvant. As of the late 1990s, it was estimated that approximately 0.8 to 1 billion doses of vaccine were produced annually worldwide (406).
As a result of a successful vaccination program in Western Europe, which resulted in a cessation of disease outbreaks after 1989, the European Union adopted a no-vaccination policy in 1992 (75). From 1992 until 2001 there were only a few limited outbreaks in this region, including Bulgaria in 1991, 1993, and 1996; Italy in 1993; Russia in 1993 and 1995; Greece in 1994, 1996, and 2000; and Albania, Macedonia, and Yugoslavia in 1996 (Fig. 1) (267). During the same period, remarkable strides in disease control were also made in South America, utilizing annual vaccination campaigns and animal culling. By the end of the 1990s, Argentina, Chile, Uruguay, the southern part of Brazil, and Guyana were recognized by the international community as being free of FMD without vaccination (107). Although FMD still occurred in the Middle East and many countries in Africa and Asia, near the end of the 20th century, it appeared that, in the developed countries and in countries that engaged in international trade of animals and animal products, FMD was under control. As a result, many of these countries discontinued vaccination entirely, and research efforts in many European and South American countries were significantly reduced.
Reemergence of FMD in Developed Countries
In 1997 an FMD outbreak was reported in Taiwan, a country that had been free of the disease for 68 years. This devastating outbreak resulted in the slaughter of more than 4 million pigs, almost 38% of the entire pig population, at a cost of approximately U.S. $6 billion (486) and reminded the international animal health community of the severe economic consequences that an FMD outbreak could have for a previously disease-free country. The outbreak, caused by a type O virus, O/Taw/97, was first reported in March 1997 and within 3 weeks spread to almost the entire island, demonstrating the ability of the virus to replicate and spread at an alarming rate (143, 486). Taiwan was declared an FMD-infected zone and lost its pork export market. The outbreak was controlled by a combination of slaughter of infected animals and vaccination. An interesting observation during this outbreak was that disease was found only in pigs and did not occur in cattle or goats that were also present on some of the infected farms (143, 486). In experiments performed at the OIE/FAO World Reference Laboratory for FMD at Pirbright, virus isolated from infected swine did not infect bovine tissue culture cells and did not cause clinical disease in cattle either by contact exposure or by direct inoculation in the tongue (143). Subsequent studies with this virus by Mason and colleagues revealed that the viral NS protein 3A has a primary role in the restricted growth of the virus in bovine cells in vitro and in vivo (47, 345) (see “Virulence factors” above).
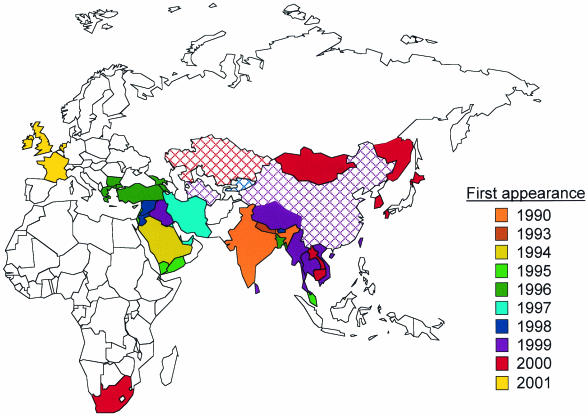
Starting in late 1999 and 2000, a series of FMD outbreaks occurred in a number of countries in East Asia. This was followed by an outbreak in South Africa and culminated in the destructive outbreak in the United Kingdom which then spread to the European continent. The World Reference Laboratory identified a serotype O PanAsia lineage virus as the causative agent of all of these outbreaks (256). This lineage had originated in India in 1990 and spread through the Middle East, Turkey, and Eastern Europe. It then moved eastward into the People's Republic of China in 1999 and then to Taiwan, South Korea, Japan, Mongolia, and far-east Russia. The virus then appeared in South Africa in late 2000 and in the United Kingdom in February 2001 (Fig. 4).
FIG. 4.
The spread of the PanAsian strain of FMDV type O from its first appearance in India in 1990 until its appearance in the United Kingdom in 2001. Solid colors, PanAsian strain present; cross-hatched colors, type O present and PanAsian strain suspected. The data and map were compiled by Nick Knowles and can be found at www.iah.bbsrc.ac.uk/virus/picornaviridae/aphthovirus.
The 1999 to 2000 outbreak in Taiwan affected cattle and goats but was more limited than the 1997 incursion. Nucleotide sequencing of virus isolated from infected animals revealed that the virus, O/Taw/99, was different than the O/Taw/97 virus but closely related to viruses circulating in the Middle East and India (217). In March 2000 a large FMD outbreak occurred in South Korea and a much more limited outbreak occurred in Japan. Both of these countries had been free of the disease for many decades (South Korea for 66 years and Japan for 92 years). The Korean outbreak was controlled by slaughter and vaccination of all cloven-hoofed animals within the affected provinces, resulting in the destruction of over 500,000 animals, mainly cattle (233). Virus isolated from infected cows was identified by the World Reference Laboratory as an O serotype, O/SKR/2000, closely related to O/Taw/99. The Japanese outbreak was limited to a few farms and was controlled by slaughter without vaccination. Sequence analysis also placed this virus, O/JPN/2000, in the same lineage as O/TAW/99 and O/SKR/2000. In September 2000 an outbreak of FMD was detected in a province of South Africa that had been free of the disease. The causative agent was identified as a member of the PanAsian type O lineage similar to O/SKR/2000 and O/JPN/2000, a serotype that had never before occurred in South Africa (Fig. 1 and 4) (421).
These outbreaks reemphasized the extreme virulence of FMDV in a variety of animal species, the vulnerability of FMD-free countries as well as countries where FMD is enzootic to new viral strains, the effects of globalization on increasing the risks of disease incursion, and hence the need for countries to more closely monitor for the presence of exotic diseases.
The 2001 United Kingdom Outbreak and Its Aftermath
On 19 February 2001, FMD was suspected in pigs at an abattoir in Essex in southeast England. The World Reference Laboratory confirmed this the next day, and the causative agent was identified as serotype O PanAsia, O/UK/2001 (256). This was the first outbreak of FMD in the United Kingdom since a 1981 disease incursion on the Isle of Wight. Subsequently it was determined that the disease had been present in the United Kingdom, but not reported, for at least 3 weeks, and by 20 February it was already present in 16 of the 23 counties that were eventually involved (430). Apparently, the first infected pigs discovered were initially exposed upon arrival at the abattoir. Further investigation determined that the index case occurred on a farm in Northumberland in northeast England. The delay in reporting the disease in Northumberland allowed sheep on neighboring farms, which had become infected (perhaps by airborne spread) but did not show clinical disease, to be moved to livestock markets and infect other susceptible animals. In fact, the frequent widespread movement of animals, especially sheep, around the United Kingdom and to other European countries significantly contributed to the severity of the outbreak. For a number of reasons, including the wide distribution of the disease by the time it was discovered and pressure from livestock producers, vaccination was not used. Instead, all infected or in-contact susceptible animals were slaughtered. In total the outbreak resulted in the slaughter of 4 million animals, mainly sheep, with an additional 2.5 million killed on welfare grounds (430). The costs to the economy of the United Kingdom were high. It has been estimated that total losses were between U.S. $12.3 billion and $13.8 billion, of which approximately 36% were losses to tourism. The government paid about U.S. $4.2 billion in compensation to the agriculture and food chain industry (454).
The outbreak subsequently spread to Northern Ireland and to France, but it only affected a few farms there and was controlled by culling. On 21 March FMD was confirmed in The Netherlands. The disease was introduced by calves that had become infected in a staging area in France where infected sheep from the United Kingdom were present (366). Soon after, the disease was detected in additional farms, and The Netherlands decided on suppressive ring vaccination, which implied that all of the vaccinated animals would be slaughtered. Approximately 200,000 animals were vaccinated.
The last case of FMD occurred in the United Kingdom on 30 September 2001, and the country regained its FMD-free status without vaccination on 22 January 2002 (430). The United Kingdom commissioned a number of inquiries to examine the government's handling of the outbreak and to determine how the country should prepare itself for and respond to future infectious disease outbreaks. Some of the key recommendations of the Royal Society's inquiry into preparation for disease outbreaks included the need to incorporate emergency vaccination as part of the control strategy from the start of any FMD outbreak (14). It specifically indicated that the policy should be vaccination-to-live; that is, meat and meat products from vaccinated animals subsequently found to be uninfected may enter the human food chain. In addition, it recommended the development of modern diagnostic methods to rapidly detect an outbreak, as well as the investment of additional funds in animal disease research to develop marker vaccines that would allow easy diagnostic distinction of vaccinated from infected or convalescent animals. The United Kingdom inquiries, as well as commissions in France and The Netherlands, indicated the need to coordinate new strategies on a regional basis within the European Union as well as at the international level through the OIE. In addition, public reaction, predominantly within The Netherlands, questioned the need for large-scale slaughter of susceptible animals, particularly the slaughter of vaccinated animals that were healthy (366).
In May 2002 at the OIE General Session, a number of amendments that addressed the world situation as a result of the unexpected reemergence of FMD were adopted. These included recognition of new diagnostic tests capable of distinguishing vaccinated from infected animals and reduction of the time, from 12 to 6 months, required for a country that vaccinated but did not slaughter these animals to regain FMD-free status. These amendments increase the likelihood that alternatives to animal culling will be utilized in the face of future FMD outbreaks.
Disease Control
Current vaccines.
The current FMD vaccine is an inactivated whole-virus preparation that is formulated with adjuvant prior to use in the field. A number of countries have established vaccine banks which contain concentrated antigen stored in the gaseous phase of liquid nitrogen (130). Antigen stored under these conditions is stable for a longer period of time than formulated vaccine (131). Banks contain antigen against a number of virus serotypes and provide member countries with an almost immediate source of vaccine. A recent report by Doel (130) provides an in-depth review of the history, production, and utilization of inactivated FMD vaccines, and we will comment only on some of the limitations of their use, especially in emergency control programs.
As already discussed (see “Outbreaks” above), the introduction of the killed FMD vaccine has been extremely successful in reducing the number of disease outbreaks in many parts of the world where the disease is enzootic. However, there are a number of concerns and limitations with its use in emergency control programs, including the following.
(i) High-containment facilities are required for the production of vaccine.
(ii) Most virus preparations used for vaccines are concentrated cell culture supernatants from FMDV-infected cells and, depending on the manufacturer, contain various amounts of contaminating viral NS proteins. Vaccinated animals develop antibody responses against the contaminating proteins, in addition to the viral structural proteins, making it difficult to reliably distinguish vaccinated from infected or convalescent animals with currently approved diagnostic tests.
(iii) The vaccine does not induce rapid protection against challenge by direct inoculation or direct contact. Thus, there is a window of susceptibility of vaccinated animals prior to the induction of the adaptive immune response.
(iv) Vaccinated animals can become long-term carriers following contact with FMDV (see “Carrier state” above).
Some of these concerns are being addressed by development of new marker vaccines that do not require infectious virus (see “Alternative vaccine strategies” below) and of diagnostic tests that are based on NS proteins that are absent or are only minor contaminants of the current vaccine (55, 122) (see “Diagnostics” below). Furthermore, efforts to understand the role of innate immunity and utilize various cytokines to both rapidly induce protection and boost the immune response are under way (see “Host response” above and “Antiviral approach” below).
Alternative vaccine strategies. (i) Proteins and peptides.
Because of the concerns described above, researchers over the past 20 to 25 years have attempted to develop alternative FMD vaccines that do not require infectious virus. Based on information concerning the FMDV capsid structure, including the prominent surface exposure of VP1 and the immunologically important VP1 G-H surface loop, a number of strategies have been employed. Initially these included use of VP1, either isolated from purified virus or produced by recombinant DNA techniques (23, 248); the use of VP1-derived peptides (441) or chemically synthesized VP1 peptides (64, 127, 160, 331, 352); the use of live vectors expressing VP1 fusion proteins (242, 246); inoculation with DNA expressing VP1 epitopes alone (478) or coadministered with DNA encoding IL-2 (479); and use of transgenic plants expressing the entire VP1-coding region or plants infected with a recombinant tobacco mosaic virus expressing VP1 (472, 473). All of these strategies present a limited subset of viral immunogens to the vaccinated animal (see “Antigenic variation” above), and although they often induce high titers of neutralizing antibodies, they do not always achieve protection against virus challenge in livestock (127, 326-328). The immunogenicity of these subunit vaccines appears to be due to their ability to present sequential epitopes from the immunologically important VP1 G-H surface loop. Although these epitopes appear to be immunodominant in many assay systems, they are not the only neutralizing epitopes on the virion (113), nor are they uniformly recognized in all host species (299). With the known variability and quasispecies nature of the FMDV genome (136), the use of a limited subset of epitopes would invite selection of antigenic variants that could cause outbreaks among animals vaccinated with these products (259, 313). In a large-scale bovine vaccination study using synthetic peptides, Taboga and colleagues found only limited protection against challenge and detected viral escape mutants that were antigenic variants of the challenge virus in vaccinated, unprotected animals (447, 448).
(ii) Live attenuated vaccines.
Reviews by Bachrach (21) and Brooksby (70) describe, in depth, the attempts to develop live attenuated FMD vaccines by the classical procedures of serial passage in nonpermissive animals or in cell culture. Attenuated strains were produced by passage in nonsusceptible species, such as mice, rabbits, and embryonated eggs, until their virulence for cattle was weakened. Field studies with these viruses were performed in Africa, the Middle East, and South America. While in some cases the attenuated vaccines induced a degree of protection, it was found that viral strains attenuated for a given host were often virulent in other susceptible species. Furthermore, it has been difficult to obtain viruses that are both attenuated and immunogenic. In addition, a major concern with live attenuated vaccines is the possibility of reversion to virulence.
More recently, attempts to develop attenuated vaccines have utilized genetic engineering to mutate regions of the genome or delete a protein-coding region. By utilizing recombinant DNA techniques, a virus was created in which the RGD receptor binding site on VP1 (see “Infectious cycle” above) was deleted. This virus was unable to bind to cells and did not cause disease in 7- to 10-day-old mice or in swine (309). Cattle inoculated with this virus, in an oil emulsion, did not develop clinical disease, developed FMDV-specific neutralizing antibodies similar to those in animals inoculated with inactivated whole-virus vaccine, and upon challenge were completely protected from clinical disease (309). However, a concern with this approach is the potential for selection of virus variants that could enter cells by utilizing nonintegrin receptors (26, 27, 447, 490) (see “Early interactions: adsorption, penetration, and uncoating” above).
The deletion of the coding region of an NS protein that is not essential for virus replication in cell culture is an alternative method of creating live attenuated vaccines. However, to be useful as a vaccine, this deletion virus must still be able to replicate in susceptible animals. The advantage of this approach, compared to the classical method of attenuation, which generally introduces mutations at a limited number of sites, is that the risk of reversion to virulence is significantly reduced.
Lpro is a virally encoded protein that is involved in the shutoff of host protein synthesis, allowing the virus to “take over” the translation machinery of the cell (see “Genome organization” above). A type A12 virus which lacked the L-coding region (leaderless) was constructed. This virus replicated in BHK cells but did not cause disease in cattle or swine (98, 296, 356) (see “Virulence factors” above). Based on its greatly reduced pathogenicity in susceptible animals, leaderless virus was examined as a live attenuated vaccine candidate. Inoculation induced an FMDV-specific neutralizing antibody response in both species. Cattle directly challenged in the tongue and swine challenged by contact exposure to an infected animal in the same room had less severe and delayed clinical disease compared to naive animals but were not fully protected (98, 296).
In contrast, a leaderless virus containing the capsid-coding region from a bovine-virulent serotype O1Campos strain in the genetic background of FMDV A12 was avirulent in cattle, as demonstrated by direct inoculation in the tongue, but caused a mild disease when inoculated into swine and was transmitted to a naive contact animal (9). These results indicate the potential for the rational design of live attenuated vaccines but underline the difficulty in designing these viruses so that they do not cause clinical disease and can replicate sufficiently to induce a protective immune response.
The leaderless A12 virus was shown to be useful as a source of antigen for traditional inactivated vaccines (98). Swine inoculated with inactivated leaderless or wild-type A12, in oil adjuvant, all developed significant FMDV-specific neutralizing antibody responses and, after virus challenge, were completely protected from clinical disease and challenge virus replication. Thus, the use of attenuated viruses as the source of antigen in traditional vaccine production could reduce the risks associated with virus escape from the production facility or incomplete virus inactivation.
(iii) Empty viral capsids.
Other experimental FMD vaccines have targeted immunogens that contain the entire repertoire of immunogenic sites present on intact virus but lack infectious nucleic acid (50, 188, 190, 269, 396). This strategy involves the molecular cloning of the regions of the viral genome necessary for the synthesis, processing, and assembly of the viral structural proteins into empty viral capsids (P1-2A- and 3Cpro-coding regions) (see “Viral translation” above). These structures are naturally produced in FMDV-infected cell culture systems, are antigenically similar to virus particles, and are as immunogenic as virions in animals (190, 400, 407). Animals inoculated with this type of vaccine could be easily distinguished from infected or convalescent animals by using currently approved technology, since the regions of the genome coding for the NS proteins used in the diagnostic assays to detect infection are not present in the empty capsid cDNA construct. In initial studies, FMDV capsid structures were expressed in Escherichia coli or in recombinant baculovirus-infected cells and inoculated into animals. Although these products did offer some protection, they did not reach the efficacy of the current inactivated whole-virus vaccine because only small amounts of antigen were obtained (50, 188, 269, 396).
To enhance the expression and delivery of empty capsid constructs, alternative vector systems that allow expression of FMDV capsid structures in infected cells and potentially induce both humoral and cell-mediated immune responses have been examined. One approach utilized a DNA inoculation-based strategy designed to produce empty capsids in inoculated animals (46, 52, 91, 95). Similar studies using cDNA that encodes the entire viral genome but contains a mutation at the cell binding site were also performed (46, 471). Initial studies in mice, inoculated either intradermally by gene gun or intramuscularly (i.m.), resulted in the induction of an FMDV-specific neutralizing antibody response that required an active viral 3Cpro in the empty capsid construct (46, 95). In swine, however, large amounts of DNA and at least two or three inoculations were needed to induce a low FMDV-specific neutralizing antibody response and variable levels of protection (46, 52, 91). In one study, coadministration of a plasmid encoding porcine granulocyte-macrophage colony-stimulating factor together with an FMDV empty capsid construct appeared to improve the FMDV-specific antibody response (91). Although the use of naked DNA vaccines has numerous advantages, its efficiency of uptake is low and the mechanism of action is still not well understood.
An alternative, and very efficient, antigen delivery system utilizes a live virus vector. Both human adenovirus and poxviruses are well-characterized vectors for foreign gene expression (182, 225, 284, 303, 349, 404) and have been used to deliver FMDV capsid proteins (1, 57, 301, 302, 320, 423, 424, 480). Human adenoviruses possess low pathogenicity in humans and animals, and wild-type virus has been safely and successfully used in oral immunization of U.S. military recruits as prevention against acute respiratory disease (109, 458). As an additional safety feature, replication-defective human Ad5 vectors, which lack a portion of the adenovirus genome, have been produced. These vectors can productively grow only in specific cell cultures that provide the missing functions (183). Despite their inability to replicate, immunization of a number of animal species with these replication-defective adenoviruses containing genes from other viruses has resulted in induction of an immune response against the foreign gene products and protection from challenge (148, 156, 234, 377, 482). An additional advantage of the adenovirus system is that human adenovirus vectors are able to bind to and internalize in cells of several animal species, including cattle and swine (375), thus ensuring rapid uptake and expression of the desired genes in these species.
We have constructed replication-defective human Ad5 vectors containing inserts coding for the FMDV serotype A12 or A24Cruzeiro structural proteins and including either wild-type (Ad5-A12 or Ad5-A24) (302, 320) or inactive (Ad5-A123CMUT) (301) 3Cpro genes. A replication-competent Ad5 containing the serotype C1Oberbayern capsid-coding region but lacking the 3Cpro-coding region (Ad5-C1) has also been constructed (424). Swine receiving two inoculations of Ad5-A12 or Ad5-A24 developed FMDV-specific neutralizing antibody responses and were protected from either contact challenge (five of six swine were protected) (302) or direct inoculation challenge (four of four swine were protected) (320). In contrast, swine inoculated with Ad5-A123CMUT (302) or swine and cattle inoculated with two doses of Ad5-C1 (423, 424) did not develop FMDV-specific neutralizing antibodies and were not protected against challenge. These results demonstrate the potential of an Ad5-vectored empty capsid vaccine approach and support the requirement of processing of the P1-2A capsid precursor protein by 3Cpro for the production of a potent and efficacious vaccine. These studies also indicated that there was no spread of recombinant virus to uninoculated swine housed in the same room, as demonstrated by the absence of both FMDV- and Ad5-specific neutralizing antibody responses (301, 302).
Similar experiments have been performed with recombinant, replication-competent vaccinia virus containing only the capsid-coding regions of either FMDV C1Oberbayern or C3Argentina85 (57, 423). Swine inoculated twice with vaccinia-C1 did not develop FMDV-specific neutralizing antibodies and were not protected from challenge (423). In contrast, mice inoculated with vaccinia-C3 developed an FMDV-specific neutralizing antibody response after the first inoculation and were protected when challenged after a second inoculation (57). To date these studies have not been extended to swine or cattle.
To be useful in emergency outbreak situations, an FMD vaccine has to be given in a single dose and induce a rapid protective response. Studies in swine with Ad5-A24 demonstrated that a single dose could induce a neutralizing antibody response and protection when the animals were challenged by direct inoculation 7, 14, or 42 days later (Table 2) (320). These animals showed no viremia or virus in nasal swabs and no evidence of virus replication. Swine inoculated with Ad5-A24 and challenged 5 days later had very low or no viremia and delayed and less severe clinical disease compared to control infected animals (319).
TABLE 2.
Rapidity of Ad5-A24 induction of protection in swine
| Vaccine | Day of challengea | Viremiab | Protectionc | Scored |
|---|---|---|---|---|
| PBSe | 14 | 6 | 0/6 | 15.8 |
| Ad5-A24f | 7 | 0 | 4/4 | 0 |
| 14 | 0 | 4/4 | 0 | |
| 42 | 0 | 4/4 | 0 |
The animals were challenged on the day indicated by direct inoculation in the heel bulb of the rear foot with 105 50% bovine infectious doses of virulent A24.
Determined by virus isolation (number of positive animals).
Number of animals protected/total number of animals.
The score is the number of digits and snout with lesions; the maximum score is 17.
PBS, phosphate-buffered saline.
Animals were inoculated i.m. with 5 × 109 PFU.
There have been few experimental vaccine trials in cattle with empty viral capsids as an immunogen. However, in a preliminary experiment, cattle inoculated with two doses of Ad5-A24 developed a very significant FMDV-specific neutralizing antibody response and were completely protected from clinical or serological evidence of FMD after direct inoculation challenge in the tongue and contact exposure to an infected animal in the same room (189).
Antiviral approach.
While both the current vaccine and the Ad5-vectored vaccine can induce complete protection by 7 days, in emergency outbreak situations it is imperative to block or reduce virus shedding as rapidly as possible to contain the outbreak. Several groups have demonstrated that in cells pretreated with IFN-α/β, productive replication of FMDV is inhibited by the IFN-induced gene products, double-stranded RNA-dependent protein kinase and 2′-5′A synthetase/RNase L (4, 96, 99, 432). IFN is one of the first lines of host cell defense against viral infection (468) and can rapidly induce a nonspecific protective response against all FMDV serotypes thus far tested (4, 96, 99, 432). Furthermore, since effective protection requires the vaccine to be matched to the outbreak strain and induce a rapid immune response, antiviral therapy could circumvent these inherent problems of FMD vaccination. Thus, IFN-α/β may be useful as an anti-FMD agent for prophylactic treatment in susceptible animals.
Clinical studies with humans have demonstrated that IFN-α/β protein is rapidly cleared; therefore, treatment requires multiple high-dose inoculations (278, 376, 422). For IFN therapy to be effective in an FMD outbreak, it must be delivered in one inoculation, in combination with vaccination, so as to induce rapid protection prior to the onset of the vaccine-induced adaptive immune response. In an attempt to meet these goals, Chinsangaram et al. (97) constructed a replication-defective Ad5 containing porcine IFN-α (Ad5-pIFN-α). Delivery via a viral vector would result in endogenous expression of IFN over a period of time, and the amount delivered could be controlled by the dosage of the recombinant virus. Swine inoculated with one dose of this recombinant virus were completely protected when challenged 1 day later by direct inoculation of FMDV (97). These animals did not develop clinical disease and had no viremia or antibody response to any viral NS protein. The degree of protection correlated with virus dose and the level of pIFN-α present in the plasma (Table 3) (97). Additional studies indicate that Ad5-pIFN-α treatment alone can protect swine from challenge for 3 to 5 days and can reduce viremia, virus shedding, and disease severity when administered 1 day postchallenge. Furthermore, a combination of pIFN-α and vaccination can provide both immediate and long-term protection (319). In recent studies with cattle, administration of Ad5-IFN-α failed to completely protect the animals from FMDV infection; however, disease was delayed and less severe compared to that in control animals (480). This lower level of protection of cattle compared to swine appears to be correlated with reduced levels of IFN detected in their plasma.
TABLE 3.
IFN-α/β can rapidly protect swine from FMDV
| Groupa | Virus dose (PFU)b | Antiviral activity at the following day postinoculationc:
|
pIFN-α (pg/ml) at the following day postinoculationd:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 0 | 1 | 2 | 3 | 4 | 5 | ||
| Ad5-Blue | 109 | <25 | <25 | <25 | <25 | <25 | <25 | 0 | 0 | 0 | 0 | 93 | 0 |
| Ad5-pIFN-α | 108 | <25 | 133 | 58 | 25 | <25 | <25 | 0 | 5,488 | 1,080 | 65 | 0 | 0 |
| 109 | <25 | 800 | 400 | 267 | 25 | 25 | 0 | 31,372 | 13,940 | 3,930 | 680 | 0 | |
Each group contained three animals.
The animals were inoculated i.m. with the indicated dose of the Ad5 vector.
Results are shown as the highest dilution that reduced the FMDV A12 plaque number by 50% and are averages for the three animals in each group.
Results (as determined by ELISA) are averages for the three animals in each group.
There have been some limited attempts to develop antiviral drug therapy that affects specific viral protein targets. Kleina and Grubman (251) demonstrated that the compound E-64 or its membrane-permeable analog E-64d, which have been identified as specific inhibitors of thiol proteases (199, 200), inhibited the proteolytic activity of Lpro both in a cell-free translation system and in cell culture. Inhibition of Lpro autocatalytic activity blocked virus capsid assembly, presumably because N-terminal myristylation of P1-2A (VP0 and VP4) and processing of P1-2A by 3Cpro were blocked. As a result there was a 1,000-fold reduction in virus yield. The recently solved crystal structure and determination of unique characteristics of Lpro by Skern and colleagues (193, 194, 260) will now allow for rational design of Lpro-specific antiviral drugs.
Finally, Sobrino and coworkers have utilized antisense technology to specifically inhibit FMDV replication in cell culture (62, 196, 397). However, this technology has not yet been extended to susceptible animals.
Diagnostics.
An essential component of any disease control strategy includes diagnostic assays to rapidly confirm the initial clinical determination of infection. For FMD this is of particular importance, since SVD, vesicular stomatitis, and vesicular exanthema of swine cause vesicular lesions in swine and cattle that cannot be distinguished from those caused by FMD (21). In addition, FMDV infection of sheep and goats can be difficult to detect clinically (138, 168). Sensitive diagnostic assays are also necessary to distinguish vaccinated from infected or convalescent animals, so that trade markets can rapidly reopen to countries that may have used vaccination as part of their disease control program and to identify carrier animals. In addition, these assays can be used for epidemiological surveillance to confirm the naive status of animals in field situations. This section examines only the development of new assays to rapidly detect FMD-infected animals as well as assays to distinguish vaccinated from infected animals.
(i) Rapid detection of virus.
Currently FMD is confirmed by antigen capture enzyme-linked immunosorbent assay (ELISA) and virus isolation. While ELISA results can be obtained in 3 to 4 h after the sample is received by the laboratory, a negative result must be confirmed by inoculation of the sample into sensitive cell cultures followed by confirmation of the virus serotype by ELISA. These assays can take up to 4 days, a time frame incompatible with the need to rapidly detect disease and initiate an appropriate disease control strategy. As a consequence, researchers have been examining alternative assay systems that allow more rapid confirmation of clinical diagnosis, which do not require a laboratory setting and may be performed “pen side.”
RT-PCR methods have been used to rapidly detect and type FMDV (83) and to detect virus infection in asymptomatic animals (289). However, this assay is often not superior in sensitivity to ELISA and virus isolation and, in addition, is labor-intensive. Most recently, real-time RT-PCR methods have been examined by a number of groups with the aim of developing portable on-site diagnosis (82, 208, 379). The assay is specific and as sensitive as virus isolation, and viral RNA could be detected in oral and nasal samples from experimentally infected animals 24 to 96 h before the onset of clinical signs (82). In addition, the assay is rapid, results can be obtained in about 2 h, and the cycler is portable. The next steps required for assessing and validating this assay are optimization of conditions with all possible field samples (i.e., blood, milk, tissue, etc.) and testing under field conditions.
(ii) Assays to differentiate infected from vaccinated animals.
In 1966 Cowan and Graves identified a highly immunogenic FMDV NS antigen, called the virus infection-associated antigen (VIAA), which reacted with sera from convalescent animals but not with sera from vaccinated animals (110). However, in later studies, investigators found that sera from multiply vaccinated animals (283, 365, 399), and even from some animals which had received a single vaccination, had antibodies to VIAA (276, 283, 320). The major reason that antibodies against an NS protein are present in sera from vaccinated animals is that FMD vaccines are not purified and, depending upon the manufacturer, contain various amounts of contaminating NS proteins (277). Nevertheless VIAA, which was subsequently identified as the viral RNA polymerase (3Dpol) (335, 369), is currently used in an agar-gel immunodiffusion test to differentiate infected from vaccinated animals.
To improve the reliability of this diagnostic assay, investigators have targeted other NS proteins as potential diagnostic reagents. Berger et al. (53) used radioimmunoprecipitation to identify a number of NS proteins that reacted with sera from convalescent animals and not with sera from vaccinated animals. Based on these results, they recommended the use of NS proteins 3AB, 2C, 3C, and 2B, or their respective peptides, as antigens in an ELISA-based assay. Bergmann et al. (54) used an enzyme-linked immunoelectrotransfer blot assay with purified NS proteins produced in E. coli by recombinant DNA techniques and found that the use of NS proteins, other than 3Dpol, could differentiate sera from infected and vaccinated animals. These workers recommended using more than one NS protein to unambiguously identify sera from infected animals. Based on this information, ELISA-based assays with various NS proteins produced by recombinant baculovirus (315, 439), in E. coli (122, 283), or with synthetically produced peptides to NS proteins (434) have been developed. Currently these assays are being validated.
With the development of new FMD marker vaccines, such as an empty viral capsid antigen, which lack portions of the viral genome coding for one or more NS proteins, the diagnostic assays described above can be used in concert to unequivocally distinguish vaccinated from infected animals. In particular, the use of the highly immunogenic 3Dpol may serve as a very sensitive diagnostic antigen to detect animals that are carriers of low levels of virus.
CONCLUDING REMARKS
The recent outbreaks of FMD in many parts of the world, particularly in developed countries, have had a profound effect on the range of disease control procedures that are currently being considered in the event of additional incursions. Thus, emergency vaccination, vaccination-to-live policies, and antiviral approaches are actively encouraged by measures recently approved by the OIE and reports from government-sponsored boards of inquiry. Furthermore, the threat of deliberate release of FMDV in the era after 11 September 2001 has caused governments, including those of the United Kingdom and the United States, to support the development of new diagnostic and disease control approaches, as well as to plan for the possibility of multiple-site release of the agent. Because of the considerable amount of information that has become available about FMDV at the molecular level and the recent, but more limited, understanding of virus-host interactions, new rapid diagnostic assays, novel vaccine candidates, and antiviral control approaches are being developed and tested.
While these additional efforts are welcome, the globalization of trade also makes it incumbent upon developed countries to consider the needs of developing countries and to design integrated FMD control strategies in an effort to eliminate or more effectively control FMD worldwide.
REFERENCES
- 1.Abrams, C. C., A. M. King, and G. J. Belsham. 1995. Assembly of foot-and-mouth disease virus empty capsids synthesized by a vaccinia virus expression system. J. Gen. Virol. 76:3089-3098. [DOI] [PubMed] [Google Scholar]
- 2.Acharya, R., E. Fry, D. Stuart, G. Fox, D. Rowlands, and F. Brown. 1989. The three-dimensional structure of foot-and-mouth disease virus at 2.9 A resolution. Nature 337:709-716. [DOI] [PubMed] [Google Scholar]
- 3.Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. [DOI] [PubMed] [Google Scholar]
- 4.Ahl, R., and A. Rump. 1976. Assay of bovine interferons in cultures of the porcine cell line IB-RS-2. Infect. Immun. 14:603-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandersen, S., I. Brotherhood, and A. I. Donaldson. 2002. Natural aerosol transmission of foot-and-mouth disease virus to pigs: minimal infectious dose for strain O1 Lausanne. Epidemiol. Infect. 128:301-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandersen, S., and A. I. Donaldson. 2002. Further studies to quantify the dose of natural aerosols of foot-and-mouth disease virus for pigs. Epidemiol. Infect. 128:313-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexandersen, S., Z. Zhang, and A. I. Donaldson. 2002. Aspects of the persistence of foot-and-mouth disease virus in animals—the carrier problem. Microbes Infect. 4:1099-1110. [DOI] [PubMed] [Google Scholar]
- 8.Alexandersen, S., Z. Zhang, A. I. Donaldson, and A. J. Garland. 2003. The pathogenesis and diagnosis of foot-and-mouth disease. J. Comp. Pathol. 129:1-36. [DOI] [PubMed] [Google Scholar]
- 9.Almeida, M. R., E. Rieder, J. Chinsangaram, G. Ward, C. Beard, M. J. Grubman, and P. W. Mason. 1998. Construction and evaluation of an attenuated vaccine for foot-and-mouth disease: difficulty adapting the leader proteinase-deleted strategy to the serotype O1 virus. Virus Res. 55:49-60. [DOI] [PubMed] [Google Scholar]
- 10.Ambros, V., and D. Baltimore. 1980. Purification and properties of a HeLa cell enzyme able to remove the 5′-terminal protein from poliovirus RNA. J. Biol. Chem. 255:6739-6744. [PubMed] [Google Scholar]
- 11.Ambros, V., R. F. Pettersson, and D. Baltimore. 1978. An enzymatic activity in uninfected cells that cleaves the linkage between poliovirion RNA and the 5′ terminal protein. Cell 15:1439-1446. [DOI] [PubMed] [Google Scholar]
- 12.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 14.Anonymous. 2002. Infectious disease in livestock. The Royal Society, London, United Kingdom.
- 15.Ansardi, D. C., Z. Moldoveanu, D. C. Porter, D. E. Walker, R. M. Conry, A. F. LoBuglio, S. McPherson, and C. D. Morrow. 1994. Characterization of poliovirus replicons encoding carcinoembryonic antigen. Cancer Res. 54:6359-6364. [PubMed] [Google Scholar]
- 16.Ansardi, D. C., D. C. Porter, and C. D. Morrow. 1992. Myristylation of poliovirus capsid precursor P1 is required for assembly of subviral particles. J. Virol. 66:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araujo, J. P., Jr., H. J. Montassier, and A. A. Pinto. 2002. Extensive antigenic and genetic variation among foot-and-mouth disease type A viruses isolated from the 1994 and 1995 foci in Sao Paulo, Brazil. Vet. Microbiol. 84:15-27. [DOI] [PubMed] [Google Scholar]
- 18.Argos, P., G. Kamer, M. J. Nicklin, and E. Wimmer. 1984. Similarity in gene organization and homology between proteins of animal picornaviruses and a plant comovirus suggest common ancestry of these virus families. Nucleic Acids Res. 12:7251-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold, E., M. Luo, G. Vriend, M. G. Rossmann, A. C. Palmenberg, G. D. Parks, M. J. Nicklin, and E. Wimmer. 1987. Implications of the picornavirus capsid structure for polyprotein processing. Proc. Natl. Acad. Sci. USA 84:21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bablanian, G. M., and M. J. Grubman. 1993. Characterization of the foot-and-mouth disease virus 3C protease expressed in Escherichia coli. Virology 197:320-327. [DOI] [PubMed] [Google Scholar]
- 21.Bachrach, H. L. 1968. Foot-and-mouth disease. Annu. Rev. Microbiol. 22:201-244. [DOI] [PubMed] [Google Scholar]
- 22.Bachrach, H. L. 1977. Foot-and-mouth disease virus: properties, molecular biology and immunogenicity. Beltsville Symp. Agric. Res. 1:3-32. [Google Scholar]
- 23.Bachrach, H. L., D. M. Moore, P. D. McKercher, and J. Polatnick. 1975. Immune and antibody responses to an isolated capsid protein of foot-and-mouth disease virus. J. Immunol. 115:1636-1641. [PubMed] [Google Scholar]
- 24.Bahnemann, H. G. 1975. Binary ethylenimine as an inactivant for foot-and-mouth disease virus and its application for vaccine production. Arch. Virol. 47:47-56. [DOI] [PubMed] [Google Scholar]
- 25.Banerjee, R., A. Echeverri, and A. Dasgupta. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71:9570-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baranowski, E., C. M. Ruiz-Jarabo, F. Lim, and E. Domingo. 2001. Foot-and-mouth disease virus lacking the VP1 G-H loop: the mutant spectrum uncovers interactions among antigenic sites for fitness gain. Virology 288:192-202. [DOI] [PubMed] [Google Scholar]
- 27.Baranowski, E., C. M. Ruiz-Jarabo, N. Sevilla, D. Andreu, E. Beck, and E. Domingo. 2000. Cell recognition by foot-and-mouth disease virus that lacks the RGD integrin-binding motif: flexibility in aphthovirus receptor usage. J. Virol. 74:1641-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barclay, W., Q. Li, G. Hutchinson, D. Moon, A. Richardson, N. Percy, J. W. Almond, and D. J. Evans. 1998. Encapsidation studies of poliovirus subgenomic replicons. J. Gen. Virol. 79:1725-1734. [DOI] [PubMed] [Google Scholar]
- 29.Barnett, P. V., S. J. Cox, N. Aggarwal, H. Gerber, and K. C. McCullough. 2002. Further studies on the early protective responses of pigs following immunisation with high potency foot and mouth disease vaccine. Vaccine 20:3197-3208. [DOI] [PubMed] [Google Scholar]
- 30.Barteling, S. J., R. H. Meloen, F. Wagenaar, and A. L. Gielkens. 1979. Isolation and characterization of trypsin-resistant O1 variants of foot-and-mouth disease virus. J. Gen. Virol. 43:383-393. [DOI] [PubMed] [Google Scholar]
- 31.Barteling, S. J., and J. Vreeswijk. 1991. Developments in foot-and-mouth disease vaccines. Vaccine 9:75-88. [DOI] [PubMed] [Google Scholar]
- 32.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basavappa, R., R. Syed, O. Flore, J. P. Icenogle, D. J. Filman, and J. M. Hogle. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 A resolution. Protein Sci. 3:1651-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastos, A. D., H. J. Bertschinger, C. Cordel, C. D. van Vuuren, D. Keet, R. G. Bengis, D. G. Grobler, and G. R. Thomson. 1999. Possibility of sexual transmission of foot-and-mouth disease from African buffalo to cattle. Vet. Rec. 145:77-79. [DOI] [PubMed] [Google Scholar]
- 36.Bautista, E. M., G. S. Ferman, and W. T. Golde. 2003. Induction of lymphopenia and inhibition of T cell function during acute infection of swine with foot and mouth disease virus (FMDV). Vet. Immunol. Immunopathol. 92:61-73. [DOI] [PubMed] [Google Scholar]
- 37.Baxt, B. 1987. Effect of lysosomotropic compounds on early events in foot-and-mouth disease virus replication. Virus Res. 7:257-271. [DOI] [PubMed] [Google Scholar]
- 38.Baxt, B., and H. L. Bachrach. 1982. The adsorption and degradation of foot-and-mouth disease virus by isolated BHK-21 cell plasma membranes. Virology 116:391-405. [DOI] [PubMed] [Google Scholar]
- 39.Baxt, B., and H. L. Bachrach. 1980. Early interactions of foot-and-mouth disease virus with cultured cells. Virology 104:42-55. [DOI] [PubMed] [Google Scholar]
- 40.Baxt, B., and Y. Becker. 1990. The effect of peptides containing the arginine-glycine-aspartic acid sequence on the adsorption of foot-and-mouth disease virus to tissue culture cells. Virus Genes 4:73-83. [DOI] [PubMed] [Google Scholar]
- 41.Baxt, B., A. E. Garmendia, and D. O. Morgan. 1989. Characterization of anti-idiotypic antibodies generated against foot-and-mouth disease virus neutralizing monoclonal antibodies. Viral Immunol. 2:103-113. [DOI] [PubMed] [Google Scholar]
- 42.Baxt, B., and P. W. Mason. 1995. Foot-and-mouth disease virus undergoes restricted replication in macrophage cell cultures following Fc receptor-mediated adsorption. Virology 207:503-509. [DOI] [PubMed] [Google Scholar]
- 43.Baxt, B., D. O. Morgan, B. H. Robertson, and C. A. Timpone. 1984. Epitopes on foot-and-mouth disease virus outer capsid protein VP1 involved in neutralization and cell attachment. J. Virol. 51:298-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baxt, B., S. Neff, E. Rieder, and P. W. Mason. 2002. Foot-and-mouth disease virus-receptor interactions: role in pathogenesis and tissue culture adaption, p. 115-123. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 45.Bazan, J. F., and R. J. Fletterick. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl. Acad. Sci. USA 85:7872-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beard, C., G. Ward, E. Rieder, J. Chinsangaram, M. J. Grubman, and P. W. Mason. 1999. Development of DNA vaccines for foot-and-mouth disease, evaluation of vaccines encoding replicating and non-replicating nucleic acids in swine. J. Biotechnol. 73:243-249. [DOI] [PubMed] [Google Scholar]
- 47.Beard, C. W., and P. W. Mason. 2000. Genetic determinants of altered virulence of Taiwanese foot-and-mouth disease virus. J. Virol. 74:987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beck, E., S. Forss, K. Strebel, R. Cattaneo, and G. Feil. 1983. Structure of the FMDV translation initiation site and of the structural proteins. Nucleic Acids Res. 11:7873-7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belnap, D. M., B. M. McDermott, Jr., D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belsham, G. J., C. C. Abrams, A. M. King, J. Roosien, and J. M. Vlak. 1991. Myristoylation of foot-and-mouth disease virus capsid protein precursors is independent of other viral proteins and occurs in both mammalian and insect cells. J. Gen. Virol. 72:747-751. [DOI] [PubMed] [Google Scholar]
- 51.Belsham, G. J., and J. K. Brangwyn. 1990. A region of the 5′ noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J. Virol. 64:5389-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benvenisti, L., A. Rogel, L. Kuznetzova, S. Bujanover, Y. Becker, and Y. Stram. 2001. Gene gun-mediated DNA vaccination against foot-and-mouth disease virus. Vaccine 19:3885-3895. [DOI] [PubMed] [Google Scholar]
- 53.Berger, H. G., O. C. Straub, R. Ahl, M. Tesar, and O. Marquardt. 1990. Identification of foot-and-mouth disease virus replication in vaccinated cattle by antibodies to non-structural virus proteins. Vaccine 8:213-216. [DOI] [PubMed] [Google Scholar]
- 54.Bergmann, I. E., P. A. de Mello, E. Neitzert, E. Beck, and I. Gomes. 1993. Diagnosis of persistent aphthovirus infection and its differentiation from vaccination response in cattle by use of enzyme-linked immunoelectrotransfer blot analysis with bioengineered nonstructural viral antigens. Am. J. Vet. Res. 54:825-831. [PubMed] [Google Scholar]
- 55.Bergmann, I. E., V. Malirat, E. Neitzert, E. Beck, N. Panizzuti, C. Sanchez, and A. Falczuk. 2000. Improvement of a serodiagnostic strategy for foot-and-mouth disease virus surveillance in cattle under systematic vaccination: a combined system of an indirect ELISA 3ABC with an enzyme-linked immunoelectrotransfer blot assay. Arch. Virol. 145:473-489. [DOI] [PubMed] [Google Scholar]
- 56.Berinstein, A., M. Roivainen, T. Hovi, P. W. Mason, and B. Baxt. 1995. Antibodies to the vitronectin receptor (integrin alpha V beta 3) inhibit binding and infection of foot-and-mouth disease virus to cultured cells. J. Virol. 69:2664-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berinstein, A., C. Tami, O. Taboga, E. Smitsaart, and E. Carrillo. 2000. Protective immunity against foot-and-mouth disease virus induced by a recombinant vaccinia virus. Vaccine 18:2231-2238. [DOI] [PubMed] [Google Scholar]
- 58.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 59.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 131:39-48. [DOI] [PubMed] [Google Scholar]
- 61.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bigeriego, P., M. F. Rosas, E. Zamora, E. Martinez-Salas, and F. Sobrino. 1999. Heterotypic inhibition of foot-and-mouth disease virus infection by combinations of RNA transcripts corresponding to the 5′ and 3′ regions. Antiviral Res. 44:133-141. [DOI] [PubMed] [Google Scholar]
- 63.Bishop, N. E., and D. A. Anderson. 1997. Hepatitis A virus subviral particles: purification, accumulation, and relative infectivity of virions, provirions and procapsids. Arch. Virol. 142:2147-2160. [DOI] [PubMed] [Google Scholar]
- 64.Bittle, J. L., R. A. Houghten, H. Alexander, T. M. Shinnick, J. G. Sutcliffe, R. A. Lerner, D. J. Rowlands, and F. Brown. 1982. Protection against foot-and-mouth disease by immunization with a chemically synthesized peptide predicted from the viral nucleotide sequence. Nature 298:30-33. [DOI] [PubMed] [Google Scholar]
- 65.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolten, R., D. Egger, R. Gosert, G. Schaub, L. Landmann, and K. Bienz. 1998. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent in situ hybridization. J. Virol. 72:8578-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolwell, C., A. L. Brown, P. V. Barnett, R. O. Campbell, B. E. Clarke, N. R. Parry, E. J. Ouldridge, F. Brown, and D. J. Rowlands. 1989. Host cell selection of antigenic variants of foot-and-mouth disease virus. J. Gen. Virol. 70:45-57. [DOI] [PubMed] [Google Scholar]
- 68.Bolwell, C., B. E. Clarke, N. R. Parry, E. J. Ouldridge, F. Brown, and D. J. Rowlands. 1989. Epitope mapping of foot-and-mouth disease virus with neutralizing monoclonal antibodies. J. Gen. Virol. 70:59-68. [DOI] [PubMed] [Google Scholar]
- 69.Borrego, B., J. A. Camarero, M. G. Mateu, and E. Domingo. 1995. A highly divergent antigenic site of foot-and-mouth disease virus retains its immunodominance. Viral Immunol. 8:11-18. [DOI] [PubMed] [Google Scholar]
- 70.Brooksby, J. B. 1982. Portraits of viruses: foot-and-mouth disease virus. Intervirology 18:1-23. [DOI] [PubMed] [Google Scholar]
- 71.Brown, C. C., J. Chinsangaram, and M. J. Grubman. 2000. Type I interferon production in cattle infected with 2 strains of foot-and-mouth disease virus, as determined by in situ hybridization. Can. J. Vet. Res. 64:130-133. [PMC free article] [PubMed] [Google Scholar]
- 72.Brown, C. C., R. F. Meyer, H. J. Olander, C. House, and C. A. Mebus. 1992. A pathogenesis study of foot-and-mouth disease in cattle, using in situ hybridization. Can. J. Vet. Res. 56:189-193. [PMC free article] [PubMed] [Google Scholar]
- 73.Brown, C. C., H. J. Olander, and R. F. Meyer. 1995. Pathogenesis of foot-and-mouth disease in swine, studied by in-situ hybridization. J. Comp. Pathol. 113:51-58. [DOI] [PubMed] [Google Scholar]
- 74.Brown, C. C., M. E. Piccone, P. W. Mason, T. S. McKenna, and M. J. Grubman. 1996. Pathogenesis of wild-type and leaderless foot-and-mouth disease virus in cattle. J. Virol. 70:5638-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown, F. 1992. New approaches to vaccination against foot-and-mouth disease. Vaccine 10:1022-1026. [DOI] [PubMed] [Google Scholar]
- 76.Brown, F., and B. Cartwright. 1961. Dissociation of foot-and-mouth disease virus into its nucleic acid and protein components. Nature 192:1163-1164. [DOI] [PubMed] [Google Scholar]
- 77.Bunch, T., E. Rieder, and P. Mason. 1994. Sequence of the S fragment of foot-and-mouth disease virus type A12. Virus Genes 8:173-175. [DOI] [PubMed] [Google Scholar]
- 78.Burrows, R. 1968. Excretion of foot-and-mouth disease prior to the development of lesions. Vet. Rec. 82:387-388. [Google Scholar]
- 79.Burrows, R., J. A. Mann, A. J. Garland, A. Greig, and D. Goodridge. 1981. The pathogenesis of natural and simulated natural foot-and-mouth disease infection in cattle. J. Comp. Pathol. 91:599-609. [DOI] [PubMed] [Google Scholar]
- 80.Butchaiah, G., and D. O. Morgan. 1997. Neutralization antigenic sites on type Asia-1 foot-and-mouth disease virus defined by monoclonal antibody-resistant variants. Virus Res. 52:183-194. [DOI] [PubMed] [Google Scholar]
- 81.Caliguiri, L. A., and I. Tamm. 1968. Action of guanidine on the replication of poliovirus RNA. Virology 35:408-417. [DOI] [PubMed] [Google Scholar]
- 82.Callahan, J. D., F. Brown, F. A. Osorio, J. H. Sur, E. Kramer, G. W. Long, J. Lubroth, S. J. Ellis, K. S. Shoulars, K. L. Gaffney, D. L. Rock, and W. M. Nelson. 2002. Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J. Am. Vet. Med. Assoc. 220:1636-1642. [DOI] [PubMed] [Google Scholar]
- 83.Callens, M., and K. De Clercq. 1997. Differentiation of the seven serotypes of foot-and-mouth disease virus by reverse transcriptase polymerase chain reaction. J. Virol. Methods 67:35-44. [DOI] [PubMed] [Google Scholar]
- 84.Cao, X., I. E. Bergmann, R. Fullkrug, and E. Beck. 1995. Functional analysis of the two alternative translation initiation sites of foot-and-mouth disease virus. J. Virol. 69:560-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao, X. M., I. E. Bergmann, and E. Beck. 1991. Comparison of the 5′ and 3′ untranslated genomic regions of virulent and attenuated foot-and-mouth disease viruses (strains O1 Campos and C3 Resende). J. Gen. Virol. 72:2821-2825. [DOI] [PubMed] [Google Scholar]
- 86.Capozzo, A. V., O. H. Periolo, B. Robiolo, C. Seki, J. L. La Torre, and P. R. Grigera. 1997. Total and isotype humoral responses in cattle vaccinated with foot and mouth disease virus (FMDV) immunogen produced either in bovine tongue tissue or in BHK-21 cell suspension cultures. Vaccine 15:624-630. [DOI] [PubMed] [Google Scholar]
- 87.Carrillo, E. C., C. Giachetti, and R. Campos. 1985. Early steps in FMDV replication: further analysis on the effects of chloroquine. Virology 147:118-125. [DOI] [PubMed] [Google Scholar]
- 88.Carrillo, E. C., C. Giachetti, and R. H. Campos. 1984. Effect of lysosomotropic agents on the foot-and-mouth disease virus replication. Virology 135:542-545. [DOI] [PubMed] [Google Scholar]
- 89.Cavanagh, D., D. J. Rowlands, and F. Brown. 1978. Early events in the interaction between foot-and mouth disease virus and primary pig kidney cells. J. Gen. Virol. 41:255-264. [DOI] [PubMed] [Google Scholar]
- 90.Cavanagh, D., D. V. Sangar, D. J. Rowlands, and F. Brown. 1977. Immunogenic and cell attachment sites of FMDV: further evidence for their location in a single capsid polypeptide. J. Gen. Virol. 35:149-158. [DOI] [PubMed] [Google Scholar]
- 91.Cedillo-Barron, L., M. Foster-Cuevas, G. J. Belsham, F. Lefevre, and R. M. Parkhouse. 2001. Induction of a protective response in swine vaccinated with DNA encoding foot-and-mouth disease virus empty capsid proteins and the 3D RNA polymerase. J. Gen. Virol. 82:1713-1724. [DOI] [PubMed] [Google Scholar]
- 92.Chang, K. H., P. Auvinen, T. Hyypia, and G. Stanway. 1989. The nucleotide sequence of coxsackievirus A9; implications for receptor binding and enterovirus classification. J. Gen. Virol. 70:3269-3280. [DOI] [PubMed] [Google Scholar]
- 93.Chang, K. H., C. Day, J. Walker, T. Hyypia, and G. Stanway. 1992. The nucleotide sequences of wild-type coxsackievirus A9 strains imply that an RGD motif in VP1 is functionally significant. J. Gen. Virol. 73:621-626. [DOI] [PubMed] [Google Scholar]
- 94.Childerstone, A. J., L. Cedillo-Baron, M. Foster-Cuevas, and R. M. Parkhouse. 1999. Demonstration of bovine CD8+ T-cell responses to foot-and-mouth disease virus. J. Gen. Virol. 80:663-669. [DOI] [PubMed] [Google Scholar]
- 95.Chinsangaram, J., C. Beard, P. W. Mason, M. K. Zellner, G. Ward, and M. J. Grubman. 1998. Antibody response in mice inoculated with DNA expressing foot-and-mouth disease virus capsid proteins. J. Virol. 72:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chinsangaram, J., M. Koster, and M. J. Grubman. 2001. Inhibition of L-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J. Virol. 75:5498-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chinsangaram, J., M. P. Moraes, M. Koster, and M. J. Grubman. 2003. Novel viral disease control strategy: adenovirus expressing alpha interferon rapidly protects swine from foot-and-mouth disease. J. Virol. 77:1621-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chinsangaram, J., P. W. Mason, and M. J. Grubman. 1998. Protection of swine by live and inactivated vaccines prepared from a leader proteinase-deficient serotype A12 foot-and-mouth disease virus. Vaccine 16:1516-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chinsangaram, J., M. E. Piccone, and M. J. Grubman. 1999. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J. Virol. 73:9891-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chow, M., J. F. Newman, D. Filman, J. M. Hogle, D. J. Rowlands, and F. Brown. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482-486. [DOI] [PubMed] [Google Scholar]
- 101.Clarke, B. E., and D. V. Sangar. 1988. Processing and assembly of foot-and-mouth disease virus proteins using subgenomic RNA. J. Gen. Virol. 69:2313-2325. [DOI] [PubMed] [Google Scholar]
- 102.Clarke, B. E., D. V. Sangar, J. N. Burroughs, S. E. Newton, A. R. Carroll, and D. J. Rowlands. 1985. Two initiation sites for foot-and-mouth disease virus polyprotein in vivo. J. Gen. Virol. 66:2615-2626. [DOI] [PubMed] [Google Scholar]
- 103.Clarke, J. B., and R. E. Spier. 1983. An investigation into causes of resistance of a cloned line of BHK cells to a strain of foot-and-mouth disease virus. Vet. Microbiol. 8:259-270. [DOI] [PubMed] [Google Scholar]
- 104.Collen, T., and T. R. Doel. 1990. Heterotypic recognition of foot-and-mouth disease virus by cattle lymphocytes. J. Gen. Virol. 71:309-315. [DOI] [PubMed] [Google Scholar]
- 105.Collen, T., L. Pullen, and T. R. Doel. 1989. T cell-dependent induction of antibody against foot-and-mouth disease virus in a mouse model. J. Gen. Virol. 70:395-403. [DOI] [PubMed] [Google Scholar]
- 106.Condy, J. B., R. S. Hedger, C. Hamblin, and I. T. Barnett. 1985. The duration of the foot-and-mouth disease virus carrier state in African buffalo (i) in the individual animal and (ii) in a free-living herd. Comp. Immunol. Microbiol. Infect. Dis. 8:259-265. [DOI] [PubMed] [Google Scholar]
- 107.Correa Melo, E., V. Saraiva, and V. Astudillo. 2002. Review of the status of foot and mouth disease in countries of South America and approaches to control and eradication. Rev. Sci. Tech. Off. Int. Epizoot. 21:429-436. [DOI] [PubMed] [Google Scholar]
- 108.Costa Giomi, M. P., I. E. Bergmann, E. A. Scodeller, P. Auge de Mello, I. Gomez, and J. L. La Torre. 1984. Heterogeneity of the polyribocytidylic acid tract in aphthovirus: biochemical and biological studies of viruses carrying polyribocytidylic acid tracts of different lengths. J. Virol. 51:799-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Couch, R. B., R. M. Chanock, T. R. Cate, D. J. Lang, V. Knight, and R. J. Heubner. 1963. Immunization with types 4 and 7 adenovirus by selective infection of the intestinal tract. Am. Rev. Intestin. Dis. 88:394-403. [DOI] [PubMed] [Google Scholar]
- 110.Cowan, K. M., and J. H. Graves. 1966. A third antigenic component associated with foot-and-mouth disease infection. Virology 30:528-540. [DOI] [PubMed] [Google Scholar]
- 111.Crawford, N. M., and D. Baltimore. 1983. Genome-linked protein VPg of poliovirus is present as free VPg and VPg-pUpU in poliovirus-infected cells. Proc. Natl. Acad. Sci. USA 80:7452-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Crowell, R. L., B. J. Landau, and J. Siak. 1981. Picornavirus receptors in pathogenesis, p. 170-180. In K. Lonberg-Holm and L. Philipson (ed.), Virus receptors, part 2. Animal viruses, series B, vol. 8. Chapman and Hall, New York, N.Y.
- 113.Crowther, J. R., S. Farias, W. C. Carpenter, and A. R. Samuel. 1993. Identification of a fifth neutralizable site on type O foot-and-mouth disease virus following characterization of single and quintuple monoclonal antibody escape mutants. J. Gen. Virol. 74:1547-1553. [DOI] [PubMed] [Google Scholar]
- 114.Cui, T., and A. G. Porter. 1995. Localization of binding site for encephalomyocarditis virus RNA polymerase in the 3′-noncoding region of the viral RNA. Nucleic Acids Res. 23:377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cui, T., S. Sankar, and A. G. Porter. 1993. Binding of encephalomyocarditis virus RNA polymerase to the 3′-noncoding region of the viral RNA is specific and requires the 3′-poly(A) tail. J. Biol. Chem. 268:26093-26098. [PubMed] [Google Scholar]
- 116.Curry, S., C. C. Abrams, E. Fry, J. C. Crowther, G. J. Belsham, D. I. Stuart, and A. M. King. 1995. Viral RNA modulates the acid sensitivity of foot-and-mouth disease virus capsids. J. Virol. 69:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Curry, S., E. Fry, W. Blakemore, R. Abu-Ghazaleh, T. Jackson, A. King, S. Lea, J. Newman, D. Rowlands, and D. Stuart. 1996. Perturbations in the surface structure of A22 Iraq foot-and-mouth disease virus accompanying coupled changes in host cell specificity and antigenicity. Structure 4:135-145. [DOI] [PubMed] [Google Scholar]
- 118.Curry, S., E. Fry, W. Blakemore, R. Abu-Ghazaleh, T. Jackson, A. King, S. Lea, J. Newman, and D. Stuart. 1997. Dissecting the roles of VP0 cleavage and RNA packaging in picornavirus capsid stabilization: the structure of empty capsids of foot-and-mouth disease virus. J. Virol. 71:9743-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dasgupta, A., P. Yalamanchili, M. Clark, S. Kliewer, L. Fradkin, S. Rubinstein, S. Das, Y. Shen, M. K. Weidman, R. Banerjee, U. Datta, M. Igo, P. Kundu, B. Barat, and A. J. Berk. 2002. Effects of picornavirus proteinases on host cell transcription, p. 321-335. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 120.Dawe, P. S., F. O. Flanagan, R. L. Madekurozwa, K. J. Sorensen, E. C. Anderson, C. M. Foggin, N. P. Ferris, and N. J. Knowles. 1994. Natural transmission of foot-and-mouth disease virus from African buffalo (Syncerus caffer) to cattle in a wildlife area of Zimbabwe. Vet. Rec. 134:230-232. [DOI] [PubMed] [Google Scholar]
- 121.Dawe, P. S., K. Sorensen, N. P. Ferris, I. T. Barnett, R. M. Armstrong, and N. J. Knowles. 1994. Experimental transmission of foot-and-mouth disease virus from carrier African buffalo (Syncerus caffer) to cattle in Zimbabwe. Vet. Rec. 134:211-215. [DOI] [PubMed] [Google Scholar]
- 122.De Diego, M., E. Brocchi, D. Mackay, and F. De Simone. 1997. The non-structural polyprotein 3ABC of foot-and-mouth disease virus as a diagnostic antigen in ELISA to differentiate infected from vaccinated cattle. Arch. Virol. 142:2021-2033. [DOI] [PubMed] [Google Scholar]
- 123.Reference deleted.
- 124.Devaney, M. A., V. N. Vakharia, R. E. Lloyd, E. Ehrenfeld, and M. J. Grubman. 1988. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J. Virol. 62:4407-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dever, T. E., M. J. Glynias, and W. C. Merrick. 1987. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc. Natl. Acad. Sci. USA 84:1814-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Diez, J., M. G. Mateu, and E. Domingo. 1989. Selection of antigenic variants of foot-and-mouth disease virus in the absence of antibodies, as revealed by an in situ assay. J. Gen. Virol. 70:3281-3289. [DOI] [PubMed] [Google Scholar]
- 127.DiMarchi, R., G. Brooke, C. Gale, V. Cracknell, T. Doel, and N. Mowat. 1986. Protection of cattle against foot-and-mouth disease by a synthetic peptide. Science 232:639-641. [DOI] [PubMed] [Google Scholar]
- 128.Dmitrieva, T. M., K. B. Norkina, and V. I. Agol. 1991. Encephalomyocarditis virus RNA polymerase preparations, with and without RNA helicase activity. J. Virol. 65:2714-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doel, T. R. 2003. FMD vaccines. Virus Res. 91:81-99. [DOI] [PubMed] [Google Scholar]
- 131.Doel, T. R., and L. Pullen. 1990. International bank for foot-and-mouth disease vaccine: stability studies with virus concentrates and vaccines prepared from them. Vaccine 8:473-478. [DOI] [PubMed] [Google Scholar]
- 132.Domingo, E., M. Davila, and J. Ortin. 1980. Nucleotide sequence heterogeneity of the RNA from a natural population of foot-and-mouth-disease virus. Gene 11:333-346. [DOI] [PubMed] [Google Scholar]
- 133.Domingo, E., J. Diez, M. A. Martinez, J. Hernandez, A. Holguin, B. Borrego, and M. G. Mateu. 1993. New observations on antigenic diversification of RNA viruses. Antigenic variation is not dependent on immune selection. J. Gen. Virol. 74:2039-2045. [DOI] [PubMed] [Google Scholar]
- 134.Domingo, E., C. Escarmis, E. Baranowski, C. M. Ruiz-Jarabo, E. Carrillo, J. I. Nunez, and F. Sobrino. 2003. Evolution of foot-and-mouth disease virus. Virus Res. 91:47-63. [DOI] [PubMed] [Google Scholar]
- 135.Domingo, E., and J. J. Holland. 1988. High error rates, population equilibrium and evolution of RNA replication systems, p. 3-36. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics, vol. III. Variability of RNA Genomes. CRC Press, Boca Raton, Fla. [Google Scholar]
- 136.Domingo, E., M. G. Mateu, M. A. Matnez, J. Dopazo, A. Moya, and F. Sobrino. 1990. Genetic variabililty and antigenic diversity of foot-and-mouth disease virus, p. 233-266. In R. G. M. E. Kurstak, F. A. Murphy, and M. H. V. Regenmortel (ed.), Virus variability, epidemiology and control, vol. 2. Plenum Publishing Corp., New York, N.Y. [Google Scholar]
- 137.Donaldson, A. I. 1987. Foot-and-mouth disease: the principal features. Irish Vet. J. 41:325-327. [Google Scholar]
- 138.Donaldson, A. I., and R. F. Sellers. 2000. Foot-and-mouth disease, p. 254-258. In W. B. Martin and I. D. Aitken (ed.), Diseases of sheep. Blackwell Science, Oxford, United Kingdom.
- 139.Donnelly, M. L., L. E. Hughes, G. Luke, H. Mendoza, E. ten Dam, D. Gani, and M. D. Ryan. 2001. The ′cleavage' activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J. Gen. Virol. 82:1027-1041. [DOI] [PubMed] [Google Scholar]
- 140.Donnelly, M. L., G. Luke, A. Mehrotra, X. Li, L. E. Hughes, D. Gani, and M. D. Ryan. 2001. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip.’ J. Gen. Virol. 82:1013-1025. [DOI] [PubMed] [Google Scholar]
- 141.Dorsch-Hasler, K., Y. Yogo, and E. Wimmer. 1975. Replication of picornaviruses. I. Evidence from in vitro RNA synthesis that poly(A) of the poliovirus genome is genetically coded. J. Virol. 16:1512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dunn, C. S., and A. I. Donaldson. 1997. Natural adaption to pigs of a Taiwanese isolate of foot-and-mouth disease virus. Vet. Rec. 141:174-175. [DOI] [PubMed] [Google Scholar]
- 144.Duque, H., and B. Baxt. 2003. Foot-and-mouth disease virus receptors: comparison of bovine alpha(V) integrin utilization by type A and O viruses. J. Virol. 77:2500-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Egger, D., R. Gosert, and K. Bienz. 2002. Role of cellular structures in viral RNA replication, p. 247-253. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 146.Eigen, M. 1971. Self organization of matter and the evolution of biological macromolecules. Naturwissenschaften 58:465-523. [DOI] [PubMed] [Google Scholar]
- 147.Ellard, F. M., J. Drew, W. E. Blakemore, D. I. Stuart, and A. M. King. 1999. Evidence for the role of His-142 of protein 1C in the acid-induced disassembly of foot-and-mouth disease virus capsids. J. Gen. Virol. 80:1911-1918. [DOI] [PubMed] [Google Scholar]
- 148.Eloit, M., P. Gilardi-Hebenstreit, B. Toma, and M. Perricaudet. 1990. Construction of a defective adenovirus vector expressing the pseudorabies virus glycoprotein gp50 and its use as a live vaccine. J. Gen. Virol. 71:2425-2431. [DOI] [PubMed] [Google Scholar]
- 149.Escarmis, C., J. Dopazo, M. Davila, E. L. Palma, and E. Domingo. 1995. Large deletions in the 5′-untranslated region of foot-and-mouth disease virus of serotype C. Virus Res. 35:155-167. [DOI] [PubMed] [Google Scholar]
- 150.Escarmis, C., M. Toja, M. Medina, and E. Domingo. 1992. Modifications of the 5′ untranslated region of foot-and-mouth disease virus after prolonged persistence in cell culture. Virus Res. 26:113-125. [DOI] [PubMed] [Google Scholar]
- 151.Evans, D. J., and J. W. Almond. 1998. Cell receptors for picornaviruses as determinants of cell tropism and pathogenesis. Trends Microbiol. 6:198-202. [DOI] [PubMed] [Google Scholar]
- 152.Evans, D. M., G. Dunn, P. D. Minor, G. C. Schild, A. J. Cann, G. Stanway, J. W. Almond, K. Currey, and J. V. Maizel, Jr. 1985. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature 314:548-550. [DOI] [PubMed] [Google Scholar]
- 153.Fares, M. A., A. Moya, C. Escarmis, E. Baranowski, E. Domingo, and E. Barrio. 2001. Evidence for positive selection in the capsid protein-coding region of the foot-and-mouth disease virus (FMDV) subjected to experimental passage regimens. Mol. Biol. Evol. 18:10-21. [DOI] [PubMed] [Google Scholar]
- 154.Feigelstock, D., M. G. Mateu, M. E. Piccone, F. De Simone, E. Brocchi, E. Domingo, and E. L. Palma. 1992. Extensive antigenic diversification of foot-and-mouth disease virus by amino acid substitutions outside the major antigenic site. J. Gen. Virol. 73:3307-3311. [DOI] [PubMed] [Google Scholar]
- 155.Fenner, F. J., P. J. Gibbs, F. A. Murphy, R. Rott, M. J. Studdert, and D. O. White. 1993. Veterinary virology, p. 403-430. Academic Press, New York, N.Y.
- 156.Fooks, A. R., D. Jeevarajah, J. Lee, A. Warnes, S. Niewiesk, V. ter Meulen, J. R. Stephenson, and J. C. Clegg. 1998. Oral or parenteral administration of replication-deficient adenoviruses expressing the measles virus haemagglutinin and fusion proteins: protective immune responses in rodents. J. Gen. Virol. 79:1027-1031. [DOI] [PubMed] [Google Scholar]
- 157.Forss, S., K. Strebel, E. Beck, and H. Schaller. 1984. Nucleotide sequence and genome organization of foot-and-mouth disease virus. Nucleic Acids Res. 12:6587-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fox, G., N. R. Parry, P. V. Barnett, B. McGinn, D. J. Rowlands, and F. Brown. 1989. The cell attachment site on foot-and-mouth disease virus includes the amino acid sequence RGD (arginine-glycine-aspartic acid). J. Gen. Virol. 70:625-637. [DOI] [PubMed] [Google Scholar]
- 159.Fracastorius, H. 1546. De sympathia et antipathia rerum liber unus. De contagione et contagiosis morbis et eorum curatione liber I, Venice, Heirs of L. A. Junta.
- 160.Francis, M. J., G. Z. Hastings, F. Brown, J. McDermed, Y. A. Lu, and J. P. Tam. 1991. Immunological evaluation of the multiple antigen peptide (MAP) system using the major immunogenic site of foot-and-mouth disease virus. Immunology 73:249-254. [PMC free article] [PubMed] [Google Scholar]
- 161.Frenkel, H. S. 1947. La culture de virus de la fievre aphteuse sur l'epithelium de la langue des bovides. Bull. Off. Int. Epiz. 28:155-162. [Google Scholar]
- 162.Fry, E. E., S. M. Lea, T. Jackson, J. W. Newman, F. M. Ellard, W. E. Blakemore, R. Abu-Ghazaleh, A. Samuel, A. M. King, and D. I. Stuart. 1999. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 18:543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gailiunas, P., and G. E. Cottral. 1966. Presence and persistence of foot-and-mouth disease virus in bovine skin. J. Bacteriol. 91:2333-2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′-noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 166.Garcia-Valcarcel, M., T. Doel, T. Collen, M. Ryan, and R. M. Parkhouse. 1996. Recognition of foot-and-mouth disease virus and its capsid protein VP1 by bovine peripheral T lymphocytes. J. Gen. Virol. 77:727-735. [DOI] [PubMed] [Google Scholar]
- 167.Gebauer, F., J. C. de la Torre, I. Gomes, M. G. Mateu, H. Barahona, B. Tiraboschi, I. Bergmann, P. A. de Mello, and E. Domingo. 1988. Rapid selection of genetic and antigenic variants of foot-and-mouth disease virus during persistence in cattle. J. Virol. 62:2041-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Geering, W. A. 1967. Foot and mouth disease in sheep. Aust. Vet. J. 43:485-489. [Google Scholar]
- 169.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2A(pro). J. Virol. 75:10979-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Giachetti, C., S. S. Hwang, and B. L. Semler. 1992. cis-acting lesions targeted to the hydrophobic domain of a poliovirus membrane protein involved in RNA replication. J. Virol. 66:6045-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gibson, C. F., A. I. Donaldson, and N. P. Ferris. 1984. Response of sheep vaccinated with large doses of vaccine to challenge by airborne foot and mouth disease virus. Vaccine 2:157-161. [DOI] [PubMed] [Google Scholar]
- 172.Giraudo, A. T., E. Beck, K. Strebel, P. A. de Mello, J. L. La Torre, E. A. Scodeller, and I. E. Bergmann. 1990. Identification of a nucleotide deletion in parts of polypeptide 3A in two independent attenuated aphthovirus strains. Virology 177:780-783. [DOI] [PubMed] [Google Scholar]
- 173.Glass, E. J., R. A. Oliver, T. Collen, T. R. Doel, R. DiMarchi, and R. L. Spooner. 1991. MHC class II restricted recognition of FMDV peptides by bovine T cells. Immunology 74:594-599. [PMC free article] [PubMed] [Google Scholar]
- 174.Golini, F., B. L. Semler, A. J. Dorner, and E. Wimmer. 1980. Protein-linked RNA of poliovirus is competent to form an initiation complex of translation in vitro. Nature 287:600-603. [DOI] [PubMed] [Google Scholar]
- 175.Gonzalez, M. J., J. C. Saiz, O. Laor, and D. M. Moore. 1991. Antigenic stability of foot-and-mouth disease virus variants on serial passage in cell culture. J. Virol. 65:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gopinath, K., J. Wellink, C. Porta, K. M. Taylor, G. P. Lomonossoff, and A. van Kammen. 2000. Engineering cowpea mosaic virus RNA-2 into a vector to express heterologous proteins in plants. Virology 267:159-173. [DOI] [PubMed] [Google Scholar]
- 178.Gorbalenya, A. E., V. M. Blinov, A. P. Donchenko, and E. V. Koonin. 1989. An NTP-binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J. Mol. Evol. 28:256-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gorbalenya, A. E., A. P. Donchenko, V. M. Blinov, and E. V. Koonin. 1989. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 243:103-114. [DOI] [PubMed] [Google Scholar]
- 180.Gorbalenya, A. E., and E. V. Koonin. 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 17:8413-8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gosert, R., D. Egger, and K. Bienz. 2000. A cytopathic and a cell culture adapted hepatitis A virus strain differ in cell killing but not in intracellular membrane rearrangements. Virology 266:157-169. [DOI] [PubMed] [Google Scholar]
- 182.Graham, F. L., and L. Prevec. 1992. Adenovirus-based expression vectors and recombinant vaccines. Bio/Technology 20:363-390. [DOI] [PubMed] [Google Scholar]
- 183.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 184.Grigera, P. R., and S. G. Tisminetzky. 1984. Histone H3 modification in BHK cells infected with foot-and-mouth disease virus. Virology 136:10-19. [DOI] [PubMed] [Google Scholar]
- 185.Groot Bramel-Verheije, M. H., P. J. Rottier, and J. J. Meulenberg. 2000. Expression of a foreign epitope by porcine reproductive and respiratory syndrome virus. Virology 278:380-389. [DOI] [PubMed] [Google Scholar]
- 186.Grubman, M. J. 1980. The 5′ end of foot-and-mouth disease virion RNA contains a protein covalently linked to the nucleotide pUp. Arch. Virol. 63:311-315. [DOI] [PubMed] [Google Scholar]
- 187.Grubman, M. J., and H. L. Bachrach. 1979. Isolation of foot-and-mouth disease virus messenger RNA from membrane-bound polyribosomes and characterization of its 5′ and 3′ termini. Virology 98:466-470. [DOI] [PubMed] [Google Scholar]
- 188.Grubman, M. J., S. A. Lewis, and D. O. Morgan. 1993. Protection of swine against foot-and-mouth disease with viral capsid proteins expressed in heterologous systems. Vaccine 11:825-829. [DOI] [PubMed] [Google Scholar]
- 189.Grubman, M. J., and P. W. Mason. 2002. Prospects, including time-frames, for improved foot and mouth disease vaccines. Rev. Sci. Tech. Off. Int. Epizoot. 21:589-600. [DOI] [PubMed] [Google Scholar]
- 190.Grubman, M. J., D. O. Morgan, J. Kendall, and B. Baxt. 1985. Capsid intermediates assembled in a foot-and-mouth disease virus genome RNA-programmed cell-free translation system and in infected cells. J. Virol. 56:120-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Grubman, M. J., B. H. Robertson, D. O. Morgan, D. M. Moore, and D. Dowbenko. 1984. Biochemical map of polypeptides specified by foot-and-mouth disease virus. J. Virol. 50:579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Grubman, M. J., M. Zellner, G. Bablanian, P. W. Mason, and M. E. Piccone. 1995. Identification of the active-site residues of the 3C proteinase of foot-and-mouth disease virus. Virology 213:581-589. [DOI] [PubMed] [Google Scholar]
- 193.Guarne, A., B. Hampoelz, W. Glaser, X. Carpena, J. Tormo, I. Fita, and T. Skern. 2000. Structural and biochemical features distinguish the foot-and-mouth disease virus leader proteinase from other papain-like enzymes. J. Mol. Biol. 302:1227-1240. [DOI] [PubMed] [Google Scholar]
- 194.Guarne, A., J. Tormo, R. Kirchweger, D. Pfistermueller, I. Fita, and T. Skern. 1998. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 17:7469-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Gutierrez, A., E. Martinez-Salas, B. Pintado, and F. Sobrino. 1994. Specific inhibition of aphthovirus infection by RNAs transcribed from both the 5′ and the 3′ noncoding regions. J. Virol. 68:7426-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gutierrez, A., A. Rodriguez, B. Pintado, and F. Sobrino. 1993. Transient inhibition of foot-and-mouth disease virus infection of BHK-21 cells by antisense oligonucleotides directed against the second functional initiator AUG. Antiviral Res. 22:1-13. [DOI] [PubMed] [Google Scholar]
- 197.Guttman, N., and D. Baltimore. 1977. Morphogenesis of poliovirus. IV. Existence of particles sedimenting at 150S and having the properties of provirion. J. Virol. 23:363-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hagino-Yamagishi, K., and A. Nomoto. 1989. In vitro construction of poliovirus defective interfering particles. J. Virol. 63:5386-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hanada, K., M. Tamai, S. Morimoto, T. Adachi, S. Ohmura, J. Sawada, and I. Tanaka. 1978. Inhibitory activities of E-64 derivatives of papain. Agric. Biol. Chem. 42:537-541. [Google Scholar]
- 200.Hanada, K., M. Tamai, S. Ohmura, J. Sawada, T. Seki, and I. Tanaka. 1978. Structure and synthesis of E-64, a new thiol protease inhibitor. Agric. Biol. Chem. 42:529-536. [Google Scholar]
- 201.Harber, J. J., J. Bradley, C. W. Anderson, and E. Wimmer. 1991. Catalysis of poliovirus VP0 maturation cleavage is not mediated by serine 10 of VP2. J. Virol. 65:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 203.Harris, T. J., and F. Brown. 1977. Biochemical analysis of a virulent and an avirulent strain of foot-and-mouth disease virus. J. Gen. Virol. 34:87-105. [DOI] [PubMed] [Google Scholar]
- 204.Haydon, D., S. Lea, L. Fry, N. Knowles, A. R. Samuel, D. Stuart, and M. E. Woolhouse. 1998. Characterizing sequence variation in the VP1 capsid proteins of foot and mouth disease virus (serotype 0) with respect to virion structure. J. Mol. Evol. 46:465-475. [DOI] [PubMed] [Google Scholar]
- 205.Haydon, D. T., A. D. Bastos, N. J. Knowles, and A. R. Samuel. 2001. Evidence for positive selection in foot-and-mouth disease virus capsid genes from field isolates. Genetics 157:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Haydon, D. T., A. R. Samuel, and N. J. Knowles. 2001. The generation and persistence of genetic variation in foot-and-mouth disease virus. Prev. Vet. Med. 51:111-124. [DOI] [PubMed] [Google Scholar]
- 207.He, Y., V. D. Bowman, S. Mueller, C. M. Bator, J. Bella, X. Peng, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hearps, A., Z. Zhang, and S. Alexandersen. 2002. Evaluation of the portable Cepheid SmartCycler real-time PCR machine for the rapid diagnosis of foot-and-mouth disease. Vet. Rec. 150:625-628. [DOI] [PubMed] [Google Scholar]
- 209.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 90:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hemadri, D., C. Tosh, R. Venkataramanan, A. Sanyal, A. R. Samuel, N. J. Knowles, and R. P. Kitching. 2000. Genetic analysis of foot-and-mouth disease virus type O isolates responsible for field outbreaks in India between 1993 and 1999. Epidemiol. Infect. 125:729-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hindiyeh, M., Q. H. Li, R. Basavappa, J. M. Hogle, and M. Chow. 1999. Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4. J. Virol. 73:9072-9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hoey, E. M., and S. J. Martin. 1974. A possible precursor containing RNA of a bovine enterovirus: the provirion 11. J. Gen. Virol. 24:515-524. [DOI] [PubMed] [Google Scholar]
- 215.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 216.Huang, C.-C., M.-H. Jong, and S.-Y. Lin. 2000. Characteristics of foot-and-mouth disease virus in Taiwan. J. Vet. Med. Sci. 62:677-679. [DOI] [PubMed] [Google Scholar]
- 217.Huang, C.-C., Y.-L. Lin, T.-S. Huang, W.-J. Tu, S.-H. Lee, M.-H. Jong, and S.-Y. Lin. 2001. Molecular characterization of foot-and-mouth disease virus isolated from ruminants in Taiwan in 1999-2000. Vet. Microbiol. 81:193-205. [DOI] [PubMed] [Google Scholar]
- 218.Hughes, G. J., V. Mioulet, R. P. Kitching, M. E. Woolhouse, S. Alexandersen, and A. I. Donaldson. 2002. Foot-and-mouth disease virus infection of sheep: implications for diagnosis and control. Vet. Rec. 150:724-727. [DOI] [PubMed] [Google Scholar]
- 219.Hyde, J. L., J. H. Blackwell, and J. J. Callis. 1975. Effect of pasteurization and evaporation on foot-and-mouth disease virus in whole milk from infected cows. Can. J. Comp. Med. 39:305-309. [PMC free article] [PubMed] [Google Scholar]
- 220.Hynes, R. O. 1999. Cell adhesion: old and new questions. Trends Cell Biol. 9:M33-M37. [PubMed] [Google Scholar]
- 221.Hynes, R. O. 1987. Integrins: a family of cell surface receptors. Cell 48:549-554. [DOI] [PubMed] [Google Scholar]
- 222.Hynes, R. O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell 110:673-687. [DOI] [PubMed] [Google Scholar]
- 223.Hynes, R. O. 1992. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69:11-25. [DOI] [PubMed] [Google Scholar]
- 224.Ilott, M. C., J. S. Salt, R. M. Gaskell, and R. P. Kitching. 1997. Dexamethasone inhibits virus production and the secretory IgA response in oesophageal-pharyngeal fluid in cattle persistently infected with foot-and-mouth disease virus. Epidemiol. Infect. 118:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Imler, J. L. 1995. Adenovirus vectors as recombinant viral vaccines. Vaccine 13:1143-1151. [DOI] [PubMed] [Google Scholar]
- 226.Jackson, T., F. M. Ellard, R. A. Ghazaleh, S. M. Brookes, W. E. Blakemore, A. H. Corteyn, D. I. Stuart, J. W. Newman, and A. M. King. 1996. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J. Virol. 70:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Jackson, T., A. M. King, D. I. Stuart, and E. Fry. 2003. Structure and receptor binding. Virus Res. 91:33-46. [DOI] [PubMed] [Google Scholar]
- 228.Jackson, T., A. P. Mould, D. Sheppard, and A. M. King. 2002. Integrin αvβ1 is a receptor for foot-and-mouth disease virus. J. Virol. 76:935-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jackson, T., D. Sheppard, M. Denyer, W. Blakemore, and A. M. King. 2000. The epithelial integrin αvβ6 is a receptor for foot-and-mouth disease virus. J. Virol. 74:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jacobson, M. F., and D. Baltimore. 1968. Polypeptide cleavages in the formation of poliovirus proteins. Proc. Natl. Acad. Sci. USA 61:77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jecht, M., C. Probst, and V. Gauss-Muller. 1998. Membrane permeability induced by hepatitis A virus proteins 2B and 2BC and proteolytic processing of HAV 2BC. Virology 252:218-227. [DOI] [PubMed] [Google Scholar]
- 233.Joo, Y.-S., S.-H. Ann, O.-K. Kim, J. Lubroth, and J.-H. Sur. 2002. Foot-and-mouth disease eradication efforts in the Republic of Korea. Can. J. Vet. Res. 66:122-124. [PMC free article] [PubMed] [Google Scholar]
- 234.Juillard, V., P. Villefroy, D. Godfrin, A. Pavirani, A. Venet, and J. G. Guillet. 1995. Long-term humoral and cellular immunity induced by a single immunization with replication-defective adenovirus recombinant vector. Eur. J. Immunol. 25:3467-3473. [DOI] [PubMed] [Google Scholar]
- 235.Kaariainen, L., and M. Ranki. 1984. Inhibition of cell functions by RNA virus infections. Annu. Rev. Microbiol. 38:91-109. [DOI] [PubMed] [Google Scholar]
- 236.Kajigaya, S., H. Arakawa, S. Kuge, T. Koi, N. Imura, and A. Nomoto. 1985. Isolation and characterization of defective-interfering particles of poliovirus Sabin 1 strain. Virology 142:307-316. [DOI] [PubMed] [Google Scholar]
- 237.Kaminski, A., S. L. Hunt, J. G. Patton, and R. J. Jackson. 1995. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1:924-938. [PMC free article] [PubMed] [Google Scholar]
- 238.Kawamura, N., M. Kohara, S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1989. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.King, A. M., D. McCahon, K. Saunders, J. W. Newman, and W. R. Slade. 1985. Multiple sites of recombination within the RNA genome of foot-and-mouth disease virus. Virus Res. 3:373-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypia, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-673. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 241.Kirchweger, R., E. Ziegler, B. J. Lamphear, D. Waters, H. D. Liebig, W. Sommergruber, F. Sobrino, C. Hohenadl, D. Blaas, R. E. Rhoads, and T. Skern. 1994. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4 gamma. J. Virol. 68:5677-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kit, M., S. Kit, S. P. Little, R. D. Di Marchi, and C. Gale. 1991. Bovine herpesvirus-1 (infectious bovine rhinotracheitis virus)-based viral vector which expresses foot-and-mouth disease epitopes. Vaccine 9:564-572. [DOI] [PubMed] [Google Scholar]
- 243.Kitching, P. 1992. Foot-and-mouth disease, p. 537-543. In A. H. Andrews, R. W. Blowey, H. Boyd, and R. G. Eddy (ed.), Bovine medicine: diseases and husbandry of cattle. Blackwell Science Inc., Oxford, United Kingdom.
- 244.Kitching, P., and S. Alexandersen. 2002. Clinical variation in foot-and-mouth disease: pigs. Rev. Sci. Tech. Off. Int. Epizoot. 21:513-518. [DOI] [PubMed] [Google Scholar]
- 245.Kitching, P., and G. H. Hughes. 2002. Clinical variation in foot-and-mouth disease: sheep and goats. Rev. Sci. Tech. Off. Int. Epizoot. 21:505-512. [DOI] [PubMed] [Google Scholar]
- 246.Kitson, J. D., K. L. Burke, L. A. Pullen, G. J. Belsham, and J. W. Almond. 1991. Chimeric polioviruses that include sequences derived from two independent antigenic sites of foot-and-mouth disease virus (FMDV) induce neutralizing antibodies against FMDV in guinea pigs. J. Virol. 65:3068-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kitson, J. D., D. McCahon, and G. J. Belsham. 1990. Sequence analysis of monoclonal antibody resistant mutants of type O foot and mouth disease virus: evidence for the involvement of the three surface exposed capsid proteins in four antigenic sites. Virology 179:26-34. [DOI] [PubMed] [Google Scholar]
- 248.Kleid, D. G., D. Yansura, B. Small, D. Dowbenko, D. M. Moore, M. J. Grubman, P. D. McKercher, D. O. Morgan, B. H. Robertson, and H. L. Bachrach. 1981. Cloned viral protein vaccine for foot-and-mouth disease: responses in cattle and swine. Science 214:1125-1129. [DOI] [PubMed] [Google Scholar]
- 249.Klein, M., H. J. Eggers, and B. Nelsen-Salz. 1999. Echovirus 9 strain Barty non-structural protein 2C has NTPase activity. Virus Res. 65:155-160. [DOI] [PubMed] [Google Scholar]
- 250.Klein, M., D. Hadaschik, H. Zimmermann, H. J. Eggers, and B. Nelsen-Salz. 2000. The picornavirus replication inhibitors HBB and guanidine in the echovirus-9 system: the significance of viral protein 2C. J. Gen. Virol. 81:895-901. [DOI] [PubMed] [Google Scholar]
- 251.Kleina, L. G., and M. J. Grubman. 1992. Antiviral effects of a thiol protease inhibitor on foot-and-mouth disease virus. J. Virol. 66:7168-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Klump, W., O. Marquardt, and P. H. Hofschneider. 1984. Biologically active protease of foot and mouth disease virus is expressed from cloned viral cDNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 81:3351-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Knipe, T., E. Rieder, B. Baxt, G. Ward, and P. W. Mason. 1997. Characterization of synthetic foot-and-mouth disease virus provirions separates acid-mediated disassembly from infectivity. J. Virol. 71:2851-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Knowles, N. J., P. R. Davies, T. Henry, V. O'Donnell, J. M. Pacheco, and P. W. Mason. 2001. Emergence in Asia of foot-and-mouth disease viruses with altered host range: characterization of alterations in the 3A protein. J. Virol. 75:1551-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Knowles, N. J., and A. R. Samuel. 2003. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 91:65-80. [DOI] [PubMed] [Google Scholar]
- 256.Knowles, N. J., A. R. Samuel, P. R. Davies, R. P. Kitching, and A. I. Donaldson. 2001. Outbreak of foot-and-mouth disease virus serotype O in the UK caused by a pandemic strain. Vet. Rec. 148:258-259. [PubMed] [Google Scholar]
- 257.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker, and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Konig, G., C. Blanco, N. J. Knowles, E. L. Palma, E. Maradei, and M. E. Piccone. 2001. Phylogenetic analysis of foot-and-mouth disease viruses isolated in Argentina. Virus Genes. 23:175-181. [DOI] [PubMed] [Google Scholar]
- 259.Krebs, O., R. Ahl, O. C. Straub, and O. Marquardt. 1993. Amino acid changes outside the G-H loop of capsid protein VP1 of type O foot-and-mouth disease virus confer resistance to neutralization by antipeptide G-H serum. Vaccine 11:359-362. [DOI] [PubMed] [Google Scholar]
- 260.Kronovetr, J., and T. Skern. 2002. Foot-and-mouth disease virus leader proteinase: a papain-like enzyme requiring an acidic environment in the active site. FEBS Lett. 528:58-62. [DOI] [PubMed] [Google Scholar]
- 261.Kuhn, R., N. Luz, and E. Beck. 1990. Functional analysis of the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 64:4625-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 263.Lea, S., R. Abu-Ghazaleh, W. Blakemore, S. Curry, E. Fry, T. Jackson, A. King, D. Logan, J. Newman, and D. Stuart. 1995. Structural comparison of two strains of foot-and-mouth disease virus subtype O1 and a laboratory antigenic variant, G67. Structure 3:571-580. [DOI] [PubMed] [Google Scholar]
- 264.Lea, S., J. Hernandez, W. Blakemore, E. Brocchi, S. Curry, E. Domingo, E. Fry, R. Abu-Ghazaleh, A. King, J. Newman, et al. 1994. The structure and antigenicity of a type C foot-and-mouth disease virus. Structure 2:123-139. [DOI] [PubMed] [Google Scholar]
- 265.Lee, W. M., S. S. Monroe, and R. R. Rueckert. 1993. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J. Virol. 67:2110-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Leforban, Y. 1999. Prevention measures against foot-and-mouth disease in Europe in recent years. Vaccine 17:1755-1759. [DOI] [PubMed] [Google Scholar]
- 267.Leforban, Y., and G. Gerbier. 2002. Review of the status of foot and mouth disease and approach to control/eradication in Europe and Central Asia. Rev. Sci. Tech. Off. Int. Epizoot. 21:477-492. [DOI] [PubMed] [Google Scholar]
- 268.Leippert, M., E. Beck, F. Weiland, and E. Pfaff. 1997. Point mutations within the βG-βH loop of foot-and-mouth disease virus O1K affect virus attachment to target cells. J. Virol. 71:1046-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lewis, S. A., D. O. Morgan, and M. J. Grubman. 1991. Expression, processing, and assembly of foot-and-mouth disease virus capsid structures in heterologous systems: induction of a neutralizing antibody response in guinea pigs. J. Virol. 65:6572-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lobert, P. E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Loeffler, F., and P. Frosch. 1897. Summarischer Bericht uber die Ergebnisse der Untersuchungen zur Erforschung der Maul- und Klauenseuche. Zentbl. Bakteriol. Parasitenkd Abt. I 22:257-259.
- 272.Logan, D., R. Abu-Ghazaleh, W. Blakemore, S. Curry, T. Jackson, A. King, S. Lea, R. Lewis, J. Newman, N. Parry, et al. 1993. Structure of a major immunogenic site on foot-and-mouth disease virus. Nature 362:566-568. [DOI] [PubMed] [Google Scholar]
- 273.Lopez de Quinto, S., and E. Martinez-Salas. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6:1380-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lopez de Quinto, S., M. Saiz, D. de la Morena, F. Sobrino, and E. Martinez-Salas. 2002. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 30:4398-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lowe, P. A., and F. Brown. 1981. Isolation of a soluble and template-dependent foot-and-mouth disease virus RNA polymerase. Virology 111:23-32. [DOI] [PubMed] [Google Scholar]
- 276.Lubroth, J., and F. Brown. 1995. Identification of native foot-and-mouth disease virus non-structural protein 2C as a serological indicator to differentiate infected from vaccinated livestock. Res. Vet. Sci. 59:70-78. [DOI] [PubMed] [Google Scholar]
- 277.Lubroth, J., M. J. Grubman, T. G. Burrage, J. F. Newman, and F. Brown. 1996. Absence of protein 2C from clarified foot-and-mouth disease virus vaccines provides the basis for distinguishing convalescent from vaccinated animals. Vaccine 14:419-427. [DOI] [PubMed] [Google Scholar]
- 278.Lukaszewski, R. A., and T. J. Brooks. 2000. Pegylated alpha interferon is an effective treatment for virulent Venezuelan equine encephalitis virus and has profound effects on the host immune response to infection. J. Virol. 74:5006-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lundquist, R. E., M. Sullivan, and J. V. Maizel, Jr. 1979. Characterization of a new isolate of poliovirus defective interfering particles. Cell 18:759-769. [DOI] [PubMed] [Google Scholar]
- 280.Luo, M., G. Vriend, G. Kamer, I. Minor, E. Arnold, M. G. Rossmann, U. Boege, D. G. Scraba, G. M. Duke, and A. C. Palmenberg. 1987. The atomic structure of Mengo virus at 3.0 A resolution. Science 235:182-191. [DOI] [PubMed] [Google Scholar]
- 281.Luz, N., and E. Beck. 1990. A cellular 57 kDa protein binds to two regions of the internal translation initiation site of foot-and-mouth disease virus. FEBS Lett. 269:311-314. [DOI] [PubMed] [Google Scholar]
- 282.Luz, N., and E. Beck. 1991. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 65:6486-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Mackay, D. K., M. A. Forsyth, P. R. Davies, A. Berlinzani, G. J. Belsham, M. Flint, and M. D. Ryan. 1998. Differentiating infection from vaccination in foot-and-mouth disease using a panel of recombinant, non-structural proteins in ELISA. Vaccine 16:446-459. [DOI] [PubMed] [Google Scholar]
- 284.Mackett, M., G. L. Smith, and B. Moss. 1982. Vaccinia virus: a selectable eukaryotic cloning and expression vector. Proc. Natl. Acad. Sci. USA 79:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Malnou, C. E., T. A. Poyry, R. J. Jackson, and K. M. Kean. 2002. Poliovirus internal ribosome entry segment structure alterations that specifically affect function in neuronal cells: molecular genetic analysis. J. Virol. 76:10617-10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Marc, D., M. Girard, and S. van der Werf. 1991. A Gly1 to Ala substitution in poliovirus capsid protein VP0 blocks its myristoylation and prevents viral assembly. J. Gen. Virol. 72:1151-1157. [DOI] [PubMed] [Google Scholar]
- 287.Marongiu, M. E., A. Pani, M. V. Corrias, M. Sau, and P. La Colla. 1981. Poliovirus morphogenesis. I. Identification of 80S dissociable particles and evidence for the artifactual production of procapsids. J. Virol. 39:341-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Marquardt, O., M. M. Rahman, and B. Freiberg. 2000. Genetic and antigenic variance of foot-and-mouth disease virus type Asia1. Arch Virol. 145:149-157. [DOI] [PubMed] [Google Scholar]
- 289.Marquardt, O., O. C. Straub, R. Ahl, and B. Haas. 1995. Detection of foot-and-mouth disease virus in nasal swabs of asymptomatic cattle by RT-PCR within 24 hours. J. Virol. Methods 53:255-261. [DOI] [PubMed] [Google Scholar]
- 290.Martinez, M. A., N. Verdaguer, M. G. Mateu, and E. Domingo. 1997. Evolution subverting essentiality: dispensability of the cell attachment Arg-Gly-Asp motif in multiply passaged foot-and-mouth disease virus. Proc. Natl. Acad. Sci. USA 94:6798-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Martinez-Salas, E., R. Ramos, E. Lafuente, and S. Lopez de Quinto. 2001. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 82:973-984. [DOI] [PubMed] [Google Scholar]
- 292.Martinez-Salas, E., J. C. Saiz, M. Davila, G. J. Belsham, and E. Domingo. 1993. A single nucleotide substitution in the internal ribosome entry site of foot-and-mouth disease virus leads to enhanced cap-independent translation in vivo. J. Virol. 67:3748-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Mason, P. W., B. Baxt, F. Brown, J. Harber, A. Murdin, and E. Wimmer. 1993. Antibody-complexed foot-and-mouth disease virus, but not poliovirus, can infect normally unsusceptible cells via the Fc receptor. Virology 192:568-577. [DOI] [PubMed] [Google Scholar]
- 294.Mason, P. W., S. V. Bezborodova, and T. M. Henry. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76:9686-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Mason, P. W., M. J. Grubman, and B. Baxt. 2003. Molecular basis of pathogenesis of FMDV. Virus Res. 91:9-32. [DOI] [PubMed] [Google Scholar]
- 296.Mason, P. W., M. E. Piccone, T. S. McKenna, J. Chinsangaram, and M. J. Grubman. 1997. Evaluation of a live-attenuated foot-and-mouth disease virus as a vaccine candidate. Virology 227:96-102. [DOI] [PubMed] [Google Scholar]
- 297.Mason, P. W., E. Rieder, and B. Baxt. 1994. RGD sequence of foot-and-mouth disease virus is essential for infecting cells via the natural receptor but can be bypassed by an antibody-dependent enhancement pathway. Proc. Natl. Acad. Sci. USA 91:1932-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Mateu, M. G. 1995. Antibody recognition of picornaviruses and escape from neutralization: a structural view. Virus Res. 38:1-24. [DOI] [PubMed] [Google Scholar]
- 299.Mateu, M. G., J. A. Camarero, E. Giralt, D. Andreu, and E. Domingo. 1995. Direct evaluation of the immunodominance of a major antigenic site of foot-and-mouth disease virus in a natural host. Virology 206:298-306. [DOI] [PubMed] [Google Scholar]
- 300.Mateu, M. G., J. Hernandez, M. A. Martinez, D. Feigelstock, S. Lea, J. J. Perez, E. Giralt, D. Stuart, E. L. Palma, and E. Domingo. 1994. Antigenic heterogeneity of a foot-and-mouth disease virus serotype in the field is mediated by very limited sequence variation at several antigenic sites. J. Virol. 68:1407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Mayr, G. A., J. Chinsangaram, and M. J. Grubman. 1999. Development of replication-defective adenovirus serotype 5 containing the capsid and 3C protease coding regions of foot-and-mouth disease virus as a vaccine candidate. Virology 263:496-506. [DOI] [PubMed] [Google Scholar]
- 302.Mayr, G. A., V. O'Donnell, J. Chinsangaram, P. W. Mason, and M. J. Grubman. 2001. Immune responses and protection against foot-and-mouth disease virus (FMDV) challenge in swine vaccinated with adenovirus-FMDV constructs. Vaccine 19:2152-2162. [DOI] [PubMed] [Google Scholar]
- 303.Mazzara, G. P., A. Destree, and A. Mahr. 1993. Generation and analysis of vaccinia virus recombinants, p. 557-581. In R. Wu (ed.), Recombinant DNA, vol. 217. Academic Press, Inc., New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 304.McCahon, D., A. M. King, D. S. Roe, W. R. Slade, J. W. Newman, and A. M. Cleary. 1985. Isolation and biochemical characterization of intertypic recombinants of foot-and-mouth disease virus. Virus Res. 3:87-100. [DOI] [PubMed] [Google Scholar]
- 305.McCullough, K. C., J. R. Crowther, W. C. Carpenter, E. Brocchi, L. Capucci, F. De Simone, Q. Xie, and D. McCahon. 1987. Epitopes on foot-and-mouth disease virus particles. I. Topology. Virology 157:516-525. [DOI] [PubMed] [Google Scholar]
- 306.McCullough, K. C., F. De Simone, E. Brocchi, L. Capucci, J. R. Crowther, and U. Kihm. 1992. Protective immune response against foot-and-mouth disease. J. Virol. 66:1835-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.McCullough, K. C., D. Parkinson, and J. R. Crowther. 1988. Opsonization-enhanced phagocytosis of foot-and-mouth disease virus. Immunology 65:187-191. [PMC free article] [PubMed] [Google Scholar]
- 308.McInerney, G. M., A. M. King, N. Ross-Smith, and G. J. Belsham. 2000. Replication-competent foot-and-mouth disease virus RNAs lacking capsid coding sequences. J. Gen. Virol. 81:1699-1702. [DOI] [PubMed] [Google Scholar]
- 309.McKenna, T. S., J. Lubroth, E. Rieder, B. Baxt, and P. W. Mason. 1995. Receptor binding site-deleted foot-and-mouth disease (FMD) virus protects cattle from FMD. J. Virol. 69:5787-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Melchers, W. J., J. G. Hoenderop, H. J. Bruins Slot, C. W. Pleij, E. V. Pilipenko, V. I. Agol, and J. M. Galama. 1997. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 71:686-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Meloen, R. H., J. I. Casal, K. Dalsgaard, and J. P. Langeveld. 1995. Synthetic peptide vaccines: success at last. Vaccine 13:885-886. [DOI] [PubMed] [Google Scholar]
- 314.Meyer, K., A. Petersen, M. Niepmann, and E. Beck. 1995. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J. Virol. 69:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Meyer, R. F., G. D. Babcock, J. F. Newman, T. G. Burrage, K. Toohey, J. Lubroth, and F. Brown. 1997. Baculovirus expressed 2C of foot-and-mouth disease virus has the potential for differentiating convalescent from vaccinated animals. J. Virol. Methods 65:33-43. [DOI] [PubMed] [Google Scholar]
- 316.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 317.Montagnier, L., and F. K. Sanders. 1963. Replicative form of encephalomyocarditis virus ribonucleic acid. Nature 199:664. [DOI] [PubMed] [Google Scholar]
- 318.Moore, D. M., and K. M. Cowan. 1978. Effect of trypsin and chymotrypsin on the polypeptides of large and small plaque variants of foot-and-mouth disease virus: relationship to specific antigenicity and infectivity. J. Gen. Virol. 41:549-562. [DOI] [PubMed] [Google Scholar]
- 319.Moraes, M. P., J. Chinsangaram, M. C. S. Brum, and M. J. Grubman. 2003. Immediate protection of swine from foot-and-mouth disease: a combination of adenoviruses expressing interferon alpha and a foot-and-mouth disease virus subunit vaccine. Vaccine 22:268-279. [DOI] [PubMed] [Google Scholar]
- 320.Moraes, M. P., G. A. Mayr, P. W. Mason, and M. J. Grubman. 2002. Early protection against homologous challenge after a single dose of replication-defective human adenovirus type 5 expressing capsid proteins of foot-and-mouth disease virus (FMDV) strain A24. Vaccine 20:1631-1639. [DOI] [PubMed] [Google Scholar]
- 321.Morasco, B. J., N. Sharma, J. Parilla, and J. B. Flanegan. 2003. Poliovirus cre(2C)-dependent synthesis of VPgpUpU is required for positive- but not negative-strand RNA synthesis. J. Virol. 77:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Morgan-Detjen, B., J. Lucas, and E. Wimmer. 1978. Poliovirus single-stranded RNA and double-stranded RNA: differential infectivity in enucleated cells. J. Virol. 27:582-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Moscufo, N., J. Simons, and M. Chow. 1991. Myristoylation is important at multiple stages in poliovirus assembly. J. Virol. 65:2372-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Moss, E. G., R. E. O'Neill, and V. R. Racaniello. 1989. Mapping of attenuating sequences of an avirulent poliovirus type 2 strain. J. Virol. 63:1884-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Muckelbauer, J. K., M. Kremer, I. Minor, G. Diana, F. J. Dutko, J. Groarke, D. C. Pevear, and M. G. Rossmann. 1995. The structure of coxsackievirus B3 at 3.5 A resolution. Structure 3:653-667. [DOI] [PubMed] [Google Scholar]
- 326.Mulcahy, G., C. Gale, P. Robertson, S. Iyisan, R. D. DiMarchi, and T. R. Doel. 1990. Isotype responses of infected, virus-vaccinated and peptide-vaccinated cattle to foot-and-mouth disease virus. Vaccine 8:249-256. [DOI] [PubMed] [Google Scholar]
- 327.Mulcahy, G., L. A. Pullen, C. Gale, R. D. DiMarchi, and T. R. Doel. 1991. Mouse protection test as a predictor of the protective capacity of synthetic foot-and-mouth disease vaccines. Vaccine 9:19-24. [DOI] [PubMed] [Google Scholar]
- 328.Mulcahy, G., E. Reid, R. D. DiMarchi, C. Gale, and T. R. Doel. 1992. Maturation of functional antibody affinity in animals immunised with synthetic foot-and-mouth disease virus. Res. Vet. Sci. 52:133-140. [DOI] [PubMed] [Google Scholar]
- 329.Murphy, M. L., M. A. Forsyth, G. J. Belsham, and J. S. Salt. 1999. Localization of foot-and-mouth disease virus RNA by in situ hybridization within bovine tissues. Virus Res. 62:67-76. [DOI] [PubMed] [Google Scholar]
- 330.Murray, K. E., and D. J. Barton. 2003. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77:4739-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Nargi, F., E. Kramer, J. Mezencio, J. Zamparo, C. Whetstone, M. H. Van Regenmortel, J. P. Briand, S. Muller, and F. Brown. 1999. Protection of swine from foot-and-mouth disease with one dose of an all-D retro peptide. Vaccine 17:2888-2893. [DOI] [PubMed] [Google Scholar]
- 332.Neff, S., and B. Baxt. 2001. The ability of integrin alpha(v)beta(3) to function as a receptor for foot-and-mouth disease virus is not dependent on the presence of complete subunit cytoplasmic domains. J. Virol. 75:527-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Neff, S., P. W. Mason, and B. Baxt. 2000. High-efficiency utilization of the bovine integrin alpha(v)beta(3) as a receptor for foot-and-mouth disease virus is dependent on the bovine beta(3) subunit. J. Virol. 74:7298-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Neff, S., D. Sa-Carvalho, E. Rieder, P. W. Mason, S. D. Blystone, E. J. Brown, and B. Baxt. 1998. Foot-and-mouth disease virus virulent for cattle utilizes the integrin alpha(v)beta3 as its receptor. J. Virol. 72:3587-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Newman, J. F., B. Cartwright, T. R. Doel, and F. Brown. 1979. Purification and identification of the RNA-dependent RNA polymerase of foot-and-mouth disease virus. J. Gen. Virol. 45:497-507. [DOI] [PubMed] [Google Scholar]
- 336.Newton, S. E., A. R. Carroll, R. O. Campbell, B. E. Clarke, and D. J. Rowlands. 1985. The sequence of foot-and-mouth disease virus RNA to the 5′ side of the poly(C) tract. Gene 40:331-336. [DOI] [PubMed] [Google Scholar]
- 337.Niepmann, M. 1996. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett. 388:39-42. [DOI] [PubMed] [Google Scholar]
- 338.Niepmann, M., A. Petersen, K. Meyer, and E. Beck. 1997. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 71:8330-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Nomoto, A., N. Kitamura, F. Golini, and E. Wimmer. 1977. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc. Natl. Acad. Sci. USA 74:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Nunez, J. I., E. Baranowski, N. Molina, C. M. Ruiz-Jarabo, C. Sanchez, E. Domingo, and F. Sobrino. 2001. A single amino acid substitution in nonstructural protein 3A can mediate adaptation of foot-and-mouth disease virus to the guinea pig. J. Virol. 75:3977-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Ochs, K., R. C. Rust, and M. Niepmann. 1999. Translation initiation factor eIF4B interacts with a picornavirus internal ribosome entry site in both 48S and 80S initiation complexes independently of initiator AUG location. J. Virol. 73:7505-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Ochs, K., A. Zeller, L. Saleh, G. Bassili, Y. Song, A. Sonntag, and M. Niepmann. 2003. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol. 77:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.O'Donnell, V. K., J. M. Pacheco, T. M. Henry, and P. W. Mason. 2001. Subcellular distribution of the foot-and-mouth disease virus 3A protein in cells infected with viruses encoding wild-type and bovine-attenuated forms of 3A. Virology 287:151-162. [DOI] [PubMed] [Google Scholar]
- 346.Oleksiewicz, M. B., A. I. Donaldson, and S. Alexandersen. 2001. Development of a novel real-time RT-PCR assay for quantitation of foot-and-mouth disease virus in diverse porcine tissues. J. Virol. Methods 92:23-35. [DOI] [PubMed] [Google Scholar]
- 347.Onodera, S., and B. A. Phillips. 1987. A novel method for obtaining poliovirus 14 S pentamers from procapsids and their self-assembly into virus-like shells. Virology 159:278-287. [DOI] [PubMed] [Google Scholar]
- 348.Palmenberg, A. C. 1990. Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 44:603-623. [DOI] [PubMed] [Google Scholar]
- 349.Panicali, D., and E. Paoletti. 1982. Construction of poxviruses as cloning vectors: insertion of the thymidine kinase gene from herpes simplex virus into the DNA of infectious vaccinia virus. Proc. Natl. Acad. Sci. USA 79:4927-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Paul, A. V. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227-246. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 351.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 352.Pfaff, E., M. Mussgay, H. O. Bohm, G. E. Schulz, and H. Schaller. 1982. Antibodies against a preselected peptide recognize and neutralize foot and mouth disease virus. EMBO J. 1:869-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Pfaff, E., H. J. Thiel, E. Beck, K. Strohmaier, and H. Schaller. 1988. Analysis of neutralizing epitopes on foot-and-mouth disease virus. J. Virol. 62:2033-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Pfister, T., K. W. Jones, and E. Wimmer. 2000. A cysteine-rich motif in poliovirus protein 2C(ATPase) is involved in RNA replication and binds zinc in vitro. J. Virol. 74:334-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Pfister, T., and E. Wimmer. 1999. Characterization of the nucleoside triphosphatase activity of poliovirus protein 2C reveals a mechanism by which guanidine inhibits poliovirus replication. J. Biol. Chem. 274:6992-7001. [DOI] [PubMed] [Google Scholar]
- 356.Piccone, M. E., E. Rieder, P. W. Mason, and M. J. Grubman. 1995. The foot-and-mouth disease virus leader proteinase gene is not required for viral replication. J. Virol. 69:5376-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Pierschbacher, M. D., and E. Ruoslahti. 1984. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 309:30-33. [DOI] [PubMed] [Google Scholar]
- 358.Pierschbacher, M. D., and E. Ruoslahti. 1984. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc. Natl. Acad. Sci. USA 81:5985-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Pilipenko, E. V., V. M. Blinov, B. K. Chernov, T. M. Dmitrieva, and V. I. Agol. 1989. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 17:5701-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Pilipenko, E. V., V. M. Blinov, L. I. Romanova, A. N. Sinyakov, S. V. Maslova, and V. I. Agol. 1989. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology 168:201-209. [DOI] [PubMed] [Google Scholar]
- 361.Pilipenko, E. V., A. P. Gmyl, S. V. Maslova, Y. V. Svitkin, A. N. Sinyakov, and V. I. Agol. 1992. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell 68:119-131. [DOI] [PubMed] [Google Scholar]
- 362.Pilipenko, E. V., S. V. Maslova, A. N. Sinyakov, and V. I. Agol. 1992. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 20:1739-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 364.Pilipenko, E. V., K. V. Poperechny, S. V. Maslova, W. J. Melchers, H. J. Slot, and V. I. Agol. 1996. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (′kissing') interactions. EMBO J. 15:5428-5436. [PMC free article] [PubMed] [Google Scholar]
- 365.Pinto, A. A., and A. J. M. Garland. 1979. Immune response to virus-infection-associated (VIA) antigen in cattle repeatedly vaccinated with foot-and-mouth disease virus inactivated by formalin or acetylethyleneimine. J. Hyg. 82:41-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Pluimers, F. H., A. M. Akkerman, P. van der Wal, A. Dekker, and A. Bianchi. 2002. Lessons from the foot and mouth disease outbreak in the Netherlands in 2001. Rev. Sci. Tech. Off. Int. Epizoot. 21:711-721. [DOI] [PubMed] [Google Scholar]
- 367.Polacino, P., G. Kaplan, and E. L. Palma. 1985. Homologous interference by a foot-and-mouth disease virus strain attenuated for cattle. Arch. Virol. 86:291-301. [DOI] [PubMed] [Google Scholar]
- 368.Polatnick, J. 1980. Isolation of a foot-and-mouth disease polyuridylic acid polymerase and its inhibition by antibody. J. Virol. 33:774-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Polatnick, J., and R. B. Arlinghaus. 1967. Foot-and-mouth disease virus-induced ribonucleic acid polymerase in baby hamster kidney cells. Virology 31:601-608. [DOI] [PubMed] [Google Scholar]
- 370.Polatnick, J., R. B. Arlinghaus, J. H. Graves, and K. M. Cowan. 1967. Inhibition of cell-free foot-and-mouth disease virus-ribonucleic acid synthesis by antibody. Virology 31:609-615. [DOI] [PubMed] [Google Scholar]
- 371.Polatnick, J., and S. H. Wool. 1983. Association of foot-and-mouth disease virus induced RNA polymerase with host cell organelles. Comp. Immunol. Microbiol. Infect. Dis. 6:265-272. [DOI] [PubMed] [Google Scholar]
- 372.Polatnick, J., and S. H. Wool. 1983. Correlation of surface and internal ultrastructural changes in cells infected with foot-and-mouth disease virus. Can. J. Comp. Med. 47:440-444. [PMC free article] [PubMed] [Google Scholar]
- 373.Porter, D. C., D. C. Ansardi, and C. D. Morrow. 1995. Encapsidation of poliovirus replicons encoding the complete human immunodeficiency virus type 1 gag gene by using a complementation system which provides the P1 capsid protein in trans. J. Virol. 69:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Porter, D. C., D. C. Ansardi, J. Wang, S. McPherson, Z. Moldoveanu, and C. D. Morrow. 1998. Demonstration of the specificity of poliovirus encapsidation using a novel replicon which encodes enzymatically active firefly luciferase. Virology 243:1-11. [DOI] [PubMed] [Google Scholar]
- 375.Prevec, L., M. Schneider, K. L. Rosenthal, L. W. Belbeck, J. B. Derbyshire, and F. L. Graham. 1989. Use of human adenovirus-based vectors for antigen expression in animals. J. Gen. Virol. 70:429-434. [DOI] [PubMed] [Google Scholar]
- 376.Qin, X. Q., N. Tao, A. Dergay, P. Moy, S. Fawell, A. Davis, J. M. Wilson, and J. Barsoum. 1998. Interferon-beta gene therapy inhibits tumor formation and causes regression of established tumors in immune-deficient mice. Proc. Natl. Acad. Sci. USA 95:14411-14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ragot, T., S. Finerty, P. E. Watkins, M. Perricaudet, and A. J. Morgan. 1993. Replication-defective recombinant adenovirus expressing the Epstein-Barr virus (EBV) envelope glycoprotein gp340/220 induces protective immunity against EBV-induced lymphomas in the cottontop tamarin. J. Gen. Virol. 74:501-507. [DOI] [PubMed] [Google Scholar]
- 378.Ray, D. K., U. K. Bhattacharyya, B. Chowdhury, P. Dasgupta, and A. K. Bhattacharyya. 1989. Studies on a severe outbreak of foot-and-mouth disease in regularly vaccinated cross-exotic dairy cattle in West Bengal (India). Indian J. Anim. Health 28:50-55. [Google Scholar]
- 379.Reid, S. M., S. S. Grierson, N. P. Ferris, G. H. Hutchings, and S. Alexandersen. 2003. Evaluation of automated RT-PCR to accelerate the laboratory diagnosis of foot-and-mouth disease virus. J. Virol. Methods 107:129-139. [DOI] [PubMed] [Google Scholar]
- 380.Rieder, E., A. Berinstein, B. Baxt, A. Kang, and P. W. Mason. 1996. Propagation of an attenuated virus by design: engineering a novel receptor for a noninfectious foot-and-mouth disease virus. Proc. Natl. Acad. Sci. USA 93:10428-10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Rieder, E., T. Bunch, F. Brown, and P. W. Mason. 1993. Genetically engineered foot-and-mouth disease viruses with poly(C) tracts of two nucleotides are virulent in mice. J. Virol. 67:5139-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Rigden, R. C., C. P. Carrasco, P. V. Barnett, A. Summerfield, and K. C. McCullough. 2003. Innate immune responses following emergency vaccination against foot-and-mouth disease virus in pigs. Vaccine 21:1466-1477. [DOI] [PubMed] [Google Scholar]
- 384.Rigden, R. C., C. P. Carrasco, A. Summerfield, and K. C. McCullough. 2002. Macrophage phagocytosis of foot-and-mouth disease virus may create infectious carriers. Immunology 106:537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Rivera, V. M., J. D. Welsh, and J. V. Maizel, Jr. 1988. Comparative sequence analysis of the 5′ noncoding region of the enteroviruses and rhinoviruses. Virology 165:42-50. [DOI] [PubMed] [Google Scholar]
- 386.Roberts, P. J., and G. J. Belsham. 1995. Identification of critical amino acids within the foot-and-mouth disease virus leader protein, a cysteine protease. Virology 213:140-146. [DOI] [PubMed] [Google Scholar]
- 387.Robertson, B. H., M. J. Grubman, G. N. Weddell, D. M. Moore, J. D. Welsh, T. Fischer, D. J. Dowbenko, D. G. Yansura, B. Small, and D. G. Kleid. 1985. Nucleotide and amino acid sequence coding for polypeptides of foot-and-mouth disease virus type A12. J. Virol. 54:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Robertson, B. H., D. M. Moore, M. J. Grubman, and D. G. Kleid. 1983. Identification of an exposed region of the immunogenic capsid polypeptide VP1 on foot-and-mouth disease virus. J. Virol. 46:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Robertson, B. H., D. O. Morgan, D. M. Moore, M. J. Grubman, J. Card, T. Fischer, G. Weddell, D. Dowbenko, and D. Yansura. 1983. Identification of amino acid and nucleotide sequence of the foot-and-mouth disease virus RNA polymerase. Virology 126:614-623. [DOI] [PubMed] [Google Scholar]
- 390.Rodriguez, P. L., and L. Carrasco. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105-8110. [PubMed] [Google Scholar]
- 391.Roehl, H. H., and B. L. Semler. 1995. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J. Virol. 69:2954-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Rohll, J. B., D. H. Moon, D. J. Evans, and J. W. Almond. 1995. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 69:7835-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Roivainen, M., T. Hyypia, L. Piirainen, N. Kalkkinen, G. Stanway, and T. Hovi. 1991. RGD-dependent entry of coxsackievirus A9 into host cells and its bypass after cleavage of VP1 protein by intestinal proteases. J. Virol. 65:4735-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Roivainen, M., L. Piirainen, T. Hovi, I. Virtanen, T. Riikonen, J. Heino, and T. Hyypia. 1994. Entry of coxsackievirus A9 into host cells: specific interactions with alpha v beta 3 integrin, the vitronectin receptor. Virology 203:357-365. [DOI] [PubMed] [Google Scholar]
- 395.Rombaut, B., A. Foriers, and A. Boeye. 1991. In vitro assembly of poliovirus 14 S subunits: identification of the assembly promoting activity of infected cell extracts. Virology 180:781-787. [DOI] [PubMed] [Google Scholar]
- 396.Roosien, J., G. J. Belsham, M. D. Ryan, A. M. King, and J. M. Vlak. 1990. Synthesis of foot-and-mouth disease virus capsid proteins in insect cells using baculovirus expression vectors. J. Gen. Virol. 71:1703-1711. [DOI] [PubMed] [Google Scholar]
- 397.Rosas, M. F., E. Martinez-Salas, and F. Sobrino. 2003. Stable expression of antisense RNAs targeted to the 5′ non-coding region confers heterotypic inhibition to foot-and-mouth disease virus infection. J. Gen. Virol. 84:393-402. [DOI] [PubMed] [Google Scholar]
- 398.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, et al. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 399.Rowlands, D. J., B. Cartwright, and F. Brown. 1969. Evidence for an internal antigen in foot-and-mouth disease virus. J. Gen. Virol. 4:479-487. [DOI] [PubMed] [Google Scholar]
- 400.Rowlands, D. J., D. V. Sangar, and F. Brown. 1974. A comparative chemical and serological study of the full and empty particles of foot-and-mouth disease virus. J. Gen. Virol. 26:227-238. [DOI] [PubMed] [Google Scholar]
- 401.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, and P. H. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 402.Rueckert, R. R., and E. Wimmer. 1984. Systematic nomenclature of picornavirus proteins. J. Virol. 50:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Ruoslahti, E. 1996. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12:697-715. [DOI] [PubMed] [Google Scholar]
- 404.Russell, W. C. 2000. Update on adenovirus and its vectors. J. Gen. Virol. 81:2573-2604. [DOI] [PubMed] [Google Scholar]
- 405.Rust, R. C., K. Ochs, K. Meyer, E. Beck, and M. Niepmann. 1999. Interaction of eukaryotic initiation factor eIF4B with the internal ribosome entry site of foot-and-mouth disease virus is independent of the polypyrimidine tract-binding protein. J. Virol. 73:6111-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Rweyemamu, M. M., and Y. Leforban. 1999. Foot-and-mouth disease and international development. Adv. Virus Res. 53:111-126. [DOI] [PubMed] [Google Scholar]
- 407.Rweyemamu, M. M., G. Terry, and T. W. Pay. 1979. Stability and immunogenicity of empty particles of foot-and-mouth disease virus. Arch. Virol. 59:69-79. [DOI] [PubMed] [Google Scholar]
- 408.Ryan, M. D., A. M. King, and G. P. Thomas. 1991. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 72:2727-2732. [DOI] [PubMed] [Google Scholar]
- 409.Sa-Carvalho, D., E. Rieder, B. Baxt, R. Rodarte, A. Tanuri, and P. W. Mason. 1997. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J. Virol. 71:5115-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Sagedahl, A., A. T. Giraudo, P. A. De Mello, I. E. Bergmann, J. L. La Torre, and E. A. Scodeller. 1987. Biochemical characterization of an aphthovirus type C3 strain Resende attenuated for cattle by serial passages in chicken embryos. Virology 157:366-374. [DOI] [PubMed] [Google Scholar]
- 411.Saiz, J. C., and E. Domingo. 1996. Virulence as a positive trait in viral persistence. J. Virol. 70:6410-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Saiz, J. C., A. Rodriguez, M. Gonzalez, F. Alonso, and F. Sobrino. 1992. Heterotypic lymphoproliferative response in pigs vaccinated with foot-and-mouth disease virus. Involvement of isolated capsid proteins. J. Gen. Virol. 73:2601-2607. [DOI] [PubMed] [Google Scholar]
- 413.Saiz, M., S. Gomez, E. Martinez-Salas, and F. Sobrino. 2001. Deletion or substitution of the aphthovirus 3′ NCR abrogates infectivity and virus replication. J. Gen. Virol. 82:93-101. [DOI] [PubMed] [Google Scholar]
- 414.Saleh, L., R. C. Rust, R. Fullkrug, E. Beck, G. Bassili, K. Ochs, and M. Niepmann. 2001. Functional interaction of translation initiation factor eIF4G with the foot-and-mouth disease virus internal ribosome entry site. J. Gen. Virol. 82:757-763. [DOI] [PubMed] [Google Scholar]
- 415.Salt, J. S. 1993. The carrier state in foot and mouth disease—an immunological review. Br. Vet. J. 149:207-223. [DOI] [PubMed] [Google Scholar]
- 416.Salt, J. S. 1998. Persistent infection with foot-and-mouth disease virus. Top. Trop. Virol. 1:77-128. [Google Scholar]
- 417.Salt, J. S., G. Mulcahy, and R. P. Kitching. 1996. Isotype-specific antibody responses to foot-and-mouth disease virus in sera and secretions of “carrier” and “non-carrier” cattle. Epidemiol. Infect. 117:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Sangar, D. V., D. N. Black, D. J. Rowlands, T. J. Harris, and F. Brown. 1980. Location of the initiation site for protein synthesis on foot-and-mouth disease virus RNA by in vitro translation of defined fragments of the RNA. J. Virol. 33:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Sangar, D. V., S. E. Newton, D. J. Rowlands, and B. E. Clarke. 1987. All foot and mouth disease virus serotypes initiate protein synthesis at two separate AUGs. Nucleic Acids Res. 15:3305-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Sangar, D. V., D. J. Rowlands, T. J. Harris, and F. Brown. 1977. Protein covalently linked to foot-and-mouth disease virus RNA. Nature 268:648-650. [DOI] [PubMed] [Google Scholar]
- 421.Sangare, O., A. D. Bastos, O. Marquardt, E. H. Venter, W. Vosloo, and G. R. Thomson. 2001. Molecular epidemiology of serotype O foot-and-mouth disease virus with emphasis on West and South Africa. Virus Genes 22:345-351. [DOI] [PubMed] [Google Scholar]
- 422.Santodonato, L., M. Ferrantini, F. Palombo, L. Aurisicchio, P. Delmastro, N. La Monica, S. Di Marco, G. Ciliberto, M. X. Du, M. W. Taylor, and F. Belardelli. 2001. Antitumor activity of recombinant adenoviral vectors expressing murine IFN-alpha in mice injected with metastatic IFN-resistant tumor cells. Cancer Gene Ther. 8:63-72. [DOI] [PubMed] [Google Scholar]
- 423.Sanz-Parra, A., M. A. Jimenez-Clavero, M. M. Garcia-Briones, E. Blanco, F. Sobrino, and V. Ley. 1999. Recombinant viruses expressing the foot-and-mouth disease virus capsid precursor polypeptide (P1) induce cellular but not humoral antiviral immunity and partial protection in pigs. Virology 259:129-134. [DOI] [PubMed] [Google Scholar]
- 424.Sanz-Parra, A., B. Vazquez, F. Sobrino, S. J. Cox, V. Ley, and J. S. Salt. 1999. Evidence of partial protection against foot-and-mouth disease in cattle immunized with a recombinant adenovirus vector expressing the precursor polypeptide (P1) of foot-and-mouth disease virus capsid proteins. J. Gen. Virol. 80:671-679. [DOI] [PubMed] [Google Scholar]
- 425.Sasaki, J., and K. Taniguchi. 2003. The 5′-end sequence of the genome of Aichi virus, a picornavirus, contains an element critical for viral RNA encapsidation. J. Virol. 77:3542-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Saunders, K., and A. M. King. 1982. Guanidine-resistant mutants of aphthovirus induce the synthesis of an altered nonstructural polypeptide, P34. J. Virol. 42:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Saunders, K., A. M. King, D. McCahon, J. W. Newman, W. R. Slade, and S. Forss. 1985. Recombination and oligonucleotide analysis of guanidine-resistant foot-and-mouth disease virus mutants. J. Virol. 56:921-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Schneider-Schaulies, J. 2000. Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 81:1413-1429. [DOI] [PubMed] [Google Scholar]
- 430.Scudamore, J. M., and D. M. Harris. 2002. Control of foot and mouth disease: lessons from the experience of the outbreak in Great Britain in 2001. Rev. Sci. Tech. Off. Int. Epizoot. 21:699-710. [DOI] [PubMed] [Google Scholar]
- 431.Sekiguchi, K., A. J. Franke, and B. Baxt. 1982. Competition for cellular receptor sites among selected aphthoviruses. Arch. Virol. 74:53-64. [DOI] [PubMed] [Google Scholar]
- 432.Sellers, R. F. 1963. Multiplication, interferon production and sensitivity of virulent and attenuated strains of the virus of foot-and-mouth disease. Nature 198:1228-1229. [DOI] [PubMed] [Google Scholar]
- 433.Sevilla, N., and E. Domingo. 1996. Evolution of a persistent aphthovirus in cytolytic infections: partial reversion of phenotypic traits accompanied by genetic diversification. J. Virol. 70:6617-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Shen, F., P. D. Chen, A. M. Walfield, J. Ye, J. House, F. Brown, and C. Y. Wang. 1999. Differentiation of convalescent animals from those vaccinated against foot-and-mouth disease by a peptide ELISA. Vaccine 17:3039-3049. [DOI] [PubMed] [Google Scholar]
- 435.Skern, T., I. Fita, and A. Guarne. 1998. A structural model of picornavirus leader proteinases based on papain and bleomycin hydrolase. J. Gen. Virol. 79:301-307. [DOI] [PubMed] [Google Scholar]
- 436.Skinner, M. A., V. R. Racaniello, G. Dunn, J. Cooper, P. D. Minor, and J. W. Almond. 1989. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 207:379-392. [DOI] [PubMed] [Google Scholar]
- 437.Sobrino, F., M. Davila, J. Ortin, and E. Domingo. 1983. Multiple genetic variants arise in the course of replication of foot-and-mouth disease virus in cell culture. Virology 128:310-318. [DOI] [PubMed] [Google Scholar]
- 438.Sorensen, J. H., D. K. Mackay, C. O. Jensen, and A. I. Donaldson. 2000. An integrated model to predict the atmospheric spread of foot-and-mouth disease virus. Epidemiol. Infect. 124:577-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Sorensen, K. J., K. G. Madsen, E. S. Madsen, J. S. Salt, J. Nqindi, and D. K. Mackay. 1998. Differentiation of infection from vaccination in foot-and-mouth disease by the detection of antibodies to the non-structural proteins 3D, 3AB and 3ABC in ELISA using antigens expressed in baculovirus. Arch. Virol. 143:1461-1476. [DOI] [PubMed] [Google Scholar]
- 440.Strebel, K., and E. Beck. 1986. A second protease of foot-and-mouth disease virus. J. Virol. 58:893-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Strohmaier, K., R. Franze, and K. H. Adam. 1982. Location and characterization of the antigenic portion of the FMDV immunizing protein. J. Gen. Virol. 59:295-306. [DOI] [PubMed] [Google Scholar]
- 442.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Sutmoller, P., S. S. Barteling, R. C. Olascoaga, and K. J. Sumption. 2003. Control and eradication of foot-and-mouth disease. Virus Res. 91:101-144. [DOI] [PubMed] [Google Scholar]
- 444.Sutmoller, P., and J. W. McVicar. 1976. Pathogenesis of foot-and-mouth disease: the lung as an additional portal of entry of the virus. J. Hyg (London). 77:235-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Sutmoller, P., J. W. McVicar, and G. E. Cottral. 1968. The epizootiological importance of foot-and-mouth disease carriers. I. Experimentally produced foot-and-mouth disease carriers in susceptible and immune cattle. Arch. Gesamte Virusforsch. 23:227-235. [DOI] [PubMed] [Google Scholar]
- 446.Svitkin, Y. V., and N. Sonenberg. 2003. Cell-free synthesis of encephalomyocarditis virus. J. Virol. 77:6551-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Taboga, O., C. Tami, E. Carrillo, J. I. Nunez, A. Rodriguez, J. C. Saiz, E. Blanco, M. L. Valero, X. Roig, J. A. Camarero, D. Andreu, M. G. Mateu, E. Giralt, E. Domingo, F. Sobrino, and E. L. Palma. 1997. A large-scale evaluation of peptide vaccines against foot-and-mouth disease: lack of solid protection in cattle and isolation of escape mutants. J. Virol. 71:2606-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Tami, C., O. Taboga, A. Berinstein, J. I. Nunez, E. L. Palma, E. Domingo, F. Sobrino, and E. Carrillo. 2003. Evidence of the coevolution of antigenicity and host cell tropism of foot-and-mouth disease virus in vivo. J. Virol. 77:1219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Tamkun, J. W., D. W. DeSimone, D. Fonda, R. S. Patel, C. Buck, A. F. Horwitz, and R. O. Hynes. 1986. Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell 46:271-282. [DOI] [PubMed] [Google Scholar]
- 450.Tesar, M., H. G. Berger, and O. Marquardt. 1989. Serological probes for some foot-and-mouth disease virus nonstructural proteins. Virus Genes 3:29-44. [DOI] [PubMed] [Google Scholar]
- 451.Tesar, M., and O. Marquardt. 1990. Foot-and-mouth disease virus protease 3C inhibits cellular transcription and mediates cleavage of histone H3. Virology 174:364-374. [DOI] [PubMed] [Google Scholar]
- 452.Teterina, N. L., K. Bienz, D. Egger, A. E. Gorbalenya, and E. Ehrenfeld. 1997. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology 237:66-77. [DOI] [PubMed] [Google Scholar]
- 453.Thomas, A. A., R. J. Woortmeijer, W. Puijk, and S. J. Barteling. 1988. Antigenic sites on foot-and-mouth disease virus type A10. J. Virol. 62:2782-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Thompson, D., P. Muriel, D. Russell, P. Osborne, A. Bromley, M. Rowland, S. Creigh-Tyte, and C. Brown. 2002. Economic costs of the foot-and-mouth disease outbreak in the United Kingdom in 2001. Rev. Sci. Tech. Off. Int. Epizoot. 21:675-687. [DOI] [PubMed] [Google Scholar]
- 455.Thomson, G. R., W. Vosloo, and A. D. Bastos. 2003. Foot and mouth disease in wildlife. Virus Res. 91:145-161. [DOI] [PubMed] [Google Scholar]
- 456.Tiley, L., A. M. King, and G. J. Belsham. 2003. The foot-and-mouth disease virus cis-acting replication element (cre) can be complemented in trans within infected cells. J. Virol. 77:2243-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Toja, M., C. Escarmis, and E. Domingo. 1999. Genomic nucleotide sequence of a foot-and-mouth disease virus clone and its persistent derivatives. Implications for the evolution of viral quasispecies during a persistent infection. Virus Res. 64:161-171. [DOI] [PubMed] [Google Scholar]
- 458.Top, F. H., E. L. Buescher, W. H. Bancroft, and P. K. Russell. 1971. Immunization with live types 7 and 4 adenovirus vaccines. II. Antibody response and protective effect against acute respiratory disease due to adenovirus type 7. J. Infect. Dis. 124:155-160. [DOI] [PubMed] [Google Scholar]
- 459.Tosh, C., D. Hemadri, and A. Sanyal. 2002. Evidence of recombination in the capsid-coding region of type A foot-and-mouth disease virus. J. Gen. Virol. 83:2455-2460. [DOI] [PubMed] [Google Scholar]
- 460.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 461.Towner, J. S., M. M. Mazanet, and B. L. Semler. 1998. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J. Virol. 72:7191-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Trautman, R., and P. Sutmoller. 1971. Detection and properties of a genomic masked viral particle consisting of foot-and-mouth disease virus nucleic acid in bovine enterovirus protein capsid. Virology 44:537-543. [DOI] [PubMed] [Google Scholar]
- 463.Vakharia, V. N., M. A. Devaney, D. M. Moore, J. J. Dunn, and M. J. Grubman. 1987. Proteolytic processing of foot-and-mouth disease virus polyproteins expressed in a cell-free system from clone-derived transcripts. J. Virol. 61:3199-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.van Bekkum, J. G., H. S. Frenkel, H. H. J. Frederiks, and S. Frenkel. 1959. Observations on the carrier state of cattle exposed to foot-and-mouth disease virus. Tijdschr. Diergeneeskd. 84:1159-1164. [Google Scholar]
- 465.van Kuppeveld, F. J., J. G. Hoenderop, R. L. Smeets, P. H. Willems, H. B. Dijkman, J. M. Galama, and W. J. Melchers. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16:3519-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.van Kuppeveld, F. J., W. J. Melchers, K. Kirkegaard, and J. R. Doedens. 1997. Structure-function analysis of coxsackie B3 virus protein 2B. Virology 227:111-118. [DOI] [PubMed] [Google Scholar]
- 467.Verlinden, Y., A. Cuconati, E. Wimmer, and B. Rombaut. 2000. Cell-free synthesis of poliovirus: 14S subunits are the key intermediates in the encapsidation of poliovirus RNA. J. Gen. Virol. 81:2751-2754. [DOI] [PubMed] [Google Scholar]
- 468.Vilcek, J., and G. C. Sen. 1996. Interferons and other cytokines, p. 375-399. In B. N. Fields, D. M. Knipe, and P. H. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 469.Waldmann, D., K. Kobe, and G. Pyl. 1937. Die aktive Immunisierung des Rindes gegen Maul- und Klauenseuche mittels Formolimpfstoff. Zentbl. Bakteriol. Parasitenkd. Infektkhrankh. 138:401-412. [Google Scholar]
- 470.Walter, B. L., T. B. Parsley, E. Ehrenfeld, and B. L. Semler. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008-12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Ward, G., E. Rieder, and P. W. Mason. 1997. Plasmid DNA encoding replicating foot-and-mouth disease virus genomes induces antiviral immune responses in swine. J. Virol. 71:7442-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Wigdorovitz, A., C. Carrillo, M. J. Dus Santos, K. Trono, A. Peralta, M. C. Gomez, R. D. Rios, P. M. Franzone, A. M. Sadir, J. M. Escribano, and M. V. Borca. 1999. Induction of a protective antibody response to foot and mouth disease virus in mice following oral or parenteral immunization with alfalfa transgenic plants expressing the viral structural protein VP1. Virology 255:347-353. [DOI] [PubMed] [Google Scholar]
- 473.Wigdorovitz, A., D. M. Perez Filgueira, N. Robertson, C. Carrillo, A. M. Sadir, T. J. Morris, and M. V. Borca. 1999. Protection of mice against challenge with foot and mouth disease virus (FMDV) by immunization with foliar extracts from plants infected with recombinant tobacco mosaic virus expressing the FMDV structural protein VP1. Virology 264:85-91. [DOI] [PubMed] [Google Scholar]
- 474.Wilson, V., P. Taylor, and U. Desselberger. 1988. Crossover regions in foot-and-mouth disease virus (FMDV) recombinants correspond to regions of high local secondary structure. Arch. Virol. 102:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Wimmer, E. 1982. Genome-linked proteins of viruses. Cell 28:199-201. [DOI] [PubMed] [Google Scholar]
- 476.Wimmer, E. 1994. Introduction, p. 1-13. In E. Wimmer (ed.), Cellular receptors for animal viruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 477.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]
- 478.Wong, H. T., S. C. Cheng, E. W. Chan, Z. T. Sheng, W. Y. Yan, Z. X. Zheng, and Y. Xie. 2000. Plasmids encoding foot-and-mouth disease virus VP1 epitopes elicited immune responses in mice and swine and protected swine against viral infection. Virology 278:27-35. [DOI] [PubMed] [Google Scholar]
- 479.Wong, H. T., S. C. Cheng, F. W. Sin, E. W. Chan, Z. T. Sheng, and Y. Xie. 2002. A DNA vaccine against foot-and-mouth disease elicits an immune response in swine which is enhanced by co-administration with interleukin-2. Vaccine 20:2641-2647. [DOI] [PubMed] [Google Scholar]
- 480.Wu, Q., M. C. S. Brum, L. Caron, M. Koster, and M. J. Grubman. 2003. Adenovirus-mediated type I interferon expression delays and reduces disease signs in cattle challenged with foot-and-mouth disease virus. J. Int. Cyt. Res. 23:359-368. [DOI] [PubMed] [Google Scholar]
- 481.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Xiang, Z. Q., Y. Yang, J. M. Wilson, and H. C. Ertl. 1996. A replication-defective human adenovirus recombinant serves as a highly efficacious vaccine carrier. Virology 219:220-227. [DOI] [PubMed] [Google Scholar]
- 483.Xiao, C., C. M. Bator, V. D. Bowman, E. Rieder, Y. He, B. Hebert, J. Bella, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2001. Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J. Virol. 75:2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Xie, Q. C., D. McCahon, J. R. Crowther, G. J. Belsham, and K. C. McCullough. 1987. Neutralization of foot-and-mouth disease virus can be mediated through any of at least three separate antigenic sites. J. Gen. Virol. 68:1637-1647. [DOI] [PubMed] [Google Scholar]
- 485.Yafal, A. G., and E. L. Palma. 1979. Morphogenesis of foot-and-mouth disease virus. I. Role of procapsids as virion precursors. J. Virol. 30:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Yang, P. C., R. M. Chu, W. B. Chung, and H. T. Sung. 1999. Epidemiological characteristics and financial costs of the 1997 foot-and-mouth disease epidemic in Taiwan. Vet. Rec. 145:731-734. [DOI] [PubMed] [Google Scholar]
- 487.Yin, J., A. V. Paul, E. Wimmer, and E. Rieder. 2003. Functional dissection of a poliovirus cis-acting replication element [PV-cre(2C)]: analysis of single- and dual-cre viral genomes and proteins that bind specifically to PV-cre RNA. J. Virol. 77:5152-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Zhang, Z. D., G. Hutching, P. Kitching, and S. Alexandersen. 2002. The effects of gamma interferon on replication of foot-and-mouth disease virus in persistently infected bovine cells. Arch. Virol. 147:2157-2167. [DOI] [PubMed] [Google Scholar]
- 489.Zhang, Z. D., and R. P. Kitching. 2001. The localization of persistent foot and mouth disease virus in the epithelial cells of the soft palate and pharynx. J. Comp. Pathol. 124:89-94. [DOI] [PubMed] [Google Scholar]
- 490.Zhao, Q., J. M. Pacheco, and P. W. Mason. 2003. Evaluation of genetically engineered derivatives of a Chinese strain of foot-and-mouth disease virus reveals a novel cell-binding site which functions in cell culture and in animals. J. Virol. 77:3269-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]