Abstract
The three-dimensional cardiac electrical imaging (3DCEI) technique was previously developed to estimate the initiation site(s) of cardiac activation and activation sequence from the noninvasively measured body surface potential maps (BSPMs). The aim of the present study was to develop and evaluate the capability of 3DCEI in mapping the transmural distribution of extracellular potentials and localizing initiation sites of ventricular activation in an in vivo animal model. A control swine model (n=10) was employed in this study. The heart-torso volume conductor model and the excitable heart model were constructed based on each animal's pre-operative MR images and a priori known physiological knowledge. Body surface potential mapping and intracavitary noncontact mapping (NCM) were simultaneously conducted during acute ventricular pacing. The 3DCEI analysis was then applied on the recorded BSPMs. The estimated initiation sites were compared to the precise pacing sites; as a subset of the mapped transmural potentials by 3DCEI, the electrograms on the left ventricular endocardium were compared to the corresponding output of the NCM system. Over the 16 LV and 48 RV pacing studies, the averaged localization error was 6.1±2.3 mm, and the averaged correlation coefficient between the estimated endocardial electrograms by 3DCEI and from the NCM system was 0.62±0.09. The present results demonstrate that the 3DCEI approach can well localize the sites of initiation of ectopic beats and can obtain physiologically reasonable transmural potentials in an in vivo setting during focal ectopic beats. This study suggests the feasibility of tomographic mapping of 3D ventricular electrograms from the body surface recordings.
Index Terms: 3D Cardiac electrical imaging, Transmural potential, Electrocardiophysiology, Body surface potential mapping, Noncontact mapping
I. Introduction
The spatial-temporal mapping of electric potentials in the heart has direct benefits on diagnosis and treatment of cardiac arrhythmias [1]–[3]. To date, endocardial mapping is routinely performed in electrophysiology (EP) labs for management of cardiac diseases. The CARTO® (Biosense Webster, Belgium) system conducts direct electro-anotomic mapping to construct electrical potential and activation maps over the endocardial surface by sequentially acquiring endocardial electrograms [4]–[5]. Alternatively, the Ensite system (St Jude Medical, Inc.) performs functional non-contact mapping (NCM) by employing a multi-electrode array (MEA) [6]–[7]. In the NCM system, the endocardial potentials are reconstructed from the electrograms recorded by the MEA via mathematical calculation.
Due to the invasive nature of the endocardial mapping techniques, the patient might be kept in the EP labs under considerable time and fluoroscopic scanning, and thus the patient’s hemodynamic instability may become a concern. Alternatively, the idea of noninvasively mapping the epicardial potentials from body surface potential maps (BSPMs) has been investigated for decades because of its potential of more conveniently guiding the management of arrhythmias. Similar to the functional mapping algorithms employed in the NCM system, by solving the so-called "inverse problem", the epicardial potentials are estimated from the BSPMs recorded by an electrode array [8]–[12]. Positive mapping results have been reported in both animal studies [11] and clinical studies [13]. Most recently, a successful human study on mapping epicardial potentials during ventricular tachycardia was reported [14].Besides mapping the endocardial/epicardial potentials, noninvasively estimating the heart surface activation sequence from the BSPMs is another popular topic regarding functional mapping [15]–[18].
Though proved successful, the mapping results using the above methods are all on the heart surfaces. An alternative approach - three-dimensional cardiac electrical imaging (3DCEI) - has been proposed by He and his coworkers, to extend the functional mapping results from the heart surfaces to the three-dimensional (3D) myocardial volume to accomplish electric tomographic imaging of cardiac electric activity [19]. For instance, ventricular arrhythmias may arise from intramural regions within the myocardial tissue, and the 3D mapping methods may better characterize and image the depth information of the initiation site(s) and the intramural delay of activation, which are hard to be characterized by heart surface mapping methods [20]. The 3DCEI technique has been previously investigated to localize the initiation of activation [21] and map the 3D activation sequence [22]–[24]. Promising validation results have been reported in a rabbit model [25]–[28] and in a swine model [29]. In the present study, we further expanded the functions of 3DCEI to noninvasively map the transmural extracellular potentials at any endocardial, epicardial or intramyocardial site. Animal studies in a control swine model were conducted to experimentally evaluate the performance of this newly proposed 3D functional tomographic mapping technique. Ventricular pacing at different locations was performed to simulate focal arrhythmogenic activities. The 3D locations of the initiation sites and the transmural electrograms were estimated by employing the 3DCEI approach and the results were evaluated with the aid of magnetic resonance imaging (MRI) and the Ensite® NCM system.
II. Methods
A. Experimental protocols
The experimental protocol was approved by the Institutional Animal Care and Use Committee at the University of Minnesota (Protocol #: 0703A04942). A control swine model (n=10) was employed in the present study. The surgical preparation of these animals for hemodynamic and electrical monitoring has been previously reported [30]. During the comparative mapping studies, each animal was intubated and anesthetized using isoflurane (1.5 minimum alveolar concentration); additionally, all were mechanically ventilated with 65% air and 35% O2 to maintain a PaCO2 of 40 ± 2 mmHg.
Right ventricular pacing was conducted with the active-fixation pacing leads implanted at either the right ventricular apex (RVA) or the RV septum (RVS) (Model 3830, Medtronic, Inc., USA). Left ventricular pacing was accomplished by a quadripolar EP catheter (MarinR, Medtronic, Inc.) at different locations on the endocardial surface.
For each animal, the pre-operative magnetic resonance imaging (MRI; ECG gated to end diastole) was acquired approximately 5–7 days before the in vivo mapping experiment so to obtain the needed anatomical geometry information. On the mapping study day, body surface potential mapping and intracavitary mapping (by the EnSite® NCM system) were simultaneously performed during the various pacing protocols. The multi-electrode array from the NCM system was put in the chamber of the left ventricle in each study. The spatial locations of the LV endocardial pacing sites were recorded on the reconstructed LV geometry acquired with the NCM system. The location of the multi-electrode array in the LV chamber was also recorded by the NCM system. For the body surface potential mapping, up to 100 disposable electrodes were placed on the shaved anterolateral chest. The 3D locations of the body surface electrodes and the fiducial points were digitized using a radio frequency localizer (Fastrak, Polhemus Inc.). The body surface potentials were referenced to that at the Wilson central terminal. After the completion of the data collection protocol, each heart was removed, perfused and fixed in formalin with the RV pacing leads remaining in place. These isolated hearts were again MRI scanned to measure the precise locations of the RV pacing sites.
B. Principles of the 3D cardiac electrical imaging
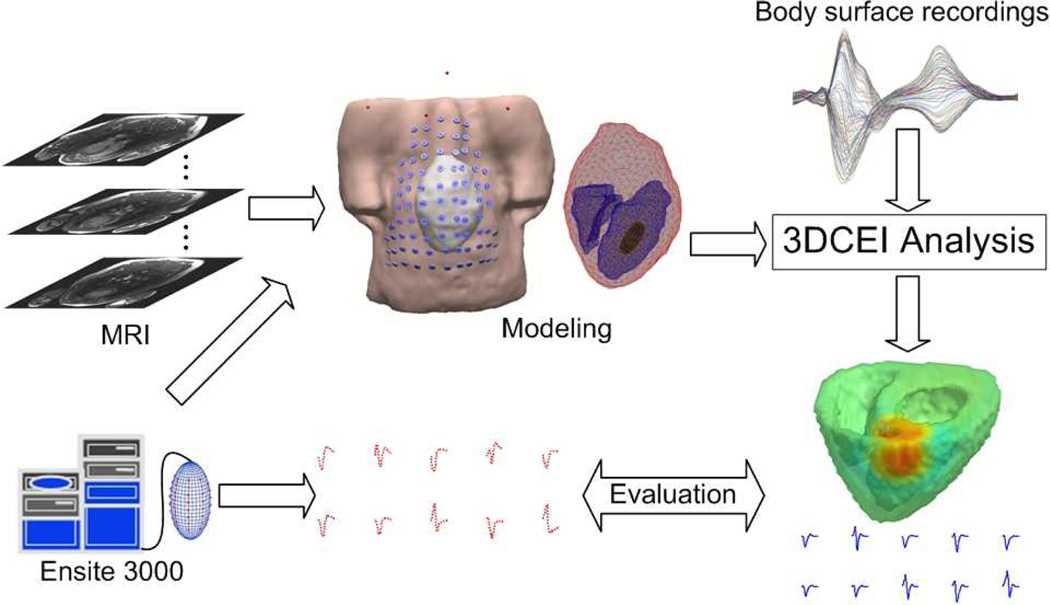
A schematic diagram of 3DCEI is shown in Fig. 1 In brief, for each animal, a heart-excitation model and a heart-torso volume conductor model were constructed based on the pre-operative MRI scans and the a priori physiological knowledge of the swine heart. The MR images were segmented to obtain the detailed cardiac geometry and the volume between the epicardium and the endocardia was discretized into over 100,000 cellular units (spatial resolution of 1.5 mm) to constitute the cellular-automaton heart model. A universal waveform of the action potential [31] was assigned to every cellular unit in the ventricles, which starts with the very steep phase 0 and followed by the transient phase 1. Since only the activation of the heart was studied in the present study, the diversity of phase 2 as well as phase 3 had little effect and so it was neglected. The other parameters of each cellular unit including cell type, conductivity tensor and conduction velocity were assigned as well. The entire heart excitation process could then be simulated and the corresponding BSPMs were calculated by applying the bidomain theory using the finite element method (FEM) [32]. In detail, once the activation time τ at location k in the ventricles and the waveform of the trans-membrane action potential at k are known, the equivalent current density at time instant t at the location k can be calculated as follows,
| (1) |
where σi is the intracellular conductivity tensor, V the trans-membrane potential. The equivalent current densities at thousands of uniformly distributed locations inside the myocardium were calculated representing the ventricular activities during activation. Based on the bidomain theory, the field potential ϕ can be calculated from those current sources employing Poisson’s equation [32],
| (2) |
where σe is the extracellular conductivity tensor. Eq. (2) was numerically solved using the finite element method (FEM). By using the variational principle and applying the Galerkin method, Eq. (2) was eventually transformed into such a set of linear equations:
| (3) |
where S is the stiffness matrix containing the information of the finite element (FE) model, G is the load matrix containing the information transformed from the equivalent current densities and ϕ is the vector of potentials at every node of the FE model. Since S is a large sparse matrix, Eq. (3) was solved with the preconditioned conjugate gradients method. The nodes corresponding to the body surface electrodes were identified and the calculated potentials at those nodes were extracted from ϕ to form the heart-model-generated BSPMs.
Fig. 1.
The schematic diagram of the noninvasive three-dimensional electrical imaging approach.
After the above forward modeling, a preliminary classification system(PCS)was employed to initialize the parameters of the heart-excitation model [21].The PCS could approximately determine the cardiac status so as to limit the searching space in the heart model. The PCS was developed based on a three-layer feed-forward artificial neural network (ANN) which was trained with a priori knowledge contained in the heart model. The measured BSPM were used as the input of the ANN and the initial heart model parameters were exported. According to the output of the PCS, the parameters of the heart computer model were initialized, and the corresponding BSPMs were calculated usingEq. (1)–(3). The model’s parameters were iteratively adjusted in an attempt to minimize the dissimilarity between the measured and the heart-model-generated BSPMs, employing a multi-objective nonlinear optimization procedure. The following equation:
| (4) |
was solved with the aid of the “Simplex Method”, where (a)ECC(x) is constructed with the average correlation coefficient (CC) between the measured and simulated BSPMs from instant T1 to instant T2 of the cardiac excitation after detection of initial activation; (b) Eint_CC(x) is constructed with the CC between the integral of the measured BSPMs and the integral of the simulated BSPMs; and (c) ENPL(x) is constructed with the relative error of the number of catheter surface recording electrodes, at which the potentials are less than a certain negative threshold, in the measured and simulated BSPMs from instant T1 to instant T2. The x is a parameter vector consisting of heart model parameters to be optimized in the process. Once the convergence criteria in Eq. (4) were satisfied, the 3D location of the initiation site of electrical activation as well as the activation sequence was determined, and then the potentials at any ventricular node during activation could be reconstructed by using the FEM as described above. The ventricles were divided into 3,000 to 4,000 small segments and each segment was represented by a regional current density, which was the source in Eq. (1).
C. Evaluation of the 3DCEI solutions
The performance of this 3DCEI approach was first validated by evaluating the localization errors of the initiation sites of activation; then the estimated 3D potentials were compared with those reconstructed by the endocardial NCM system on the LV endocardial surface.
The locations of the initiation sites of activation were inversely estimated in the heart-excitation models, which were obtained from the pre-operative MR images. The precise locations of the pacing sites in the RV were obtained by locating the distal end of the pacing leads in the post-operative MR images of the isolated hearts; note that each heart was fixed in an end diastolic state. For the purpose of determining the precise 3D locations of the initiation sites in the resultant heart-excitation models, an image registration procedure between the isolated hearts and the beating hearts were performed as described in [29]. In brief, the registration included two steps. First, initial transformation parameters were obtained by “landmark registration”, during which 3 to 4 anatomical endocardial locations in the two sets of endocardial surfaces were manually fitted. Second, “surface registration” was performed. The two surfaces were registered by applying a multidimensional registration approach, which aims to minimize the Euclidean distance between the two surfaces. After registration, the locations of the initiation sites of activation in the RV were determined in the heart-excitation model and could be directly compared with the estimated locations.
Each heart was also paced at endocardial locations within the LV with an EP catheter. The endocardial geometries of the LV and the locations of the endocardial pacing sites were recorded by the NCM system. In order to determine the locations of the pacing sites reflected in the resultant heart-excitation models, the constructed endocardial surface was extracted from the NCM recordings and then registered with the corresponding surface of the beating heart segmented from the pre-operative MR images. The registration approach used here was described in [29]. We defined the locations of the endocardial pacing sites within each heart-excitation model as the closest point on the endocardial surface of the beating heart to the corresponding pacing sites on the endocardial surface recorded by the NCM system.
The reconstructed transmural extracellular potentials were evaluated with the output of the NCM system over the endocardium. The NCM system can reconstruct and export the electrograms at up to 2,048 sites uniformly distributed on the endocardium. It has been reported that when the distance of an endocardial site to the balloon catheter is less than 40 mm, the reconstructed potentials by the NCM system at this site is more trustable [33]. So the electrograms at the endocardial sites which satisfied the above criterion were extracted from the NCM system's output, and the corresponding electrograms estimated by the 3DCEI approach were also extracted from the 3D results. These two sets of endocardial electrograms were then quantitatively compared. Furthermore, the 3D iso-potential maps estimated by 3DCEI were also presented.
III. Results
A. Localization of initiation site
Ten animals were studied. In each animal, RV pacing was performed at RVA and RVS. After the mapping studies, the RV pacing leads were kept in place when the heart was cut off and fixed. In our studies, some of the RVS leads slipped away from the original sites when isolating the hearts and thus the precise pacing locations could not be determined. Those data had to be excluded. All available RV pacing sites and the corresponding localization results in each of the animals are shown in Table I The LV endocardial pacing was tried in the following regions by moving an EP catheter: LV basal anterior (LVBA), LV basal lateral (LVBL), LV basal septum (LVBS), LV middle anterior (LVMA), LV middle posterior (LVMP), LV middle lateral (LVML), LV middle septum (LVMS), LV apical anterior (LVAA), LV apical posterior (LVAP), LV apical lateral (LVAL), LV apical septum (LVAS) and LV apex (LVA). All available LV pacing sites and the corresponding localization results in each of the animals are shown in Table I In summary, over the 16 RV pacing sites and 48 LV pacing sites in ten animals, the averaged localization error was 6.1±2.3 mm (mean ± standard deviation).
TABLE I.
Localization Errors (Unit MM)
| Pig # | Pacing site | RVA | RVS | LVBA | LVBL | LVBS | LAMA | LVMP | LVML | LVMS | LVAA | LVAP | LVAL | LVAS | LVA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 9.2 | 6.7 | 6.7 | 3.4 | |||||||||||
| 2 | 6.2 | 4.5 | 5.2 | 5.6 | 10.6 | 6.0 | 7.6 | 2.1 | |||||||
| 3 | 4.7 | 6.4 | 9.0 | 2.1 | 7.6 | 3.0 | 4.2 | 7.5 | 3.0 | 4.2 | |||||
| 4 | 10.1 | 8.9 | 6.2 | 4.5 | 4.5 | 2.1 | 6.2 | 3.0 | 7.5 | 4.8 | 6.7 | ||||
| 5 | 7.8 | 4.5 | 6.0 | 9.6 | 2.6 | 10.1 | 7.0 | 3.0 | 6.4 | ||||||
| 6 | 6.2 | 9.2 | 6.0 | 3.0 | 9.2 | ||||||||||
| 7 | 92 | 9.0 | 9.2 | 4.2 | 5.6 | 6.9 | 2.1 | ||||||||
| 8 | 5.4 | 5.6 | 6.2 | ||||||||||||
| 9 | 7.7 | 7.7 | 5.4 | 5.4 | 8.6 | ||||||||||
| 10 | 7.5 | 5.0 | |||||||||||||
RVA: RV apex; RVS: RV septum; LVBA: LV basal anterior; LVBL: LV basal lateral; LVBS: LV basal septum; LVMA: LV middle anterior; LVMP: LV middle posterior; LVML: LV middle lateral; LVMS: LV middle septum; LVAA: LV apical anterior; LVAP: LV apical posterior; LVAL: LV apical lateral; LVAS: LV apical septum; LVA: LV apex.
B. Estimation of transmural extracellular potentials
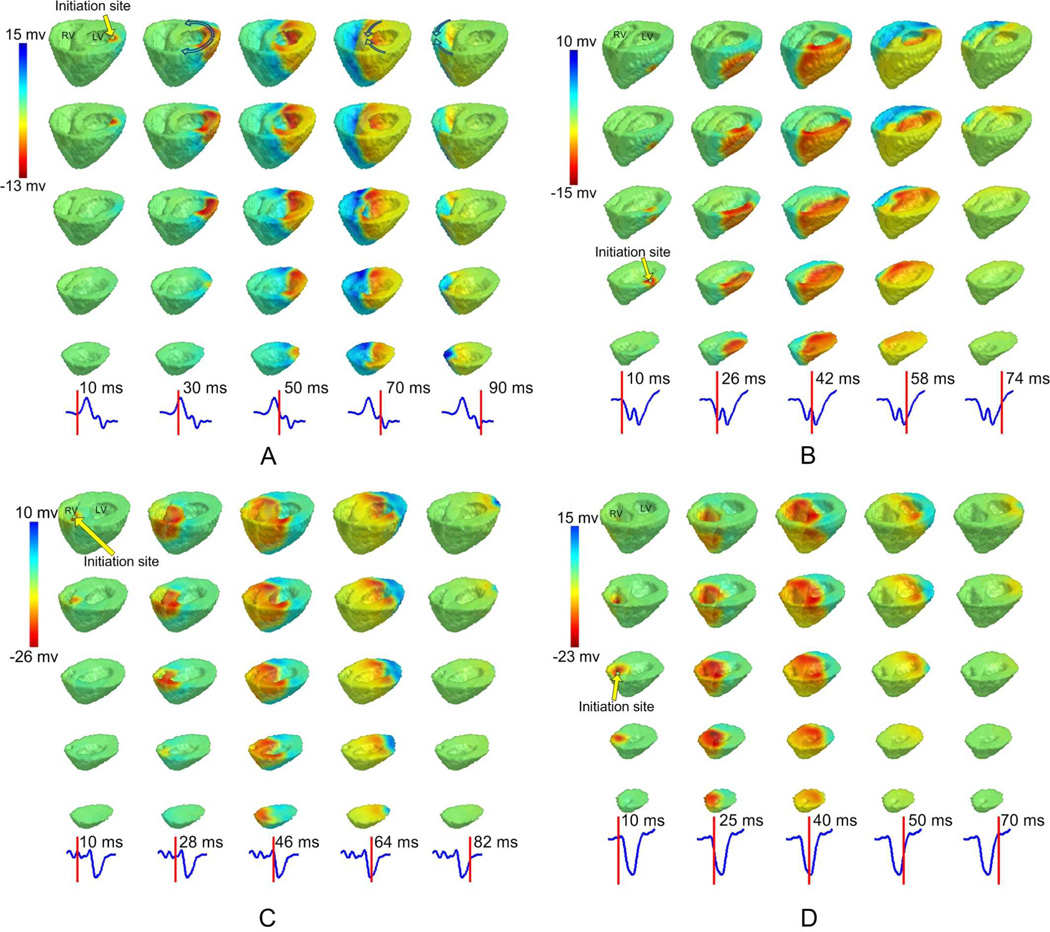
The transmural extracellular potentials throughout the ventricles during activation were reconstructed. The performance of the new function of estimating transmural potentials in the ventricles were first evaluated in simulation. As shown in the supplementary file, the results were promising with high accuracy. Then analysis was conducted with realistic animal data. Fig. 2A depicts the detailed iso-potential maps during LV pacing. The transmural wavefronts of propagation at different time instants were clearly illustrated. First, the initiation site of activation was located on the endocardium of the LV basal lateral region as shown in the first column of Fig. 2A. The breakthrough on the LV epicardium occurred about 15 ms after pacing stimulation. Then as shown with arrows in the second column of Fig. 2A, the excitation propagated towards the LV anterior wall and posterior wall, respectively. The two wavefronts collided at the septal region as shown in the fourth column, and eventually annihilated each other at the RV lateral wall. The activation at both heart surfaces and intramural regions was characterized by the iso-potential maps in informative detail. It was found in the reconstructed iso-potential maps that the extracellular potential at a myocardial site decreased dramatically when the propagation wavefront arrived. This phenomenon was consistent with the electrophysiological nature of depolarization. Fig. 2B depicts the reconstructed iso-potential maps when pig #3 was paced at LV middle anterior. Examples of reconstructed results by 3DCEI when the heart was paced at the RV region were illustrated in Fig. 2C and Fig. 2D. Similar-quality iso-potential maps were presented in all examples. In summary, physiologically reasonable and high-resolution transmural potentials can be reconstructed in all pacing studies in the ten animals studied. Examples of the 3D iso-potential maps over the entire activation can be found in the supplementary video clips.
Fig. 2.
The reconstructed transmural extracellular potentials reconstructed by 3DCEI.Three-dimensional iso-potential maps throughout the ventricles are shown when A: pig #1 is paced at LV basal lateral; B: pig #3 is paced at LV middle lateral; C: pig #5 is paced at RV septum; D: pig #7 is paced at RV apex. In each panel, the iso-potential maps are shown on different horizontal levels from base to apex (each column), and in time sequence. The time instant after pacing corresponding to each column is indicated on the lead II ECG waveform. The initiation site of activation is indicated with an arrow.
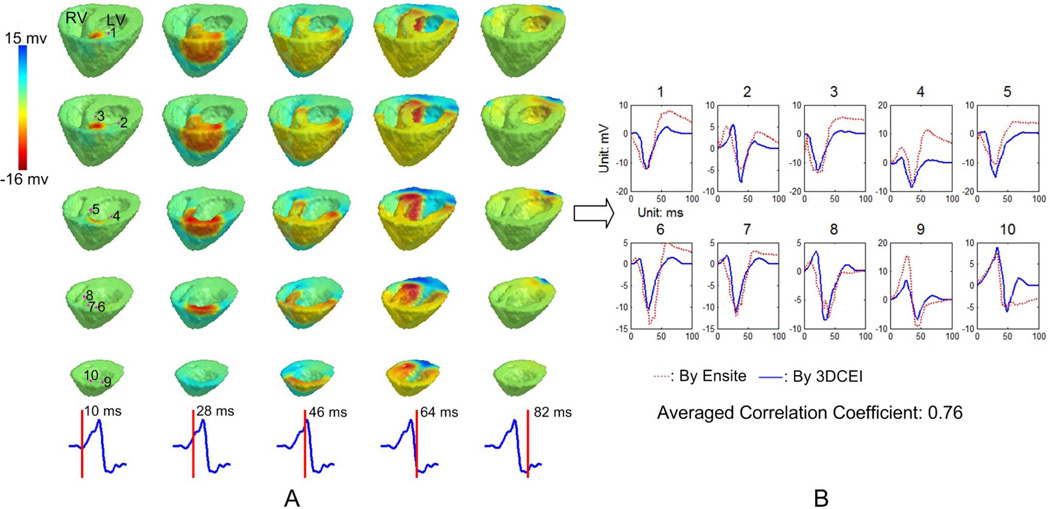
The reconstructed potentials were further evaluated with the aid of the NCM system. Since the NCM system only exported the potentials on the LV endocardial surface, the corresponding reconstructed electrograms by 3DCEI were extracted and the correlation coefficients (CC) between the two sets of electrograms were calculated. In Fig. 3, the estimated 3D iso-potential maps when the heart was paced at LV middle anterior are first shown in panel A. Then ten LV endocardial sites whose distances to the MEA are less than 40 mm are selected and the eletrograms at those locations are shown in panel B. Consistency between the estimated electrograms by 3DCEI and the output of the NCM system was observed in the figure and demonstrated by the averaged CC of 0.76 over all of the ten selected electrograms.
Fig. 3.
A: The reconstructed transmural extracellular potentials by 3DCEI when pig #1 is paced at LV middle anterior. The iso-potential maps are shown on different horizontal levels from base to apex (each column), and in time sequence. The time instant after pacing corresponding to each column is indicated on the lead II ECG waveform. B: The samples of the estimated endocardial electrograms compared with the corresponding output of the NCM system. The endocardial locations where the estimated electrograms are displayed are indicated in the first column of panel A.
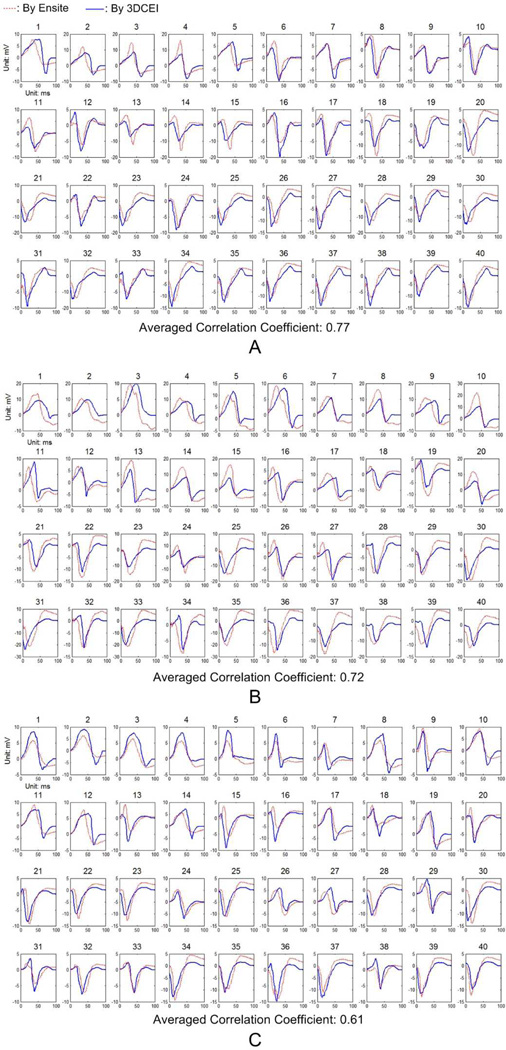
The averaged CC between the endocardial electrograms estimated by 3DCEI and the corresponding output of the NCM system was calculated for each of the 64 pacing studies and the results were summarized in Table II Over all the studies, the averaged "averaged CC" was 0.62±0.09 (mean ± standard deviation). Illustrative comparisons are depicted in Fig. 4 with three pacing studies from different animals. For example, electrograms at 40 spatial endocardial locations are shown and compared in Fig. 4A when pig #2 was paced at LV apical lateral. Those locations were selected from the early activated region (20, 23, 25, 27, 36, etc), late activated region (1, 2, 3, 4, 5, etc) and regions between them (6, 8, 10, 12, 17, etc) to cover the activation sequence on the endocardium. Good consistency between the two sets of electrograms could be observed in most of the samples, but discrepancies also existed, such as the temporal shift (20, 28, 32, etc) and the difference on morphology (1, 14, 31, etc). Similar consistency and discrepancy were also observed in panel B and C. The reasons of discrepancy will be discussed in the following section.
TABLE II.
Correlation Coefficients of Endocardial Electrograms
| Pig # | Pacing site | RVA | RVS | LVBA | LVBL | LVBS | LVMA | LVMP | LVML | LVMS | LVAA | LVAP | LVAL | LVAS | LVA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.68 | 058 | 0.76 | 0.51 | |||||||||||
| 2 | 0.52 | 0.40 | 0.74 | 0.62 | 0.53 | 0.77 | 0.57 | 0.64 | |||||||
| 3 | 0.58 | 0.60 | 0.57 | 0.69 | 0.75 | 0.67 | 0.60 | 0.62 | 0.57 | 0.68 | |||||
| 4 | 0.46 | 0.51 | 0.41 | 0.57 | 0.65 | 0.64 | 0.70 | 0,75 | 0.70 | 0.72 | 0.74 | ||||
| 5 | 0.59 | 0.55 | 0.71 | 0.50 | 0.69 | 0.56 | 0.68 | 0.71 | 0.71 | ||||||
| 6 | 0.61 | 0.37 | 0.49 | 0.68 | 0.60 | ||||||||||
| 7 | 0.50 | 0.61 | 0.56 | 0.62 | 0.69 | 0.68 | 0.73 | ||||||||
| 8 | 0.69 | 0.63 | 0.64 | ||||||||||||
| 9 | 0.58 | 0.70 | 0.71 | 0.67 | 0.73 | ||||||||||
| 10 | 0.61 | 0.65 | |||||||||||||
RVA: RV apex; RVS: RV septum; LVBA: LV basal anterior; LVBL: LV basal lateral; LVBS: LV basal septum; LVMA: LV middle anterior; LVMP: LV middle posterior; LVML: LV middle lateral; LVMS: LV middle septum; LVAA: LV apical anterior; LVAP: LV apical posterior; LVAL: LV apical lateral; LVAS: LV apical septum; LVA: LV apex.
Fig. 4.
The comparisons between the estimated endocardial electrograms by 3DCEI and the output of the NCM system when A: pig #2 is paced at LV apical lateral; B: pig #4 is paced at LV apical septum; C: pig #6 is paced at RV apex.
IV. Discussion
In the present study, the heart-model-based 3DCEI approach was evaluated with a large number of employed animals in an environment mimicking the clinical setting. The performance in localizing the origin of the focal cardiac events was rigorously validated with 16 RV and 48 LV pacing studies and the averaged localization error of 6.1 mm suggests capability of the 3DCEI approach to precisely localize the origins of ectopic beats. Furthermore, estimating the transmural potentials from the noninvasive body surface potentials was for the first time reported and then evaluated with the aid of the NCM system. The comparison between the 3DCEI-estimated electrograms and the NCM system's output on the LV endocardial surface suggested that the 3DCEI's results were consistent with the well accepted mapping results in clinical applications over the endocardium but provided additional transmural information.
Localizing the origins of the abnormal cardiac events has direct benefits in diagnosing and treating cardiac arrhythmias. Electroanatomic (CARTO) and noncontact (Ensite) endocardial mapping systems have been widely used for determining ectopic foci. On the other hand, noninvasive methods which serve this purpose by analyzing the body surface ECGs or the intracavitary electrograms have been investigated, including tracing the moving dipole(s) [34]–[36], reconstructing the epicardial or endocardial potentials [7], [13]–[14] and estimating the heart surface activation sequence [37]. Localizing the origin of the ectopic foci by estimating the 3D cardiac activation was previously proposed by our group [21]–[23] and the 3D inverse approaches have also been reported by other colleagues [31], [38]–[39].We have preliminarily evaluated this approach in animal models [25], [29]. In the present study, we report our results in a larger number of animals as well as a larger number of successful pacing studies for localization, in addition to trasnmural potential imaging.
Mapping the transmural potentials is of importance for both diagnosing cardiac arrhythmias and investigating the intrinsic mechanisms of cardiac activities. Mapping studies using invasive needle electrodes were performed in both animal [40] and human [1], [41] hearts for further understanding the abnormal ventricular activities. Due to the invasive nature, diagnosing arrhythmias in patients by mapping the intramural potentials with needle electrodes is not realistic. In the present study, with the functional mapping of transmural extracellular potentials from the noninvasive body surface potentials, we have proposed an alternative way to serve the purpose. The electrograms during activation at any ventricular location can be obtained without using heart surface or intramural electrodes. This new function incorporated into the 3DCEI approach has the potential of characterizing cardiac arrhythmias in a more efficient way, because the presented intramural potentials could straightforwardly provide the intrinsic information behind the myocardial walls, which may be hard to be captured by heart surface mapping. On the other hand, this technique could become a useful tool in the research of the electrical characteristics of cardiac activities, especially for the arrhythmias with intramural origins.
The new function of mapping transmural potentials was evaluated with the aid of the NCM system by comparing the estimated electrograms on the endocardial surface with the corresponding output from the NCM system. Although good consistency between the two sets of data was observed from the majority of our pacing studies, the following limitations are noteworthy. First, only the estimated results on the LV endocardium were compared and the intramural potentials throughout the ventricles were not quantitatively validated. Second, the output of the NCM system is not a real "gold standard" because those results are reconstructed from the recordings of the noncontact multi-electrode array, which also implies that the discrepancy between our results and the NCM system's output does not necessarily mean either of the mapping approach has better or worse performance on the endocardium. Nevertheless, the NCM system's output has been widely accepted in clinical applications and represents a valuable reference. Thus the comparison in the present study suggests that the estimated potentials by the 3DCEI approach should also be valuable for the applications in EP labs. Furthermore, the illustrated 3D iso-potential maps (Fig. 2, 3) and the supplementary video clips are self-consistent and physiologically reasonable, suggesting the validity of the 3DCEI imaging results which overcome the limitations of heart surface results and provide more informative and straightforward data. In order to rigorously validate the estimated transmural potentials, studies with the aid of plunge needle electrode mapping are required in future studies.
The employed heart models in the present study were assumed isotropic and healthy. In reality, the heart is anisotropic due to the existence of myocardial fibers, and structural abnormalities may exist when ectopic beats occur. The effects of cardiac anisotropy in solving inverse problems are still an open question. In our practice, when isotropy was assumed, reasonably good results was still obtained regarding both localization of the origin of activation (Table I) and estimation of the transmural potentials (Table II, Fig. 2, 3 and 4, supplementary videos). On the other hand, the anisotropy does affect the mapping results. For instance, the discrepancy of the reconstructed endocardial electrograms shown in Fig. 3 and Fig. 4 may be related to the absence of anisotropy. In theory, the assumption of isotropy also reduces the bidomain problem in Eq. (2) into a monodomain one. Incorporating the cardiac anisotropy into the heart models employed in the 3DCEI approach is in our future plan, and the conductivity tensor characterizing the anisotropy may be obtained with the aid of diffusion tensor magnetic resonance imaging (DTMRI) [42].Furthermore, while beyond the scope of this proof of concept study, incorporating pathologic information into the functional mapping approach is of importance for imaging diseased hearts [39].
In conclusion, the feasibility of mapping the transmural potentials in the ventricles from the noninvasive body surface potentials employing the 3DCEI approach have been experimentally demonstrated; the performance of the 3DCEI approach on localizing the initiation site of activation has been evaluated with a large number of studies and the results are promising. The present study suggests that the 3DCEI approach has the potential of enhancing our ability of understanding the electrical characteristics of cardiac activation and guiding the management of cardiac arrhythmias.
Supplementary Material
Acknowledgment
The authors would like to thank Jason Quill and Chengzong Han for technical assistance of the data acquisition.
This work was supported in part by NIH R01HL080093, NSF CBET-0756331, and a grant from the Institute of Engineering in Medicine of the University of Minnesota. Chenguang Liu was supported in part by a Doctoral Dissertation Fellowship from the Graduate School of the University of Minnesota.
References
- 1.Attin M, Ideker RE, Pogwizd SM. Mechanistic insights into ventricular arrhythmias from mapping studies in humans. Heart Rhythm. 2008;vol. 5(6 Suppl):S53–S58. doi: 10.1016/j.hrthm.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeppenfeld K, Schalij MJ, Bartelings MM, Tedrow UB, Koplan BA, Soejima K, Stevenson WG. Catheter ablation of ventricular tachycardia after repair of congenital heart disease: electroanatomic identification of the critical right ventricular isthmus. Circulation. 2007;vol. 116:2241–2252. doi: 10.1161/CIRCULATIONAHA.107.723551. [DOI] [PubMed] [Google Scholar]
- 3.Helm RH, Byrne M, Helm PA, Daya SK, Osman NF, Tunin R, Halperin HR, Berger RD, Kass DA, Lardo AC. Three-dimensional mapping of optimal left ventricular pacing site for cardiac resynchronization. Circulation. 2007;vol. 115:953–961. doi: 10.1161/CIRCULATIONAHA.106.643718. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Haim SA, Osadchy D, Schuster I, Gepstein L, Hayam G, Josephson ME. Nonfluoroscopic, in vivo navigation and mapping technology. Nature Med. 1996;2:1393–1395. doi: 10.1038/nm1296-1393. [DOI] [PubMed] [Google Scholar]
- 5.Gepstein L, Hayam G, Ben-Haim SA. A novel method for nonfluoroscopic catheter-based electroanatomical mapping of the heart: in vitro and in vivo accuracy results. Circulation. 1997;vol. 95:1611–1622. doi: 10.1161/01.cir.95.6.1611. [DOI] [PubMed] [Google Scholar]
- 6.Schilling RJ, Peters NS, Davies DW. Simultaneous endocardial mapping in the human left ventricle using a noncontact catheter: comparison of contact and reconstructed electrograms during sinus rhythm. Circulation. 1998;vol. 98:887–898. doi: 10.1161/01.cir.98.9.887. [DOI] [PubMed] [Google Scholar]
- 7.Gornick CC, Adler SW, Pederson B, Hauck J, Budd J, Schweitzer J. Validation of a new noncontact catheter system for electroanatomic mapping of left ventricular endocardium. Circulation. 1999;vol. 99:829–835. doi: 10.1161/01.cir.99.6.829. [DOI] [PubMed] [Google Scholar]
- 8.Barr RC, Spach MS. Inverse calculation of QRS-T epicardial potentials from normal and ectopic beats in the dog. Circ. Res. 1978;vol. 42:661–675. doi: 10.1161/01.res.42.5.661. [DOI] [PubMed] [Google Scholar]
- 9.Shahidi AV, Savard P, Nadeau R. Forward and inverse problems of electrocardiography: modeling and recovery of epicardial potentials in humans. IEEE Trans. Biomed. Eng. 1994;vol. 41:249–256. doi: 10.1109/10.284943. [DOI] [PubMed] [Google Scholar]
- 10.He B, Wu D. A bioelectric inverse imaging technique based on surface Laplacians. IEEE Trans. Biomed. Eng. 1997;vol. 44:529–538. doi: 10.1109/10.594893. [DOI] [PubMed] [Google Scholar]
- 11.Oster HS, Taccardi B, Lux RL, Ershler PR, Rudy Y. Noninvasive electrocardiographic imaging: reconstruction of epicardial potentials, electrograms, and isochrones and localization of single and multiple electrocardiac events. Circulation. 1997;vol. 96:1012–1024. doi: 10.1161/01.cir.96.3.1012. [DOI] [PubMed] [Google Scholar]
- 12.Greensite F, Huiskamp G. An improved method for estimating epicardial potentials from the body surface. IEEE Trans. Biomed. Eng. 1998;vol. 45:98–104. doi: 10.1109/10.650360. [DOI] [PubMed] [Google Scholar]
- 13.Ramanathan C, Ghanem RN, Jia P, Ryu K, Rudy Y. Noninvasive electrocardiographic imaging for cardiac electrophysiology and arrhythmia. Nature Med. 2004;vol. 10:422–428. doi: 10.1038/nm1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Cuculich PS, Zhang J, Desouza KA, Vijayakumar R, Chen J, Faddis MN, Lindsay BD, Smith TW, Rudy Y. Noninvasive electroanatomic mapping of human ventricular arrhythmias with electrocardiographic imaging. Sci. Transl. Med. 2011;vol. 3:98ra84. doi: 10.1126/scitranslmed.3002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huiskamp G, Greensite F. A new method for myocardial activation imaging. IEEE Trans. Biomed. Eng. 1997;vol. 44:433–446. doi: 10.1109/10.581930. [DOI] [PubMed] [Google Scholar]
- 16.Berger T, Fischer G, Pfeifer B, Modre R, Hanser F, Trieb T, Roithinger FX, Stuehlinger M, Pachinger O, Tilg B, Hintringer F. Single-beat noninvasive imaging of cardiac electrophysiology of ventricular pre-excitation. J. Am. Coll. Cardiol. 2006;vol. 48:2045–2052. doi: 10.1016/j.jacc.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Berger T, Pfeifer B, Hanser FF, Hintringer F, Fischer G, Netzer M, Trieb T, Stuehlinger M, Dichtl W, Baumgartner C, Pachinger O, Seger M. Single-beat noninvasive imaging of ventricular endocardial and epicardial activation in patients undergoing CRT. PLoS ONE. 2011;vol. 6:e16255. doi: 10.1371/journal.pone.0016255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dam PM, Oostendorp TF, Linnenbank AC, van Oosterom A. Non-invasive imaging of cardiac activation and recovery. Ann. Biomed. Eng. 2009;vol. 37:1739–1756. doi: 10.1007/s10439-009-9747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He B, Liu C. Cardiac electrophysiological imaging—solving the inverse problem of electrocardiography. In: Sigg D, editor. Cardiac Electrophysiology Methods and Models. Springer; 2010. [Google Scholar]
- 20.Avari JN, Rhee EK. Cardiac resynchronization therapy for pediatric heart failure. Heart Rhythm. 2008;vol. 5:1476–1478. doi: 10.1016/j.hrthm.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 21.Li G, He B. Localization of the site of origin of cardiac activation by means of a heart-model-based electrocardiographic imaging approach. IEEE Trans. Biomed. Eng. 2001;vol. 48:660–669. doi: 10.1109/10.923784. [DOI] [PubMed] [Google Scholar]
- 22.He B, Li G, Zhang X. Noninvasive three-dimensional activation time imaging of ventricular excitation by means of a heart-excitation model. Phys. Med. Bio. 2002;vol. 47:4063–4078. doi: 10.1088/0031-9155/47/22/310. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, Liu C, He B. Noninvasive reconstruction of three-dimensional ventricular activation sequence from the inverse solution of distributed equivalent current density. IEEE Trans. Med. Imaging. 2006;vol. 25:1307–1318. doi: 10.1109/tmi.2006.882140. [DOI] [PubMed] [Google Scholar]
- 24.Liu C, He B. Noninvasive estimation of the cardiac activation sequence using the extended Kalman Filter. IEEE Trans. Biomed. Eng. 2011;vol. 58:541–549. doi: 10.1109/TBME.2010.2066564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Ramachandra I, Liu Z, Muneer B, Pogwizd SM, He B. Noninvasive three-dimensional electrocardiographic imaging of ventricular activation sequence. Am. J. Physiol. Heart. Circ. Physiol. 2005;vol. 289:H2724–H2732. doi: 10.1152/ajpheart.00639.2005. [DOI] [PubMed] [Google Scholar]
- 26.Han C, Liu Z, Zhang X, Pogwizd SM, He B. Noninvasive three-dimensional cardiac activation imaging from body surface potential maps: a computational and experimental study on a rabbit model. IEEE Trans. Med. Imaging. 2008;vol. 27:1622–1630. doi: 10.1109/TMI.2008.929094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han C, Pogwizd SM, Killingsworth CR, He B. Noninvasive imaging of three-dimensional cardiac activation sequence during pacing and ventricular tachycardia. Heart Rhythm. 2011;vol. 8:1266–1272. doi: 10.1016/j.hrthm.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han C, Pogwizd SM, Killingsworth CR, He B. Noninvasive reconstruction of the three-dimensional ventricular activation sequence during pacing and ventricular tachycardia in the canine heart. Am. J. Physiol. Heart. Circ. Physiol. 2012;vol. 302:H244–H252. doi: 10.1152/ajpheart.00618.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Skadsberg N, Ahlberg S, Swingen C, Iaizzo PA, He B. Estimation of global ventricular activation sequences by noninvasive 3-dimensional electrical imaging: validation studies in a swine model during pacing. J. Cardiovasc. Electrophysiol. 2008;vol. 19:535–540. doi: 10.1111/j.1540-8167.2007.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laske TG, Skadsberg ND, Hill AJ, Klein GJ, Iaizzo PA. Excitation of the intrinsic conduction system through his and intraventricular septal pacing. Pacing Clin. Electrophysiol. 2006;vol. 29:397–405. doi: 10.1111/j.1540-8159.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- 31.Ohyu S, Okamoto Y, Kuriki S. Use of ventricular propagated excitation model in the magnetocardiographic inverse problem for reconstruction of electrophysiological properties. IEEE Trans. Biomed. Eng. 2002;vol. 49:509–519. doi: 10.1109/TBME.2002.1001964. [DOI] [PubMed] [Google Scholar]
- 32.He B, Liu C, Zhang Y. Three-dimensional cardiac electrical imaging from intracavity recordings. IEEE Trans. Biomed. Eng. 2007;vol. 54:1454–1460. doi: 10.1109/TBME.2007.891932. [DOI] [PubMed] [Google Scholar]
- 33.Abrams DJ, Earley MJ, Sporton SC, Kistler PM, Gatzoulis MA, Mullen MJ, Till JA, Cullen S, Walker F, Lowe MD, Deanfield JE, Schilling RJ. Comparison of noncontact and electroanatomic mapping to identify scar and arrhythmia late after the Fontan procedure. Circulation. 2007;vol. 115:1738–1746. doi: 10.1161/CIRCULATIONAHA.106.633982. [DOI] [PubMed] [Google Scholar]
- 34.Ideker RE, Bandura JP, Larsen RA, Cox JW, Keller JW, Brody DA. Localization of heart vectors produced by epicardial burns and ectopic stimuli: validation of a dipole ranging method. Circ. Res. 1975;vol. 36:105–112. doi: 10.1161/01.res.36.1.105. [DOI] [PubMed] [Google Scholar]
- 35.Armoundas AA, Feldman AB, Mukkamala R, Cohen RJ. A single equivalent moving dipole model: an efficient approach for localizing sites of origin of ventricular electrical activation. Ann. Biomed. Eng. 2003;vol. 31:564–576. doi: 10.1114/1.1567281. [DOI] [PubMed] [Google Scholar]
- 36.Lai D, Liu C, Eggen M, Iaizzo P, He B. Equivalent moving dipole localization of cardiac ectopic activity in a swine model during pacing. IEEE Trans. Inf. Technol. Biomed. 2010;vol. 14:1318–1325. doi: 10.1109/TITB.2010.2051448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tilg B, Fischer G, Modre R, Hanser F, Messnarz B, Schocke M, Kremser C, Berger T, Hintringer F, Roithinger FX. Model-based imaging of cardiac electrical excitation in humans. IEEE Trans. Med. Imaging. 2002;vol. 21:1031–1039. doi: 10.1109/TMI.2002.804438. [DOI] [PubMed] [Google Scholar]
- 38.Dössel O, Bauer WR, Farina D, Kaltwasser C, Skipa O. Imaging of bioelectric sources in the heart using a cellular automaton model. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2005;Vol. 2:1067–1070. doi: 10.1109/IEMBS.2005.1616603. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Zhang H, Wong K, Liu H, Shi P. Noninvasive computational imaging of cardiac electrophysiology for 3-d infarct. IEEE Trans. Biomed. Eng. 2011;vol. 58:1033–1043. doi: 10.1109/TBME.2010.2099226. [DOI] [PubMed] [Google Scholar]
- 40.Pogwizd SM. Nonreentrant mechanisms underlying spontaneous ventricular arrhythmias in a model of nonischemic heart failure in rabbits. Circulation. 1995;vol. 92:1034–1048. doi: 10.1161/01.cir.92.4.1034. [DOI] [PubMed] [Google Scholar]
- 41.Chung MK, Pogwizd SM, Miller DP, Cain ME. Three-dimensional mapping of the initiation of nonsustained ventricular tachycardia in the human heart. Circulation. 1997;vol. 95:2517–2527. doi: 10.1161/01.cir.95.11.2517. [DOI] [PubMed] [Google Scholar]
- 42.Helm P, Beg MF, Miller MI, Winslow RL. Measuring and mapping cardiac fiber and laminar architecture using diffusion tensor MR imaging. Ann. N. Y. Acad. Sci. 2005;vol. 1047:296–307. doi: 10.1196/annals.1341.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.