Abstract
Frequent exposure to nickel compounds has been considered as one of the potential causes of human lung cancer. However, the molecular mechanism of nickel-induced lung carcinogenesis remains obscure. In the current study, slight S-phase increase, significant G2/M cell cycle arrest, and proliferation blockage were observed in human bronchial epithelial cells (Beas-2B) upon nickel exposure. Moreover, the induction of cyclin D1 and cyclin E by nickel was shown for the first time in human pulmonary cells, which may be involved in nickel-triggered G1/S transition and cell transformation. In addition, we verified that hypoxiainducible factor-1α, an important transcription factor of nickel response, was not required for the cyclin D1 or cyclin E induction. The role of p53 in nickel-induced G2/M arrest was excluded, respecting that its protein level, ser15 phosphorylation, and transcriptional activity were not changed in nickel response. Further study revealed that cyclin A was not activated in nickel response, and cyclin B1, which not only promotes G2/M transition but also prevents M-phase exit of cells if not degraded in time, was up-regulated by nickel through a manner independent of hypoxia-inducible factor. More importantly, our results verified that overexpressed cyclin B1, veiling the effect of cyclin D1 or cyclin E, mediated nickel-caused M-phase blockage and cell growth inhibition, which may render pulmonary cells more sensitive to DNA damage and facilitates cancer initiation. These results will not only deepen our understanding of the molecular mechanism involved in nickel carcinogenecity, but also lead to the further study on chemoprevention of nickel-associated human cancer.
Introduction
Exposure to nickel(II) has largely increased in industrial societies due to the environmental pollution by heavy metals at all stages of production, use, and disposal (1, 2). Epidemiologic studies have shown the close correlation between the incidence of respiratory cancer and nickel exposure. Due to the workplace exposure and the nonoccupational exposure in surrounding environments, the average daily exposure to nickel by inhalation has been estimated at 0.2 and 0.4 μg for rural and urban dwellers, respectively (3, 4).The levels found in the lungs of autopsied U.S. subjects with no known occupational exposure to nickel, ranged between 1.8 μg/cm2 and 2.1 μg/cm2 of lung surface area, and nickel refinery workers had as high as 15 μg/cm2 of nickel (3, 4). Several types of cellular damage, including DNA damage and DNA repair inhibition, have been identified to contribute to nickel-triggered carcinogenesis (5). The hypoxic signing cascade caused by nickel(II) ions and the subsequent gene expression silence located near heterochromatin caused by a loss of histone H4 and H3 acetylation and DNA hypermethylation was reported to be relevant with nickel carcinogenicity (6, 7). Moreover, nickel can stimulate signaling pathways that increase the expression of numerous inflammatory cytokines, profibrotic proteins, and hypoxic response proteins, such as plasminogen activator inhibitor-1, interleukin (IL)-8, IL-6, cyclooxygenase-2, vascular endothelial growth factor (VEGF), and CAP43 (NDRG1; refs. 8-12). Induction of these genes may contribute to the pathologic effects of nickel, including cancers.
Most of the genes whose transcription is regulated by nickel exposure were identified as targets of the hypoxia-signaling cascade mediated by hypoxia-inducible factor-1α (HIF-1α; ref. 13). In this pathway, nickel(II) facilitates continuous oxidation of intracellular ascorbate by ambient oxygen, and then it may lead to the inhibition of hydroxylases. Therefore HIF-1α becomes more stable due to the weakness of oxygen-involved hydroxylation and subsequent degradation (14-16). The accumulated HIF-1α subsequently modulates the expression of downstream genes involved in proliferation, survival, metabolism, and tumor-igenesis. In addition to HIF-dependent pathway, other activated pathways by nickel such as κB kinase 2/nuclear factor-κB (17-19) and Phosphoinositide 3′ kinases/Akt (20), mitogen-activated protein kinase/activator protein (18, 19), and Nuclear factor of activated T cells (21, 22) are also believed to associate with its carcinogenic activities.
Aberrant cell cycle progression is one of the most important cellular events during the initiation and promotion stages of carcinogenesis, and overgrowth of genetic mutated cells is indispensable in tumor development. It is believed that enhancement of cell cycle transition plays an essential role in tumor promotion, whereas the prolonged mitosis facilitates tumor initiation in some cases (23, 24). Therefore, one question that has been raised is whether metal ions, including nickel(II), induce cancer by interfering cell cycle progression. Microarray analysis of nickel(II)-transformed mouse fibroblasts displaying a malignant phenotype elucidated the overexpression of β-catenin and its downstream target cyclin D1 (25). Cyclin D1 and cyclin E, as important regulators of G1-S phase progression, have been associated with the development of many kinds of cancer, such as the tumors in the breast, lung, and bladder (26, 27). In addition, Chinese hamster ovary cells exposed to nickel were reported to present an increase of G2/M phase proportion (28), in which cyclin A and cyclin B are usually involved. Therefore, it is important to investigate the overall regulation of cyclins, including cyclin D, cyclin E, cyclin B, and cyclin A, as well as p53, which is an important cell cycle regulator, in the cells upon nickel exposure and their biological functions in nickel carcinogenesis. Considering the lung is the primary target organ of nickel compounds in vivo, human bronchial epithelial cells Beas-2B were used to explore the carcinogenicity of nickel in lung cancer.
Materials and Methods
Reagents and Cell Lines
Human bronchial epithelial cells Beas-2B were cultured in DMEM (GIBCO BRL) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin G, and 100 μg/mL streptomycin at 37°C in 5% CO2 atmosphere. Nickel compounds were purchased from Sigma. The substrate for the luciferase assay was purchased from Promega. The anti-β-actin antibody was obtained from Santa Cruz Biotechnology, Inc. Anti-VEGF antibody was purchased from Upstate Biotechnology. Anti-cyclin D1, -cyclin E, -p53, -p53ser15, -HIF-1α, -HIF-1β, -cyclin B1, and -cyclin A2 antibodies were purchased from Cell Signaling Technology.
Plasmids
VEGF-luciferase reporter plasmid containing human VEGF promoter was constructed by inserting a 2.65-kb KpnI-BssHII fragment of the human VEGF promoter sequence from -2274 to +379 relative to the transcription initiation site into the pGL2-basic vector (Promega) as described previously (20). The cyclin D1-luciferase reporter was constructed by inserting the sequence of promoter regions of human gene CCND1 into luciferase reporter vector pGL3-basic. The ΔHRE(-559 to -392) cyclin D1 reporter, which contains a cyclin D1 promoter sequence lacking the HIF-1α binding site, was derived from cyclin D1(-962)-luciferase reporter by overlap extension PCR as shown in Fig. 4E using the following primers: A 5′-ggggtaccgagctcttacgcgtg-3′; B 5′-aaggccggcaggccagtaaattgcaagaa-3′; C 5′-tggcctgccggccttccta-3′ and D 5′-cccaagcttctggggagggctgtg-3′. The p53 and HRE-luciferase reporter were constructed as described previously (29). DN-HIF is a dominant-negative mutant of HIF-1, which inhibits the transcription activity of HIF-1α (30). The cyclin B1 expression plasmid (pCMX cyclin B1) was a kind gift from Carlos Perez-Stable (VA Medical Center, Miami, FL; ref. 31).
Figure 4.
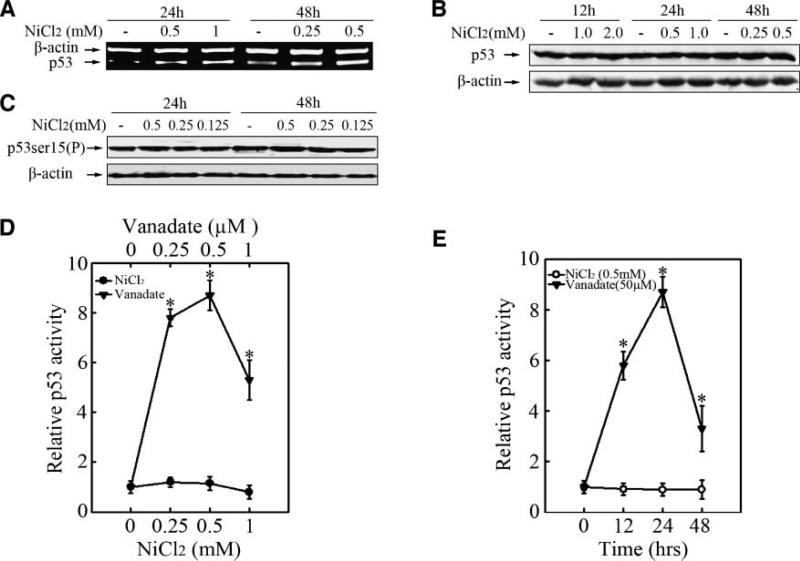
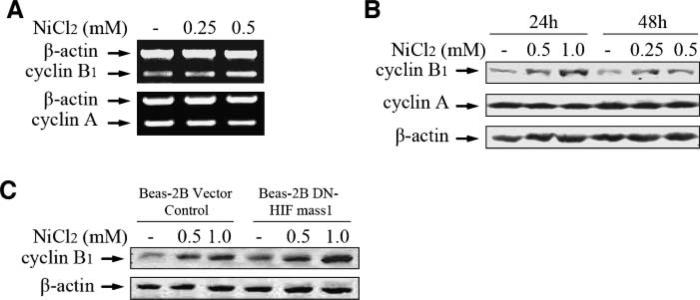
P53 was not involved in the nickel-induced G2/M arrest. Beas-2B cells (2.5 × 105) were seeded into each well of 6-well plates. After being cultured at 37°C overnight, they were treated with 0.25 mmol/L, 0.5 mmol/L, or 1.0 mmol/L NiCl2 for 24 or 48 h as indicated. RT-PCR (A) and Western blot were done as described above (B, C). For p53 transactivation assay (D, E), Beas-2B p53-luc mass1 cells were seeded into each well of 96-well plates and cultured overnight. The cells were treated with vanadate or nickel at the indicated dosages for various time periods. The luciferase activity was determined and the results were presented as relative p53 activity.
Small Interfering RNA Constructs
The procedure for preparing constructs coding for small interfering RNA was described previously (32). Four CCNB1 target sequences were tested for cyclin B1 knockdown by small hairpin RNAs (shRNA), including one 21-nucleotide and three 19-nucleotide gene-specific sequences spanning respectively from nucleotides No. 1, 340 to 360 (33); No. 2, 827 to 845; No. 3, 556 to 574; and No. 4, 1131 to 1149 downstream of the gene transcription start site. The selected sequence was inserted into a BglII/HindIII-cut pSuper vector separately to generate the pSuper/sicyclin B1 plasmid. After identification, the vector carrying the No. 4 sequence was selected to suppress cyclin B1 gene expression, due to its high efficiency and specificity.
Establishment of Stable Transfectants
Beas-2B cells were cultured in a 6-well plate until they reached 85% to 90% confluence. Four and a half micrograms of VEGF-luciferase, cyclin D1-luciferase, cyclin E-luciferase, HRE-luciferase, p53-luciferase, DN-HIF expression plasmid or empty vector control and 0.5 μg hygromycine resistant plasmid were mixed respectively with 10 μL of Lipofectamine 2000 reagent (Invitrogen). These mixtures were used to transfect in each well without penicillin-streptomycin and serum. After 5 to 6 h, the medium was replaced with 10% FBS DMEM. Approximately 36 to 48 h after the beginning of the transfection, the medium was replaced with 10% FBS DMEM containing 400 μg/mL hygromycine. After selection for 14 to 21 d with hygromycine, the stable transfectants Beas-2B VEGF-luc mass1 cells, Beas-2B cyclin D1-luc mass1 cells, Beas-2B cyclin E-luc mass1 cells, Beas-2B HRE-luc mass1 cells, Beas-2B p53-luc mass1 cells, Beas-2B DN-HIF mass1 cells, and Beas-2B Vector Control mass1 were identified by measuring the basal level of luciferase activity. Stable transfectant Beas-2B DN-HIF-VEGF-luc mass1 was established by transfecting Beas-2B using DN-HIF expressing plasmid together with VEGF-luciferase reporter plasmid.
Luciferase Reporter Gene Assays
Confluent monolayer of stable luciferase reporter transfectants were trypsinized, and 8 × 103 viable cells suspended in 100 μL of 10% FBS DMEM were seeded into each well of 96-well plates. The cells were incubated at 37°C in a humidified atmosphere of 5% CO2 in air. After the cell density reached 80% to 90% confluence, the cells were treated with nickel at various concentrations. Cells were lysed with 50 μL lysis buffer, and the luciferase activity was measured using Promega luciferase assay reagent with a luminometer (Wallac 1420 Victor2 multipliable counter system). The results are expressed as relative HIF-1α or p53 activation, and VEGF or cyclin D1 induction relative to medium control. Student's t-test was used to determine the significance of the differences, and the differences were considered significant at P < 0.05.
Reverse Transcription-PCR
Beas-2B cells were cultured in the 6-well plates respectively until they reached 85% to 90% confluence. The cells were then exposed to nickel compounds for indicated time periods. Cells were washed once with ice-cold PBS and extracted for whole RNA with TRIzol reagent following the manufacturer's instructions (Invitrogen). The cDNA was synthesized from 1 μg RNA by using First-Strand Synthesis System for RT-PCR (Invitrogen). The PCR was done by using 2 μL synthesized cDNA and specific human VEGF primers (sense: 5′-ccttgctgctctacctccac-3′, antisense: 5′-atctgcatggtgatgttgga-3′), human wide type p53 primers (sense: 5′-gaacccttgcttgcaatagg-3′, antisense: 5′-gtgaggtaggtgcaaatgcc-3′), human cyclin A2 primers (sense: 5′-gccattagtttacctggacccaga-3′, antisense: 5′-cactgacatggaagacaggaacct-3′), human cyclin B1 primers (sense: 5′-aagagctt taaactttggtctggg-3′, antisense: 5′-ctttgtaagtccttgatttaccatg-3′), human cyclin D1 primers (sense: 5′-agctcctgtgctgcgaagtggaaac-3′, antisense: 5′- agtgttcaatgaaatcgtgcggggt-3′), human cyclin E1 primers (sense: 5′-atacagacccacagagacag-3′, antisense: 5′-tgccatccacagaaatactt-3′) or β-actin primers (sense: 5′- gcgagaagatgacccagatcat -3′, antisense: 5′- gctcaggaggagcaatgatctt -3′). The PCR products were separated on 1.5% agarose gels. The relative mRNA levels of target genes were normalized to the internal reference β-actin that was coamplified in the same reaction for each sample. The reverse transcription-PCR was conducted within the linear ranges of PCR cycles and RNA input.
Western Blot Assays
Beas-2B cells (2 × 105) were cultured in each well of 6-well plates to 70% to 80% confluence. After being cultured for 24 h, the cells were exposed to nickel for 12, 24, 36, or 48 h. The cells were then washed once with ice-cold PBS and extracted with SDS-sample buffer. The cell extracts were separated on polyacryl-amide-SDS gels, transferred, and probed with rabbit-specific antibody against β-actin, HIF-1α, HIF-1β, cyclin D1, or p53, mouse-specific antibody against cyclin E, cyclin B1, cyclin A, or phospho-p53ser15. The protein band, specifically bound to the primary antibody, was detected with antimouse or antirabbit Alexa-800 secondary antibodies, and scanned and analyzed with the Odyssey Infrared Imaging System (Li-Cor Biosciences).
Cell Proliferation Assay
Beas-2B cells were cultured in 96-well culture plates at 8 × 103 cells per well for 24 h. The cells were then exposed to nickel compound. The Cell Counting Kit-8 (CCK-8; Dojindo) was used to assay cell proliferation according to the manufacturer's instructions. Cell proliferation of each group (six wells of each group) was qualified by measuring the absorbance at 450 nm using a microplate reader at the time points of 0, 12, 24, 36, 48, and 72 h after nickel treatment.
Cell Cycle Analysis
Beas-2B cells (2 × 105) were cultured in each well of 6-well plates to 70% to 80% confluence with normal culture medium. The cell culture medium was replaced with 0.1% FBS DMEM with 2 mmol/L L-glutamine, 100 U/mL penicillin G, and 100 μg/mL streptomycin, and cultured for 24 h and then exposed to nickel compounds. The cells were harvested and fixed with 5 mL of ice-cold 70% ethanol overnight. The fixed cells were washed twice with PBS, and then suspended in 500 μL propidium iodide staining solution (propidium iodide 50 μg/mL, RNase A 10 mg/mL, and 0.1% Triton X-100; Sigma Chemical) for at least 1 h at 4°C. The DNA content was determined by flow cytometry using the MoFlo XDP Cell Sorter (Beckman Coulter) and Summit Software v5.0.
Dual measurement of cyclin A2 expression and DNA content was made on MoFlo XDP Cell Sorter (Beckman Coulter). After ethanol fixation and propidium iodide staining, cells (2 × 105) were then incubated with 10 μL FITC-conjugated anti-cyclin A (Beckman Coulter) for 10 min and then subjected to cytometry assay. Immunofluorescence data were analyzed with Summit Software v5.0 (Beckman Coulter). In these analyses, G2/M cells were sorted by the fluorescence intensity of chromatin stained with propidium iodide and mitotic cluster was distinguished from G2/M cells as cyclin A2–negative events.
Results
Nickel-triggered G2/M Cell Cycle Arrest and Proliferation Blockage
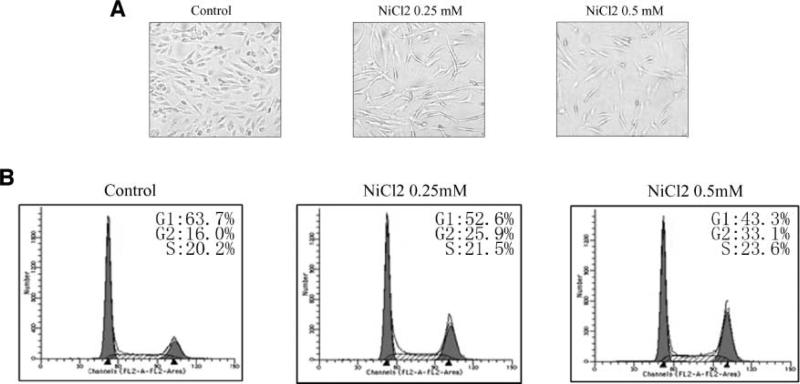
To explore the molecular mechanism implicated in nickel-induced human lung cancer, Beas-2B cells were utilized in the present study. The cells were exposed to various dosages of NiCl2 according to the previous studies (12). We observed that the growth of Beas-2B cells was significantly inhibited after 24 hours of NiCl2 treatment (Fig. 1A). Moreover, the cell cycle distribution of Beas-2B cells exposed to NiCl2 was evaluated by fluorescence-activated cell sorter analysis. We found that exposure of Beas-2B cells to 0.25 mmol/L or 0.5 mmol/L NiCl2 resulted in a slightly increased S-phase population and a significant G2/M cell cycle arrest (Fig. 1B). To further confirm the effect of nickel compounds on cell proliferation, HEK293 and A549 cells were utilized and the results were consistent with that from Beas-2B cells (supplementary data 1a to d).
Figure 1.
Nickel-triggered G2/M cell cycle arrest and proliferation blockage. 1 × 103 of Beas-2B cells were seeded into each well of 96-well plates, cultured in 10% FBS DMEM overnight, and then exposed to NiCl2. The cells were photographed under microscopy after 36 h exposure (A). Beas-2B cells were seeded into each well of 6-well plates and cultured in DMEM containing 10% FBS. After the cell density reached 70% to 80%, the cells were exposed to 0.25 mmol/L or 0.5 mmol/L NiCl2 for 36 h, and then were fixed and stained with propidium iodide as described previously. Cell cycle distribution was determined by flow cytometry (B). Each experiment was repeated for at least three times.
Nickel Up-regulated Cyclin D1 and Cyclin E Expression
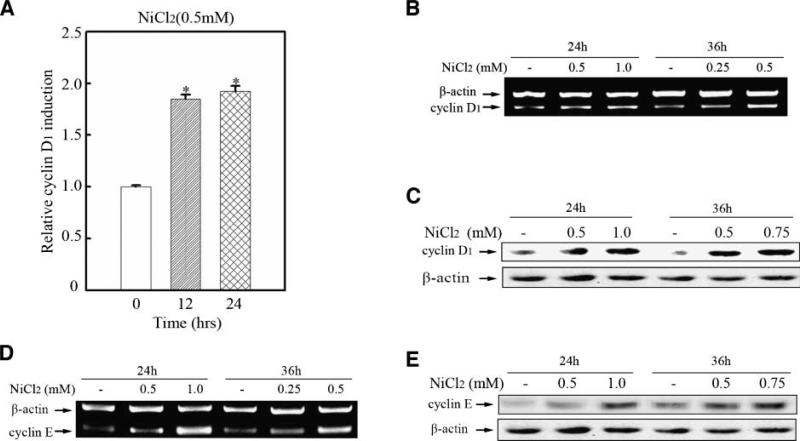
Overexpression of cyclin D1 has been shown in numerous carcinomas, and it is well accepted that cyclin D1 overexpression–rendered deregulated cell cycle progression is closely related to carcinogenesis. In the present study, our results showed that cyclin D1 expression was increased by nickel compounds in bronchial epithelial cells. Figure 2A shows that nickel exposure triggered cyclin D1 promoter activation. The up-regulation of cyclin D1 at the transcription and protein levels was further confirmed by reverse transcription-PCR and Western blot assay (Fig. 2B and C).
Figure 2.
Nickel up-regulated cyclin D1 and cyclin E expression. 8 × 103 Beas-2B cyclin D1-luc mass1 cells (A) were seeded into each well of a 96-well plate. After being cultured at 37°C overnight, the cells were treated with 0.5 mmol/L NiCl2 for various time periods as indicated. The cells were then extracted with lysis buffer, and the luciferase activity was measured as described in Materials and Methods. Bar, mean and SD of the triplicate wells; (*), a significant increase from medium control cells (P < 0.05). Beas-2B cells (B, D) were seeded into 100-mm dishes. After being cultured at 37°C overnight, they were treated with NiCl2 for 24 or 36 h as indicated. RNA isolation and reverse transcription-PCR (RT-PCR) were carried out as described in Materials and Methods. 2.5 × 105 Beas-2B cells (C, E) were seeded into each well of 6-well plate and cultured in 10 % FBS DMEM. Twenty-four hours later, the cells were exposed to NiCl2 at various dosages for 24 or 36 h as indicated and then Western blot assay was done.
Overexpression of cyclin in carcinogenesis can be linked to the premature S-phase entrance and disturbance of the DNA-replication (34-36). In the present study, we showed, for the first time, that cyclin E was up-regulated at both RNA and protein levels in bronchial epithelia cells with nickel treatment (Fig. 2D and E).
HIF-1 was not Involved in Nickel-induced Cyclin D1 or Cyclin E Expression
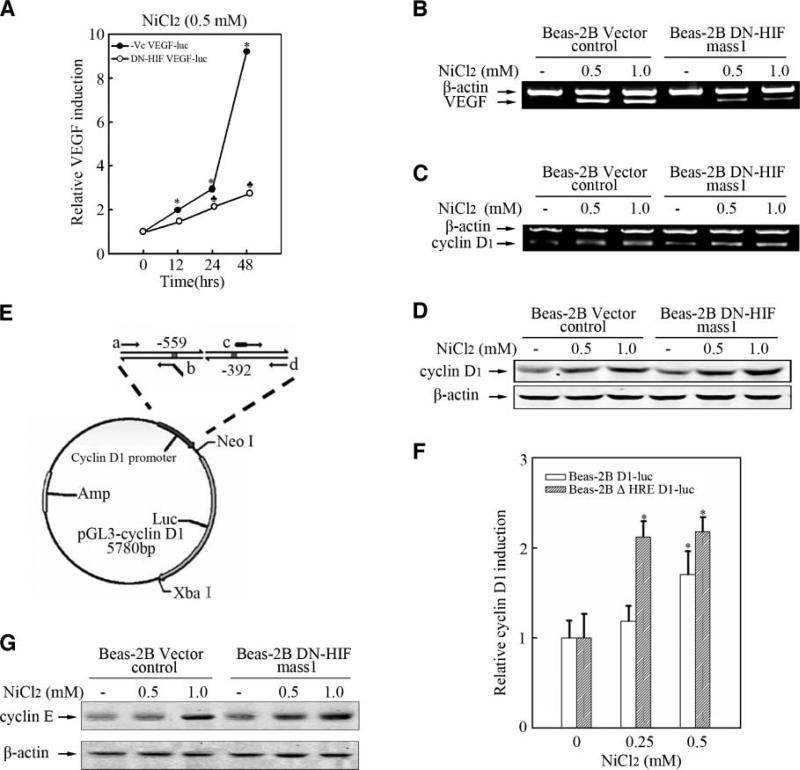
HIF-1, a important transcription factor that regulates multiple downstream target genes expression, plays a key role in mediating the biological response of nickel compounds (20). Bioinformation analysis revealed that cyclin D1 promoter region contains HRE sequence that is the canonical binding site of HIF-1. Therefore, it is important to explore the role of HIF-1 in nickel-induced cyclin D1 expression. We found that nickel can induce HIF-1α activation and VEGF expression in Beas-2B cells (supplemental data 2a to f) and overexpression of DN-HIF, which is a dominant negative mutant of HIF-1α, dramatically blocked VEGF induction by nickel compound (Fig. 3A to C). However, cyclin D1 or cyclin E up-regulation by nickel was not impaired in DN-HIF stable transfectants (Fig. 3D and G). In further study, cyclin D1 luciferase reporter plasmid and ΔHRE cyclin D1-luciferase reporter plasmid, which was lack of HIF binding sequence, were transiently transfected into Beas-2B cells respectively. As shown in Fig. 3E and F, nickel-induced cyclin D1 promoter activation was not impaired in ΔHRE cyclin D1-luciferase reporter plasmid–transfected Beas-2B cells compared with that transfected with cyclin D1-luciferase reporter plasmid. Taken together, these data strongly suggest HIF-1 is not required in nickel-induced cyclin D1 or cyclin E expression in bronchial epithelia cells.
Figure 3.
HIF-1 was not involved in nickel-induced cyclin D1 or cyclin E expression. 8 × 103 Beas-2B-Vc VEGF-luc mass1 and Beas-2B DN-HIF VEGF-luc mass1 cells (A) were seeded into each well of a 96-well plate. After being cultured at 37°C overnight, the cells were treated with NiCl2 for various dosages as indicated. The cells were then extracted and luciferase activity was determined as described previously. (♣), a significant decrease as compared with nickel-treated Beas-2B VEGF-Luc mass1 cells (P < 0.05). Beas-2B Vector control cells, Beas-2B DN-HIF mass1 cells (B, C, D, and G) were seeded into 100-mm dishes. After being cultured at 37°C overnight, the cells were treated with NiCl2 for 24 h. RT-PCR and Western blot were carried out as described previously. E. The graphic representation of ΔHRE cyclin D1 reporter plasmid construction. 2 × 104 cyclin D1-luc plasmid transiently transfected Beas-2B cells and ΔHRE cyclin D1-luc transiently transfected Beas-2B cells (F) were seeded into each well of a 48-well plates respectively. After being cultured at 37°C overnight, the cells were treated with 0.25 mmol/L or 0.5 mmol/L NiCl2 for 12 h. The results of relative cyclin D1 induction were analyzed as described above.
P53 was not Involved in Nickel-induced G2/M Arrest
Considering the essential role of p53 in G2/M arrest has been widely accepted (37), expression and activation of p53 upon nickel response were investigated in the present study. Our results showed the dose-dependent increase of p53 mRNA in Beas-2B cells with nickel treatment (Fig. 4A). However, p53 total protein and phosphorylated p53 at Ser15, which is important for p53 activation in G2/M checkpoint, were not obviously changed (Fig. 4B and C). In addition, we tested the transcriptional activity of p53 in nickel response by gene reporter assay. Vanadate, which has been verified to induce p53 activation, was used as positive control. As shown in Fig. 4D and E, vanadate treatment triggered a near 10-fold increase of p53 transcriptional activation, but there was no significant increase in p53 transcription activity in Beas-2B cells exposed to NiCl2 for various time periods. Taken together, our results showed that p53 was not involved in nickel-induced G2/M arrest.
Expression of Cyclin B1, but not Cyclin A, was Enhanced by Nickel Treatment
It has been well established that cyclin A and cyclin B are both important regulators in mitosis progression. The destruction of cyclin A and cyclin B is reported to be required for anaphase onset (escape from mitosis; ref. 38). Therefore, to elucidate the mechanism underling nickel-induced G2/M cell cycle arrest, it is essential to explore the potential regulation of cyclin A and cyclin B1 by nickel compound. As shown in Fig. 5A and B, cyclin B1 expression was notably up-regulated by nickel at both mRNA and protein levels, but not cyclin A, which suggests cyclin B1 might be required in nickel-induced G2/M arrest. Although the cyclin B1 promoter region contains HRE-like sequence, overexpression of DN-HIF did not impair nickel-triggered cyclin B1 expression, showing that cyclin B1 induction by nickel is via HIF-1-independent pathway (Fig. 5C).
Figure 5.
Expression of cyclin B1, but not cyclin A, was enhanced by nickel treatment. 2.5 × 105 Beas-2B cells were seeded into well of 6-well plates. After being cultured at 37°C overnight, they were treated with 0.25 mmol/L or 0.5 mmol/L NiCl2 for 48 h. RNA isolation and RT-PCR were carried out (A). 2.5 × 105 of Beas-2B cells (B), Beas-2B vector control cells, and Beas-2B DN-HIF mass1 (C) were seeded into each well of 6-well plate respectively and cultured in 10% FBS DMEM overnight. The cells were then exposed to various dosage of NiCl2 as indicated for 24 or 48 h and subjected to Western blot assay.
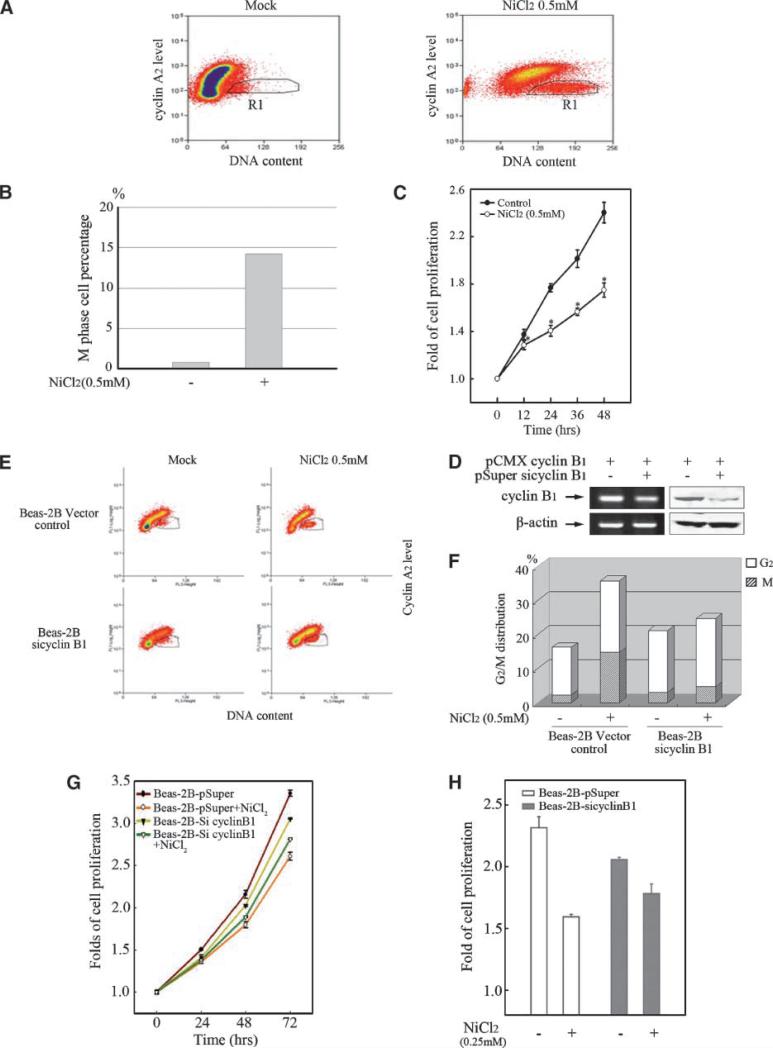
Cyclin B1 is Responsible for the Nickel-induced M-phase Arrest and Cell Growth Inhibition
To figure out the precise machinery of nickel-induced G2/M cell cycle arrest, the M-phase cells were gated from the whole cell population after nickel treatment regarding their DNA content values and cyclin A2 level. As shown in Fig. 6A, M-phase cells (black-line thresholds in the right-bottom figure) were sorted from the cells with a 4N DNA content (G2/M cluster) as cyclin A2–negative events, because cyclin A2 is abruptly degraded in the early phase of mitosis (before metaphase). In this result, the proportion of M-phase cells was proved to be largely increased by nickel treatment corresponding with the G2/M blockage (Fig. 6B). Excessive cyclin B1 expression is regarded as one of the most deleterious cellular events which prevent cells from mitosis exit and lead to aberrant M-phase blockage. Thus, proliferation of Beas-2B cells was inhibited by nickel treatment, which is shown in Fig. 6C.
Figure 6.
Cyclin B1 is responsible for the nickel-induced M-phase arrest and cell growth inhibition. Beas-2B cells (A, B) were seeded into each well of 6-well plates and cultured in DMEM containing 10% FBS until the cell density reached 70% to 80%. After the treatment of 0.5 mmol/L NiCl2 for 48 h, the cells were stained with propidium iodide and then incubated with 10 μL FITC-conjugated anti-cyclin A2. M-phase cell population was sorted by flow cytometry as described in Materials and Methods. D. Beas-2B cells were transiently transfected with pCMX cyclin B1 expression plasmid together with pSuper/sicylin B1 or Vector control respectively using Lipofectamine 2000 reagent. RT-PCR and Western blot were carried out as described above. Beas-2B cells transiently transfected by pSuper/siCyclin B1 or vector control (E, F) were seeded respectively into each well of 6-well plates and cultured in DMEM containing 10% FBS. After the cell density reached 70% to 80%, the cells were exposed to 0.5 mmol/L NiCl2 for 48 h. Cell cycle distribution was determined by flow cytometry as described above. Every result was from one representative of three independent experiments. 1 × 103 of Beas-2B cells (C) or Beas-2B cells transiently transfected by pSuper/siCyclin B1 or vector control (G and H) were seeded into each well of 96-well plates, cultured in 10% FBS DMEM overnight, and then exposed to 0.5 mmol/L (C) or 0.25 mmol/L (G) NiCl2 for various time periods. The cell proliferation was evaluated as described in Materials and Methods. The comparison of cell growth between the Beas-2B cells transiently transfected by pSuper/siCyclin B1 and vector control was presented (H). Each result was presented as an arithmetical average of three independent assays.
To further confirm that up-regulated cyclin B1 is responsible for nickel-induced cell cycle arrest and cell growth inhibition, pSuper/siCyclin B1 was constructed and identified (Fig. 6D). As shown in Fig. 6E and F, the M-phase blockage induced by nickel treatment was markedly impaired by cyclin B1 silence in fluorescence-activated cell sorter assay, indicating that nickel-induced G2/M blockage was mainly due to the deficient mitosis exit which was mediated by excessive cyclin B1. In addition, the slightly increased G2-phase proportion by nickel was supposed to be a subsequent event following the interfered G2/M transition caused by M-phase blockage. Moreover, we found that silence of cyclin B1 not only led to a notable cell growth inhibition as reported previously, but also dramatically impaired nickel-trigged cell growth inhibition in Beas-2B cells (Fig. 6G and H).
Discussion
The correlation between nickel compound exposure and increased risk of human respiratory cancer has been widely reported in epidemiologic studies (39, 40). However, the molecular mechanism of nickel-induced carcinogenesis is not yet fully understood. In the current study, we showed that nickel compound was able to induce G2/M arrest in Beas-2B cells and consequently caused significant cell growth inhibition. The expressions of cyclin D1 and cyclin E were up-regulated by nickel, but only a slight increase of S-phase cell population was observed. Further study showed that the expression of cyclin B1, an important regulator of G2/M and M-phase progression, was also enhanced by nickel compound. And overexpression of cyclin B1 was responsible for the nickel-triggered mitosis exit blockage, which veiled the effects of cyclin D1 and cyclin E, and resulted in the significant inhibition of cell proliferation.
One of the most important features that distinguish cancer cells from normal cells is aberrant cell division, usually resulting from the uncontrolled expression of cyclins (23, 24). Overexpression of cyclins has been proved to be implicated in the malignant transformation of a variety of human cancers (26, 27). Cyclin D1 has been reported to distinguish benign and premalignant human breast lesions from any form of breast carcinoma (41). Recently, Ouyang W and his colleagues reported that exposure to soluble nickel compounds caused a significant inhibition of cell growth and G1/G0 cell cycle arrest in human A549 cancer cells, which was concomitant with a down-regulation of cyclin D1 (42). In the current study, the nickel-induced cyclin D1 expression was first found at both RNA and protein levels in human bronchial epithelia Beas-2B cells. Cyclin E is the other critical activator in G1/S transition (43). High level of cyclin E protein in cancer has been also linked to poor prognosis and several aggressive features of cancer such as estrogen receptor negativity, poor differentiation, p53-mutations, pRb-inactivation, and p27Kip1 down-regulation (27, 44). Our results showed that expression of cyclin E was increased in nickel-treated bronchial epithelia cells. It was observed that Beas-2B cell population at the S phase can be marginally increased by nickel exposure. This observation was partially consistent with another report that CHO cell population at the S phase was gradually increased in cultures exposed to rising quantities of nickel(II) (45). Therefore, influence of up-regulated cyclin D1 and/or cyclin E on uncontrolled cell cycle progression, undifferentiation, and cell transformation should be involved, at least partially, in nickel carcinogenecity.
The role of p53 in mediation of G2/M cell cycle arrest has been widely accepted, but whether p53 participates in nickel-induced G2/M arrest remains obscure. Salnikow et al. reported that acute nickel exposure activates p53 transcription based on the result from GeneChip analysis (46), and the induction of wild-type p53 by nickel compound was observed in MCF-7 and A549 cells (47). Huang C et al. documented that exposure of cells to either Ni3S2 or NiCl2 did not show any influence of p53 activity (21). Lee SH and Shiao YH documented that the level of p53 protein was not changed in nickel(II)-treated normal rat kidney cells, and p53 was not invovled nickel(II)-induced apoptosis and G2/M arrest either (28, 48). In the current study, although the dose-dependent increase of p53 mRNA in Beas-2B cells upon nickel exposure was observed, p53 total protein and phosphorylated p53Ser15 were not obviously changed by nickel treatment. Furthermore, the absence of p53 activation in nickel response was revealed by p53 gene report assay. Our results were consistent with the epidemiologic report that expression of p53 in cells from nasal biopsies is not different between nickel workers and control population (49).
Cyclin B1, which is essential for cell cycle progression through mitosis, is overexpressed in a variety of cancers (50, 51). The deregulated expression of cyclin B1 seems to be closely associated with early events in neoplastic transformation (52) and poor prognosis (53). Therefore, deregulation of cyclin B1 has been considered as a key event in carcinogenesis and tumor therapy.
As a partner of Cdk1, the biofunction of cyclin B1 in cell cycle progression consists of G2/M transition and M-phase exit. When cyclin B1-Cdk1 is fully activated, cells are committed to mitosis. Almost at the same time, phosphorylation of anaphase-promoting complex/cyclo-some (APC/C), the most important regulator of the ubiquitin-mediated proteolysis in mitosis progress control, was triggered by cyclin B1-Cdk1 to generate the active form of APC/C bound by Cdc20 (APC/CCdc20). Then APC/CCdc20 mediates the degradation of securin and cyclin B, which acts as the inhibitor of anaphase and cytokinesis, to render exit from mitosis when spindle checkpoint is inactivated (38, 50, 54). In the current study, the accumulation of cyclin B1 by nickel may result from the deregulation of APC/C activation or some other mechanism such as sustained activation of cyclin B1 expression. Because cyclin B1 remains at high level, instead of being eliminated upon nickel exposure, the cells are unable to exit from mitosis and are blocked in anaphase.
Owning to the cell cycle check point, numerous DNA damage produced during interphase or prophase of cell division can be repaired, but not those during anaphase when the chromosomes are condensed. Because gene transcription is silenced during anaphase, prolonging division leads to decreased DNA repair because the cell cannot carry out its vital functions. In view of the deleterious effects of nickel exposure, including genome toxicity and epigenetic alteration, the prolonged anaphase will lead to an extended duration of cells in fragile state upon nickel exposure, thus rendering the cells more susceptible to nickel carcinogenecity. The significance of cyclin B1 in carcinogen-esis and cancer therapy still needs to be further investigated.
HIF-1 heterodimer has been reported to be a key transcription factor among the nickel-responsive genes. Genetic studies comparing the growth of tumors with and without HIF-1 have elucidated the function of HIF-1 in tumor vascularization and growth (55). We have also reported that HIF-1α plays an essential role in VEGF induction by nickel in mouse epidermal Cl 41 cells (20, 56). In the present study, the accumulation of HIF-1α, but not activator protein (data not shown), another important transcription factor for VEGF expression, was found to be required in VEGF induction in Beas-2B cells with nickel treatment. Given that VEGF has been well documented in inducing microvascular permeability as well as mediating angiogenesis and vasculogenesis (57), there is also growing evidence indicating that it also plays a crucial role in tumor promotion during the multistep process of chemically induced carcinogenesis (58, 59). Although there locates HRE sequence in the promoter region of both cyclin D1 and cyclin B1, our results showed HIF was not involved in nickel-induced cyclin D1 and cyclin B1 expression, the detailed regulatory machinery of which is still under investigation in our lab.
In summary, we show here, for the first time, that nickel exposure is able to facilitate cyclin D1 and cyclin E expression, which may promote G1/S transition and cell transformation of pulmonary cells. More importantly, our results verified that nickel-triggered cyclin B1 accumulation and subsequent M-phase arrest is responsible for the inhibitory effect of nickel on cell proliferation in bronchial epithelia cells. Because the malfunction of G1/S transition and M exit promotion in the cell microenvironment are both important for cancer initiation and promotion, these results will not only broaden our knowledge of the molecular mechanism involved in nickel-induced human lung cancer, but also lead to the further study on chemoprevention and therapy of nickel-associated cancer.
Supplementary Material
Acknowledgments
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We thank professor Binghua Jiang from the Mary Babb Randolph Cancer Center, West Virginia University for the generous gift of the HRE-luciferase construct and DN-HIF expression plasmid (pCEP4/HIF-1αΔNBΔAB), and professor Perez-Stable Geriatric from Veterans Affairs Medical Center for the kind gift of pCMX/cyclin B1 plasmid.
Grant support: NIH/National Cancer Institute CA112557 and National Natural Science Foundation of China grants 30530790, 30800576, and 30620130434. Shanghai Key Basic Science Foundation grant 07DJ14006, 06DJ14009, International collaboration program 08400701000 and Shanghai Pujiang program.
Footnotes
Note: Supplementary data for this article are available at Cancer Epidemiology, Biomarkers & Prevention Online (http://cebp.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Bennett BG. Exposure of man to environmental nickel-an exposure commitment assessment. Sci Total Environ. 1982;22:203–12. doi: 10.1016/0048-9697(82)90065-1. [DOI] [PubMed] [Google Scholar]
- 2.Doll R, Mathews JD, Morgan LG. Cancers of the lung and nasal sinuses in nickel workers: a reassessment of the period of risk. Br J Ind Med. 1977;34:102–5. doi: 10.1136/oem.34.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doll R, Morgan LG, Speizer FE. Cancers of the lung and nasal sinuses in nickel workers. Br J Cancer. 1970;24:623–32. doi: 10.1038/bjc.1970.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett BG. Environmental nickel pathways to man. IARC Sci Publ. 1984:487–95. [PubMed] [Google Scholar]
- 5.Hartwig A, Asmuss M, Ehleben I, et al. Interference by toxic metal ions with DNA repair processes and cell cycle control: molecular mechanisms. Environ Health Perspect. 2002;110(Suppl 5):797–9. doi: 10.1289/ehp.02110s5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zoroddu MA, Schinocca L, Kowalik-Jankowska T, Kozlowski H, Salnikow K, Costa M. Molecular mechanisms in nickel carcinogenesis: modeling Ni(II) binding site in histone H4. Environ Health Perspect. 2002;110(Suppl 5):719–23. doi: 10.1289/ehp.02110s5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa M, Davidson TL, Chen H, et al. Nickel carcinogenesis: epigenetics and hypoxia signaling. Mutat Res. 2005;592:79–88. doi: 10.1016/j.mrfmmm.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Andrew A, Barchowsky A. Nickel-induced plasminogen activator inhibitor-1 expression inhibits the fibrinolytic activity of human airway epithelial cells. Toxicol Appl Pharmacol. 2000;168:50–7. doi: 10.1006/taap.2000.9009. [DOI] [PubMed] [Google Scholar]
- 9.Barchowsky A, Soucy NV, O'Hara KA, Hwa J, Noreault TL, Andrew AS. A novel pathway for nickel-induced interleukin-8 expression. J Biol Chem. 2002;277:24225–31. doi: 10.1074/jbc.M202941200. [DOI] [PubMed] [Google Scholar]
- 10.Namiki A, Brogi E, Kearney M, et al. Hypoxia induces vascular endothelial growth factor in cultured human endothelial cells. J Biol Chem. 1995;270:31189–95. doi: 10.1074/jbc.270.52.31189. [DOI] [PubMed] [Google Scholar]
- 11.Zhou D, Salnikow K, Costa M. Cap43, a novel gene specifically induced by Ni2+ compounds. Cancer Res. 1998;58:2182–9. [PubMed] [Google Scholar]
- 12.Ding J, Zhang X, Li J, et al. Nickel compounds render anti-apoptotic effect to human bronchial epithelial Beas-2B cells by induction of cyclooxygenase-2 through an IKKβ/p65-dependent and IKKα- and p50-independent pathway. J Biol Chem. 2006;281:39022–32. doi: 10.1074/jbc.M604798200. [DOI] [PubMed] [Google Scholar]
- 13.Viemann D, Schmidt M, Tenbrock K, et al. The contact allergen nickel triggers a unique inflammatory and proangiogenic gene expression pattern via activation of NF-κB and hypoxia-inducible factor-1α. J Immunol. 2007;178:3198–207. doi: 10.4049/jimmunol.178.5.3198. [DOI] [PubMed] [Google Scholar]
- 14.Kaczmarek M, Timofeeva OA, Karaczyn A, Malyguine A, Kasprzak KS, Salnikow K. The role of ascorbate in the modulation of HIF-1α protein and HIF-dependent transcription by chromium(VI) and nickel(II). Free Radic Biol Med. 2007;42:1246–57. doi: 10.1016/j.freeradbiomed.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang GS, Li Q, Chen H, Costa M. Effect of metal ions on HIF-1α and Fe homeostasis in human A549 cells. Mutat Res. 2006;610:48–55. doi: 10.1016/j.mrgentox.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Davidson TL, Chen H, Di Toro DM, D'Angelo G, Costa M. Soluble nickel inhibits HIF-prolyl-hydroxylases creating persistent hypoxic signaling in A549 cells. Mol Carcinog. 2006;45:479–89. doi: 10.1002/mc.20176. [DOI] [PubMed] [Google Scholar]
- 17.Chen F, Ding M, Castranova V, Shi X. Carcinogenic metals and NF-κB activation. Mol Cell Biochem. 2001;222:159–71. [PubMed] [Google Scholar]
- 18.Huang Y, Davidson G, Li J, et al. Activation of nuclear factor-κB and not activator protein-1 in cellular response to nickel compounds. Environ Health Perspect. 2002;110(Suppl 5):835–9. doi: 10.1289/ehp.02110s5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz MT, Goncalo M, Figueiredo A, Carvalho AP, Duarte CB, Lopes MC. Contact sensitizer nickel sulfate activates the transcription factors NF-κB and AP-1 and increases the expression of nitric oxide synthase in a skin dendritic cell line. Exp Dermatol. 2004;13:18–26. doi: 10.1111/j.0906-6705.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Davidson G, Huang Y, et al. Nickel compounds act through phosphatidylinositol-3-kinase/Akt-dependent, p70(S6k)-independent pathway to induce hypoxia inducible factor transactivation and Cap43 expression in mouse epidermal Cl41 cells. Cancer Res. 2004;64:94–101. doi: 10.1158/0008-5472.can-03-0737. [DOI] [PubMed] [Google Scholar]
- 21.Huang C, Li J, Costa M, et al. Hydrogen peroxide mediates activation of nuclear factor of activated T cells (NFAT) by nickel subsulfide. Cancer Res. 2001;61:8051–7. [PubMed] [Google Scholar]
- 22.Lu H, Shi X, Costa M, Huang C. Carcinogenic effect of nickel compounds. Mol Cell Biochem. 2005;279:45–67. doi: 10.1007/s11010-005-8215-2. [DOI] [PubMed] [Google Scholar]
- 23.Cohen SM, Ellwein LB. Cell growth dynamics in long-term bladder carcinogenesis. Toxicol Lett. 1988;43:151–73. doi: 10.1016/0378-4274(88)90026-4. [DOI] [PubMed] [Google Scholar]
- 24.Charnley G, Wilson JD. Evaluation of the form of the cell growth rate function of the two-stage model for carcinogenesis. Prog Clin Biol Res. 1991;369:291–301. [PubMed] [Google Scholar]
- 25.Kowara R, Karaczyn A, Cheng RY, Salnikow K, Kasprzak KS. Micro-array analysis of altered gene expression in murine fibroblasts transformed by nickel(II) to nickel(II)-resistant malignant phenotype. Toxicol Appl Pharmacol. 2005;205:1–10. doi: 10.1016/j.taap.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Alao JP. The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foulkes WD, Brunet JS, Stefansson IM, et al. The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloidmicrovascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res. 2004;64:830–5. doi: 10.1158/0008-5472.can-03-2970. [DOI] [PubMed] [Google Scholar]
- 28.Shiao YH, Lee SH, Kasprzak KS. Cell cycle arrest, apoptosis and p53 expression in nickel(II) acetate-treated Chinese hamster ovary cells. Carcinogenesis. 1998;19:1203–7. doi: 10.1093/carcin/19.7.1203. [DOI] [PubMed] [Google Scholar]
- 29.Huang C, Ma WY, Li J, Hecht SS, Dong Z. Essential role of p53 in phenethyl isothiocyanate-induced apoptosis. Cancer Res. 1998;58:4102–6. [PubMed] [Google Scholar]
- 30.Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Phosphatidy linositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001;12:363–9. [PubMed] [Google Scholar]
- 31.Gomez LA, de Las Pozas A, Reiner T, Burnstein K, Perez-Stable C. Increased expression of cyclin B1 sensitizes prostate cancer cells to apoptosis induced by chemotherapy. Mol Cancer Ther. 2007;6:1534–43. doi: 10.1158/1535-7163.MCT-06-0727. [DOI] [PubMed] [Google Scholar]
- 32.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 33.Soni DV, Sramkoski RM, Lam M, Stefan T, Jacobberger JW. Cyclin B1 is rate limiting but not essential for mitotic entry and progression in mammalian somatic cells. Cell Cycle. 2008;7:1285–300. doi: 10.4161/cc.7.9.5711. [DOI] [PubMed] [Google Scholar]
- 34.Sharma PS, Sharma R, Tyagi R. Inhibitors of cyclin dependent kinases: useful targets for cancer treatment. Curr Cancer Drug Targets. 2008;8:53–75. doi: 10.2174/156800908783497131. [DOI] [PubMed] [Google Scholar]
- 35.Ekholm-Reed S, Mendez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawamura K, Izumi H, Ma Z, et al. Induction of centrosome amplification and chromosome instability in human bladder cancer cells by p53 mutation and cyclin E overexpression. Cancer Res. 2004;64:4800–9. doi: 10.1158/0008-5472.CAN-03-3908. [DOI] [PubMed] [Google Scholar]
- 37.Meulmeester E, Jochemsen AG. p53: a guide to apoptosis. Curr Cancer Drug Targets. 2008;8:87–97. doi: 10.2174/156800908783769337. [DOI] [PubMed] [Google Scholar]
- 38.Yu H, Yao X. Cyclin B1: conductor of mitotic symphony orchestra. Cell Res. 2008;18:218–20. doi: 10.1038/cr.2008.20. [DOI] [PubMed] [Google Scholar]
- 39.Langard S. Nickel-related cancer in welders. Sci Total Environ. 1994;148:303–9. doi: 10.1016/0048-9697(94)90408-1. [DOI] [PubMed] [Google Scholar]
- 40.Shen HM, Zhang QF. Risk assessment of nickel carcinogenicity and occupational lung cancer. Environ Health Perspect. 1994;102(Suppl 1):275–82. doi: 10.1289/ehp.94102s1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinstat-Saslow D, Merino MJ, Manrow RE, et al. Overexpression of cyclin D mRNA distinguishes invasive and in situ breast carcinomas from non-malignant lesions. Nat Med. 1995;1:1257–60. doi: 10.1038/nm1295-1257. [DOI] [PubMed] [Google Scholar]
- 42.Ouyang W, Zhang D, Li J, Verma UN, Costa M, Huang C. Soluble and insoluble nickel compounds exert a differential inhibitory effect on cell growth through IKKα-dependent cyclin D1 down-regulation. J Cell Physiol. 2009;218:205–14. doi: 10.1002/jcp.21590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berglund P, Landberg G. Cyclin e overexpression reduces infiltrative growth in breast cancer: yet another link between proliferation control and tumor invasion. Cell Cycle. 2006;5:606–9. doi: 10.4161/cc.5.6.2569. [DOI] [PubMed] [Google Scholar]
- 44.Mazumder S, DuPree EL, Almasan A. A dual role of cyclin E in cell proliferation and apoptosis may provide a target for cancer therapy. Curr Cancer Drug Targets. 2004;4:65–75. doi: 10.2174/1568009043481669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costa M, Cantoni O, de Mars M, Swartzendruber DE. Toxic metals produce an S-phase-specific cell cycle block. Res Commun Chem Pathol Pharmacol. 1982;38:405–19. [PubMed] [Google Scholar]
- 46.Salnikow K, Davidson T, Kluz T, Chen H, Zhou D, Costa M. GeneChip analysis of signaling pathways effected by nickel. J Environ Monit. 2003;5:206–9. doi: 10.1039/b210262p. [DOI] [PubMed] [Google Scholar]
- 47.Salnikow K, An WG, Melillo G, Blagosklonny MV, Costa M. Nickel-induced transformation shifts the balance between HIF-1 and p53 transcription factors. Carcinogenesis. 1999;20:1819–23. doi: 10.1093/carcin/20.9.1819. [DOI] [PubMed] [Google Scholar]
- 48.Lee SH, Choi JG, Cho MH. Apoptosis, bcl2 expression, and cell cycle analyses in nickel(II)-treated normal rat kidney cells. J Korean Med Sci. 2001;16:165–8. doi: 10.3346/jkms.2001.16.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Z, Suo Z, Sudbo J, Holm R, Boysen M, Reith A. Diagnostic implications of p53 protein reactivity in nasal mucosa of nickel workers. Anal Quant Cytol Histol. 1997;19:345–50. [PubMed] [Google Scholar]
- 50.Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 51.Mashal RD, Lester S, Corless C, et al. Expression of cell cycle-regulated proteins in prostate cancer. Cancer Res. 1996;56:4159–63. [PubMed] [Google Scholar]
- 52.Chang TH, Schlegel R. SV40 T antigen increases the expression and activities of p34cdc2, cyclin A, and cyclin B prior to immortalization of human diploid fibroblasts. J Cell Biochem. 1996;60:161–72. doi: 10.1002/(sici)1097-4644(19960201)60:2<161::aid-jcb1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 53.Soria JC, Jang SJ, Khuri FR, et al. Overexpression of cyclin B1 in early-stage non-small cell lung cancer and its clinical implication. Cancer Res. 2000;60:4000–4. [PubMed] [Google Scholar]
- 54.Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–51. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 55.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 56.Ouyang W, Li J, Shi X, Costa M, Huang C. Essential role of PI-3K, ERKs and calcium signal pathways in nickel-induced VEGF expression. Mol Cell Biochem. 2005;279:35–43. doi: 10.1007/s11010-005-8214-3. [DOI] [PubMed] [Google Scholar]
- 57.Robinson CJ, Stringer SE. The splice variants of vascular endothelial growth factor (VEGF) and their receptors. J Cell Sci. 2001;114:853–65. doi: 10.1242/jcs.114.5.853. [DOI] [PubMed] [Google Scholar]
- 58.Blagosklonny MV. Antiangiogenic therapy and tumor progression. Cancer Cell. 2004;5:13–7. doi: 10.1016/s1535-6108(03)00336-2. [DOI] [PubMed] [Google Scholar]
- 59.Larcher F, Murillas R, Bolontrade M, Conti CJ, Jorcano JL. VEGF/VPF overexpression in skin of transgenic mice induces angiogenesis, vascular hyperpermeability and accelerated tumor development. Oncogene. 1998;17:303–11. doi: 10.1038/sj.onc.1201928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.