Abstract
Mammalian oxygen homeostasis is dependent on the HIF family of transcription factors. The CSN subunit, CSN5, binds both the CODD of HIF-1α and the pVHL tumor suppressor. High CSN5 expression generates a pVHL-independent form of CSN5 that stabilizes HIF-1α aerobically by inhibiting HIF-1α prolyl-564 hydroxylation. Aerobic CSN5 association with HIF-1α occurs independently of the CSN holocomplex, leading to HIF-1α stabilization independent of Cullin 2 deneddylation. CSN5 weakly associates with HIF-1α under hypoxia, but is required for optimal hypoxia-mediated HIF-1α stabilization. These results indicate that CSN5 regulates aerobic as well as hypoxic HIF-1α stability by different mechanisms during oncogenesis.
Keywords: Von Hippel-Lindau, COP9 signalosome, hypoxia inducible factor-1α
Adaptation to hypoxia in humans requires the hypoxia-inducible factor (HIF) family of transcriptional regulators (for review, see Kim and Kaelin 2003). Various isoforms of both HIF-α and HIF-β exist, with HIF-1α and HIF-2α protein stability being regulated by dioxygen levels (Kim and Kaelin 2003). HIF-1α is targeted for ubiquitylation and degradation under aerobic conditions by trans-4-hydroxylation of highly conserved LXXLAP motifs in oxygen-dependent degradation domains (ODDs) at P564 (CODD) and P402 (NODD; Kim and Kaelin 2003). The 4-prolyl-hydroxylation is performed by a conserved family of O2-, Fe(II)-, and 2-oxoglutarate-dependent prolyl-hydroxylase isozymes termed prolyl hydroxylase domain enzymes (PHD1-3; Kim and Kaelin 2003). Prolyl-hydroxylation allows recognition of HIF-1α by the von Hippel-Lindau tumor suppressor protein (pVHL), which serves as the recognition component of an E3 ubiquitin ligase complex (pVHL/Elongin C/Elongin B [VCB], Cullin 2 [Cul2], and Rbx-1). Limiting oxygen availability or direct inhibition of the PHD enzymes by large divalent metal ions, iron chelators, certain 2-oxoglutarate analogs, or the absence of ascorbate allows HIF-α to escape hydroxylation and recognition by pVHL. Aerobic HIF-1α stabilization can also be induced by a variety of growth factors and oncogenes (Kim and Kaelin 2003).
The COP9 signalosome (CSN) holocomplex is composed of eight subunits designated CSN1–CSN8 (for review, see Wolf et al. 2003). Until recently, the function of the CSN was obscure, although it appeared to be involved in regulating protein stability. Mutational analysis in Saccharomyces pombe revealed that CSN disruption resulted in the accumulation of neddylated cullins (Wolf et al. 2003). The conjugation of the ubiquitin-like protein Nedd8 to cullins is thought to be required for ubiquitin E2 recruitment. CSN5 is found both in the nucleus and cytoplasm and contains a JAB-1/MPN domain metalloenzyme motif (JAMM) that forms the catalytic region of an isopeptidase (Wolf et al. 2003). CSN5 requires assembly into the CSN holocomplex to function as a Nedd8 isopeptidase. The CSN5 JAMM domain is responsible for the cleavage of Nedd8 from cullins, resulting in an inability of cullin complexes to catalyze ubiquitin ligation. Ongoing Cul2 neddylation is required for pVHL-mediated HIF-α degradation, and inhibition of Nedd8 conjugation to Cul2 results in HIF-1α protein accumulation under aerobic conditions (Ohh et al. 2002). Evidence exists, however, that suggests the deneddylating function of the CSN holoenzyme is not the sole function of this protein complex and that smaller CSN-containing complexes exist that do not possess Cullindeneddylating functions (Wolf et al. 2003). Therefore, the CSN complexes are thought to function in the regulation of protein stability in a variety of ways.
Cullin-containing E3 ubiquitin ligases control the degradation of multiple proteins linked to oncogenic pheno-types such as those involved in cell cycle control, apoptosis, vascular morphogenesis, meiosis-to-mitosis transition, DNA-damage responses, and oxygen homeostasis (Wolf et al. 2003). Thus, regulators of these ligases could potentially be mutated or exhibit altered expression in cancer patients. This study examines the role of CSN5 in the aerobic and hypoxic regulation of HIF-1α protein stabilization. Our data suggest that the high CSN5 expression seen in some tumors is sufficient for aerobic HIF-1α stability by directly preventing HIF-1α proline 564 hydroxylation and VHL-mediated recognition and ubiquitylation.
Results and Discussion
CSN5 binds HIF-1α and regulates its transcriptional activity under aerobic conditions
Retroviral cDNA libraries were screened by using a cell line stably expressing an HIF-inducible (3xHRE) promoter driving hnGFP expression by FACS to isolate clones that possessed gain-of-function aerobic HIF activity. One of the cDNAs possessing the ability to activate the 3xHRE reporter under aerobic conditions was CSN5. Expression of CSN5 led to the activation of the 3xHRE-hnGFP reporter (Fig. 1A), stabilization of HIF-1α protein, and expression of the HIF-responsive VEGF gene (Fig. 1B). Expression of other CSN subunits (CSN6) or CSN5 paralogs (Rpn11) are insufficient for HIF-1α stabilization or HIF-dependent gene expression.
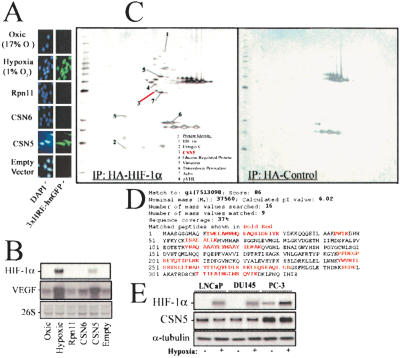
Figure 1.
(A) Functional screen for activators of HIF activity. U-251 cells stably expressing three tandomly linked hypoxia-response elements (3xHRE) driving hnGFP gene expression were used to screen retroviral expression libraries. Highly fluorescent clones (aerobic) were FACS sorted and expanded, and the expressed cDNA was isolated. CSN5 was identified in 3 of 13 clones expressing aerobic hnGFP. Shown is the positive control of HIF-mediated transcription (hypoxia 1% O2) and negative control (17% O2). U-251 (3xHRE) cell lines containing Rpn11 (CSN5 paralog and negative control), CSN6 (CSN cofactor and negative control), or CSN5 are shown in the lower three panels. (B) Protein and mRNA from U-251 cellular lysates were detected by Western blot (anti-HIF-1α, top panel) or HIF-regulated gene expression assessed by increased VEGF in Northern blots (lower panel). (C) Proteomic identification of CSN5 as an HIF-1α binding protein. LNCaP cells were retrovirally infected with pLXSN-HA-HIF-1α or the empty pLXSN-HA vector. Forty-eight hours postinfection, HA-HIF-1α and HA-complexed proteins were isolated by using anti-HA affinity chromatography under native lysis conditions. Protein complexes were washed, separated by two-dimensional electrophoresis, and silver stained. Proteins differentially associated with HA-HIF-1α and not the HA-epitope alone were excised, proteolytically digested, and subjected to MALDI-MS for mass peptide fingerprinting. CSN5 was one of the most prominently stained spots (red arrow). (D) The MASCOT Search results for the CSN5 MS spectrum. (E) CSN5 is expressed at higher levels in prostate cancer cells containing deregulated (aerobic) HIF-1α stabilization. LNCaP, DU145, and PC-3 cell lines were subjected to 17% O2 (-) or 1% O2 (+) for 12 h, and the proteins blotted for HIF-1α or CSN5.
An affinity purification of aerobic HA-tagged HIF-1α protein complexes was performed concurrent with the functional genetic screen by using mass spectrometry to identify proteins that directly interact with HIF-1α. CSN5 was found to specifically associate with HA-HIF-1α but not the empty vector (Fig. 1C). Thus, CSN5 was found to positively regulate HIF function and to be physically associated with the HIF-1α complex, supporting a previous CSN5–HIF-1α interaction found in a yeast two-hybrid study (Bae et al. 2002). However, that study suggested that the mechanism underlying CSN5-induced HIF-1α protein stabilization was related to CSN5 competition with p53 for HIF-1α. Yet CSN5 stabilizes HIF-1α at higher pO2 concentrations (1%–17% O2) than hypoxia-stabilized (0.1%–0% O2) p53, suggesting CSN5 mediates aerobic and hypoxic HIF-1α stabilization independent of p53.
CSN5 is overexpressed and required for aerobic HIF-1α stability in PC-3
HIF-1α has been shown to be aerobically stabilized in the prostate bone metastasis PC-3, but not DU145 or LNCaP prostate lymph node metastases, by an undetermined mechanism (Jiang et al. 2001). CSN5 is expressed at a higher level in PC-3 than in DU145 and LNCaP (Fig. 1E). Although CSN5 expression is altered by only several fold in PC-3, CSN5 basal protein expression is already very high; thus, relatively small changes in CSN5 expression could shift homeostatic balance of pVHL-associated CSN5 to the target of this ligase (HIF-1α), which is present in far lower cellular concentrations. In this regard, it is interesting to note that the CSN5 gene is found on chromosome 8q, which has been found to possess multiple and frequent amplifications in prostate cancer (Schulz et al. 2002).
Aerobic CSN5 expression is necessary and sufficient to stabilize HIF-α
CSN5 significantly stabilized HIF-1α under aerobic conditions (Fig. 2A). Inhibition of 26S proteosome function by MG262 stabilized HIF-1α to a greater degree than did CSN5 alone, although the combination of CSN5 and MG262 was not additive, suggesting that CSN5 stabilizes HIF-1α through inhibition of a 26S-dependent degradation pathway. Interestingly, CSN5 overexpression in RCC4 cells having aerobic HIF-1α stability due to loss of pVHL does not synergize with pVHL loss, indicating that CSN5 and pVHL likely regulate HIF-1α stability by functioning within the same pathway (Fig. 2B).
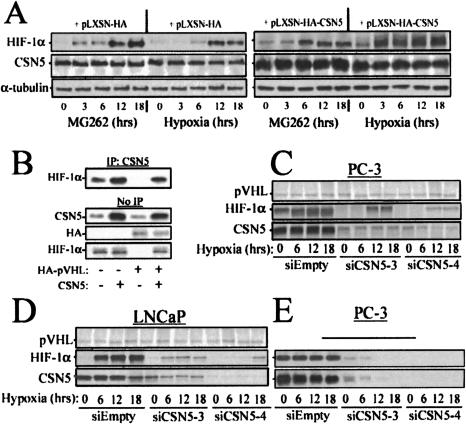
Figure 2.
CSN5 mediates HIF-1α stabilization upstream of the 26S proteosome and is additive to hypoxia-induced HIF-1α stabilization. (A) LNCaP were infected with HA-CSN5 or an HA-vector retrovirus, incubated 36 h, and placed under hypoxia (1% pO2) or treatment with 500 nM MG262 for the indicated durations. The cells were lysed at the indicated times and the protein blotted with anti-HIF-1α, CSN5, and α-tubulin. (B) Aerobic CSN5-induced HIF-1α stabilization is not significantly enhanced by loss of pVHL. RCC4 cells lacking wild-type pVHL and expressing aerobic HIF-1α were stably transfected with HA-pVHL and/or CSN5. Cell lysates were prepared and immunoblots performed to examine the levels of total CSN5, HA (wild-type pVHL), and HIF-1α (bottom three panels). Alternatively, cell lysates were immunoprecipitated by using anti-CSN5 and immunoblots performed to examine HIF-1α associated with the CSN5 immunoprecipitates (top panel). Aerobic HIF-1α stabilization is prevented and hypoxic HIF-1α stabilization is delayed by CSN5 RNAi knockdown. PC-3 (C,E) and LNCaP (D) cells were infected with siRNA-CSN5 or control retroviruses. The cell lines were incubated for 36 h before incubation under hypoxia (1% pO2; C,D) or aerobic conditions (E). The cells were lysed at the indicated times and the protein blotted with anti-HIF-1α, pVHL, and CSN5.
RNAi inhibition of CSN5 expression indicated that decreased CSN5 resulted in a delay in the kinetics of hypoxia-induced HIF-1α stabilization but not complete blockade of protein stabilization (Fig. 2C,D), whereas CSN5 expression is required to maintain aerobic HIF-1α expression (Fig. 2E). This suggests that CSN5 overexpression in certain tumors may be sufficient to maintain constitutive HIF-1α stabilization. Previous studies have also indicated that the CSN exhibits dual or paradoxical roles both in the degradation and stabilization of E3-mediated protein stability (Wolf et al. 2003). Thus, these results indicate that CSN5 is necessary for aerobic HIF-1α stability and mediates the kinetics and amplitude of hypoxia-inducible HIF-1α stability. This also implies that the regulation of CSN5 expression is a key event in determining whether E3 targets are stabilized or targeted for degradation.
CSN5 binds pVHL and the HIF-1α CODD irrespective of proline 564 hydroxylation
To examine if HIF-1α was physically associated with CSN5 independent of any associated proteins or posttranslational modifications, we incubated recombinant CSN5 with either HIF-1α, VCB, or p27 recombinant proteins, as shown in Figure 3A and B. CSN5 interacts specifically with the CODD and the positive control p27, but not the bHLH-PAS domain of HIF-1α, the NODD, Elongin C, or Elongin B. Interestingly, CSN5 bound the pVHL/Elongin C/Elongin B complex, suggesting that CSN5 interacts with both the E3 ligase and its substrate. Analysis of conserved identities among CSN5-binding proteins (CODD, p27, and pVHL) revealed limited homology, although certain D, E, and L residues could be aligned. Interestingly, CSN5 binds both the hydroxylated CODD as well as the nonhydroxylated CODD, suggesting it can recognize a form of HIF-1α that pVHL cannot (Fig. 3A). Although mutation of the critical HIF-1α CODD proline (564) greatly diminished CSN5 binding to the CODD, the rigid structural constraints conferred by prolyl residues on protein ternary structure makes it unclear whether decreased binding of CSN5 is due to direct contact with Pro564 or to radical structural alterations induced by loss of the prolyl residue in this small peptide.
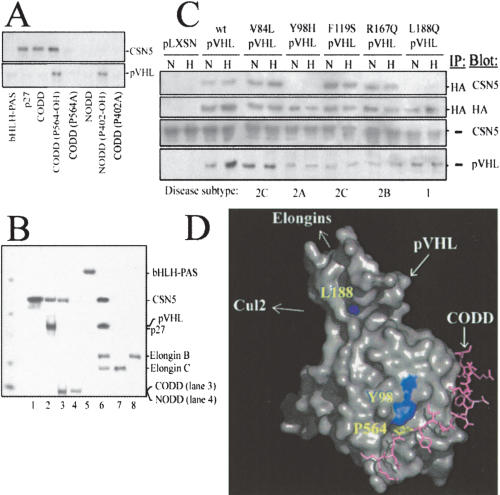
Figure 3.
CSN5 binds wild-type pVHL (but not specific clinically manifested mutants) in addition to the CODD, irrespective of P564-hydroxylation. (A) Recombinant His6-tagged bHLH-PAS, p27, or His6-tagged synthesized CODD, CODD(P564A), CODD(hydroxyl-P564), NODD, NODD (P402A), and NODD(hydroxyl-P402) were bound to individual NTA columns and incubated with either recombinant CSN5 (upper panel) or VCB (lower panel; 4 h at 4°C). Elutions were separated by SDS-PAGE and visualized by Coomassie stain. (B) Recombinant His6-tagged proteins (CSN5 [1], p27 [2], CODD [3], NODD [4], bHLH-PAS [5], “VCB” [6], Elongin C [7], Elongin B [8]) were bound to individual NTA columns. Recombinant CSN5 was loaded and incubated on the columns (4 h at 4°C) and washed thoroughly, and the His6-fusion proteins were proteolytically eluted. Elutions were separated by SDS-PAGE and visualized by Coomassie stain. (C) 786-0 cells expressing wild-type HA-pVHL or indicated mutants under hypoxic (H) or aerobic (N) conditions were immunoprecipitated with anti-HA and blotted as indicated with anti-HA, anti-pVHL, and anti-CSN5. (D) Crystal structure of the HIF-1α CODD (pink) complexed to pVHL (gray; Hon et al. 2002). pVHL residues that alter CSN5 binding are highlighted in yellow (pVHL L188 and Y98; and HIF-1α P564).
Clinical VHL mutations alter CSN5–pVHL interaction
To determine if clinically manifested pVHL mutations affect CSN5 interaction, we stably expressed several mutants representing the various phenotype/genotype pVHL classifications in the 786-0 VHL-/- cell line under aerobic or hypoxic conditions, and immunoprecipitated and immunoblotted them as shown (Fig. 3C). Hypoxia had no effect on the CSN5–pVHL interactions. Some mutations (Y98H and L188Q) but not others (V84L, F119S, R167Q) that lose the ability to degrade HIF-1α also prevent CSN5:pVHL interaction (Hoffman et al. 2001). This presents the possibility that a subset of VHL mutants could possess deregulated HIF-α degradation due to an inability to interact with CSN5. The HIF-1α (P564) and pVHL residues (Y98 and L188) that alter CSN5 binding when mutated are localized either in the HIF-1α–pVHL interface (P564, Y98) or in a region near the Elongin–pVHL contact interface (L188; Fig. 3D) and reside on a linear face of pVHL (Hon et al. 2002). If CSN5 and the rest of the CSN bind along this surface, the CSN holocomplex (>450–600 kD) could easily be juxtaposed within the proposed Cullin–Rbx1 interface, which is the known site of action for the CSN. Because CSN2 and CSN6 have previously been shown to interact with cullins and Rbx1, this model can explain how CSN5 can interact with both HIF-1α and the VCB–Cul2 E3 ligase complex.
Expression of CSN5 mutants incapable of cullin deneddylation still stabilize HIF-1α
The “neddylation” of Cul2 has been reported to be required for pVHL-mediated degradation of HIF-1α (Ohh et al. 2002). Expression of CSN5 could therefore potentially stimulate CSN-mediated Cul2 deneddylation, resulting in HIF-1α stabilization. Because the CSN5 JAMM motif binds metal ions that are required for its proteolytic activity, mutations in the metal-chelating residues (D151N) result in a nonfunctional peptidase. To test if CSN5-mediated HIF-1α stabilization is dependent on proteolytic activity and associated Cul2 deneddylation, we expressed either wild-type CSN5 or CSN5(D151N) and examined Cul2 neddylation and HIF-1α stabilization (Fig. 4A). CSN5 (D151N) expression led to HIF-1α stabilization to the same extent as wild-type CSN5 despite Cul2 remaining in a partially neddylated state. This indicates that CSN5 can mediate HIF-1α stability in a manner that is at least partially independent of its proteolytic activity.
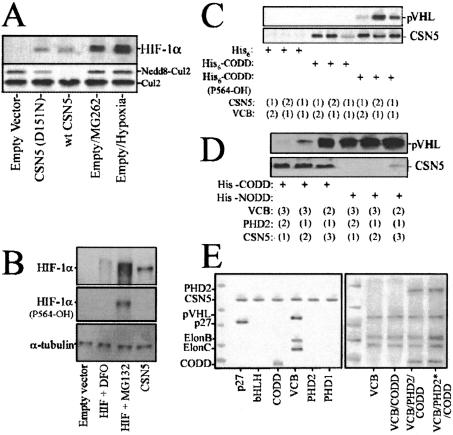
Figure 4.
(A) Expression of CSN5 mutants incapable of cullin deneddylation stabilize HIF-1α. LNCaP were retrovirally infected with empty vector (lanes 1,4,5), pLXSN-HA-CSN5 (lane 2), or pLXSN-HA-CSN5(D151N; lane 3). At 36 h postinfection, some of the cells were treated with 500 nM MG262 for 6 h (lane 4) or placed under hypoxia (1% pO2 for 12 h, lane 5). The cellular lysates were blotted for HIF-1α, Cul2, and pVHL. (B) CSN5 expression results in an inhibition of HIF-1α proline 564 hydroxylation. HKO cells were transiently transfected with full-length human HIF-1α, CSN5, or empty vector and treated with DFO (100 μM; 4 h) or MG132 (10 μM; 1 h). Total cell lysates were prepared and immunoblots were performed to examine the levels of total HIF-1α, hydroxylated proline 564 of HIF-1α, and α-tubulin. (C) CSN5 sterically prevents PHD hydroxylation of HIF-1α (P564) and pVHL binding. Synthesized His6-tagged CODD or CODD(hydroxyl-P564) were bound to individual NTA columns and incubated with either recombinant CSN5 or VCB in the order indicated for 4 h at 4°C [(1)/(1) denotes coincubation]. Elutions were blotted as indicated. (D) Recombinant His6-tagged CODD or NODD were bound to individual NTA columns and incubated with recombinant PHD2, CSN5, or VCB in the order indicated for 4 h at 4°C. Elutions were blotted as indicated. (E) CSN5 does not directly bind PHD1 or PHD2 or modulate their activity. In the left panel, recombinant His6-tagged CSN5 was bound to individual NTA columns and incubated with recombinant p27, bHLH, CODD, VCB, PHD2, or PHD1 for 4 h at 4°C. In the right panel, recombinant His6-tagged VCB complexes were bound to individual NTA columns (lanes 2–5). Recombinant HA-tagged PHD2 bound to anti-HA agarose was incubated with recombinant CSN5 (PHD2*) for 4 h at 4°C, washed, and eluted by using HA peptide. Recombinant HIF-1α CODD was incubated in buffer alone, untreated PHD2, or PHD2* for 4 h at 4°C and added to the His6-tagged VCB columns as indicated (lanes 3–5, respectively) and incubated for 2 h at 4°C. The columns were extensively washed, and the proteolytic elutions were separated by SDS-PAGE and visualized by Coomassie (left panel) or Fast Blue (right panel) stain.
CSN5 expression results in an inhibition of HIF-1α P564 hydroxylation
One mechanism by which CSN5 could alter HIF-1α stability would be to alter HIF-1α hydroxylation at P564. To test this possibility, we expressed HIF-1α in HIF-1α null fibroblasts (HKO) and treated them with DFO or MG132, or transfected them with CSN5 or an empty vector. The cell lysates were probed with an antibody that specifically recognizes the 4-hydroxylated P564 (Fig. 4B). DFO and CSN5 prevented HIF-1α P564 hydroxylation, whereas MG132 had no effect on the HIF-1α that was hydroxylated on P564. This indicates that CSN5 and DFO exert HIF-1α stabilization at or upstream of PHD-mediated HIF-1α hydroxylation prior to 26S-mediated degradation.
CSN5 sterically prevents HIF-1α P564 hydroxylation and pVHL recognition without affecting the activity of the PHDs
Synthetic His6-CODD or a hydroxylated P564 variant were bound to Ni2+-sepharose columns and CSN5 or the VCB complex added in the indicated sequence (Fig. 4C). CSN5 could in large part prevent pVHL association with the hydroxylated P564 CODD, suggesting that CSN5 directly prevents pVHL association with HIF-1α. The CODD-complexed CSN5 did not bind pVHL, indicating that the CSN5–pVHL interaction interface was masked or structurally altered, preventing this interaction. However, CSN5 was able to interact with the pVHL–CODD (OH-P564) as well as the pVHL–NODD (OH-P564) complexes, suggesting the possibility for multiple levels of CSN5-mediated regulation of the VCB E3 complex (Fig. 4D). CSN5 could potentially be altering CODD prolyl-hydroxylation by binding to or by proteolytically altering the PHDs into inactive states. We have found, however, that CSN5 does not bind either PHD1 or PHD2 directly (Fig. 4E, left panel) nor does it alter PHD2 activity when preincubated in in vitro interaction studies (Fig. 4E, right panel).
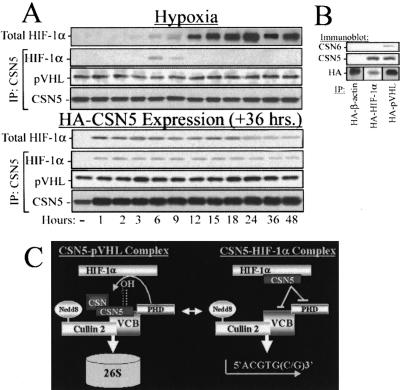
Endogenous CSN5 associates constitutively with pVHL and transiently with HIF-1α under hypoxic conditions
To more fully understand the CSN5–HIF-1α interaction under hypoxia, we conducted time courses by using LNCaP at 1% O2 (Fig. 5A, top) or LNCaP retrovirally infected with HA-HIF-1α under aerobic conditions (Fig. 5A, bottom). CSN5 associates with hypoxia-induced HIF-1α transiently and only during the initial phase of HIF-1α protein stabilization (Fig. 5A, top) but constitutively with aerobically stabilized HA-HIF-1α (Fig. 5A, bottom). To determine if the VCB complex associated with CSN5 with similar kinetics and transience to HIF-1α, we analyzed the CSN5 time courses shown in Figure 5A for pVHL. This indicates that pVHL is constitutively associated with CSN5, presenting the potential for dynamic antagonism between pVHL and CSN5 in the regulation of HIF-1α stability.
Figure 5.
CSN5 is transiently associated with HIF-1α at the onset of hypoxia-induced HIF-1α stabilization. (A) LNCaP were treated for the indicated durations at 1% O2 (top panels) or by using transient retroviral HA-HIF-1α expression (bottom panels) followed by native lysis; CSN5 IP; and blotting with anti-CSN5, anti-HIF-1α, or anti-pVHL. Whole lysates were blotted with HIF-1α as well to indicate HIF-1α stabilization (top panel). CSN-independent CSN5 is associated with HIF-1α, whereas the CSN holocomplex is associated with the pVHL E3 ligase. (B) 786-0 cells lacking wild-type pVHL or wild-type HIF-1α expression were retrovirally infected with HA-β-actin (lane 1), HA-HIF-1α (lane 2), or HA-pVHL (lane 3). At 36 h postinfection, the cells were lysed using native protocols, immunoprecipitated, and subjected to Western blotting as indicated. (C) A model of CSN5 regulation of HIF-1α stability when in differential CSN5–HIF-1α or CSN5–VCB complexes.
CSN5 has been proposed to have multiple and largely undefined roles in the cell, depending on whether it is associated with the other CSN subunits or performing a CSN-independent function (Wolf et al. 2003) We therefore isolated HA-HIF-1α or HA-pVHL bound CSN5 fractions and examined if other CSN subunits were associated with these complexes (Fig. 5B). Although HA-HIF-1α contained CSN5 but not other CSN subunits, HA-pVHL fractions contained CSN subunits associated with the CSN holoenzyme. This leads us to propose the following model (Fig. 5C), in which the CSN5 monomer functions to block PHD2 and pVHL-mediated recognition of HIF-1α and the pVHL-associated CSN holoenzyme assists HIF-1α protein degradation by facilitating Cul2 neddylation/deneddylation.
The data presented in this communication demonstrate that CSN5 interacts directly with both the CODD (independent of prolyl-hydroxylation state) and the pVHL E3 ligase. Because CSN5, pVHL, and the PHDs interact with the same region of HIF-1α, the potential for CSN5 altering pVHL or PHD affinity for HIF-1α seems likely. This poses the possibility that CSN5-associated pVHL and PHD-associated pVHL could have reciprocal and antagonistic association/dissociation mechanisms, leading to the dramatic switch-like mechanism seen in vivo. This is supported by the finding that CSN5 expression leads to an inhibition of HIF-1α hydroxylation at P564, signifying that CSN5 sterically prevents PHD association with HIF-1α but does not directly interact with or alter PHD enzymatic activity.
In conclusion, CSN5 expression is sufficient and necessary for aerobic HIF-1α stabilization and is dependent on CSN5 expression level. CSN5 expression also affects the kinetics and amplitude of hypoxia-induced HIF-1α stabilization. Overexpression of CSN5 has been associated with tumor progression and could reflect the deregulation of aerobic HIF stability, which is thought to be an important transition in tumor progression. Recently, CSN5 mRNA expression has been shown to be down-regulated by prolonged and severe hypoxia (Denko et al. 2003). Under these same conditions, HIF-1α stability is also decreased, further supporting the functional relationship between the two molecules. Because other known regulators of HIF stability or function (PHD2, ARD1) are also regulated at the message level, CSN5 regulation by hypoxia is consistent with the tight control of HIF expression controlled by these feedback mechanisms.
Materials and methods
Antibodies and plasmids
MG262 was purchased from Affiniti. The antibodies used were as follows: anti-Cul2 (abcam), anti-α-tubulin (Research Diagnostic, Inc.), anti-CSN5 and anti-CSN6 (Affiniti), anti-HIF-1α (USB), anti-pVHL (Transduction Labs), and anti-HA-agarose (Roche). Anti-hydroxylated HIF-1α (P564) was described previously (Chan et al. 2002). HA-VHL was cloned into pcDNA 3.1. The VHL and Elongin B genes were cloned into pGEX/KG. Elongin C, Elongin B, PHD2, CSN5, p27, and the HIF-1α-bHLH-PAS (residues 1–295) were expressed in pXA-BN. The CODD fragment (residues 543–583) and NODD fragments (residues 380–440) were expressed in pGEX/KG. The cDNAs encoding CSN5, CSN6, and Rpn11 were expressed in pLXSN-HA. Mutations in VHL (V84L, Y98H, F119S, R167Q, and L188Q) and CSN5 (D151N) were introduced by PCR-mediated mutagenesis.
Protein binding assays and structural studies
Protein tertiary structures were elaborated by using Molscript and Pymd programs. The fusion proteins were induced with IPTG and disrupted by sonication on ice in 50 mM Tris-HCl (pH 8.0), 0.25 M NaCl, and 0.2% inhibitor cocktail (Sigma P8340). The supernatant was separated by IMAC (Ni2+; Amersham Pharmacia); washed; eluted with imidazole, thrombin, or the addition of SDS gel loading dye; boiled for 5 min; separated on a 4%–12% SDS-PAGE gel; and visualized by Coomassie staining. For preparation of the VCB complex, pGEX/KG–VHL–ElonB and pXA–ElonC were cotransformed and expression was carried out as described earlier and purified using a glutathione Sepharose 4B column. The CODD and NODD were hydroxylated by PHD2 in vivo when both proteins were coexpressed in Escherichia coli with the addition of 2 mM 2-oxoglutarate, 2 mM ascorbate, and 250 μM FeSO4. All affinity-purified proteins were further purified by gel-filtration chromatography.
Functional expression library screen
The model is described in the legend for Figure 1. Clones that had low aerobic (basal) hnGFP and significant hypoxia hnGFP inducibility were isolated. A single clone was expanded and used to screen human prostate and brain retroviral libraries (BD Biosciences Clontech). At 36 h postinfection, aerobic clones were sorted by FACS for the 0.002% most highly fluorescent cells. The individual cells were expanded in 96 well plates and the retrovirally expressed cDNA for each clone was isolated.
Two-dimensional electrophoresis and mass spectrometry
LNCaP cells were infected with pLXSN-HA-HIF-1α or pLXSN-HA. Infected cells were lysed in “native” lysis buffer (0.1% NP-40, 150 mM NaCl, 10 mM Tris-HCl at pH 7.4, 10 mM PMSF, 10 mM sodium orthovanadate, 0.2% protease inhibitor cocktail). One milligram of protein was immunoprecipitated with 30 μL anti-HA/agarose overnight at 4°C. Beads were washed five times and proteins eluted in 500 μL rehydration buffer (Bio-Rad). ReadyStrip 17-cm IPG strips (pH 3–10; Bio-Rad) were actively rehydrated with sample followed by isoelectric focusing. Focused strips were equilibrated (Bio-Rad) for 10 min, separated on 8%–16% SDS-PAGE gels, and then silver stained by using a low-fixation procedure. Spots were excised, equilibrated in 200 μL of 25 mM (NH4)2CO3 for 10 min, and destained in 200 μL of 100 mM Na2O3S3 and 30 mM FeK3(CN)6 for 10 min. The gel pieces were washed two times with 200 μL of 25 mM (NH4)2CO3, dehydrated in 50% CH3CN/50% 25 mM (NH4)2CO3 for 10 min, then in 100% CH3CN for 5 min, followed by vacuum centrifugation. Proteins were digested by trypsin at room temperature overnight. Peptides (0.5 μL) were loaded onto a MALDI-TOF plate, allowed to dry, and 0.5 μL of matrix (80% CH3CN/20% 25 mM [NH4]2CO3) added. Peptides were separated by MALDI-TOF (Voyager DE-STR, Applied Biosystems), and generated spectra were calibrated to trypsin autolysis products and searched against protein databases by using MASCOT.
Immunoprecipitations (IPs) and Westerns
Following treatment as described, the cells were washed (two times PBS), lysed as described earlier, and cleared using IgG-Protein G Plus-agarose (Bio-Rad). One milligram of protein was incubated overnight at 4°C with 10 μg of the indicated antibody followed by 30 μL Protein G agarose (1 h). IPs were washed five times with lysis buffer and eluted by boiling for 5 min in 30 μL SDS loading dye. Proteins were separated on a 12% SDS-PAGE and immunoblotted with the indicated antibody.
Cell culture and gene expression
All cells were grown in DMEM supplemented with 10% FBS in 15-cm tissue culture plates until confluent. Hypoxia treatment was carried out in a Bactron II hypoxia chamber (1% O2, 5% CO2, balance N2; Sheldon). Transfections and retroviral infections were carried out as previously described (Chan et al. 2002).
Retroviral CSN5 siRNA construction and expression
The corresponding sequences are siCSN5-3 (5′-GCUCAGAGUAUC GAUGAAAUUCAAGAGAUUUCAUCGAUACUCUGAGCUU-3′) and siCSN5-4 (5′-CAUGCAGGAAGCUCAGAGUUUCAAGAGAACUCU GAGCUUCCUGCAUGUU-3′). The reverse complement was synthesized and the two strands annealed, cut at the 3′ and 5′ ends, and directionally inserted into the recently described pMSCV-U6 retroviral cassette (Devroe and Silver 2002). The siRNA vectors were transfected in the AMPHO-Phoenix retroviral packaging cell lines. Retroviruses were infected into cells and incubated for a further 36 h.
Acknowledgments
We regret that few primary references could be cited due to space limitations. We acknowledge the generous gifts of reagents from J. Gleadle and P. Ratcliffe (PHD cDNAs), J. Conaway (pVHL, Elongin C and Elongin B vectors), and P. Silver (pMSCV-U6). We also thank the excellent administrative and technical assistance from Debra Becker, Laura Casias, Barbara Frederick, and Alyssa DiGiacomo. This research was supported by grants from Pfizer Pharmaceuticals and NCI CA102301 (W.Z), the American Heart Association 0225388Z (C.V.F.), and NCI CA67166 (A.J.G.)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1180104.
Corresponding author.
References
- Bae M.K., Ahn, M.Y., Jeong, J.W., Bae, M.H., Lee, Y.M., Bae, S.K., Park, J.W., Kim, K.R., and Kim, K.W. 2002. Jab1 interacts directly with HIF-1alpha and regulates its stability. J. Biol. Chem. 277: 9-12. [DOI] [PubMed] [Google Scholar]
- Chan D.A., Sutphin, P.D., Denko, N.C., and Giaccia, A.J. 2002. Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1alpha. J. Biol. Chem. 277: 40112-40117. [DOI] [PubMed] [Google Scholar]
- Denko N.C., Fontana, L.A., Hudson, K.M., Sutphin, P.D., Raychaudhuri, S., Altman, R., and Giaccia, A.J. 2003. Investigating hypoxic tumor physiology through gene expression patterns. Oncogene 22: 5907-5914. [DOI] [PubMed] [Google Scholar]
- Devroe E. and Silver, P.A. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M.A., Ohh, M., Yang, H., Klco, J.M., Ivan, M., and Kaelin Jr., W.G. 2001. von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum. Mol. Genet. 10: 1019-1027. [DOI] [PubMed] [Google Scholar]
- Hon W.C., Wilson, M.I., Harlos, K., Claridge, T.D.W., Schofield, C.J., Pugh, C.W., Maxwell, P.H., Ratcliffe, P.J., Stuart, D.I., and Jones, E.Y. 2002. Structural basis for the recognition of hydroxyproline in HIF-1α by pVHL. Nature 417: 975-978. [DOI] [PubMed] [Google Scholar]
- Jiang B.H., Jiang, G., Zheng, J.Z., Lu, Z., Hunter, T., and Vogt, P.K. 2001. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 12: 363-369. [PubMed] [Google Scholar]
- Kim W. and Kaelin Jr., W.G. 2003. The von Hippel-Lindau tumor suppressor protein: New insights into oxygen sensing and cancer. Curr. Opin. Genet. Dev. 13: 55-60. [DOI] [PubMed] [Google Scholar]
- Ohh M., Kim, W.Y., Moslehi, J.J., Chen, Y., Chau, V., Read, M.A., and Kaelin Jr., W.G. 2002. An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 3: 177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz W.A., Elo, J.P., Florl, A.R., Pennanen, S., Santourlidis, S., Engers, R., Buchardt, M., Seifert, H.H., and Visakorpi, T. 2002. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer 35: 58-65. [DOI] [PubMed] [Google Scholar]
- Wolf D.A., Zhou, C., and Wee, S. 2003. The COP9 signalosome: An assembly and maintenance platform for cullin ubiquitin ligases? Nat. Cell Biol. 5: 1029-1033. [DOI] [PubMed] [Google Scholar]