Abstract
The bulk of cellular proteins derive from the translation of eukaryotic translation initiation factor (eIF)4E-bound mRNA. However, recent studies of nonsense-mediated mRNA decay (NMD) indicate that cap-binding protein (CBP)80-bound mRNA, which is a precursor to eIF4E-bound mRNA, can also be translated during a pioneer round of translation. Here, we report that the pioneer round, which can be assessed by measuring NMD, is not inhibited by 4E-BP1, which is known to inhibit steady-state translation by competing with eIF4G for binding to eIF4E. Therefore, at least in this way, the pioneer round of translation is distinct from steady-state translation. eIF4GI, poly(A)-binding protein (PABP)1, eIF3, eIF4AI, and eIF2α coimmunopurify with both CBP80 and eIF4E, which suggests that each factor functions in both modes of translation. Consistent with roles for PABP1 and eIF2α in the pioneer round of translation, PABP-interacting protein 2, which is known to destabilize PABP1 binding to poly(A) and inhibit steady-state translation, as well as inactive eIF2α, which is also known to inhibit steady-state translation, also inhibit NMD. Polysome profiles indicate that CBP80-bound mRNAs are translated less efficiently than their eIF4E-bound counterparts.
Keywords: Pioneer translation initiation complex, steady-state translation initiation complex, nonsense-mediated mRNA decay
Nonsense-mediated mRNA decay (NMD) is a quality-control mechanism that eliminates mRNAs that prematurely terminate translation (Li and Wilkinson 1998; Hentze and Kulozik 1999; Maquat 2003, 2004a,b; Singh and Lykke-Andersen 2003). As such, NMD functions to eliminate the production of potentially deleterious truncated proteins. In mammalian cells, NMD is restricted to newly synthesized mRNA (Belgrader et al. 1994; Cheng et al. 1994). Consistent with this, data indicate that NMD targets mRNA bound by the cap-binding protein (CBP)80-CBP20 heterodimer during what has been called a pioneer round of translation (Ishigaki et al. 2001; Lejeune et al. 2002). The pioneer round precedes the exchange of CBP80-CBP20 by eukaryotic initiation factor (eIF)4E (Lejeune et al. 2002), which supports the bulk of cellular protein synthesis (Hershey and Merrick 2000).
Evidence for a pioneer round of translation consists of the following. First, nonsense-containing CBP80-bound mRNA is reduced in abundance to an extent that is comparable to the corresponding nonsense-containing eIF4E-bound mRNA (Ishigaki et al. 2001). Considering that eIF4E-bound mRNA derives from CBP80-bound mRNA (Lejeune et al. 2002), this finding indicates that NMD is restricted to CBP80-bound mRNA. Second, inhibition of translation and, thus, nonsense codon recognition using either cycloheximide or suppressor tRNA increases the level of nonsense-containing CBP80-bound mRNA (Ishigaki et al. 2001; Lejeune et al. 2002). Third, α-CBP80 antibody but not α-eIF4E antibody immunopurifies Upf2 and Upf3/3X (also called Upf3a/b) NMD factors (Ishigaki et al. 2001; Lejeune et al. 2002). Fourth, α-Upf3/3X antibody immunopurifies CBP80 but not eIF4E (Ishigaki et al. 2001). These findings also indicate that CBP80-bound mRNA but not eIF4E-bound mRNA is targeted for NMD.
Upf2 and Upf3/3X factors are recruited to mRNA by the exon junction complex (EJC) of proteins that is deposited ∼20-24 nt upstream of exon-exon junctions as a consequence of pre-mRNA splicing (Dreyfuss et al. 2002; Jurica and Moore 2003; Maquat 2003, 2004a; Reed 2003). This explains the observation that intron position within pre-mRNA generally determines which nonsense codons elicit NMD (Nagy and Maquat 1998). Considering all data, the pioneer translation initiation complex is envisioned to consist of newly synthesized, polyadenylated mRNA bound by CBP80-CBP20 at the 5′-cap, the EJC and associated Upf2 and Upf3/3X NMD factors at exon-exon junctions, and poly(A)-binding protein (PABP)2 at the 3′-poly(A) tail (Maquat 2003, 2004a; Singh and Lykke-Andersen 2003).
In this communication, we validate the existence of the pioneer round of translation in mammalian cells and continue to characterize constituents of the pioneer translation initiation complex. We demonstrate that the pioneer round of translation is distinct from steady-state translation by demonstrating that 4E-BP1, which is known to inhibit steady-state translation by competing with eIF4G for binding to eIF4E (Haghighat et al. 1995; Mader et al. 1995; Marcotrigiano et al. 1999), inhibits the production of luciferase activity as well as a secreted protein but does not inhibit NMD. We also demonstrate that eIF4GI, PAPB1, eIF3, eIF4AI, and eIF2α, which are known to function in steady-state translation, coimmunopurify not only with eIF4E but also with CBP80. At least eIF2α and PABP1 are integral components of the pioneer translation initiation complex based on functional assays in which an inhibitor of each protein was found to inhibit NMD. Therefore, despite the pioneer and steady-state modes of translation using distinct mRNP substrates, they involve several of the same translation initiation factors. An additional difference between the two modes of translation is evident with our finding that eIF4AIII, which may or may not be a translation initiation factor (see Discussion), coimmunopurifies with CBP80 but not with eIF4E. Finally, we use polysome gradients to show that CBP80-bound β-globin, glutathione peroxidase 1, and SMG7 mRNAs are translated less efficiently than their eIF4E-bound counterparts.
Results
4E-BP1 inhibits steady-state translation but not the pioneer round of translation
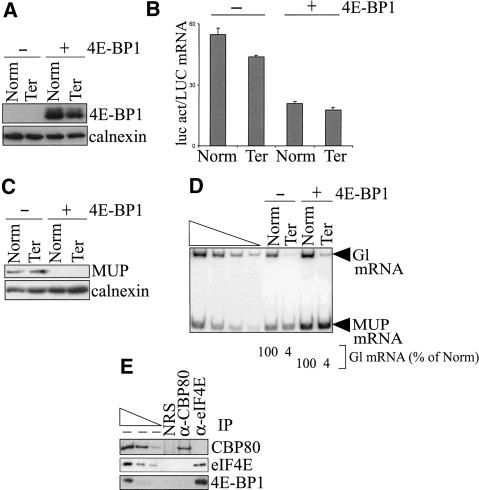
4E-BP1 inhibits steady-state translation by competing with eIF4G for binding to eIF4E (Haghighat et al. 1995; Mader et al. 1995; Marcotrigiano et al. 1999). In search of a difference between the pioneer round and steady-state translation to validate the existence of the pioneer round, we tested if 4E-BP1 specifically targets steady-state translation but not the pioneer round of translation. To this end, COS cells were transiently transfected with four plasmids: (1) one of two pmCMV-Gl test plasmids that encode either nonsense-free (Norm) or nonsense-containing (Ter) β-globin (Gl) mRNA (Zhang et al. 1998); (2) the phCMV-MUP reference plasmid (Belgrader and Maquat 1994); (3) the pGL2 reporter plasmid that produces luciferase (luc); and (4) one of two pACTAG2 plasmids that encode either nothing (-) or 4E-BP1 (+; Gingras et al. 1999). Two days after transfection, total COS-cell lysates were generated, and protein was purified from half. The levels of 4E-BP1 and calnexin were quantitated using Western blotting and α-4E-BP1 and α-calnexin antibodies. The levels of calnexin were used to control for variations in protein loading. Steady-state translation was measured by assaying luc activity and, subsequently, normalizing the level of activity to the level of LUC mRNA. Steady-state translation was also measured by assaying the level of MUP, which is a secreted protein unlike calnexin or luc. We rationalized that measuring the level of MUP, which is secreted within 30 min (H. Baumann, pers. comm.), would provide a more sensitive assay for the 4E-BP1-mediated block in steady-state translation than would measuring firefly luc, which has a half-life of 3 h (Thompson et al. 1991). Additionally, the association of 4E-BP1 with eIF4E and CBP80 was tested in immunopurifications (IPs) that used α-CBP80 antibody, α-eIF4E antibody, or normal rabbit serum (NRS), the latter of which controlled for nonspecific IP. RNA was purified from the other half of the lysate, and the pioneer round of translation was measured by quantitating the levels of Gl and MUP mRNAs using RT-PCR (Ishigaki et al. 2001; Lejeune et al. 2002, 2003). For each transfection, the level of Gl mRNA was normalized to the level of MUP mRNA to control for variations in transfection efficiencies and RNA recovery.
Expression of plasmid-encoded 4E-BP1 was demonstrated by Western blotting (Fig. 1A). Luc activity assays, which provided a measure of steady-state translation, demonstrated that 4E-BP1 expression reduced the production of luc activity threefold (Fig. 1B). 4E-BP1 expression also reduced to undetectable the amount of MUP produced from phCMV-MUP, which provided another measure of steady-state translation (Fig. 1C). In contrast, however, RT-PCR demonstrated that 4E-BP1 expression did not affect the relative levels of Gl Ter mRNA and Gl Norm mRNA, that is, did not abrogate NMD, which provided a measure of the pioneer round of translation (Fig. 1D). Importantly, a comparable (threefold) reduction of luc activity by a different inhibitor did abrogate NMD (see below), indicating that 4E-BP1 does not target the pioneer round of translation. The affinity of 4E-BP1 for eIF4E is regulated by phosphorylation: Hypophosphorylated 4E-BP1 binds efficiently to eIF4E, whereas hyperphosphorylated 4E-BP1 does not (Gingras et al. 1999). Expression of 4E-BP1 that harbored two amino acid changes (T37A and T46A) that preclude phosphorylation and support constitutive eIF4E binding (Gingras et al. 1999) also reduced the production of luc activity but did not affect NMD (data not shown). Consistent with an effect of 4E-BP1 on the production of luc activity but not NMD, 4E-BP1 was immunopurified with α-eIF4E antibody but not with α-CBP80 antibody or NRS (Fig. 1E).
Figure 1.
4E-BP1 inhibits the production of luc activity without abrogating NMD. COS cells were transiently transfected with pmCMV-Gl (Norm or Ter), phCMV-MUP, pGL2, and either pACTAG2 empty vector (-) or expressing 4E-BP1 (+). (A) 4E-BP1 is detected in cells transfected with a 4E-BP1 expression vector (+) but not in cells transfected with empty vector (-) as determined by Western blotting, where the level of endogenous calnexin controlled for variations in the amount of cellular protein analyzed. (B) Luc activity (act), which was normalized to the level of LUC mRNA, is inhibited in cells expressing 4E-BP1. (C) Production of the secreted protein MUP, which was measured relative to the level of calnexin, is inhibited in cells expressing 4E-BP1. (D) The level of nonsense-containing Gl mRNA (Ter) is the same percentage of the level of nonsense-free Gl (Norm) in cells regardless of 4E-BP1 expression. For each lane, the level of Gl mRNA was normalized to the level of MUP mRNA, and the normalized level of Norm was defined as 100. Serial dilutions of RNA in the left four lanes demonstrate that the RT-PCR analysis is semiquantitative. (E) α-eIF4E antibody but not α-CBP80 antibody, or, as a control for nonspecific IP, normal rabbit serum (NRS), immunopurifies 4E-BP1. Serial dilutions of protein in the left three lanes demonstrate that the Western blot analyses of immunopurified proteins using antibodies against CBP80, eIF4E, or 4E-BP1 are semiquantitative. All results are representative of at least two independently performed experiments.
Taken together, these results indicate that 4E-BP1 inhibits steady-state translation but not the pioneer round of translation. This finding supports our initial proposal that a pioneer round of translation does, in fact, take place and is distinct from steady-state translation (Ishigaki et al. 2001). Notably, the finding that inhibiting steady-state translation does not abrogate NMD is consistent with our previous conclusion that NMD does not target steady-state mRNA, that is, eIF4E-bound mRNA, but is restricted to newly synthesized mRNA, namely, CBP80-bound mRNA (Belgrader et al. 1994; Cheng et al. 1994; Ishigaki et al. 2001; Lejeune et al. 2002).
eIF4GI, PABP1, eIF3, eIF4AI, and eIF2α coimmunopurify with both CBP80 and eIF4E, whereas eIF4AIII coimmunopurifies with only CBP80
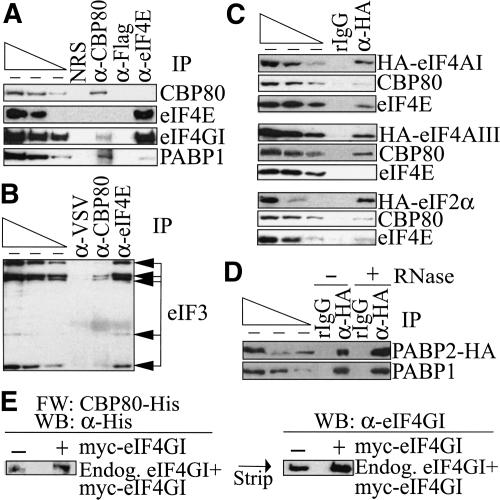
In previous characterizations of the pioneer translation initiation complex, CBP80 was shown to coimmunopurify not only with CBP20 in an RNase-insensitive manner but also with components of the EJC, NMD factors Upf2 and Upf3X, and with PABP2 in an RNase-sensitive manner (Ishigaki et al. 2001; Lejeune et al. 2002). CBP80 was also shown to coimmunopurify with eIF4GI and ribosomal protein L10 (Ishigaki et al. 2001; McKendrick et al. 2001). These results are consistent with the existence of a pioneer translation initiation complex that consists of mRNA bound by CBP80-CBP20 at the cap, EJCs and associated Upf2 and Upf3/3X proteins at exon-exon junctions, and PABP2 at the poly(A) tail. To further characterize the pioneer complex, total COS-cell lysates were immunopurified using α-CBP80 or α-eIF4E antibody. α-VSV and α-Flag antibody were used to control for the IP specificity of α-CBP80 antibody and α-eIF4E antibody, respectively. Subsequently, the presence of CBP80, eIF4E, and different steady-state translation initiation factors was analyzed using Western blotting.
CBP80 but not eIF4E was detected when protein was immunopurified with α-CBP80 antibody; eIF4E but not CBP80 was detected when protein was immunopurified with α-eIF4E antibody; and neither cap-binding protein was detected when protein was immunopurified using either α-VSV or α-Flag antibody, which demonstrated the specificity of each IP (Fig. 2A). eIF4GI and PABP1 were detected in both CBP80 and eIF4E IPs (Fig. 2A). Although the association of eIF4GI with CBP80 was shown previously (Ishigaki et al. 2001), the association of PABP1 with CBP80 was surprising considering that PABP2 had previously been shown to coimmunopurify with CBP80 as well as Upf3/3X (Ishigaki et al. 2001). The higher abundance of PABP1 in the CBP80 IP relative to the eIF4E IP may reflect differences in the stability of the interactions in cells, after cell lysis, or both. In theory, eIF4GI, PABP1, or any protein that coimmunopurifies with CBP80 may be an integral component of the pioneer translation initiation complex or involved in remodeling of the pioneer to the steady-state translation initiation complex. At least PABP1 appears to be an integral component based on results of functional studies (see below). Five subunits of eIF3 were also detected in both CBP80 and eIF4E IPs (Fig. 2B), where α-VSV served to control for the specificity of both IPs because both antibodies derived from rabbit.
Figure 2.
Steady-state translation initiation factors coimmunopurify not only with eIF4E but also with CBP80. (A) α-eIF4E antibody as well as α-CBP80 antibody immunopurify eIF4GI and PABP1. COS cells were lysed, and proteins were immunopurified using rabbit α-CBP80 or mouse α-eIF4E antibody, where rabbit α-VSV or mouse α-Flag antibody was used to control for the IP specificity, respectively. Immunopurified proteins were analyzed by Western blotting using antibodies against CBP80, eIF4E, eIF4GI, and PABP1. (B) α-eIF4E antibody as well as α-CBP80 antibody immunopurify subunits of eIF3. As in A except that rabbit α-VSV antibody was used to control for IP specificity of rabbit α-eIF4E, and immunopurified proteins were analyzed by Western blotting using antibody against eIF3. (C) eIF4AI and eIF2α coimmunopurify with CBP80 and eIF4E, whereas eIF4AIII coimmunopurifies with CBP80 and not eIF4E. COS cells were transfected with a vector expressing HA-eIF4AI, HA-eIF4AIII, or HA-eIF2α, and cell lysates were immunopurified using α-HA antibody or, as a control for IP specificity, rIgG. Immunopurified proteins were analyzed by Western blotting using antibody against HA, CBP80, or eIF4E. (D) PABP1 and PABP2 coimmunopurify in an RNase-insensitive manner. COS cells were transfected with a vector expressing PABP2-HA, and cell lysates were immunopurified using α-HA antibody or, as a control for the IP specificity, rIgG. Lysates were (+) or were not (-) exposed to RNase A prior to IP and analyzed by Western blotting using antibody against HA or PABP1. (E) Purified CBP80-His interacts directly with cellular eIF4GI and exogenously produced myc-eIF4GI using Far-Western analysis. Using untransfected cells (-) or cells transfected with an expression vector encoding myc-eIF4GI (+) at a level that was 2.5-fold above the level of endogenous eIF4GI, total-cell protein was subjected to Far-Western blotting (FW) using bacculovirus-produced CBP80-His. CBP80-His was subsequently detected by Western blotting (WB) using α-His antibody (left). Reactivity was removed, and blots were subjected to Western blotting using α-eIF4GI antibody (right). For all panels except E, serial dilutions of protein in the leftmost three lanes, which start with different amounts of protein based on antibody reactivity, demonstrate that the Western blot analyses are semiquantitative.
Neither eIF4AI nor eIF2α was detected in CBP80 or eIF4E IPs (data not shown). To increase the sensitivity of detecting either protein, COS cells were transiently transfected with a vector expressing either eIF4AI or eIF2α that harbored an N-terminal HA tag. Considering data indicating that eIF4AIII is a component of the EJC and is required for NMD (N. Sonenberg, pers. comm.), COS cells were also transiently transfected with a vector expressing eIF4AIII that harbored an N-terminal HA tag. IPs were performed using α-HA antibody or rIgG, the latter of which controlled for nonspecific IP using α-HA antibody. Western blotting demonstrated that CBP80 and eIF4E were detected in HA-eIF1AI or HA-eIF2α IPs (Fig. 2C). In contrast, CBP80 but not eIF4E was detected in the HA-eIF4AIII IP (Fig. 2C).
To further examine the significance of the association of PABP1 with the pioneer translation initiation complex, lysates were generated from COS cells that had been transiently transfected with a vector expressing PABP2 that harbored a C-terminal HA tag and immunopurified using α-HA antibody or rIgG. Prior to IP, half of the sample was exposed to RNase A. RT-PCR analysis of GAPDH mRNA in each IP demonstrated that digestion was complete in those samples exposed to RNase A (data not shown). Western blotting demonstrated that PABP1 was detected in the HA IP either with or without exposure to RNase A (Fig. 2D). These data suggest that PABP1 and PABP2 can be present on the same mRNA molecule and, thus, may coexist in the pioneer translation initiation complex (see below).
Taken together, results from the CBP80 and HA IPs indicate that the pioneer translation initiation complex appears to share several components with the steady-state translation initiation complex and also differs from the steady-state complex by its association with eIF4AIII (see Discussion).
Evidence that eIF4GI interacts directly with CBP80
In theory, CBP80 may function analogously to eIF4GI in the pioneer initiation complex, especially if eIF4GI is not an integral component of the complex. This possibility exists considering that (1) CBP80 contains sequences that are similar to the middle domain of eIF4GI that interact with eIF4AI, eIF4AII, eIF4AIII, and eIF3 (Aravind and Koonin 2000; Ponting 2000); (2) eIF4AIII is required for NMD (N. Sonenberg, pers. comm.); and (3) CBP80 coimmunopurifies with HA-eIF4AIII (Fig. 2C).
To test for a direct interaction between the CBP80 and eIF4GI, lysates from COS cells that were untransfected (-) or transfected (+) with a vector that encoded eIF4GI harboring an N-terminal myc tag (myc-eIF4GI) were generated and analyzed by Far-Western blotting for binding to purified recombinant CBP80 that harbored a C-terminal His tag (CBP80-His). CBP80-His interacted directly with endogenous eIF4GI as well as myc-eIF4GI in total-cell lysate (Fig. 2E, left). CBP80-His binding was shown to be to eIF4GI because binding at the same position was obtained when the blot was stripped and probed with α-eIF4GI antibody (Fig. 2E, right). These results indicate that CBP80 binds directly to eIF4GI independently of other cellular proteins. Although these data suggest that eIF4GI is an integral component of the pioneer translation initiation complex, we cannot rule out the possibility that the CBP80-eIF4GI interaction reflects an intermediate in the transition from the pioneer to the steady-state complex.
eIF2α functions in NMD
During steady-state translation, eIF2 forms a ternary complex with GTP and Met-tRNAiMet for the loading of GTP-Met-tRNAiMet that is required onto the 40S ribosomal subunit in the process of steady-state translation initiation (Hershey and Merrick 2000). eIF2α is the smallest subunit of eIF2 and a phosphoprotein (Kaufman et al. 1989). It is possible that eIF2α is an integral component of the pioneer translation initiation complex because HA-eIF2α coimmunopurified with CBP80 (Fig. 2C).
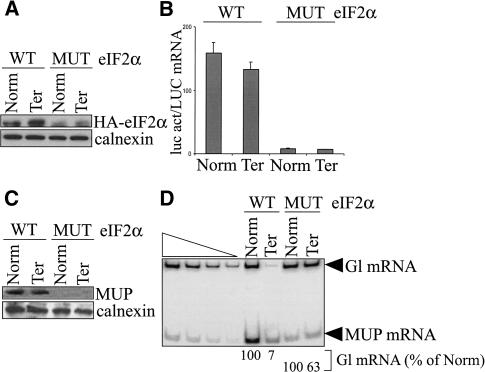
To test this possibility, eIF2α was assayed for a role in NMD. COS cells were transiently transfected with a pmCMV-Gl test plasmid (either Norm or Ter), the phCMV-MUP reference plasmid, the pGL2 reporter plasmid that produces luc, and one of two pMT2-HA-eIF2α plasmids. The latter encoded HA-eIF2α WT (wild type) or HA-eIF2α S51D MUT, which is a phosphomimetic version that is constitutively inactive (Srivastava et al. 1998). Total COS-cell lysates were generated, and the effects of each eIF2α protein were determined as were the effects of 4E-BP1 (Fig. 1).
Western blotting using α-HA antibody demonstrated that eIF2α WT and eIF2α MUT were expressed (Fig. 3A). Luc activity assays demonstrated that eIF2α MUT inhibited steady-state translation 19-fold relative to eIF2α WT (Fig. 3B). Consistent with this finding, eIF2α MUT also reduced the amount of MUP produced from phCMV-MUP to undetectable (Fig. 3C). Furthermore, RT-PCR analyses demonstrated that eIF2α MUT inhibited NMD ninefold relative to eIF2α WT (Fig. 3D). This result indicates that eIF2α is an integral component of the pioneer translation initiation complex.
Figure 3.
eIF2α MUT abrogates NMD, which indicates that eIF2α functions in NMD. COS cells were transiently transfected with pmCMV-Gl (Norm or Ter), phCMV-MUP, pGL2, and pMT2-HA-eIF2α, either WT (wild type) or S51D MUT. (A) Western blotting of total-cell protein using antibodies against HA and calnexin demonstrates that HA-eIF2α WT and HA-eIF2α MUT are expressed. (B) Assays of luc activity, where activity in total-cell protein was normalized to the level of LUC mRNA, demonstrate that eIF2α MUT inhibits activity relative to eIF2α WT. (C) Production of the secreted protein MUP, which was measured relative to the level of calnexin, is inhibited in cells expressing eIF2α MUT. (D) RT-PCR analysis of total-cell RNA using primers specific for Gl and MUP mRNAs demonstrates that eIF2α MUT inhibits NMD relative to eIF2α WT. Analyses were as in Figure 1D.
PABP1 functions in NMD
To determine if PABP1 is an integral component of the pioneer translation initiation complex together with PABP2 or if PABP1 is only detected in the process of replacing PABP2 during remodeling of the pioneer complex to the steady-state translation initiation complex, the effect of Paip2 on NMD was tested. Whereas Paip1 binds to PABP1 and enhances cellular translation as measured by luc activity (Craig et al. 1998), Paip2 competes with Paip1 for PABP1 so as to destabilize PABP1 binding to poly(A) and inhibit cellular translation (Khaleghpour et al. 2001).
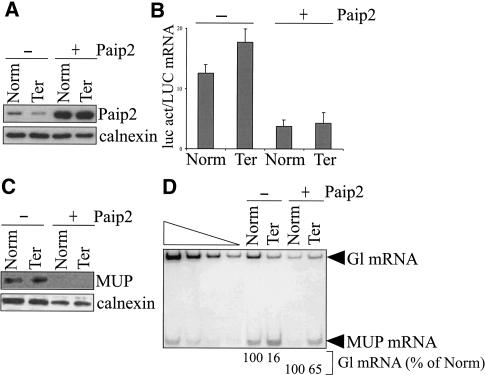
COS cells were transiently transfected with the pmCMV-Gl Norm or Ter test plasmid, the phCMV-MUP reference plasmid, the pGL2 reporter plasmid that produces luc, and either pcDNA3 or pcDNA3-HA-Paip2 (Khaleghpour et al. 2001). Total COS-cell lysates were generated, and the effects of Paip2 were determined. Western blotting using α-Paip2 antibody demonstrated that Paip2 was expressed at a level that was sevenfold above the level of endogenous Paip2 (Fig. 4A). Luc activity assays demonstrated that Paip2 expression inhibited the production of luc activity threefold (Fig. 4B). Consistent with this finding, Paip2 expression also reduced the amount of MUP produced from phCMV-MUP to undetectable (Fig. 4C). Finally, RT-PCR demonstrated that Paip2 inhibited NMD (Fig. 4D). Importantly, a comparable reduction of luc activity by 4E-BP1 did not abrogate NMD (Fig. 1), indicating that the Paip2-mediated inhibition of NMD is not caused by a general malaise of the cells or an indirect effect due to down-regulated global protein synthesis. Data obtained using Paip2, together with the finding that the interaction of PABP1 and PABP2 is stable after RNase A treatment (Fig. 2C), indicate that PABP1 functions in NMD and, therefore, is likely to be an integral component of the pioneer translation initiation complex along with PABP2. In view of data demonstrating that both PAPB2 and PABP1 shuttle between the nucleus and cytoplasm (Afonina et al. 1998; Calado et al. 2000), these data raise the important issue of when during mRNA maturation PABP1 joins the poly(A) tail.
Figure 4.
Paip2 abrogates NMD, which indicates that PABP1 functions in NMD. COS cells were transiently transfected with pmCMV-Gl (Norm or Ter), phCMV-MUP, pGL2, and either pcDNA3 (-) or pcDNA3-HA-Paip2 (+). (A) Western blotting of total-cell protein using antibodies against Paip2 and calnexin demonstrates that plasmid-encoded Paip2 is sevenfold more abundant than endogenous Paip2. (B) Assays of luc activity, where activity in total-cell protein was normalized to the level of LUC mRNA, demonstrate that Paip2 inhibits activity. (C) Production of the secreted protein MUP, which was measured relative to the level of calnexin, is inhibited in cells expressing Paip2. (D) RT-PCR analysis of total-cell RNA using primers specific for Gl and MUP mRNAs demonstrates that Paip2 inhibits NMD. Analyses were as in Figure 1D.
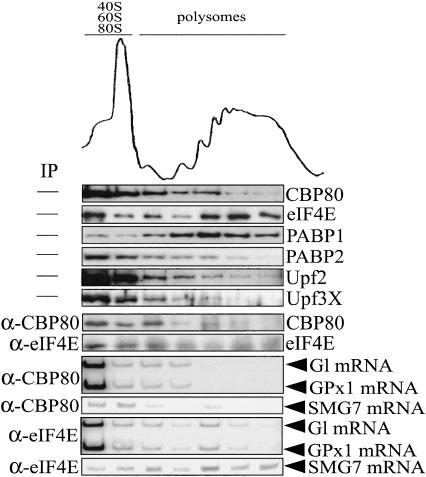
CBP80-bound Gl, GPx1, and SMG7 mRNAs are associated with fewer ribosomes than the respective eIF4E-bound mRNAs
In search of additional differences between the pioneer round of translation and steady-state translation, we determined if the translation initiation efficiency of CBP80-bound mRNA differs from that of eIF4E-bound mRNA. To this end, 293T cells were transiently transfected with nonsense-free pmCMV-Gl (i.e., Norm) and nonsense-free pmCMV-GPx1, the latter of which encodes glutathione peroxidase (GPx) 1 mRNA (Moriarty et al. 1998). Cells were lysed 2 d after transfection, and polysomes were fractionated according to size in a 10%-50% sucrose gradient. Fractions were collected. Proteins and RNAs in each fraction were analyzed using, respectively, Western blotting and RT-PCR before or after IP with α-CBP80 or α-eIF4E antibody.
eIF4E and PABP1, which are involved in steady-state translation (Borman et al. 2000), were detected in all fractions prior to IP and were concentrated in polysomes (Fig. 5). The polysome association of these proteins was deemed to be real because dissociating polysomes by adding EDTA prior to generating the gradient shifted protein migration to subpolysomal fractions (data not shown). CBP80, PABP2, and the NMD factors Upf2 and Upf3X, all of which are confined to the pioneer round of translation (Ishigaki et al. 2001; Lejeune et al. 2002), were also detected in all fractions but were concentrated in subpolysomal fractions (Fig. 5; endogenous CBP20 could not be assayed because of the low avidity of the α-CBP20 antibody). These results provide the first indication that CBP80-bound mRNAs are less efficiently translated than eIF4E-bound mRNAs. They are also consistent with the idea that at least some proteins that are associated with the pioneer round of translation are removed by translating ribosomes, as has been demonstrated for the EJC component Y14 (Dostie and Dreyfuss 2002).
Figure 5.
Evidence that CBP80-bound mRNAs are translated less efficiently than the corresponding eIF4E-bound mRNAs. 293T cells were transiently transfected with pmCMV-Gl and pmCMV-GPx1. Cells were lysed, and polysomes were separated according to size in a 10%-50% sucrose gradient. Each gradient fraction was analyzed using rabbit α-CBP80 or mouse α-eIF4E antibody. Proteins were analyzed using Western blotting and antibodies against CBP80, eIF4E, PABP1, PABP2, Upf2, or Upf3/3X. After IP using antibody against eIF4E or CBP80, CBP80 and eIF4E were analyzed using Western blotting, and Gl, GPx1, and SMG7 mRNAs were analyzed using RT-PCR. Results are representative of at least three independently performed experiments.
Analysis of the distribution of eIF4E after IP using α-eIF4E antibody and of CBP80 after IP using α-CBP80 antibody indicated that eIF4E relative to CBP80 is enriched in polysomes, as expected from data obtained before IP, although the efficiency of eIF4E IP at higher sucrose concentrations was less efficient than at lower sucrose concentrations (Fig. 5; data not shown for three other independently performed experiments). Consistent with the enrichment of eIF4E relative to CBP80 in polysomal fractions, Gl and GPx1 mRNAs as well as endogenous SMG7 mRNA in IPs using α-eIF4E antibody were enriched in polysomal fractions relative to the corresponding mRNAs in IPs using α-CBP80 antibody (Fig. 5). We do not understand why Gl and GPx1 mRNAs are relatively enriched in fractions that are smaller than 80S. Regardless, all of these data are consistent with the conclusion that CBP80-bound Gl and GPx1 mRNAs are translated less efficiently than their eIF4E-bound counterparts.
Notably, Gl mRNA is subject to nucleus-associated NMD, whereas GPx1 mRNA is subject to cytoplasmic NMD (Moriarty et al. 1998; Zhang et al. 1998). Therefore, the finding that both CBP80-bound Gl mRNA and CBP80-bound GPx1 mRNA are associated with monosomes and small polysomes suggests that (1) CBP80 by itself is inadequate to determine susceptibility to NMD; and (2) Gl mRNA, like GPx1 mRNA, acquires eIF4E primarily in the cytoplasm.
Discussion
Our earlier work offered support for the concept that NMD in mammalian cells occurs during a pioneer round of translation that targets CBP80-bound mRNA (Ishigaki et al. 2001; Lejeune et al. 2002). Consistent with this, kinetic analyses indicate that NMD is restricted to newly synthesized mRNA (Belgrader et al. 1994; Cheng et al. 1994; Lejeune et al. 2003). Here, we validate the existence of the pioneer round of translation and continue to characterize its constituent proteins. We demonstrate that the pioneer round of translation is insensitive to inhibition by 4E-BP1 and, thus, is distinct from steady-state translation (Fig. 1). We also demonstrate that CBP80-bound Gl, GPx1, and SMG7 mRNAs are associated with ribosomes, although a fewer number of ribosomes than their eIF4E-bound counterparts (Fig. 5). Additionally, we show that eIF4AIII copurifies with CBP80 but not with eIF4E (Fig. 2). This finding is consistent with two papers that were published while our work was under review. In the first paper, eIF4AIII was shown to constitute a component of the EJC of proteins that is deposited upstream of exon-exon junctions as a consequence of pre-mRNA splicing (Chan et al. 2004). In the second paper, eIF4AIII was shown to be required for NMD (Palacios et al. 2004). Notably, despite the 65% sequence identity of eIF4AIII and eIF4AI, and despite data indicating that eIF4AIII binds to a region of eIF4GI that also binds eIF4AI (Li et al. 1999), it is unknown if eIF4AIII functions in the pioneer round of translation, the subsequent steps of mRNA decay that typify NMD, or both.
Although there are differences between the pioneer round of translation and steady-state translation, there are also several similarities. At least some mechanistic conservation makes sense given the complexity of the protein synthetic machinery and the large fraction of cellular resources that are devoted to mRNA translation (Hershey and Merrick 2000). We demonstrate that eIF4GI, PABP1, eIF3, eIF4AI, and eIF2α coimmunopurify with CBP80 as well as eIF4E (Fig. 2). This suggests that each protein either functions in both pioneer and steady-state translation initiation complexes or is involved in remodeling of the pioneer complex to the steady-state complex so as to function only in the steady-state complex. Functional analyses indicate that at least PAPB1 and eIF2α function in both pioneer and steady-state translation initiation complexes (Figs. 3, 4). Notably, the significance of our finding that eIF4AI coimmunopurifies with CBP80 remains uncertain in light of evidence that eIF4AI and eIF4AIII have similar binding sites within eIF4G (Li et al. 1999) and eIF4AIII not only inhibits steady-state translation in rabbit reticulocyte lysates (Li et al. 1999) but also is required for NMD (Palacios et al. 2004). We conclude that the pioneer and steady-state translation initiation complexes involve several of the same translation initiation factors despite involving distinct mRNP substrates.
Despite support for the existence of a pioneer translation initiation complex and a pioneer round of translation in mammalian cells, an alternative role for CBP80-CBP20 in RNA metabolism was recently put forth based on studies of Saccharomyces cerevisiae. In yeast, eIF4GI can simultaneously interact with CBP80-CBP20 and eIF4E, but in a competitive manner (Fortes et al. 2000). Furthermore, CBP80 added to yeast extracts stimulates translation 2.5-fold provided that the extracts derive from strains that harbor a mutated eIF4GI that interacts only weakly with eIF4E and poly(A)-binding protein (Fortes et al. 2000). Although these results indicate that CBP80 together with eIF4GI mediate translation initiation, the results of more recent studies indicate that cells expressing eIF4GI that harbors a point mutation that abolishes binding to CBP80 fail to manifest a detectable defect in growth or composition of the transcriptome (Baron-Benhamou et al. 2003). It was concluded that the interaction of CBP80 with eIF4GI is not essential for translation, that this interaction at most reflects eIF4GI function in remodeling CBP80-CBP20-bound mRNA to eIF4E-bound mRNA, and that the current proposal for a pioneer round of translation should be questioned (Baron-Benhamou et al. 2003). However, it was also noted that CBP80 is less conserved between yeast and mammals than, for example, CBP20, which opens up the possibility that CBP80 functions differently in the two types of organisms (Baron-Benhamou et al. 2003). Furthermore, CBP80 is not required for cell viability in S. cerevisiae (Das et al. 2000) and, thus, must not be required for translation.
Whether or not the CBP80-eIF4GI interaction in mammalian cells reflects a functional association within the pioneer translation initiation complex or the process of remodeling this complex to the steady-state translation initiation complex remains unknown. Future studies will undoubtedly offer insight into mechanistic similarities and differences between the translation of newly synthesized mRNAs in S. cerevisiae and mammalian cells.
Materials and methods
Cell culture and transfection
COS-7 and 293T cells were cultured in DMEM (GIBCO-BRL) that was supplemented with 10% fetal calf serum (GIBCO-BRL). Transient transfections were performed essentially following Ishigaki et al. (2001).
Cell fractionation, lysis, and immunopurification
COS cells (1-4 × 107) were lysed, and proteins and RNA were purified either before or after immunopurification (IP; Ishigaki et al. 2001; Lejeune et al. 2002). Immunopurified proteins and RNAs were then analyzed using Western blotting and RT-PCR, respectively.
Western blotting
Proteins before or after IP were electrophoresed in polyacrylamide (5%-12%), transferred to HyBond ECL nitrocellulose (Amersham), and probed with antibody against CBP80, eIF4E (Santa Cruz), eIF4GI (Bushell et al. 2000), eIF2α (Santa Cruz), eIF3 (Etchison et al. 1982), PABP1 (Gorlach et al. 1994), PABP2 (Krause et al. 1994), 4E-BP1 (Gingras et al. 1998), one of the Upf proteins (Ishigaki et al. 2001), HA (Roche), Penta-His (QIAGEN), calnexin (StressGen), or MUP (Berger and Szoka 1981). Reactivity to each antibody was detected using horseradish peroxidase-conjugated donkey α-rabbit (Amersham), sheep α-mouse (Amersham), donkey α-goat (Amersham), or rabbit α-rat antibody (Sigma). Reactivity of the secondary antibody was visualized using SuperSignal West Pico or Femto solution (Pierce).
Luciferase activity assays
Assays were performed as previously described (Lejeune et al. 2003).
Far-Western blotting
An XhoI-NotI fragment that encodes C-terminal His-tagged CBP80 was inserted into the corresponding sites of pBacPAK8, and recombinant baculovirus was prepared and used to infect Sf21 cells according to the manufacturer's instructions (BD Biosciences). Cells were lysed in 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 2.5 mM β-mercaptoethanol, 1% NP-40, and complete EDTA-free protease inhibitor cocktail (Roche Diagnostics), and CBP80-His was purified using Ni-NTA resin (QIAGEN). Wash buffer was supplemented with 2.5 mM β-mercaptoethanol, and elution buffer was supplemented with 2.5 mM β-mercaptoethanol and 10% glycerol. CBP80-His was then dialyzed for 2-3 h in 100 mM Tris (pH 7.5), 100 mM potassium acetate, 2 mM magnesium acetate, 1 mM DTT, 0.1 M EDTA, 10% glycerol, 1 mM PMSF, 1 mM benzamidine, and complete EDTA-free protease inhibitor cocktail.
Total protein from untransfected cells and cells transfected with pmyc-eIF4GI (Coldwell et al. 2004) was resolved in 6% SDS-polyacrylamide and transferred to nitrocellulose. Membranes were incubated for 24 h at 4°C in blocking buffer (dialysis buffer minus DTT and supplemented with 0.05% Tween-20) containing 5% milk, and then incubated overnight at 4°C in blocking buffer containing 5 μg of purified CBP80-His. Membranes were then washed once in blocking buffer and twice in Tris-buffered saline containing 0.05% Tween-20. Complexes were detected using Western blotting.
RT-PCR
Gl (Norm and 39Ter), GPx1, MUP, LUC, and SMG7 mRNAs were analyzed by RT-PCR as previously described (Ishigaki et al. 2001; Lejeune et al. 2003). Primers for the amplification of SMG7 mRNA were 5′-CCAAAGGAGACCATCTGACC-3′ (sense) and 5′-CCTCATCTCGGCTTTCC-3′ (antisense). The simultaneous analysis of serial dilutions of RNA ensured that RT-PCR was quantitative. RT-PCR products were quantitated by PhosphorImaging (Molecular Dynamics).
Polysome fractionation
293T cells (1.2 × 107) were transiently transfected with nonsense-free pmCMV-Gl and pmCMV-GPx1 and lysed with polysome extraction buffer (Johannes and Sarnow 1998). After centrifugation at 13,000 × g for 10 min at 4°C, the lysate was layered onto a 10%-50% sucrose gradient and centrifuged in a SW41 rotor (36,000 rpm) for 2 h at 4°C. Fractions were collected from the top using an ISCO fraction collection system, with concomitant and continuous measurement of absorbance at 254 nm.
Acknowledgments
We thank E. Izaurralde for α-CBP80 antibody; S. Morley for α-eIF4GI antibody; N. Sonenberg for pcDNA3-HA-Paip2, α-Paip2 antibody, pACTAG2, pACTAG2-4E-BP1, α-4E-BP1, pcDNA3-HA-eIF4AI, and pcDNA3-HA-eIF4AIII; J. Hershey for α-eIF3 antibody; G. Dreyfuss for α-PABP1 antibody; E. Wahle for pcDNA3-PABP2-HA and α-PABP2 antibody; M. Coldwell and S. Morley for pcDNA3.1-myc-eIF4GI; P. Anderson for pMT2-HA-eIF2α plasmids; and H. Baumann and W. Held for α-MUP antibody. We also thank N. Sonenberg for communicating unpublished results, and Y.-T. Yu and Y.K. Kim for comments on the manuscript. This work was supported by NIH grants to L.E.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1170204.
Corresponding author.
References
- Afonina E., Stauber, R., and Pavlakis, G.N. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 273: 13015-13021. [DOI] [PubMed] [Google Scholar]
- Aravind L. and Koonin, E.V. 2000. Eukaryote-specific domains in translation initiation factors: Implications for translation regulation and evolution of the translation system. Genome Res. 10: 1172-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron-Benhamou J., Fortes, P., Inada, T., Preiss, T., and Hentze, M.W. 2003. The interaction of the cap-binding complex (CBC) with eIF4G is dispensable for translation in yeast. RNA 9: 654-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P. and Maquat, L.E. 1994. Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol. Cell. Biol. 14: 6326-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Cheng, J., Zhou, X., Stephenson, L.S., and Maquat, L.E. 1994. Mammalian nonsense codons can be cis effectors of nuclear mRNA half-life. Mol. Cell. Biol. 14: 8219-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F.G. and Szoka, P. 1981. Biosynthesis of the major urinary proteins in mouse liver: A biochemical genetic study. Biochem. Genet. 19: 1261-1273. [DOI] [PubMed] [Google Scholar]
- Borman A.M., Michel, Y.M., and Kean, K.M. 2000. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: The eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res. 2821: 4068-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M., Wood, W., Clemens, M.J., and Morley, S.J. 2000. Changes in integrity and association of eukaryotic protein synthesis initiation factors during apoptosis. Eur. J. Biochem. 267: 1083-1091. [DOI] [PubMed] [Google Scholar]
- Calado A., Kutay, U., Kuhn, U., Wahle, E., and Carmo-Fonseca, M. 2000. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA 6: 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.C., Dostie, J., Diem, M.D., Feng, W., Mann, M., Rappsilber, J., and Dreyfuss, G. 2004. eIF4A3 is a novel component of the exon junction complex. RNA 10: 200-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Belgrader, P., Zhou, X., and Maquat, L.E. 1994. Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell. Biol. 14: 6317-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldwell M.J., Hashemzadeh-Bonehi, L., Hinton, T.M., Morley, S.J., and Pain, V.M. 2004. Expression of fragments of translation initiation factor eIF4GI reveals a nuclear localisation signal within the N-terminal apoptotic cleavage fragment N-FAG. J. Cell. Science (in press). [DOI] [PubMed]
- Craig A.W., Haghighat, A., Yu, A.T., and Sonenberg, N. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392: 520-523. [DOI] [PubMed] [Google Scholar]
- Das B., Guo, Z., Russo, P., Chartrand, P., and Sherman, F. 2000. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 20: 2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J. and Dreyfuss, G. 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 12: 1060-1067. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3: 195-205. [DOI] [PubMed] [Google Scholar]
- Etchison D., Milburn, S.C., Edery, I., Sonenberg, N., and Hershey, J.W. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257: 14806-14810. [PubMed] [Google Scholar]
- Fortes P., Inada, T., Preiss, T., Hentze, M.W., Mattaj, I.W., and Sachs, A.B. 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6: 191-196. [PubMed] [Google Scholar]
- Gingras A.C., Kennedy, S.G., O'Leary, M.A., Sonenberg, N., and Hay, N. 1998. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes & Dev. 12: 502-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Gygi, S.P., Raught, B., Polakiewicz, R.D., Abraham, R.T., Hoekstra, M.F., Aebersold, R., and Sonenberg, N. 1999. Regulation of 4E-BP1 phosphorylation: A novel twostep mechanism. Genes & Dev. 13: 1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlach M., Burd, C.G., and Dreyfuss, G. 1994. The mRNA poly(A)-binding protein: Localization, abundance, and RNA-binding specificity. Exp. Cell Res. 211: 400-407. [DOI] [PubMed] [Google Scholar]
- Haghighat A., Mader, S., Pause, A., and Sonenberg, N. 1995. Repression of cap-dependent translation by 4E-binding protein 1: Competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14: 5701-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik, A.E. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96: 307-310. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B. and Merrick, W.C. 2000. The pathway and mechanism of initiation of protein synthesis. In Translational control of gene expression (ed. M.B. Mathews), pp. 33-88. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Ishigaki Y., Li, X., Serin, G., and Maquat, L.E. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607-617. [DOI] [PubMed] [Google Scholar]
- Johannes G. and Sarnow, P. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4: 1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M.S. and Moore, M.J. 2003. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell 12: 5-14. [DOI] [PubMed] [Google Scholar]
- Kaufman R.J., Davies, M.V., Pathak, V.K., and Hershey, J.W. 1989. The phosphorylation state of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol. Cell. Biol. 9: 946-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K., Svitkin, Y.V., Craig, A.W., DeMaria, C.T., Deo, R.C., Burley, S.K., and Sonenberg, N. 2001. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell 7: 205-216. [DOI] [PubMed] [Google Scholar]
- Krause S., Fakan, S., Weis, K., and Wahle, E. 1994. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp. Cell Res. 214: 75-82. [DOI] [PubMed] [Google Scholar]
- Lejeune F., Ishigaki, Y., Li, X., and Maquat, L.E. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 21: 3536-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F., Li, X., and Maquat, L.E. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12: 675-687. [DOI] [PubMed] [Google Scholar]
- Li S. and Wilkinson, M.F. 1998. Nonsense surveillance in lymphocytes? Immunity 8: 135-141. [DOI] [PubMed] [Google Scholar]
- Li Q., Imataka, H., Morino, S., Rogers Jr., G.W., Richter-Cook, N.J., Merrick, W.C., and Sonenberg, N. 1999. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol. Cell. Biol. 19: 7336-7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader S., Lee, H., Pause, A., and Sonenberg, N. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15: 4990-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E. 2003. Nonsense-mediated mRNA decay in mammalian cells: From pre-mRNA processing to mRNA translation and degradation. In Translation mechanisms (ed. L. Brakier-Gingras), pp. 210-222. Landes Bioscience, Austin, TX.
- ____. 2004a. Nonsense-mediated mRNA decay: Splicing, translation, and mRNP dynamics. Nat. Rev. Mol. Cell. Biol. 5: 89-99. [DOI] [PubMed] [Google Scholar]
- ____. 2004b. Nonsense-mediated mRNA decay: A comparative analysis of different species. Curr. Genomics 5: 175-190. [Google Scholar]
- Marcotrigiano J., Gingras, A.C., Sonenberg, N., and Burley, S.K. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol. Cell 3: 707-716. [DOI] [PubMed] [Google Scholar]
- McKendrick L., Thompson, E., Ferreira, J., Morley, S.J., and Lewis, J.D. 2001. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 21: 3632-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty P.M., Reddy, C.C., and Maquat, L.E. 1998. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 18: 2932-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E. and Maquat, L.E. 1998. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 23: 198-199. [DOI] [PubMed] [Google Scholar]
- Palacios I.M., Gatfield, D., St Johnston, D., and Izaurralde, E. 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753-757. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. 2000. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem. Sci. 25: 423-426. [DOI] [PubMed] [Google Scholar]
- Reed R. 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15: 326-331. [DOI] [PubMed] [Google Scholar]
- Singh G. and Lykke-Andersen, J. 2003. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 28: 464-466. [DOI] [PubMed] [Google Scholar]
- Srivastava S.P., Kumar, K.U., and Kaufman, R.J. 1998. Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273: 2416-2423. [DOI] [PubMed] [Google Scholar]
- Thompson J.F., Hayes, L.S., and Lloyd, D.B. 1991. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene 103: 171-177. [DOI] [PubMed] [Google Scholar]
- Zhang J., Sun, X., Qian, Y., and Maquat, L.E. 1998. Intron function in the nonsense-mediated decay of β-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 4: 801-815. [DOI] [PMC free article] [PubMed] [Google Scholar]