Abstract
The Ser-Arg-rich (SR) proteins comprise a large family of nuclear phosphoproteins that are required for constitutive and alternative splicing. A subset of SR proteins shuttles continuously between the nucleus and the cytoplasm, suggesting that the role of shuttling SR proteins in gene expression may not be limited to nuclear pre-mRNA splicing, but may also include unknown cytoplasmic functions. Here, we show that shuttling SR proteins, in particular SF2/ASF, associate with translating ribosomes and stimulate translation when tethered to a reporter mRNA in Xenopus oocytes. Moreover, SF2/ASF enhances translation of reporter mRNAs in HeLa cells, and this activity is dependent on its ability to shuttle from the nucleus to the cytoplasm and is increased by the presence of an exonic-splicing enhancer. Furthermore, SF2/ASF can stimulate translation in vitro using a HeLa cell-free translation system. Thus, the association of SR proteins with translating ribosomes, as well as the stimulation of translation both in vivo and in vitro, strongly suggest a role for shuttling SR proteins in translation. We propose that shuttling SR proteins play multiple roles in the posttranscriptional expression of eukaryotic genes and illustrate how they may couple splicing and translation.
Keywords: pre-mRNA splicing, SR proteins, nucleocytoplasmic shuttling, ribosomes, translation
Pre-mRNA splicing, an essential step in gene expression, is catalyzed by a large ribonucleoprotein complex, termed the spliceosome. This macromolecular machine consists of small nuclear ribonucleoproteins particles (snRNPs) U1, U2, U4, U5, and U6 and a multitude of non-snRNP splicing factors that includes Ser-Arg-rich (SR) proteins (for review, see Kramer 1996; Will and Luhrmann 1997; Jurica and Moore 2003). The SR proteins constitute a family of structurally and functionally related proteins, playing dual roles in both constitutive and alternative pre-mRNA splicing (Fu 1995; Valcarcel and Green 1996; Graveley 2000). SR proteins are characterized by their modular domain structure, with one or two N-terminal RNA recognition motifs (RRMs) and a C-terminal domain rich in arginine and serine residues, termed the RS domain. The RRMs determine RNA-binding specificity, whereas the RS domain acts as a splicing activator domain by mediating protein-protein interactions with other components of the splicing machinery (Wu and Maniatis 1993; Graveley and Maniatis 1998). The RS domain also directs subcellular localization and nucleocytoplasmic shuttling of individual SR proteins (Hedley et al. 1995; Caceres et al. 1997, 1998).
SR proteins play numerous roles in pre-mRNA splicing and spliceosome assembly, but perhaps their most significant function is in splice-site recognition and selection (Tacke and Manley 1999; Sanford et al. 2003). Metazoan splice sites contain low information content and are far more degenerate than those found in simpler eukaryotes (Burge et al. 1998); thus, splice-site specificity is conferred by the presence of additional sequences that facilitate splice-site selection, such as exonic-splicing enhancers and silencers (ESEs and ESSs, respectively). These elements recruit trans-acting factors that function as adapters between the pre-mRNA and the basal-splicing machinery. For instance, SR family proteins bound to ESEs can promote U2AF recruitment to the polypyrimidine tract and activate an adjacent 3′ splice site. In certain cases, SR proteins may act to antagonize the negative activity of hnRNP proteins recognizing ESS elements (for review, see Blencowe 2000; Hastings and Krainer 2001; Caceres and Kornblihtt 2002). The SR proteins are not only required for constitutive splicing, but also influence regulation of alternative splicing, and this activity is antagonized by hnRNP A/B proteins in a concentration-dependent manner (Mayeda and Krainer 1992; Caceres et al. 1994; Yang et al. 1994). SF2/ASF and hnRNP A1 competitive binding to pre-mRNA underlies their functional antagonism in splice-site selection (for review, see Hastings and Krainer 2001).
Initially, SR proteins were found to be functionally redundant in constitutive splicing assays; however, differences in alternative splicing regulation, as well as genetic analysis of SR protein function suggested that not all SR proteins are functionally redundant. For example, SF2/ASF is essential for cell viability in the DT40 chicken cell line, and its depletion cannot be rescued by overexpression of other SR proteins (Wang et al. 1996, 1998). Genetic disruption of SRp55/B52 in Drosophila results in lethality during development, and individual SR proteins were able to complement the loss of B52 in most tissues, except in the brain, where B52 is the predominant protein (Ring and Lis 1994; Peng and Mount 1995; Hoffman and Lis 2000). In the mouse, SRp20 was shown to be essential for early development (Jumaa et al. 1999), whereas conditional deletion of the SR protein SC35 in the thymus causes a defect in T-cell maturation (Wang et al. 2001). RNA interference (RNAi) experiments with Caenorhabditis elegans SR proteins showed that, whereas CeSF2/ASF is an essential gene, functional knockouts of other SR genes resulted in no obvious phenotype, which is indicative of functional redundancy (Kawano et al. 2000; Longman et al. 2000). These results suggest that at least some SR proteins are functionally redundant, and that the requirement for a particular SR protein may be due to specific functions in the tissue or developmental stage in which a particular SR protein is predominant.
At steady state, SR proteins are localized in the nucleus and are distributed both in the nucleoplasm and in interchromatin granule clusters (IGCs) or speckles (Spector et al. 1991; for review, see Lamond and Spector 2003). However, a subset of SR proteins shuttle continuously between the nucleus and the cytoplasm, reminiscent of what was observed for several hnRNP proteins (Pinol-Roma and Dreyfuss 1992; Caceres et al. 1998). The shuttling behavior of this subset of SR proteins argues against their function being limited to the nucleus, and allows for the possibility that shuttling SR proteins may have additional roles in mRNA transport and/or in cytoplasmic events such as mRNA localization, stability, or regulation of translation. Despite the extensive characterization of the activities of SR proteins in nuclear pre-mRNA splicing, little is known regarding any potential cytoplasmic functions of shuttling SR proteins. In contrast, cytoplasmic activities for a number of shuttling hnRNP proteins have been characterized and include the roles for hnRNP A2/B1 and squid/hrp40 in mRNA localization (Hoek et al. 1998; Lall et al. 1999) and for hnRNP D in regulation of mRNA stability (Loflin et al. 1999; Xu et al. 2001; for review, see Shyu and Wilkinson 2000; Dreyfuss et al. 2002). In addition, polypyrimidine tract-binding protein (hnRNP I/PTB) is essential for internal initiation of translation of viral RNAs (Kaminski et al. 1995), whereas hnRNP K and E1 mediate translational silencing (Ostareck et al. 1997; Habelhah et al. 2001). Interestingly, two shuttling SR proteins, SRp20 and 9G8, function to promote mRNA export of intronless RNAs (Huang and Steitz 2001) and also act as adapter proteins for TAP-dependent mRNA export (Huang et al. 2003). Moreover, SF2/ASF has been shown to control mRNA stability in the cytoplasm for a specific mRNA (Lemaire et al. 2002).
Here, we have used a variety of assays to elucidate putative cytoplasmic functions for shuttling SR proteins. We show by sucrose gradient centrifugation of HeLa cytoplasmic extracts that two shuttling SR proteins, SF2/ASF and SRp20, cosediment with the 80S ribosome particle, and in the case of SF2/ASF, also with polysomes. A functional role for shuttling SR proteins in translation is confirmed by three lines of evidence. First, we show that SR proteins are able to stimulate translation when tethered to a luciferase reporter in Xenopus oocytes. Second, SF2/ASF is also able to stimulate translation of a reporter in HeLa cells in an enhancer-dependent manner. Finally, SF2/ASF can stimulate translation in vitro in a HeLa cell-free translation system. These findings establish a novel cytoplasmic role for shuttling SR proteins and demonstrate that SR proteins are important determinants of the fate of an mRNA from the early nuclear RNA processing events until its translation in the cytoplasm.
Results
Shuttling SR proteins cosediment with ribosomal particles
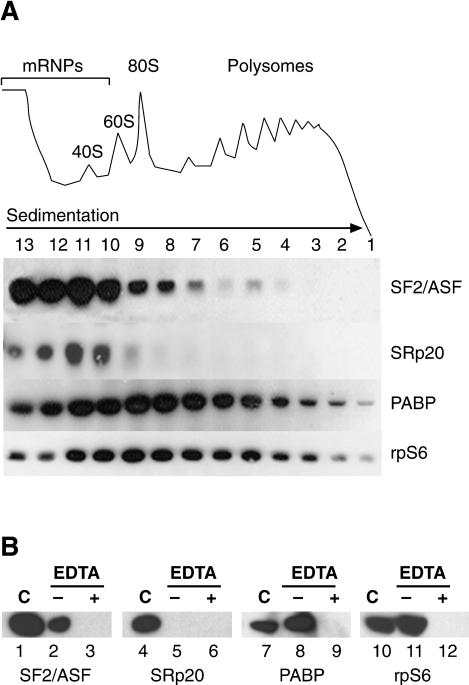
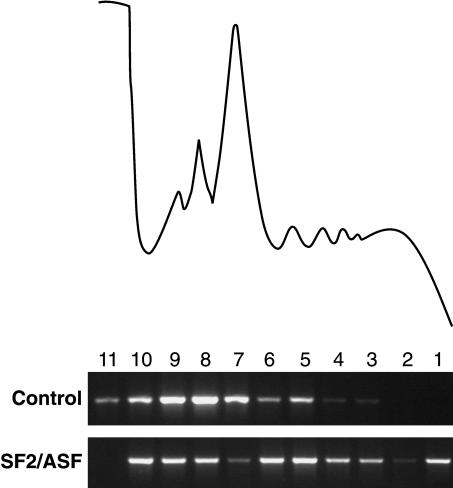
As an initial approach toward elucidating the cytoplasmic functions of shuttling SR proteins, HeLa cell cytoplasm was fractionated across 10%-50% sucrose gradients, and the distribution of SR proteins was analyzed by Western blotting. Two shuttling SR proteins, SF2/ASF and SRp20, were broadly distributed between the mRNP complexes and the ribosomal subunits. Interestingly, SF2/ASF, and to a lesser extent, SRp20, also cosedimented with the 80S ribosome, as was the case for the ribosomal protein S6, an integral component of the small ribosomal subunit (rpS6; Fig. 1A). In contrast, the nonshuttling SR protein, SRp40, was present only in trace amounts in HeLa cytosolic extract and was never observed cosedimenting with ribosomal complexes (data not shown). Interestingly, a fraction of SF2/ASF was also detected in the lighter polysomal fractions, whereas SRp20 was absent from polysomes. Treatment of cytoplasmic extracts with EDTA induces dissociation of mono- and polyribosomes into ribosomal subunits, and under these conditions, SF2/ASF is redistributed to the top of the gradient (Supplementary Fig. S1). Analysis of pelleted polysomes showed that SF2/ASF and Poly(A)-binding protein (PABP), a bona fide translational regulator, cosedimented with polyribosomes as marked by the presence of rpS6, a ribosomal subunit protein; however, little, if any, SRp20 appeared to associate with polysomes (Fig. 1B). Extracts treated with EDTA, which compromises the integrity of ribosomes, failed to sediment rpS6, PABP, or SF2/ASF (Fig. 1B). Thus, we conclude that SF2/ASF, but not SRp20, is associated with polysomes. Taken together, these results suggest that shuttling SR proteins can interact with the translation machinery and may play a role in translational regulation.
Figure 1.
Shuttling SR proteins are associated with the translation machinery. (A) HeLa cell cytosolic extracts were fractionated across a 10%-50% sucrose gradients and analyzed by Western blotting with antibodies against SF2/ASF, SRp20, poly(A)-binding protein (PABP), and the ribosomal protein, rpS6. (Top) UV absorbance (254 nm) profile of cytosolic ribonucleoprotein complexes. (B) Association of SF2/ASF with polyribosomes requires intact ribosomal particles. Western blot analysis of RNA-binding proteins associated with polyribosomes, purified by pelleting through a 10%-50% sucrose gradient from mock treated (-) or EDTA-treated (+) HeLa cytosolic extracts. Unfractionated cytosolic extract is shown in lanes designated by C.
Tethered SR proteins activate translation of a reporter in Xenopus oocytes
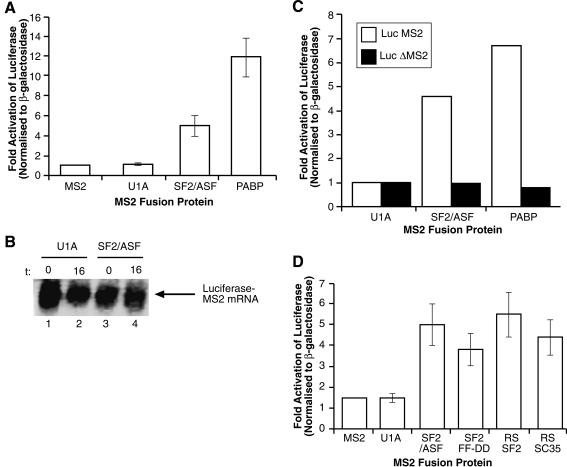
Cosedimentation of SF2/ASF with the translation machinery suggests that SF2/ASF, and possibly other shuttling SR proteins, may play a role in translational regulation. This hypothesis was directly tested using a variety of functional assays. First, the well-established tethered function assay in Xenopus oocytes was used (Gray et al. 2000). Briefly, mRNAs encoding fusion proteins between the bacteriophage RNA-binding protein MS2 and shuttling SR proteins, or bona fide translational regulators such as PABP, were injected into Xenopus oocytes. Following a 6-h incubation to allow time for protein production, oocytes were subsequently coinjected with a luciferase reporter mRNA containing cognate MS2-binding sites in the 3′UTR (or lacking MS2 sites as a control). A β-galactosidase mRNA lacking MS2-binding sites was coinjected to control for nonspecific effects of the MS2 fusion proteins on translation of the luciferase reporter. Translational activity was quantified as luciferase activity normalized to β-galactosidase activity as previously described (Gray et al. 2000). Figure 2A shows that like MS2 alone, the fusion of MS2 to the U1 snRNP-specific protein U1A (MS2-U1A) was unable to activate translation. This was not due to lack of MS2-U1A fusion protein, as the expression of the protein can be seen in 35S-labeled oocytes (Supplementary Fig. S2). In contrast, PABP strongly activates translation of the reporter mRNA. Interestingly, MS2-SF2/ASF also activated translation when tethered to the luciferase mRNA reporter. Synthesis of the MS2 fusion proteins was confirmed by 35S-met labeling of the injected oocytes (Supplementary Fig. S2). Once corrected for the number of methionines, translation of the MS2-SF2 fusion protein seems to be reduced compared with that of U1A or PABP; thus, the activity of SF2/ASF could be higher than apparent from the luciferase assays. Therefore, a shuttling SR protein can enhance the translation of reporter mRNA in a heterologous in vivo translation assay (Fig. 2A). It should be noted that in this experimental system, mRNA export becomes irrelevant, as the reporter mRNAs (both luciferase and β-galactosidase) are coinjected in the cytoplasm. Importantly, stimulation of luciferase expression mediated by MS2-SF2/ASF was observed in the absence of an increase in the stability of reporter mRNA, as measured by Northern blots, indicating that the fusion proteins function at the level of translation (Fig. 2B). Furthermore, the effect of MS2-SF2/ASF on luciferase translation was specific and occurred only in cis, as no stimulation was observed in the absence of MS2-binding sites in the 3′UTR of the luciferase reporter (Fig. 2C).
Figure 2.
Tethered SR proteins stimulate translation in Xenopus oocytes. (A) mRNAs encoding either the MS2 protein alone, or fusions between MS2-U1A, MS2-SF2/ASF, and MS2-PABP were coinjected into the cytoplasm of Xenopus oocyctes with a luciferase reporter mRNA containing MS2p-binding sites within its 3′UTR and a β-galactosidase mRNA lacking MS2-binding sites. The injected fusion proteins did not significantly affect β-galactosidase levels and nonspecific effects of the fusion proteins on translation were taken into account by normalizing luciferase activity to β-galactosidase activity. These data represent the average stimulation from three independent experiments. (B) Northern blot analysis showing constant levels of mRNA reporter upon injection of MS2-U1A (lanes 1,2) and MS2-SF2/ASF (lanes 3,4). (t) Time after injection (0 and 16 h, respectively). (C) The effect of the MS2-SF2/ASF fusion protein is specific to reporter mRNAs containing cognate MS2-binding sites. Microinjection experiments were performed using a luciferase reporter mRNA containing (white bars) or lacking (black bars) functional MS2-binding sites. (D) The RS domain of SR proteins is sufficient to stimulate translation in vivo. mRNAs encoding MS2, MS2-U1A, MS2-SF2/ASF, MS2-SF2 FF-DD, MS2-RS SF2/ASF, or MS2-RS SC35 were injected into the cytoplasm of Xenopus oocytes along with reporter mRNAs as described above. These data represents the average stimulation from three independent experiments.
Next, we asked whether the RNA-binding activity of SF2/ASF was required for its function as a translational regulator in the tethering system. Interestingly, we found that an RNA-binding-deficient mutant of SF2/ASF harboring a double point mutation in its first RRM that severely affects sequence-specific RNA binding (Caceres and Krainer 1993) was able to stimulate translation when fused to the MS2 protein (MS2 FF-DD, Fig. 2D). This experiment demonstrates that RNA binding is most likely required to recruit SF2/ASF to the mRNA, rather than to promote an interaction with ribosomal RNAs (Fig. 2D). The RS domain of SR proteins has been shown to function as a splicing-activator domain, presumably by promoting protein-protein interactions with essential components of the splicing machinery (Wu and Maniatis 1993; Zuo and Maniatis 1996; Graveley and Maniatis 1998). Therefore, we asked whether the RS domains of both shuttling and nonshuttling SR proteins (SF2/ASF and SC35, respectively) could function as translational activators in vivo when fused to the MS2 protein. Figure 2D shows that the RS domain of either SR protein was sufficient to stimulate translation in vivo. It should be noted that the shuttling activity of SR proteins becomes irrelevant in this assay, as the mRNAs coding for the particular SR fusion protein were injected in the cytoplasm. Fusion proteins between MS2 and full-length SF2/ASF and SC35 (shuttling and nonshuttling SR proteins, respectively) stimulated translation to similar extents in Xenopus oocytes (data not shown). Thus, these results suggest that shuttling SR proteins are capable of functionally interacting with the translation machinery in vivo and that the RS domain is sufficient to mediate this effect.
SF2/ASF activates translation of a reporter in HeLa cells in an enhancer-dependent manner
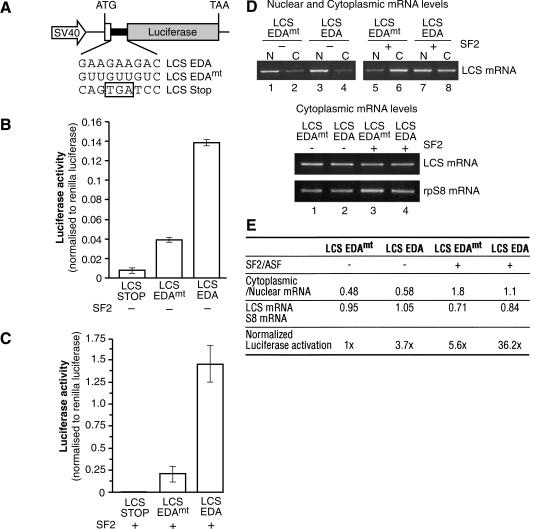
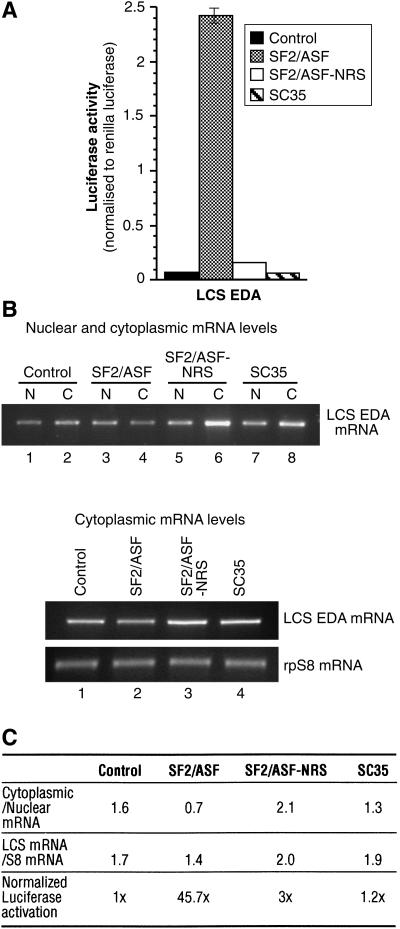
Next, we investigated the role of the shuttling SR protein SF2/ASF in translation in mammalian cells. We developed a luciferase-based reporter system that encodes the C-terminal 33 amino acids from β-alactosidase fused in-frame to a Firefly luciferase gene via a short synthetic linker sequence. Restriction enzyme sites within this linker sequence were used to insert an exonic-splicing enhancer (ESE), derived from the EDA alternative exon of the fibronectin gene (Caputi et al. 1994), which is known to recruit SF2/ASF as well as 9G8 (Lavigueur et al. 1993; Cramer et al. 1999). This ESE sequence was inserted either in a wild-type or in a mutant version that lacked binding sites for SF2/ASF (pLCS-EDA or pLCS-EDAmt, Fig. 3A). As a control, we generated a construct containing a stop codon within the linker (pLCS-stop). This is an ideal strategy to incorporate SR protein-binding sites into an ORF without perturbing the sequence or structure of the reporter. After transfection into HeLa cells, this reporter gene system is transcribed under the control of the SV40 promoter, and translation initiating at the unique ATG continues through the linker region and leads to the synthesis of the luciferase reporter. Thus, we were able to test the effect of an SR protein-binding site with endogenous or transiently overexpressed SR proteins on translation of the reporter construct by assaying luciferase levels. A Renilla luciferase reporter driven by a thymidine kinase promoter was cotransfected as a control for transfection efficiency, and data are expressed as a ratio between the enzymatic activities of the two reporters. The luciferase activity values were corrected by the effects on mRNA export induced by the presence of the ESE-binding site or overexpression of SF2/ASF (see below). We found that the presence of the EDA exonic enhancer sequence, which is recognized by SF2/ASF, stimulated luciferase activity by nearly fourfold (Fig. 3B, cf. activity of pLCS EDA vs. pLCS EDAmt). This effect most likely reflects the activity of endogenous SF2/ASF protein that is recruited by the EDA enhancer, resulting in an increased translation of the reporter mRNA. If this hypothesis were correct, then we would predict that overexpression of SF2/ASF should further stimulate luciferase activity. This is the case, as transfected SF2/ASF strongly induces luciferase activity of the reporter harboring the ESE sequence (Fig. 3C,E). An increase in the activity of the luciferase reporter lacking an ESE sequence was also observed upon SF2/ASF overexpression, suggesting the presence of additional binding sites (other than the inserted ESE sequence) for SF2/ASF within the luciferase ORF. Software designed to predict the presence of SF2/ASF-binding sites detects several additional potential binding sites within the luciferase ORF, two of which show a strong resemblance to the EDA ESE (Cartegni et al. 2003). Interestingly, under these conditions, the levels of the control reporter, Renilla luciferase, which contains far fewer putative SF2/ASF-binding sites, remained unchanged (data not shown). More importantly, the effect of the EDA ESE sequence is maintained under SF2/ASF overexpression (Fig. 3C,E).
Figure 3.
SF2/ASF stimulates translation of a luciferase reporter mRNA in HeLa cells. (A) Schematic diagrams of the pLCS reporter system. The fibronectin EDA ESE or a mutant version were inserted in frame and upstream of the Firefly luciferase ORF. After cotransfection into HeLa cells with a Renilla luciferase reporter (driven by the thymidine kinase promoter), the pLCS reporters are transcribed under the control of the SV40 promoter and translation leads to the synthesis of the reporter enzymes. The promega Dual Luciferase Reaction (DLR) system is used to assay levels of the Firefly (translational reporter) and Renilla (to control for transfection efficiency) luciferase levels. (B) The presence of an ESE stimulates expression of a Firefly Luciferase reporter mRNA. HeLa cells were transiently transfected with pLCS-Stop, pLCS-EDA, or pLCS-EDAmt, which contains a mutation in the EDA sequence along with the nonspecific Renilla luciferase reporter. The data are expressed as a ratio of Firefly luciferase activity to Renilla luciferase activity, and have been normalized to changes in nuclear and cytoplasmic distribution of the reporter mRNAs. (C) Overexpression of SF2/ASF stimulates expression of reporter mRNAs in vivo. HeLa cells were transiently transfected with pLCS-Stop, pLCS-EDA, or pLCS-EDAmt along with the nonspecific Renilla luciferase reporter and pCGT7-SF2/ASF and analyzed as described above. (D) Analysis of the nucleocytoplasmic distribution of reporter mRNAs (top) and of the cytosolic levels of reporter mRNA relative to the endogenous rpS8 mRNA (bottom) by RT-PCR. (E) Table summarizing the effects of the EDA ESE and overexpression of SF2/ASF on LCS reporter mRNA distribution, cytoplasmic accumulation, and reporter enzyme expression.
We assayed nuclear and cytoplasmic reporter mRNA levels to control that any increase in translation efficiency may be due to an increase in mRNA export. RT-PCR analysis was used to assay the amount of luciferase mRNA reporter in nuclear and cytoplasmic total RNA preps from cells cotransfected with SF2/ASF or a control plasmid. The ratio of cytoplasmic to nuclear luciferase mRNA remained relatively constant for both the LCS-EDA and LCS-EDAmt (Fig. 3D, top, cf. lanes 1,2 and 3,4). Although overexpression of SF2/ASF increased the proportion of mRNAs in the cytoplasm, this effect was similar for both LCS-EDA and LCS-EDAmt and did not result in significant increases in the steady-state levels of these RNAs in the cytoplasm when compared with an endogenous mRNA (ribosomal protein rpS8; Fig. 3D, bottom). Consequently, the results presented in Figure 3B and C have been normalized to reflect differences in nuclear and cytoplasmic reporter mRNA levels. These experiments demonstrate that overexpression of SF2/ASF does not dramatically affect the extent of mRNA export and/or stability, and further supports the hypothesis that the primary effect of SF2/ASF in this assay is at the level of translation (Fig. 3D,E).
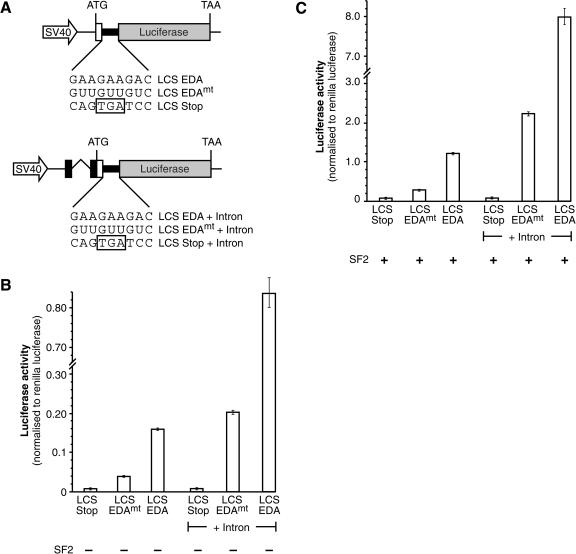
The luciferase-based reporter mRNAs described above lack introns, suggesting the possibility that SF2/ASF may enhance the translation of intronless mRNAs. To determine whether SF2/ASF could enhance the translation of spliced mRNAs, we generated a related series of constructs that contain an intron in their 5′UTR. Figure 4 clearly shows that SF2/ASF positively affects translation of both intron-containing and intronless reporter mRNAs. The presence of an intron in these luciferase-based reporters causes a general stimulation of gene expression; however, it does not substantially alter the positive effect of translation mediated by an SF2/ASF-binding site or by overexpressed SF2/ASF protein (Fig. 4). Thus, the effect of SF2/ASF on translational regulation does not require pre-mRNA splicing, raising the possibility that it could affect translation of both intronless and spliced mRNAs.
Figure 4.
SF2/ASF stimulates the translation of both intronless and intron-containing reporter mRNAs. (A) Schematic diagrams of the intronless and intron-containing LCS reporter mRNAs. The fibronectin EDA ESE or a mutant version were inserted in-frame and upstream of the Firefly luciferase ORFs. (Right) Note the presence of an heterologous intron in the 5′UTR. (B) The presence of an ESE (EDA ESE) stimulates translation of both intronless and intron-containing Firefly Luciferase reporter mRNAs. HeLa cells were transiently transfected with pLCS-Stop, pLCS-EDA, or pLCS-EDAmt, which contains a mutation in the EDA sequence along, lacking or containing an intron, with the nonspecific Renilla luciferase reporter. The data are expressed as a ratio of Firefly luciferase activity to Renilla luciferase activity. (C) Overexpression of SF2/ASF dramatically increases the expression of both intronless and intron-containing LCS reporter mRNAs in HeLa cells. HeLa cells were transiently transfected with pLCS-Stop, pLCS-EDA, or pLCS-EDAmt with or without an intron along with the nonspecific Renilla luciferase reporter and pCGT7-SF2/ASF and analyzed as described above.
Nucleocytoplasmic shuttling of SR proteins is required for their activity in translational activation
Our previous studies determined that the RS domain of specific SR proteins determines the nucleocytoplasmic properties of shuttling SR proteins (Caceres et al. 1998). We have also previously shown that SC35 is actively retained in the nucleus, due to the presence of a nuclear retention sequence (NRS) in the C terminus of its RS domain, and that fusion of this NRS C-terminal of SF2/ASF led to nuclear retention of this chimeric protein (SF2/ASF-NRS; Cazalla et al. 2002). Thus, we asked if the nucleocytoplasmic shuttling activity of SF2/ASF was required to stimulate the expression of ESE containing reporters in vivo. Whereas overexpression of wild-type SF2/ASF activated translation of the luciferase reporter harboring an SF2/ASF-binding site (pLCS-EDA), as shown above, SC35 failed to activate translation of this reporter (Fig. 5). Interestingly, the presence of a nuclear retention signal in SF2/ASF dramatically reduces activation of the luciferase reporter (SF2/ASF-NRS; Fig. 5). As in Figure 3, the luciferase activity was normalized to reflect slight differences in the nuclear and cytoplamic distribution of the luciferase reporter. Thus, compromising the shuttling ability of SR proteins causes a significant reduction in the ability of SF2/ASF to activate translation in this assay.
Figure 5.
Nucleocytoplasmic shuttling of SF2/ASF is important for stimulation of translation in vivo. (A) HeLa cells were cotransfected with pLCS-EDA and empty vector (black bar), pCGT7-SF2/ASF (gray bar), pCGT7-SF2/ASF-NRS (white bar), or pCGT7-SC35 (hatched bar). Dual Luciferase assays measured the effect of shuttling on the translation of the reporter enzymes. These data represent the average stimulation from three independent experiments. (B) Analysis of the nucleocytoplasmic distribution of the reporter mRNAs (top) or of the cytosolic levels of reporter mRNA relative to the endogenous rpS8 mRNA (bottom) by RT-PCR. (C) Table summarizing the effects of overexpression of SF2/ASF, SF2/ASF-NRS, and SC35 on LCS reporter mRNA distribution, cytoplasmic accumulation, and reporter enzyme expression.
Translational regulation by other shuttling SR proteins in HeLa cells
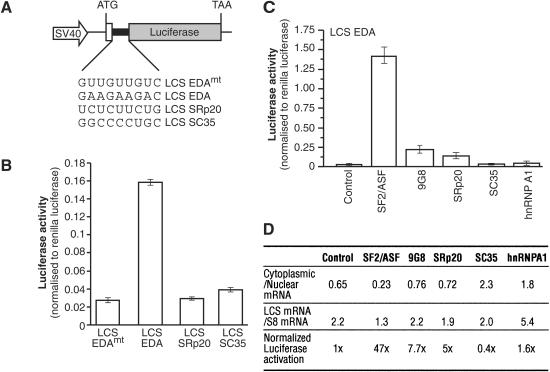
The previous experiments suggest that SF2/ASF can stimulate translation in vivo. To determine whether this activity was common to other shuttling SR proteins, we generated two additional LCS reporter constructs containing binding sites for SRp20 and SC35 (Fig. 6A). The motif chosen for SRp20 is an ESE from exon 4 of the SRp20 gene, which is involved in the autoregulation of SRp20 alternative splicing (Jumaa and Nielsen 1997), whereas the SC35 motif was derived from a functional SELEX approach (Liu et al. 2000). We found that the presence of the EDA exonic enhancer sequence, which is recognized by SF2/ASF and 9G8, stimulated luciferase activity by nearly fourfold as described above for Figure 3. In contrast, the presence of binding sites for either SRp20 or SC35 does not enhance translation of the respective reporter mRNAs (Fig. 6B). This experiment most likely reflects the activity of the different endogenous SR proteins that are recruited by the different ESE sequences. Thus, we conclude that endogenous SRp20 and SC35 fail to activate translation of reporters harboring their respective binding sites.
Figure 6.
Effect of shuttling SR proteins on the activation of translation of a luciferase reporter mRNA in HeLa cells. (A) Schematic diagrams of the pLCS reporter constructs containing in-frame ESEs recognized by SF2/ASF and 9G8, SRp20 and SC35. (B) The presence of the EDA ESE, but not the SRp20 ESE nor the SC35 ESE, stimulates expression of a Firefly Luciferase reporter mRNA. HeLa cells were transiently transfected with pLCS-EDA, with pLCS-EDAmt, which contains a mutation in the EDA sequence, or with pLCS SRp20 ESE and pLCS SC35 ESE along with the nonspecific Renilla luciferase reporter. The data are expressed as a ratio of Firefly luciferase activity to Renilla luciferase activity. (C) Overexpression of SF2/ASF, but not of 9G8, SRp20, SC35, or hnRNP A1, stimulate expression of the LCS EDA reporter mRNA in vivo. HeLa cells were cotransfected with pLCS-EDA and empty vector (control), pCGT7-SF2/ASF, pCGT7-9G8, pCGT7-SRp20, pCGT7-SC35, and pCGT7-hnRNP A1 along with the nonspecific Renilla luciferase reporter and pCGT7-SF2/ASF and analyzed as described above. (D) Table summarizing the effects of overexpression of SR proteins and hnRNP A1 on LCS reporter mRNA distribution, cytoplasmic accumulation, and reporter enzyme expression.
Next, we asked whether overexpression of different SR proteins was able to activate translation of LCS reporter mRNAs harboring binding sites for different SR proteins. Western blot analysis demonstrated that each protein was overexpressed to comparable levels (data not shown). As expected, SF2/ASF strongly activated the EDA ESE-containing reporter (Fig. 6C). Interestingly, overexpression of 9G8, which is known to regulate the EDA ESE (Cramer et al. 1999) only marginally activated expression of this reporter mRNA. Although 9G8 is not a potent activator of the luciferase reporter harboring the EDA ESE in this assay, we cannot rule out a positive role for this shuttling SR protein in translation of mRNAs harboring 9G8 high-affinitiy binding sites (Cavaloc et al. 1999). Furthermore, overexpression of SRp20 only weakly activated expression of the reporter mRNA. As described for Figures 3 and 5, the luciferase activity was normalized for any effects of the overexpressed proteins on mRNA nuclear/cytoplasmic distribution. In contrast to SF2/ASF, overexpression of SC35 and the shuttling RNA-binding protein hnRNP A1 failed to significantly activate expression of the LCS EDA reporter. Moreover, cotransfection of either SRp20 with the LCS SRp20 reporter construct or SC35 with the LCS SC35 reporter construct failed to significantly increase the expression of these reporter mRNAs (data not shown). Importantly, overexpression of SR proteins does not dramatically affect the extent of mRNA export and/or stability and further supports the hypothesis that the primary effect of SF2/ASF in this assay is at the level of translation (Fig. 6D)
SF2/ASF increases the utilization of reporter mRNA by the translation machinery
The data presented above suggest that SF2/ASF plays a role in translation. If this hypothesis is correct, then we would predict that overexpression of SF2/ASF will increase the utilization of the reporter mRNA by the translation machinery. We followed the association of the reporter mRNA with ribosomal complexes across sucrose gradients in the presence or absence of SF2/ASF. A direct role for SF2/ASF in the stimulation of translation should result in a shift of the more efficiently translated reporter mRNA being enriched in the polyribosome fractions of sucrose gradients. This was indeed the case, as the distribution of the pLCS-EDA reporter shifted to the polysomes upon overexpression of SF2/ASF (Fig. 7). Taken together, these data, along with those of the preceding experiments, strongly suggest that the predominant effect of SF2/ASF on luciferase expression is at the level of translation.
Figure 7.
Overexpression of SF2/ASF enhances translation of reporter mRNAs. RT-PCR analysis of LCS EDA reporter mRNA levels from HeLa cells transfected with pLCS EDA and either empty expression vector (control) or pCGT7-SF2/ASF (SF2/ASF) fractionated across a 10%-50% sucrose gradient as described in Figure 1. Each transfection also contained the Renilla luciferase reporter.
SF2/ASF enhances translation in a HeLa cell-free translation system
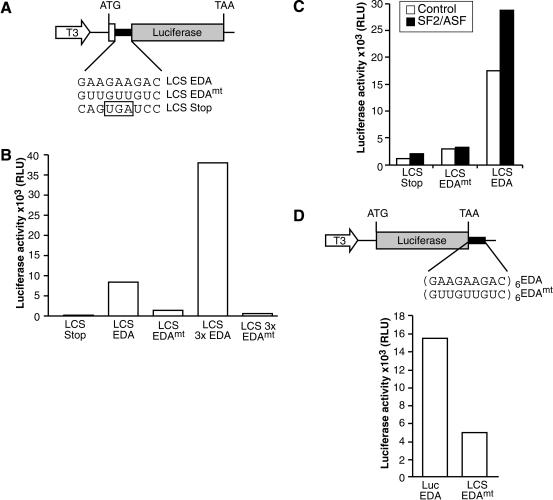
Finally, a translationally competent HeLa cell-free extract was utilized to directly test whether SF2/ASF could enhance translation in vitro (Bergamini et al. 2000). Reporter mRNAs analogous to those described in Figure 3 were synthesized in vitro and are shown schematically in Figure 8A and D. We show that extracts programmed with equal amounts of reporter mRNA constructs containing an in-frame EDA ESE are translated more efficiently than those carrying a mutated ESE. This demonstrates that the presence of an heterologous SF2/ASF-binding site within the ORF can dramatically enhance translation (Fig. 8B). Moreover, reporter mRNAs containing three copies of the EDA ESE were expressed nearly three times greater than those containing a single ESE, indicating that, as is the case in pre-mRNA splicing (Graveley et al. 1998), ESE function additively rather than synergistically in translation. These data suggest that endogenous SF2/ASF present in the extract can stimulate the translation of reporter mRNAs. Furthermore, addition of recombinant SF2/ASF protein enhances this translational activation (Fig. 8C). Interestingly, heterologous EDA ESEs could also stimulate translation of the luciferase reporter mRNA from the 3′UTR, strongly suggesting that the effect of the ESE on translation is independent of changing the coding capacity of the reporter mRNA (Fig. 8D). Importantly, stimulation of translation by SF2/ASF was observed in the absence of an increase in the stability of reporter mRNA, as measured by Northern blots (data not shown). These data indicate that SF2/ASF plays a direct role in translation in a cell-free system, programmed by equal amounts of mRNA templates.
Figure 8.
ESEs and recombinant SF2/ASF stimulate translation of reporter mRNAs in vitro. (A,B) Insertion of the EDA ESE into the luciferase ORF stimulates translation in vitro. Constructs described in Figure 3A were transcribed in vitro. Reporter mRNAs (200 ng) containing either one or three copies of a wild-type or mutant version of the EDA ESE were incubated in a HeLa translation extract. Following incubation at 37°C, luciferase assays were performed (Promega). (C) Recombinant SF2/ASF stimulates expression of LCS reporter mRNAs in vitro. In vitro translation reactions were performed in the presence (black bars) or absence (white bars) of recombinant SF2/ASF (200 ng), as described above. (D) Insertion of the EDA ESE into the 3′UTR enhances translation of the luciferase reporter mRNA in vitro. Reporter mRNAs (100 ng) containing either six copies of a wild-type or mutant EDA ESE in the 3′UTR were incubated in HeLa translation extract. Following incubation at 37°C, luciferase assays were performed.
In summary, we have identified a novel cytoplasmic function for shuttling SR proteins in translation. Although the precise role of SR proteins in translation is yet to be determined, our findings suggest that SR proteins play numerous roles in the metabolism of mRNA and posttranscriptional gene expression in both the nuclear and cytoplasmic compartments of the cell.
Discussion
The nuclear roles of SR proteins in pre-mRNA splicing have been extensively studied. In contrast, until only recently, the cytosolic functions of shuttling SR proteins have remained enigmatic. Cosedimentation of cytoplasmic SR proteins with the translation machinery suggested that SR proteins might play a role in translational regulation. This hypothesis was directly tested using three different functional assays. First, we showed that shuttling SR proteins could stimulate translation of reporter mRNAs when tethered via the MS2 protein in Xenopus oocytes. This experimental approach provided a direct assay of the ability of SF2/ASF to stimulate translation independently of its RNA-binding specificity. Additionally, we developed a complementary mammalian cellular system to assay the activity of SR proteins in translation regulation, which not only allowed us to independently confirm the results obtained with the tethering system in Xenopus oocytes, but also to test the requirement for nucleocytoplasmic shuttling in translational activity. We found that the presence of the fibronectin EDA ESE (a binding site for SF2/ASF) can stimulate the expression of luciferase both in vivo and in vitro. We also demonstrated that the ability of SR proteins to shuttle from the nucleus to the cytoplasm plays a major role in their ability to stimulate translation in vivo (Fig. 5). Thus, the regulation of the nucleocytoplasmic shuttling activity may control the levels of SF2/ASF in the cytoplasm and therefore modulate translational regulation of mRNA targets of SF2/ASF.
It is possible that the role of SF2/ASF in translation regulation may be associated with an early step in translation. We have clearly shown that overexpression of SF2/ASF markedly induces a shift on the ESE-containing reporter toward the polysomal fractions, strongly suggesting that SF2/ASF is affecting translation initiation (Fig. 7). Moreover, our finding that the EDA ESE can stimulate the expression of a luciferase reporter mRNA in vitro when present either in the ORF or the 3′UTR, strongly suggest that the effect of the ESE is at the level of translation rather than the unlikely possibility that the specific activity of the reporter enzyme encoded by the LCS EDA and LCS EDAmt mRNAs differs dramatically.
Recent work from several laboratories has demonstrated functional coupling of different steps in the gene expression pathway. It is widely thought that coordination of pre-mRNA synthesis with the RNA processing machinery is important for regulating and optimizing expression of eukaryotic genes and, that shuttling RNA-binding proteins may play important roles in this process. There is substantial evidence showing a clear effect for an intron on gene expression. Two recent studies showed that the presence of an intron not only influences 3′ end processing, but also significantly enhances the translational utilization of cytoplasmic mRNAs (Lu and Cullen 2003; Nott et al. 2003). This stimulatory effect of introns on translation may, in part, be attributable to the exon junction complex (EJC; Wiegand et al. 2003; Nott et al. 2004).
Here, we have shown that splicing is not required for the stimulatory role of shuttling SR proteins in translation, as SF2/ASF positively affects translation of both intron-containing and intronless reporter mRNAs (Fig. 4). We have also shown that SF2/ASF is associated with polyribosomes in cytoplasmic extracts and can enhance translation both in vivo and in vitro. One plausible hypothesis may be that shuttling SR proteins are involved in regulating the translation of specific mRNA targets. Elucidation of mRNA targets of shuttling SR proteins will play a key role in understanding how shuttling SR proteins influence cytoplasmic steps in gene expression.
Finally, our results underscore the impact of alternative pre-mRNA splicing in increasing the complexity of the proteome. Most typically, alternative splicing decisions lead to the generation of more than one mRNA isoform. We have shown that the presence of an ESE in an mRNA can stimulate its translation in vivo. These data suggest the intriguing possibility that the ratio of exon-included versus skipped mRNA isoforms would be substantially amplified at the protein level by the enhanced translation of the ESE containing isoforms. Thus, shuttling SR proteins may provide an unexpected level of regulation and coupling between alternative pre-mRNA splicing and mRNA translation.
In summary, we have shown that shuttling SR proteins associate with the translational machinery and stimulate translation in Xenopus oocytes and in mammalian cells in vivo, and also in a HeLa cell-free extract system. Although the precise role of SR proteins in translation is yet to be determined, our findings suggest that SR proteins play numerous roles in the metabolism of mRNA in both the nuclear and cytoplamic compartments of the cell. Taken together, these results strongly suggest that SR proteins are multifunctional regulators of mRNA metabolism with diverse roles that couple the processes of splicing, mRNA export, and translation.
Materials and methods
Cell fractionation and sucrose gradient centrifugation
HeLa cells and HeLa Cell cytoplasmic extracts were purchased from 4C Biotech. Cytoplasmic translation extracts were prepared as described below. The salt concentration of the 4C biotech cytoplasmic extract was adjusted to 20 mM Tris (pH 7.5), 5 mM MgCl2, 100 mM KCl, 0.3% NP-40. The extracts were incubated for 10 min on ice, and insoluble material was pelleted by centrifugation at 10,000 rpm for 10 min in a cold microfuge. The resulting supernatant and/or HeLa cytoplasmic translation extracts were then loaded onto a 10%-50% sucrose gradient containing 20 mM Tris (pH 7.5), 5 mM MgCl2, 100 mM KCl and centrifuged for 2 h at 38,000 rpm in a Sorval TH-641 rotor. Following centrifugation, the gradients were fractionated using a Pharmacia Superfrac fraction collector and the absorbance of cytosolic RNA was at 254 nm and was recorded by an inline UV monitor (Pharmacia). For transfection experiments, HeLa cells were removed from a single T-75 flask with trypsin and washed four times with 10 mL of ice-cold PBS. The cell pellet was then resuspended in ice-cold lysis buffer (as described above) and incubated on ice for 10 min. Nuclei and insoluble material were then pelleted at 10,000 rpm for 10 min in a cold microfuge. Sucrose gradient fractionation was carried out as described above.
Antibodies
The following antibodies were used for Western blot analyses: mAb anti-SF2/ASF (clone 96; Hanamura et al. 1998), mAb anti-SRp20 (Neugebauer and Roth 1997), anti-PABP (Gorlach et al. 1994), anti-SRp40 (Snow et al. 1997), anti-rpS6 (Cell Signalling), and anti-T7 monoclonal antibody (Novagen).
Western blot analysis
Protein samples isolated from sucrose gradient fractions, pelleted polysomes, or HeLa nuclear and cytosolic extracts were resolved by SDS-PAGE. Proteins were then transferred to Hybond P membranes (Amersham Pharmacia Biotech). Nonspecific binding sites were blocked by incubation of the membrane with 5% nonfat dry milk in TBST (20 mM Tris at pH 7.5, 137 mM NaCl, and 0.1% Tween 20). Proteins were detected using the following primary antibodies (described above) diluted in 5% nonfat dry milk in TBST: mouse monoclonal anti-SF2/ASF (1:500), mouse monoclonal anti-SRp20 (1:10), rabbit polyclonal anti-PABP (1:2000), rabbit polyclonal anti-rpS6 (1:500), rabbit polyclonal anti-SRp40 (1:1000), and mouse monoclonal anti-T7 (1:10,000). Following washing in TBST, blots were incubated with the appropriate secondary antibodies conjugated to horse-radish peroxidase (Pierce) and detected with Super Signal West Pico detection reagent (Pierce). For Figure 1, the membrane was stripped (100 mM glycine at pH 2.5 and 50 mM NaCl), equilibrated in TBST, blocked in 5% nonfat dry milk, and reprobed as described above.
Microinjection of Xenopus oocytes
In vitro transcription, microinjection of Xenopus oocytes, and luciferase assays were performed as previously described (Gray et al. 2000).
Luciferase reporter system in HeLa cells
The parental plasmid used for the construction of the luciferase reporter systems, pBPLUGA, was a generous gift of Dr. Ian Eperon and has been described elsewhere (Kollmus et al. 1996). pBPLUGA encodes a cassette containing β-galactosidase, a short linker sequence followed by the Firefly luciferase gene. Oligonucleotides encoding an SF2/ASF ESE, or a mutated version, were cloned into the BamHI and SalI sites between the β-galactosidase and luciferase genes, maintaining the ORF. Oligonucleotides encoding an SRp20 or SC35 ESE were cloned into the BamHI and SalI sites between the β-galactosidase and luciferase genes, maintaining the ORF. A total of 10 nM of complementary oligonucleotides were annealed in a 100-μL reaction containing 1 M NaCl, 25 mM EDTA, 50 mM Tris (pH 7.5). The solution was heated to 90°C for 5 min, then cooled to 50°C and incubated for 1 h. The annealed oligos were then desalted by two sequential G25 spin columns. The desalted annealed oligos were then ligated directly into the pBPLUGA. The sequence of each oligo is listed as follows: EDA forward, TCGAGAAGA AGACG; EDA reverse, GATCCGTCTTCTTC; EDA mut forward, TCGAGTTGTTGTCG; EDA mut reverse, GATCCGAC AACAAC; SRp20 forward, TCGATCTCTTCTCG; SRp20 feverse, GATCCGAGAAGAGA; SC35 forward, TCGAGGCCCC TGCG; SC35 reverse, GATCCGCAGGGGCC; 3xEDA forward, TCGAGAAGAAGACGAAGAAGACGAAGAAGAC; 3xEDA reverse, GATCCGTCTTCTTCGTCTTCTTCGTCTTCTTC; 3xEDA mut forward, TCGAGTTGTTGTCGGTTGTTGTCG GTTGTTGTCG; 3xEDA mut reverse, GATCCGACAACAAC GACAACAACGACAACAAC.
The pLCS and pLuc plasmids (Figs. 3, 4, 8, respectively) were generated by PCR amplification from the pBPLUGA plasmids containing the ESE sequences. For pLCS, the 5′ primer annealed 46 codons upstream from the luciferase ORF. For pLuc, the 5′ primer annealed two codons upstream of the luciferase ATG. Amplification with both primer sets yielded a full-length Firefly luciferase ORF, preceded by either a 5′ leader sequence derived from the last 33 codons of β-galactosidase, followed by the SF2/ASF ESE (pLCS series) or by the 5′ end of luciferase, lacking any leader sequence (oligo sequences shown as follows): pLCS 5′ (HindIII), GGGAAGCTTCAACAGATGGGGATTGGTG GC; pLuc 5′ (HindIII), GGGAAGCTTTTCCTCAGATGTCCC GAGGATCC; pLuc 3′(BamHI), GGGGGATCCTTACAATTT GGACTTTCC.
The PCR products were digested with HindIII and EcoRV, then cloned into pBPLUGA, and digested by HindIII and EcoRV, resulting in the deletion of the β-galactosidase sequence present in the parental plasmid pBPLUGA. Intron-containing versions of the pLCS series were generated by amplifying the chimeric intron from the TK-Renilla luciferase expression plasmid by PCR (Promega) using the following primers: 5′ Intron (XhoI), GGGCTCGAGGATTCTTCTGACACAACAG; 3′ Intron (HindIII), GGGAAGCTTCTATAGTGAGTCGTATTAA.
The resulting PCR product was digested with XhoI and HindIII and cloned directionally into the XhoI and HindIII sites in the 5′UTR of the pLCS plasmids.
The in vitro transcription constructs were generated by excising the complete pLCS or pLuc cassettes using HindIII and KpnI and cloning into the same sites in pBlueScript (Stratagene). pLuc 6xEDA and pLuc 6xEDAmt were constructed by cloning oligos (sequence below) into the unique BglII site in the 3′UTR of pLuc. Transformants were screened by sequencing; due to the nondirectional nature of this cloning strategy, multiple insertion events were obtained. The 6xEDA and 6xEDAmt resulted from ligation of two copies of the oligo dimers into either the sense or antisense orientation: 3xEDA forward, GATCGAA GAAGACGAAGAAGACGAAGAAGAC; 3xEDA reverse, GATCGTCTTCTTCGTCTTCTTCGTCTTCTTC.
Cell transfection and dual luciferase assays
HeLa cells were grown to 90% confluence in 24-well plates. A mixture of 1.6 μg of plasmid DNA consisting of 400 ng pLCS reporter, 200 ng TK-Renilla Luciferase (Promega) and 1.0 μg of the pCGT7 expression vector, and Lipofectamine 2000 (Invitrogen) was used to transfect cells following the manufacturers' instructions. The transfection medium was replaced with fresh medium after a 5-h incubation, and cells were then incubated for another 42 h. For dual luciferase assays HeLa cells were lysed using passive lysis buffer (Promega) and the levels of Firefly and Renilla luciferase were assayed using Promega's Dual Luciferase Assay Kit. The luminescence was measured with a Monolight 3010 luminometer (Pharmingen).
RNA isolation and RT-PCR
For mRNA analysis, HeLa cells were grown to 90% confluence in 6-well plates and transfected with pLCS constructs, tk-Renilla Luciferase and pCGT7 SF2/ASF or empty vector, as described above. Cells were fractionated as described for the sucrose gradient analyses, and total RNA was isolated from each fraction using the TRI Reagent as described by the manufacturer's instructions (Sigma). Following digestion with RQ DNAse (Promega), the mRNAs for the pLCS reporter constructs and the ribosomal protein rpS8 were amplified using the One Step RT-PCR kit (Invitrogen) and the following primers: LCS forward, CGGAATTCCAGCTGAGCG; LCS reverse, CAGGGCGTAT CTCTTCATA; rpS8 forward, GGGTCTAGAATGGGCATCT CTCGGGACA; rpS8 reverse, GGGGGATCCTTATTTGCCT TTGCGGCC.
RT-PCR was carried out for 18, 22, 24, 28, 30, and 34 cycles to determine the linear range of amplification. Densitometry was performed using the software suite provided with the QuantityOne imaging system (Bio-Rad). For RT-PCR analysis of cytosolic reporter mRNA levels, equal amounts (1 μg) of total RNA was reverse transcribed using oligo dT. The luciferase reporter mRNA and rpS8 mRNA were then amplified by PCR using the primers described above. For RT-PCR analysis of luciferase reporters across sucrose gradients, one-third of each fraction was extracted with TRI Reagent. Purified RNA was digested with RQ DNAse, and cDNA was synthesised using the LCS Reverse primer and Super Script Reverse Transcriptase (Invitrogen). One-quarter of the cDNA from each fraction was amplified by 40 cycles of PCR with LCS Forward and Reverse primers and Ampli-taq (Perkin Elmer). Northern blot analysis was carried out are previously described (Gray et al. 2000).
Preparation of HeLa in vitro translation extracts and in vitro translation reactions
HeLa Cell free extracts were prepared from 2.5 × 109 HeLa Cells (4Cbiotech) as previously described (Bergamini et al. 2000). Translation reactions were programmed with 200 ng of reporter mRNA in the presence or absence of 200 ng of recombinant SF2/ASF (ProteinOne) or BC100 (20 mM Tris HCl at pH 7.5, 100 mM KCl, 0.2 mM EDTA, and 20% glycerol) and incubated at 37°C for 30 min. Reactions were stopped by dilution into 50 μL passive lysis buffer (Promega). Luciferase activity was assayed using Promega Luciferase Activating Reagent.
Acknowledgments
We are grateful to Juan Valcárcel for comments and critical reading of the manuscript. We thank Tom Misteli and Nick Hastie for helpful discussions, Brent Graveley and Ian Eperon for constructs, and Bill Richardson for technical assistance. This work was supported by the MRC (N.K.G. and J.F.C.), DAAD (K.B.), and nonconcurrent postdoctoral fellowships from EMBO and the Caledonian Research Foundation (J.R.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.286404.
Corresponding author.
References
- Bergamini G., Preiss, T., and Hentze, M.W. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6: 1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J. 2000. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25: 106-110. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Padgett, R.A., and Sharp, P.A. 1998. Evolutionary fates and origins of U12-type introns. Mol. Cell 2: 773-785. [DOI] [PubMed] [Google Scholar]
- Caceres J.F. and Kornblihtt, A.R. 2002. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 18: 186-193. [DOI] [PubMed] [Google Scholar]
- Caceres J.F. and Krainer, A.R. 1993. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 12: 4715-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J.F., Stamm, S., Helfman, D.M., and Krainer, A.R. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265: 1706-1709. [DOI] [PubMed] [Google Scholar]
- Caceres J.F., Misteli, T., Screaton, G.R., Spector, D.L., and Krainer, A.R. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell. Biol. 138: 225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres J.F., Screaton, G.R., and Krainer, A.R. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes & Dev. 12: 55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M., Casari, G., Guenzi, S., Tagliabue, R., Sidoli, A., Melo, C.A., and Baralle, F.E. 1994. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 22: 1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., Wang, J., Zhu, Z., Zhang, M.Q., and Krainer, A.R. 2003. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31: 3568-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y., Bourgeois, C.F., Kister, L., and Stevenin, J. 1999. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA 5: 468-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D., Zhu, J., Manche, L., Huber, E., Krainer, A.R., and Caceres, J.F. 2002. Nuclear export and retention signals in the RS domain of SR proteins. Mol. Cell Biol. 22: 6871-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P., Caceres, J.F., Cazalla, D., Kadener, S., Muro, A.F., Baralle, F.E., and Kornblihtt, A.R. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4: 251-258. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3: 195-205. [DOI] [PubMed] [Google Scholar]
- Fu X.D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1: 663-680. [PMC free article] [PubMed] [Google Scholar]
- Gorlach M., Burd, C.G., and Dreyfuss, G. 1994. The mRNA poly(A)-binding protein: Localization, abundance, and RNA-binding specificity. Exp. Cell Res. 211: 400-407. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B.R. and Maniatis, T. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1: 765-771. [DOI] [PubMed] [Google Scholar]
- Graveley B.R., Hertel, K.J., and Maniatis, T. 1998. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 17: 6747-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.K., Coller, J.M., Dickson, K.S., and Wickens, M. 2000. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19: 4723-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelhah H., Shah, K., Huang, L., Ostareck-Lederer, A., Burlingame, A.L., Shokat, K.M., Hentze, M.W., and Ronai, Z. 2001. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell. Biol. 3: 325-330. [DOI] [PubMed] [Google Scholar]
- Hanamura A., Caceres, J.F., Mayeda, A., Franza, B.R., and Krainer, A.R. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4: 430-444. [PMC free article] [PubMed] [Google Scholar]
- Hastings M.L. and Krainer, A.R. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell. Biol. 13: 302-309. [DOI] [PubMed] [Google Scholar]
- Hedley M.L., Amrein, H., and Maniatis, T. 1995. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl. Acad. Sci. 92: 11524-11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek K.S., Kidd, G.J., Carson, J.H., and Smith, R. 1998. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry 37: 7021-7029. [DOI] [PubMed] [Google Scholar]
- Hoffman B.E. and Lis, J.T. 2000. Pre-mRNA splicing by the essential Drosophila protein B52: Tissue and target specificity. Mol. Cell. Biol. 20: 181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. and Steitz, J.A. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7: 899-905. [DOI] [PubMed] [Google Scholar]
- Huang Y., Gattoni, R., Stevenin, J., and Steitz, J.A. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11: 837-843. [DOI] [PubMed] [Google Scholar]
- Jumaa H. and Nielsen, P.J. 1997. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 16: 5077-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H., Wei, G., and Nielsen, P.J. 1999. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 9: 899-902. [DOI] [PubMed] [Google Scholar]
- Jurica M.S. and Moore, M.J. 2003. Pre-mRNA splicing. Awash in a sea of proteins. Mol. Cell 12: 5-14. [DOI] [PubMed] [Google Scholar]
- Kaminski A., Hunt, S.L., Patton, J.G., and Jackson, R.J. 1995. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1: 924-938. [PMC free article] [PubMed] [Google Scholar]
- Kawano T., Fujita, M., and Sakamoto, H. 2000. Unique and redundant functions of SR proteins, a conserved family of splicing factors, in Caenorhabditis elegans development. Mech. Dev. 95: 67-76. [DOI] [PubMed] [Google Scholar]
- Kollmus H., Flohe, L., and McCarthy, J.E. 1996. Analysis of eukaryotic mRNA structures directing cotranslational incorporation of selenocysteine. Nucleic Acids Res. 24: 1195-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65: 367-409. [DOI] [PubMed] [Google Scholar]
- Lall S., Francis-Lang, H., Flament, A., Norvell, A., Schupbach, T., and Ish-Horowicz, D. 1999. Squid hnRNP protein promotes apical cytoplasmic transport and localization of Drosophila pair-rule transcripts. Cell 98: 171-180. [DOI] [PubMed] [Google Scholar]
- Lamond A.I. and Spector, D.L. 2003. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4: 605-612. [DOI] [PubMed] [Google Scholar]
- Lavigueur A., La Branch, H., Kornblihtt, A.R., and Chabot, B. 1993. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes & Dev. 7: 2405-2417. [DOI] [PubMed] [Google Scholar]
- Lemaire R., Prasad, J., Kashima, T., Gustafson, J., Manley, J.L., and Lafyatis, R. 2002. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: A novel function for SR proteins. Genes & Dev. 16: 594-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.X., Chew, S.L., Cartegni, L., Zhang, M.Q., and Krainer, A.R. 2000. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol. 20: 1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loflin P., Chen, C.Y., and Shyu, A.B. 1999. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes & Dev. 13: 1884-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., Johnstone, I.L., and Caceres, J.F. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19: 1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. and Cullen, B.R. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A. and Krainer, A.R. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68: 365-375. [DOI] [PubMed] [Google Scholar]
- Neugebauer K.M. and Roth, M.B. 1997. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes & Dev. 11: 1148-1159. [DOI] [PubMed] [Google Scholar]
- Nott A., Meislin, S.H., and Moore, M.J. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9: 607-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Le Hir, H., and Moore, M.J. 2004. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes & Dev. 18: 210-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D.H., Ostareck-Lederer, A., Wilm, M., Thiele, B.J., Mann, M., and Hentze, M.W. 1997. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89: 597-606. [DOI] [PubMed] [Google Scholar]
- Peng X. and Mount, S.M. 1995. Genetic enhancement of RNA-processing defects by a dominant mutation in B52, the Drosophila gene for an SR protein splicing factor. Mol. Cell. Biol. 15: 6273-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S. and Dreyfuss, G. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355: 730-732. [DOI] [PubMed] [Google Scholar]
- Ring H.Z. and Lis, J.T. 1994. The SR protein B52/SRp55 is essential for Drosophila development. Mol. Cell. Biol. 14: 7499-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Longman, D., and Caceres, J.F. 2003. Multiple roles of the SR protein family in splicing regulation. Prog. Mol. Subcell. Biol. 31: 33-58. [DOI] [PubMed] [Google Scholar]
- Shyu A.B. and Wilkinson, M.F. 2000. The double lives of shuttling mRNA binding proteins. Cell 102: 135-138. [DOI] [PubMed] [Google Scholar]
- Snow B.E., Heng, H.H., Shi, X.M., Zhou, Y., Du, K., Taub, R., Tsui, L.C., and McInnes, R.R. 1997. Expression analysis and chromosomal assignment of the human SFRS5/SRp40 gene. Genomics 43: 165-170. [DOI] [PubMed] [Google Scholar]
- Spector D.L., Fu, X.D., and Maniatis, T. 1991. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10: 3467-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R. and Manley, J.L. 1999. Determinants of SR protein specificity. Curr. Opin. Cell. Biol. 11: 358-362. [DOI] [PubMed] [Google Scholar]
- Valcarcel J. and Green, M.R. 1996. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 21: 296-301. [PubMed] [Google Scholar]
- Wang J., Takagaki, Y., and Manley, J.L. 1996. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes & Dev. 10: 2588-2599. [DOI] [PubMed] [Google Scholar]
- Wang J., Xiao, S.H., and Manley, J.L. 1998. Genetic analysis of the SR protein ASF/SF2: Interchangeability of RS domains and negative control of splicing. Genes & Dev. 12: 2222-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Y., Xu, X., Ding, J.H., Bermingham Jr., J.R., and Fu, X.D. 2001. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell 7: 331-342. [DOI] [PubMed] [Google Scholar]
- Wiegand H.L., Lu, S., and Cullen, B.R. 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. 100: 11327-11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Luhrmann, R. 1997. Protein functions in pre-mRNA splicing. Curr. Opin. Cell. Biol. 9: 320-328. [DOI] [PubMed] [Google Scholar]
- Wu J.Y. and Maniatis, T. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75: 1061-1070. [DOI] [PubMed] [Google Scholar]
- Xu N., Chen, C.Y., and Shyu, A.B. 2001. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol. Cell. Biol. 21: 6960-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Bani, M.R., Lu, S.J., Rowan, S., Ben David, Y., and Chabot, B. 1994. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl. Acad. Sci. 91: 6924-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo P. and Maniatis, T. 1996. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes & Dev. 10: 1356-1368. [DOI] [PubMed] [Google Scholar]