Abstract
The discovery of diverse codon reassignment events has demonstrated that the canonical genetic code is not universal. Studying coding reassignment at the molecular level is critical for understanding genetic code evolution, and provides clues to genetic code manipulation in synthetic biology. Here we report a novel reassignment event in the mitochondria of Ashbya (Eremothecium) gossypii, a filamentous-growing plant pathogen related to yeast (Saccharomycetaceae). Bioinformatics studies of conserved positions in mitochondrial DNA-encoded proteins suggest that CUU and CUA codons correspond to alanine in A. gossypii, instead of leucine in the standard code or threonine in yeast mitochondria. Reassignment of CUA to Ala was confirmed at the protein level by mass spectrometry. We further demonstrate that a predicted 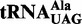 is transcribed and accurately processed in vivo, and is responsible for Ala reassignment. Enzymatic studies reveal that
is transcribed and accurately processed in vivo, and is responsible for Ala reassignment. Enzymatic studies reveal that 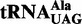 is efficiently recognized by A. gossypii mitochondrial alanyl-tRNA synthetase (AgAlaRS). AlaRS typically recognizes the G3:U70 base pair of tRNAAla; a G3A change in Ashbya
is efficiently recognized by A. gossypii mitochondrial alanyl-tRNA synthetase (AgAlaRS). AlaRS typically recognizes the G3:U70 base pair of tRNAAla; a G3A change in Ashbya
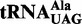 abolishes its recognition by AgAlaRS. Conversely, an A3G mutation in Saccharomyces cerevisiae
abolishes its recognition by AgAlaRS. Conversely, an A3G mutation in Saccharomyces cerevisiae
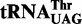 confers tRNA recognition by AgAlaRS. Our work highlights the dynamic feature of natural genetic codes in mitochondria, and the relative simplicity by which tRNA identity may be switched.
confers tRNA recognition by AgAlaRS. Our work highlights the dynamic feature of natural genetic codes in mitochondria, and the relative simplicity by which tRNA identity may be switched.
INTRODUCTION
When the genetic code was first deciphered in the 1960s, it was considered to be universal, with all organisms using the same standard code. Later it was shown that several codons have been recoded with different amino acids, thus creating non-standard genetic codes that are present in modern organisms (1). To date, 11 codon reassignment events have been reported in the nuclear genomes of bacteria, archaea and eukaryotes, and 16 have been found in mitochondria [reviewed in (1–4)]. Most recently, single-cell sequencing and biochemical analyses identified yet another, UGA tryptophan-to-glycine reassignment event in SR1 bacteria (5). These dogma-breaking discoveries suggest that the genetic code is evolvable in nature, and that it could be engineered in synthetic organisms. Enabled by genome engineering technologies, such as multiplex automated genome engineering (6), conjugative assembly genome engineering (7) and de novo genome synthesis (8), editing and rewriting the genetic code have emerged as an exciting topic in synthetic biology. Multiplex automated genome engineering, and conjugative assembly genome engineering have been used to change all 321 known UAG stop codons to the synonymous UAA stop codon. This will enable the abolition of UAG function, thereby permitting subsequent reassignment from ‘stop’ to any natural or non-natural amino acid (7).
Natural codon reassignment events may be explained by different evolutionary scenarios, depending on special circumstances. The codon capture mechanism (9) evokes that a specific set of codons and the corresponding tRNA completely disappear from a genome before a novel tRNA evolves to read such codons with a different specificity. In contrast, the ambiguous intermediate mechanism (10) posits that a codon does not need to disappear before reassignments and that it is ambiguously translated into distinct amino acids. More recently, reassignment scenarios have been discussed within an extended gain–loss framework of codons and tRNAs, pointing out that it is necessary to consider the details of each case carefully (11).
Codon reassignment is facilitated in mitochondrial DNAs (mtDNAs) because they encode only a small set of proteins, and tend to be A + T rich, which introduces a strong codon bias. For instance, UAA stop codons are highly preferred over UGA and UAG, making the latter available for reassignment from ‘stop’ to ‘sense’. The use of UGA (tryptophan) has evolved several times independently in the mitochondria of unrelated eukaryotic lineages, so has the introduction of UAG (Leu or Ala) [for a review see (12)]. Not only may stop codons be assigned to amino acids, but sense codons [e.g. UCA in Scenedesmus obliquus (13,14), and UUA, UUG in Pycnococcus provasolii (15)] may also convert to stop. It is interesting that UCA codons are absent in mitochondrial genes of close relatives of S. obliquus [i.e. Chlamydomonas and Pedinomonas (14)], in support of a codon capture mechanism in this case. Finally, sense codons may switch identity from one amino acid to another. The principle is similar to stop codon reassignment and facilitated when amino acids are encoded by alternative codon families, Leu (CUN or UUA/UUG in the standard code; N denotes A, U, G or C), Arg (CGN or AGA/AGG) and Ser (UCN or AGU/AGC). The less frequently used codon family may disappear, either completely or with only few remaining codons in non-critical sites of proteins, to allow switching of the aminoacyl-tRNA synthetase (aaRS) recognition site of a given tRNA to a different identity. Alternatively, extended phases of codon ambiguity may exist. For example, mutation of the tRNASer anticodon to CAG and an m1G37 modification confer recognition of this tRNA by both seryl- and leucyl–tRNA synthetases, which is responsible for the ambiguous decoding of CUG by Ser and Leu in Candida albicans (16,17).
In this context, mitochondria are a special case. Their genes tend to be A + T rich with A or U preferred in third codon positions, and with genomes encoding small gene sets, which together simplifies liberation of codons for subsequent identity switch. However, an unmodified U in the wobble position of a tRNA’s anticodon is able to accommodate any of the four standard nucleotides in the codon’s third position (18,19), reducing the number of tRNAs that are required to recognize all codons to only 25 or even less [reviewed in (12,20)]. Consequently, switching identity of mitochondrial four-codon families by a codon capture mechanism requires liberation of the complete codon family before codon reassignment, which explains why so far only one such case has been identified. This example is the reassignment of CUN codons from Leu to Thr in the mitochondria of Saccharomyces cerevisiae (21,22), which belongs to a group of yeast species (Saccharomycetaceae) that lost all seven standard mitochondrial genes encoding subunits of the NADH dehydrogenase complex. Responsible for the reassignment event in the remaining eight protein-coding genes of S. cerevisiae is an unusual 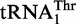 with a UAG anticodon (23), and an 8- rather than 7-nt-long anticodon loop, which has evolved from a tRNAHis ancestor (21). Here, we report a second instance in which a tRNA closely related to
with a UAG anticodon (23), and an 8- rather than 7-nt-long anticodon loop, which has evolved from a tRNAHis ancestor (21). Here, we report a second instance in which a tRNA closely related to 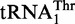 reassigns CUU and CUA codons in the mitochondrial genome of the yeast A. gossypii to Ala. We further provide preliminary evidence for a third event in another yeast species (Nakaseomyces bacillisporus), in which CGA is read as histidine rather than arginine.
reassigns CUU and CUA codons in the mitochondrial genome of the yeast A. gossypii to Ala. We further provide preliminary evidence for a third event in another yeast species (Nakaseomyces bacillisporus), in which CGA is read as histidine rather than arginine.
MATERIALS AND METHODS
Cloning, mutagenesis and general methods
The A. gossypii AlaRS gene was cloned into pET28a expression vector (Novagen) with an N-terminal six-His tag. Expression of recombinant proteins was induced at 37°C for 4 hours with 0.5 mM isopropyl β-D-1-thiogalactopyranoside in Escherichia coli strain BL21-codon plus in Luria–Bertani media. His-tagged proteins were purified according to the standard procedures. Mitochondrial tRNA genes were cloned into pUC18 vector (GenScript), and mutations were introduced using QuikChange Site-Directed Mutagenesis Kit (Stratagene).
In vitro assays with tRNAs
In vitro tRNA transcripts were obtained using the T7 RNA polymerase runoff procedure as described (24). Aminoacylation experiments were performed as described (25) in the presence of 100 mM Na-HEPES, pH 7.2, 30 mM KCl, 10 mM MgCl2, 2 mM ATP, 25 µM [14C] Ala or [14C] Thr, 5 µM tRNA transcripts and 30-3000 nM aaRSs.
Identification of mitochondrial tRNAs in RNA-Seq data
A. gossypii cells (ATCC 10895) were grown to an optical density of ∼2.5 in a medium containing 1% yeast extract, 0.5% glucose and 3% glycerol, pH 5.5. Purification of a crude mitochondrial fraction followed previously published procedures (26), and RNA-Seq sequences (Illumina MiSeq; provided by the Genome Quebec Innovation Center) were generated from total, Trizol-extracted mitochondrial RNA, without size selection. Sequences in fastq format were quality-trimmed (phred 20, minimum sequence length 20 nt) and adapter-clipped using Seqtrimnext (http://rubygems.org/gems/seqtrimnext). tRNA sequences were analyzed by scanning the trimmed sequences with a 15-nt-long sequence window (using the basic Linux tools grep and wc).
Identification of mitochondrial proteins by mass spectrometry
Purified mitochondria were solubilized in a buffer containing 20 mM Hepes/KOH, pH 7.4, 60 mM NH4Cl, 10 mM MgCl2, 0.5 mM ethylenediaminetetraacetic acid, 1 mM phenylmethylsulfonyl fluoride and digitonin (2 g/g of protein), followed by incubation on ice for 30 min and homogenization in a Potter homogenizer. After centrifugation at 18 000 g for 15 min, the supernatant was collected, and a small fraction (∼150 µg protein) was separated for 30 min (4–14% Blue Native Poly-Acrylamide Gel Electrophoresis (BN-PAGE); Hoefer apparatus with an 18 × 16 cm electrophoresis chamber; 140 V and 9 mA). The preparation of BN-PAGE gels, electrophoresis buffer and samples followed previously published procedures (27). The protein-containing zone was cut from the gel and submitted to liquid chromatography tandem mass spectrometry analysis (28,29), provided by a service platform at the Université de Montréal (Institute for Research in Immunology and Cancer). It includes destaining, reduction, alkylation, tryptic digestion and functional annotation by Mascot (30).
Sequence alignments and identification of mitochondrial tRNAs
Derived mitochondrial protein sequences were aligned with Muscle version 3.6 (31). Mitochondrial tRNAs were identified with MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl). It uses tool components of the Infernal package v1.1rc2 (32,33), notably cmbuild and cmcalibrate, to build a covariance search model from aligned tRNA training sets, followed by cmsearch to screen genomic sequences. The tRNA sequences shown in Figures 2 and 6 were aligned with cmsearch and the –A switch, using the standard tRNA model of MFannot. For visualization, editing and reformatting of sequence alignments, we used the Genetic Data Environment (GDE) sequence editor (34). A modified GDE version that functions with current 64-bit Linux versions, together with the appropriate libraries, is available on request.
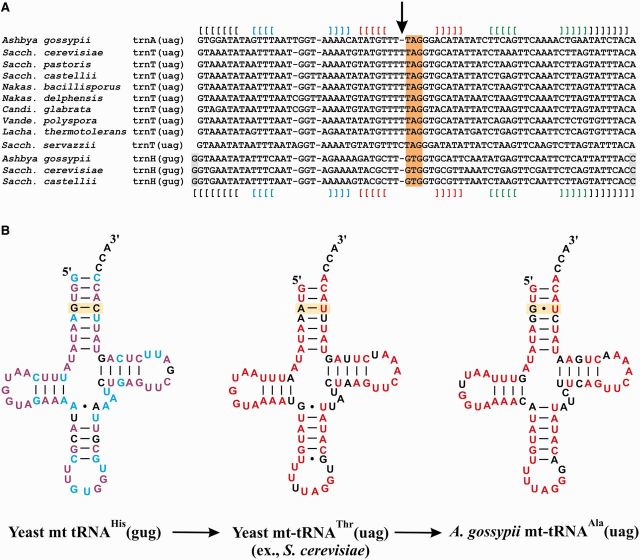
Figure 2.
Primary and secondary structures of yeast mitochondrial 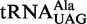 ,
, 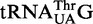 and
and 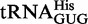 . (A) Selected yeast mitochondrial tRNAs with UAG or GUG anticodons (anticodon marked orange) are aligned (upper and lower blocks of sequences, respectively). Square brackets indicate the four standard helical regions in tRNAs. The arrow points to a nucleotide insertion that leads to a characteristic 8-nt anticodon loop in
. (A) Selected yeast mitochondrial tRNAs with UAG or GUG anticodons (anticodon marked orange) are aligned (upper and lower blocks of sequences, respectively). Square brackets indicate the four standard helical regions in tRNAs. The arrow points to a nucleotide insertion that leads to a characteristic 8-nt anticodon loop in 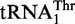 (
(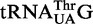 ). In A. gossypii, the anticodon loop has 7 nucleotides, and this tRNA reads CUN codons as Ala but not Thr. As
). In A. gossypii, the anticodon loop has 7 nucleotides, and this tRNA reads CUN codons as Ala but not Thr. As 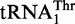 is most likely derived from tRNAHis by duplication, tRNAHis is included in the alignment for comparison [see also (21)]. Note the characteristic G residue at position −1 (constituting the 5′-terminus of tRNAHis) that pairs with the C at the 3′-discriminator position (both positions marked in gray). (B) Secondary structures of tRNAs that illustrate a possible tRNA reassignment scenario, S. cerevisiae
is most likely derived from tRNAHis by duplication, tRNAHis is included in the alignment for comparison [see also (21)]. Note the characteristic G residue at position −1 (constituting the 5′-terminus of tRNAHis) that pairs with the C at the 3′-discriminator position (both positions marked in gray). (B) Secondary structures of tRNAs that illustrate a possible tRNA reassignment scenario, S. cerevisiae
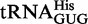 (left), A. gossypii
(left), A. gossypii
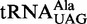 (right) and S. cerevisiae
(right) and S. cerevisiae
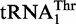 (middle). Nucleotide identity is coded as follows: red, A. gossypii
(middle). Nucleotide identity is coded as follows: red, A. gossypii
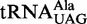 versus S. cerevisiae
versus S. cerevisiae
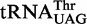 ; magenta, identity across all three tRNAs; and blue, additional identity between A. gossypii and S. cerevisiae
; magenta, identity across all three tRNAs; and blue, additional identity between A. gossypii and S. cerevisiae
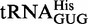 . Following a codon capture mechanism,
. Following a codon capture mechanism, 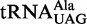 and
and 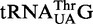 would have evolved from a
would have evolved from a 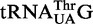 because the latter is present in the sister lineage Lachancea species (for a species phylogeny, see Figure 5), and absent in K. lactis. For abbreviations, see Figure 1.
because the latter is present in the sister lineage Lachancea species (for a species phylogeny, see Figure 5), and absent in K. lactis. For abbreviations, see Figure 1.
Figure 6.
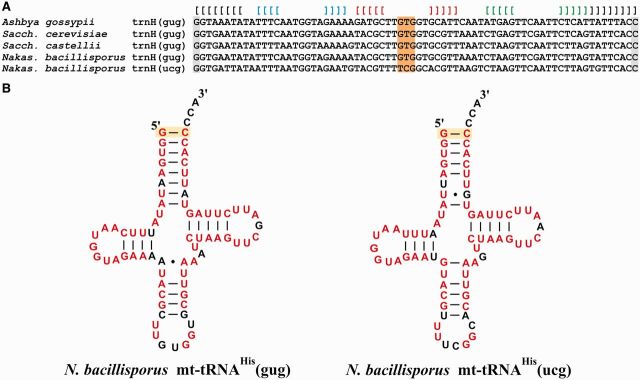
Primary and secondary structures of N. bacillisporus mitochondrial 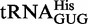 and putative
and putative 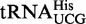 . (A) Selected yeast mitochondrial tRNAsHis with GUG anticodons (anticodon marked orange) are aligned in comparison with an unusual, predicted N. bacillisporus mitochondria tRNAHis with a UCG anticodon (that according to the standard genetic code would recognize CGN arginine). Square brackets indicate the four typical helical regions in tRNAs. Note the characteristic G residue at position −1 (constituting the 5′ terminus of tRNA histidine) that pairs with the C at the 3′ discriminator position (both positions marked in gray). (B) Secondary structures of the two mitochondrial N. bacillisporus histidine tRNAs. Nucleotide identity is coded in red, and the recognition sequence for HisRS (G–C base pair at position −1) is marked yellow.
. (A) Selected yeast mitochondrial tRNAsHis with GUG anticodons (anticodon marked orange) are aligned in comparison with an unusual, predicted N. bacillisporus mitochondria tRNAHis with a UCG anticodon (that according to the standard genetic code would recognize CGN arginine). Square brackets indicate the four typical helical regions in tRNAs. Note the characteristic G residue at position −1 (constituting the 5′ terminus of tRNA histidine) that pairs with the C at the 3′ discriminator position (both positions marked in gray). (B) Secondary structures of the two mitochondrial N. bacillisporus histidine tRNAs. Nucleotide identity is coded in red, and the recognition sequence for HisRS (G–C base pair at position −1) is marked yellow.
Phylogeny of yeast species based on mtDNA-encoded protein sequences
Thirteen standard, mtDNA-encoded derived protein sequences (Cob, Cox1,2,3, Atp6,9 and Nad1,2,3,4,4L,5,6) were aligned in two steps. Briefly, sequences were pre-aligned with Muscle (31) and then refined with HMMalign (S. Eddy; http://hmmer.janelia.org). Sequence positions that were not aligned with a posterior probability value of 1.0 are discarded, and alignments were concatenated. The final dataset includes 40 species and has 3583 amino acid positions. The phylogenetic analysis was performed with PhyloBayes (35), the CAT/GTR model, six discrete categories, four independent chains, 14 000 cycles (corresponding to ∼770 000 generations) and the –dc parameter to remove constant sites. The first 10 000 cycles were discarded as burn-in.
Phylogeny of yeast mitochondrial tRNAs
The phylogenetic analysis with PhyloBayes (CAT, GTR, six categories; four independent chains, 11 000 cycles corresponding to ∼1 800 000 generations) contained all tRNA sequences from the species presented in Figure 5. Only the section of the tRNA phylogeny covering the related 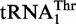 and tRNAHis clusters is shown in Figure 3.
and tRNAHis clusters is shown in Figure 3.
Figure 5.
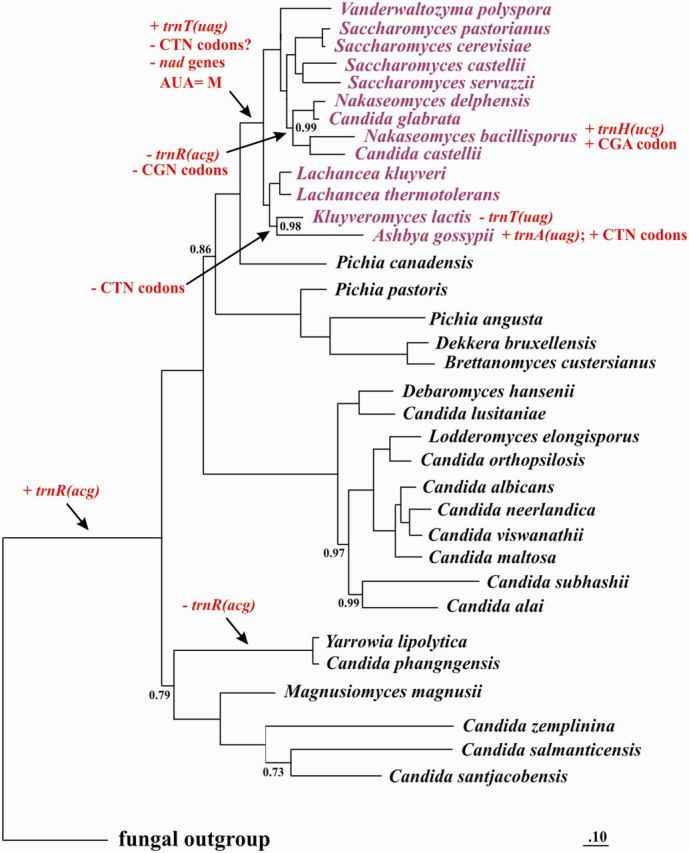
Evolution of yeast species and codon reassignment events. The phylogenetic analysis with PhyloBayes and the CAT/GTR model is based on 13 mtDNA-encoded proteins. All divergence points are supported by posterior probability values of 1.0, except where indicated; Saccharomycetaceae are in magenta. The fungal outgroup consisted of Rhizopus oryzae, Aspergillus niger, Podospora anserina, Fusarium oxysporum, Cantharellus cibarius and Ustilago maydis. All mitochondrial sequences were downloaded from the ‘Organelle Genome Resources’ Web site of NCBI. The red arrow points to the concomitant loss of all seven nad genes (subunits of NADH dehydrogenase complex) and the start of major mitochondrial codon reassignments, including AUA methionine, CUN threonine, CUN alanine and (most unusual) CGN histidine.
Figure 3.
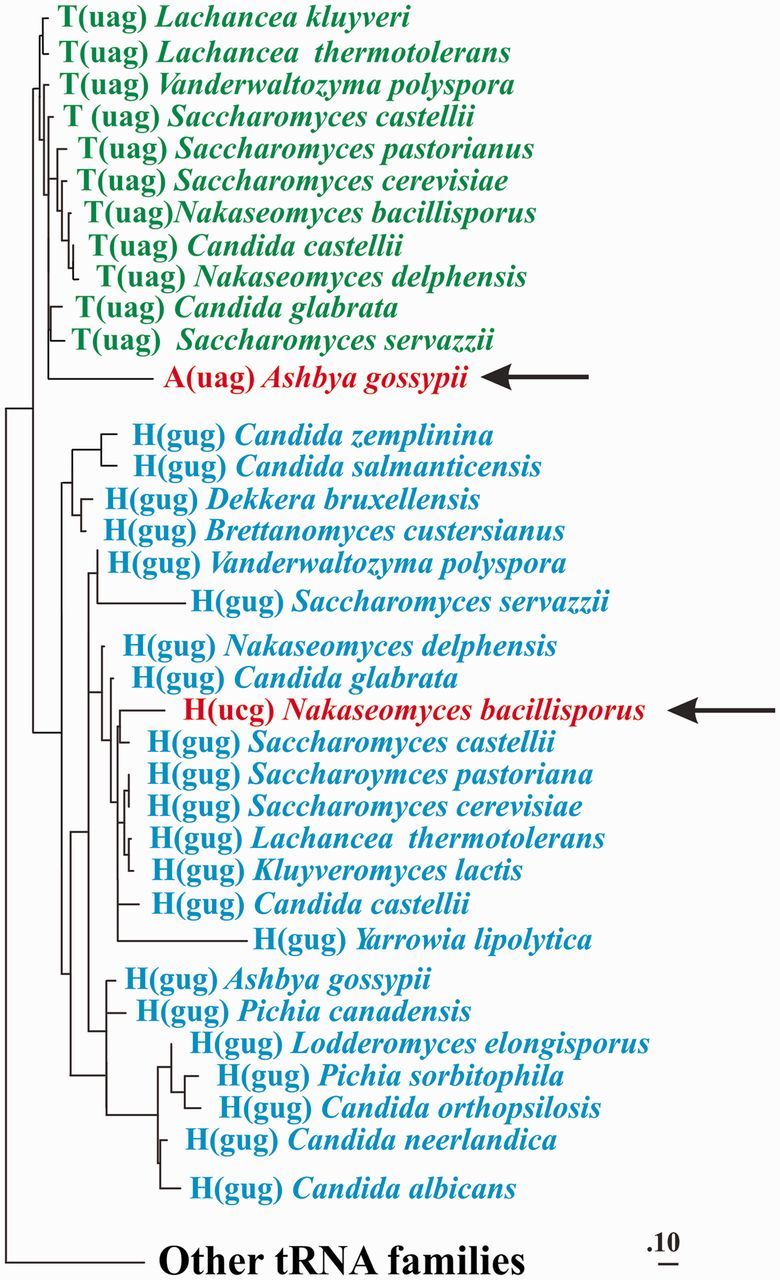
Phylogeny of yeast mitochondrial tRNAs. The phylogenetic analysis contains representatives from a broad selection of yeast species. Only the section of the tree in which relevant 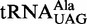 ,
, 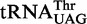 and
and 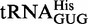 cluster together is shown. The posterior probability support for the two tRNA groups is indicated. Green and blue indicate
cluster together is shown. The posterior probability support for the two tRNA groups is indicated. Green and blue indicate 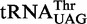 , and
, and 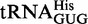 families, respectively. Red indicates reassigned tRNAs. Note that phylogenetic analysis with tRNA sequences depends on only a few informative nucleotide positions, which does not allow resolving the branching order within these groups.
families, respectively. Red indicates reassigned tRNAs. Note that phylogenetic analysis with tRNA sequences depends on only a few informative nucleotide positions, which does not allow resolving the branching order within these groups.
RESULTS
Bioinformatic analyses suggest that CUU and CUA codons are reassigned from Leu to Ala in mitochondria of A. gossypii
Multiple sequence alignment of derived mitochondrial protein sequences reveals numerous positions where Ashbya does not conform to otherwise highly conserved, or even invariant, amino acids. For instance, in the given example of a cytochrome oxidase Cox2 sequence alignment (Figure 1), three columns containing CUN-encoded amino acids (amino acid shown in lower case) are highlighted. In the first two marked sequence columns, the pattern of conservation is consistent with translation of CUN to Thr (indicated by a lower-case t) for the given species, as experimentally confirmed in S. cerevisiae (21,22). In the third column, CUN corresponds to Ala in A. gossypii (shown as a lower-case a). This trend applies to all eight regular mtDNA-encoded proteins in Ashbya (49 CUA and 32 CUU codons; Supplementary Table S1), with most deviations in less well-conserved amino acid positions.
Figure 1.

Alignment of derived Cox2 protein sequences and mass spectrometry at the protein level indicate that CUN decodes alanine in A. gossypii. The alignment corresponding to amino acids 141–195 of A. gossypii Cox2 is shown. Amino acids corresponding to CUN codons are in lower case, and translated into either Thr (most Saccharomycetaceae) or Ala (A. gossypii, confirmed by mass spectrometry of mitochondrial proteins. The sequence of the respective tryptic peptide is FIVTAaDVIHDFAVPSLGIK). Lower case m corresponds to AUA codons. This conservation pattern is valid for all mtDNA-encoded proteins of the shown species. Abbreviations: Sacch., Saccharomyces; Candi., Candida; Kluyv., Kluyveromyces; Lacha., Lachancea; Nakas., Nakaseomyces; and Vande., Vanderwaltozyma.
In the context of codon reassignment, the strict avoidance of CUG and CUC codons seems noteworthy. An analysis of overall mitochondrial codon usage reveals that Ashbya is as biased in other codon families, with 24 sense codons unused, most of which with a C or G in the third position (Supplementary Table S1). In other yeast species such as S. cerevisiae, overall codon bias is similar, although somewhat less extreme (Supplementary Table S1). A notable exception is in Kluyveromyces lactis, a close relative of Ashbya. This species avoids CUN codons altogether (Supplementary Table S1), and has no corresponding mtDNA-encoded tRNA with a UAG anticodon that would allow the recognition of this codon family.
Mass spectrometry confirms the identity of CUU and CUA codons in A. gossypii mitochondrial proteins
To confirm the predicted CUA/CUU codon identity in A. gossypii as Ala, tryptic digests of proteins extracted from purified mitochondria were analyzed by mass spectrometry. To identify potential translation variants, results were analyzed based on three sets of inferred proteins in which CUA/CUU was translated as Leu, Thr or Ala. Among 475 identified proteins, two (Cox1 and Cox2) were mtDNA-encoded, with peptides covering gene regions with one CUA codon each. In both cases (experiment repeated three times), CUA was translated as Ala but not Thr or Leu (for mass spectrometry data of identified Cox2 peptides, see Supplementary Figure S1). Protein regions translated with CUU codons were not identified.
In silico identification of the A. gossypii tRNA decoding CUU and CUA codons
Identification of yeast mitochondrial tRNAs is highly sensitive and without false positives when using covariance search models (32), which are used by our annotation tool MFannot (12). All known tRNA structures with a UAG anticodon (tRNAUAG) were identified throughout yeast species (e.g. Figure 2A), with the notable exception of K. lactis, as mentioned earlier in the text. In distinction to the 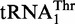 in S. cerevisiae and most other Saccharomycetaceae, the A. gossypii tRNAUAG has a standard 7-nt anticodon loop (Figure 2A). A comparison of tRNAUAG across yeast species further reveals >70% sequence identity (e.g. Figure 2A and B), pointing to a recent common ancestry.A phylogenetic analysis of mtDNA-encoded tRNAs confirms this view, clustering
in S. cerevisiae and most other Saccharomycetaceae, the A. gossypii tRNAUAG has a standard 7-nt anticodon loop (Figure 2A). A comparison of tRNAUAG across yeast species further reveals >70% sequence identity (e.g. Figure 2A and B), pointing to a recent common ancestry.A phylogenetic analysis of mtDNA-encoded tRNAs confirms this view, clustering 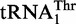 of Saccharomycetaceae with A. gossypii tRNAUAG (Figure 3). This analysis further confirms that tRNAUAG from yeast mitochondria was originally derived from a duplication of a histidine tRNA, as previously proposed (21). The tRNA phylogeny further reveals an unexpected clustering of N. bacillisporus mitochondrial tRNA(UCG) within
of Saccharomycetaceae with A. gossypii tRNAUAG (Figure 3). This analysis further confirms that tRNAUAG from yeast mitochondria was originally derived from a duplication of a histidine tRNA, as previously proposed (21). The tRNA phylogeny further reveals an unexpected clustering of N. bacillisporus mitochondrial tRNA(UCG) within 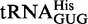 (Figure 3). A more detailed comparison with histidine tRNAs (Figure 6) reveals a shared recognition signal for histidyl-tRNA synthetase (HisRS) (17), a G residue at position −1 (the 5′ terminus of tRNA histidine) that pairs with a C at the 3′ discriminator position. This suggests that tRNA (UCG) might recognize the CGN codon family not as arginine (see Discussion).
(Figure 3). A more detailed comparison with histidine tRNAs (Figure 6) reveals a shared recognition signal for histidyl-tRNA synthetase (HisRS) (17), a G residue at position −1 (the 5′ terminus of tRNA histidine) that pairs with a C at the 3′ discriminator position. This suggests that tRNA (UCG) might recognize the CGN codon family not as arginine (see Discussion).
To demonstrate that A. gossypii tRNAUAG is expressed, properly processed and matured with a 3′-CCA terminus, RNA-Seq data from total RNA were produced and analyzed. This tRNA is expressed at about the same level as other mitochondrial tRNA species. The 5′- and 3′-processing intermediates and mature tRNA (with CCA addition at the 3′) were confirmed. The sequence of tRNAUAG is identical to that of the genomic DNA (including the anticodon), suggesting that the tRNA is not edited post-transcriptionally (36).
CUU/CUA-decoding tRNA in A. gossypii mitochondria is recognized by the mitochondrial alanyl-tRNA synthetase
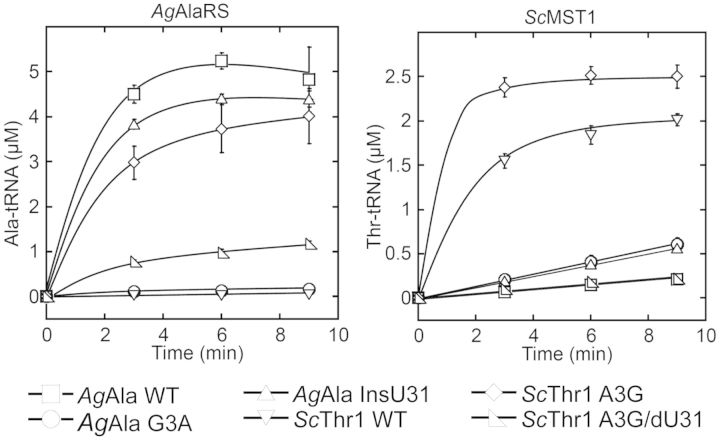
The tRNA with a predicted UAG anticodon in A. gossypii mitochondria is closely related to S. cerevisiae mitochondrial 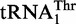 (Figures 2B and 3). Therefore, we tested whether the A. gossypii mitochondrial tRNAUAG is a substrate for ScMST1, which is the enzyme responsible for attaching Thr onto S. cerevisiae mitochondrial
(Figures 2B and 3). Therefore, we tested whether the A. gossypii mitochondrial tRNAUAG is a substrate for ScMST1, which is the enzyme responsible for attaching Thr onto S. cerevisiae mitochondrial 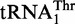 . In vitro aminoacylation experiments show that the wild-type (WT) A. gossypii mitochondrial tRNAUAG is not recognized by ScMST1 (Figure 4). Previously we showed that the enlarged
. In vitro aminoacylation experiments show that the wild-type (WT) A. gossypii mitochondrial tRNAUAG is not recognized by ScMST1 (Figure 4). Previously we showed that the enlarged 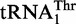 anticodon loop is an important identity element for ScMST1 (21,37). In line with that, a U insertion (InsU31) in the anticodon loop of A. gossypii mitochondrial tRNAUAG converts it to a moderate substrate for ScMST1 in an aminoacylation experiment with Thr (Figure 4). To further validate the identity of this tRNAUAG species, we purified the recombinant A. gossypii mitochondrial AlaRS (AgAlaRS) and tested Ala acylation. The WT tRNAUAG from A. gossypii mitochondria turned out to be a good substrate for AgAlaRS, and was therefore named
anticodon loop is an important identity element for ScMST1 (21,37). In line with that, a U insertion (InsU31) in the anticodon loop of A. gossypii mitochondrial tRNAUAG converts it to a moderate substrate for ScMST1 in an aminoacylation experiment with Thr (Figure 4). To further validate the identity of this tRNAUAG species, we purified the recombinant A. gossypii mitochondrial AlaRS (AgAlaRS) and tested Ala acylation. The WT tRNAUAG from A. gossypii mitochondria turned out to be a good substrate for AgAlaRS, and was therefore named 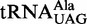 .
.
Figure 4.
Aminoacylation of mitochondrial tRNAAla and tRNAThr variants by AgAlaRS and ScMST1. The reaction was performed at 37°C in the presence of 3 µM AgAlaRS or ScMST1, 5 µM tRNA transcript and 25 µM [14C]Ala or [14C]Thr.
The G3:U70 pair of 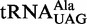 is critical for reassignment of CUU and CUA codons as Ala
is critical for reassignment of CUU and CUA codons as Ala
The major identity element for AlaRS enzymes is the G3:U70 base pair (38,39). This pair is also present in A. gossypii mitochondrial 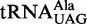 . A G3A change in
. A G3A change in 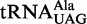 abolishes its recognition by AgAlaRS (Figure 4), suggesting that AgAlaRS recognizes its tRNA substrate in a similar manner as the characterized AlaRSs. As expected, the WT S. cerevisiae mitochondrial
abolishes its recognition by AgAlaRS (Figure 4), suggesting that AgAlaRS recognizes its tRNA substrate in a similar manner as the characterized AlaRSs. As expected, the WT S. cerevisiae mitochondrial 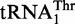 , which contains an A3:U70 pair, is not a substrate for AgAlaRS. However, an A3G mutant of S. cerevisiae mitochondrial
, which contains an A3:U70 pair, is not a substrate for AgAlaRS. However, an A3G mutant of S. cerevisiae mitochondrial 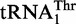 gains 12% alanylation efficiency compared with the WT A. gossypii mitochondrial
gains 12% alanylation efficiency compared with the WT A. gossypii mitochondrial 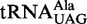 (Figure 4 and Table 1). Further, deletion of U31 in the A3G/dU31 mutant prevented the tRNA from recognition by MST1. These results suggest that A. gossypii mitochondrial
(Figure 4 and Table 1). Further, deletion of U31 in the A3G/dU31 mutant prevented the tRNA from recognition by MST1. These results suggest that A. gossypii mitochondrial 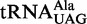 has evolved to an orthogonal Ala tRNA not recognized by ThrRS, either via duplication of
has evolved to an orthogonal Ala tRNA not recognized by ThrRS, either via duplication of 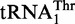 or its evolutionary precursor tRNAHis (21). The emergence of a G3:U70 pair would be the defining evolutionary step during this reassignment process.
or its evolutionary precursor tRNAHis (21). The emergence of a G3:U70 pair would be the defining evolutionary step during this reassignment process.
Table 1.
Aminoacylation efficiency of tRNA variants by A. gossypii AlaRS
| tRNA | kcat (min−1) | Km (µM) | kcat/Km (µM−1 min−1) | Relative kcat/Km |
|---|---|---|---|---|
| tRNAAla WT | 2.1 ± 0.4 | 0.53 ± 0.35 | 5.3 ± 3.0 | 100 |
| tRNAAla InsU31 | 2.1 ± 0.2 | 1.0 ± 0.4 | 2.2 ± 0.8 | 42 |
| tRNAThr A3G | 1.0 ± 0.04 | 1.6 ± 0.08 | 0.65 ± 0.03 | 12 |
DISCUSSION
Reassignment of CUN codons in yeast mitochondria
The mitochondrial genetic code has been rapidly evolving across eukaryotes, including a relatively recent CUN codon reassignment close to the divergence of the Saccharomycetaceae family (Figure 5). This group of yeast species has reduced mitochondrial gene numbers substantially (no nad genes, only 7–8 protein-coding genes are left in Saccharomycetaceae), which implies that codon families can be even more easily eliminated than in other mitochondrial systems. For example, K. lactis (a close relative of Ashbya) does not use CUN codons (Supplementary Table S1) and has no mtDNA-encoded tRNA with a UAG anticodon that would allow the recognition of this codon family. Based on the combined evidence, we favor the interpretation that CUN codon reassignment followed a codon capture mechanism, where CUN codons and the corresponding tRNA first vanished in the mitochondrial genome (completely, or more likely, reduced to a few codons that have little impact on protein structure and function), followed by the emergence of a tRNA with UAG anticodon that decodes CUN as alanine and the reappearance of more CUN codons (9,21). A short period of coding ambiguity cannot be ruled out if this new tRNA arose by duplication. However, a single mutation at the aaRS recognition site would suffice for an identity switch from threonine to alanine, without codon ambiguity. We favor this latter interpretation over other alternatives that may rather apply to more complex genetic systems [e.g. the nuclear genome of C. albicans (3)] than to yeast mitochondria with much fewer protein-coding genes.
Our previous work reveals that in Saccharomyces, 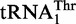 , which is responsible for CUN codon reassignment from Leu to Thr, was derived from a mitochondrial tRNAHis (21). Loss of CUN codons (accompanied by loss of nad genes) and evolution of
, which is responsible for CUN codon reassignment from Leu to Thr, was derived from a mitochondrial tRNAHis (21). Loss of CUN codons (accompanied by loss of nad genes) and evolution of 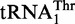 from tRNAHis
via gene duplication presumably occurred in a common ancestor of Saccharomycetaceae (Figure 5). Gene duplications are common in mtDNA. For example, among extant yeast species, the C. albicans strain SC5314 mitochondrial genome encodes two copies of tRNAHis within duplicated genome segments—the basis for neo-functionalization of one of the gene copies (40).
from tRNAHis
via gene duplication presumably occurred in a common ancestor of Saccharomycetaceae (Figure 5). Gene duplications are common in mtDNA. For example, among extant yeast species, the C. albicans strain SC5314 mitochondrial genome encodes two copies of tRNAHis within duplicated genome segments—the basis for neo-functionalization of one of the gene copies (40).
In the current work, we show that A. gossypii mitochondria use CUA and CUU to decode Ala instead of Leu or Thr. 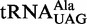 clearly clusters with yeast mitochondrial
clearly clusters with yeast mitochondrial 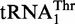 , from which it most likely also derived via gene duplication. This reassignment implies a reduction of the unusual 8-nt anticodon loop (the recognition site of threonyl–tRNA synthetase) to only seven nucleotides, and introduction of a G3:U70 base pair, which allows efficient recognition of CUN codons by A. gossypii mitochondrial alanyl-tRNA synthetase (Figure 2).
, from which it most likely also derived via gene duplication. This reassignment implies a reduction of the unusual 8-nt anticodon loop (the recognition site of threonyl–tRNA synthetase) to only seven nucleotides, and introduction of a G3:U70 base pair, which allows efficient recognition of CUN codons by A. gossypii mitochondrial alanyl-tRNA synthetase (Figure 2).
Evolution of mitochondrial 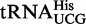 from
from 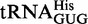 in N. bacillisporus
in N. bacillisporus
The tRNA phylogeny further reveals a divergence of a N. bacillisporus mitochondrial tRNA(UCG) from within the 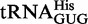 cluster (Figure 3). According to the standard translation code, a tRNA with this anticodon is expected to recognize the CGN arginine codon family. Yet, based on the following considerations, we predict instead that it most likely translates CGA as His: (i)
cluster (Figure 3). According to the standard translation code, a tRNA with this anticodon is expected to recognize the CGN arginine codon family. Yet, based on the following considerations, we predict instead that it most likely translates CGA as His: (i) 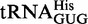 and
and 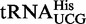 in this species are 86% identical at the primary sequence level (Figure 6), (ii) both tRNAs have a gene-encoded G at position −1, and a C in the discriminator position, forming a base pair that is the known recognition signal for HisRS (21), (iii) CGN codons do not exist in neighbor species of N. bacillisporus (Nakaseomyces delphensis, Candida glabrata and Candida castellii; Figure 5), preparing the way for CGN codon capture and (iv) N. bacillisporus has a single CGA codon in the cox1 gene, at an overall poorly conserved amino acid position. Remarkably, its closest neighbor C. castellii has a histidine in this sequence position that is overall not well conserved. An identity switch of the unique CGA codon from arginine to histidine would therefore have little, if any, functional bearing, but as N. bacillisporus HisRS has to recognize the GUG anticodon of
in this species are 86% identical at the primary sequence level (Figure 6), (ii) both tRNAs have a gene-encoded G at position −1, and a C in the discriminator position, forming a base pair that is the known recognition signal for HisRS (21), (iii) CGN codons do not exist in neighbor species of N. bacillisporus (Nakaseomyces delphensis, Candida glabrata and Candida castellii; Figure 5), preparing the way for CGN codon capture and (iv) N. bacillisporus has a single CGA codon in the cox1 gene, at an overall poorly conserved amino acid position. Remarkably, its closest neighbor C. castellii has a histidine in this sequence position that is overall not well conserved. An identity switch of the unique CGA codon from arginine to histidine would therefore have little, if any, functional bearing, but as N. bacillisporus HisRS has to recognize the GUG anticodon of 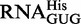 (21), a specificity change of HisRS is required to also recognize CGA (or alternatively, by a duplicated and modified HisRS). Whether this implies a transition period with ambiguous codon recognition depends on the facility with which mutations become established. Further experimental evidence is needed to validate our prediction that CGA is reassigned from Arg to His in N. bacillisporus mitochondria.
(21), a specificity change of HisRS is required to also recognize CGA (or alternatively, by a duplicated and modified HisRS). Whether this implies a transition period with ambiguous codon recognition depends on the facility with which mutations become established. Further experimental evidence is needed to validate our prediction that CGA is reassigned from Arg to His in N. bacillisporus mitochondria.
Natural evolution of orthogonal tRNAs
According to our interpretation, CUN codon identity in yeast mitochondria has evolved in the order Leu to Thr to Ala, with corresponding specific tRNAs for Thr and Ala deriving from duplicates of 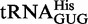 and
and 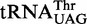 , respectively (Figure 2B). The suggested complete reassignment of these codons requires that tRNAs are orthogonal to the corresponding aaRS (recognized by a single cognate aaRS). In other words, recognition of a tRNA by more than one aaRSs would result in ambiguous decoding, as observed in the nuclear genome of several Candida species, where CUG is read by both Leu and Ser (16,41).
, respectively (Figure 2B). The suggested complete reassignment of these codons requires that tRNAs are orthogonal to the corresponding aaRS (recognized by a single cognate aaRS). In other words, recognition of a tRNA by more than one aaRSs would result in ambiguous decoding, as observed in the nuclear genome of several Candida species, where CUG is read by both Leu and Ser (16,41).
Experimental data and the presence of known tRNA identity signatures are consistent with our hypothesis. Mass spectrometry identified Ala (not Leu, His or Thr) in highly conserved positions of A. gossypii mtDNA-encoded proteins, corresponding to CUA at the codon level and suggesting that 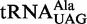 is orthogonal to AlaRS. We further show experimentally that
is orthogonal to AlaRS. We further show experimentally that 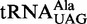 is not recognized by S. cerevisiae MST1, in line with the previous observation that this enzyme recognizes the extended 8-nt-long anticodon loop of
is not recognized by S. cerevisiae MST1, in line with the previous observation that this enzyme recognizes the extended 8-nt-long anticodon loop of 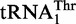 (37). Most decisively, A. gossypii
(37). Most decisively, A. gossypii
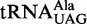 has a G3:U70 base pair, which is a known identity signature for AlaRS (38,42). Recognition of this tRNA by mitochondrial HisRS can be excluded, as it is known to recognize G−1 (a nucleotide at position −1 according to standard nomenclature that is not present in other tRNAs), the anticodon GUG and the discriminator base C73 (21). A. gossypii
has a G3:U70 base pair, which is a known identity signature for AlaRS (38,42). Recognition of this tRNA by mitochondrial HisRS can be excluded, as it is known to recognize G−1 (a nucleotide at position −1 according to standard nomenclature that is not present in other tRNAs), the anticodon GUG and the discriminator base C73 (21). A. gossypii
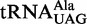 has all of these identity elements altered, and is therefore unlikely to be a substrate for HisRS. Finally, this tRNA has the same anticodon as mitochondrial tRNALeu outside Saccharomycetaceae. However, leucyl–tRNA synthetases typically recognize A73 (not present in A. gossypii
has all of these identity elements altered, and is therefore unlikely to be a substrate for HisRS. Finally, this tRNA has the same anticodon as mitochondrial tRNALeu outside Saccharomycetaceae. However, leucyl–tRNA synthetases typically recognize A73 (not present in A. gossypii
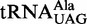 ) and not the anticodon (37). Thus, we conclude that CUA and CUU are decoded solely by Ala in A. gossypii mitochondria.
) and not the anticodon (37). Thus, we conclude that CUA and CUU are decoded solely by Ala in A. gossypii mitochondria.
Studying sense codon recoding is not only important for understanding the evolution of life, but also provides valuable insights into manipulation of the genetic code to create synthetic organisms. One roadblock of sense codon recoding is the orthogonality of the synthetic tRNA (Krishnakumar et al., submitted for publication), which requires that the synthetic tRNA is not recognized by any endogenous aaRS. Identifying naturally evolved orthogonal tRNAs would thus offer insights into the principles that have to be followed, and the translational components that need to be adapted, for recoding the translation machinery of synthetic organisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [43].
FUNDING
The National Institute of General Medical Sciences [GM022854 to D.S.], FQRNT and the National Research Council [NSERC to B.F.L.], the Canadian Research Chair Program (to B.F.L.) and the Department of Energy [DE-FG02-02ER63445 to G.C.]. Funding for open access charge: NIH [GM022854].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Chris Cho (Yale University) and Lise Forget (Université de Montréal) for technical support and Miljan Simonović (University of Illinois at Chicago) for insightful discussion and comments.
REFERENCES
- 1.Ambrogelly A, Palioura S, Söll D. Natural expansion of the genetic code. Nat. Chem. Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 2.Santos MA, Moura G, Massey SE, Tuite MF. Driving change: the evolution of alternative genetic codes. Trends Genet. 2004;20:95–102. doi: 10.1016/j.tig.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Moura GR, Paredes JA, Santos MA. Development of the genetic code: insights from a fungal codon reassignment. FEBS Lett. 2010;584:334–341. doi: 10.1016/j.febslet.2009.11.066. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe K, Yokobori S. tRNA modification and genetic code variations in animal mitochondria. J. Nucleic Acids. 2011;2011:623095. doi: 10.4061/2011/623095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell JH, O'Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T, Söll D, Podar M. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc. Natl Acad. Sci. USA. 2013;110:5540–5545. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 9.Osawa S, Jukes TH. Codon reassignment (codon capture) in evolution. J. Mol. Evol. 1989;28:271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- 10.Schultz DW, Yarus M. Transfer RNA mutation and the malleability of the genetic code. J. Mol. Biol. 1994;235:1377–1380. doi: 10.1006/jmbi.1994.1094. [DOI] [PubMed] [Google Scholar]
- 11.Sengupta S, Yang X, Higgs PG. The mechanisms of codon reassignments in mitochondrial genetic codes. J. Mol. Evol. 2007;64:662–688. doi: 10.1007/s00239-006-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang BF, Lavrov D, Beck N, Steinberg V. In: Organelle Genetics. Bullerwell CE, editor. Heidelberg: Springer, Berlin; 2012. pp. 431–474. [Google Scholar]
- 13.Kück U, Jekosch K, Holzamer P. DNA sequence analysis of the complete mitochondrial genome of the green alga Scenedesmus obliquus: evidence for UAG being a leucine and UCA being a non-sense codon. Gene. 2000;253:13–18. doi: 10.1016/s0378-1119(00)00228-6. [DOI] [PubMed] [Google Scholar]
- 14.Nedelcu AM, Lee RW, Lemieux C, Gray MW, Burger G. The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome. Genome Res. 2000;10:819–831. doi: 10.1101/gr.10.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turmel M, Otis C, Lemieux C. A deviant genetic code in the reduced mitochondrial genome of the picoplanktonic green alga Pycnococcus provasolii. J. Mol. Evol. 2010;70:203–214. doi: 10.1007/s00239-010-9322-6. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki T, Ueda T, Watanabe K. The ‘polysemous' codon–a codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 1997;16:1122–1134. doi: 10.1093/emboj/16.5.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massey SE, Moura G, Beltrao P, Almeida R, Garey JR, Tuite MF, Santos MA. Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Res. 2003;13:544–557. doi: 10.1101/gr.811003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heckman J, Sarnoff J, Alzner-DeWeerd B, Yin S, RajBhandary U. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc. Natl Acad. Sci. USA. 1980;77:3159–3163. doi: 10.1073/pnas.77.6.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogalski M, Karcher D, Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 2008;15:192–198. doi: 10.1038/nsmb.1370. [DOI] [PubMed] [Google Scholar]
- 20.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su D, Lieberman A, Lang BF, Simonovic M, Söll D, Ling J. An unusual tRNAThr derived from tRNAHis reassigns in yeast mitochondria the CUN codons to threonine. Nucleic Acids Res. 2011;39:4866–4874. doi: 10.1093/nar/gkr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Tzagoloff A. Assembly of the mitochondrial membrane system: sequences of yeast mitochondrial valine and an unusual threonine tRNA gene. Cell. 1979;18:47–53. doi: 10.1016/0092-8674(79)90352-0. [DOI] [PubMed] [Google Scholar]
- 23.Sibler AP, Dirheimer G, Martin RP. Nucleotide sequence of a yeast mitochondrial threonine-tRNA able to decode the CUN leucine codons. FEBS Lett. 1981;132:344–348. doi: 10.1016/0014-5793(81)81194-5. [DOI] [PubMed] [Google Scholar]
- 24.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang BF, Burger G. Purification of mitochondrial and plastid DNA. Nat. Protoc. 2007;2:652–660. doi: 10.1038/nprot.2007.58. [DOI] [PubMed] [Google Scholar]
- 27.Daoud R, Forget L, Lang BF. Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res. 2011;40:1728–1736. doi: 10.1093/nar/gkr941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wessels HJ, Vogel RO, van den Heuvel L, Smeitink JA, Rodenburg RJ, Nijtmans LG, Farhoud MH. LC-MS/MS as an alternative for SDS-PAGE in blue native analysis of protein complexes. Proteomics. 2009;9:4221–4228. doi: 10.1002/pmic.200900157. [DOI] [PubMed] [Google Scholar]
- 29.Fandino AS, Rais I, Vollmer M, Elgass H, Schagger H, Karas M. LC-nanospray-MS/MS analysis of hydrophobic proteins from membrane protein complexes isolated by blue-native electrophoresis. J. Mass. Spectrom. 2005;40:1223–1231. doi: 10.1002/jms.903. [DOI] [PubMed] [Google Scholar]
- 30.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eddy S. 2013. Infernal website. http://infernal.janelia.org. (26 June 2013, date last accessed)
- 34.Smith SW, Overbeek R, Woese CR, Gilbert W, Gillevet PM. The genetic data environment an expandable GUI for multiple sequence analysis. Comput. Appl. Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- 35.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 36.Alfonzo JD, Thiemann O, Simpson L. The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 1997;25:3751–3759. doi: 10.1093/nar/25.19.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling J, Peterson KM, Simonovic I, Cho C, Söll D, Simonovic M. Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment. Proc. Natl Acad. Sci. USA. 2012;109:3281–3286. doi: 10.1073/pnas.1200109109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musier-Forsyth K, Usman N, Scaringe S, Doudna J, Green R, Schimmel P. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991;253:784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- 39.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- 41.Miranda I, Silva R, Santos MA. Evolution of the genetic code in yeasts. Yeast. 2006;23:203–213. doi: 10.1002/yea.1350. [DOI] [PubMed] [Google Scholar]
- 42.Hou YM, Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 43.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.