Abstract
Cdc13, the telomere end-binding protein from Saccharomyces cerevisiae, is a multidomain protein that specifically binds telomeric single-stranded DNA (ssDNA) with exquisitely high affinity to coordinate telomere maintenance. Recent structural and genetic data have led to the proposal that Cdc13 is the paralog of RPA70 within a telomere-specific RPA complex. Our understanding of Cdc13 structure and biochemistry has been largely restricted to studies of individual domains, precluding analysis of how each domain influences the activity of the others. To better facilitate a comparison to RPA70, we evaluated the ssDNA binding of full-length S. cerevisiae Cdc13 to its minimal substrate, Tel11. We found that, unlike RPA70 and the other known telomere end-binding proteins, the core Cdc13 ssDNA-binding activity is wholly contained within a single tight-binding oligosaccharide/oligonucleotide/oligopeptide binding (OB)-fold. Because two OB-folds are implicated in dimerization, we also evaluated the relationship between dimerization and ssDNA-binding activity and found that the two activities are independent. We also find that Cdc13 binding exhibits positive cooperativity that is independent of dimerization. This study reveals that, while Cdc13 and RPA70 share similar domain topologies, the corresponding domains have evolved different and specialized functions.
INTRODUCTION
In Saccharomyces cerevisiae, the Cdc13 protein is an essential regulator of telomere maintenance, mediating both the vital end-protection activity and orchestrating the access of telomerase to telomere ends (1–7). Together with the essential proteins Stn1 and Ten1, Cdc13 protects the ends of the chromosome from catastrophic resection and activation of the DNA damage response (1–4). Cdc13 is also the key positive regulator of telomerase action, recruiting telomerase to the chromosomal end through a direct interaction with the telomerase subunit Est1 to facilitate telomerase-mediated extension of the G-strand (2,6,8). Conversely, the C-terminus of Cdc13 interacts with Stn1 to inhibit telomerase and coordinate the synthesis of the C-strand following telomerase extension (1–7). All of these activities depend on the ability of Cdc13 to bind to yeast telomeric single-stranded DNA (ssDNA) with high affinity and precise specificity (1–4,9,10).
High-resolution structural studies of several protein fragments have revealed that the multidomain 924-residue Cdc13 protein is composed of four distinct oligosaccharide/oligonucleotide/oligopeptide binding (OB)-folds (Figure 1). The first OB-fold domain to be structurally characterized, spanning residues 497–694, was found to contain robust DNA-binding activity and was therefore assigned as the ‘DNA-binding domain’ (DBD) (2,8,11,18). This OB-fold interacts with telomeric ssDNA with an apparent dissociation constant (KD,app) of 3 ± 1 pM by specifically recognizing a GxGT motif in the 5′-end of the ligand, which confers the ability to recognize the degenerate S. cerevisiae telomeric sequence (12,13). This was followed by the recent structures of the N-terminal, middle and C-terminal domains, all of which also adopt OB-folds (14–17,19) (Figure 1). The N-terminal OB-fold (NOB, residues 1–227) mediates Cdc13 homodimerization, which is disrupted by the L91R point mutation (15,19). The roles of the middle OB-fold (MOB; residues 344–497) and C-terminal OB-fold (COB; residues 694–924) domains are less well understood. MOB has been reported to form a homodimer in isolation, as well as interact with the telomere-associated protein Stn1 (17), while COB has been genetically demonstrated to interact with Stn1 and its partner Ten1 to exert end-protection function (7).
Figure 1.
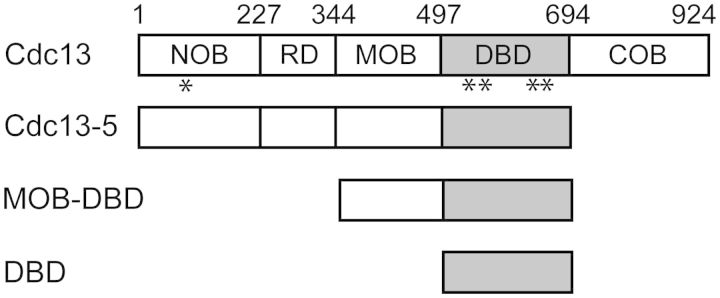
Cdc13 protein constructs. Cdc13 and derivative proteins were recombinantly expressed and purified. NOB, N-terminal OB-fold; RD, recruitment domain; MOB, middle OB-fold domain; DBD, DNA-binding domain; COB, C-terminal OB-fold. Locations of the point mutations studied are indicated by asterisks.
Together with the structures of domains of Stn1 and the Schizosaccharomyces pombe Ten1, these comprehensive domain studies revealed a structural topology that strongly suggests that Cdc13, Stn1 and Ten1 form a telomere-specific RPA complex (t-RPA) (15,21–23). However, there are significant differences between the Cdc13 and RPA70, raising the question of how far the RPA analogy extends in defining the functions of Cdc13. Specifically, in addition to a telomerase recruitment domain between NOB and MOB, the OB-fold structures of Cdc13 domains are distinctly different from the RPA70 paralogous domains (15–17,19,24–26). Also, in contrast to Cdc13, there is no evidence to date indicating that RPA70 homodimerizes (27,28). While the individual domain structures provide critical insight into biochemical functions of Cdc13, the structures do not address how these four OB-folds function within the context of the full-length protein to execute the many regulatory functions of Cdc13.
In vivo, the essential function of Cdc13 is dependent on its ssDNA-binding activity, as the disruption of ssDNA binding is lethal (2,9). Cdc13-DBD binds to Tel11 with a tight affinity (KD,app = 3 pM) (13,18), while we and others have reported that full-length Cdc13 binds with significantly reduced affinity, between 300 and 500 pM (4,9,15,18,20). This difference suggested the modulation of intrinsic ssDNA-binding activity of the DBD by other domains within the full-length protein. Additionally, other telomeric end-binding proteins, including S. pombe Pot1 (SpPot1) and Sterkiella nova TEBP, use multiple domains to bind telomeric ssDNA (29–31,35–39). Similarly, RPA uses multiple OB-fold domains to bind ssDNAs in two distinct modes. Minimally, the large subunit of RPA, RPA70, binds an 8-nt ssDNA with a KD,app of 50 nM using tandem OB-fold domains, DBD-A and DBD-B, that are centrally located within the protein (25,32). Individually, these domains each bind to ssDNA with KD,app > 100 mM, forming unstable complexes that are only detectable using cross-linking, indicating that both OB-folds are integral to proper binding activity (33).
To address the question of how well the single OB-fold domain of Cdc13 recapitulates the activity of the intact protein, we have thoroughly evaluated the ssDNA-binding activity of full-length Cdc13 from S. cerevisiae using the same strategies used on the Cdc13-DBD. First, we compared the affinity and specificity of Cdc13 to Cdc13-DBD. Second, we evaluated whether the OB-folds of Cdc13 impact its ssDNA-binding activity. Finally, we evaluated the role of dimerization in Cdc13 binding of ssDNA. We find that Cdc13 uses the single OB-fold of the DBD to bind to ssDNA and that dimerization has no regulatory impact on ssDNA-binding activity. These biochemical features distinguish Cdc13 from the other known telomere end-binding proteins as well as from RPA70, suggesting evolutionary specialization of each OB-fold within the Cdc13 protein.
MATERIALS AND METHODS
Protein expression and purification
Cdc13 protein was expressed with an N-terminal His6 tag in Sf9 cells and purified as described previously with minor modification (20,18). Following lysis by Dounce homogenization and Ni2+ affinity purification, the protein was separated on a HiLoad 16/600 Sephadex 200 (S200) gel filtration column (GE Healthcare) in 50 mM potassium phosphate, pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol (βME) and 5% glycerol. The eluted protein was then buffer-exchanged into the same buffer with 10% glycerol. In a critical departure from previous protocols, polysorbate-20 (Tween-20) was fully omitted, as its presence disrupted the solution dimer of Cdc13 as evaluated by SEC-MALS (data not shown). Point mutations were introduced into pHTB-Cdc13 via site-directed mutagenesis using QuikChange (Stratagene), and the mutant proteins expressed and purified as described above. Cdc13-DBD proteins were expressed with a C-terminal His6-tag and purified as described, including elution over a S75 gel filtration column (GE Healthcare) in storage buffer (20). Cdc13-MOB-DBD and Cdc13-MOB were cloned into the pTXB1 bacterial expression vector (New England BioLabs) and expressed as fusions to an intein-chitin–binding domain (CBD) tag in Escherichia coli BL21 (DE3) Rosetta cells. Cells were lysed by sonication in lysis buffer [20 mM Tris–HCl, pH 8.5, at 4°C, 500 mM NaCl, 1 mM ethylenediaminetetracetic acid (EDTA), 1 mM phenylmethylsulfonyl fluoride (PMSF) and a Complete EDTA-free protease inhibitor tablet (Roche)]. The proteins were affinity-purified by flowing the clarified cell extract over 10 ml bed volume of chitin beads (New England BioLabs) per 1 l cell culture at 0.5 ml/min, followed by a 20-column-volume wash with wash buffer (20 mM Tris–HCl, pH 8.5 at 4°C, 500 mM NaCl at 2 ml/min). Self-cleavage of the intein protein was induced by static overnight incubation at 4°C in one column volume of cleavage buffer (20 mM Tris–HCl, pH 8.5, 500 mM NaCl, 50 mM βME). The flowthrough was concentrated and subjected to size exclusion chromatography using appropriate columns (Cdc13-MOB-DBD, S200; Cdc13-MOB, S75) (GE Healthcare) in storage buffer (50 mM Tris, pH 7.5, 300 mM NaCl, 50 mM Na2SO4 and 5 mM βME). The purified protein was pooled and polished over a second chitin column (2–5 ml bed volume) in storage buffer to remove residual intein-CBD, and the flowthrough was concentrated and stored at 4°C. For all recombinant proteins, the concentration of active protein was determined by both electrophoretic mobility shift assay (EMSA) and double-filter binding assay apparent equilibrium binding constant using ssDNA ligand concentrations well above the KD,app. For Cdc13 proteins, activity correction values were defined as the ratio of protein:ssDNA required to fully shift 200 nM of the cognate Tel11 oligo (d(GTGTGGGTGTG)). For Cdc13-DBD and Cdc13-MOB-DBD, the activity correction values were defined as the ratio of protein:ssDNA required to fully shift 100 pM of Tel11.
Binding assays
Binding activity was evaluated by both EMSA and double-filter binding as described previously, with significant modifications to the binding buffer conditions (12,13,18,20). For EMSA, all Cdc13 proteins were assayed in 50 mM Tris–HCl, pH 7.8 at 4°C, 75 mM KCl, 75 mM NaCl, 1 mM dithiothreitol (DTT), 0.1 mM EDTA, 0.1 mg/ml bovine serum albumin and 15% glycerol. ssDNA oligonucleotides (Integrated DNA Technologies) were resuspended in nuclease-free water and 5′-end-labeled using γ-32 P-ATP with T4 polynucleotide kinase (New England Biolabs). Free ATP was removed with G25 spin columns (GE Healthcare), and labeled ssDNA ligands were diluted to 2× final concentration in binding buffer. Final ssDNA concentrations in the binding reactions were held between 0.75 and 2 pM, well below the KD,app of the interaction being assayed. The reactions were electrophoresed for 20 min (Cdc13-DBD and Cdc13-MOB-DBD) or 35 min (Cdc13) at 200 V through 6.7% acrylamide (19:1 acrylamide/bisacrylamide) Tris-borate-EDTA (TBE) gels containing 5% glycerol in a running buffer of 1× TBE + 5% glycerol. Double-filter binding assays were carried out essentially as described, with the following modifications (13). All buffers again used Tris–HCl instead of HEPES, and binding reactions were carried out in high-salt binding buffer (50 mM Tris–HCl, pH 7.8, at 4°C, 750 mM KCl, 1 mM DTT, 0.1 mM EDTA, 0.1 mg/ml bovine serum albumin and 15% glycerol). Wash buffer (50 mM Tris–HCl, pH 7.8, at 4°C, 1 mM DTT) was used to presoak all filters and to prepare and wash the wells before and after the application of the binding reactions. For both EMSA and double-filter binding, a minimum of three independent replicates was performed. Unless noted, all errors are reported as standard error of the mean.
The binding studies to determine the specificity requirements for Cdc13 binding were carried out using the same nucleotide substitution library approach previously used to determine the specificity profile for Cdc13-DBD (13). Oligonucleotides randomized individually for the three non-cognate bases at each position in Tel11 (Integrated DNA Technologies) were labeled and diluted as described for single oligonucleotides above.
Dried gels and filters were exposed to phosphor screens and imaged on a Typhoon XL phosphorimager (GE Healthcare). Data were quantitated using ImageQuantTL (GE Healthcare) and processed using Microsoft Excel. Kaleidagraph (Synergy Software) was used to plot and fit the data to a modified binding isotherm for KD,app determination as described (12). For the evaluation of cooperativity, nonlinear least-squares analysis was performed in MatLab (MathWorks). We used the equations for a three-state system as described by Senear and Brenowitz (41), with modification to include two additional parameters to accommodate incomplete saturation and background signal, similar to the previously described binding isotherm modification (12). Cooperativity was evaluated without making any assumptions about the relationship between the two binding sites (41). The macroscopic binding constants were solved using the following equations:
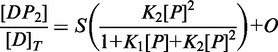 |
where [DP0]/[D]T, [DP1]/[D]T, [DP2]/[D]T are the fractions of ssDNA that are unbound, singly bound and doubly bound by protein, respectively; K1 and K2 are the macroscopic binding constants; [P] is total protein concentration; S is a saturation offset; and O is a baseline offset.
RESULTS
Optimized protocol for Cdc13 binding assays reveals a tight KD,app to Tel11
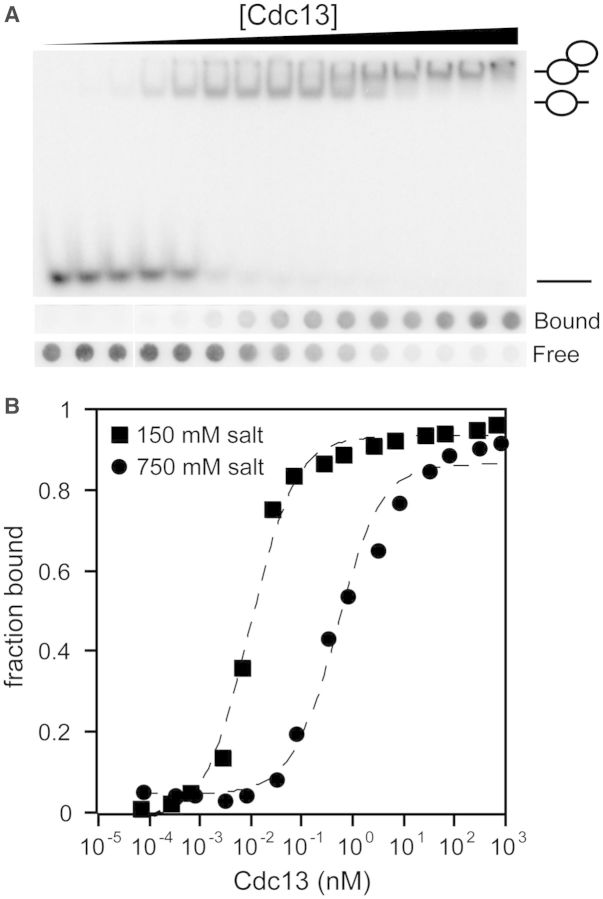
We have improved the purification and binding assay protocols for S. cerevisiae Cdc13. Critically, we omitted Tween-20 from all steps of purification. We found that while the inclusion of Tween-20 in storage buffer allows Cdc13 to be concentrated without forming aggregates, it also prevents the formation of the Cdc13 homodimer. When purified in the absence of Tween-20, Cdc13 forms a homodimer as analyzed by tandem size exclusion chromatography and multiangle light scattering (Mn = 206 kDa, 1.7% polydispersity; Supplementary Figure S1), consistent with previous reports (15,19). Further optimization of binding assay conditions allows several important advances in the analysis of Cdc13 ssDNA binding to a telomeric oligonucleotide of 11 bases [Tel11; d(GTGTGGGTGTG)] (4,9,12,18), eliminating the presence of protein retained in the loading wells of EMSA gels at intermediate protein concentrations, while also improving the band resolution of species that migrate in the gel (Figure 2 and data not shown) (9,20). These optimized conditions yielded KD,app values significantly tighter than previously reported, 13.4 ± 0.5 pM, under physiological salt conditions. We obtain a KD,app value of 3.4 ± 0.5 pM for Cdc13-DBD at the same condition, in good agreement with the previously reported value of 3 ± 1 pM (2,6,8,11,18).
Figure 2.
Cdc13 binds to Tel11 tightly at physiological salt concentrations. (A) Representative samples of EMSA (top) and double-filter binding assay (bottom) data obtained using improved experimental conditions and gel filtration-purified recombinant Cdc13 as described in the ‘Materials and Methods’ section. Schematic figures to the right of the gel indicate species composition from bottom to top as free, singly and doubly bound Tel11. Filter labels: ‘bound’, nitrocellulose filter; ‘free’, HyBond filter. EMSA experiments were conducted in binding buffer containing 150 mM salt (75 mM KCl + 75 mM NaCl). Filter binding experiments were conducted in binding buffer containing 750 mM KCl. (B) Data from (A) were plotted as a function of fraction bound versus protein concentration and fit to a two-state binding model. Square, EMSA data acquired at 150 mM salt (75 mM KCl + 75 mM NaCl); circle, double-filter binding data acquired at 750 mM KCl. Dashed lines are the least-squares fits of the binding isotherms for each set of data. Mean KD,app values from at least three independent experiments are reported in Table 1. The background was subtracted from the raw counts before calculating the fraction bound.
The affinity of Cdc13-DBD for Tel11 is dependent on ionic strength in a log-linear relationship (12,13). A buffer containing 750 mM KCl attenuated the binding affinity by ∼1000-fold to 1 nM, and thus allowed the use of a more experimentally tractable amount of radiolabeled ligand (12,13). Therefore, in addition to EMSA experiments at physiological salt concentrations, we also performed high-salt double-filter binding assays with Cdc13 (Figure 2). Cdc13 exhibits a KD,app = 410 ± 40 pM under these conditions, similar to that of 210 ± 20 pM obtained for Cdc13-DBD at identical conditions (Table 1). The KD,app values obtained at both high and low salt reflect considerably tighter binding behavior than previously reported, which we ascribe to the improvement in protein quality and binding conditions (4,12,13,18,20). Thus, Cdc13 binds to Tel11 with an affinity only ∼2- to 4-fold weaker than Cdc13-DBD (Table 1). These apparent binding affinities are in close accord and strongly suggest that the DBD comprises the entirety of the ssDNA-binding activity of the full-length protein.
Table 1.
Apparent KD values for Cdc13 proteins
| Protein | KD,app,LS (pM) | KD,rel,LS | KD,app,HS (pM) | KD,rel,HS |
|---|---|---|---|---|
| Cdc13 | 13.4 ± 0.4 | 1.0 | 410 ± 40 | 1 |
| Cdc13-DBD | 3.4 ± 0.5 | 0.25 | 210 ± 20 | 0.5 |
| Cdc13-MOB-DBD | 2.3 ± 0.2 | 0.17 | 180 ± 30 | 0.4 |
| Cdc13-F539A | 140 ± 20 | 10 | 12 000 ± 2000 | 30 |
| Cdc13-Y561A | 17 ± 1 | 1.3 | 28 000 ± 6300 | 70 |
| Cdc13-K622A | 50 ± 3 | 3.7 | 3800 ± 680 | 9 |
| Cdc13-R635A | 600 ± 80 | 45 | 140 000 ± 33 000 | 300 |
| Cdc13-L91R | 5.0 ± 0.6 | 0.37 | 220 ± 50 | 0.5 |
LS, low-salt EMSA conditions; HS, high-salt filter binding conditions.
KD,rel is relative to Cdc13 for the corresponding binding assay. Errors are standard error of the mean, n ≥ 3.
The improved band resolution of the EMSA clearly reveals the presence of two Cdc13-Tel11 bound species, which was not previously resolved on native gels (Figure 2A). The apparent constant KD of the transition between these two bands is 5.1 ± 0.8 nM. This value for Cdc13 dimerization is consistent with the 9 nM KD of dimerization independently measured for Cdc13-NOB using analytical ultracentrifugation in slightly different buffer conditions (19), suggesting that Cdc13 homodimerization is not affected by ssDNA binding.
Binding specificity and affinity features of the DBD translate to the full-length protein
To determine whether Cdc13 makes the same contacts with the ssDNA ligand as identified in Cdc13-DBD (12,13), we measured the specificity profile for the Tel11 cognate ligand using the same strategy that was used for Cdc13-DBD, using oligonucleotide pools under high-salt binding conditions (13). Each pool was composed of three oligonucleotides in which a single position was substituted with each of the noncognate bases (Supplementary Table S1). The difference between the free energy of binding to the noncognate pool and the free energy of binding to the cognate Tel11 ligand reveals the specifically recognized positions within the ligand. The overall specificity profile of Cdc13 determined using this strategy is similar to that obtained for Cdc13-DBD (Figure 3, Supplementary Figure S2). In particular, G1, G3 and T4 define the same 5′-GxGT motif observed for Cdc13-DBD, contributing the majority of the sequence specificity. Similarly, positions T2, G7 and G9 all make lesser, but significant, contributions to the free energy of binding for both Cdc13 and Cdc13-DBD, suggesting that Cdc13 recognizes the Tel11 ssDNA ligand by the same mechanism as established for Cdc13-DBD.
Figure 3.
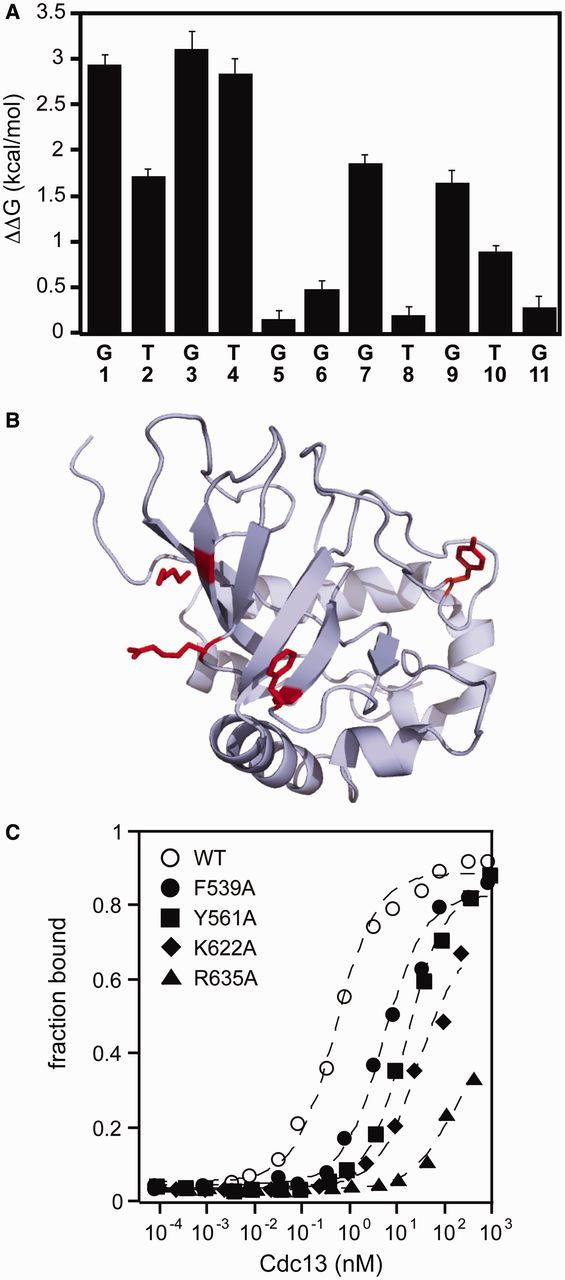
Cdc13 binding specificity recapitulates Cdc13-DBD specificity. (A) The specificity profile of Cdc13 was determined using a panel of singly substituted ssDNA oligonucleotides (Supplementary Table S2). The bar graph plots the ΔΔG°′ of binding, defined as ΔΔG = RT ln(KD1/KD2), where KD1 is the apparent binding constant for the substituted oligonucleotide and KD2 is the apparent binding constant for the cognate Tel11. Errors are reported as the standard error of the mean (n = 3 independent experiments). (B) Structure of Cdc13-DBD (PDB: 1KXL), with the four residues mutated in alanine represented as red sticks. The figure was generated using Mac PyMol (Delano Scientific) (C) Representative binding data of the DBD point mutants within Cdc13 as evaluated by high-salt filter binding. Open circle, wild-type Cdc13; closed circle, Cdc13-F539A; closed square, Cdc13-Y561A; closed diamond, Cdc13-K622A; closed triangle, Cdc13-R635A. Dashed lines are the least-squares fits of the binding isotherm for each set of data.
The details of the protein–nucleic acid interface can be probed from the protein perspective using alanine-scanning mutagenesis. Four point mutations within the DBD were introduced into Cdc13: F539A, Y561A, K622A and R635A. Three of these four mutations (F539A, Y561A and R635A) were previously assessed in Cdc13-DBD (in the sequence numbering used in Anderson et al. (12), these are F44A, Y66A and R140A, respectively). In Cdc13, these point mutations reduce binding affinity by at least an order of magnitude (Figure 3 and Table 1), closely tracking the effect observed in the isolated DBD (12) and support the notion that the amino acid contacts between Tel11 and Cdc13-DBD are conserved in the Tel11/Cdc13 interaction. Together, the differences in the free energy of Cdc13 binding we observed in both the specificity profile and the alanine mutagenesis are similar to the changes to the free energy of binding by the isolated DBD (Supplementary Figure S2).
The domains flanking the DBD do not participate in ssDNA binding
The DBD is flanked by two OB-folds, MOB and COB. To directly determine whether either or both of these domains are responsible for the modest difference in apparent ssDNA affinity between Cdc13 and Cdc13-DBD, we tested binding using two constructs: MOB-DBD and Cdc13-5, which lacks the COB (i.e., is composed of NOB-RD-MOB-DBD) (Figure 1). Independently of a recent report (17), we used multiple sequence alignment and secondary structure prediction to identify boundaries for the predicted OB-fold in the middle of Cdc13 (14,15,43) (Supplementary Figure S3). Cdc13-MOB-DBD exhibits ssDNA-binding affinity that is indistinguishable from that of Cdc13-DBD (Table 1; Supplementary Figure S3). We also purified the isolated Cdc13-MOB (residues 344–496), which eluted from a SEC-MALS column as a mixture of monomer and dimer (Supplementary Figure S4). Cdc13-MOB exhibited no intrinsic ssDNA-binding activity to Tel11, further supporting the notion that MOB does not contribute to Cdc13 ssDNA-binding activity (Supplementary Figure S4). Similarly, we found that the KD,app and specificity requirements of Cdc13-MOB-DBD were indistinguishable from that of Cdc13 (Supplementary Figure S5). Thus, the flanking OB-folds do not impact the activity of the DBD.
ssDNA-binding affinity is independent of dimerization
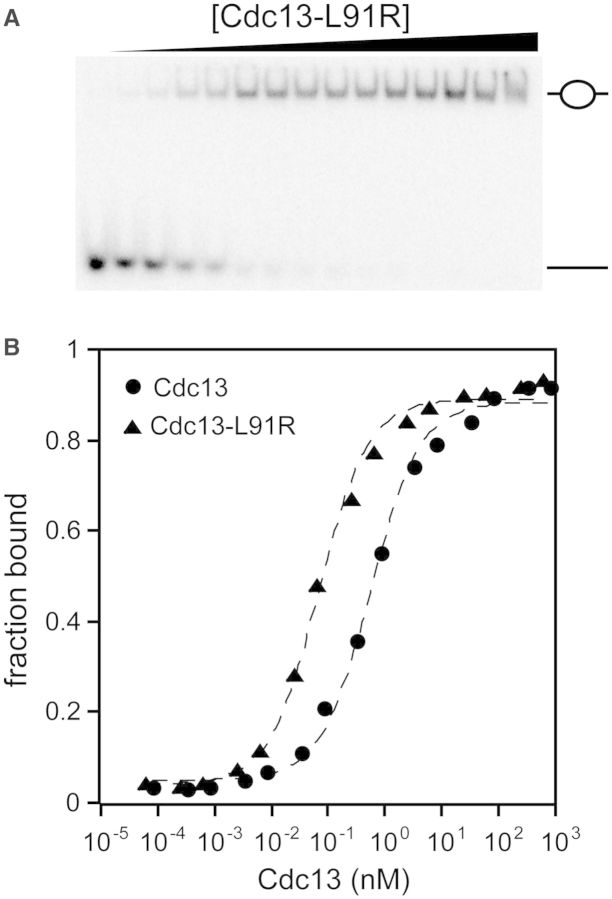
The observation that Cdc13 forms a homodimer distinguishes it from both RPA70 and other telomere end-binding proteins, and this dimerization could impact its ssDNA activity. While the EMSAs provided evidence that ssDNA binding does not disrupt dimerization, we were also interested in whether the ability to dimerize affected the ssDNA-binding activity. Dimerization of both Cdc13-NOB and Cdc13 is fully disrupted by a single point mutation, L91R (15,19). The dimerization-deficient Cdc13-L91R was confirmed to be a monomer in solution by SEC-MALS (Mn = 105 kDa, 2.9% polydispersity; Supplementary Figure S6), and demonstrated that the protein no longer dimerizes when bound to Tel11 in gel shift assays (Figure 4). This strictly monomeric form of Cdc13 bound to Tel11 similarly to wild type, with ∼2- to 3-fold tighter affinity than Cdc13, with a KD,app comparable with Cdc13-DBD (Table 1).
Figure 4.
Cdc13-L91R binds to Tel11 with higher affinity than wild-type Cdc13. (A) Representative low-salt EMSA gel showing a single bound species. As in Figure 2, the schematic figures to the right of the gel indicate whether the respective species in the gel is composed of zero or one Cdc13 molecule bound to Tel11. (B) Representative binding data of Cdc13 and Cdc13-L91R as measured by either low-salt EMSA or high-salt filter binding. Closed circle, wild-type Cdc13; closed triangle, Cdc13-L91R. Dashed lines are the least-squares fits of the binding isotherm for each set of data.
Cdc13 cooperatively binds to long ssDNA substrates
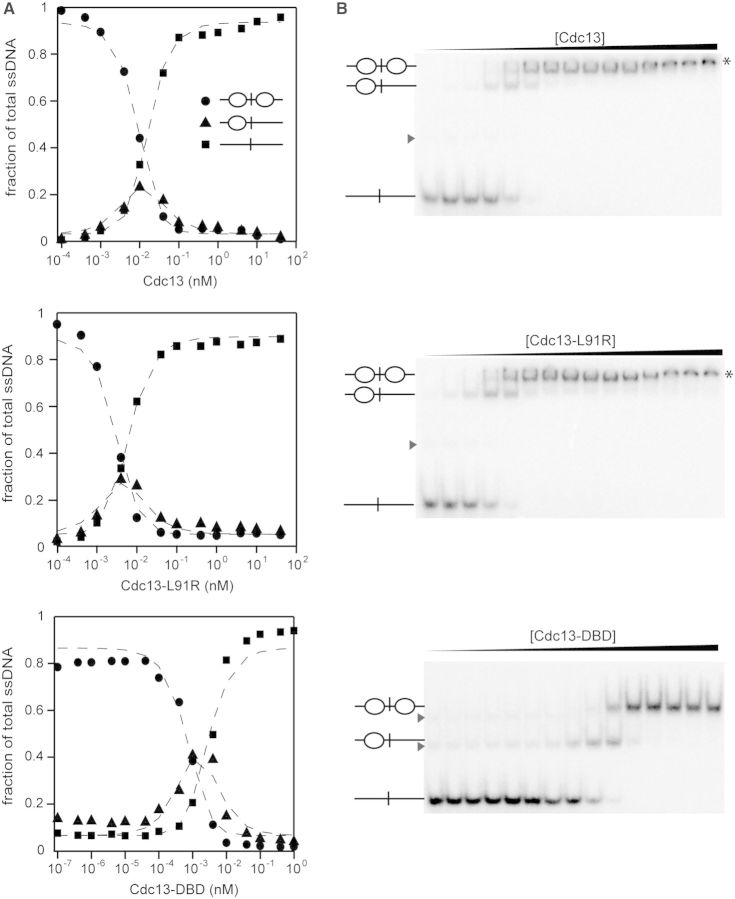
During G-phase, the single-stranded telomeric 3′ overhangs are <16 nt, but they are elongated to >30 nt during mid- to late-S phase (44,45). Because Cdc13 is a dimer in the cell, we were curious whether Cdc13 dimerization facilitated cooperativity in ssDNA binding. Therefore, we also asked whether Cdc13 exhibited any cooperative binding behavior to adjacent ssDNA sites. Positive cooperativity can be identified in a two-site system using EMSAs (41). Because we are able to resolve singly and doubly bound species by EMSA, we used this approach to evaluate the binding of Cdc13 proteins to a substrate containing two Tel11 binding sites separated by six adenosines (5′-Tel11-A6-Tel11-3′) (Figure 5). First, we assessed the apparent binding affinity by quantitating the unbound and bound species, then fitting to the binding isotherm as described above. As expected, for a ligand containing two Cdc13 binding sites, the KD,app values of all three proteins for Tel11-A6-Tel11 were half the KD,app measured for Tel11 owing to the doubling of the on-rate in this two-site system (46).
Figure 5.
Cdc13 proteins exhibit cooperativity when binding adjacent sites. (A) The two-site ligand Tel11-A6-Tel11 was incubated with titrations of Cdc13, Cdc13-L91R or Cdc13-DBD under low-salt conditions (75 mM KCl + 75 mM NaCl) and analyzed by EMSA. Three species are formed, as indicated by the inset schematics. The signal of each species is plotted as a fraction of total ssDNA signal: closed circle, free oligo; closed triangle, singly bound oligo; closed square, doubly bound oligo. Three independent data sets were collected and used to solve the cooperativity constants listed in Table 2. Dashed lines are the least-squares fits of the binding isotherm for each set of data. (B) Representative EMSA gels for the data plotted in (A). Asterisks indicate a third binding event to a low-affinity cryptic binding site that occurred at high concentrations of protein. Gray arrowheads indicate lower-mobility DNA conformations.
Next, we used the statistical mechanical approach described by Senear and Brenowitz to define the fraction of DNA bound by zero, one or two proteins in terms of two macroscopic binding constants, K1 and K2 (Equations 1, 2 and 3; Figure 5; Table 2) (41). Notably, this approach requires no assumption about the equivalency of the two binding sites in the ssDNA. The data were simultaneously fitted with the equations described in ‘Materials and Methods’ section to determine values for K1 and K2. The relationship between K2 and 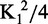 defines whether positive cooperativity is present; if K2 >
defines whether positive cooperativity is present; if K2 > 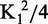 , then positive cooperativity exists between the two binding events (41). We found modest positive cooperativity in Cdc13 binding to Tel11-A6-Tel11 (Table 2). Surprisingly, this positive cooperativity was not owing to homodimerization, as similar cooperativity is observed with the dimerization-defective Cdc13-L91R protein. An alternative explanation is that Cdc13 binding remodels the G-rich structures present in the longer substrate, thus making the second binding site more accessible to the second Cdc13 molecule. To test this hypothesis, we also assessed Tel11-A6-Tel11 binding by Cdc13-DBD, which does not dimerize even at millimolar concentrations (11). The observation of similar modest positive cooperativity (Table 2) in binding by Cdc13-DBD supports the idea that the cooperativity is owing to substrate remodeling rather than protein–protein interactions.
, then positive cooperativity exists between the two binding events (41). We found modest positive cooperativity in Cdc13 binding to Tel11-A6-Tel11 (Table 2). Surprisingly, this positive cooperativity was not owing to homodimerization, as similar cooperativity is observed with the dimerization-defective Cdc13-L91R protein. An alternative explanation is that Cdc13 binding remodels the G-rich structures present in the longer substrate, thus making the second binding site more accessible to the second Cdc13 molecule. To test this hypothesis, we also assessed Tel11-A6-Tel11 binding by Cdc13-DBD, which does not dimerize even at millimolar concentrations (11). The observation of similar modest positive cooperativity (Table 2) in binding by Cdc13-DBD supports the idea that the cooperativity is owing to substrate remodeling rather than protein–protein interactions.
Table 2.
Cooperativity constants for binding Tel11-A6-Tel11
| Protein | K2 (×1018 M−2) | K1 (×109 M−1) | 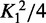 |
ΔGcoop (kcal/mol) |
|---|---|---|---|---|
| Cdc13 | 7350 | 52 | 671 | −1.3 |
| Cdc13-L91R | 48 350 | 171 | 7310 | −1.1 |
| Cdc13-DBD | 478 540 | 960 | 230 400 | −0.4 |
K1 and K2 are the macroscopic binding constants as described by Senear and Brenowitz (41).
Positive cooperativity is indicated by K2 > 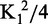 . ΔGcoop is the free energy of cooperativity, calculated with the assumption that the two binding sites are equivalent. The numbers shown are the mean of values solved for three independent data sets.
. ΔGcoop is the free energy of cooperativity, calculated with the assumption that the two binding sites are equivalent. The numbers shown are the mean of values solved for three independent data sets.
DISCUSSION
Cdc13 is a central regulator of telomere maintenance in yeast. Through a finely tuned network of interactions with other proteins, Cdc13 exerts its essential end-capping function in concert with Stn1 and Ten1, and controls telomerase-mediated extension of the telomere by directly interacting with Est1 and Stn1 (2,6,7). As is the case with many large multifunctional proteins, our understanding of Cdc13 structure and biochemistry has been mostly restricted to studies of individual domains owing to the relative ease of working with structurally defined small domains. The Cdc13-DBD exhibited remarkably tight affinity and specificity that are well-tuned to the enigmatic requirements of yeast telomere recognition (12,13). However, owing to the difficulties of obtaining well-behaved large proteins, the biochemical features of the DBD within the full-length protein are less understood, even though auxiliary domains provide an apt route for the modulation of activity.
Our comprehensive characterization of the ssDNA-binding activity of full-length S. cerevisiae Cdc13 reveals that its ssDNA-binding activity is in fact much more similar to that observed for Cdc13-DBD than previously reported (13,18,20,42). We find that affinity, specificity and impact of alanine mutants are similar between the Cdc13-DBD and full-length proteins. Within Cdc13, the restriction of ssDNA-binding activity so completely to one domain frees the others to participate in more elaborate protein–protein interactions (7,17). Indeed, both the lethal deletion of Cdc13 as well as defective telomerase recruitment have been rescued by fusing the structurally defined Cdc13-DBD to proteins that need to be localized to the telomere, including Est1, Est2, Est3 and Stn1 (2,8,47–50). These results also bring the relationship between Cdc13 and Cdc13-DBD more in line with other telomeric ssDNA-binding proteins, such as Pot1 (29), where the DBDs fully recapitulate the ssDNA activity. Furthermore, it is now known that the telomere end-binding proteins generally exhibit specificity for the 5′-end of their minimal ssDNA ligand (29,38,40). While the underlying reason for this is not readily apparent, this skew in specificity may contribute to the ability of the end-binding proteins to fully protect telomeric ssDNA overhang by accommodating degenerate telomeric sequences found in some organisms while simultaneously allowing telomerase access to the 3′-end (29).
In contrast to other members of the telomere end-binding protein family, Cdc13 forms a homodimer through its N-terminal domain. This dimer is relevant in vivo, as the calculated nuclear concentration of Cdc13 is ∼200 nM, which is well above the KD of dimerization, and disruption of dimerization leads to short telomeres (15,19). The modest impact of disruption of dimerization on ssDNA-binding affinity indicates that the cellular consequences observed with the L91R mutation can be ascribed to impacts other than a significant change in the fraction of cellular Cdc13 bound to telomeric ssDNA. This is in contrast to what was observed for Candida tropicalis Cdc13, where mutation of its putative dimerization interfaces produced significant changes in ssDNA binding, including an apparent reduction in affinity and the formation of bound species with different mobilities (16). Interestingly, the C. tropicalis Cdc13 dimerizes through its COB domain (16), and the respective phylogenetic clades of these Cdc13 proteins exhibit a wide spectrum of sequence specificity and binding affinity (34). Together, these data suggest that some mechanisms of Cdc13 function may be different between S. cerevisiae and C. tropicalis, and these differences could be manifested in the functional relationship between the dimerization domain and the DBD.
Cdc13 binds to adjacent sites on a longer ssDNA ligand (Figure 5) with modest cooperativity, which was retained by both the L91R mutant and the DBD alone, suggesting that something other than protein–protein interactions is responsible for this effect. Similar positive cooperativity at adjacent binding sites has been previously observed for the N-terminal OB-fold of S. pombe Pot1 (Pot1pN) (35) Long GT-rich ssDNA is highly susceptible to forming secondary structure conformations (51), which can be observed as lower-mobility species visible in native gels (Figure 5). We propose that the positive cooperativity in binding adjacent sites arises from the ability of one Cdc13 molecule to remodel the ssDNA substrate into a conformation more easily bound by a second Cdc13 protein. Consistent with this hypothesis, Cdc13-induced changes in ssDNA structure were recently observed using a 48-mer ssDNA oligonucleotide (52). The ability to alter ssDNA conformations may be important for telomere maintenance in vivo, as the 3′ telomeric single-stranded overhang is longer in S-phase when the telomere association of Cdc13 is highest (44,53).
The coincidence of biochemical features exhibited by Cdc13 and just one of its four OB-folds renders this protein unique within this family, as the other telomere end-binding proteins, and indeed many other proteins, all use multiple OB-folds to bind their ligand (54). While the domain topologies of Cdc13, Stn1 and Ten1 suggest that these proteins function as a t-RPA, the differences in the purposing of the OB-folds in Cdc13 suggest some divergence of roles. Such an evolution would not be unexpected for a protein that is part of the highly specialized nucleoprotein structures that comprise the telomere. A comparison of the ssDNA-binding activities of the putative Cdc13/Stn1/Ten1 heterotrimer and RPA will provide valuable insight to the critical end-protection function exerted by Cdc13 as well as its interaction with Stn1 and Ten1. Additionally, it will be important to determine how the protein–protein interactions involving the other OB-fold domains of Cdc13 cooperate to function as an integrated whole that regulates chromosome integrity and telomere maintenance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute for General Medical Studies of the National Institutes of Health [GM059414 to D.S.W., Ruth L. Kirschstein National Research Service Award GM093528 to K.A.L. and institutional training appointments GM008732 to D.A.P. and GM065103 to S.E.A. and J.N.E.]; National Science Foundation [MCB1121842 to D.S.W.]; University of Colorado Cancer Center [to D.S.W.]. Funding for open access charge: National Institute for General Medical Studies of the National Institutes of Health [GM059414].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Vicki Lundblad for helpful comments and discussion, Eric Lee and Brian Gleeson for assistance with MatLab programming and Thayne Dickey and Robert Hom for helpful comments on the manuscript. Baculovirus production and insect cell culture were carried out by the Protein Production/MoAB/Tissue Culture Core Facility at the University of Colorado Cancer Center (University of Colorado Anschutz Medical Campus, Aurora, CO).
REFERENCES
- 1.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 2.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 3.Lundblad V, Szostak J. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 4.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 5.Evans S, Lundblad V. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 2000;113(Pt 19):3357–3364. doi: 10.1242/jcs.113.19.3357. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y, Zakian VA. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc. Natl Acad. Sci. USA. 2011;108:20362–20369. doi: 10.1073/pnas.1100281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 9.Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl Acad. Sci. USA. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Durocher D. De novo telomere formation is suppressed by the Mec1-dependent inhibition of Cdc13 accumulation at DNA breaks. Genes Dev. 2010;24:502–515. doi: 10.1101/gad.1869110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke D. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 12.Anderson EM, Halsey WA, Wuttke DS. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry. 2003;42:3751–3758. doi: 10.1021/bi027047c. [DOI] [PubMed] [Google Scholar]
- 13.Eldridge A, Halsey W, Wuttke D. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry. 2006;45:871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theobald DL, Wuttke DS. Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Sun J, Yang Y, Wan K, Mao N, Yu TY, Lin YC, Dezwaan DC, Freeman BC, Lin JJ, Lue NF, et al. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase α. Cell Res. 2011;21:258–274. doi: 10.1038/cr.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu EY, Sun J, Lei M, Lue NF. Analyses of Candida Cdc13 orthologues revealed a novel OB fold dimer arrangement, dimerization-assisted DNA-binding, and substantial structural differences between Cdc13 and RPA70. Mol. Cell. Biol. 2012;32:186–198. doi: 10.1128/MCB.05875-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason M, Wanat JJ, Harper S, Schultz DC, Speicher DW, Johnson FB, Skordalakes E. Cdc13 OB2 dimerization required for productive Stn1 binding and efficient telomere maintenance. Structure. 2012;21:109–120. doi: 10.1016/j.str.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson EM, Halsey WA, Wuttke DS. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell MT, Smith JS, Mason M, Harper S, Speicher DW, Johnson FB, Skordalakes E. Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation. Mol. Cell. Biol. 2010;30:5325–5334. doi: 10.1128/MCB.00515-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zappulla DC, Roberts JN, Goodrich KJ, Cech TR, Wuttke DS. Inhibition of yeast telomerase action by the telomeric ssDNA-binding protein, Cdc13p. Nucleic Acids Res. 2009;37:354–367. doi: 10.1093/nar/gkn830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelinas AD, Paschini M, Reyes FE, Héroux A, Batey RT, Lundblad V, Wuttke DS. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl Acad. Sci. USA. 2009;106:19298–19303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs DM, Lipton AS, Isern NG, Daughdrill GW, Lowry DF, Gomes X, Wold MS. Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker. J. Biomol. NMR. 1999;14:321–331. doi: 10.1023/a:1008373009786. [DOI] [PubMed] [Google Scholar]
- 25.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 26.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–1863. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brosey CA, Chagot ME, Ehrhardt M, Pretto DI, Weiner BE, Chazin WJ. NMR analysis of the architecture and functional remodeling of a modular multidomain protein, RPA. J. Am. Chem. Soc. 2009;131:6346–6347. doi: 10.1021/ja9013634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brosey CA, Yan C, Tsutakawa SE, Heller WT, Rambo RP, Tainer JA, Ivanov I, Chazin WJ. A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res. 2013;41:2313–2327. doi: 10.1093/nar/gks1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschuler SE, Dickey TH, Wuttke DS. Schizosaccharomyces pombe protection of telomeres 1 utilizes alternate binding modes to accommodate different telomeric sequences. Biochemistry. 2011;50:7503–7513. doi: 10.1021/bi200826a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croy JE, Podell ER, Wuttke DS. A new model for Schizosaccharomyces pombe telomere recognition: the telomeric single-stranded DNA-binding activity of Pot11-389. J. Mol. Biol. 2006;361:80–93. doi: 10.1016/j.jmb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Croy JE, Altschuler SE, Grimm NE, Wuttke DS. Nonadditivity in the recognition of single-stranded DNA by the Schizosaccharomyces pombe protection of telomeres 1 DNA-binding domain, Pot1-DBD. Biochemistry. 2009;48:6864–6875. doi: 10.1021/bi900307x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bastin-Shanower SA, Brill SJ. Functional analysis of the four DNA binding domains of replication protein A. The role of RPA2 in ssDNA binding. J. Biol. Chem. 2001;276:36446–36453. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyka IM, Dhar K, Binz SK, Wold MS. Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry. 2003;42:12909–12918. doi: 10.1021/bi034930h. [DOI] [PubMed] [Google Scholar]
- 34.Mandell EK, Gelinas AD, Wuttke DS, Lundblad V. Sequence-specific binding to telomeric DNA is not a conserved property of the Cdc13 DNA binding domain. Biochemistry. 2011;50:6289–6291. doi: 10.1021/bi2005448. [DOI] [PubMed] [Google Scholar]
- 35.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe. Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 36.Peersen OB, Ruggles JA, Schultz SC. Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA. Nat. Struct. Biol. 2002;9:182–187. doi: 10.1038/nsb761. [DOI] [PubMed] [Google Scholar]
- 37.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 38.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 39.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 40.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 41.Senear DF, Brenowitz M. Determination of binding constants for cooperative site-specific protein-DNA interactions using the gel mobility-shift assay. J. Biol. Chem. 1991;266:13661–13671. [PubMed] [Google Scholar]
- 42.Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 43.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 45.Larrivée M, LeBel C, Wellinger RJ. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss JN. The Hill equation revisited: uses and misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- 47.Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124:1155–1168. doi: 10.1016/j.cell.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 48.Förstemann K, Zaug AJ, Cech TR, Lingner J. Yeast telomerase is specialized for C/A-rich RNA templates. Nucleic Acids Res. 2003;31:1646–1655. doi: 10.1093/nar/gkg261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes TR, Evans SK, Weilbaecher RG, Lundblad V. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- 50.Grandin N, Damon C, Charbonneau M. Cdc13 cooperates with the yeast Ku proteins and Stn1 to regulate telomerase recruitment. Mol. Cell. Biol. 2000;20:8397–8408. doi: 10.1128/mcb.20.22.8397-8408.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giraldo R, Suzuki M, Chapman L, Rhodes D. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: a circular dichroism study. Proc. Natl Acad. Sci. USA. 1994;91:7658–7662. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lue NF, Zhou R, Chico L, Mao N, Steinberg-Neifach O, Ha T. The telomere capping complex CST has an unusual stoichiometry, makes multipartite interaction with G-Tails, and unfolds higher-order G-Tail structures. PLoS Genet. 2013;9:e1003145. doi: 10.1371/journal.pgen.1003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taggart AKP, Teng SC, Zakian VA. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 54.Dickey TH, Altschuler SE, Wuttke DS. Single-stranded DNA-binding proteins: multiple domains for multiple functions. Structure. 2013;21:1074–1084. doi: 10.1016/j.str.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.