Abstract
In addition to silencing specific genes, small interfering RNA (siRNA) transfection is also associated with the non-specific induction of inflammatory cytokines and type I interferon. Those so-called “off-target” effects have considerable implications for the interpretation of in vitro studies and clinical application of siRNA. The present study attempted to develop a better understanding of the mechanism involved in these off target effects. Synthesized siRNA significantly enhances DNA-mediated interferon lambda-1 response (IFN-λ1/IL-29), a newly characterized antiviral interferon in non-immune or primary immune cells. This enhancement was most pronounced by double-stranded siRNA with at least a 2-nucleotide overhang at one 3′ terminus in a dose-dependent manner, while the presence of DNA was indispensable. A pull-down assay using biotinylated siRNA- or DNA-conjugated beads indicated that retinoic acid-inducible gene I (RIG-I) and interferon gamma-inducible protein 16 (IFI16) were involved in the sensing of siRNA and DNA, respectively. Co-immunoprecipitation analysis further revealed that RIG-I and IFI16 formed a complex via siRNA, and the dissociation of IFI16 from this complex in the presence of DNA activated the downstream STING-TBK1-IRF3 (stimulator of interferon genes – tank-binding kinase 1 – interferon regulatory factor 3) pathway, shedding light on a new physiological signalling pathway to activate innate immunity. Collectively, these findings may provide rational information for siRNA-induced innate immunity, with important implications for developing siRNA-based reagents to control human diseases.
INTRODUCTION
RNA interference is a post-transcriptional gene-silencing process by which double-stranded RNA directly degrades sequence-specific mRNA (1–3). In mammalian cells, RNA interference can be triggered by 21–25 nucleotide (nt) lengths of synthetic RNA duplexes, referred to as small interfering RNA (siRNA) (4,5). Typically, siRNAs are chemically synthesized with a central 19-bp duplex region and symmetric 2-base 3′ overhangs on the termini, with each strand having a 5′ phosphate group and a 3′ hydroxyl group. siRNAs can be exogenously introduced into cells by various transfection methods to knockdown a specific gene of interest. The potential of 21-mer siRNAs for use as therapeutic agents to reduce the activity of specific gene products has received considerable attention, and successful knockdown of target gene expression in mice has been demonstrated by several groups (6–10).
In the innate immune system, pattern recognition receptors (PRRs) in host cells recognize conserved pathogen-associated molecular patterns expressed by microbes and then activate immune responses (11,12). Many nucleic acids, including double- and single-stranded RNA and DNA, can stimulate innate immune responses (13–15). siRNAs were originally thought incapable of inducing immune responses because they are short and designed to mimic the natural product of Dicer in vivo. However, a number of recent studies have shown that siRNA can induce strong innate immunity (16–19). Examples include the production of interferon (IFN)-α, tumour necrosis factor alpha, interleukin (IL)-6 and IL-12. Previous studies indicated that transfection of siRNA could activate double-stranded RNA-activated protein kinase R (PKR) or toll-like receptor 3 (TLR3) in a sequence-independent pattern (16,19) or sequence-dependently recognized by TLR7 in mice and TLR7/8 in humans (20). siRNA can be chemically synthesized, but it can also be delivered into cells through viral vector-transcripted short-hairpin RNA (shRNA) (21). Kenworthy et al. reported that shRNA delivered by a lentiviral vector triggers RIG-I-mediated IFN activation. This IFN activation depends on the sequence, a 5′ triphosphate and correct processing of the RNA hairpin by Dicer (22).
The mechanisms that drive the immunostimulatory properties of the siRNA as well as the recognition pathway and signalling components involved in sensing siRNA are still incompletely understood. Of all three subtypes of IFNs, type III IFNs are the most recently discovered (23,24). In humans, type III IFNs include the three members IFN-λ1, IFN-λ2 and IFN-λ3 (also known as IL-29, IL-28A and IL-28B, respectively), which can also exert broad antiviral activity, yet they use a distinct heterodimeric receptor complex (IFN–λR1/IL–10R2) compared with type I IFNs (23,24). To our knowledge, there is no report describing whether transfection of siRNA induces type III IFNs or has any effect on other reagent-mediated type III IFNs. Investigation of this question will provide a comprehensive evaluation of the immunological functions of siRNA and shed light on potential clinical application of siRNA-based reagents.
We have previously reported that transfection of non-coding DNA plasmids or infection by a DNA virus, herpes simplex virus (HSV) type-2, led to a robust and selective induction of IFN-λ1 in several cell types. Ku70, a protein involved in DNA repair, has been identified as a double-stranded DNA binding protein to initiate type III IFN using IRF1 and IRF7 (25). Based on these findings, the present study was initially sought to elucidate the mechanism of DNA-mediated type III immune activation by knocking down potential signal mediators using siRNAs. Unexpectedly, we found that siRNA had a profound enhancing effect on the DNA-mediated induction. The results of this study show for the first time that siRNA significantly enhances DNA-mediated IFN-λ1 production. Further investigation revealed that the induction of type III IFN by siRNA occurs via a crosstalk between siRNA sensor, RIG-I and DNA sensor, IFI16 signalling pathway.
MATERIALS AND METHODS
Cells, antibodies, viruses and siRNAs
Human cervical cancer (HeLa), human embryonic kidney 293 (HEK293), human rhabdomyosarcoma (RD), SV40 T-antigen transformed HEK293 (293T) and acute monocytic leukaemia (THP-1) cell lines were obtained from American Type Culture Collection (Rockville, MD, USA) and maintained following manufacturer’s instructions. To obtain monocyte-derived immature dendritic cells (iDC), CD14+ monocytes were isolated from peripheral blood mononuclear cells (PBMCs) of healthy donors as previously described (26), and monocytes were cultured at 0.5 × 106 cells/ml in RPMI1640 (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) (Hyclone, Logan, UT, USA); 50 ng/ml granulocyte macrophage colony-stimulating factor (GM-CSF) (R&D Systems, Minneapolis, MN, USA); 50 ng/ml IL-4 (R&D Systems); 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (Quality Biology, Gaithersburg, MD, USA) and 5 µg/ml Gentamicin (Invitrogen) for 7 days. Non-adherent cells were harvested as iDCs for use in the subsequent experiments. Antibodies used in this study were as follows: anti-RIG-I (Cell Signalling Technology, Danvers, MA, USA); anti-IFI16 (Santa Cruz, Santa Cruz, CA, USA); anti-STING (LifeSpan BioSciences, Seattle, WA, USA); anti-TBK1 and anti-Flag (Cell Signalling Technology) and anti-IRF3 (Santa Cruz). For virus infection, HSV-1 (MacIntyre strain; Advanced Biotechnologies, Columbia, MD, USA) was used. All double-stranded or single-stranded siRNAs were chemically synthesized by Applied Biosystems (Foster City, CA, USA) or Integrated DNA Technologies (Coralville, IA, USA). The sequences or siRNA ID# is listed in the Supplementary Table S1.
Gel electrophoresis and western blot
Cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer (Thermo scientific, Rockford, IL, USA) in the presence of protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA) and Halt phosphatase inhibitor cocktail (Thermo scientific). Samples for native gel electrophoresis were prepared in an NP40-containing lysis buffer (50 mM HEPES, pH7.4, 125 mM NaCl, 0.2% NP40) with protease and phosphatase inhibitor cocktails. Protein concentrations of the cell lysates were quantified using a BCA protein assay (Thermo Scientific) to ensure equal amounts of total protein were loaded in each well of NuPAGE 4–12% Bis-Tris Gel (Invitrogen). Samples for the IRF-3 dimerization assay were run on a polyacrylamide gel under non-denaturing conditions. Proteins were transferred onto a nitrocellulose membrane and blotted with the appropriate antibodies, followed by the horseradish peroxidase (HRP)-conjugated secondary antibodies, and the detection was performed using ECL plus western blotting detection reagents (GE Healthcare, Piscataway, NJ, USA). The intensity of the band was analysed by NIH Image J (http://rsbweb.nih.gov/ij/).
Transfections
siRNAs were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocols. DNA transfections were conducted with Lipofectamine 2000 (Invitrogen) for HeLa cells or TransIT-293 (Mirus Bio LLC, Madison, WI, USA) for 293T cells, according to the manufacturers’ protocols.
Plasmids
Plasmid DNA, used as a stimulator, was the empty pCR2.1 vector (Invitrogen). The His-tagged IFI16 expression vector was constructed as follows: IFI16 cDNA was synthesized from the total cellular RNA of IFN-α–treated HeLa cells using the Superscript First-Strand Synthesis System for reverse transcriptase-polymerase chain reaction (RT-PCR) (Invitrogen). The reverse-transcripted cDNA encoding IFI16 was cloned into pEF1/V5-His (Invitrogen). The insertion was confirmed by DNA sequencing using BigDye version 3.0 (Applied Biosystems). Other expression vectors used in this study were constructed using a similar method. The cDNA encoding RIG-I, STING and TBK1 were cloned into pEF1-Flag, pEF6/Myc-His and pEF1/V5-His (all vectors were obtained from Invitrogen), respectively. The DNA encoding the IFN-λ1 promoter region amplified from HeLa genomic DNA was cloned into the pGL4 luciferase vector (Promega, Madison, WI, USA) and named as pGL4-IFN-λ1. A plasmid pTK-RL, which expresses Renilla luciferase under the control of HSV thymidine kinase promoter was obtained from Promega and was used to normalize experimental variations such as transfection efficiency.
Luciferase assay for IFN-λ1 promoter activity
293T cells (30 × 103 cells/well) were seeded in six-well plates and co-transfected with 100 ng of pGL4-IFN-λ1, 10 ng of pRL-TK (Promega) and designated amounts of RIG-I, IFI16, STING or TBK1 expression vectors. The cells were transfected with 10 nM siRNA on day 2 and further transfected with 1 µg of pCR2.1 plasmid DNA on day 3. On day 4, the cells were collected for the luciferase assay. This assay was performed using the Dual-Glo luciferase assay system reagents (Promega) and relative luciferase activity calculated based on the ratio of the luminescence of firefly luciferase to Renilla luciferase.
RNA extraction and real-time reverse transcription PCR
Total cellular RNA was isolated from cells using the RNeasy isolation kit (Qiagen, Valencia, CA, USA). The cDNA was synthesized from total RNA using Taqman reverse transcription reagents (Applied Biosystems) with random hexamer primer, according to the manufacturer’s instructions. IFN-λ1 mRNA expression levels were measured using quantitative RT-PCR on a CFX96 real-time system (BioRad, Hercules, CA, USA); the two-temperature cycle of 95°C for 15 s and 60°C for 1 min (repeated for 40 cycles) was used. Relative quantities of IFN-λ1 transcripts were calculated using the ΔΔCt method with GAPDH as a reference. Normalized samples were expressed relative to the average ΔCt value for controls (untreated cells or cells without siRNA transfection) to obtain relative fold change in expression levels. Probes specific for IFN-λ1 (Hs01050642_gH) and GAPDH (Hs99999905_m1) were purchased from Applied Biosystems.
Oligonucleotide pull-down assay and mass spectrometry
Cytoplasmic extracts were generated by disrupting IFN-α–treated HeLa cells using hypotonic buffer (Active Motif, Carlsbad, CA, USA), and pelleting the cell debris by centrifugation at 14 000g for 10 min at 4°C. A total of 1 mg of cytoplasmic proteins were incubated with 100 µl of Dynabeads M-280 streptavidin (Invitrogen) conjugated with 2 nmol 5′ biotinylated siRNA or 100 bp linearized DNA. Precipitated oligonucletide–protein complexes were washed extensively in protein binding buffer (20 mM HEPES, 50 mM KCL, 10% glycerol, 5 mM MgCL2 and 1 mM dithiothreitol (DTT)). Proteins were eluted from the beads by boiling in NuPAGE lithium dodecyl sulfate (LDS) sample loading buffer (Invitrogen) and were separated on NuPAGE 4–12% Bis-Tris gel (Invitrogen) and stained using Silver Quest (Invitrogen). Individual bands were excised from the gel, digested using trypsin and analysed by liquid chromatography and mass spectrometry (MS; LTQ XP, ThermoElectron, San Jose, CA, USA) (27).
Co-immunoprecipitation
HeLa cells (1 × 106) were seeded on 100-mm dishes and transfected with a FLAG-tagged RIG-I expression vector using a TransIT-HeLaMONSTER Transfection Kit (Mirus Bio LLC). The cells were further transfected with 10 nM siRNA and 10 µg of plasmid DNA at day 2 and day 3, respectively. The cells were lysed in Pierce immunoprecipitation (IP) lysis buffer (Thermo Scientific) with 10% glycerol (Sigma-Aldrich). The lysates were centrifuged at 10 000g for 10 min at 4°C to remove cell debris. The supernatants were immunoprecipitated with anti-Flag agarose (Sigma-Aldrich), and analysed by immunoblotting with anti-FLAG (Cell Signalling Technology) or anti-IFI16 antibody (Santa Cruz).
For an siRNA cleavage assay, HeLa cells in a 100-mm culture dish were transfected with 10 µg of FLAG-tagged RIG-I vector (pRIG-I-FLAG) followed by 10 nM siRNA transfection as described above. The cells were then harvested 24 h after siRNA transfection and lysed using Pierce IP lysis buffer, and then treated with or without 0.01 U/ml of RNase V1 (Invitrogen) at 25°C for 10 min. The treated samples were immunoprecipitated with anti-FLAG agarose as described above.
Enzyme-linked immunosorbent assay
The level of IFN-λ1 or IFN-β protein in culture supernatants was measured using Duoset human IFN-λ1 enzyme-linked immunosorbent assay (ELISA) kit or human IFN-β ELISA kit (R&D Systems) according to the manufacturer’s protocol.
Statistical analysis
Results were representative of at least three independent experiments. The values were expressed as mean and standard deviation (SD) of individual samples. Statistical significance was determined by the Student’s t-test. A P < 0.05 was considered a significant difference between the experimental groups.
RESULTS
Chemically synthesized siRNA unexpectedly enhances DNA-mediated IFN-λ1 induction
We previously reported that DNA transfection induces IFN-λ1 (also called IL-29), a type III IFN rather than type I IFNs, in HEK293 cells. Of interest, this DNA-mediated type III IFN induction was little, if any, observed in HeLa cells (25), and a further study demonstrated that the IFN-λ1 gene could be induced when HeLa cells were treated with IFN-α (Figure 1A). These results suggest that IFN-inducible factors may be required for the induction of IFN-λ1 in HeLa cells. Therefore, we treated cells with siRNA targeting different IFN-α–inducible genes, and the cells transfected with non-targeting siRNAs as a negative control. As shown in Figure 1B, we did not identify any factor that dramatically inhibited IFN-λ1 production, although the target genes were knocked down. Conversely, and unexpectedly, many of these siRNAs, with or without knocking down targets, induced IFN-λ1 production. We also noticed that two siRNAs with the same target showed different effects on IFN-λ1 induction. For example, both siTRIM21-1 and siTRIM21-2 knocked down the TRIM21 gene; however, the cells treated with siTRIM21-1 produced higher levels of IFN-λ1 production. Of interest, Non-human Ctrl siRNA that was used as a Non-targeting siRNA induced a significantly high level of IFN-λ1 production. It was speculated that diversity in the IFN-λ1 induction among different siRNAs was owing to the difference in siRNA stability. To compare the stability among siRNAs, an siRNA stability assay was performed using Non-human Ctrl siRNA and three other siRNAs, siGFP, siDENND2D-2 and siTRIM21-1: siGFP (a non-human targeting siRNA with lower level of IFN-λ1 induction compared with Non-human Ctrl siRNA), siDENND2D-2 (a targeting siRNA with a higher IFN-λ1 gene induction than Non-human Ctrl siRNA) and siTRIM21-1 (a targeting siRNA with a similar level of the gene induction as Non-human Ctrl siRNA). The results demonstrated that all siRNAs remain the similar level within 48 h after transfection (Supplementary Figure S1), indicating that the capabilities of the siRNA to induce IFN-λ1 were not correlated with their stabilities in HeLa cells. Based on this unexpected observation, we concluded that siRNA significantly induced type III IFN in our experimental settings, and this non-specific activity was induced via a target-independent manner and with no correlation with their stability. Non-human Ctrl siRNA was used as siRNA stimulant in the subsequent experiments.
Figure 1.
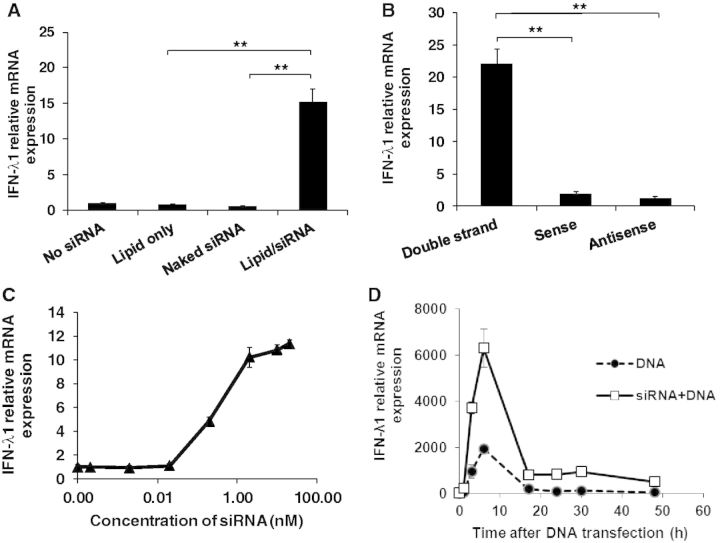
siRNA unexpectedly enhances DNA-mediated IFN-λ1 induction. (A) HeLa cells were treated with or without IFN-α (1000 U/ml) and plasmid DNA (1 µg) transfection on day 1 and day 2, respectively. Total RNA was extracted and relative IFN-λ1 expression was measured by real-time RT-PCR, and the gene expression was compared with untreated cells. (B) HeLa cells were transfected with or without 100 nM siRNA targeting different genes on day 1, and then treated with IFN-α on day 2 followed by plasmid DNA transfection on day 3. The total RNA was extracted 24 h after DNA transfection. Relative IFN-λ1 mRNA expression was measured by real-time RT-PCR. The level of gene expression in each well was compared with that of the control cells (without siRNA transfection) after normalization of the expression level of GAPDH. (C) Different cell types, RD, HEK293, THP-1 and iDC, were transfected with or without 10 nM Non-human Ctrl siRNA followed by DNA transfection. IFN-λ1 gene expression was measured by real-time RT-PCR. The expression level was compared with that of the cells without siRNA transfection. (D and E) HeLa cells were transfected with 10 nM siRNA followed by 1000 U/ml IFN-α treatment, and then transfected with or without 1 µg of DNA. (D) Relative IFN-λ1 mRNA expression levels was compared with the untreated HeLa cells, and (E) protein level of IFN-λ1 and IFN-β in the culture supernatants was measured by real-time RT-PCR and ELISA, respectively. UD represents undetectable level of IFN-λ1 or IFN-β protein, the detection limit of this assay is 62.5 pg/ml for IFN-λ1 and 25 pg/ml for IFN-β. All results represent the mean of three independent experiments. Error bars show SD. *P < 0.05; **P < 0.001 (Student’s t-test).
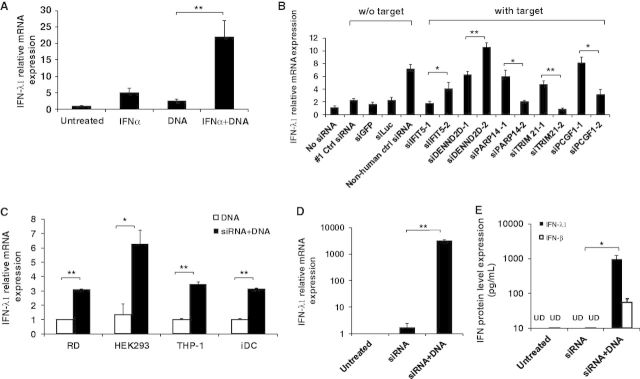
To further confirm this observation, RD, HEK293, THP-1 and iDC cells were examined using similar experimental conditions (Figure 1C). Compared with those without siRNA transfection before DNA transfection, the cells sequentially transfected with siRNA and DNA showed significantly higher levels of IFN-λ1 gene production. These results in RD, HEK293, THP-1 and iDC cells were consistent with those observed in HeLa cells. Therefore, siRNA-induced type III IFN is not unique to HeLa cells; siRNA can also induce IFN-λ1 production in other cell lines and even in primary cells.
Because siRNA-induced IFN-λ1 production was based on the sequential transfection of siRNA and DNA, we further tested whether siRNA transfection alone can induce similar levels of IFN-λ1. We treated HeLa cells with Non-human Ctrl siRNA and further transfected them with or without DNA. The result showed that siRNA transfection alone induced low levels of IFN-λ1; however, the cells that were transfected with siRNA followed by DNA transfection induced IFN-λ1 at a level 1000-fold greater than untreated cells, indicating that DNA transfection is required for siRNA to induce a high level of type III IFN response (Figure 1D). Quantification of IFN-λ1 in culture supernatants using ELISA showed that only cells transfected with a combination of siRNA and DNA induced IFN-λ1 up to a concentration of 1000 pg/ml. The protein levels in untreated cells and cells transfected with siRNA alone were below the level of detection (62.5 pg/ml) (Figure 1E). We also measured the protein amounts of IFN-β in the transfection supernatants. IFN-β was also induced by the transfection, but the induced amounts are much lower than that of IFN-λ1 (Figure 1E). To evaluate biological relevance of IFN-λ1 induction, we tested whether the amounts of the induced IFN-λ1 is high enough to inhibit human immunodeficiency virus (HIV)-1 replication in primary macrophages and dendritic cells. In consistence with another study (28), >1000 pg/ml of IFN-λ1 was sufficient to inhibit HIV-1 replication in macrophages and also in iDCs (Supplementary Figure S2), suggesting that the off-target effect by siRNAs may have an impact on antiviral effect.
Characterization of non-specific activities of siRNA
To determine whether excluding the transfection lipid has an impact on IFN-λ1 activity, HeLa cells were transfected with lipid, naked siRNA or the siRNA–lipid complex. Only siRNA delivered by lipid, rather than lipid itself or naked siRNA, induced high levels of IFN-λ1 production (Figure 2A). Because chemically synthesized siRNA is a double-stranded RNA, we tested whether sense or antisense single-stranded siRNA was able to induce IFN-λ1. The results indicated that only double-stranded siRNA induced type III IFN response (Figure 2B). To further characterize the immunostimulatory capacity of siRNAs, HeLa cells were transfected with Non-human Ctrl siRNA at a concentration ranging from 0.001 to 100 nM, followed by DNA transfection. As shown in Figure 2C, the Non-human Ctrl siRNA induced IFN-λ1 at concentrations as low as 1 nM and plateaued at 10 nM. Taken together, siRNA-induced type III IFN appeared to respond in a dose-dependent manner. Next, we performed a kinetic analysis of siRNA-induced type III IFN response. The results shown in Figure 2D clearly indicated that IFN-λ1 production reached its peak at 6 h after DNA transfection regardless of whether siRNA was transfected. These results consistently indicated that siRNA highly enhanced DNA-mediated IFN-λ1 production. Our results demonstrated that IFN-α treatment was required for HeLa cells but not for other cell types to induce siRNA effect on the DNA-mediated type III IFN response (Figure 1C). To further define the role of IFN-α treatment in HeLa cells, the cells were treated with IFN-α at different concentrations and then the relative IFN-λ1 mRNA expression was compared (Supplementary Figure S3). The data indicated that IFN-α treatment dose dependently enhances siRNA and DNA-mediated IFN-λ1 production, and in the absence of IFN-α, siRNA had no significant impact on the DNA-mediated gene activation in HeLa cells.
Figure 2.
Characterization of siRNA-induced IFN-λ1 production. (A) HeLa cells were transfected with a transfection lipid reagent Lipofectamine RNAiMAX, naked siRNA or siRNA with the lipid, followed by IFN-α treatment and DNA transfection on day 2 and day 3, respectively. (B) HeLa cells were transfected with double-stranded siRNA, sense or antisense single-stranded siRNA at 10 nM, followed by IFN-α treatment and DNA transfection. (C) HeLa cells were transfected with various concentrations of Non-human Ctrl siRNA (0.0001–100 nM), followed by 1000 U/ml IFN-α treatment and 1 µg of DNA transfection as described above. For experiments (A), (B) and (C), total RNA was extracted for relative mRNA expression measurements using real-time RT-PCR. The gene expression level was compared with the cells treated with IFN-α and DNA but not siRNA. (D) HeLa cells were transfected with or without Non-human Ctrl siRNA, followed by IFN-α treatment and DNA transfection. Total RNA was extracted at different time points after plasmid DNA transfection. Gene expression was compared with untreated cells at time zero. **P < 0.001 (Student’s t-test).
At least one TT overhang at the 3′ terminus is required for siRNA to highly enhance DNA-mediated IFN-λ1 induction
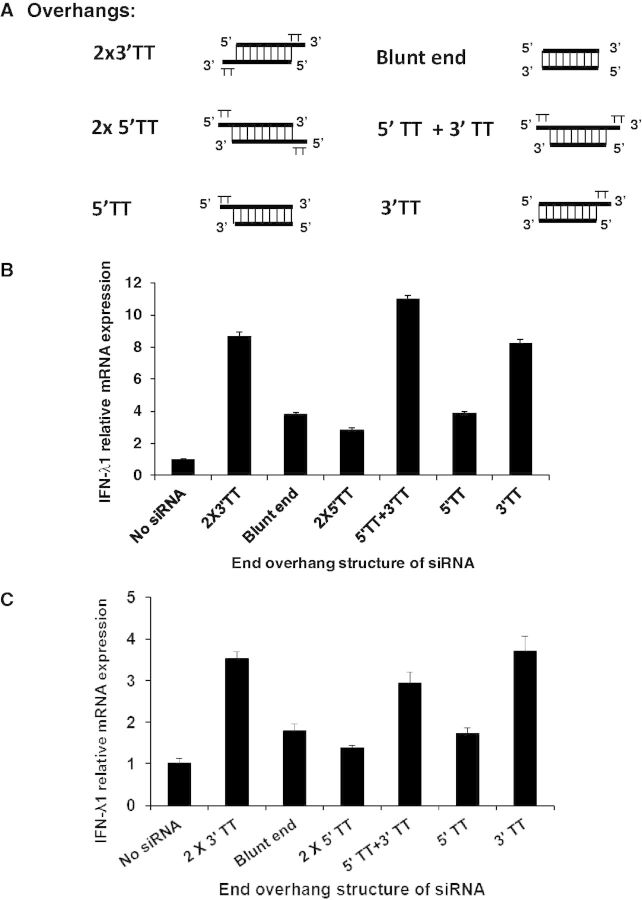
It has been reported that blunt ends at the termini are an essential structure for siRNA to induce type I IFN (29). In this study, all chemically synthesized siRNAs are composed of 19 nt double-stranded RNA with a two thymidine base (TT) overhang at the 3′ terminus of each strand (Figure 3A). We examined whether this structure was essential for type III IFN induction. The siRNA with two 3′-end TT overhangs (2 × 3′ TT) represents general siRNA used for RNA interference. Other overhang structures, blunt end, two 5′-end overhang (2 × 5′ TT), a combination of 5′-end and 3′-end overhangs (5′ TT + 3′ TT), one 5′-end overhang (5′ TT) and one 3′-end overhang (3′ TT), were tested in this study (Figure 3A). The result showed that the siRNA with 2 × 3′ TT, 5′ TT + 3′ TT or 3′ TT induced higher levels of IFN-λ1 production by >8-fold. In contrast, siRNA with a blunt end, 2 × 5′ TT or 5′ TT overhangs had, on average, only a 4-fold IFN-λ1 production compared with the cells without siRNA transfection (Figure 3B). To define cell specificity, we performed the same experiment using THP-1 cells. A similar trend in the IFN-λ1–inducing activity was observed in THP-1 cells (Figure 3C). These results suggested that, unlike the blunt end siRNA-mediated type I IFN response (30), at least one TT overhang at the 3′ terminus is required for chemically synthesized siRNA to significantly enhance DNA-mediated type III IFN activity. It was speculated that the overhang structure may change the susceptibility of the siRNA to nuclease digestion, therefore led to the differences in the IFN-λ1 induction. To compare the stability of the siRNAs, real-time RT-PCR was performed to quantitate the remaining intact siRNA in the cells. The result indicated that siRNA with 2 × 3′ TT overhang is less stable than siRNA with other type of overhang. However, the variation in stability of siRNA does not correlate with their IFN-λ1 induction (Supplementary Figure S4).
Figure 3.
A minimum TT overhang at one 3′ terminus of siRNA is required to highly induce IFN-λ1 production. (A) Schematic of chemically synthesized double-stranded Non-human Ctrl siRNA (19 nt) with different TT overhangs. (B) HeLa cells were transfected with 10 nM siRNA containing different TT overhangs, followed by IFN-α treatment (1000 U/ml) and plasmid DNA (1 µg) transfection on day 2 and day 3, respectively. The gene expression levels were measured using real-time RT-PCR. Cells without siRNA transfection were used as a control. (C) THP-1 cells were transfected with 10 nM siRNA containing different TT overhangs, followed by plasmid DNA (1 µg) transfection on day 2. The gene expression levels were measured using real-time RT-PCR and compared with cells without siRNA transfection.
Type III IFN is stimulated through crosstalk between RIG-I and IFI16 signalling pathway
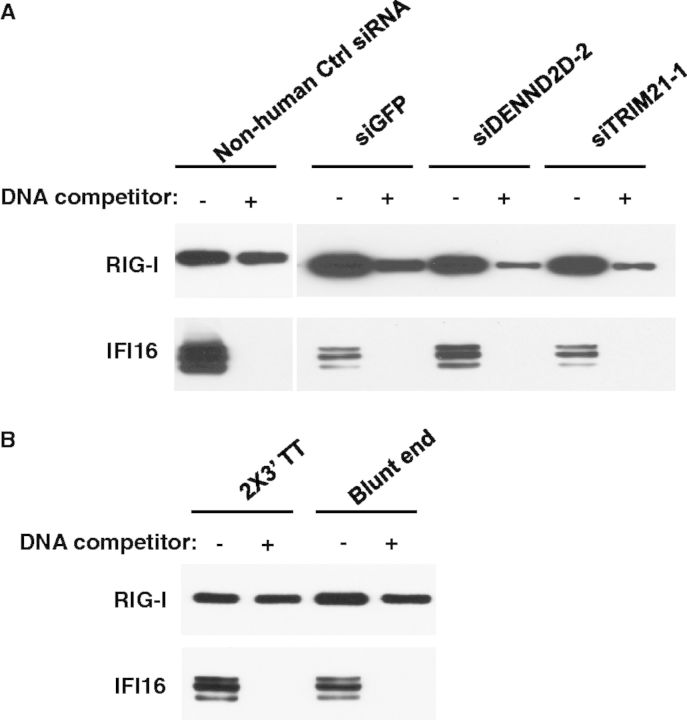
To investigate the molecular mechanism by which siRNA induces type III IFN, a series of pull-down assays using siRNA- or DNA-conjugated magnetic beads were conducted to identify siRNA and DNA sensor proteins in IFN-α–treated HeLa cells. Cytosolic fractions from IFN-α–treated HeLa cells were incubated with siRNA-conjugated magnetic beads with or without 10-fold excess amounts of Non-human Ctrl siRNA or plasmid DNA as competitors. The proteins bound on the beads with or without competitors were separated using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and identified by MS analysis. The addition of the siRNA competitor led to the disappearance of the specific siRNA binding protein. The MS analysis identified RIG-I as the siRNA binding protein, which was further confirmed using western blot (Figure 4A). When a 10-fold amount of siRNA was added as a competitor, the band corresponding to RIG-I completely disappeared. In contrast, when plasmid DNA was added as a competitor, the RIG-I protein band persisted, suggesting RIG-I specifically bound to siRNA. To elucidate the DNA-binding protein in the cytosol, a similar set of studies were performed using DNA-conjugated beads. The result indicated that IFI16 was a DNA binding protein (Figure 4B). RIG-I activation after ligand binding is thought to involve multiple steps that include the activation of its ATPase activity, conformational changes and oligomerization (31,32). To determine whether siRNA induces RIG-I oligomerization, we performed a size-exclusion chromatography using cytosolic fraction from 293T transfected by RIG-I expression vector with or without siRNA. The result demonstrated that, in the presence of siRNA, RIG-I was eluted in higher molecular mass fractions (Supplementary Figure S5), indicating that chemically synthesized 19-nt siRNA induces oligomerization of RIG-I. To precisely demonstrate the interaction of siRNA and RIG-I, the AlphaScreen assay was performed using purified Flag-tagged RIG-I protein and siRNA in a cell-free assay system. As shown in the Supplementary Figure S6, a short 19-nt RNA is able to bind with RIG-I, which is previously reported usually be activated by triphosphate RNA (5′ PPP-RNA). Therefore furthermore, we compared IFN-λ1 and IFN-β inducing activity between 5′ PPP-RNA and siRNA (non-5′ PPP) in this DNA-mediated IFN induction. The data indicated that 5′ PPP-RNA also induced IFN-λ1 or IFN-β; however, the induced level by 5′ PPP-RNA was significantly lower than that by non-5′ PPP siRNA (Supplementary Figure S7A). To verify the IFN-β induction capability of the 5′ PPP-RNA as a valid control, another control experiment was performed. In the absence of DNA transfection, 5′ PPP-RNA induced a higher level of IFN-β than IFN-λ1 in IFN-α–treated HeLa cells. While non-5′ PPP siRNA had no significant impact on the induction of both IFNs (Supplementary Figure S7B), taken together, the results supported that 5′ PPP-RNA is a valid control in this study and 5′ PPP structure may not play a key role in the DNA-mediated IFN induction.
Figure 4.
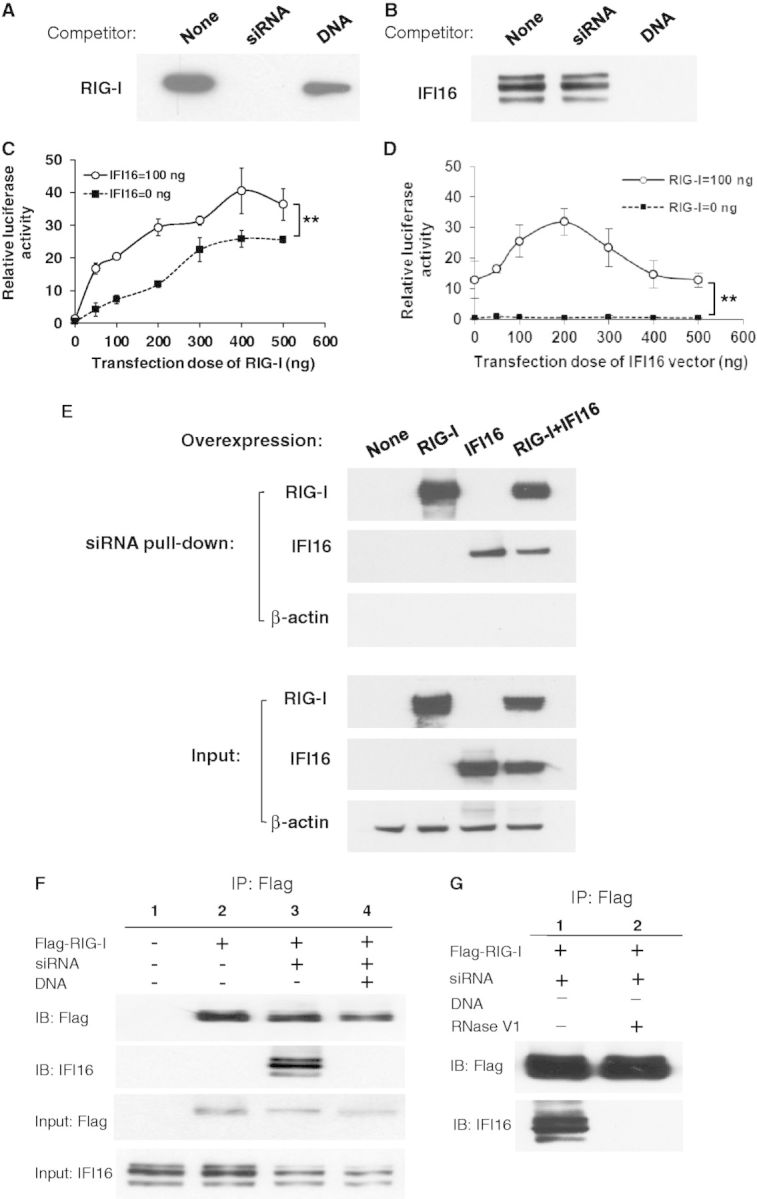
siRNA enhances DNA-mediated IFN-λ1 production through a RIG-I and IFI16 crosstalk signalling pathway. Cytoplasmic extracts of IFN-α–treated HeLa cells were incubated with (A) siRNA-conjugated or (B) DNA-conjugated magnetic beads in the absence or presence of siRNA or DNA as competitors. Bound proteins on the beads were separated by SDS-PAGE gel, and western blot was performed using (A) anti-RIG-I antibody or (B) anti-IFI16 antibody. (C and D) 293T cells were co-transfected with IFN-λ1 reporter plasmid (pGL4-IFN-λ1) and Renilla luciferase plasmid (pRL-TK) with different amounts of IFI16 or RIG-I expression vectors. Cells were then stimulated with 10 nM siRNA and 1 µg of plasmid DNA on day 2 and 3, respectively. On day 4, cells were collected for luciferase assay. (E) 293T cells were transfected with the RIG-I or IFI16 expression vector, and then the cell lysate was subjected for siRNA pull-down assay using siRNA-conjugated beads. The siRNA bound proteins were separated on SDS-PAGE gels, western blot was performed to detect RIG-I and IFI16. (F) HeLa cells were transfected with flag-tagged RIG-I vector and further stimulated with siRNA and DNA transfection on day 2 or day 3, respectively. Cytosolic lysates were immunoprecipitated using anti-flag antibody. Precipitated proteins were analysed by western blot using anti-flag and anti-IFI16 antibodies. Input controls were also included. (G) HeLa cells were transfected with FLAG-RIG-I vector followed by siRNA transfection. The cells were lysed by Pierce IP lysis buffer and treated with or without RNase V1. The treated samples were then immunoprecipitated by anti-FLAG conjugated agarose and analysed by western blot using anti-FLAG and anti-IFI16 antibodies.
To determine whether RIG-I and IFI16 are involved in the siRNA-enhanced DNA-mediated IFN-λ1 production, we performed an IFN-λ1 reporter assay. In our previous work, we demonstrated that transfection with any DNA-induced IFN-λ1 in HEK293 cells; however, transfection of DNA into 293T did not induce the gene activation (25). To compare the difference in DNA-mediated IFN-λ1 activation between HEK293 and 293T cells, a GFP expression vector, pCMV-GFP, was transfected into both cells and gene expression was compared. Both cells expressed a high level of GFP on transfection, suggesting that both cells were sufficiently transfected by plasmid DNA (Supplementary Figure S8A). The transfection, however, did not induce IFN-λ1 gene activation in 293T cells (Supplementary Figure S8B). To compare the endogenous level of RIG-I and IFI16 expression in HEK293 and 293T cells, western blot analysis was performed. The result indicated that the level of RIG-I expression was low or below the level of detection in both cell lines. However, HEK293 expresses a higher basal level of IFI16 compared with that of 293T cells (Supplementary Figure S8C). Meanwhile, untreated HeLa cells express endogenous IFI16 but not RIG-I (Supplementary Figure S8D). These results indicated that 293T cells are appropriate for studying the IFN-λ1 promoter function using a reporter plasmid with a series of expression plasmids. 293T cells were co-transfected with pGL4-IFN-λ1 and different amounts of RIG-I plasmid with 0 or 100 ng of IFI16 expression plasmid and then stimulated with siRNA and DNA (Figure 4C). The results indicated that overexpression of RIG-I dose-dependently activated IFN-λ1 promoter activity. In the presence of the IFI16 expression plasmid, RIG-I significantly enhanced the promoter activity. We next studied the impact of IFI16 expression plasmid on IFN-λ1 promoter activity. 293T cells were co-transfected with pGL4-IFN-λ1 and various amounts of the IFI16 expression vector with 0 or 100 ng of RIG-I plasmid followed by stimulation of siRNA and DNA (Figure 4D). However, IFI16 alone was not sufficient to activate the IFN-λ1 promoter. Only in the presence of RIG-I, IFI16 was able to activate the IFN-λ1 promoter in a bell-shaped dose-dependent manner. A combination of co-transfection of 100 ng of RIG-I expression vector and 200 ng of IFI16 expression plasmid induced the maximum level of luciferase activity. To confirm whether the IFN-λ1 promoter activity is induced by siRNA or DNA transfection alone, a control experiment was performed using 293T cells co-transfected with RIG-I and IFI16 expression vector. The results illustrated that with the co-transfection of RIG-I and IFI16 vector, siRNA alone or DNA alone is not able to induce a high level of the IFN-λ1 promoter activation (Supplementary Figure S9). Collectively, these data indicated that activation of the IFN-λ1 promoter was enhanced by siRNA and DNA co-stimulation via a RIG-I and IFI16 crosstalk pathway.
To define the mechanism of the crosstalk between RIG-I and IFI16, we performed a set of siRNA pull-down assay using RIG-I or/and IFI16-overexpressed 293T cell lysate. The result shown in the Figure 4E indicated that both RIG-I and IFI16 can independently bind siRNA, and the binding was persistent even in the presence of both RIG-I and IFI16, suggesting that RIG-I and IFI16 bind with siRNA at different binding sites. As shown in Fig 4D, overexpression of IFI16 alone had no impact on IFN-λ1 promoter activation. Therefore, the physical binding between siRNA and IFI16 is not sufficient to trigger IFN-λ1 transduction.
To further define how RIG-I and IFI16 crosstalk and initiate downstream signalling, we designed a co-immunoprecipitation assay to determine whether IFI16 interacts with RIG-I in the context of siRNA and DNA stimulation (Figure 4F). The results showed that IFI16 was co-immunoprecipitated with RIG-I only on siRNA transfection (Figure 4F, Lane 3). Interestingly, IFI16 dissociated from the RIG-I–siRNA–IFI16 complex when cells were further transfected with DNA (Figure 4F, Lane 4). It appears that siRNA stimulation leads to the formation of a complex between RIG-I and IFI16. On DNA transfection, this complex dissociates owing to a possible conformational change. Because a sequential transfection of siRNA followed by DNA is the experimental condition for the cells to induce high levels of IFN-λ1, and our pull-down assay suggested that RIG-I and IFI16 are the sensors to bind with transfected siRNA and DNA, respectively. Therefore, the co-immunoprecipitation assay results indicated that IFI16 first formed a complex with RIG-I and siRNA, and then released from the complex on the subsequent transfection with DNA and formed a new complex with the DNA. These results suggested that the dissociation of IFI16 switches on the downstream signalling pathway.
To precisely demonstrate whether the association of RIG-I and IFI16 is mediated through siRNA, we collected the cell lysate from FLAG-RIG-I overexpressed and siRNA transfected cells and the cell lysate was treated with RNase V1, which cleaves the base-paired nt residues, and then a co-immunoprecipitation assay was performed (Figure 4G). The results illustrated that without RNase V1 treatment, RIG-I formed the complex with the IFI16 (Lane 1). However, after the cell lysate was treated with RNase V1, the band of IFI16 was diminished (Lane 2). Those results further supported that IFI16 forms a complex with RIG-I in the presence of siRNA, thus siRNA is an essential factor for making the complex.
The association of RIG-I and IFI16 through siRNA is not related with the sequence or DNA overhang of the siRNA
We have demonstrated that non-human Ctrl siRNA enhanced DNA-mediated IFN-λ1 induction through a crosstalk signalling pathway between RIG-I and IFI16. To define whether the association of RIG-I and IFI16 via siRNA is specific to Non-human Ctrl siRNA, a series of siRNA pull-down assays were performed using siGFP, siDENND2D-2 and siTRIM21-1 (Figure 5A). Those siRNAs possess the similar stability but with different levels of IFN-λ1 induction (Supplementary Figure S1). In consistence with the results showed in the Figure 4E, the data from this assay demonstrated that all siRNAs did pull down RIG-I along with IFI16, and the addition of the DNA competitor induced a dissociation of IFI16, therefore the interaction of RIG-I and IFI16 via siRNA is caused in a siRNA sequence independent manner.
Figure 5.
The association of RIG-I and IFI16 through siRNA are not related with the sequence or DNA overhang of the siRNA. (A) A set of siRNA pull-down assay was performed in the absence or presence of DNA as competitor. Bound proteins on the beads were separated by SDS-PAGE, and western blot was performed using anti-RIG-I and anti-IFI16 antibodies. The results using other siRNAs, siGFP, siDENND2D-2, siTRIM21-1 were compared with the results from Non-human Ctrl siRNA. (B) The results of siRNA pull-down assay with or without DNA as competitor were compared between siRNA with a TT overhang and a blunt end siRNA.
The siRNA we used in this study possesses 2-nt DNA overhang at their 3′ terminus. IFI16 is previously known as a DNA binding protein (33), and so it is possible that the binding of IFI16 with siRNA was through the DNA overhang of the siRNA. To determine whether the overhanged DNA played a role in the association, we performed a pull-down assay using a blunt end siRNA. The result demonstrated that RIG-I is still associated with IFI16 through the blunt end siRNA, and the DNA competitor induced a dissociation of IFI16 from the complex (Figure 5B).
IFN-λ1 is induced through the STING-TBK1-IRF3 pathway
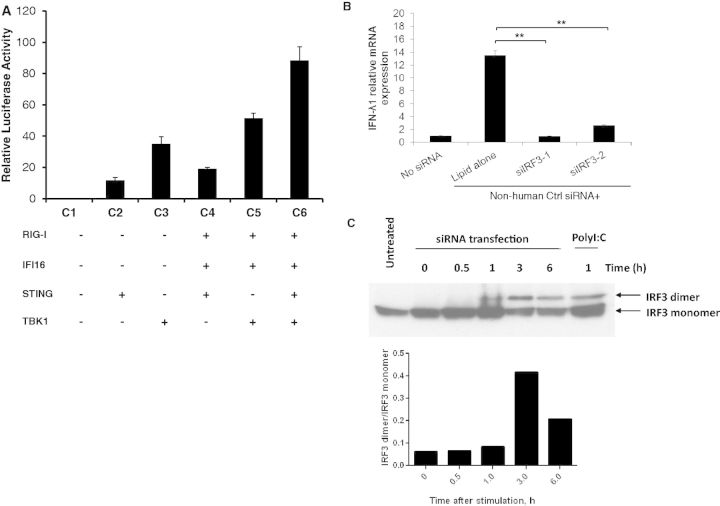
We next assessed what downstream signal mediators were involved in the induction of IFN-λ1. STING (TMEM173) is a signalling protein required for TBK1-dependent IFN-β responses to virus infection and DNA transfection (34,35). IFI16 senses intracellular DNA, which induces IFN-β induction via the STING, TBK1, IRF3 pathway (33). It is possible that the induction of IFN-λ1 uses a similar pathway. Therefore, an IFN-λ1 reporter assay was conducted by overexpression of the downstream components STING and TBK1 in 293T cells (Figure 6A). Indeed, the overexpression of STING or TBK1 was sufficient to enhance the activity of IFN-λ1 promoter (Figure 6A: lanes C2 and C3). The highest level of IFN-λ1 induction was obtained when TBK1 and STING were co-transfected with RIG-I and IFI16 (Figure 6A: lanes C4, C5 and C6). These results suggested that both STING and TBK1 were involved in the siRNA-induced type III IFN pathway, and that the activation of downstream STING and TBK1 was required for induction of the type III IFN response.
Figure 6.
STING, TBK1 and IRF3 activation are involved in siRNA-induced IFN-λ1 production. (A) 293T cells were co-transfected with pGL4-IFN-λ1 and pRL-TK, together with expression vectors of RIG-I, IFI16, STING and TBK1 as indicated combinations. The cells in the condition of C1–C5 were transfected with an empty vector to ensure each well has the same amounts of DNA transfection as the condition C6. Cells were then stimulated with siRNA and plasmid DNA transfection on day 2 and day 3, respectively. On day 4, cells were collected for luciferase assay. Relative luciferase activity was determined by the ratio of firefly to Renilla readings. (B) HeLa cells were transfected with or without 10 nM Non-human Ctrl siRNA along with two different siRNA targeting IRF3 (siIRF3-1 and siIRF3-2). Cells were then treated with IFN-α (1000 U/ml) followed by DNA transfection on day 2 and day 3, respectively. RNA was extracted 6 h after DNA transfection. Relative IFN-λ1 mRNA expression level was measured by real-time RT-PCR and compared with the cells treated with IFN-α and DNA transfection but without siRNA transfection. **P < 0.001 (Student’s t-test). (C) Top: HeLa cells were stimulated with 10 nM siRNA and then treated with IFN-α (1000 U/ml) and plasmid DNA transfection (1 µg). Cells were lysed at different time points after DNA transfection. HeLa cells stimulated by Poly(I:C) for 1 h were set as a positive control for IRF3 activation. The collected cell lysates were separated on native PAGE and analysed by western blot using anti-IRF3 antibody. Bottom: band intensity of IRF3 dimer relative to that of IRF3 monomer was assessed by NIH Image J.
It has been reported that STING is an indispensable signal mediator that promotes the phosphorylation of IRF3 by TBK1 in the cytosolic DNA signalling pathway (36). Having confirmed that both STING and TBK1 are involved in the siRNA-induced IFN-λ1 production, we next examined whether IRF3 was also activated in this signalling transduction pathway. Two different siRNAs targeting IRF3 were used to knock down IRF3 and tested whether the siRNA-enhanced IFN-λ1 induction was affected. The results indicated that IFN-λ1 gene expression was significantly decreased when IRF3 was knocked down, supporting that IRF3 has a direct role in activating type III IFN response (Figure 6B). Furthermore, A Native PAGE was used to detect dimerization of IRF3 (an active form of IRF3 to transduce signalling). HeLa cells were transfected with siRNA and DNA, and the cells were lysed at different time points after DNA transfection, as indicated in Figure 6C. IRF3 dimerization occurred within 1 h after DNA transfection and reached its peak at 3 h after DNA transfection. The ratio of IRF3 dimer to IRF3 monomer was calculated by NIH Image J software (data shown in the lower panel of Figure 6C). The results indicated that the activation of IRF3 is also involved in the type III IFN induction pathway. The kinetic process of IRF3 dimerization is consistent with the time course of the siRNA-induced IFN-λ1 response (Figure 2D).
siRNA enhances HSV-1 infection–mediated type III IFN
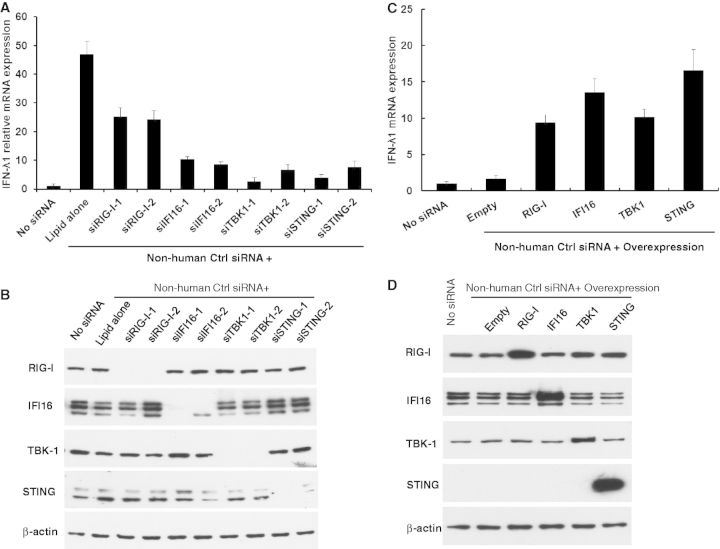
To ascertain the physiological relevance of our findings, we examined whether siRNA could induce a potent IFN-λ1 induction even in the presence of DNA virus infection. It was shown that infection with HSV-1, but not HSV-2, induced IFN-λ1 gene activation by ∼20-folds compared with untreated cells. The HSV-1–induced IFN-λ1 activation was further enhanced by Non-human Ctrl siRNA transfection (Figure 7A). To determine the roles of RIG-I, IFI16, TBK1 and STING in the DNA virus–mediated type III innate immunity, we also delivered siRNA targeting RIG-I (siRIG-I-1 and siRIG-I-2), IFI16 (siIFI16-1 and siIFI16-2), TBK1 (siTBK1-1 and siTBK1-2) and STING (siSTING-1 and siSTING-2). IFN-λ1 production was significantly reduced when RIG-I, IFI16, TBK1 and STING were knocked down. The knockdown efficiency of the siRNA was confirmed by western blot (Figure 7B). We also knocked down mitochondrial antiviral signalling protein (MAVS), another important signalling mediator that functions downstream of RIG-I (37). Compared with the result from knocking down of STING, knocking down of MAVS suppressed the IFN-λ1 gene induction by ∼20–50% (Supplementary Figure S10), suggesting that MAVS is partially involved, and with less contribution as that of STING in the RIG-I and IFI16 crosstalk signalling pathway. Taken together, our data indicated that siRNA also enhanced a DNA virus, HSV-1 infection-mediated type III IFN response and RIG-I, IFI16, STING and TBK1 were involved in the DNA virus infection–mediated IFN-λ1 induction. We also assessed the effect of the overexpression of RIG-I, IFI16, STING and TBK1 in the siRNA- and HSV-1-mediated IFN-λ1 induction. To define the impact of overexpressed proteins, we treated cells with a suboptimal concentration of IFN-α (100 U/ml) with a short incubation time (6 h). This treatment conferred a lower background of IFN-λ1 induction. The results indicated that the virus-mediated IFN-λ1 induction was highly increased when the cells were overexpressed with RIG-I, IFI16, STING or TBK1 (Figure 7C and D). It is noted that the overexpression of RIG-I, IFI16, STING or TBK1 alone could induce a certain level of IFN-λ1; however, this level was much lower than that induced by the cells with overexpression and siRNA stimulation. Taken together, RIG-I, IFI16, STING and TBK1 play an important role in the siRNA-enhanced HSV-1 infection-mediated IFN-λ1 induction pathway.
Figure 7.
siRNA also enhances HSV-1–mediated IFN-λ1 production. (A) HeLa cells were transfected with or without 10 nM Non-human Ctrl siRNA along with siRNA targeting RIG-I (siRIG-I-1 and siRIG-I-2), IFI16 (siIFI16-1 and siIFI16-2), TBK1 (siTBK1-1 and siTBK1-2) or STING (siSTING-1 and siSTING-2). Cells were then treated with IFN-α (1000 U/ml) followed by HSV-1 infection at a MOI of 5. RNA was extracted 6 h after virus infection. Relative IFN-λ1 mRNA expression level was measured by real-time RT-PCR and compared with the cells treated with IFN-α and HSV-1 infection (no siRNA in the figure, the IFN-λ1 induction under this condition has ∼20-folds increase compared with that of uninfected cells). (B) Whole-cell lysates were collected, and an equal amount of total proteins were separated using SDS-PAGE and subjected to western blot to detect RIG-I, IFI16, TBK1 and STING expression. β-actin was used as a loading control. (C) Impact of the overexpression of RIG-I, IFI16, TBK1 and STING on the siRNA-enhanced HSV-1–mediated IFN-λ1 production. HeLa cells were transfected with RIG-I, IFI16, STING or TBK1 expression vector and followed by a transfection of Non-human Ctrl siRNA. An empty vector was transfected to the cells without RIG-I, IFI16, STING or TBK1 overexpression to ensure each condition with the same amount of DNA transfection. The transfected cells were then treated with IFN-α (100 U/ml) for 6 h (a minor IFN-α treatment to confer a lower IFN-λ1 induction) followed by HSV-1 infection at a MOI of 5. RNA was extracted 6 h after HSV-1 infection. Relative IFN-λ1 mRNA expression level was measured by real-time RT-PCR and compared with the cells without siRNA transfection. (D) Whole-cell lysates of the cells were collected, and an equal amount of total proteins was loaded on SDS-PAGE and then subjected to western blot to detect RIG-I, IFI16, TBK1 and STING.
DISCUSSION
It has been reported that chemically synthesized siRNAs can non-specifically induce innate immune response and produce off-target effects through PRRs, including TLR3, TLR7/8 and PKR (20,38). However, knowledge of the immunostimulatory effects of siRNA is still limited. It is not clear whether siRNA induces, or has any effects on the response of type III IFN, a newly characterized IFN with type I IFN–like antiviral activity. Results from the current study demonstrate that siRNA can synergistically enhance DNA-mediated type III IFN response through a novel crosstalk signalling pathway between RNA and DNA sensor.
The current study demonstrated that siRNA induces type III IFN response in a dose-dependent manner. When double-stranded, sense or antisense single-stranded siRNA was transfected into the cells, only double-stranded siRNA induced higher levels of type III IFN. It is reported that sense single-stranded siRNA is usually able to induce type I IFN or proinflammatory cytokines at the similar level as that induced by double-stranded siRNA through the TLRs (17,20). This discrepancy is likely due to a difference in receptors: the siRNA-sensing protein identified in our study is RIG-I, rather than endosomal TLRs. Single-stranded siRNA is probably not sufficient to activate RIG-I activity. In this regard, our data support the view that recognition of non-self RNA by RIG-I requires the presence of double-stranded RNA, at least of short double-stranded RNA (30). Innate immunity is a rapid response in which invading pathogens are efficiently detected by host PRRs. The type III IFN response triggered by siRNAs reaches its peak 6 h after stimulation, a response that is considered rapid, and is the cell’s first line of defence against pathogen. Furthermore, this response is not restricted to HeLa cells but also exists in other cell lines and primary dendritic cells, suggesting that siRNA-induced type III IFN response is a common immune response. And we further proved that the IFN-λ1 produced in this system is high enough to inhibit HIV-1 virus replication in primary macrophages and dendritic cells, supporting a more physiological relevance to the antiviral effect of IFN-λ1. Taken together, these findings provide a novel basic profile for siRNA-induced type III IFN response.
Activation of RIG-I by 5′-end triphosphate RNA leads to its association with MAVS and activation of downstream cytosolic kinases, which in turn activate the transcription factors IRF3 and NF-κB, respectively (39,40). In the current study, we have demonstrated that a 19-nt siRNA also has the capability to bind with RIG-I and further induce its oligomerization. It has been reported that siRNAs produced by bacteriophage T7 RNA polymerase induce potent IFN-α and IFN-β in a variety of cell lines, and the 5′-end triphosphate is required for this induction (18). However, in this study, we have demonstrated that 5′-end triphosphate is not required for DNA-mediated IFN response, suggesting that the mechanisms of the IFN induction are different between the absence and presence of DNA in signalling triggering. Furthermore, our findings have shown that at least a 2-nt overhang at one 3′ terminus is required for siRNA to highly enhance DNA-mediated IFN-λ1 production, which is confirmed in HeLa cells and THP-1 cells, suggesting that the end structure of siRNA may contribute to RIG-I recognition and IFN-λ1 triggering. Of note, Marques et al. reported that having a blunt-end structure is essential for siRNA to induce type I IFN; therefore, the 2-nt overhang is added to evade the type-I IFN response (29). This discrepancy further suggested that the mechanism of type III IFN induction probably differs from that of type I IFN induction.
5′ GUCCUUCCAA 3′ and 5′ UGUGU 3′ have been previously defined as IFN- or cytokine-activating motifs (20,38). None of the siRNAs in our study contains any of these sequences. We included 40 siRNAs to evaluate their IFN-λ1 immunostimulatory properties, and the results suggested that different siRNAs displayed various abilities to induce IFN-λ1 production with a target-independent manner, and further study is required to discover its sequence dependency. In addition, results from siRNA stability assay and the evaluation of siRNA binding affinity to RIG-I showed that the diversity in the IFN-λ1 inducing activity may not be correlated with their stability and affinity to RIG-I. Therefore, we propose that, as the ligand of RIG-I, the molecular or structural features for siRNA to trigger type III IFN are different from those required for type I IFN induction.
As we have noted above, DNA transfection is indispensable for siRNA-induced type III IFN response. Based on this observation, we hypothesized that the involvement of a DNA sensor may play an important role in initiating the type III IFN signalling pathway. The results indeed suggest an siRNA and DNA sensor crosstalk pathway. The dissociation of DNA sensor protein triggers the downstream STING-TBK1-IRF3 pathway as illustrated in Figure 8. Although whether a type I IFN–inducing pathway or other potential co-activators are required for initiating the formation of RIG-I and IFI16 complex remains unclear, we demonstrated for the first time that in the presence of siRNA and DNA, a crosstalk pathway between RIG-I and IFI16 to induce a type III IFN response. In addition, we confirmed that in an HSV-1 (DNA virus) infection model, siRNA-induced type III IFN occurs through a RIG-I and IFI16 crosstalk and STING-TBK1-IRF3 signalling pathway, conferring a physiological relevance to our findings. We have previously identified that Ku70, a DNA repair protein, serves as a novel DNA sensor protein in cytosol and induces type III IFN response (25). In this study, however, we demonstrated that IFI16 but not Ku70 acts as a DNA sensor to initiate downstream signalling in the presence of siRNA. A further study investigating the mechanism of regulation of the DNA sensing among Ku70, IFI16 and RIG-I is currently being conducted. Preliminary data suggested that in the presence of siRNA, RIG-I preferentially exerted a synergistic effect in the IFN-λ1 activation with IFI16 rather than Ku70 (data not shown). We hypothesized that in the presence of siRNA, the DNA sensing system may be altered, and cells differentially responded on siRNA and DNA stimulation. Another alternative interpretation would be that the transfected siRNA is recognized by RIG-I, and this RIG-I sensing pathway may prime some factor(s), which is an initiator for DNA sensing by IFI16. Therefore, this uncharacterized factor probably plays an important role to transduce signalling from RIG-I pathway to IFI16 pathway. Further study is required to precisely define this crosstalk between RIG-I and IFI16 signalling pathway. IFI16 was previously identified as a DNA sensor that induces type I IFN (33). However, our study reveals it to be a DNA sensor that initiates type III IFN response. Our findings detailing the downstream signalling pathway was consistent with these previous studies (33,34,41), supporting that type III and type I IFN responses share a partially similar signalling pathway.
Figure 8.
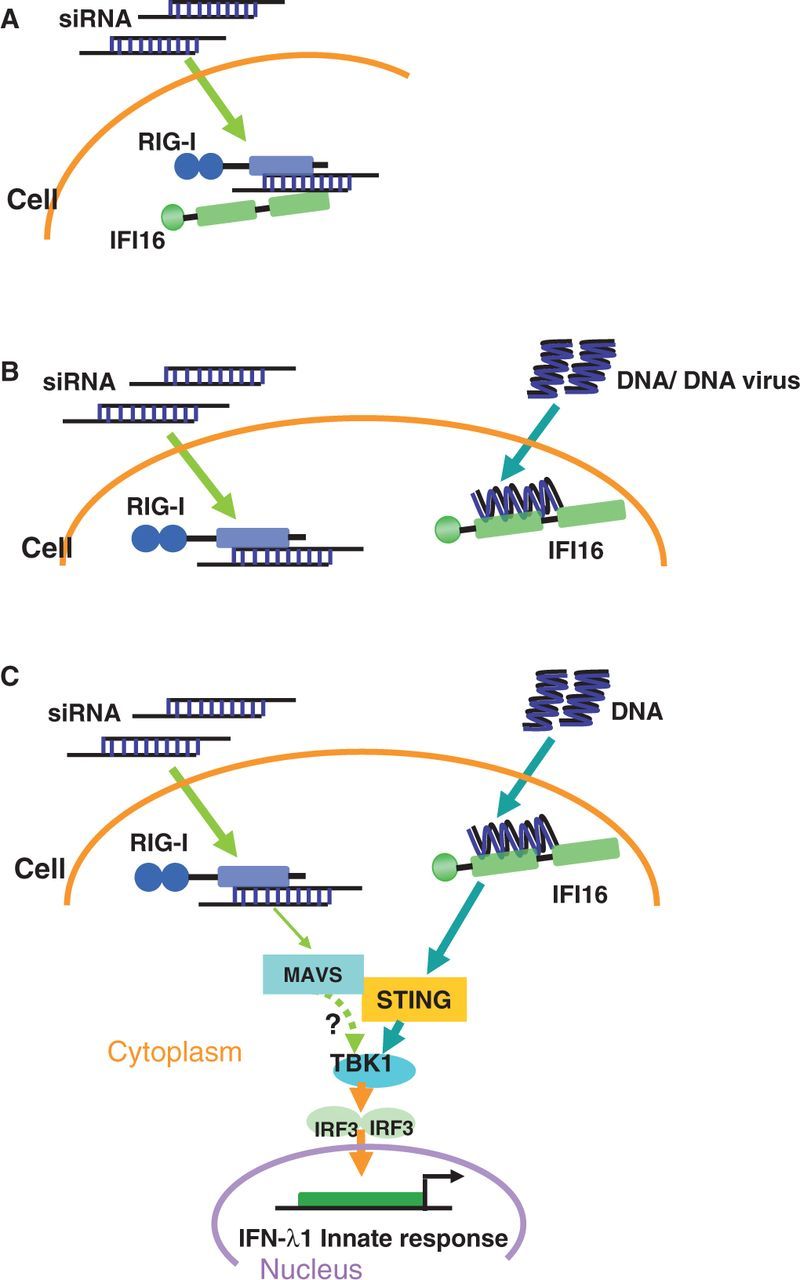
A simplified view of siRNA-enhanced DNA-mediated IFN-λ1 signalling pathway. (A) RIG-I and IFI16 formed a protein complex in the presence of siRNA. (B) IFI16 dissociated with the RIG-I–siRNA–IFI16 complex and bound with invaded DNA or DNA virus. (C) The dissociation of IFI16 initiated the downstream STING-TBK1-IRF3 signalling pathway. RIG-I/MAVS pathway also partially contributed the type III IFN induction pathway, but how MAVS interact with downstream TBK1 or STING remains unclear, where is indicated by a question mark and a dashed line.
This study also indicated that IFI16 is able to bind to siRNA even in the absence of RIG-I. And either RIG-I or IFI16 can independently bind with siRNA. Co-immunoprecipitation assay and RNase V1 cleavage experiment further delineated that RIG-I and IFI16 form a complex in the presence of siRNA. This association is not related with the sequence and the 2-nt DNA overhang of the siRNA. Additionally, this is the first time to demonstrate that IFI16 also bind with 19-nt small double-stranded RNA; however, the physical interaction between IFI16 and siRNA was not sufficient to trigger IFN-λ1 in the absence of RIG-I. At this moment, it is still unclear that how IFI16 interacts with siRNA to form the RIG-I-siRNA-IFI16 and how the siRNA-IFI16 interaction is involved in the IFN-λ1 signal induction, and a further molecular study is needed.
The immunostimulatory ‘side effects’ of siRNA must be taken into account when considering possible application of siRNA for in vivo therapy. Mice injected with immunostimulatory siRNAs present signs of toxicity, including elevated levels of serum alanine and aspartate aminotransferases, and a reduced number of lymphocytes and platelets (42,43). However, in some scenarios, activation of the immune system combined with siRNA-mediated gene silencing might actually be desired, leading to enhanced therapeutic effects, suggesting that siRNA containing potent immunostimulatory molecular features could prove useful in the treatment of viral infections and tumours (44). Gene silencing and immunostimulation, therefore, represent a double-edged sword in the use of siRNA in vivo (45). Our findings demonstrate that chemically synthesized siRNA can enhance DNA-mediated type III IFN response in non-immune or immune cells, thus identifying a new immunostimulatory characteristic of siRNAs. Induction of IFN response, including not only type I IFNs but also type III IFNs, needs to be considered in interpreting the results of in vitro and in vivo siRNA-based gene silencing. The molecular or structural features and immunorecognition mechanism identified in this study will enable the development of methods to avoid immunostimulation of siRNA or the design of siRNA to take advantage of immunostimulation, depending on the aim.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [46,47].
FUNDING
Federal funds from the National Cancer Institute, National Institutes of Health [contract No. HHSN261200800001E] (whole or in part). The content of this publication does not necessarily reflect the reviews or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US government. Finding for open access: Federal funds from the National Cancer Institute, National Institutes of Health [contract No. HHSN261200800001E] (whole or in part).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Richard Lempicki, Dawei Huang, Jun Yang, Hiromi Imamichi and Howard Young for discussion, Demitra Perlegas for technical support, Raphael Oguariri, Lue Dai, Sanjay Swaminathan and Nancy Parrish for critical reading of the manuscript; Alicia Gussio for assistance with supplies.
REFERENCES
- 1.Vaucheret H, Béclin C, Fagard M. Post-transcriptional gene silencing in plants. J. Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- 2.Sharp PA. RNA interference—2001. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 3.Brantl S. Antisense-RNA regulation and RNA interference. Biochim. Biophys. Acta. 2002;1575:15–25. doi: 10.1016/s0167-4781(02)00280-4. [DOI] [PubMed] [Google Scholar]
- 4.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 5.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 6.Sørensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J. Mol. Biol. 2003;327:761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 7.Hamar P, Song E, Kökény G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc. Natl Acad. Sci. USA. 2004;101:14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 9.Ge Q, Filip L, Bai A, Nguyen T, Eisen HN, Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc. Natl Acad. Sci. USA. 2004;101: 8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song E, Lee S-K, Wang J, Ince N, Ouyang N, Min J, Chen J, Shankar P, Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 11.Parkin J, Cohen B. An overview of the immune system. Lancet. 2001;357:1777–1789. doi: 10.1016/S0140-6736(00)04904-7. [DOI] [PubMed] [Google Scholar]
- 12.Janeway CA, Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 13.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-[kappa]B by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 14.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via Ttoll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 15.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl Acad. Sci. USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BRG. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 17.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J. Mol. Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Kim D-H, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 19.Kariko K, Bhuyan P, Capodici J, Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J. Immunol. 2004;172:6545 – 6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- 20.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 21.Sui H-Y, Zhao G-Y, Huang J-D, Jin D-Y, Yuen K-Y, Zheng B-J. Small interfering RNA targeting M2 gene induces effective and long term inhibition of influenza A virus replication. PLoS One. 2009;4:e5671. doi: 10.1371/journal.pone.0005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenworthy R, Lambert D, Yang F, Wang N, Chen Z, Zhu H, Zhu F, Liu C, Li K, Tang H. Short-hairpin RNAs delivered by lentiviral vector transduction trigger RIG-I-mediated IFN activation. Nucleic Acids Res. 2009;37:6587–6599. doi: 10.1093/nar/gkp714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003;4:63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 24.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003;4:69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang D-W, Lempicki RA, Baseler MW, et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fakruddin JM, Lempicki RA, Gorelick RJ, Yang J, Adelsberger JW, Garcia-Pineres AJ, Pinto LA, Lane HC, Imamichi T. Noninfectious papilloma virus-like particles inhibit HIV-1 replication: implications for immune control of HIV-1 infection by IL-27. Blood. 2007;109:1841–1849. doi: 10.1182/blood-2006-02-001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, Grewal SIS. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–422. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou W, Wang X, Ye L, Zhou L, Yang Z-Q, Riedel E, Ho W-Z. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J. Virol. 2009;83:3834–3842. doi: 10.1128/JVI.01773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BRG. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 30.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 32.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 33.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl Acad. Sci. USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seth RB, Sun L, Ea C-K, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Judge A, Sood V, Shaw J, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 39.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M, et al. 5'-triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 40.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 41.Barber GN. STING-dependent signaling. Nat. Immunol. 2011;12:929–930. doi: 10.1038/ni.2118. [DOI] [PubMed] [Google Scholar]
- 42.Marques JT, Williams BRG. Activation of the mammalian immune system by siRNAs. Nat. Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 43.Medzhitov R, Janeway CA. Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 44.Gantier MP, Tong S, Behlke MA, Irving AT, Lappas M, Nilsson UW, Latz E, McMillan NAJ, Williams BRG. Rational design of immunostimulatory siRNAs. Mol. Ther. 2010;18:785–795. doi: 10.1038/mt.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlee M, Hornung V, Hartmann G. siRNA and isRNA: two edges of one sword. Mol. Ther. 2006;14: 463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Kenakin T. Principles: receptor theory in pharmacology. Trends Pharmacol. Sci. 2004;25:186–192. doi: 10.1016/j.tips.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Chen Q, Swaminathan S, Yang D, Dai L, Sui H, Yang J, Hornung RL, Wang Y, Huang DW, Hu X, et al. Interleukin-27 is a potent inhibitor of cis HIV-1 replication in monocyte-derived dendritic cells via a type I interferon-independent pathway. PLoS One. 2013;8:e59194. doi: 10.1371/journal.pone.0059194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.