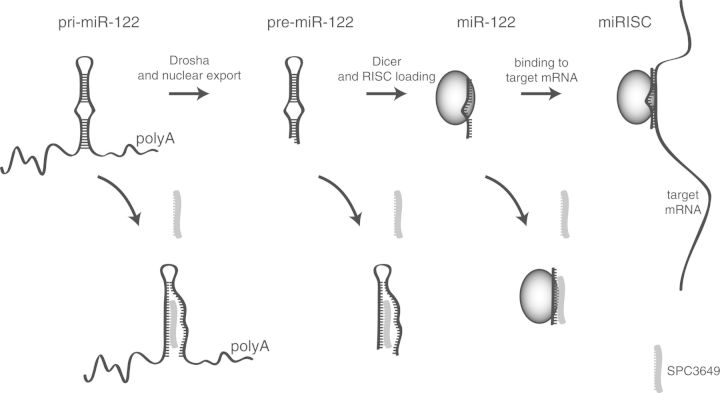
Abstract
MicroRNAs (miRNAs) are short noncoding RNAs, which bind to messenger RNAs and regulate protein expression. The biosynthesis of miRNAs includes two precursors, a primary miRNA transcript (pri-miRNA) and a shorter pre-miRNA, both of which carry a common stem-loop bearing the mature miRNA. MiR-122 is a liver-specific miRNA with an important role in the life cycle of hepatitis C virus (HCV). It is the target of miravirsen (SPC3649), an antimiR drug candidate currently in clinical testing for treatment of HCV infections. Miravirsen is composed of locked nucleic acid (LNAs) ribonucleotides interspaced throughout a DNA phosphorothioate sequence complementary to mature miR-122. The LNA modifications endow the drug with high affinity for its target and provide resistance to nuclease degradation. While miravirsen is thought to work mainly by hybridizing to mature miR-122 and blocking its interaction with HCV RNA, its target sequence is also present in pri- and pre-miR-122. Using new in vitro and cellular assays specifically developed to discover ligands that suppress biogenesis of miR-122, we show that miravirsen binds to the stem-loop structure of pri- and pre-miR-122 with nanomolar affinity, and inhibits both Dicer- and Drosha-mediated processing of miR-122 precursors. This inhibition may contribute to the pharmacological activity of the drug in man.
INTRODUCTION
MicroRNAs (miRNAs) are the products of a multistep biogenesis (1). Most primary miRNA transcripts (pri-miRNA) are produced from Pol II-mediated transcription. They are cleaved by the nuclear Microprocessor complex to yield a pre-miRNA stem-loop with a short overhang at its 3′-terminus (2–4). The pre-miRNA is transported to the cytoplasm where Dicer and its partner proteins excise the loop, creating a double-stranded RNA composed of mature miRNA strands (5–7). Both arms of the stem may be functional and may be used in a cell-context–dependent fashion (8,9). Mature miRNAs are bound by Argonaute proteins and associated factors to form the miRNA-induced silencing complex (miRISC) (10). MiRISC is guided to target sites in the 3′-untranslated regions (UTRs) of mRNAs where it inhibits protein translation or induces mRNA degradation (11–14).
MiR-122 is transcribed from a single locus and is conserved among vertebrates (15). It is highly and selectively expressed in liver where it regulates lipid metabolism (16). In addition to its natural function, miR-122 plays a pivotal role in the in the life cycle of the liver-tropic hepatitis C virus (HCV) (17,18). MiRNAs can be inhibited from binding their targets by complementary oligonucleotides (antimiRs) (19). MiR-122 was first inhibited in cultured cells using a complementary 2′-O-methyl RNA (2′-O-Me RNA) (20), and in mice using phosphorothioated, and cholesterol-linked-2′-O-Me RNA (16). Since then, a wide array of modified antimiRs has been investigated, the most prominent of which is a mixed locked nucleic acid (LNA)/DNA phosphorothioated sequence SPC3649. SPC3649 has been tested in mice (21,22), African green monkeys (22), as well as chimpanzee models of HCV infection (23). In mid-stage clinical trials, SPC3649 (miravirsen) potently suppresses viral load when administered to HCV patients (24).
As our knowledge of miRNA biogenesis grows, miRNA precursors are increasingly appreciated as functional RNAs with their own regulatory elements (25). Responding to a need for chemical tools to investigate the individual steps of biogenesis, we launched a program to characterize ligands binding selectively to miRNA precursors. Such ligands may eventually find use as antagonists in therapeutic applications where the new synthesis of miRNAs is limiting. We have established a variety of novel assays to characterize miR-122 precursor/ligand interactions including a minimalistic Dicer enzymatic assay, a surface plasmon resonance (SPR) binding assay to quantify binding kinetics and a cellular reporter assay for Microprocessor cleavage of pri-miRNAs. In the course of this work, we discovered that SPC3649, the active component of miravirsen, suppresses the biogenesis of miR-122 at the pri- and pre-miRNA levels. This suppression may contribute to the pharmacological activity of this candidate drug.
MATERIALS AND METHODS
Olignoucleotide sequences
HPLC-MS binding assay
Synthetic pre-miR-122 (final concentration 0.25 µM), the four test antimiRs (final concentration 2.5 µM) and their mixtures were incubated in H2O (Millipore) for 15 min at room temperature, then analyzed on a high pressure liquid chromatography - mass spectrometer (HPLC-MS) apparatus (Agilent 1200/6130) fitted with a C-18 reverse phase (RP) column (Waters Acquity OST C18, 2.1 × 50 mm, 1.7 μm) equilibrated at 40°C to limit denaturation of the putative duplex. Elution conditions were MeOH (18–30% in 10 min, then up to 90% in 5 min) in aqueous hexafluoroisopropanol (HFIP) (0.4 M)/triethylammonium (TEA) (16.2 mM) buffer.
HPLC-MS Dicer assay
Synthetic pre-miR-122 (final concentration 2.5 µM) was incubated alone, with 1 unit of commercial Dicer (T520002, Genlantis, San Diego) or with commercial Dicer after preincubation with oligonucleotides (final concentration 5 µM) in a buffered aqueous solution (Tris–HCl 30 mM, MgCl2 3 mM, NaCl 50 mM, DTT 1 mM) for 2 h at 37°C. The reactions were stopped by addition of 0.5 µl of 0.5 M EDTA (pH 8) and kept at 4°C before analysis using a C18 RP column equilibrated at 70°C with detection set to 260 nm. Elution conditions were 12–25% MeOH in aqueous HFIP (0.4 M)/TEA (16.2 mM) buffer in 10 min. Observed mass of miR-122-5p: 7115 Da; calculated mass: 7116 Da; observed mass of 5′-phosphorylated miR-122-3p: 7016 Da; calculated mass: 7017 Da; observed mass of loop sequence 5′-phos-UGUCUAAACUAUCA: 4455 Da; calculated mass: 4456 Da; observed mass of extended miR-122-5p: 8684 Da; calculated mass of 5′-OH miR-122-5p elongated by 5 nt: 8685 Da.
Cell cultures and transfections
HeLa and Huh-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) GlutaMAXTM-I (31966-021, Gibco®, Life Technologies) supplemented with 10% of fetal bovine serum. Transfections were performed according to the manufacturer's protocol with Lipofectamine 2000 (11668-019, Invitrogen, Life Technologies, Carlsbad) for antimiRs, Oligofectamine (12252-011, Life Technologies, Carlsbad) for pre-miRNA and JetPEI (101-10, Polyplus transfections, Illkirch) for plasmid DNA.
Luciferase assays
Cells were seeded in opaque white 96-well plates (136 101, Nunc, Roskilde) in 80-µl medium per well, and transfected according to the experimental setup with the reagents described above. Reporter plasmids were transfected at 10 ng [Glycogen Synthase 1 (GYS1) reporter, HeLa cells], 15 ng (Drosha reporter and control reporter, HeLa cells; miR-122-5p sensor reporter, Huh-7 cells) or 20 ng (miR-122-3p reporter, Huh-7 cells). The assay was performed with the Dual-Glo® Luciferase Assay System (E2980, Promega, Fitchburg). The cell culture medium was removed and 30 μl of a solution composed of luciferase buffer with dissolved luciferase substrate and water in a ratio of 1:1 were added to each well. After 10 min, the luminescence was measured on a Mithras LB 940 plate reader (Berthold Technologies, Bad Wildbad). Fifteen microliters of a 1% solution of stop & glo substrate in stop & glo buffer were added to each well and luminescence was again measured after 10 min. Each time luminescence was measured for 1 s per well. For each well, Renilla luminescence counts were normalized on firefly luminescence counts, then average values were computed.
psiCHECK-2 reporter constructs
The target sequences were either cloned from HeLa genomic DNA (pri-miR-122 Drosha reporter) by polymerase chain reaction (PCR) with primers containing suitable restriction sites in a two-step process, amplified from synthetic DNA (miR-122-5p reporter, miR-122-3p reporter) or produced by overlap extension PCR (26,27) (Drosha reporter control), using oligonucleotides (Supplementary Table S1) from Microsynth (Balgach) and Finnzymes Phusion High Fidelity DNA Polymerase (F530-S, Finnzymes, Vantaa).
After amplification, the samples were analyzed by agarose gel electrophoresis (1.5%) in a field of 100 V for 1 h. The PCR reactions corresponding to bands of the correct size on the gel were purified, digested and ligated into the previously digested and dephosphorylated psiCHECK-2 plasmid (C8021, Promega, Fitchburg). The ligated fragment was transformed into Subcloning Efficiency™ DH5α™ Competent Escherichia coli (18265-017, Life technologies, Carlsbad), plated on LB agar with 100 µg/ml Amp and incubated for at least 16 h at 37°C. The single colonies (or in the case of a florid growth, pools of colonies) were picked and resuspended in water. An aliquot of the suspension was cracked by heating to 95°C for 15 min and analyzed by PCR using two primers designed to anneal at 50 bp from the restriction sites of psiCHECK-2 vector using an annealing temperature of 64°C. Aliquots of clones yielding positive bands were used to make glycerol stocks and DNA minipreps and were sequenced by Microsynth (Balgach). All the PCR purifications were performed following the protocol of MinElute® PCR Purification Kit (250) (28006, Qiagen, Venlo). Miniprep were performed following the protocol of QIAprep® Spin Miniprep Kit (250) (27106, Qiagen, Venlo). Digestions were performed following the guideline of the NEB Web site (http://www.neb.com) using NotI and XhoI according to the supplied protocol (NEB, Ipswich). The ligations were performed with ratios of 1:3, 1:5, 1:10 (vector to insert) using the dephosphorylated psiCHECK-2 vector with T4 DNA Ligase (M1804, Promega, Fitchburg).
qRT-PCR
Except for the experiments with murine primary hepatocytes, total RNA was extracted using the RNeasy kit (74104, Qiagen, Venlo). TaqMan® quantitative reverse transcription (RT) PCR (qRT-PCR) was performed using standard reagents from Applied Biosystems (TaqMan® MicroRNA Assays: miR-122-5p: 002245; miR-122-3p: 002130; miR-16-5p: 000391; miR-191-5p: 002299). The pre-miR-122 TaqMan® probe was made by Applied Biosystems with the pre-miR-122 sequence used as input in the online custom TaqMan® small RNA assay design tool (4398987). The RT was performed using the TaqMan® primers from MicroRNA Assays and the TaqMan® MicroRNA Reverse Transcription Kit (4366596, Life Technologies, Carlsbad) with 25 ng of total RNA. The PCR was performed in a LightCycler 480 instrument (Roche, Penzberg) with TaqMan® Unversal PCR Master Mix. No AmpErase UNG (4324020, Invitrogen, Life Technologies, Carlsbad) according to the manufacturer's protocol. Each reaction was carried out in four technical replicates. Ct values were calculated for each and averaged.
SYBR Green qRT-PCR was performed with total RNA extracted in the same way as above. RT was performed with M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant (M3681, Promega, Fitchburg) and equal amounts of oligo-dT (C1101, Promega, Fitchburg) and random hexamer primers (SO142, Thermo Scientific, Waltham). SYBR Green PCR was performed in a LightCycler 480 instrument (Roche, Penzberg) with FastStart Universal SYBR Green Master (Rox) (04913914001, Roche, Penzberg). Each reaction was carried out in three technical replicates. Ct values were calculated for each and averaged.
SPR measurements
The SPR measurements of SPC3649, AMO-122 and controls against the hairpin of pre-miR-122 were performed using a SPR2 machine (Sierra Sensors GmbH, Hamburg, Germany). The measurements were performed at flow rates of 25 µl/min at 25°C, and 37°C, respectively. For the analysis, a matrix-free carboxymethylated amine chip (C1 type) was used. During the immobilization process, a phosphate buffered saline (PBS) buffer (200 mg/l KCl, 200 mg/l KH2PO4, 8 g/l NaCl and 1.15 g/l Na2HPO4, 0.05% TWEEN20, pH 7.4) was used at a flow rate of 25 µl/min. The chip was activated by the injection of 150 µl of a EDC/NHS solution (containing 50 mM N-hydroxysuccinimide and 200 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (Sierra Sensors) in desalted, sterile water) at a flow rate of 25 µl/min at 37°C (this yields high immobilization levels). After activation, the chip was coated with Streptavidin (Jackson ImmunoResearch Laboratories, Inc., PA, USA). For the analysis, we used two different chips of the same chemistry. The immobilizations were done with injections of 100 µg/ml streptavidin in NaAc, 10 mM, pH 5.5 (170 µl, 25 µl/min), over both flow cells yielding 1850 RU for both chips. Subsequent injection of 100 µl of a 1 M ethanolamine/NaOH solution, pH 8.5, was used for blocking of unreacted sites. For the interaction analysis, flow cell 1 was left unreacted (containing only streptavidin) and served as reference cell. After changing to a Tris-buffer (trishydroxymethylaminomethane 20 mM, MgCl2.6H2O 5 mM, CaCl2.6H2O 2 mM, KCl 2 mM, NaCl 140 mM and TWEEN 20 0.05% in sterile, desalted and rigorously degassed water), the biotinylated ligands (pre-miR-122) were bound onto flow cell 2 of the streptavidin-coated surface: 110 RU for the first chip (30 nM solution of biotinylated ligand, 100 µl at 25 µl/min, Tris-buffer) and 165 RU for the second chip (same conditions). The sensorgrams of the analytes were obtained by injections of 300 µl of 342 nM solutions with two-fold dilutions in Tris-buffer followed by a dissociation time of 330 s. As a regeneration solution, we used a 1:1:1 mixture of water, an acid regeneration solution (equal volumes of oxalic acid, H3PO4, formic acid and malonic acid, each at 0.15 M, mixed and adjusted to pH 3.0 with HCl) and an ionic regeneration solution [KSCN (0.46 M), MgCl2 (1.83 M), urea (0.92 M) and guanidine HCl (1.83 M)]. The pH-value of this solution was ∼2. The fitting of the sensorgrams was performed using Scrubber2.0 (BioLogic Software, http://www.biologic.com.au) using a 1:1 binding model with accounting for mass-transport limitation.
Primary hepatocytes isolation and transfection
Eight- to twelve-week-old male C57BL/6 mice were anesthetized by intraperitoneal injection of 150 µl of pentobarbital (Esconarkon US vet) prediluted 1:5 in PBS. The liver was perfused by cannulation of the caudal vena cava with the portal vein as a drain. The liver was perfused with prewarmed Hank's Balanced Salt Solution (14175-053, Invitrogen, Life Technologies, Carlsbad) containing 0.5 mM ethylene glycol tetraacetic acid (EGTA) followed by digestion of the extracellular matrix with prewarmed digestion media [DMEM, 1 g/l glucose (31885-049, Invitrogen,), 1% Penicillin-Streptomycin (Invitrogen, 15140-122), 15 mM HEPES (Invitrogen, 15630-056, Life Technologies, Carlsbad) and 30 µg/ml Liberase (05401119001, Roche, Penzberg)] each for 4 min at a flow rate of 5 ml per min. The liver was surgically removed, hepatocytes released into 5 ml digestion media by shaking and supplemented with 20 ml ice cold low glucose media [DMEM, 1 g/l glucose (31885-049, Invitrogen, Life Technologies, Carlsbad), 1% Penicillin-Streptomycin (15140-122, Invitrogen, Life Technologies, Carlsbad), 10% (v/v) fetal bovine serum (F7524, Sigma, St. Louis) and 1% Glutamax (35050, Invitrogen, Life Technologies, Carlsbad)] and filtered through a 100 µm Cell Strainer (352360, BD, Franklin Lakes). The suspension was then washed three times with 25 ml of ice cold low glucose media (50 g at 4°C for 2 min). Hepatocytes were counted and plated at 300 000 cells/well in precoated six-well plates (353846, BD Franklin Lakes) in low glucose media and incubated at 37°C in a humidified atmosphere containing 5% CO2. Four hours after plating, cells were transfected using Dharmafect1 (T-2001-03, Thermo Scientific, Waltham) using the manufacturer’s instructions. The transfection was performed in Hepatozyme media [HepatoZYME-SFM (17705-021, Invitrogen, Life Technologies, Carlsbad) containing 1% Penicillin-Streptomycin (15140-122, Invitrogen, Life Technologies, Carlsbad), 1% Glutamax (35050-061, Invitrogen, Life Technologies, Carlsbad)] with 5 µl of Dharmafect1/well and an end volume of 1.25 ml. Twenty-four hours after transfection, the media was changed to 2 ml Hepatozyme media per well. At the end, the cells were washed twice with ice cold PBS, 1 ml of Trizol reagent (15596-018, Invitrogen, Life Technologies, Carlsbad) was added and frozen at −80°C for later RNA extraction. RNA was extracted using the Trizol according to manufacturer's instructions, except for the isopropanol precipitation of RNA, which was done for half an hour at −20°C.
RESULTS
Synthesis of pre-miRNAs
Pre-miRNAs can be prepared by in vitro transcription from DNA templates or by solid-phase synthesis. They are typically 50–90 nt in length, which likely represents the upper limit for chemical synthesis using conventional reagents. Pre-miR-122 is 58 nt long and carries a 2 nt overhang at the 3′-terminus (www.mirbase.org). We synthesized pre-miR-122 with terminal 5′- and 3′-OH groups for solution phase binding assays and cell assays. It was purified by RP-HPLC (see Figure 1a) and was characterized by mass spectrometry (MS). For immobilization of pre-miR-122 for SPR studies, we extended pre-miR-122 with a 3′-oligo-dT bearing a terminal biotin moiety.
Figure 1.
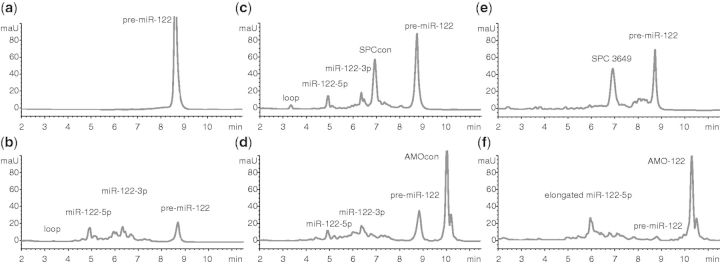
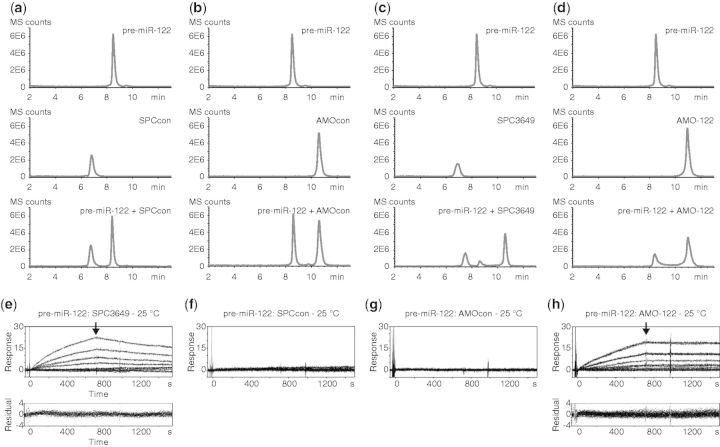
SPC3649 inhibits Dicer processing of pre-miR-122 in vitro. AntimiRs (5 µM) were added to synthetic pre-miR-122 in buffer (2.5 µM) before addition of Dicer. Reactions were quenched after 2 h and analyzed by HPLC-MS. (a) Pre-miR-122 alone is stable in buffer. (b) Dicer cleaves pre-miR-122 into miR-122-5p, miR-122-3p and the loop. SPCcon (c) and AMOcon (d) do not affect processing, whereas SPC3649 (e) inhibits processing. (f) AMO-122 shifts the cleavage site to yield miR-122-5p elongated by 5 nt.
SPC3649 inhibits Dicer processing of pre-miR-122 in vitro
We have optimized an enzymatic assay to characterize ligands that will modulate the Dicer processing of pre-miRNAs. The assay uses HPLC to separate substrates and products, and coupled MS to determine their mass and their identity. Previous accounts of Dicer assays typically used lysates or immunoprecipitated Dicer. Our assay uses a commercially available Dicer, which is reportedly optimized for the production of short interfering RNAs from double-stranded RNA. It has previously been used together with a fluorescently labeled blunt-ended pre-miRNA analog (28,29). Use of the authentic pre-miRNA in our assay ensured processing to the bona fide miR-122-5p- and 3p- strands, thus simulating closely Dicer processing of pre-miR-122 in the cell.
Incubation of pre-miR-122 with Dicer resulted in its cleavage into well-separated products, which were unambiguously identified by mass as the miR-122-5p and the 5′-phosphorylated miR-122-3p miRNAs. These were sequence-identical with the most abundant miRNAs from hsa-mir-122 registered in miRBase and from deep-sequencing, respectively (30) (Figure 1b). The 5′-phosphorylated 14-nt apical loop of pre-miR-122 was also identified. Although the efficiency and precision of the cleavage varied with batches of the enzyme, these three products were consistently among the most abundant. Taken together, the experiments showed that this enzyme recognizes and cleaves chemically synthesized pre-miR-122 in vitro into the correct products under the chosen conditions, in the absence of cofactors or protein binding partners such as TAR (HIV-1) RNA binding protein 1 (TRBP) (31).
During optimization of this assay we tested four antimiRs (Supplementary Table S1): SPC3649 (miravirsen), a 15-nt LNA/DNA phosphorothioate sequence complementary to nt 2–16 of miR-122-5p; SPCcon, identical in sequence to SPC3649 except for two mismatched nt in the center and close to the terminus (21); AMO-122, a previously described (16), partially phosphorothioated 2'-O-Me RNA antimiR fully complementary to miR-122-5p; AMOcon: a scrambled sequence of AMO-122 bearing no significant complementarity to pre-miR-122. These controls were intended to guide optimization of assay parameters (concentrations, time points, buffers, etc.), as well as to indicate any nonspecific inhibition of Dicer. Incubation of pre-miR-122 with SPCcon (Figure 1c) or AMOcon (Figure 1d) did not affect significantly its processing by Dicer. For both oligonucleotides the amounts of 5p and 3p products were comparable with those seen in the control reaction (Figure 1b). In contrast, SPC3649 (Figure 1e) slowed the reaction significantly, decreasing the yields of miRNAs to barely detectable levels. Surprisingly, AMO-122 also appeared to inhibit the course of the processing by shifting the cleavage event into the loop region of the hairpin (Figure 1f). A single dominant product was visible in the chromatogram, and mass analysis suggested that it was an elongated miR-122-5p strand. In summary, both of the antimiRs affected Dicer processing of pre-miR-122, whereas the noncomplementary control sequences did not. This implied that SPC3649 and AMO-122 invade the stem of pre-miR-122 by binding to the 5p-strand and preventing correct and efficient cleavage by Dicer. In the case of the former, Dicer processing was halted; for the latter, AMO-122 may have acted as a surrogate passenger strand (32), shifting the cleavage to produce an extended miRNA.
SPC3649 invades the hairpin structure of pre-miR-122
Invasion of structured RNAs by oligonucleotides to induce misfolding of the RNA has been described earlier. For example, short LNA oligonucleotides induced misfolding of group I introns from rRNA genes in pathogenic organisms and inhibited self-splicing (33); similarly, LNAs were used to inhibit the RNA component of telomerase (34). In the miRNA field, morpholino-modified oligonucleotides were shown to inhibit biogenesis of zebra fish miRNAs (35).
We used two assays to demonstrate that SPC3649 invades the pre-miR-122 hairpin consistent with its inhibition of Dicer processing. In one, incubation of SPCcon or AMOcon separately with pre-miR-122 and injection into an HPLC column showed distinct separated peaks of oligoribonucleotides in the MS chromatograms (Figures 2a and b, respectively). In contrast, a short incubation of SPC3649 with pre-miR-122 gave attenuation of both components and the appearance of a sharp new slower-running peak containing the masses of both pre-miR-122 and SPC3649, consistent with an RNA hybrid structure comprising SPC3649 and pre-miR-122 (Figure 2c). Incubation of pre-miR-122 with AMO-122 showed no new slower-running peaks, but did result in some broadening of the pre-miR-122 peak (Figure 2d), possibly indicating a dynamic equilibrium between species with different binding affinities to the column.
Figure 2.
SPC3649 binds pre-miR-122 with nanomolar affinity in vitro. Overlaid HPLC-MS chromatograms of synthetic pre-miR-122 (0.25 µM) incubated with antimiRs (2.5 µM) in water: pre-miR-122 alone produces a single peak (top trace); antimiRs were analyzed alone (center traces) and then mixed with pre-miR-122 hairpin in solution and injected (lower traces). No effects on the peak of pre-miR-122 were observed on incubation with (a) SPCcon or (b) AMOcon. (c) The addition of SPC3649 resulted in loss of pre-miR-122 and the appearance of a new peak, containing both SPC3649 and pre-miR-122. (d) Addition of AMO-122 to pre-miR-122 resulted in a broadening of the pre-miR-122 peak. SPR sensorgrams of antimiRs bound to immobilized pre-miR-122 at four concentrations (342 nM; 2-fold dilutions) and 25°C: each trace represents a different concentration of antimiR; the highest binding (Response) is seen with higher concentrations of antimiR; the arrow indicates a switch to running buffer and the beginning of the dissociation phase: (e) SPC3649 binds concentration dependently to pre-miR-122 with calculated KD of 22 nM: ka was 17000 M−1s−1, and kd was 3.8 × 10−4 s−1. (f) SPCcon and (g) AMOcon show no association with pre-miR-122. (h) AMO-122 binds concentration dependently to pre-miR-122 with calculated KD of 18 nM (ka 3900 M−1 s−1, kd 7 × 10−5 s−1). Where binding data was fitted (e and h), the plotted residuals of the fit are shown in lower traces.
To quantify the affinity of SPC3649 for pre-miR-122, we used SPR spectroscopy. SPR is an established technique for the precise measurement of affinities and kinetics of intermolecular interactions (36). It is commonly used to characterize interactions involving proteins, but its use with folded RNAs is growing (37,38). Pre-miR-122 and mature miR-122-5p-strand (as control) were each synthesized with an extended biotinylated 3′-dT6 spacer group for binding to the streptavidin surface of an SPR chip. We then determined the KD of SPC3649 for the surface-bound pre-miR-122 as well as for single-stranded mature miR-122-5p from sensorgrams generated under a range of conditions. For SPC3649, we identified one binding site on pre-miR-122 at 25 (Figure 2e) and 37°C from which we calculated KDs of 22 and 62 nM, respectively. No measurable binding was observed for either SPCcon or AMOcon to pre-miR-122 under these conditions (Figures 2f, 2g, resp.). We also attempted to quantify the interaction of the antimiRs with mature miR-122-5p. Under similar experimental conditions, we observed a rapid and irreversible association of SPC3649 with miR-122-5p (Supplementary Figure S1). As no dissociation of the duplex was seen on switching to a buffer wash, we were unable to calculate a KD for this irreversible binding, consistent with the expected higher affinity of the antimiR for the unstructured mature miRNA compared with the hairpin. The 23 nt 2’-O-Me RNA AMO-122 also associated equally strongly with pre-miR-122 (KD 18 nM) (Figure 2h), in accordance with its modulation of pre-miR-122 processing by Dicer (Figure 1f). Furthermore, the rate of dissociation (kd) of AMO-122 was 5-fold slower than that of SPC3649, suggesting that once in place the longer duplex formed by AMO-122 has an extended half-life, and may be more difficult to displace by the dangling 3p strand of the pre-miRNA. The combined data from these in vitro experiments suggested that SPC3649 might invade the stem structure of both pri- and pre-miR-122 precursors in vivo and disrupt their processing to mature miRNAs.
SPC3649 inhibits biogenesis of miR-122 in cells
We investigated the ability of SPC3649 to interfere with the individual steps of miR-122 biogenesis using combinations of oligonucleotides and luciferase reporter constructs in HeLa cells, which do not express miR-122 (20), in Huh-7 cells expressing moderate levels of endogenous miR-122-5p (15) and in highly expressing primary hepatocytes (15). Both of the precursors of miR-122 contain the same hairpin secondary structure. However, pre-miR-122 is typically present at much lower levels in cells compared with pri-miRNAs or mature miRNAs (39). Therefore, we examined the capacity of SPC3649 to inhibit Dicer processing of synthetic pre-miR-122 delivered exogenously into HeLa cells.
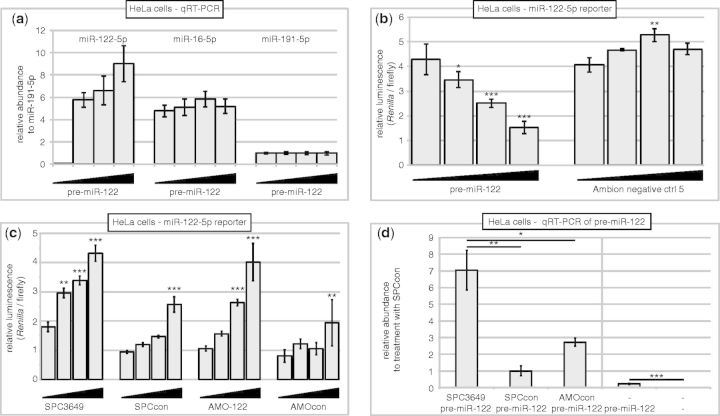
We first confirmed that pre-miR-122 is efficiently transfected into HeLa cells and is processed by endogenous Dicer. Consistent with the data shown in Figure 1b, we detected a dose-dependent production of miR-122-5p (Figure 3a) demonstrating that endogenous Dicer recognizes the synthetic pre-miR-122. To confirm that this miR-122-5p was functional, we cotransfected the RNA hairpin with a dual reporter plasmid expressing Renilla and firefly luciferase. A previously characterized binding site for miR-122-5p from the GYS1 3′-UTR (40) was inserted into the 3′-UTR of the Renilla mRNA, whereas the firefly luciferase was used to normalize for differences in transfection efficiency of the plasmid. Dose-dependent downregulation of Renilla luciferase activity was observed consistent with suppression of the 3′-UTR by the 5p strand: a control short interfering RNA had no effect (Figure 3b). When the experiment was performed in the presence of SPC3649 or AMO-122, a dose-dependent derepression of Renilla luciferase activity was observed (Figure 3c). SPCcon showed a greatly reduced effect, but some inhibition at the highest dose consistent with previous observations that two mismatches in the sequence of a high affinity antimiR would not necessarily remove all of its inhibitory activity (32). AMOcon—a scrambled sequence—showed no significant effect. This assay does not distinguish, as the source of derepression of luciferase activity, the inhibition of Dicer processing of pre-miR-122 from a sequestration of the mature miR-122-5p. Measuring the abundance of mature miR-122 by PCR or northern blot could theoretically differentiate between these two mechanisms as inhibition of Dicer would result in decreased mature miRNA. However, both of these methods are reportedly prone to artifacts caused by the presence and interference of the antimiR in samples with the assayed miR-122-5p (16,41). Therefore, we asked instead whether the antimiR treatment of cells processing synthetic pre-miR-122 would show an accumulation of pre-miR-122 levels in line with a strand invasion of the hairpin and a disruption of Dicer activity. For this we used PCR primers specific for the precursor rather than the mature miRNAs. To confirm correct functioning of the PCR assay, pre-miR-122 was transfected alone into HeLa cells, and after 24 h, RNA was isolated: intact pre-miR-122 was detected in treated but not untreated control cells (Figure 3d, right panel). Next, in a double transfection experiment, cells were treated first with SPC3649, SPCcon or AMOcon, followed by transfection with pre-miR-122 (Figure 3d, left panel). After 24 h, AMOcon treatment showed intact pre-miR-122, albeit at higher levels than observed in the single transfection (Figure 3d, right panel): this might have been due to sequence-unspecific effects or a negative influence of the second transfection on processing. SPCcon showed a similar level of intact pre-miR-122 as AMOcon (Figure 3d, left panel). However, the highest accumulation of pre-miR-122 was seen from treatment with SPC3649, consistent with inhibited processing of exogenously delivered pre-miR-122 by the SPC3649 and in agreement with experiments performed in vitro (Figures 1 and 2).
Figure 3.
Inhibition of Dicer processing of pre-miR-122 by SPC3649 and AMO-122 (a) HeLa cells were treated with synthetic pre-miR-122 (0, 2.5, 5, 10 nM). RNA was extracted 24 h later and analyzed by qPCR. The threshold cycle (Ct) values were normalized to miR-191-5p and transformed to generate relative abundance values. Error bars are SEM of four qPCR replicates. (b) Synthetic pre-miR-122 (0, 2.5, 5, 10 nM) silences GYS1 3′-UTR luciferase reporter in HeLa cells dose dependently. Renilla and firefly luciferase luminescence were measured after 72 h, and Renilla activity was normalized to that of firefly. Error bars are SD of three transfections. Data were analyzed by ANOVA using Dunnett’s post-test in Graphpad Prism 6, comparing against the lowest dose in each group (*P < 0.05, **P < 0.01, ***P < 0.001). (c) AntimiRs inhibit miR-122–mediated suppression of GYS1 3′-UTR reporter. Cells were transfected with reporter plasmid and treated with antimiRs (0.6, 2.5, 10, 40 nM), before transfection with pre-miR-122 (10 nM). Normalized Renilla activity was determined as in (b). Data were analyzed by ANOVA using Dunnett’s post-test in Graphpad Prism 6, comparing against the lowest dose in each group (**P < 0.01, ***P < 0.001). (d) SPC3649 accumulates intact synthetic pre-miR-122 in HeLa cells. Cells were transfected with 50 nM SPC3649, SPCcon or AMOcon, respectively, and 6 h later, 10 nM pre-miR-122 (left panel); cells were transfected with 10 nM pre-miR-122 alone or left untreated (right panel). After 24 h, qPCR was performed for pre-miR-122 and miR-191 expression. Ct values were calculated and transformed into relative abundance, using miR-191 as internal standard and normalizing on SPCcon. Error bars are SEM of four qPCR replicates. P-values were calculated using the two-tailed unpaired Student’s t-test with equal variances in Graphpad Prism 6 (*P < 0.05, **P < 0.01, ***P < 0.001).
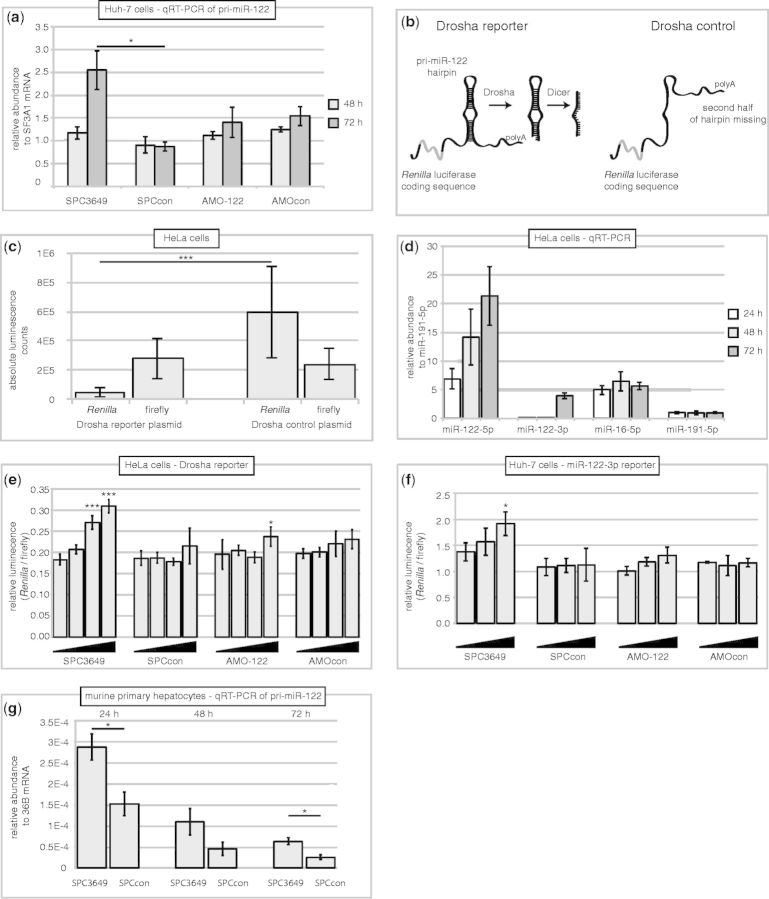
In liver cells, pre-miR-122 is expressed at 400-fold lower levels than mature miR-122 (39). Therefore, we turned to the potential effects of SPC3649 on the Drosha processing of endogenous pri-miR-122 in the nucleus of Huh-7 cells: single-stranded oligonucleotides are active in the nucleus after transfection. We transfected Huh-7 cells separately with the four antimiRs and then studied residual endogenous pri-miR-122 levels using qRT-PCR. A significant accumulation of pri-miR-122 at 72 h after transfection was seen for treatment with SPC3649, but not for AMO-122 or the negative controls (Figure 4a). This result would be consistent with invasion of pri-miR-122 by SPC3649, and the inhibition of Drosha processing in the nucleus. To exclude that the accumulation of endogenous pri-miR-122 was not solely due an indirect effect, for example, from enhanced transcription of the gene, we developed a Drosha reporter assay similar to one recently described (42). A sequence containing the pri-miR-122 hairpin was cloned into the 3′-UTR of Renilla luciferase in a standard psiCHECK-2 dual luciferase vector. We anticipated that the corresponding reporter mRNA would fold into the same hairpin structure as in the pri-miRNA (Figure 4b) and therefore would be recognized and processed by the Microprocessor; we have shown previously that artificially constructed fusion mRNAs usually maintain their local folding structures (43). Thus, Drosha was expected to excise pre-miR-122 from this UTR, and in so doing would cleave the polyA tail of the reporter mRNA, subjecting it to degradation by 3′-exonucleases with a concomitant loss of Renilla activity. In parallel, a control vector comprising the same pri-miR-122 insert, but lacking the 3p-strand of the stem-loop was also constructed. The mRNA produced from this construct was not expected to be a substrate for Drosha, and therefore should not undergo cleavage-mediated degradation. We first compared the two reporters by measuring their firefly and Renilla luminescence counts after transfection of equal amounts into HeLa cells. As expected, the firefly luciferase counts from both reporters were comparable. However, the Renilla counts from the Drosha reporter were a fraction of those of the control vector, consistent with degradation of Renilla mRNA (Figure 4c). To confirm the processing of this mRNA by Drosha, we assayed for the presence of mature miR-122 miRNAs using qRT-PCR. After 24, 48 and 72 h, clearly detectable levels of miR-122-5p were present (Figure 4d), consistent with a correct excision and processing of miR-122 from the reporter 3′-UTR by both Drosha and Dicer. We then cotransfected the antimiRs with the Drosha reporter plasmid into HeLa cells and measured Renilla luciferase levels. A clear dose-dependent derepression effect was observed in the case of SPC3649, with possibly a small effect from the AMO-122. In contrast, neither of the negative controls showed significant activity (Figure 4e). These observations are consistent with a sequence-specific inhibition of Drosha processing of the pri-miR-122 reporter by SPC3649, in accordance with strand invasion of the hairpin structure.
Figure 4.
SPC3649 inhibits Microprocessor-mediated processing of pri-miR-122. (a) Huh-7 cells were transfected with antimiRs (40 nM). RNA was isolated after 48 h and 72 h. qPCR was performed for expression of pri-miR-122 and for SF3A1 mRNA (housekeeping gene). Ct values were calculated and transformed into relative abundance. Error bars indicate SEM of three qPCR replicates. P-values were calculated using the two-tailed unpaired Student’s t-test with equal variances in Graphpad Prism 6 (*P < 0.05). (b) Principal function of a Drosha reporter mRNA and its control. The pri-miR-122 stem-loop and 100 nt flanking sequence were cloned into the 3′-UTR of the Renilla luciferase in the psiCHECK-2 reporter plasmid (left). The expressed mRNA forms a stem-loop, which is recognized by the Microprocessor and is processed to miR-122. The control plasmid (right) expresses an mRNA that lacks the 3p sequence of the pri-miR-122. (c) Plasmids from (b) were transfected separately into HeLa cells. Levels of firefly and Renilla activity were measured after 24 h. Error bars are SD of 12 transfections. P-values were calculated using the two-tailed unpaired Student’s t-test with equal variances in Graphpad Prism 6 (***P < 0.001). (d) HeLa cells were transfected with 200 ng of pri-miR-122 Drosha reporter. RNA was isolated after 24, 48 and 72 h, respectively. qPCR was performed for miR-122-5p, miR-122-3p, miR-16 and miR-191. MiR-191 was used for normalization. Error bars are SEM of four qPCR replicates. (e) SPC3649 inhibits Drosha processing of a pri-miR-122 reporter in HeLa cells. Cells were transfected with the Drosha reporter and after 6 h, antimiRs (6.25, 12.5, 25, 50 nM). Renilla and firefly luciferase were assayed after further 36 h. Error bars are SD of four transfections. Data were analyzed by ANOVA using Dunnett’s post-test in Graphpad Prism 6, comparing against the lowest dose in each group (*P < 0.05, ***P < 0.001). (f) Huh-7 cells were transfected with antimiRs (0.6, 2.5, 10 nM). After 3 days, a psiCHECK-2 miR-122-3p sensor reporter was transfected. Renilla and firefly activity were assayed 24 h later. Error bars indicate SD of three transfections. Data were analyzed by ANOVA using Dunnett’s post-test in Graphpad Prism 6, comparing against the lowest dose in each group (*P < 0.05). (g) Primary hepatocytes were extracted from a mouse, cultured and transfected with SPC3649 and SPCcon (50 nM). RNA was isolated after 24, 48 and 72 h. qPCR (SYBR Green) was performed for the pri-miR-122 sequence, using 36B mRNA for normalization. Error bars are SEM of three qPCR replicates. P-values were calculated using the two-tailed unpaired Student’s t-test with equal variances in Graphpad Prism 6 (*P < 0.001).
As explained above, we did not try to distinguish between inhibition of miR-122 biogenesis and sequestration of mature miR-122-5p by antimiRs by assaying the functional activity of endogenous miR-122-5p. However, an inhibition of pri-miRNA processing will not only suppress new synthesis of the mature 5p strand, but also any 3p miRNA. MiR-122-3p has been detected in Huh-7 by sequencing although counts were low (30) and we were unable to quantify it reproducibly in experiments using qPCR. Nevertheless, we attempted to examine the effects of the antimiRs on the activity of endogenous miR-122-3p using a highly sensitive miR-122-3p luciferase reporter containing a fully complementary target sequence to miR-122-3p. The reporter and SPC3649 were cotransfected into Huh-7 cells. After 3 days, we detected a small but significant derepression of Renilla luminescence by SPC3649, with possibly some effect from AMO-122, but no effects from the negative controls (Figure 4f). As SPC3649 and AMO-122 have no significant complementarity to miR-122-3p, these results support the notion that SPC3649 and possibly AMO-122 invade the stem structure of pri-miR-122 precursor in the nucleus and suppress the Drosha-mediated production of the 3p mature miRNA.
In clinical trials, patients with HCV infections are treated subcutaneously with miravirsen (SPC3649), which accumulates in liver and sequesters mature miR-122. Liver hepatocytes express high levels of miR-122. Murine pre-miR-122 is highly homologous to its human ortholog: the sequences of the 5p strands are identical and the murine 3p strand is shorter by 3 nt. Freshly prepared primary murine hepatocytes were treated separately with two doses of SPC3649 and SPCcon. Isolation of RNA and measurement of pri-miR-122 levels on three successive days showed significantly higher levels (2- to 3-fold) of pri-miR-122 after the SPC3649 treatment compared with SPCcon (Figure 4g). This contrasts with the data obtained from Huh-7 cells in which an accumulation was only seen on day 3 (Figure 4a). The data suggest that inhibition of miR-122 biogenesis in the nucleus and the cytoplasm of liver hepatocytes occurs and hence may contribute to the clinical activity of miravirsen.
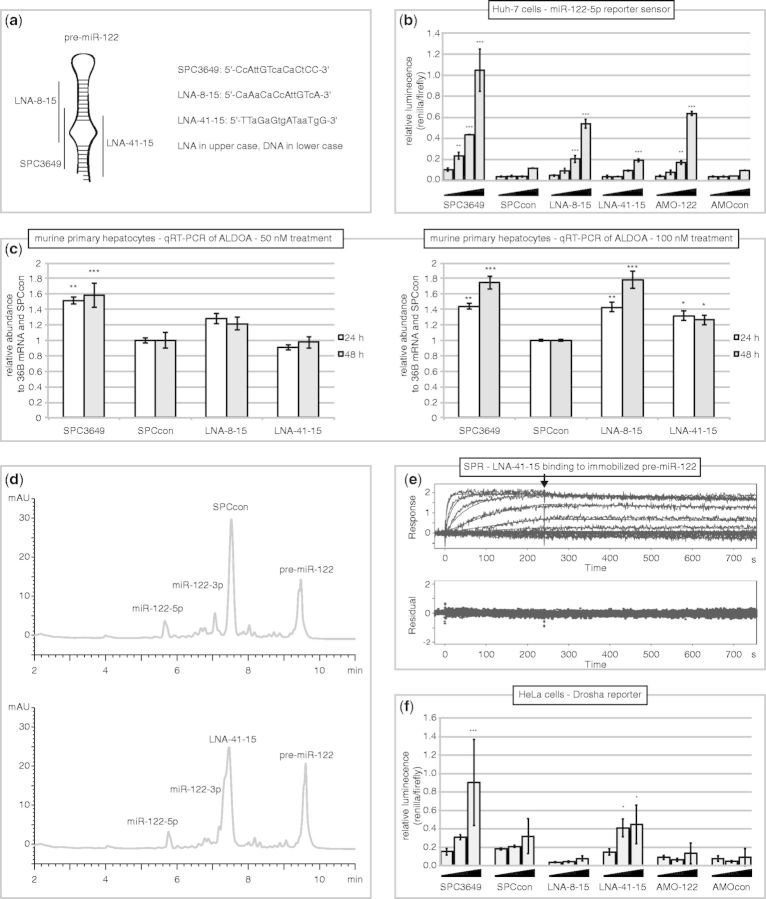
Taken together, we have shown that SPC3649 binds to the stem-loop structures of pri- and pre-miR-122 and inhibits miR-122 biogenesis. However, it is not known if and by how much inhibition of miR-122 processing contributes to the pharmacological activity of miravirsen. We therefore investigated additional oligonucleotides complementary to other regions of the miR-122 hairpin to consider the properties of SPC3649 in a wider context. Two 15-nt LNA/DNA chimera sequences were acquired (Figure 5a): LNA-8-15, which overlaps with SPC3649 and is complementary to the 3′-end of miR-122-5p, and LNA-41-15, which is complementary to the 3′-end of miR-122-3p.
Figure 5.
Targeting of miR-122-3p by LNA-41-15 inhibits biogenesis of miR-122. (a) Representative binding sites of the three LNA/DNA chimera antimiRs on pre-miR-122. (b) LNA-8-15 and LNA-41-15 derepress a miR-122-5p sensor construct in cells expressing miR-122-5p. Huh-7 cells were transfected with a miR-122-5p sensor construct based on the psiCHECK-2 reporter vector. Six hours later, the cells were transfected with increasing concentrations (0.6, 2.5, 10, 40 nM) of antimiRs. Renilla and firefly luciferase were assayed after further 48 h. Error bars are SD of three transfections. Data were analyzed by ANOVA using Dunnett’s post-test in Graphpad Prism 6, comparing against the lowest dose in each group (**P < 0.01, ***P < 0.001). (c) SPC3649, LNA-8-15 and LNA 41-15 derepress ALDOA expression. Murine primary hepatocytes were cultured and transfected with 50 nM (left) or 100 nM (right) of antimiRs. Cells were lysed after 24, 48 and 72 h, and total RNA was isolated. qPCR (SYBR Green) was performed for the ALDOA mRNA, using 36B4 mRNA for normalization. Error bars are SEM of three transfections (50 nM experiment) or two transfections (100 nM experiment). Data were analyzed by ANOVA using Dunnett’s post-test in Graphpad Prism 6, comparing against SPCcon for each time point (*P < 0.05, **P < 0.01, ***P < 0.001). (d) Pre-miR-122 was incubated with SPCcon or LNA-41-15 and 1 U of recombinant commercial Dicer was added. The reactions were incubated at 37°C for 2 h, quenched with EDTA and analyzed by LCMS apparatus. LNA-41-15 slows the cleavage of pre-miR-122 by Dicer compared with SPCcon by ∼15%. (e) SPR sensorgrams of LNA-41-15 bound to immobilized pre-miR-122 at six concentrations (166.7 nM; 3-fold dilutions) and 25°C: increased binding (response) is seen with higher concentrations of antimir; the arrow indicates a switch to running buffer and the beginning of the dissociation phase. For LNA-41-15 a KD of ∼240 pm (ka 1 × 106 M−1 s−1, kd 2.4 x 10−4 s−1) was determined. The plotted residuals of the fit are shown in the lower panel. (f) LNA-41-15 inhibits Drosha processing of a pri-miR-122 reporter in HeLa cells. Cells were transfected with the Drosha reporter plasmid and after 6 h, antimiRs (6.25, 12.5, 25 nM). Renilla and firefly luciferase were assayed after further 36 h. Error bars are SD of three transfections. Data were analyzed by ANOVA using Dunnett’s post-test in Graphpad Prism 6, comparing against the lowest dose in each group (* P < 0.05, *** P < 0.001).
First, we assayed all the antimiRs against a luciferase reporter sensor bearing a fully complementary target site to miR-122-5p in miR-122-expressing Huh-7 cells. This assay shows the consequences of inhibiting mature miR-122-5p, as well as the processing of endogenous pri- and pre-miR-122. As expected, the miR-122-5p antimiRs (SPC3649, LNA-8-15, AMO122) showed a strong concentration-dependent derepression of luciferase activity, with the greatest effect from SPC3649 (Figure 5b). LNA-41-15, which was inactive against an unrelated control reporter (data not shown), was also active against the miR-122-5p sensor. It showed ∼20% of the activity of SPC3649 at the two highest concentrations. As LNA-41-15 is complementary to the 3p arm of miR-122, this outcome was consistent with inhibition of pri- and/or pre-miR-122 processing. We consolidated these observations by examining the effects of the antimiRs on aldolase A, fructose-bisphosphate (ALDOA) (16) and GYS1 (40), two natural targets of miR-122-5p in primary murine hepatocytes. Indeed, in comparison with treatment with SPCcon, higher levels of ALDOA mRNA were found 1 and 2 days after transfection of hepatocytes with the two 5p antimiRs (SPC3649, LNA-8-15) (Figure 5c). In contrast, but consistent with the data from the reporter assay, the 3 p antimiR LNA-41-15 showed activity only at the 100 nM treatment. Similar results were obtained on GYS1 mRNA at 100 nM (data not shown).
We reran three assays in an effort to derive additional evidence that LNA-41-15 inhibited miR-122 biogenesis via binding to the 3p arm of the precursors: the in vitro Dicer assay, the SPR binding assay and the Drosha reporter assay. Incubation of pre-miR-122 with recombinant Dicer and SPCcon (Figure 5d, upper panel) yielded a similar result to that shown previously (Figure 1c). Assaying LNA-41-15 under identical conditions resulted reproducibly in visibly less cleavage of the substrate consistent with invasion of pre-miR-122 and inhibition of Dicer (Figure 5d, lower panel). We then measured the affinity of LNA-41-15 for immobilized pre-miR-122 on an SPR biosensor: LNA-41-15 bound strongly to pre-miR-122 (Figure 5e): it did not bind to miR-122-5p (data not shown). In addition, LNA-8-15 was unable to invade pre-miR-122 under the same conditions (data not shown). Finally, we used the Drosha reporter assay in HeLa cells to provide insight as to whether LNA-41-15 could inhibit processing of pri-miR-122. Consistent with the results of the biosensor binding assays, SPC3649 and LNA-41-15 inhibited sequence-specifically cleavage of this reporter mRNA, whereas LNA-8-15 showed no activity (Figure 5f).
In summary, we have shown that invasion of the hairpin structure of miR-122 precursors by two independent antimiRs targeting separately the 5p and 3p arms (SPC3659 and LNA-41-15, respectively) leads to its inhibition of processing by Drosha and Dicer. As LNA-41-15 does not bind to mature miR-122-5p, its inhibition of the function of the mature miRNA (miR-122-5p) (Figure 5b and c) likely derives from inhibition of the precursors. Extrapolating these observations to SPC3649 it appears likely that part of the pharmacological effects of miravirsen is attributable to its suppression of miR-122 biogenesis, in addition to its sequestration of mature miR-122-5p.
DISCUSSION
To date, any discussion of the mechanism of action of miravirsen centers upon its hybridization with mature miR-122-5p. Indeed, mature miR-122 5p is more abundant than its precursors in hepatocytes and has a long half-life (44). However, the target sequence of miravirsen is also present in the stem-loop structures of pre-miR-122 and pri-miR-122. As part of a program to discover ligands that bind to miRNA stem-loops, we have developed new assays that allow us to study regulation of the individual steps of miR-122 biogenesis (45). These include: a minimal in vitro enzymatic/HPLC-MS assay, which gauges semiquantitatively inhibition of Dicer cleavage of pre-miRNAs; an SPR assay to measure the kinetics of ligand interactions with a surface-bound pre-miRNA and a cell reporter assay to monitor Microprocessor-mediated cleavage of pri-miRNAs. Using these assays we discovered that SPC3649 (miravirsen) invades the stem structure of miR-122 precursors and inhibits Drosha and Dicer processing of pri-miR-122 and pre-miR-122, respectively (Figure 6).
Figure 6.
SPC3649 modulates the biogenesis of miR-122. Pri-miR-122 is processed by the Microprocessor to pre-miR-122, which is in turn processed by Dicer and passed to miRISC. SPC3649 invades the stem-loop structures of the primary transcript (pri-miRNA) as well as the pre-miRNA, and also sequesters the mature miRNA.
We demonstrated invasion and sequence-specific binding of pre-miR-122 by SPC3649 in two in vitro assays (Figure 2). This nanomolar interaction was shown to be functional in an enzymatic assay in which the cleavage of synthetic pre-miR-122 by Dicer was halted by SPC3649 (Figure 1). The biophysical properties of SPC3649 in vitro also correlated with its cellular activity. Using exogenously delivered pre-miR-122, which was processed into functional 5p and 3p strands in HeLa cells, we showed that cotransfection with SPC3649 led to its attenuated processing, causing accumulation of pre-miR-122 (Figure 3d). Similarly, the processing of pri-miR-122 in the nucleus of Huh-7 and primary murine hepatocytes by the Microprocessor was also inhibited by SPC3649, leading to accumulation of endogenous pri-miR-122 (Figure 4). To show that these effects were due to inhibited Drosha activity, we used a pri-miR-122 reporter in which the pri-miR-122 was inserted into the 3′-UTR of luciferase. In non-miR-122-expressing HeLa cells, the reporter mRNA was processed to mature miR-122-5p and this was accompanied by loss of luciferase activity. Treatment with SPC3649 led to derepression of luciferase consistent with strand invasion of pri-miR-122 and inhibition of processing (Figure 4).
Taken together, our assays demonstrated that miRNA biogenesis can be inhibited by high-affinity binding antimiRs (Figure 6). High affinity, however, is not sufficient for activity against miRNA precursors. SPC3649 and AMO-122 have similar affinities for pre-miR-122, yet the considerably longer AMO-122 was less active in cell assays. Furthermore, the slower rate of dissociation of AMO-122 from pre-miR-122 infers a longer lifetime of the interaction (i.e. drug residence time), at least in vitro. Consequently, the elevated activity of SPC3649 against miR-122 precursors may partly derive from better pharmacokinetics, e.g. superior cellular uptake or nuclear localization.
It is interesting to speculate whether inhibition of miR-122 biogenesis by SPC3649/miravirsen contributes to its pharmacological activity. Some insight on this was provided by data from experiments with LNA-41-15, an LNA/DNA chimera complementary to miR-122-3p. The activity of SPC3649 derives from its binding to mature miR-122-5p as well as its inhibition of miR-122 biogenesis. On the other hand, LNA-41-15 selectively inhibits miR-122 biogenesis alone, i.e. it does not bind to miR-122-5p. In a sensitive reporter assay for miR-122-5p activity, LNA-41-15 showed ∼20% of the activity of SPC3649 (Figure 5b), leading us to conclude that inhibition of biogenesis can contribute to the overall activity of an antimir, if it is able to invade the stem-loop structure of the precursors. However, our experimental design does not permit us to quantify such a contribution because SPC3649 and LNA-41-15 are independent sequences with their own pharmacokinetics and pharmacodynamics properties. Thus, it cannot be assumed that they inhibit miR-122 biogenesis to the same degree.
Today, it is accepted that regulation of several prominent miRNAs is controlled at the precursor level, for example, the LIN28/pre-let-7 interaction (46). Concerning miR-122, its precursors (not mature miR-122) cycle with the circadian rhythm, and one of the models proposed to explain this phenomenon had a key role for the precursors in regulation of circadian gene expression (39). The importance of targeting miRNA precursors pharmacologically awaits the development of new classes of high-affinity binding ligands.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [CRS133_130576 to M.S.; 205321_124720 to J.H] (in part). Funding for open access charge: ETH Zurich.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The biotinylated hairpin of pre-miR-122 was kindly provided by A. Brunschweiger. We are grateful to H. Towbin for reading the manuscript. We thank M. Zimmermann for synthesis of oligonucleotides and J. Zagalak for primers for the Huh-7 experiments.
REFERENCES
- 1.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 4.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 7.Gurtan AM, Lu V, Bhutkar A, Sharp PA. In vivo structure–function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA. 2012;18:1116–1122. doi: 10.1261/rna.032680.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 11.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 12.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selbach M, Schwanhaeusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 14.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 16.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 17.Jopling CL. Regulation of hepatitis C virus by microRNA-122. Biochem. Soc. Trans. 2008;36:1220–1223. doi: 10.1042/BST0361220. [DOI] [PubMed] [Google Scholar]
- 18.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm. Res. 2010;27:1788–1799. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 20.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 21.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 23.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 25.Choudhury NR, Michlewski G. Terminal loop-mediated control of microRNA biogenesis. Biochem. Soc. Trans. 2012;40:789–793. doi: 10.1042/BST20120053. [DOI] [PubMed] [Google Scholar]
- 26.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 27.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 28.Davies BP, Arenz C. A homogenous assay for micro RNA maturation. Angew. Chem. Int. Ed. Engl. 2006;45:5550–5552. doi: 10.1002/anie.200601332. [DOI] [PubMed] [Google Scholar]
- 29.Davies BP, Arenz C. A fluorescence probe for assaying micro RNA maturation. Bioorg. Med. Chem. 2008;16:49–55. doi: 10.1016/j.bmc.2007.04.055. [DOI] [PubMed] [Google Scholar]
- 30.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HY, Doudna JA. TRBP alters human precursor microRNA processing in vitro. RNA. 2012;18:2012–2019. doi: 10.1261/rna.035501.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Childs JL, Disney MD, Turner DH. Oligonucleotide directed misfolding of RNA inhibits Candida albicans group I intron splicing. Proc. Natl Acad. Sci. USA. 2002;99:11091–11096. doi: 10.1073/pnas.172391199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elayadi AN, Braasch DA, Corey DR. Implications of high-affinity hybridization by locked nucleic acid oligomers for inhibition of human telomerase. Biochemistry. 2002;41:9973–9981. doi: 10.1021/bi025907j. [DOI] [PubMed] [Google Scholar]
- 35.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geschwindner S, Carlsson JF, Knecht W. Application of optical biosensors in small-molecule screening activities. Sensors. 2012;12:4311–4323. doi: 10.3390/s120404311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darfeuille F, Hansen JB, Orum H, Di Primo C, Toulme JJ. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res. 2004;32:3101–3107. doi: 10.1093/nar/gkh636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Agata R, Spoto G. Artificial DNA and surface plasmon resonance. Artif. DNA PNA XNA. 2012;3:45–52. doi: 10.4161/adna.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatfield D, Le Martelot G, Vejnar CE, Gerlach D, Schaad O, Fleury-Olela F, Ruskeepaa AL, Oresic M, Esau CC, Zdobnov EM, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, Bhat B, Swayze EE, Bennett CF, Esau C. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37:70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allegra D, Mertens D. In-vivo quantification of primary microRNA processing by Drosha with a luciferase based system. Biochem. Biophys. Res. Commun. 2011;406:501–505. doi: 10.1016/j.bbrc.2011.02.055. [DOI] [PubMed] [Google Scholar]
- 43.Husken D, Asselbergs F, Kinzel B, Natt F, Weiler J, Martin P, Haner R, Hall J. mRNA fusion constructs serve in a general cell-based assay to profile oligonucleotide activity. Nucleic Acids Res. 2003;31:e102. doi: 10.1093/nar/gng103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011;39:5692–5703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Towbin H, Wenter P, Guennewig B, Imig J, Zagalak JA, Gerber AP, Hall J. Systematic screens of proteins binding to synthetic microRNA precursors. Nucleic Acids Res. 2013;41:e47. doi: 10.1093/nar/gks1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.