Abstract
The establishment of DNA synthesis during the S phase is a multistep process that occurs in several stages beginning in late mitosis. The first step is the formation of a large prereplicative complex (pre-RC) at individual replication origins and occurs during exit from mitosis and entry into G1 phase. To better understand the genetic requirements for pre-RC formation, we selected chromosomal suppressors of a temperature-sensitive cdc6-4 mutant defective for pre-RC assembly. Loss-of-function mutations in the chromatin-modifying genes SIR2, and to a lesser extent in SIR3 and SIR4, suppressed the cdc6-4 temperature-sensitive lethality. This suppression was independent of the well-known silencing roles for the SIR proteins at the HM loci, at telomeres, or at the rDNA locus. A deletion of SIR2 uniquely rescued both the DNA synthesis defect of the cdc6-4 mutant and its severe plasmid instability phenotype for many origins. A SIR2 deletion suppressed additional initiation mutants affecting pre-RC assembly but not mutants that act subsequently. These findings suggest that Sir2p negatively regulates the initiation of DNA replication through a novel mechanism and reveal another connection between proteins that initiate DNA synthesis and those that establish silent heterochromatin in budding yeast.
Keywords: DNA replication, SIR2, deacetylase
DNA replication occurs during the S phase of each cell cycle and initiates at discrete sites called origins of replication (Newlon and Theis 1993). Although DNA synthesis is restricted to the S phase, the ability to initiate replication is determined by molecular events at each origin beginning during anaphase of the previous cell cycle and continuing into early G1 phase. During this period, the assembly of prereplicative complexes (pre-RCs) directly at origins establishes replication competence. Pre-RCs contain the proteins necessary but not sufficient to initiate DNA replication (for review, see Diffley 1996; Bell and Dutta 2002). Subsequent events during G1 lead to origin unwinding and the recruitment of polymerases that initiate bidirectional DNA synthesis. Importantly, the ability to reinitiate replication from any one origin is not possible once cells have entered S phase because pre-RC assembly is prevented during the S, G2, and M phases. The inhibition of pre-RC assembly occurs through a variety of mechanisms controlling the availability and nuclear localization of key pre-RC components (Labib et al. 1999; Nguyen et al. 2001). This temporal determination of replication is necessary to prevent reinitiation of DNA replication within a single cell cycle that, if allowed to occur, would cause increases in ploidy and promote genomic instability.
In Saccharomyces cerevisiae, the origin recognition complex (ORC) determines the sites that initiate DNA replication by binding to a bipartite consensus sequence within the origin (Bell and Stillman 1992; Rao and Stillman 1995). ORC then serves as a “landing pad” for the assembly of the multiple initiation proteins at origins prior to DNA synthesis (Stillman 1996). The ORC–origin interaction occurs in a nucleosome-free region for the ARS1 origin, and both ORC binding and the initiation of replication are inhibited by nucleosomes occupying this binding site (Simpson 1990; Lipford and Bell 2001). Positioning nucleosomes distal to the origin also inhibits initiation but not ORC binding, additionally suggesting that protein–nucleosomal contacts or chromatin structure is important for pre-RC assembly (Lipford and Bell 2001).
The first step in pre-RC assembly occurs when the Cdc6 protein binds to ORC and promotes MCM (minichromosome maintenance) helicase loading (Liang et al. 1995; Cocker et al. 1996; Aparicio et al. 1997; Tanaka et al. 1997). Cdc6p is a member of the large AAA+ family of ATP-binding proteins (Neuwald et al. 1999; Davey et al. 2002). This family includes proteins that use ATP binding or hydrolysis to promote conformational changes in nucleic acids or protein substrates. An X-ray crystallographic structure of an archaebacterial Cdc6-like protein bound to ADP (Liu et al. 2000) has revealed that it shares striking structural similarity with other members of this family, including the bacterial initiator protein DnaA (Erzberger et al. 2002), the pentameric γ–δ clamp loader (an analog of eukaryotic RFC; Jeruzalmi et al. 2001), and the human NSF-d2 hexamer that is involved in intracellular protein trafficking (Lenzen et al. 1998; Yu et al. 1998). Molecular genetic evidence indicates that ATP binding and/or hydrolysis is required for Cdc6p activity because mutation of conserved residues within the ATP-binding motifs of Cdc6p homologs either impairs or eliminates cellular growth and the ability to load the MCM helicase (Perkins and Diffley 1998; DeRyckere et al. 1999; Weinreich et al. 1999; Frolova et al. 2002). We had shown previously that substitutions of a conserved lysine residue in the Cdc6p Walker A box, which is predicted to contact β–γ phosphates of ATP, compromised or eliminated CDC6 function (Weinreich et al. 1999). Although Cdc6p may act as a direct loader of the MCM helicase, there are no mechanistic data demonstrating how Cdc6p performs its essential function during initiation.
In addition to its essential role in DNA replication, ORC is involved in the formation of heterochromatin (Bell et al. 1993; Foss et al. 1993). ORC is required for heterochromatin assembly at the two silent mating-type HM loci in budding yeast, it influences heterochromatin structure at telomeres (Fox et al. 1997), and is present at heterochromatic regions in Drosophila (Pak et al. 1997). ORC has been shown to interact with both the Drosophila and Xenopus heterochromatin protein 1 (HP1). Budding yeast Orc1p recruits Sir1p to the HM loci (Triolo and Sternglanz 1996; Fox et al. 1997), which is required to help establish the silent chromatin state. However, the Orc1p domain that recruits Sir1p to the silencers is not required for ORC's role in DNA replication (Bell et al. 1995).
There are three additional silent information regulator genes (SIR2–4; Rine and Herskowitz 1987) that have well-established roles for formation of an alternative chromatin structure at the heterochromatic HM loci (Rusche et al. 2003) and at telomeres (Gottschling et al. 1990). SIR2 is the founding member of a conserved family of NAD+-dependent histone and protein deacetylases (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000). SIR2 is required for transcriptional silencing at the HM loci and telomeres as well as suppression of recombination between the 100–200 directly repeated copies of the ribosomal DNA (rDNA) genes (Gottlieb and Esposito 1989). This latter function of Sir2p promotes longevity in yeast (Guarente 2000; Sinclair 2002). Sir2p deacetylates specific acetyl-lysine residues within the N-terminal tails of histones H3 and H4 in S. cerevisiae, and this catalytic activity is required for its ability to function as a silencing protein. Mammalian SIR2 homologs have been shown to deacetylate nonhistone proteins including p53 (Luo et al. 2001; Vaziri et al. 2001), tubulin (North et al. 2003), and also the TAFI68 subunit of RNA polymerase I (Muth et al. 2001), raising the possibility that histones may not be the only targets of the budding yeast Sir2p. The closest murine SIR2 homolog, SIRT1, is essential for normal development, and few homozygous mice survive to birth (Cheng et al. 2003). SIR3 and SIR4 encode chromatin-binding proteins unique to budding yeast. Sir3p and Sir4p interact with hypoacetylated histone H3 and H4 N-terminal tails (Hecht et al. 1995; Carmen et al. 2002) and are essential for heterochromatin formation at the silent-mating-type loci and at telomeres. Sir4p is thought to initiate the formation of heterochromatin through its ability to interact with multiple proteins, including the DNA-binding protein Rap1p (Luo et al. 2002), which is present at telomeres and silencers; histones H3 and H4 (Hecht et al. 1995); and Sir1p, Sir2p, and Sir3p (Gasser and Cockell 2001).
To further understand the process of pre-RC assembly, we isolated chromosomal suppressors of a cdc6 temperature-sensitive allele that altered a conserved lysine residue (K114A) in the ATP-binding pocket. Some of these suppressors mapped to (and inactivated) the heterochromatin genes SIR2, SIR3, and SIR4. Our findings established that inactivation of the SIR2–4 genes suppress an initiation mutant, although likely by different genetic pathways, and that this suppression is independent of the known roles for the SIRs in the cell. Because a deletion of SIR2 (but not of SIR3 or SIR4) suppressed additional pre-RC mutants and reversed many of the replication defects of cdc6-4, we suggest that Sir2p is acting to inhibit pre-RC assembly through its enzymatic activity as a protein deacetylase.
Results
Deletion of SIR2, SIR3, or SIR4 suppresses a replication initiation mutant
A strain containing the cdc6-4 mutation, which changes the conserved lysine at position 114 in the Walker A motif to an alanine, was plated at 37°C to select spontaneous suppressors of its temperature-sensitive lethality. We isolated multiple independent clones that could grow at 37°C but observed that many of the suppressor strains were sterile because they could not mate with a strain of the opposite mating type. There are four SIR genes in yeast required for full transcriptional repression at the silent-mating-type cassettes, HMR and HML, and mutations within these chromatin-modifying genes lead to defects in mating (Rine and Herskowitz 1987). Therefore, we tested whether wild-type plasmid copies of the SIR genes complemented the sterility of the suppressors. The mating defects of two strains were complemented by SIR2, four were complemented by SIR3, and one was complemented by SIR4. None of the mating-defective strains was complemented by SIR1. We subsequently confirmed that in each case the suppressor of temperature sensitivity was linked to the same SIR gene that complemented the mating deficiency (see Materials and Methods), suggesting that a loss of SIR2–4 function was likely also responsible for the suppression of the cdc6-4 mutant.
We tested whether null mutations in the SIR genes would suppress the cdc6-4 temperature-sensitive defect. ORC also has a role in silencing by binding to the HM silencers and recruiting Sir1p through an Orc1p N-terminal domain that is dispensable for its role in DNA replication (Bell et al. 1995; Triolo and Sternglanz 1996; Gardner et al. 1999). Deleting SIR2, SIR3, or SIR4 (but not SIR1 or amino acids 2–235 of Orc1p) suppressed the temperature sensitivity of cdc6-4. We examined the degree of suppression by spotting serial 10-fold dilutions of each double mutant strain both at the permissive temperature of 25°C and also at the nonpermissive temperature of 37°C to solid media (Fig. 1A). Deleting SIR2 almost fully restored wild-type growth at 37°C. Deletions of SIR3 and SIR4 were less efficient suppressors than a SIR2 deletion, by 10-fold and 100-fold, respectively. However, deletion of SIR1 or of the ORC1 N terminus had little or no ability to suppress cdc6-4. The deletions of SIR2, SIR3, or SIR4 were required to suppress the cdc6-4 temperature sensitivity because transformation of the corresponding wild-type SIR gene into the sirΔ cdc6-4 double-mutant strains reversed both the TS+ phenotype and the sterility (data not shown). Because deletion of SIR2 (but not of SIR3 or SIR4) rescued the plasmid stability and S-phase defects of the cdc6-4 mutant (see below), the SIR3 and SIR4 deletions are likely partially rescuing the growth of cdc6-4 by a different mechanism than the loss of SIR2, which is the focus of this report.
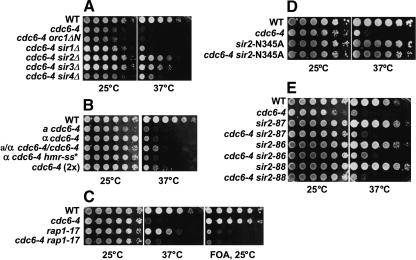
Figure 1.
Tenfold serial dilutions of strains were spotted onto plates and incubated at 25°C (3 d) and 37°C (2 d). (A) Deletion of SIR2-4 suppresses the temperature-sensitivity of cdc6-4. M138 (W303-1A), M386 (cdc6-4), M636 (cdc6-4 orc1ΔN6–235), M638 (cdc6-4 sir1Δ), M922 (cdc6-4 sir2Δ), M971 (cdc6-4 sir3Δ), and M974 (cdc6-4 sir4Δ). (B) Simultaneous MATa and MATα expression does not suppress cdc6-4. M138 (WT), M386 (MATa cdc6-4), M599 (MATα cdc6–4), M1101 (MATa/MATα cdc6-4/cdc6-4), M1102 (MATα cdc6-4 hmra-ss*), and M576 (MATα cdc6–4[2×]). (C) Disruption of telomeric silencing does not suppress cdc6-4. M1020 (WT, VIIL::URA3-tel), M1021 (cdc6-4 VIIL::URA3-tel), AJL369-4d (rap1-17 VIIL::URA3-tel), M1010 (cdc6-4 rap1-17 VIIL::URA3-tel). (D) Loss of Sir2p deacetylase activity suppresses cdc6-4. M138 (WT), M386 (cdc6-4), M795 (sir2-N345A), and M1100 (cdc6-4 sir2-N345A). (E) Loss of Sir2p rDNA localization does not suppress cdc6-4. M138 (WT), M386 (cdc6-4), M1117 (sir2-87), M1118 (cdc6-4 sir2-87), M1155 (sir2-86), M1164 (cdc6-4 sir2-86), M1156 (sir2-88), and M1166 (cdc6-4 sir2-88).
The loss of mating-type or telomeric silencing does not indirectly suppress the cdc6-4 mutant
Because SIR2 is required for transcriptional silencing at the HM loci and telomeres, we first tested whether a deletion of SIR2 could be indirectly affecting replication in the cdc6-4 mutant through its roles in these processes. However, a priori, it seemed very unlikely that a loss of HM or telomeric silencing alone suppressed the cdc6-4 mutant because the disruption of SIR2, SIR3, or SIR4 completely abolished both TPE and HM silencing, but these same deletions had a 100-fold differential effect on the growth of cdc6-4 at 37°C (Fig. 1A).
Disruption of SIR2, SIR3, or SIR4 in a haploid strain causes expression of the genes at both the HMRa and HMLα silent mating-type loci with accompanying transcriptional changes at the mating-type-responsive genes (Rusche et al. 2003). We, therefore, compared the cdc6-4 temperature sensitivity in cells expressing a, α, or both a and α information. The MATa, MATα, and the MATa/MATα cdc6-4 diploid were all temperature sensitive (Fig. 1B). We also constructed a MATα cdc6-4 HMRa-ss* haploid strain that was completely defective for silencing at HMRa because of a cis mutation of the Rap1p-binding site in the synthetic silencer (McNally and Rine 1991). The a and α gene expression caused by a loss of silencing at HMRa in the haploid also did not suppress the cdc6-4 temperature sensitivity (Fig. 1B). These data indicate that expression of a, α, or both a and α mating-type genes does not suppress the temperature sensitivity of the cdc6-4 mutant. Neither did a duplication (tandem integration) of cdc6-4 rescue the temperature-sensitive phenotype (Fig. 1B), even though this strain increased cdc6-4 mRNA expression 7.8-fold (data not shown). Therefore, increasing the number of cdc6-4 copies either in the diploid or with a double integration also does not suppress its temperature sensitivity.
We next examined whether a loss of telomeric silencing could suppress the cdc6-4 mutant because the Sir2–4 proteins are required for the transcriptional silencing of genes near the telomeres, termed the telomere position effect (TPE; Gottschling et al. 1990; Aparicio et al. 1991). The specialized telomeric chromatin structure requiring the SIR proteins also contributes to the late replication timing of origins located within the subtelomeric domain (Stevenson and Gottschling 1999; Cosgrove et al. 2002), and it was possible that a loss of TPE might be affecting more origins than expected. Rap1p is required for formation of the telomeric and subtelomeric chromatin structure in part through recruiting Sir3p and Sir4p to the telomeres (Lustig et al. 1990; Kyrion et al. 1993; Cockell et al. 1995; Strahl-Bolsinger et al. 1997; Moretti and Shore 2001). Therefore, we crossed cdc6-4 to the TS+ rap1-17 allele that is known to abolish telomeric silencing and the Rap1p–Sir4p interaction. The cdc6-4 rap1-17 double mutant lost TPE as evidenced by the derepression of the URA3 reporter near telomere VII-L (Fig. 1C). However, all of the double mutants we recovered were still temperature sensitive, indicating that the loss of telomeric silencing or of the specialized telomeric chromatin structure per se is not capable of suppressing the cdc6-4 initiation mutant.
The loss of Sir2p deacetylase activity but not its rDNA localization suppresses the growth of the cdc6-4 initiation mutant
SIR2 encodes an NAD+-dependent histone deacetylase that targets acetyl lysines at the histone H3 and H4 N termini. An N345A mutation in the catalytic domain of Sir2p abolishes its deacetylase activity together with its ability to transcriptionally silence its targets (Imai et al. 2000), although the N345A and additional Sir2p catalytic mutant proteins still bind in a spatially restricted pattern at the HMR-E silencer and the rDNA locus (Hoppe et al. 2002; Rusche et al. 2002). We isolated cdc6-4 sir2-N345A double mutants and observed that this catalytically inactive sir2 mutant suppressed the cdc6-4 temperature sensitivity similarly to a deletion of SIR2 (Fig. 1D). Therefore, a loss of Sir2p enzymatic activity suppressed the cdc6-4 mutant.
Sir2p has a unique role within the nucleolus independent of the other SIR proteins. Sir2p inhibits recombination between the 100–200 copies of the directly repeated rDNA gene cassettes (Gottlieb and Esposito 1989) and also silences Pol II transcribed genes placed directly adjacent to or within the rDNA locus (Bryk et al. 1997; Smith and Boeke 1997; Buck et al. 2002). Sir2p, Net1p, and Cdc14p are subunits of the nucleolar RENT complex (regulator of nucleolar silencing and telophase exit), and Sir2p localization to the nucleolus requires Net1p (Shou et al. 1999; Straight et al. 1999; Visintin et al. 1999). It was therefore possible that a loss of the nucleolar function of Sir2p suppressed the cdc6-4 mutant in an indirect manner. We examined whether separation-of-function sir2 mutations that cause a loss of Sir2p localization to the nucleolus (but still allow silencing at the HM loci and at telomeres) suppressed the cdc6-4 mutant. A sir2-87 mutant that deletes the last 15 amino acids of Sir2p is specifically defective for rDNA silencing (Cuperus et al. 2000). The Sir2-87 protein does not localize to the nucleolus because of an impaired interaction with Net1p. We verified that the sir2-87 mutant was defective for rDNA silencing and crossed it to the cdc6-4 mutant. However, because the double-mutant sir2-87 cdc6-4 strains were still temperature sensitive (Fig. 1E), a loss of rDNA localization in this mutant did not suppress the cdc6-4 mutant. Two additional rDNA silencing alleles (sir2-86 and sir2-88), that we confirmed were defective for rDNA silencing (Cuperus et al. 2000), also did not suppress the temperature sensitivity of the cdc6-4 mutant (Fig. 1E). Taken together, these data indicate that a loss of Sir2p function at the rDNA locus is not responsible for the cdc6-4 suppression seen in the sir2-null mutant.
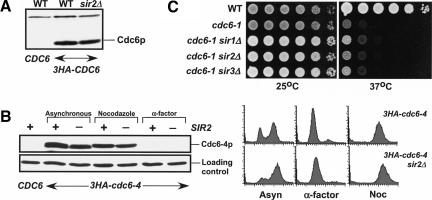
Cdc6p is an unstable protein that has a half-life of ∼5 min (Drury et al. 2000). This instability varies during the cell cycle and is mediated by ubiquitin-mediated proteolysis of Cdc6p. Although SIR2 is not known to regulate ubiquitin-mediated protein stability, we tested whether deletion of SIR2 had an effect on Cdc6p protein levels. We compared total cell extracts of wild-type and sir2Δ strains expressing 3HA-Cdc6p as well as 3HA-Cdc6-4p at the permissive temperature. Because the levels of wild-type and Cdc6-4p are the same in a wild-type strain and in the strains deleted for SIR2 (Fig. 2A,B), a deletion of SIR2 is not causing a significant stabilization of Cdc6p. We also compared Cdc6-4p levels in SIR2WT and sir2Δ strains using G2/M and G1 synchronized cells (Cdc6p is highly unstable in α-factor-arrested cells) and again saw no significant differences in Cdc6-4p abundance (Fig. 2B). In addition, deletion of SIR1, SIR2, or SIR3 does not suppress the cdc6-1 temperature sensitivity (Fig. 2C), indicating that the loss of the SIR genes is not bypassing the requirement for CDC6.
Figure 2.
Deletion of SIR2 does not alter the abundance of the wild-type or Cdc6-4 proteins. (A) 12CA5 Western blot for Cdc6p from asynchronous total cell extracts of W303-1A (untagged), M276 (3HA-CDC6), and M1065 (3HA-CDC6 sir2Δ). (B) 12CA5 Western blot of Cdc6-4p and FACS samples from asynchronous cells and cells completely arrested in G2/M phase using 15 μg/mL nocodazole or in G1 phase using 10 μg/mL α-factor; W303-1A (untagged), M1257 (3HA-cdc6-4), M1274 (3HA-cdc6-4 sir2Δ), M1299 (3HA-cdc6-4 sir2Δ hmlΔ). (C) Deletion of SIR1, SIR2, or SIR3 does not suppress the cdc6-1 temperature sensitivity. W303-1A (WT), M379 (cdc6-1), M1103 (cdc6-1 sir1Δ), M1104 (cdc6-1 sir2Δ), M1105 (cdc6-1 sir3Δ).
Deletion of SIR2 rescues the DNA replication defect of cdc6-4
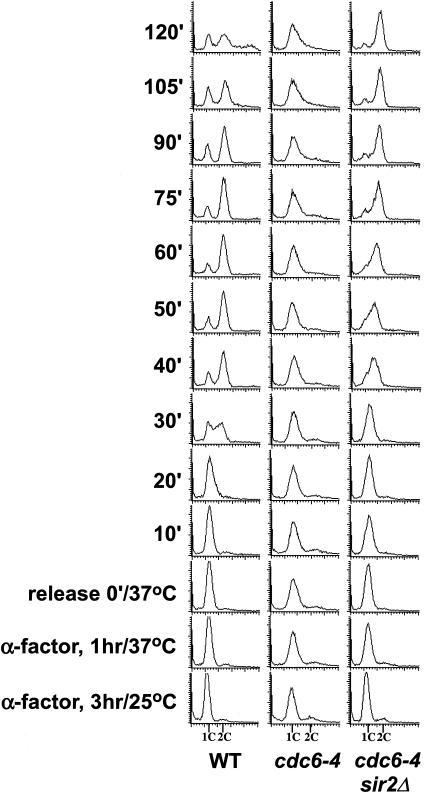
If deletion of SIR2 is suppressing the initiation defect of the cdc6-4 mutant, the double-mutant strain should progress through S phase when released from a G1 block at the restrictive temperature. The cdc6-4 mutant progresses through S phase very slowly even at the permissive temperature and is defective in loading the MCM proteins to chromatin (Weinreich et al. 1999). The wild-type, cdc6-4, and cdc6-4 sir2Δ strains were arrested in G1 phase, shifted to the restrictive temperature at the G 1-arrest point, and then released into the cell cycle at the restrictive temperature for cdc6-4 and examined for DNA content by flow cytometry (Fig. 3). The wild-type strain entered S phase within 30 min of the G1 release and had largely completed S phase by 60 min as evidenced by the subsequent reappearance of cells with 1C DNA content. The cdc6-4 strain, however, remained arrested at the G1 stage with largely a 1C DNA content and did not progress through S phase. In contrast to the cdc6-4 mutant, the sir2Δ cdc6-4 double mutant exhibited a nearly wild-type S phase because the cells begin S phase between 30 and 40 min. These cells do not re-enter G1 phase with the same timing as the wild-type cells, perhaps indicating that DNA replication is not completed as accurately as in wild type or that additional functions of Cdc6p during G2/M are not effectively bypassed by a deletion of SIR2. Asynchronous cultures of cdc6-4 sir2Δ grown at 25°C also exhibit a substantial portion of cells in the G2/M phase, indicative of a cell cycle delay (data not shown). Importantly, the deletion of SIR2 allowed S phase to occur in the cdc6-4 mutant in a manner reflecting its nearly wild-type growth at 37°C, indicating that the loss of SIR2 is suppressing the initiation defect of cdc6-4.
Figure 3.
Deletion of SIR2 promotes entry into S phase in the cdc6-4 mutant. M138 (WT), M386 (cdc6-4), and M940 (cdc6-4 sir2Δ hmlΔ) were arrested with α-factor for 3 h at 25°C, shifted for 1 h to 37°C,andthenreleasedfrom thearrestinto fresh YPD medium at 37°C. Cell cycle progression was monitored by flow cytometry as described (Weinreich et al. 1999).
Loss of SIR2 rescues additional initiation mutants, but not mutants that act after pre-RC assembly
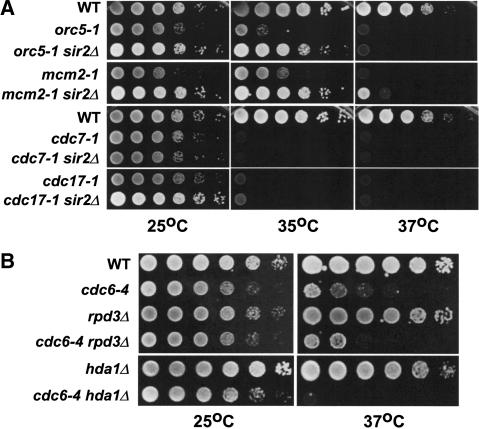
Given that the loss of SIR2 suppressed the cdc6-4 initiation mutant, we tested whether temperature-sensitive mutations in other initiation mutants could be suppressed by deletion of SIR2. Deletion of SIR2 almost completely suppressed the temperature-sensitive growth of the orc5-1 and mcm2-1 pre-RC mutants at 35°C, but not at 37°C (Fig. 4A). In contrast, deletion of SIR2 did not suppress the temperature-sensitive phenotypes of the cdc7-1 (protein kinase) or the cdc17-1 (DNA polymerase α) mutants that act in steps subsequent to pre-RC assembly at temperatures ranging from 37°C to 30°C (Fig. 4A). Because the ORC and MCM genes are required for pre-RC assembly together with CDC6, this suggested that SIR2 negatively regulates initiation at the level of pre-RC assembly but not at stages subsequent to this step.
Figure 4.
(A) Deletion of SIR2 can partially rescue additional pre-RC mutants. M138 (WT), M198 (orc5-1), M1096 (orc5-1 sir2Δ), M359 (mcm2-1), M1097 (mcm2-1 sir2Δ), M444 (cdc7-1), M1098 (cdc7-1 sir2Δ), M354 (cdc17-1), and M1099 (cdc17-1 sir2Δ). (B) Deletion of the histone deacetylases RPD3 or HDA1 will not suppress cdc6-4. M138 (WT), M386 (cdc6-4), WJY140 (rpd3Δ), M1080 (cdc6-4 rpd3Δ), M1161 (hda1Δ), and M1174 (cdc6-4 hda1Δ).
We also examined the specificity of the sir2 suppression by testing whether additional histone deacetylase mutants could suppress the cdc6-4 temperature sensitivity. Based on sequence comparisons among organisms, RPD3, HDA1, and SIR2 form three distinct classes of histone deacetylases; class I, II, and III, respectively (Marks et al. 2001; Kurdistani and Grunstein 2003). RPD3 encodes a global histone deacetylase that negatively regulates the expression of many genes and, importantly, a deletion of RPD3 has been shown to advance the replication time of both early and late origins in a manner correlated with the loss of histone acetylation near origins (Vogelauer et al. 2002). Rpd3p deacetylates all four histones in vitro, but interestingly, it does not deacetylate H4 K16, an important Sir2p target (Rusche et al. 2003). The Hda1p histone deacetylase also negatively regulates transcription of many genes throughout the genome, but it preferentially acts in a subtelomeric region termed the HAST domain (Robyr et al. 2002) and is thought to deacetylate only histones H3 and H2A. We constructed double mutants of rpd3Δ or hda1Δ with cdc6-4 and found that deletions of these histone deacetylases would not suppress the temperature sensitivity of the cdc6-4 mutant (Fig. 4B). This indicates that loss of a SIR2 specific deacetylase function is required to suppress cdc6-4.
Deletion of SIR2 rescues the plasmid instability phenotype of cdc6-4
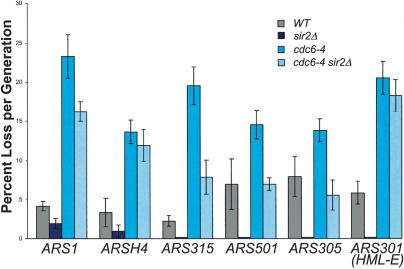
As a third test of the effect of SIR2 on replication initiation, we compared the plasmid loss rates of wild-type, cdc6-4, and cdc6-4 sir2Δ strains. Initiation mutants exhibit a high plasmid loss rate because they fail to initiate DNA replication from the plasmid origin in every cell cycle, and this is also true for cdc6 mutants (Hogan and Koshland 1992). The high plasmid loss rate of an initiation mutant can be rescued by increasing the frequency at which the plasmid origin fires or by adding additional origins to the plasmid, which increases the probability that any one origin will fire. If deleting SIR2 is causing an increase in initiation frequencies at specific origins, then the sir2Δ should also reverse the plasmid instability phenotype of the cdc6-4 mutant. We therefore measured plasmid loss rates in wild-type, sir2Δ, cdc6-4, and the cdc6-4 sir2Δ strains at the permissive temperature for six different origin sequences. Wild-type strains typically lose plasmids at rates between 3% and 6% per generation under nonselective conditions, and this was true for all of the origins we tested (Fig. 5). In the wild-type background, deletion of SIR2 slightly improved the plasmid loss rates for the ARS1 and ARSH4 plasmids but substantially improved loss rates for the remaining origins. The cdc6-4 mutant had a highly elevated plasmid loss rate that varied from 15% to 25% per generation for these six origins. Significantly, deletion of SIR2 rescued the high plasmid loss rate of cdc6-4 but only for some of the origins we tested, suggesting that SIR2 does not negatively regulate all origins with the same efficiency. A sir2Δ completely reversed the instability phenotype for the ARS305 and ARS501 plasmids and substantially reversed the loss rate of the ARS315 plasmid. However, a sir2Δ had a partial effect on the ARS1 plasmid and no effect on the loss rates of the ARSH4 and HML-E (ARS301) plasmids. Because ARS1, ARS301, ARS305, and ARS315 were present within an otherwise identical plasmid context, the SIR2-dependent variation in plasmid stability in the cdc6-4 strain was caused by the origin sequences present on the plasmids.
Figure 5.
Deletion of SIR2 rescues the plasmid instability defect conferred by cdc6-4 at certain origins and improves plasmid stability in wild-type cells. Values were determined as in Dani and Zakian (1983) and are reported are percentage plasmid loss rate per generation in nonselective medium at 25°C. They represent the average of at least four measurements. The ARS501 plasmid is pR151 (Ferguson et al. 1991), ARSH4 is pRS416, and the remaining plasmids have ∼300–500-bp origin fragments replacing the ARS1 origin on pARS1-WT (Marahrens and Stillman 1992).
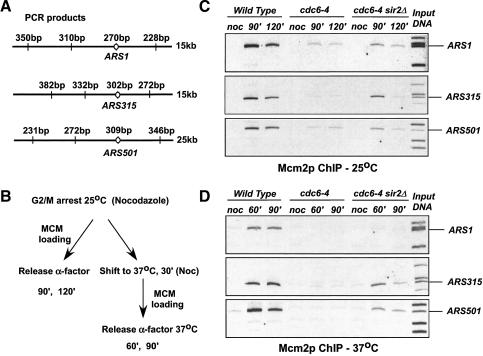
Deletion of SIR2 promotes MCM origin binding in the cdc6-4 mutant
Cdc6p is required to load MCM proteins at origins of replication (Cocker et al. 1996; Aparicio et al. 1997; Tanaka et al. 1997). We therefore used chromatin immunoprecipitation (ChIP) to address whether MCM protein could be loaded in the cdc6-4 and cdc6-4 sir2Δ mutants at several of the origins that were differentially affected by deletion of SIR2 in the plasmid instability assay. We used PCR primers to amplify short sequences that contained the ARS1, ARS315, and ARS501 origins following ChIP. Adjacent non-origin sequences within several kilobases of the ARS elements were amplified as negative controls (Fig. 6A). Wild-type, cdc6-4, and cdc6-4 sir2Δ cells were arrested at the permissive temperature in G2/M and then either released at the permissive temperature or raised to the nonpermissive temperature of 37°C prior to release into G1. During progression into G1, MCM proteins are loaded at origins in a CDC6-dependent manner (outlined in Fig. 6B). We used a monoclonal antibody against Mcm2p to identify MCM protein loading. In all cases, no Mcm2p was associated with the origins in G2/M-arrested cells (Fig. 6C,D). In the wild type, Mcm2p specifically associated with origin-containing fragments for these three origins at 25°C and 37°C during release to the G1 arrest. For cdc6-4, we observed a similar, weak recovery for all of the origins at 25°C, but the cdc6-4 mutant was completely defective for loading Mcm2p at 37°C. Deletion of SIR2 in the cdc6-4 background had a differential effect for the origins we tested. We saw a partial recovery of ARS1 Mcm2p loading at 25°C but very little at 37°C. For the ARS315 and ARS501 origins, deletion of SIR2 allowed more efficient Mcm2p loading at both the permissive and nonpermissive temperatures. These data correlate well with the plasmid instability assays (Fig. 5) and also the flow cytometry data (Fig. 3), showing that deleting SIR2 promoted S phase in the cdc6-4 mutant.
Figure 6.
Deletion of SIR2 restores Mcm2p loading at origins. ChIP assays were performed as described in Materials and Methods using cells grown in YPD medium to mid-log phase, arrested in G2/M with 15 μg/mL nocodazole, and then released into YPD medium containing 5 μg/mL α-factor for 60, 90, or 120 min, so that 95% of cells had a 1C DNA content as determined by flow cytometry. W303-1A (WT), M386 (cdc6-4), and M940 (cdc6-4 sir2Δ hmlΔ). A representative input DNA PCR sample is shown for each origin examined.
Discussion
We have uncovered a negative regulatory role for Sir2p in DNA replication using a cdc6-4 mutant defective for initiation. Because a deletion of SIR2 promoted a nearly wild-type S phase in the cdc6-4 mutant background, a loss of Sir2p function is restoring the ability to replicate DNA (and therefore initiate DNA replication) on a genome-wide level. A SIR2 deletion also partially rescued the temperature-sensitive growth of two other pre-RC mutants, orc5-1 and mcm2-1, but not of replication mutants that act after pre-RC assembly. This suggests that Sir2p is affecting some early step in pre-RC formation and also that the loss of SIR2 is not enhancing DNA replication in a generalized or nonspecific manner.
SIR2 is required for silent heterochromatin formation in yeast; however, the loss of SIR2 is not indirectly suppressing the initiation defect of cdc6-4 through its known functions in the genome, either by transcriptional silencing at the HM loci, at telomere proximal genes, or by some indirect mechanism affected by increased recombination or a loss of silencing within the rDNA repeats. Although we initially isolated the sir2 suppressor using the cdc6-4 allele integrated at the LEU2 locus, a deletion of SIR2 also suppresses the temperature-sensitive phenotype of cdc6-4 when it is present at its normal chromosomal location (D.L. Pappas Jr. and M. Weinreich, unpubl.). Importantly, we have shown that both Cdc6p and Cdc6-4p abundance is the same in a wild-type and in a SIR2 deletion strain. Thus, a SIR2 deletion suppresses cdc6-4 in a context-independent fashion that is also not explained by an increase in cdc6-4 expression. Taken together, these data suggest that SIR2 has additional targets in the cell distinct from the silent heterochromatic loci.
Both the plasmid stability and ChIP assays indicated that loss of SIR2 did not affect all origins equally. A SIR2 deletion substantially reversed the cdc6-4 plasmid instability phenotype for the ARS305, ARS315, and ARS501 origin plasmids but only partially rescued ARS1 plasmid instability and had no effect for the ARSH4 and ARS301 plasmids. In a wild-type strain, the SIR2 deletion caused similar effects with the exception of the ARS301 origin plasmid, which was now very efficiently maintained. The fact that the high plasmid loss rate of some origins was rescued by deletion of SIR2 but others were unaffected or minimally affected, suggests that Sir2p is having specific effects at particular origins or that some origins are less sensitive to Sir2p inhibition is this assay. The different effects on plasmid stability seen by deletion of SIR2 could also reflect different chromatin structures surrounding the origins, differences in origin sequence, or efficiency of origin usage. The observation that a sir2Δ in an otherwise wild-type background brings some plasmid retention rates to nearly 100% suggests that SIR2 inhibits origin usage in wild-type cells.
A SIR2 deletion had a differential effect on MCM loading at origins in the cdc6-4 mutant. To examine pre-RC formation, we tested Mcm2p binding to three efficient origins (by ChIP) in the wild-type, cdc6-4, and cdc6-4 sir2Δ mutant strains. These experiments showed that the cdc6-4 mutant was defective for Mcm2p origin loading at the permissive temperature and inactive for Mcm2p loading at the nonpermissive temperature at all three origins, as suggested previously (Weinreich et al. 1999). Deletion of SIR2 promoted Mcm2p binding to the ARS315 and ARS501 chromosomal origins at 25°C and 37°C; however, a sir2Δ had only a minimal effect at the ARS1 origin at the permissive temperature and did not promote Mcm2p binding to ARS1 at the nonpermissive temperature. These results correlated well with the plasmid instability data and further suggested that the loss of SIR2 differentially affects pre-RC formation at these origins. Because ARS1, ARS315, and ARS501 are efficient chromosomal origins that fire at different times during S phase (Raghuraman et al. 2001), it appears that the rescue of origin activity is not strictly correlated with their time of activation but more properly with pre-RC assembly.
Consistent with our finding that Sir2p is a negative regulator of chromosomal DNA replication, a recent report found that Sir2p negatively regulated initiation events within the rDNA locus using the technique of molecular combing (Pasero et al. 2002). Each 9.1-kb rDNA repeat contains an origin of replication, although only ∼20%–25% of these origins are used during the cell cycle. A SIR2 deletion resulted in a twofold increase in origin initiation within the rDNA locus on Chromosome XII. Another systematic ChIP study across the rDNA repeat has shown that Sir2p is enriched at two positions: at the NTS1 (non-transcribed 1) spacer and also at the NTS2 near the Pol I 35S gene promoter and extending within the 35S gene (Huang and Moazed 2003). These experiments suggest that the replication fork barrier protein Fob1 and RNA polymerase I target Sir2p to the NTS1 and NTS2, respectively. Thus, the current understanding of Sir2p localization in the cell suggests that potentially three strategies are used to bring Sir2p to its targets: Sir4p recruits Sir2p to the HM loci and telomeres; Fob1p and Pol I recruit Sir2p (i.e., RENT) to the rDNA locus. It is possible that Sir2p has additional chromosomal targets in the cell that have not been seen because of their transient nature or that Sir2p can interact with an initiation protein. At the rDNA locus, because Sir2p deacetylates nucleosomes throughout the entire region, a loss of Sir2p would likely also affect the nucleosomal acetylation state near the origin, and this could contribute to the increased initiation frequencies observed.
Another precedent indicating that Sir2p affects chromosome maintenance comes from the finding that increased expression of Sir2p from the regulated GAL1 promoter caused lethality in budding yeast, and a transient increase in Sir2p levels (that did not result in lethality) substantially increased chromosome loss rates (Holmes et al. 1997). Although the precise mechanism of the chromosome loss was not known, increased minichromosome loss also occurred for plasmids that did not contain telomeric sequences, suggesting that this effect was independent of Sir2p binding near telomeres. Both the lethality and the increased chromosome loss rates upon Sir2p induction could certainly be explained by the negative regulation of DNA replication that we have uncovered, because suppression of initiation frequencies throughout the genome would lead to chromosome loss and cell death.
It is interesting to note that temperature-sensitive mutations within the ORC genes were first isolated as silencing-defective mutants using the HMRa synthetic silencer (Foss et al. 1993), and we have found that deletion of SIR2 restores growth to cdc6-4 and orc5-1 mutants. It is well established that ORC is required for silencing at the HM loci. However, ORC promotes silencing through a replication-independent mechanism whereby Orc1p recruits Sir1p to the silencer via the ORC-binding sites within the silencer DNA elements (Triolo and Sternglanz 1996; Fox et al. 1997). Although there is currently no evidence for this, it is not inconceivable that nonsilencer origins could also recruit one or more SIR proteins, because Sir1p interacts with ORC and the Sir2–4 proteins are present on the chromatin throughout an ∼3–4-kb region surrounding the silencer that naturally includes the ORC-binding sites (Rusche et al. 2002; Zhang et al. 2002). Because a deletion of SIR1 or the Orc1p interaction domain with Sir1p will not suppress the cdc6-4 initiation mutant, it is very unlikely that the effect of Sir2p on replication is mediated through a known heterochromatic role for ORC. How then might Sir2p negatively affect DNA replication?
Sir2p could directly inhibit origin usage by deacetylating origin-proximal nucleosomes, which either hinders recruitment of a key initiation factor or promotes binding of an inhibitory factor. Sir2p is unique among the three classes of deacetylases in that it requires NAD+ as a cofactor for its enzymatic activity (Moazed 2001), and it has a distinct substrate specificity from both the Rpd3p and Hda1p deacetylases. Because deleting neither RPD3 nor HDA1 restored growth to the cdc6-4 mutant, this suggests that either Sir2p-specific histone modifications are influencing the initiation of DNA replication or that an unknown function of Sir2p not shared by Rpd3p or Hda1p is negatively affecting initiation frequencies. A recent report has shown that histone acetylation near origins promotes earlier firing for both early and late origins. A deletion of the RPD3 deacetylase was shown to advance replication timing for many individual origins and was correlated with earlier binding of Cdc45p to origins (Vogelauer et al. 2002). Cdc45p associates with origins after pre-RC formation in a manner correlated with their time of activation and thus is a temporal marker for the initiation of DNA synthesis (Aparicio et al. 1999). The authors also targeted the histone acetyltransferase Gcn5p to the internal late origin ARS1412, which caused its earlier activation and increased histone acetylation near ARS1412. These data established that histone acetylation promotes replication initiation and that one consequence of histone acetylation is the earlier recruitment of Cdc45p to origins. Thus, if SIR2 is directly targeted to some origins in the cell, localized histone deacetylation could inhibit initiation.
Although Sir2p is a histone deacetylase, it could also deacetylate a nonhistone protein such as a pre-RC component or regulator of initiation and thereby inhibit DNA replication. As mentioned earlier, mammalian SIR2 orthologs have been shown to deacetylate nonhistone proteins such as p53 (Luo et al. 2001; Vaziri et al. 2001) and tubulin (North et al. 2003). Although there is no evidence that Sir2p normally silences genes apart from the silent-mating-type cassettes, the rDNA locus, or near telomeres, it is also possible that Sir2p regulates the transcription of an unknown gene that is limiting for initiation. Whatever the exact mechanism, Sir2p inhibits a subset of origins, and therefore some particular aspect of origin sequence or structure not shared among all origins makes them sensitive to SIR2 inhibition. Because initiation from all origins is not restored in the cdc6-4 sir2Δ mutant, the G2/M delay seen in this strain could be caused by slowed or incomplete DNA replication.
SIR2 is the only SIR gene that is conserved in metazoans (Brachmann et al. 1995), and human cells alone contain seven SIR2 orthologs, SIRT1–SIRT7 (Frye 2000). Because Sir2p is required for heterochromatin formation and also negatively regulates DNA replication in the budding yeast, we speculate that a conservation of this function in metazoans could afford one mechanism to link replication domains with heritable transcriptional states during development or in response to cell-autonomous signals.
Materials and methods
Construction of yeast strains, growth media, and genetic methods
The yeast strains used in this work are listed in Supplementary Table 1. Genetic manipulations were performed according to standard techniques. YPD denotes rich medium, and FOA denotes synthetic complete medium containing 1 mg/mL 5-fluoro-orotic acid.
SIR2 was PCR-amplified from wild-type genomic DNA with forward oligonucleotide SIR2-Sal and reverse oligonucleotide SIR2-Xba (Supplementary Table 2). The PCR reaction contained 10 ng of genomic DNA, 200 μM dNTP, 50 pmoles of each oligonucleotide, and 2.5 units of PfuTurbo (Stratagene) in a 50-μL reaction volume. The PCR product was digested with SalI and XbaI and cloned into the respective sites in pRS416 to generate pDP56. pDP56 served as a template for SIR2 mutagenesis. Mutagenesis of SIR2 was performed using the corresponding oligonucleotides in Supplementary Table 2 with the QuikChange method (Stratagene) according to manufacturer's specifications. In each case, the entire SIR2 gene was sequenced to verify the presence of only a single mutation.
sir2 mutations were integrated at the SIR2 locus in W303-1A as outlined below. For sir2-N345A and sir2-87, a 1.4-kb BsrGI/XbaI fragment was cloned into the Acc65I/XbaI sites of YIplac211 (Gietz and Sugino 1988). For sir2-88, a 1.4-kb BsrGI/XbaI fragment was cloned into the Acc65I/XbaI sites of YIplac204 (Gietz and Sugino 1988). For sir2-86, a 1.6-kb SphI/StuI fragment was cloned into the SphI/SmaI sites of YIplac204. The resulting plasmids, pDP135 (sir2-N345A), pDP139 (sir2-87), pDP309 (sir2-88), and pDP307 (sir2-86), respectively, were linearized with BglII to direct integration. All integrations were confirmed by PCR of genomic DNA. PCR reactions were performed with 5 ng of genomic DNA, 15 pmoles of oligonucleotides SIR2-Sal (SIR2-specific) and M13-reverse sequencing primer (vector-specific), and 1.5 units of Taq DNA polymerase (Invitrogen) in a 25-μL reaction volume. In the case of sir2-86, reaction conditions were the same except that 15 pmoles of SIR2-Xba was used as the SIR2-specific oligonucleotide. PCR products were visualized by agarose gel electrophoresis followed by ethidium bromide staining (0.5 μg/mL). PCR products were also subjected to automated DNA sequencing using an ABI 3700 Genetic Analyzer (Applied Biosystems). Individual sequencing reactions contained 50 ng of PCR product and 3.2 pmoles of a SIR2-specific sequencing oligonucleotide (SIR2-SP1, SP2, SP3 or SP4; Supplementary Table 2).
To construct M1161, hda1::kanMX was PCR-amplified from the Research Genetics strain 5347 using primers HDA1-1F and HDA1-1R (Supplementary Table 2) and integrated into W303-1A in a single step. The correct insertion was verified by PCR using the original primers and a second set of primers (HDA1-2F and HDA1-2R; Supplementary Table 2) flanking these and the HDA1 ORF.
Isolation and characterization of extragenic suppressors of cdc6-4
Strain M386 (W303-1A cdc6-4) was grown at 25°C in YPD until stationary phase. Approximately 2 × 106 cells each were plated onto multiple YPD plates containing 1% formamide and incubated for 3 d at 37°C. This concentration of fomamide did not affect the growth of the wild-type strain at 37°C but did tighten up the ts phenotype of the cdc6-4 strain. Multiple independent spontaneous Ts+ revertants (rgc mutants [restores growth of cdc6-4]) were isolated that had a secondary nonmating phenotype. Plasmids containing SIR1, SIR2, SIR3, or SIR4 (from C. Fox, UW-Madison) were introduced into suppressor strains and then assayed for mating proficiency. To confirm that suppression of cdc6-4 was conferred by a single gene mutation, strains transformed with their respective complementing SIR plasmid were crossed to M599 (MATα cdc6-4). Diploids were selected, cured of the wild-type SIR plasmid, and sporulated. In all cases, the tetrad analysis showed a 2:2 ratio of Ts+:Ts- demonstrating the suppression of cdc6-4 in each strain was caused by a single gene mutation. The sterile phenotype cosegregated with suppression of cdc6-4, indicating that it is a pleiotropic phenotype associated with the suppressor mutation. SIR2, SIR3, and SIR4 were tested to determine if they were, indeed, allelic to the original mutations. A TRP1 nutritional marker was integrated adjacent to the SIR2, SIR3, and SIR4 loci individually in strain M599 (MATα cdc6-4). The three resulting integrant strains were crossed to rgc strains transformed with the respective complementing SIR plasmid. Diploids were selected and sporulated as above. In all cases, Ts+:Ts- segregated 2:2 and the Trp+ phenotype (wild-type SIR) segregated opposite the suppressor. These data indicate that the rgc mutants are bona fide suppressors of cdc6-4 and are allelic to SIR2, SIR3, and SIR4.
To quantitate growth of strains, cells were 10-fold serially diluted and spotted onto YPD at 25°C and 37°C or YPD +1% formamide at 37°C for the cdc6-4-containing strains shown in Figures 1 and 4. Formamide diminished the appearance of sporadic cdc6-4 revertants, which made scoring difficult. However, the same quantitative suppression could be seen for the cdc6-4 sir mutants in the absence of formamide.
FACS analysis
Strains M138 (WT), M386 (cdc6-4), and M940 (cdc6-4 sir2Δ hmlΔ) were grown in YPD medium at 25°C to OD600 = 0.25 and synchronized in G1 by the addition of 10 μg/mL α-factor for 3 h. Cultures were shifted for 1 h to 37°C in the presence of α-factor and then released into fresh medium in the absence of α-factor at 37°C. Aliquots of cells were harvested at the indicated times, processed for flow cytometry as described (Weinreich et al. 1999), and analyzed using a Becton-Dickenson FACScalibur machine.
Cloning of ARS elements from Chromosome III
ARS elements for Chromosome III were defined in Poloumienko et al. (2001). ARS305 and ARS315 were PCR-amplified from genomic DNA with EcoRI and HindIII linkers (Supplementary Table 2), whereas ARS301 was PCR-amplified from genomic DNA with SacI and HindIII. All were then cloned in place of the 192-bp ARS1-containing fragment from pARS1-WT (Marahrens and Stillman 1992). The wild-type DNA sequence was verified for each ARS element, and each recombinant plasmid was shown to contain a functional origin by the high-frequency transformation assay. Chromosomal coordinates contained on the plasmids are for ARS301 (11,045–11,532), ARS305 (39,382–39,724), and ARS315 (224,804–225,318).
Plasmid instability assays
Plasmids pARS1-WT, pRS416 (ARSH4), pRF21 (ARS315), pR151 (ARS501), pRF4 (ARS305), and pRF12 (ARS301) were introduced into strains M138 (WT), CFY366 (sir2Δ), M386 (cdc6-4), and M922 (cdc6-4 sir2Δ). Plasmid instability was performed as previously described (Dani and Zakian 1983). Results are reported as the average of four to six measurements with accompanying standard errors.
ChIP
ChIP was performed as described (Strahl-Bolsinger et al. 1997) with the following modifications: the lysis buffer contained 300 mM NaCl and the immunoprecipitation (IP) was performed using an Mcm2p monoclonal antibody (kindly provided by B. Stillman) cross-linked to protein A Sepharose beads. 1/25-th of the IP DNA and 1/500-th of the input DNA were subjected to 30 cycles of multiplex PCR for ARS1 and 28 cycles for ARS315 and ARS501 using the indicated primers (Supplementary Table 2). PCR products were separated on 5% polyacrylamide gels.
Acknowledgments
We thank Catherine Fox, Art Lustig, Dan Gottschling, David Shore, Carol Newlon, Jeff Smith, Michael Grunstein, Steve Bell, and Bruce Stillman for generously providing yeast strains and reagents. We also thank Catherine Fox for encouragement and critical reading of the manuscript, and Marleah Russo and Kelli VanDussen for expert technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1173204.
Corresponding author.
References
- Aparicio O.M., Billington, B.L., and Gottschling, D.E. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279-1287. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Weinstein, D.M., and Bell, S.P. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59-69. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Stout, A.M., and Bell, S.P. 1999. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl. Acad. Sci. 96: 9130-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P. and Dutta, A. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333-374. [DOI] [PubMed] [Google Scholar]
- Bell S.P. and Stillman, B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128-134. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Kobayashi, R., and Stillman, B. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262: 1844-1849. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Mitchell, J., Leber, J., Kobayashi, R., and Stillman, B. 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83: 563-568. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Sherman, J.M., Devine, S.E., Cameron, E.E., Pillus, L., and Boeke, J.D. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes & Dev. 9: 2888-2902. [DOI] [PubMed] [Google Scholar]
- Bryk M., Banerjee, M., Murphy, M., Knudsen, K.E., Garfinkel, D.J., and Curcio, M.J. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & Dev. 11: 255-269. [DOI] [PubMed] [Google Scholar]
- Buck S.W., Sandmeier, J.J., and Smith, J.S. 2002. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111: 1003-1014. [DOI] [PubMed] [Google Scholar]
- Carmen A.A., Milne, L., and Grunstein, M. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 277: 4778-4781. [DOI] [PubMed] [Google Scholar]
- Cheng H.L., Mostoslavsky, R., Saito, S., Manis, J.P., Gu, Y., Patel, P., Bronson, R., Appella, E., Alt, F.W., and Chua, K.F. 2003. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. 100: 10794-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M., Palladino, F., Laroche, T., Kyrion, G., Liu, C., Lustig, A.J., and Gasser, S.M. 1995. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: Evidence for a multicomponent complex required for yeast telomeric silencing. J. Cell Biol. 129: 909-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker J.H., Piatti, S., Santocanale, C., Nasmyth, K., and Diffley, J.F. 1996. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379: 180-182. [DOI] [PubMed] [Google Scholar]
- Cosgrove A.J., Nieduszynski, C.A., and Donaldson, A.D. 2002. Ku complex controls the replication time of DNA in telomere regions. Genes & Dev. 16: 2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus G., Shafaatian, R., and Shore, D. 2000. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 19: 2641-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani G.M. and Zakian, V.A. 1983. Mitotic and meiotic stability of linear plasmids in yeast. Proc. Natl. Acad. Sci. 80: 3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey M.J., Jeruzalmi, D., Kuriyan, J., and O'Donnell, M. 2002. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell. Biol. 3: 826-835. [DOI] [PubMed] [Google Scholar]
- DeRyckere D., Smith, C.L., and Martin, G.S. 1999. The role of nucleotide binding and hydrolysis in the function of the fission yeast cdc18+ gene product. Genetics 151: 1445-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F.X. 1996. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 10: 2819-2830. [DOI] [PubMed] [Google Scholar]
- Drury L.S., Perkins, G., and Diffley, J.F. 2000. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 10: 231-240. [DOI] [PubMed] [Google Scholar]
- Erzberger J.P., Pirruccello, M.M., and Berger, J.M. 2002. The structure of bacterial DnaA: Implications for general mechanisms underlying DNA replication initiation. EMBO J. 21: 4763-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.M., Brewer, B.J., Reynolds, A.E., and Fangman, W.L. 1991. A yeast origin of replication is activated late in S phase. Cell 65: 507-515. [DOI] [PubMed] [Google Scholar]
- Foss M., McNally, F.J., Laurenson, P., and Rine, J. 1993. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science 262: 1838-1844. [DOI] [PubMed] [Google Scholar]
- Fox C.A., Ehrenhofer-Murray, A.E., Loo, S., and Rine, J. 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276: 1547-1551. [DOI] [PubMed] [Google Scholar]
- Frolova N.S., Schek, N., Tikhmyanova, N., and Coleman, T.R. 2002. Xenopus Cdc6 performs separate functions in initiating DNA replication. Mol. Biol. Cell 13: 1298-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R.A. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273: 793-798. [DOI] [PubMed] [Google Scholar]
- Gardner K.A., Rine, J., and Fox, C.A. 1999. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics 151: 31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S.M. and Cockell, M.M. 2001. The molecular biology of the SIR proteins. Gene 279: 1-16. [DOI] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino, A. 1988. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527-534. [DOI] [PubMed] [Google Scholar]
- Gottlieb S. and Esposito, R.E. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56: 771-776. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio, O.M., Billington, B.L., and Zakian, V.A. 1990. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell 63: 751-762. [DOI] [PubMed] [Google Scholar]
- Guarente L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes & Dev. 14: 1021-1026. [PubMed] [Google Scholar]
- Hecht A., Laroche, T., Strahl-Bolsinger, S., Gasser, S.M., and Grunstein, M. 1995. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell 80: 583-592. [DOI] [PubMed] [Google Scholar]
- Hogan E. and Koshland, D. 1992. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 89: 3098-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes S.G., Rose, A.B., Steuerle, K., Saez, E., Sayegh, S., Lee, Y.M., and Broach, J.R. 1997. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics 145: 605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe G.J., Tanny, J.C., Rudner, A.D., Gerber, S.A., Danaie, S., Gygi, S.P., and Moazed, D. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22: 4167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. and Moazed, D. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes & Dev. 17: 2162-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S., Armstrong, C.M., Kaeberlein, M., and Guarente, L. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795-800. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D., O'Donnell, M., and Kuriyan, J. 2001. Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell 106: 429-441. [DOI] [PubMed] [Google Scholar]
- Kurdistani S.K. and Grunstein, M. 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell. Biol. 4: 276-284. [DOI] [PubMed] [Google Scholar]
- Kyrion G., Liu, K., Liu, C., and Lustig, A.J. 1993. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes & Dev. 7: 1146-1159. [DOI] [PubMed] [Google Scholar]
- Labib K., Diffley, J.F., and Kearsey, S.E. 1999. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1: 415-422. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton, A., Tafrov, S.T., Heller, R.C., Stebbins, J., Pillus, L., and Sternglanz, R. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. 97: 5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzen C.U., Steinmann, D., Whiteheart, S.W., and Weis, W.I. 1998. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell 94: 525-536. [DOI] [PubMed] [Google Scholar]
- Liang C., Weinreich, M., and Stillman, B. 1995. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81: 667-676. [DOI] [PubMed] [Google Scholar]
- Lipford J.R. and Bell, S.P. 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7: 21-30. [DOI] [PubMed] [Google Scholar]
- Liu J., Smith, C.L., DeRyckere, D., DeAngelis, K., Martin, G.S., and Berger, J.M. 2000. Structure and function of Cdc6/Cdc18: Implications for origin recognition and checkpoint control. Mol. Cell 6: 637-648. [DOI] [PubMed] [Google Scholar]
- Luo J., Nikolaev, A.Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L., and Gu, W. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107: 137-148. [DOI] [PubMed] [Google Scholar]
- Luo K., Vega-Palas, M.A., and Grunstein, M. 2002. Rap1–Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes & Dev. 16: 1528-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig A.J., Kurtz, S., and Shore, D. 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250: 549-553. [DOI] [PubMed] [Google Scholar]
- Marahrens Y. and Stillman, B. 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817-823. [DOI] [PubMed] [Google Scholar]
- Marks P., Rifkind, R.A., Richon, V.M., Breslow, R., Miller, T., and Kelly, W.K. 2001. Histone deacetylases and cancer: Causes and therapies. Nat. Rev. Cancer 1: 194-202. [DOI] [PubMed] [Google Scholar]
- McNally F.J. and Rine, J. 1991. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 5648-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. 2001. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13: 232-238. [DOI] [PubMed] [Google Scholar]
- Moretti P. and Shore, D. 2001. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 21: 8082-8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth V., Nadaud, S., Grummt, I., and Voit, R. 2001. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 20: 1353-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind, L., Spouge, J.L., and Koonin, E.V. 1999. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9: 27-43. [PubMed] [Google Scholar]
- Newlon C.S. and Theis, J.F. 1993. The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev. 3: 752-758. [DOI] [PubMed] [Google Scholar]
- Nguyen V.Q., Co, C., and Li, J.J. 2001. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411: 1068-1073. [DOI] [PubMed] [Google Scholar]
- North B.J., Marshall, B.L., Borra, M.T., Denu, J.M., and Verdin, E. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11: 437-444. [DOI] [PubMed] [Google Scholar]
- Pak D.T., Pflumm, M., Chesnokov, I., Huang, D.W., Kellum, R., Marr, J., Romanowski, P., and Botchan, M.R. 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91: 311-323. [DOI] [PubMed] [Google Scholar]
- Pasero P., Bensimon, A., and Schwob, E. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes & Dev. 16: 2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G. and Diffley, J.F. 1998. Nucleotide-dependent prereplicative complex assembly by Cdc6p, a homolog of eukaryotic and prokaryotic clamp-loaders. Mol. Cell 2: 23-32. [DOI] [PubMed] [Google Scholar]
- Poloumienko A., Dershowitz, A., De, J., and Newlon, C.S. 2001. Completion of replication map of Saccharomyces cerevisiae chromosome III. Mol. Biol. Cell 12: 3317-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman M.K., Winzeler, E.A., Collingwood, D., Hunt, S., Wodicka, L., Conway, A., Lockhart, D.J., Davis, R.W., Brewer, B.J., and Fangman, W.L. 2001. Replication dynamics of the yeast genome Science 294: 115-121. [DOI] [PubMed] [Google Scholar]
- Rao H. and Stillman, B. 1995. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl. Acad. Sci. 92: 2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J. and Herskowitz, I. 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116: 9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D., Suka, Y., Xenarios, I., Kurdistani, S.K., Wang, A., Suka, N., and Grunstein, M. 2002. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437-446. [DOI] [PubMed] [Google Scholar]
- Rusche L.N., Kirchmaier, A.L., and Rine, J. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481-516. [DOI] [PubMed] [Google Scholar]
- Shou W., Seol, J.H., Shevchenko, A., Baskerville, C., Moazed, D., Chen, Z.W., Jang, J., Charbonneau, H., and Deshaies, R.J. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233-244. [DOI] [PubMed] [Google Scholar]
- Simpson R.T. 1990. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature 343: 387-389. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A. 2002. Paradigms and pitfalls of yeast longevity research. Mech. Ageing Dev. 123: 857-867. [DOI] [PubMed] [Google Scholar]
- Smith J.S. and Boeke, J.D. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & Dev. 11: 241-254. [DOI] [PubMed] [Google Scholar]
- Smith J.S., Brachmann, C.B., Celic, I., Kenna, M.A., Muhammad, S., Starai, V.J., Avalos, J.L., Escalante-Semerena, J.C., Grubmeyer, C., Wolberger, C., et al. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. 97: 6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J.B. and Gottschling, D.E. 1999. Telomeric chromatin modulates replication timing near chromosome ends. Genes & Dev. 13: 146-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. 1996. Cell cycle control of DNA replication. Science 274: 1659-1663. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht, A., Luo, K., and Grunstein, M. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 11: 83-93. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Shou, W., Dowd, G.J., Turck, C.W., Deshaies, R.J., Johnson, A.D., and Moazed, D. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97: 245-256. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp, D., and Nasmyth, K. 1997. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90: 649-660. [DOI] [PubMed] [Google Scholar]
- Triolo T. and Sternglanz, R. 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381: 251-253. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Dessain, S.K., Ng Eaton, E., Imai, S.I., Frye, R.A., Pandita, T.K., Guarente, L., and Weinberg, R.A. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107: 149-159. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang, E.S., and Amon, A. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398: 818-823. [DOI] [PubMed] [Google Scholar]
- Vogelauer M., Rubbi, L., Lucas, I., Brewer, B.J., and Grunstein, M. 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10: 1223-1233. [DOI] [PubMed] [Google Scholar]
- Weinreich M., Liang, C., and Stillman, B. 1999. The Cdc6p nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc. Natl. Acad. Sci. 96: 441-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R.C., Hanson, P.I., Jahn, R., and Brunger, A.T. 1998. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat. Struct. Biol. 5: 803-811. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hayashi, M.K., Merkel, O., Stillman, B., and Xu, R.M. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21: 4600-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]