Abstract
DNA repair associated with DNA replication is important for the conservation of genomic sequence information, whereas reconstitution of chromatin after replication sustains epigenetic information. We have isolated and characterized mutations in the BRU1 gene of Arabidopsis that suggest a novel link between these underlying maintenance mechanisms. Bru1 plants are highly sensitive to genotoxic stress and show stochastic release of transcriptional gene silencing. They also show increased intrachromosomal homologous recombination and constitutively activated expression of poly (ADP-ribose) polymerase-2 (AtPARP-2), the induction of which is associated with elevated DNA damage. Bru1 mutations affect the stability of heterochromatin organization but do not interfere with genome-wide DNA methylation. BRU1 encodes a novel nuclear protein with two predicted protein–protein interaction domains. The developmental abnormalities characteristic of bru1 mutant plants resemble those triggered by mutations in genes encoding subunits of chromatin assembly factor (CAF-1), the condensin complex, or MRE11. Comparison of bru1 with these mutants indicates cooperative roles in the replication and stabilization of chromatin structure, providing a novel link between chromatin replication, epigenetic inheritance, S-phase DNA damage checkpoints, and the regulation of meristem development.
Keywords: Arabidopsis thaliana, DNA repair, epigenetic inheritance, gene silencing, homologous recombination
A dynamic chromatin structure contributes to the regulation of repair and transcription of DNA templates. Chromatin components involved in both processes have been described that imply shared molecular mechanisms modulating DNA accessibility for repair and transcription (Green and Almouzni 2002). The first molecular link between transcription and DNA repair was revealed during characterization of transcription factor IIH (TFIIH), which is required for initiation of RNA synthesis by RNA polymerase II and for efficient repair of DNA through nucleotide excision (Feaver et al. 1993; Schaeffer et al. 1993; Drapkin et al. 1994; Wang et al. 1994).
Accessibility is determined by compaction of chromatin, which consists of loosely packaged, transcriptionally active euchromatin, and heterochromatin, which is condensed and transcriptionally silent and consists mainly of transposable elements and repetitive sequences. Chromatin states are inherited during DNA replication, providing a scaffold for epigenetic information that influences transcriptional gene regulation.
Several chromatin components determining heritable features of chromatin also have an influence on epigenetic regulation of gene activity and efficiency of DNA repair or genome stability. For example, SIR proteins in yeast mediate formation of a compact chromatin structure similar to heterochromatin in multicellular eukaryotes (Gross 2001) and are required for transcriptional gene silencing (TGS) and for suppression of homologous recombination of rDNA repeats (Guarente 2000). They are also involved in repair of DNA double-strand breaks (DSBs) by nonhomologous end joining (NHEJ; Tsukamoto et al. 1997). Thus, SIR mutations result in both release of TGS (Gross 2001) and increased sensitivity to DNA-damaging agents (Tsukamoto et al. 1997; Critchlow and Jackson 1998; Martin et al. 1999; Mills et al. 1999). A further requisite for accurate regulation of TGS and proper responses to genotoxic stress is the chromatin assembly factor 1 (CAF-1), which is conserved throughout eukaryotes (Kaufman and Almouzni 2000). CAF-1 interacting with PCNA (Shibahara and Stillman 1999) facilitates incorporation of histones H3 and H4 into newly synthesized DNA during S phase (Smith and Stillman 1989) and unscheduled DNA-repair synthesis (Gaillard et al. 1996). Yeast mutants with defective CAF-1 subunits (cac mutants) are hypersensitive to UV irradiation (Kaufman et al. 1997) and unable to maintain telomeric silencing (Kaufman et al. 1997; Tchenio et al. 2001).
The replication-coupling assembly factor (RCAF; Tyler et al. 1999) also facilitates the assembly of nucleosomes onto newly replicated DNA. RCAF is a protein complex of a Drosophila ortholog of the anti-silencing function-1 protein (ASF1) and histones H3 and H4 (Tyler et al. 1999) that acts synergistically with CAF-1 in chromatin assembly. In yeast, both CAF-1 and ASF1/RCAF are involved in chromatin reconstitution after DNA replication and repair-coupled DNA synthesis, but their functions do not fully overlap: cac1 and asf1 mutations have dissimilar effects on nucleotide excision repair and homologous recombination and also differ in silencing at subtelomeric regions or mating-type loci (Tyler et al. 1999). Nevertheless, they clearly have cooperative functions, as asf1/cac1 double mutants exhibit synergistic loss of silencing and deficiencies in DNA repair (Tyler et al. 1999).
A genetic link between DNA repair and TGS has been documented for unicellular green algae (Chlamydomonas reinhardtii). Transcriptionally silenced transgenes and transposable elements were activated in mutants mut-9 and mut-11, both of which were hypersensitive to conditions provoking DSBs (Jeong et al. 2002). MUT-9 encodes a putative serine/threonine protein kinase and MUT-11 a novel protein containing a WD-40 repeat (Jeong et al. 2002). Their precise molecular functions are unknown.
Although genes involved in DNA repair or epigenetic regulation of transcription have been studied extensively in multicellular eukaryotes (Gorbunova and Levy 1999; Mittelsten Scheid and Paszkowski 2000; Hoeijmakers 2001; Martienssen and Colot 2001; Li 2002; Grewal and Moazed 2003), evidence for components linking DNA repair and epigenetic inheritance is scarce. It is reasonable to predict that such shared components exist also in higher plants and may be revealed by forward genetic approaches. Here we describe the results of two parallel searches in two different laboratories for Arabidopsis mutants affected in DNA-damage responses and for mutants affected in maintenance of TGS. This led to the isolation of different mutant alleles of BRU1, which encodes a novel nuclear protein. Bru1 mutants have phenotypic similarities to Arabidopsis mutants in CAF-1 subunits (fas1 and fas2; Kaya et al. 2001), MRE11 (Bundock and Hooykaas 2002), and condensin genes (Siddiqui et al. 2003).
Phenotypic, genetic, and molecular characterization of bru1, fas1, fas2, mre11, and mutants in subunits of the condensin complex indicates that BRU1 is involved in epigenetic inheritance during DNA replication. Moreover, mutations in Arabidopsis MRE11 or condensin subunits genes, in addition to defects in an S-phase checkpoint (Aono et al. 2002; D'Amours and Jackson 2002; Hagstrom and Meyer 2003), also compromise epigenetic regulation, as shown by release of heterochromatin-mediated TGS. Thus, our results imply a tight link between chromatin replication, postreplicative DNA repair, and an S-phase DNA-damage checkpoint.
Results
Isolation of bru1 mutants
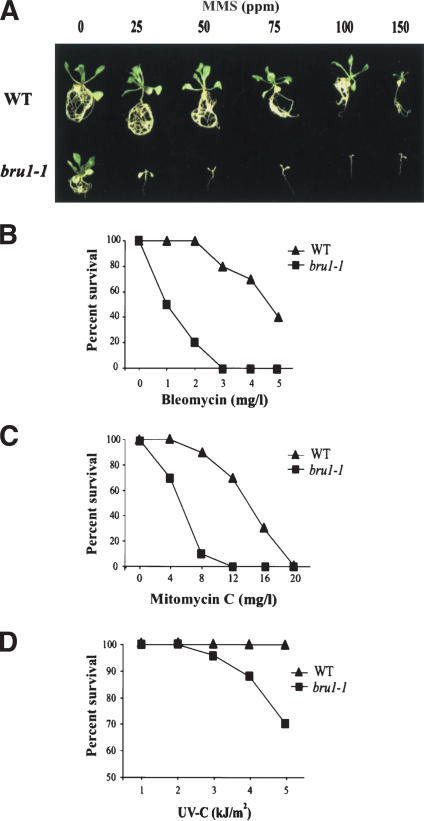
In a search for plant components involved in the recognition and repair of DNA damage, we screened 2500 Arabidopsis T3 families mutagenized by random insertion of foreign DNA (T-DNA) for individuals with elevated sensitivity to methyl methane sulfonate (MMS). MMS alkylates DNA and is considered to mimic DNA DSB damage (Schwartz 1989). One transgenic family segregated individuals unable to withstand 25 ppm of MMS, whereas wild-type plants tolerated 100 ppm (Fig. 1A). This sensitivity threshold of 25 ppm is significantly (two- to threefold) lower than for previously characterized Arabidopsis mutants hypersensitive to MMS (Mengiste et al. 1999; Ulm et al. 2001; Bundock and Hooykaas 2002; Garcia et al. 2003). The sensitivity segregates as a recessive monogenic trait. The homozygous mutant plants are also hypersensitive to bleomycin (inducer of DSBs; Fig. 1B), mitomycin C (cross-linking of DNA strands; Fig. 1C), and UV-C irradiation (Fig. 1D). Importantly, the sensitivity of the mutant seems to be restricted to genotoxic stresses. Mutant plants do not differ from wild-type plants in sensitivity to abiotic stresses such as high salinity, elevated osmolarity, or oxidative stress (data not shown). The mutant was named brushy1 (bru1) because of its morphological abnormalities, and this particular mutant allele was designated bru1-1. The most prominent morphological alterations of plants homozygous for bru1-1 are retarded growth of primary roots, distorted phyllotaxy (resulting in an irregular branching pattern), and fasciation (thick and flattened stems and fused organs; Fig. 2A–F). The degree of fasciation varies between individual bru1-1 mutant plants and even between neighboring branches of the same plant (Fig. 2D). Fasciation affects stems, lateral shoots, and even siliques, which arise close together and form brush-like structures (Fig. 2C,F). There is no progression in severity of the phenotypes over subsequent generations. Flowering time also varies among individuals, some of which show a prolonged vegetative phase, particularly under short-day conditions (data not shown).
Figure 1.
Sensitivity of bru1 to genotoxic stress. Representative seedlings (A) and the survival rate (B–D) after each treatment are displayed. Levels of resistance of wild-type Ws (WT) and bru1-1 to MMS (A), bleomycin (B), mitomycin C (C), and UV-C (D). Ten (A–C) or fifty (D) seedlings were tested in each treatment.
Figure 2.
Developmental aberrations in bru1. Branching pattern of wild-type (A) and bru1-1 (B,C) plants. (D) Fasciated (white arrow) and normal (black arrow) stems from the same bru1-1 plant. (E) Root growth of seedlings of wild-type Ws (left) and bru1-1 (right). (F) Fused siliques in bru1-1. (G) bru1-3 mutant allele (left) and wild type (right).
An independent screen after ethyl methane sulfonate (EMS) mutagenesis for mutations that release TGS at a transgenic locus encoding the luciferase (Luc) marker gene yielded a mutant that morphologically closely resembles bru1-1. Genetic analysis and sequencing of the affected BRU1 gene in the two mutants showed them to be allelic, and the second mutant allele was designated bru1-2.
A third mutant allele (bru1-3) was identified in a collection of T-DNA insertion lines from the Torrey Mesa Research Institute (http://www.tmri.org). Although some aspects of the altered morphology of bru1-3 resemble bru1-1 and bru1-2 phenotypes, this mutant allele is further characterized by severe dwarfing and very low seed set (Fig. 2G). This strong phenotype of bru1-3, which is not observed for the other two alleles, suggests that bru1-1 and bru1-2 are likely partial loss-of-function alleles. Extreme sensitivity to various genotoxic stresses, as initially observed for bru1-1, is also characteristic of bru1-2 and bru1-3 mutant plants (data not shown). The segregation of all observed mutant traits indicates that bru1-2 and bru1-3 are also recessive.
The BRU1 gene encodes a novel nuclear protein
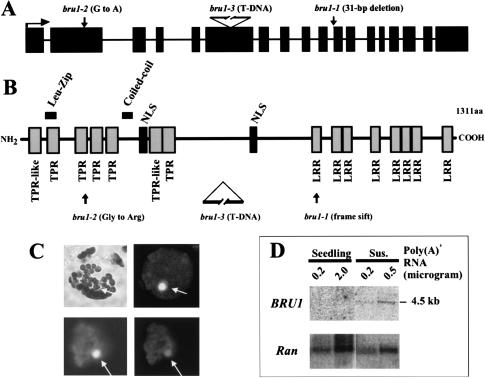
Because the T-DNA residing in the bru1-1 strain was not genetically linked with the mutation, the BRU1 gene was identified by map-based cloning (data not shown). Sequence analysis of bru1-2 and phenotypic characterization of the putative insertion allele bru1-3 assigned all three alleles to the BRU1 gene (Fig. 3A,B). BRU1 is located in the upper arm of chromosome 3 and encodes a putative open reading frame of 1311 amino acids (At3g18730; GenBank accession no. AY560347). The predicted BRU1 protein shows no obvious similarity to any protein with an assigned function present in the public databases. However, BRU1 contains two conserved domains involved in protein–protein interaction, namely tetratricopeptide repeats (TPRs; Lamb et al. 1995; Blatch and Lassle 1999) and leucine-rich repeats (LRRs; Kobe and Deisenhofer 1994), located in the N- and C-terminal parts of the protein, respectively (Fig. 3B). BRU1 also includes a predicted coiled-coil region and a leucine zipper motif that can potentially interact with DNA (Fig. 3B). There is no other gene related to BRU1 present in Arabidopsis. There is a putative BRU1 homolog in rice (chromosome 2, BAC clone; AP004095) that shows 45% identity and 62% similarity over the entire coding region. However, there are no obvious BRU1 homologs in animals or fungi.
Figure 3.
The BRU1 gene encodes a novel nuclear protein. (A) Structure of the BRU1 gene and its mutant alleles (exons are marked as black rectangles, introns as lines, and translation start as horizontal arrow). Positions of mutations are indicated. (B) Structure of the BRU1 protein. Predicted functional motifs and positions of mutations are indicated. (C) Nuclear localization of BRU1–GFP fusion protein, showing a bright-field image of a transformed proto-plast (top left) and a dark-field image of GFP fluorescence (top right, no Triton X-100 treatment). Below images of DAPI-stained nuclei (bottom left) and GFP fluorescence (bottom right) in the presence of Triton X-100. Nuclei are indicated by arrows. (D) Northern blot analysis of BRU1 mRNA. Lanes with poly(A)+ RNA from seedlings and suspension culture cells (Sus.) are marked. RNA was hybridized with probes for BRU1 and AtRanBP1a (Ran; Haizel et al. 1997) as a loading control.
The bru1-1 allele has a 31-bp deletion removing the junction between the 10th intron and the 11th exon, causing mis-splicing and thereby a frame-shift, which results in truncation of the C-terminal LRR domain (Fig. 3A,B). The bru1-2 mutation produces a single nucleotide substitution (G to A) and consequent amino acid exchange (Gly into Arg) in the N-terminal TPR region. Interestingly, the change in bru1-2 is at the amino acid in position 27 that is crucial for the antiparallel helical structure of the 34-amino acid-long TPR unit (Blatch and Lassle 1999). In bru1-3, the T-DNA insertion disrupts the linker section between the TPR and LRR regions (Fig. 3B).
As mutations in BRU1 render the plant extremely sensitive to DNA-damaging treatment, it was to be expected that BRU1 performs its function in the nucleus. Indeed, BRU1 contains two predicted nuclear localization signals (Fig. 3B). To verify the subcellular localization of BRU1, we constructed an N-terminal protein fusion of BRU1 with the green fluorescent protein (GFP). The BRU1–GFP fusion protein, when expressed in Nicotiana plumbaginifolia protoplasts, was found exclusively in the nucleus (Fig. 3C).
The BRU1 mRNA (4.5 kb) is not readily detectable by Northern blot analysis of various plant tissues, but it is clearly more abundant in suspension culture cells (Fig. 3D). Moreover, a tobacco homolog of BRU1 is expressed preferentially in S phase (T. Suzuki, pers. comm.).
Frequency of intra-chromosomal homologous recombination is elevated in bru1
Because bru1 mutants are sensitive to treatments that increase levels of DSBs, and DSBs are repaired by either homologous recombination (HR) or nonhomologous end joining (NHEJ), we examined whether bru1 is altered in either mechanism. To determine the level of HR, bru1-1 was crossed to a transgenic line containing a chromosomally integrated recombination substrate consisting of two overlapping parts of the β-glucuronidase (GUS) transgene (Fig. 4A; Swoboda et al. 1994). An intrachromosomal recombination event between the homologous regions of the incomplete transgenes should restore the structure of the GUS gene and its expression. Thus, the frequency of HR can be estimated by histochemical staining and determination of the number of blue tissue sectors expressing GUS. Compared with the wild type, bru1-1 plants had approximately fourfold higher levels of HR (Fig. 4A). Elevated HR was apparent under standard growth conditions without any artificial increase in DNA damage by application of genotoxins. Thus, bru1-1 carries out HR with an efficiency actually exceeding that of the wild type.
Figure 4.
Frequency of intrachromosomal homologous recombination, DSB-repair proficiency, and constitutive induction of a genotoxic stress responsive gene in bru1. (A, top) The recombination trap containing defective but overlapping sequences of the GUS reporter gene separated by a hygromycin phosphotransferase (HPT) marker gene as indicated. Homologous recombination generates a functional GUS reporter gene. The recombination trap was introduced into a bru1-1 heterozygous plant by crossing, and its F2 progeny was genotyped for individuals homozygous for bru1-1 or BRU1 and homozygous for GUS transgene. (Bottom) Progenies of three mutant (columns a–c) and two wild-type (columns a and b) parental lines were examined for recombination frequency based on average numbers of GUS sectors per plant. The average scores in all bru1-1 and wild-type lines (mean) and the numbers of individuals tested from each parental line (n) are indicated. (B) Time course of DSB repair after bleomycin treatment in bru1. DSBs induced by bleomycin were detected in a comet assay. Levels of DSBs are given as percent DNA in a tail of comet images. Each experimental point is represented by the mean value and standard error from at least three independent experiments in which 100 comets on four gels were evaluated. (NT) not treated with bleomycin. (C) RNA blot analysis of AtPARP-2 mRNA. Aliquots (20 μg) of total RNA from wild type (WT) and bru1-1 were loaded and hybridized with AtPARP-2- and AtRanBP1a-specific probes as indicated.
Bru1 is proficient in NHEJ
To investigate whether the sensitivity of bru1 to high levels of DSBs is due to a deficiency in NHEJ, we assessed repair proficiency of DSBs induced by bleomycin using the comet assay (Fig. 4B). Surprisingly, bleomycin-induced DSBs were repaired in bru1-1 as rapidly as in the wild type (Fig. 4B). This is in contrast to the mim1 mutant deficient in a protein related to structural maintenance of chromosome (SMC) proteins (Mengiste et al. 1999) and affected in both HR (Mengiste et al. 1999) and NHEJ (Fig. 4B). These results suggest that bru1 is proficient in both DSB repair pathways, despite its hypersensitivity to agents inducing additional DNA damage.
Bru1 has constitutively activated genotoxic stress responses
As increased levels of DSBs result in elevated levels of HR in the wild type (Lebel et al. 1993; Puchta et al. 1993, 1996), we hypothesized that bru1 has naturally increased levels of DSBs and that additional genotoxic treatment with cumulative levels of DSBs would simply exceed the capacity of the repair pathways, possibly leading to an apoptotic-like response. As a consequence, constitutively increased levels of DSBs in bru1 should be reflected, even under normal growth conditions, by constitutive induction of genotoxic stress responses. To test this hypothesis, we examined the expression of the Arabidopsis poly (ADP-ribose) polymerase-2 gene (AtPARP-2), a transcriptionally induced marker gene associated with elevated levels of DSBs (Doucet-Chabeaud et al. 2001; Chen et al. 2003). Consistent with the hypothesis, expression of AtPARP-2 is significantly (two- to three-fold) up-regulated in both bru1-1 and bru1-2 plants compared with the wild type and under normal growth conditions (Fig. 4C; data not shown).
Release of transcriptional gene silencing in bru1 mutants
The bru1-2 allele was isolated in a screen for mutations interfering with the maintenance of TGS at a silent LUC locus (Fig. 5A). In bru1-2, silencing of LUC is released in leaves of the mutant plants; however, this release is not uniform and luciferase activation can be observed in sectors of tissue (Fig. 5A). These sectors probably reflect stochastic deregulation of silencing followed by mitotic transmission of the newly acquired active state. TGS mutants, such as mom1 (Amedeo et al. 2000), met1 (Saze et al. 2003), and ddm1 (Jeddeloh et al. 1998) release TGS rather uniformly throughout the entire plant (Fig. 5B). To examine whether patchy release of silencing is characteristic of bru1 mutations and not simply a peculiarity of the transgenic LUC locus, we crossed the bru1-1 mutant allele to a line carrying transcriptionally silent GUS transgene (line 6B5; Morel et al. 2000). In subsequent analysis of the segregating F2 progeny for release of GUS expression, only bru1-1 homozygotes released silencing at the GUS transgene (Fig. 5B; data not shown). Importantly, activation of the GUS locus occurred in a stochastic fashion (Fig. 5B). Therefore, the patchy release of silencing seems to be an idiosyncrasy of bru1 mutations. Such epigenetic variegation has not been reported for any other silencing mutant, but fasciata (fas) mutants show a similar phenotype (H. Kaya, T. Araki, and K. Shibahara, unpubl.). In addition, bru1-1 plants were examined for the release of silencing at pericentromeric repeats (Fig. 5C), which are transcriptionally silent in wild-type Arabidopsis but activated in a number of mutants affected in TGS maintenance (TSI [transcriptionally silent information]; Steimer et al. 2000; Jackson et al. 2002; Saze et al. 2003). Again, only bru1-1 homozygotes, but not heterozygous or wild-type segregants, released silencing of TSI. Interestingly, the level of TSI expression varied between homozygous bru1-1 plants, likely reflecting the stochastic nature of silencing release. Variable release of TSI silencing was also observed in homozygous mutants of the other two bru1 alleles (data not shown).
Figure 5.
Maintenance of TGS is compromised in bru1. (A) bru1-2 releases silencing of a transcriptionally repressed LUC locus. Expression of LUC was examined in wild-type (Ctl) and bru1-2 backgrounds (left column shows bright-light image, right column shows luciferase signals in false-color image). (B) Release of silencing at a transcriptionally silent GUS locus in bru1-1 or mom1-1 homozygous background and wild-type control. Expression of GUS is apparent as blue staining. (C) bru1 activates transcriptionally silent pericentromeric repeats (TSI). (Upper panel) Segregated homozygous bru1-1 (bb, lanes 4,6,7), heterozygous (Bb, lanes 2,5), and wild-type (BB, lanes 1,3,8) individuals were tested for expression of TSI (Steimer et al. 2000) by RNA blot analysis. (Lower panel) Ethidium bromide (EtBr)-stained gel before blotting as loading control. (D) bru1-1 does not affect HpaII and MspI cytosine methylation at the 180-bp centromeric repeats nor at reactivated TSI loci. Genomic DNA was digested with HpaII and MspI and examined by Southern blot analysis with a probe specific for 180-bp centromeric repeats (Vongs et al. 1993) or with a TSI cDNA (Steimer et al. 2000). The mutant ddm1-7 (Vongs et al. 1993) served as a control. (E) Expression of TSI RNA in seedlings of the double mutant heterozygous for a mutation in the AtCAP-E1 gene and homozygous for a mutation in the AtCAP-E2 gene (Siddiqui et al. 2003), and of the homozygous mre11-1 mutant (Bundock and Hooykaas 2002), analyzed by Northern blot as in C (left), and by RT–PCR (right), with (+) or without (-) reverse transcriptase (RT). RNA levels were compared with the corresponding wild-type and bru1-1 seedlings. EtBr staining of ribosomal RNAs (rRNA) and RT–PCR of Actin2 mRNA were used as standards.
The bru1 mutation does not affect global DNA methylation
Because TGS in plants is often associated with DNA hypermethylation and its release with a decrease in methylation (Martienssen and Colot 2001), we examined the effect of bru1 mutations on the normally hypermethylated 180-bp centromere repeats (Vongs et al. 1993). We performed Southern blot analysis after restriction with HpaII and MspI, which both recognize the sequence CCGG. HpaII restriction is inhibited by methylation of either of the two cytosines of the recognition site, whereas MspI is inhibited only by methylation of the first cytosine. As shown in Figure 5D, no differences in digestion patterns were detected between wild type and bru1-1, indicating that DNA methylation in either CpG or CpNpG sequences is not significantly affected by the bru1 mutation. This is in sharp contrast to the ddm1 mutation, which clearly affects DNA methylation levels at the centromeric repeats (Fig. 5D; Vongs et al. 1993; Mittelsten Scheid et al. 1998). Therefore, bru1 mutations release TGS via a mechanism not associated with global changes in DNA methylation levels. Moreover, it is possible that TGS release is also not accompanied by local changes in DNA methylation, because HpaII and MspI site methylation of TSI loci was not detectably altered despite their transcriptional activation in bru1-1 (Fig. 5D).
Relationship of bru1 to other mutants with similar changes in plant morphology
The morphological alterations such as fasciation that are characteristic of bru1 alleles are reminiscent of fas1 and fas2 (Leyser and Furner 1992) or clavata (clv) mutants (Clark et al. 1993; Carles and Fletcher 2003). However, the FAS and CLV genes are thought to function in different pathways (Leyser and Furner 1992; Kaya et al. 2001). fas1 and fas2 are loss-of-function alleles in genes encoding the p150 and p60 subunits of the CAF-1 complex, respectively (Kaya et al. 2001), whereas CLV1, CLV2, and CLV3 seem to encode components of a signal transduction pathway, namely a heterodimeric receptor kinase and its ligand, involved in intercellular communication (Carles and Fletcher 2003). As BRU1 is a nuclear protein involved in genome stability, its function is more likely to be related to the function of FAS than to the CLV complex located at the plasmalemma. In addition, whereas CLV genes are specific regulators of shoot apical meristem organization, both BRU1 and FAS also affect root development. Interestingly, mutations in MRE11 (AtMRE11-1; Bundock and Hooykaas 2002) or SMC2 (AtCAP-E1 and AtCAP-E2, encoding core subunits of the condensin complex; Siddiqui et al. 2003) were also reported to produce fasciation in Arabidopsis. Both MRE11 and condensin complexes are likely to be involved in chromatin/DNA replication and S-phase DNA-damage checkpoints of the cell cycle (D'Amours and Jackson 2002; Hagstrom and Meyer 2003). Therefore, we investigated possible links between chromatin/DNA replication, DNA-damage response, TGS and meristem maintenance, by focusing on the relationship between bru1 and fas, as well as mre11 and condensin mutants.
We tested whether mre11 and the condensin mutant also affect epigenetic regulation in heterochromatin. Plants homozygous for mutant alleles of AtMRE11 gene have reduced vigor and exhibit fasciation in later developmental stages (Bundock and Hooykaas 2002). AtCAP-E1 and AtCAP-E2 are functionally redundant, and individuals homozygous for both mutant alleles as well as plants with genotype AtCAP-E1-/-, AtCAP-E2+/- die during embryo development (Siddiqui et al. 2003). However, AtCAP-E1+/-, AtCAP-E2-/- plants are viable but exhibit fasciation due to meristem disorganization (Siddiqui et al. 2003). Importantly, the AtCAP-E1+/-, AtCAP-E2-/- double mutants express TSI at levels similar to bru1 (Fig. 5E). Furthermore, seedlings homozygous for the AtMRE11-1 mutation also express TSI, but at a level lower than in bru1 (Fig. 5E). These results suggest that defects in the S-phase DNA-damage checkpoint or inaccuracies during chromatin/DNA replication cause instability of epigenetic states of newly replicated chromatin, and that this is reflected by the fasciation phenotypes common to the mutants examined. These possibilities were further supported by defects in fas mutants, strikingly similar to those observed in bru1, as described below.
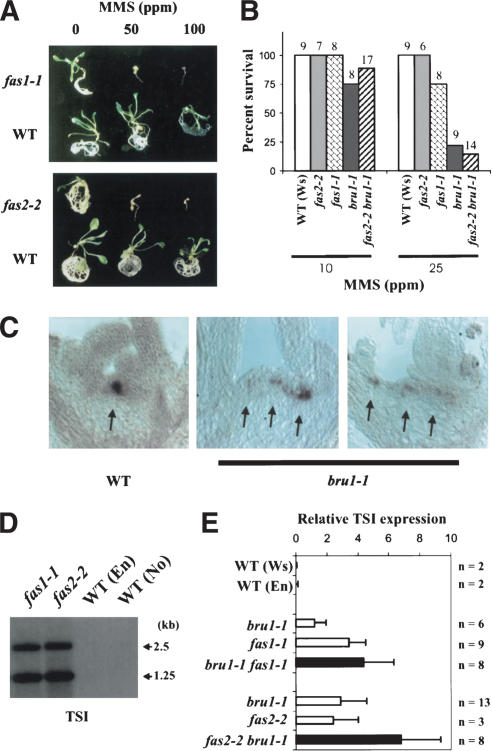
With regard to the sensitivity to genotoxic stress, both fas1 and fas2 were sensitive to 50 ppm of MMS, whereas wild-type plants tolerated 100 ppm (Fig. 6A). Thus, fas1 and fas2 are also hypersensitive to genotoxic stress, but to a lower extent than bru1 (Fig. 6B).
Figure 6.
Functional similarity between bru1 and fas1 or fas2 mutants. (A) Sensitivity of fas1-1 and fas2-2 and their respective wild types En and No (WT) to MMS. Ten seedlings were tested in each treatment, and representative seedlings are displayed. (B) Sensitivity of mutants to low concentrations of MMS. The number of seedlings used for each treatment is indicated above the bars. The survival rates after each treatment are shown. (C) Distorted shoot apical meristem and misexpression of the WUSCHEL gene in bru1-1 (middle and right) compared with wild type (left). Localization of WUS mRNA in shoot apical meristems of wild type and bru1-1 is visible as dark purple signals (arrows). (D) fas1-1 and fas2-2 activate transcriptionally silent pericentromeric repeats (TSI). An RNA blot with equal loading of total cellular RNA (as verified by EtBr-staining of rRNAs) was hybridized with the TSI-specific probe as in Figure 5C. (E) Quantification of TSI expression in bru/fas double mutants. Samples of total RNA of wild type (WT), bru1-1, fas1-1, fas2-2, and their double mutants were analyzed by RNA blotting with a probe specific for TSI and AtRanBP1a. Means and standard deviations of TSI RNA levels relative to AtRanBP1a mRNA are shown. The numbers of plants tested (n) are indicated. Expression levels in the double mutants were compared to single mutants segregated from two parental lines (middle three and bottom three).
To investigate whether fas mutations interfere with TGS, we examined expression of TSI and reactivation of the GUS marker, which is silenced within the line 6b5, in the fas1 and fas2 background. Similarly to bru1-1, TSI was expressed in both fas1 and fas2 (Fig. 6D), and crosses between fas1 or fas2 and line 6b5 segregated fas mutant plants stochastically expressing GUS (data not shown). Moreover, transcription of the CACTA transposable element (Miura et al. 2001), which is silent in wild-type plants but activated in the ddm1 mutant, was also found to be stochastically up-regulated in a small proportion of cells of fas mutants (H. Kaya, T. Araki, and K. Shibahara, unpubl.). As in bru1, release of TGS in fas1 and fas2 was not accompanied by global changes in DNA methylation (data not shown).
In both fas mutants, shoot apical meristems are disorganized, and this is reflected by distorted meristematic localization of WUSCHEL (WUS) mRNA (Kaya et al. 2001). WUS is required for maintenance of stem cell identity in the shoot apical meristem (Mayer et al. 1998). Similar to fas1 and fas2, disorganized meristems and an abnormal, dispersed expression pattern of WUS mRNA were observed in bru1 (Fig. 6C).
We also examined a potential epistatic relationship between bru1-1; fas1-1 and bru1-1; fas2-2 alleles by constructing double mutant lines. Both double mutants were viable and showed a range of phenotypes similar to those of either of the single parental mutants (Fig. 2; data not shown). Moreover, double mutants showed a level of sensitivity to MMS similar to that of single bru1-1 mutant plants (Fig. 6B). Thus, by these criteria, bru1 is most likely to be epistatic to fas1 and fas2. To test whether BRU1 interacts directly with CAF-1, an in vitro protein interaction assay was employed, using a Baculovirus-expressed BRU1 protein tagged with GST and in vitro-translated subunits of CAF-1, which were previously shown to form an active complex (Kaya et al. 2001). However, we were unable to detect specific interaction between BRU1 and the CAF-1 subunits (data not shown). Surprisingly, double mutants exhibit an additive effect on the release of TGS, reflected by elevated levels of TSI expression (Fig. 6E). Taken together, these results suggest that BRU1 is involved in chromatin assembly, and that BRU1 and CAF-1 contribute additively to postreplicative stability of epigenetic states.
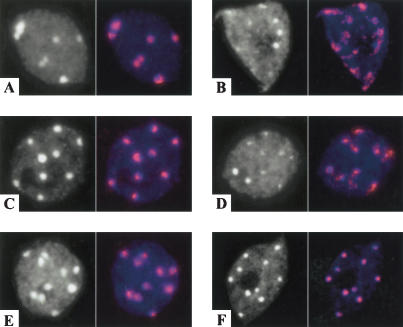
We examined the effects of the bru1, fas1, and fas2 mutations on heterochromatin organization in interphase nuclei by fluorescence in situ hybridization (FISH) with a probe specific for centromeric repetitive DNA (Vongs et al. 1993). The analysis revealed altered heterochromatin distribution patterns in bru1. Although the organization of centromeric heterochromatin in the majority of bru1-1 nuclei is similar to wild type (Fig. 7A,C), some nuclei showed a significant diffusion of chromocenters (Fig. 7B). Similarly, decondensation of centromeric heterochromatin was observed in some but not all nuclei in bru1-2 (Fig. 7D) and bru1-3 (data not shown). The frequency of nuclei with decondensed chromocenters varies between 2% and 8% and is correlated with the severity of the developmental aberrations. Thus, BRU1 is involved in heterochromatin condensation. In contrast, there were no changes in heterochromatin condensation in fas1 (data not shown) or fas2 (Fig. 7F) compared with wild type (Fig. 7E). Double mutants for bru1 and fas1 were not different in centromeric heterochromatin organization relative to the bru1 single mutant (data not shown).
Figure 7.
Chromatin organization in interphase nuclei of bru1 and fas2 mutants. DAPI-stained nuclei (left panels) and FISH with a probe specific for 180-bp centromeric repeats (right panels) visualizing the compaction of the centromeric DNA (red) in wild-type Ws (A), bru1-1 (B), wild-type Col (C), bru1-2 (D), wildtype No (E), and fas2-2 (F).
Discussion
We have isolated and characterized an allelic series of mutations in the BRU1 gene of Arabidopsis that result in a drastic increase in sensitivity to genotoxic stress, elevated levels of intrachromosomal homologous recombination, and instability in the maintenance of TGS. Considering that other previous Arabidopsis TGS mutants, such as ddm1 (Schaeffer et al. 1993; Vongs et al. 1993; Mittelsten Scheid et al. 1998), mom1 (Amedeo et al. 2000), and met1 (Kankel et al. 2003; Saze et al. 2003), have unchanged resistance to DNA-damaging treatments (data not shown), BRU1 seems to play a dual function as guardian of both genetic and epigenetic information.
BRU1 encodes a novel protein. Clues to BRU1 function were provided by comparative analysis of bru1 with mutants in CAF-1 subunits, MRE11, or condensin subunits. These mutations result in developmental alterations similar to bru1, and the corresponding genes have been functionally related to DNA and chromatin replication. Interestingly, these complexes are thought to be important for the replication of chromosomal regions containing repeated, transcriptionally silent DNA packaged into heterochromatin (Enomoto and Berman 1998; D'Amours and Jackson 2002; Lobachev et al. 2002; Hagstrom and Meyer 2003), whereas bru1 mutations release gene silencing and alter chromatin structure in the area of pericentromeric heterochromatin.
We substantiated the phenotypic correlations by demonstrating that fas1 and fas2 mutants were also hypersensitive to MMS, although less so than bru1. This may reflect functional redundancy between CAF-1 (Kaya et al. 2001) and the putative ASF1 chromatin-assembling subunits encoded by genes At5G38110 and At1G66740 in Arabidopsis, whereas there is no obvious redundancy for BRU1. It is also possible that BRU1 has further roles in the prevention or the repair of DNA damage.
In the apical meristems of fas1 and fas2 mutants, the maintenance of spatial expression patterns of the WUS gene is disturbed, and it has been proposed that CAF-1 secures the organization of meristems by stabilization of the epigenetic inheritance of gene expression patterns (Kaya et al. 2001). In bru1, WUS is also mis-expressed, and the shoot apical meristem structure is disrupted similarly to fas1 and fas2. A common role of BRU1 and CAF-1 subunits in epigenetic inheritance is further supported by the activation of transcriptionally silent pericentromeric repeats in all these mutants. Moreover, this TGS release occurs without obvious changes in DNA methylation and in a variegated fashion, implying related mechanisms of silencing control. Importantly, epistatic analyses suggested that these proteins serve both overlapping and nonoverlapping functions.
Although no direct interaction was detected between BRU1 and the CAF-1 subunits or histones, BRU1 may be indirectly involved in histone chaperoning mediated by CAF-1 or associated with CAF-1 through additional protein(s). In yeast, CAF-1-mediated nucleosome assembly following DNA replication facilitates stabilization of newly reconstituted chromatin (Enomoto and Berman 1998). Considering the alterations in centromeric heterochromatin observed in bru1, BRU1 possibly plays a role in chromatin replication and may also be involved in postreplicative stabilization of chromatin structure.
As with the fas mutants, bru1 shares several features beyond phenotypic similarity with mutations in MRE11 and condensin genes (Bundock and Hooykaas 2002; Siddiqui et al. 2003). MRE11 is part of the MRE11/RAD50/NBS complex that is thought to be involved in DSB repair by NHEJ. Mutations in the individual subunits produce largely divergent phenotypes. Both mre11 and rad50 mutants are hypersensitive to treatments provoking DSBs (although not to the extent of the bru1 mutation; Gallego et al. 2001; Bundock and Hooykaas 2002) and show elevated levels of homologous recombination (in Arabidopsis documented only for rad50; Gherbi et al. 2001). However, only mre11 mutants, and not rad50 mutants, exhibit fasciated phenotypes similar to bru1. Thus, deficiency in NHEJ does not inevitably result in developmental abnormalities characteristic of bru1 and mre11. This implies that BRU1 function is unlikely to be linked to NHEJ. Indeed, bru1 mutants are proficient in NHEJ, as shown by the comet assay. Depletion of the MRE11 complex in Xenopus leads to spontaneous accumulation of DSBs (Costanzo et al. 2001). Considering the similarity of bru1 and mre11 mutants in Arabidopsis, it is possible that hypersensitivity of bru1 to genotoxic treatments is in part a result of constitutively elevated levels of DSBs, prior to induction of additional DNA DSBs by exogenously applied genotoxins. This is in agreement with the constantly elevated expression level in bru1 of AtPARP-2, a marker gene associated with elevated levels of DNA damage (Doucet-Chabeaud et al. 2001; Chen et al. 2003). In addition, consistent with the spontaneous up-regulation of genotoxic stress response, we observed less damage in bru1 than in wild type shortly after bleomycin treatment (Fig. 4B; data not shown). Importantly, defects in chromatin assembly in human cells induced by a dominant-negative mutation of the CAF-1 p150 subunit result in stalled replication forks that are inappropriately processed and lead to increased DSBs (Ye et al. 2003). This is accompanied by activation of the S-phase checkpoint, resulting in S-phase arrest (Ye et al. 2003). RNAi depletion of CAF-1 also causes S-phase checkpoint activation and accumulation of cells in early and mid-S phase. In this case, the arrest seemed to be a consequence of a perturbation in chromatin replication (Hoek and Stillman 2003), because the ATR (ATM/Rad3-related) signaling kinase but not the ATM (ataxia-telangiectasia-mutated) pathway was activated. The existence of a plant homolog of the mammalian ATM kinase (Garcia et al. 2003), a key mediator of S-phase checkpoints and activation of concomitant DNA-repair responses, suggests that a similar genome surveillance system could operate in plants. It is possible that chromatin replication in the absence of functional BRU1 protein causes instability of the replication forks, leading either to elevated DSBs or to deficiencies in chromatin replication. In both cases, this results in activation of the S-phase checkpoint control.
Mutations in condensin and mre11, but not in rad50 genes, result in developmental abnormalities similar to bru1. Noticeably, both condensin and MRE11 are involved in S-phase checkpoint activation (Aono et al. 2002; D'Amours and Jackson 2002; Hagstrom and Meyer 2003). Thus, it is plausible that BRU1 also plays a role in this process together with the CAF-1 complex, acting upstream of the checkpoint activation. Interestingly, the most crucial roles assigned to MRE11 and CAF-1 are for the smooth and high-fidelity replication of DNA repeat regions, which are usually packaged into compact heterochromatin (Lobachev et al. 2002; Hoek and Stillman 2003). In the present study, we demonstrate that the silencing of TSI is also compromised in mre11 and condensin mutants. The similar developmental abnormalities and destabilization of the genetic and epigenetic inheritance observed in all mutants examined here emphasize the tight link between chromatin assembly and the S-phase checkpoint. One possibility is that inaccurate chromatin replication and/or checkpoint defects can trigger downstream events such as accumulation of DNA damage, aberrant chromatin structures, and release of epigenetic gene silencing. Obviously, all of these events can also take place during unscheduled DNA synthesis accompanying DNA repair, and deficiencies in the components involved may contribute to the postreplicative increase of DNA damage, thus exhausting its efficient repair.
In summary, we propose that the novel BRU1 protein is involved in structural and functional stabilization of chromatin. Complex and multiple facets of bru1 mutants and the overlapping roles of BRU1, CAF-1, MRE11, and condensin strongly suggest that BRU1 is a new molecular link between maintenance of both genetic and epigenetic information and the control of development.
Materials and methods
Plant material and growth conditions
For isolation of bru1-1 and bru1-2, we screened 2500 T-DNA-mutagenized Arabidopsis (ecotype Wassilevskija [Ws]) lines, generated at the Institute de la Recherche Agronomique (INRA), Versailles, France, and 50,000 lines of Arabidopsis (ecotype Columbia [Col] containing a transcriptionally silent LUC locus) mutagenized by EMS. bru1-3 (ecotype Col) was obtained from the Torrey Mesa Research Institute (http://www.tmri.org). fas1-1 (ecotype Enkheim [En]) and fas2-2 (ecotype Nossen [No]) were described previously (Leyser and Furner 1992; Kaya et al. 2001). The genotypes of bru1 and fas mutants were determined by PCR with primers differentiating between wild-type and mutated loci. The line 6b5 (ecotype Col) containing transcriptionally silent, multiple copies of a GUS transgene (Morel et al. 2000) was kindly provided by H. Vaucheret (INRA, Versailles, France). Plants were grown in 12 h light/12 h dark cycles (short day) or 16 h light/8 h dark cycles (long day) at 21°C during the day and 16°C during the night. Seedlings of mutants homozygous for AtMRE11-1 (ecotype Ws) and the double mutant for AtCAP-E1+/- (ecotype Ws); AtCAP-E2-/- (ecotype Col) were grown and selected on half-strength MS medium supplemented with 10 g/L sucrose, 1.5% agar, and 50 mg/L kanamycin in long-day conditions as described (Bundock and Hooykaas 2002; Siddiqui et al. 2003).
Tests for sensitivity to genotoxic stresses
Three-day-old seedlings grown under aseptic conditions were tested for sensitivity to MMS (Fluka) as described (Revenkova et al. 1999). Sensitivity to bleomycin, mitomycin C, and UV-C were examined as described (Mengiste et al. 1999).
Map-based cloning of the BRU1 gene
bru1- was crossed to Col and Landsberg erecta (Ler) wild-type plants. DNA from 1400 F2 plants was analyzed for SSLP and CAPS markers, generated, and evaluated based on the polymorphism data provided by CEREON (http://www.arabidopsis.org/Cereon). Genes at the mapped interval were amplified by PCR and sequenced. A BRU1 cDNA was amplified by PCR from cDNA mixtures, using primers designed to contain sequences corresponding to 5′- and 3′-ends of the annotated ORF, respectively (The Arabidopsis Information Resource, http://arabidopsis.org; forward, 5′-TAGGAATTCCTGATGGGTCGA TTAGATGTAGCTGCGGCG-3′, and reverse, 5′-TAGGAATT CCTGGTCCTTTCTGCAGCATGATTTGACTCCGC-3′).
Subcellular localization of BRU1–GFP fusion protein
The BRU1 cDNA was inserted after sequence determination into a plant expression vector (derivative of pUC-based vector) containing a reporter gene encoding enhanced GFP (F64L, S65C) and driven by a modified CaMV 35S promoter. The resulting plasmids were introduced into Nicotiana plumbaginifolia protoplasts by PEG-mediated transformation, and the protoplasts were incubated at 28°C in the dark for 8 h. GFP signals were analyzed using a Leitz DMR fluorescence microscope, and images were captured with a SPOT RT camera (Diagnostic Instruments) using the SPOT advanced software. For visualization of nuclei, protoplasts were stained with 10 mg/mL DAPI in the presence of 0.5% Triton X-100.
Southern and Northern blot analyses, RT–PCR, fluorescence in situ hybridization, and in situ mRNA localization
Southern and Northern blot analyses were performed and specific probes for TSI and AtRanBP1a were prepared as described (Steimer et al. 2000). A cDNA probe for AtPARP-2 was obtained by RT–PCR as described (Doucet-Chabeaud et al. 2001). RT–PCR with poly-dT primer for cDNA synthesis and with Actin2- and TSI-specific primers for cDNA amplification was performed as described (Saze et al. 2003). For fluorescence in situ hybridization, the biotin-labeled 180-bp centromeric repeat (Vongs et al. 1993) was hybridized to nuclear spreads prepared from ethanol-acetic acid-fixed rosette leaves, detected with Texas Red conjugated avidin (Mittelsten Scheid et al. 2002; Probst et al. 2003), and analyzed using a Leitz DMR fluorescence microscope. For each genotype, at least five plants were examined and 150–350 nuclei per plant were scored. In situ localization of WUS mRNA was performed as described (Kaya et al. 2001).
GUS staining and intrachromosomal recombination assay
bru1-1 was crossed to line 651 (ecotype C24) carrying a reporter construct for intrachromosomal recombination (Swoboda et al. 1994). Four-week-old F3 plants homozygous for the mutant bru1-1 allele or wild-type BRU1 gene and homozygous for the GUS transgene were stained histochemically as described (Amedeo et al. 2000).
Comet assay
A single-cell gel electrophoresis (comet) assay to detect DSBs (N/N protocol) was performed as described (Angelis et al. 1999, 2000), using cells from aerial tissues of 10-day-old seedlings. DSB repair was measured after induction of DSBs by treatment with bleomycin (30 μg/mL) for 1 h.
Luciferase assay
Plants carrying the transcriptionally silent LUC locus were sprayed with a 1-mM aqueous solution of the substrate luciferin. Luciferin (Molecular Probes) was dissolved in sterile water and stored frozen as a 25-mM stock solution. In vivo imaging of luciferase activity was performed with a CCD camera system employing Argus 50 Software (Hamamatsu Photonic Deutschland).
Note added in proof
While this manuscript was under review, Guyomarc'h et al. (2004) and Suzuki et al. (2004) reported isolation of additional mutant alleles of the BRU1 gene causing similar developmental abnormalities as described here. These data complement and confirm the phenotypic description of bru1 mutants given here.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Acknowledgments
We thank B. Hohn for the reporter line for the HR assay, Y. Habu for the pGFPEx-ENS plasmid, D. Riggs for seeds of the AtCAP-E1+/-; AtCAP-E2-/- double mutants, P. Bundock for seeds of the mre11-1 mutant, E. Richards for the 180-bp repeat probe, H. Vaucheret for the Arabidopsis thaliana 6b5 line, INRA for the mutant collection, TMRI for the bru1-3 mutant, and K. Afsar and S. Lienhard for technical assistance. We thank S. Adams and P. King for comments on the manuscript. This work was supported by grants #521/01/1418 from Grant Agency of the Czech Republic, #A6038201 from the Grant Agency of the Academy of Sciences of the Czech Republic, by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan (#14036219 to T.A.), and by the Deutsche Forschungsgemeinschaft (SFB 363, project #14). H.K. was supported by a Japan Society for the Promotion of Science Research Fellowship for Young Scientists. K.S. was supported by grant-in-aid 15023259 for scientific research on priority area in cancer research, 20263098 for young scientists (A), and 14GS0321 for creative scientific research from MEXT of Japan, and by grants from HFSP. S.T. was supported by EU Grant QLK3-CT-2000-00365/BBW00.0187-2.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.295404.
Corresponding author.
References
- Amedeo P., Habu, Y., Afsar, K., Scheid, O.M., and Paszkowski, J. 2000. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405: 203-206. [DOI] [PubMed] [Google Scholar]
- Angelis K.J., Dusinska, M., and Collins, A.R. 1999. Single cell gel electrophoresis: Detection of DNA damage at different levels of sensitivity. Electrophoresis 20: 2133-2138. [DOI] [PubMed] [Google Scholar]
- Angelis K.J., McGuffie, M., Menke, M., and Schubert, I. 2000. Adaptation to alkylation damage in DNA measured by the comet assay. Environ. Mol. Mutagen. 36: 146-150. [DOI] [PubMed] [Google Scholar]
- Aono N., Sutani, T., Tomonaga, T., Mochida, S., and Yanagida, M. 2002. Cnd2 has dual roles in mitotic condensation and interphase. Nature 417: 197-202. [DOI] [PubMed] [Google Scholar]
- Blatch G.L. and Lassle, M. 1999. The tetratricopeptide repeat: A structural motif mediating protein–protein interactions. Bioessays 21: 932-939. [DOI] [PubMed] [Google Scholar]
- Bundock P. and Hooykaas, P. 2002. Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14: 2451-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C.C. and Fletcher, J.C. 2003. Shoot apical meristem maintenance: The art of a dynamic balance. Trends Plant Sci. 8: 394-401. [DOI] [PubMed] [Google Scholar]
- Chen I.P., Haehnel, U., Altschmied, L., Schubert, I., and Puchta, H. 2003. The transcriptional response of Arabidopsis to genotoxic stress—A high-density colony array study (HDCA). Plant J. 35: 771-786. [DOI] [PubMed] [Google Scholar]
- Clark S.E., Running, M.P., and Meyerowitz, E.M. 1993. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119: 397-418. [DOI] [PubMed] [Google Scholar]
- Costanzo V., Robertson, K., Bibikova, M., Kim, E., Grieco, D., Gottesman, M., Carroll, D., and Gautier, J. 2001. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8: 137-147. [DOI] [PubMed] [Google Scholar]
- Critchlow S.E. and Jackson, S.P. 1998. DNA end-joining: From yeast to man. Trends Biochem Sci. 23: 394-398. [DOI] [PubMed] [Google Scholar]
- D'Amours D. and Jackson, S.P. 2002. The Mre11 complex: At the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3: 317-327. [DOI] [PubMed] [Google Scholar]
- Doucet-Chabeaud G., Godon, C., Brutesco, C., de Murcia, G., and Kazmaier, M. 2001. Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol. Genet. Genomics 265: 954-963. [DOI] [PubMed] [Google Scholar]
- Drapkin R., Reardon, J.T., Ansari, A., Huang, J.C., Zawel, L., Ahn, K., Sancar, A., and Reinberg, D. 1994. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 368: 769-772. [DOI] [PubMed] [Google Scholar]
- Enomoto S. and Berman, J. 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes & Dev. 12: 219-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver W.J., Svejstrup, J.Q., Bardwell, L., Bardwell, A.J., Buratowski, S., Gulyas, K.D., Donahue, T.F., Friedberg, E.C., and Kornberg, R.D. 1993. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell 75: 1379-1387. [DOI] [PubMed] [Google Scholar]
- Gaillard P.H., Martini, E.M., Kaufman, P.D., Stillman, B., Moustacchi, E., and Almouzni, G. 1996. Chromatin assembly coupled to DNA repair: A new role for chromatin assembly factor I. Cell 86: 887-896. [DOI] [PubMed] [Google Scholar]
- Gallego M.E., Jeanneau, M., Granier, F., Bouchez, D., Bechtold, N., and White, C.I. 2001. Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant J. 25: 31-41. [DOI] [PubMed] [Google Scholar]
- Garcia V., Bruchet, H., Camescasse, D., Granier, F., Bouchez, D., and Tissier, A. 2003. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherbi H., Gallego, M.E., Jalut, N., Lucht, J.M., Hohn, B., and White, C.I. 2001. Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2: 287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V.V. and Levy, A.A. 1999. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 4: 263-269. [DOI] [PubMed] [Google Scholar]
- Green C.M. and Almouzni, G. 2002. When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep. 3: 28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I. and Moazed, D. 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798-802. [DOI] [PubMed] [Google Scholar]
- Gross D.S. 2001. Sir proteins as transcriptional silencers. Trends Biochem. Sci. 26: 685-686. [DOI] [PubMed] [Google Scholar]
- Guarente L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes & Dev. 14: 1021-1026. [PubMed] [Google Scholar]
- Guyomarc'h S., Vernoux, T., Traas, J., Zhou D.X., and Delarue, M. 2004. MGOUN3, an Arabidopsis gene with Tetratrico-Peptide-Repeat-related motifs, regulates meristem cellular organization. J. Exp. Bot. 55: 673-684. [DOI] [PubMed] [Google Scholar]
- Hagstrom K.A. and Meyer, B.J. 2003. Condensin and cohesin: More than chromosome compactor and glue. Nat. Rev. Genet. 4: 520-534. [DOI] [PubMed] [Google Scholar]
- Haizel T., Merkle, T., Pay, A., Fejes, E., and Nagy, F. 1997. Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J. 11: 93-103. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411: 366-374. [DOI] [PubMed] [Google Scholar]
- Hoek M. and Stillman, B. 2003. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. 100: 12183-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth, A.M., Cao, X., and Jacobsen, S.E. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556-560. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Bender, J., and Richards, E.J. 1998. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes & Dev. 12: 1714-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong B.R., Wu-Scharf, D., Zhang, C., and Cerutti, H. 2002. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. Proc. Natl. Acad. Sci. 99: 1076-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel M.W., Ramsey, D.E., Stokes, T.L., Flowers, S.K., Haag, J.R., Jeddeloh, J.A., Riddle, N.C., Verbsky, M.L., and Richards, E.J. 2003. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P. and Almouzni, G. 2000. DNA replication, nucleotide excision repair and nucleosome assembly. In Chromatin structure and gene expression, 2nd ed. (eds. S.C.R Elgin and J.L. Workman), pp. 24-48. Oxford University Press, Oxford.
- Kaufman P.D., Kobayashi, R., and Stillman, B. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes & Dev. 11: 345-357. [DOI] [PubMed] [Google Scholar]
- Kaya H., Shibahara, K.I., Taoka, K.I., Iwabuchi, M., Stillman, B., and Araki, T. 2001. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131-142. [DOI] [PubMed] [Google Scholar]
- Kobe B. and Deisenhofer, J. 1994. The leucine-rich repeat: A versatile binding motif. Trends Biochem Sci. 19: 415-421. [DOI] [PubMed] [Google Scholar]
- Lamb J.R., Tugendreich, S., and Hieter, P. 1995. Tetratrico peptide repeat interactions: To TPR or not to TPR? Trends Biochem Sci. 20: 257-259. [DOI] [PubMed] [Google Scholar]
- Lebel E.G., Masson, J., Bogucki, A., and Paszkowski, J. 1993. Stress-induced intrachromosomal recombination in plant somatic cells. Proc. Natl. Acad. Sci. 90: 422-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser H.M.O. and Furner, I.J. 1992. Characterization of three shoot apical meristem mutants of Arabidopsis thaliana. Development 116: 397-403. [Google Scholar]
- Li E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3: 662-673. [DOI] [PubMed] [Google Scholar]
- Lobachev K.S., Gordenin, D.A., and Resnick, M.A. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183-193. [DOI] [PubMed] [Google Scholar]
- Martienssen R.A. and Colot, V. 2001. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293: 1070-1074. [DOI] [PubMed] [Google Scholar]
- Martin S.G., Laroche, T., Suka, N., Grunstein, M., and Gasser, S.M. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621-633. [DOI] [PubMed] [Google Scholar]
- Mayer K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805-815. [DOI] [PubMed] [Google Scholar]
- Mengiste T., Revenkova, E., Bechtold, N., and Paszkowski, J. 1999. An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 18: 4505-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills K.D., Sinclair, D.A., and Guarente, L. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97: 609-620. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O. and Paszkowski, J. 2000. Transcriptional gene silencing mutants. Plant Mol. Biol. 43: 235-241. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O., Afsar, K., and Paszkowski, J. 1998. Release of epigenetic gene silencing by trans-acting mutations in Arabidopsis. Proc. Natl. Acad. Sci. 95: 632-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelsten Scheid O., Probst, A.V., Afsar, K., and Paszkowski, J. 2002. Two regulatory levels of transcriptional gene silencing in Arabidopsis. Proc. Natl. Acad. Sci. 99: 13659-13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H., and Kakutani, T. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212-214. [DOI] [PubMed] [Google Scholar]
- Morel J.B., Mourrain, P., Beclin, C., and Vaucheret, H. 2000. DNA methylation and chromatin structure affect transcriptional and posttranscriptional transgene silencing in Arabidopsis. Curr. Biol. 10: 1591-1594. [DOI] [PubMed] [Google Scholar]
- Probst A.V., Fransz, P.F., Paszkowski, J., and Scheid, O.M. 2003. Two means of transcriptional reactivation within heterochromatin. Plant J. 33: 743-749. [DOI] [PubMed] [Google Scholar]
- Puchta H., Dujon, B., and Hohn, B. 1993. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 21: 5034-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1996. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. 93: 5055-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E., Masson, J., Koncz, C., Afsar, K., Jakovleva, L., and Paszkowski, J. 1999. Involvement of Arabidopsis thaliana ribosomal protein S27 in mRNA degradation triggered by genotoxic stress. EMBO J. 18: 490-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Scheid, O.M., and Paszkowski, J. 2003. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34: 65-69. [DOI] [PubMed] [Google Scholar]
- Schaeffer L., Roy, R., Humbert, S., Moncollin, V., Vermeulen, W., Hoeijmakers, J.H., Chambon, P., and Egly, J.M. 1993. DNA repair helicase: A component of BTF2 (TFIIH) basic transcription factor. Science 260: 58-63. [DOI] [PubMed] [Google Scholar]
- Schwartz J.L. 1989. Monofunctional alkylating agent-induced S-phase-dependent DNA damage. Mutat. Res. 216: 111-118. [DOI] [PubMed] [Google Scholar]
- Shibahara K. and Stillman, B. 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575-585. [DOI] [PubMed] [Google Scholar]
- Siddiqui N.U., Stronghill, P.E., Dengler, R.E., Hasenkampf, C.A., and Riggs, C.D. 2003. Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130: 3283-3295. [DOI] [PubMed] [Google Scholar]
- Smith S. and Stillman, B. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58: 15-25. [DOI] [PubMed] [Google Scholar]
- Steimer A., Amedeo, P., Afsar, K., Fransz, P., Scheid, O.M., and Paszkowski, J. 2000. Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12: 1165-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Inagaki, S., Nakajima, S., Akashi, T., Ohto, M., Kobayashi, M., Seki, M., Shinozaki, K., Kato, T., Tabata, S., et al. 2004. A novel Arabidopsis gene TONSOKU is required for proper cell arrangement in root and shoot apical meristems. Plant J. (in press). [DOI] [PubMed]
- Swoboda P., Gal, S., Hohn, B., and Puchta, H. 1994. Intrachromosomal homologous recombination in whole plants. EMBO J. 13: 484-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchenio T., Casella, J.F., and Heidmann, T. 2001. A truncated form of the human CAF-1 p150 subunit impairs the maintenance of transcriptional gene silencing in mammalian cells. Mol. Cell Biol. 21: 1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y., Kato, J., and Ikeda, H. 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388: 900-903. [DOI] [PubMed] [Google Scholar]
- Tyler J.K., Adams, C.R., Chen, S.R., Kobayashi, R., Kamakaka, R.T., and Kadonaga, J.T. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402: 555-560. [DOI] [PubMed] [Google Scholar]
- Ulm R., Revenkova, E., di Sansebastiano, G.P., Bechtold, N., and Paszkowski, J. 2001. Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes & Dev. 15: 699-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A., Kakutani, T., Martienssen, R.A., and Richards, E.J. 1993. Arabidopsis thaliana DNA methylation mutants. Science 260: 1926-1928. [DOI] [PubMed] [Google Scholar]
- Wang Z., Svejstrup, J.Q., Feaver, W.J., Wu, X., Kornberg, R.D., and Friedberg, E.C. 1994. Transcription factor b (TFIIH) is required during nucleotide-excision repair in yeast. Nature 368: 74-76. [DOI] [PubMed] [Google Scholar]
- Ye X., Franco, A.A., Santos, H., Nelson, D.M., Kaufman, P.D., and Adams, P.D. 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell 11: 341-351. [DOI] [PubMed] [Google Scholar]