Abstract
The nucleotide sequence around the translational initiation site is an important cis-acting element for post-transcriptional regulation. However, it has not been fully understood how the sequence context at the 5′-untranslated region (5′-UTR) affects the translational efficiency of individual mRNAs. In this study, we provide evidence that the 5′-UTRs of Arabidopsis genes showing a great difference in the nucleotide sequence vary greatly in translational efficiency with more than a 200-fold difference. Of the four types of nucleotides, the A residue was the most favourable nucleotide from positions −1 to −21 of the 5′-UTRs in Arabidopsis genes. In particular, the A residue in the 5′-UTR from positions −1 to −5 was required for a high-level translational efficiency. In contrast, the T residue in the 5′-UTR from positions −1 to −5 was the least favourable nucleotide in translational efficiency. Furthermore, the effect of the sequence context in the −1 to −21 region of the 5′-UTR was conserved in different plant species. Based on these observations, we propose that the sequence context immediately upstream of the AUG initiation codon plays a crucial role in determining the translational efficiency of plant genes.
INTRODUCTION
In the cell, one of the most essential life processes is the production of cellular proteins. This is largely regulated at the transcriptional level, but recent studies show that translational regulation also plays a critical role in determining protein levels. Translation is the process by which cells produce their constituent proteins from messenger RNAs (mRNAs) and thus is one of the most essential cellular processes in living organisms. Translation is a complex process involving a large number of components including mRNAs, tRNAs, ribosomes and many ribosome-associated protein factors. All these factors contribute to the translational efficiency in various ways. In addition, transport of mRNAs from the nucleus to the cytosol (1), and subcellular localization and stability of mRNAs also contribute to the translational efficiency (2,3).
Of this large number of components involved in the translation, mRNA itself has many features that are involved in controlling the translational efficiency. Most of the control elements are located in the 5′- and 3′-untranslated regions (5′- and 3′-UTRs) (4). In the 5′-UTR, the m7G cap, secondary structure, length and nucleotide sequence context around the AUG start codon are known to affect the translational efficiency (5,6). Among these factors, the secondary structure of mRNAs is regarded as the most important element for controlling translational efficiency mostly in a negative fashion (7,8). Computational calculation using the RNA base-pairing simulation method has commonly been used to determine the secondary structure of mRNAs (9). Moreover, a recent study employing deep sequencing of mRNA fragments generated by structure-specific enzymes has provided evidence that the secondary structure around the start codon reduces translational efficiency (10). In addition, specific sequence motifs present in the 5′-UTR are involved in controlling translational efficiency in either positive or negative manners depending on their nature (11,12). An example is the internal ribosomal entry sites (IRESs) found in many viral mRNAs (13). These motifs direct the cap-independent translation. Another is the upstream open reading frames (uORFs) in the 5′-UTR that can modulate the translational efficiency (14,15). In addition, the 3′-UTR also contains many sequence motifs that affect the translational efficiency (16). Of these motifs, the poly(A) tail and its binding proteins have been most extensively characterized at the molecular level (4,17).
In addition to the specific sequence motifs in the 5′- or 3′-UTRs, the length and sequence context of the 5′-UTR of an mRNA also affect translational efficiency. The effect of the sequence context of the 5′-UTR on translational efficiency has been extensively studied in a variety of organisms including plants by mainly focusing on the sequence context around the translation initiation site (18–22). In many vertebrate genes, a sequence, CC(A/G)CCAUGG, known as the Kozak sequence, is preferentially detected and this ‘consensus’ sequence is crucial for efficient translation initiation (18). In plants, similar approaches have shown that A residues at positions −1 to −4 of the 5′-UTR are crucial for high translational efficiency (19,23,24). In addition, high-throughput analysis using polysome fractionation and microarrays has identified a consensus sequence around the AUG start codon of Arabidopsis genes (20). These results raise the possibility that the favourable sequence for high translational efficiency in plants does not conform to the Kozak sequence found in animal systems. Moreover, later studies using the whole genome sequence databases from various eukaryote species have revealed that preferred nucleotide sequences around the AUG codon are quite diverse among different species (25).
Despite a great deal of advances in understanding the role of the 5′-UTR in translational efficiency in many different organisms, it is still not fully understood how the sequence context of the 5′-UTR affects translational efficiency at the single nucleotide level. The whole genome sequencing of a large number of organisms including Arabidopsis has revealed that the 5′-UTRs show a great diversity in their nucleotide sequence. However, it is not fully understood how the sequence diversity affects the translational efficiency. Previous studies have mainly focused on the effect of the nucleotide sequence immediately upstream (from nucleotide positions −1 to −4) or downstream (+4) of the AUG start codon on the translational efficiency (20,23,25).
To expand our understanding of the effect of the 5′-UTR on translational efficiency, we obtained the 5′-UTR sequences from 25 Arabidopsis genes that show a great diversity in their 5′-UTR sequences, and compared their translational efficiency at the single nucleotide level by generating nucleotide substitution mutants. In this analysis, we decided to use the 21 nucleotide (nt) region from positions −1 to −21 of the 5′-UTR so as to get closer to represent the full-length sequence context of the 5′-UTR on the translational efficiency and at the same time to minimize the effect of the secondary structure on the translational efficiency. Here, we provide evidence that the translational efficiency of various Arabidopsis genes varies over 200-fold depending on their 5′-UTRs and that the immediate upstream region from the nucleotide positions −5 to −1 is most critical in determining translational efficiency although the further upstream region of the 5′-UTR also plays an important role depending on the sequence context. Moreover, we provide evidence that of the four different types of nucleotides, nucleotides A and T are the most favourable and unfavourable, respectively, in translation.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis plants (ecotype Columbia-0) were grown on B5 plates in a growth chamber at 20–22°C under a 16-h/8-h light/dark cycle and 70% relative humidity. Leaf tissues from 2.5 week-old plants were used for protoplast isolation.
Vector construction
To generate the reporter construct, the 5′-UTRs were fused to the coding region of green fluorescent protein (GFP). The RbcS1A 5′-UTR::GFP construct was generated by two rounds of PCR. First, 10 nt and 21 nt-long 5′-UTRs of the RbcS1A::GFP construct were amplified with primers P1 and P2 using GFP as template. Subsequently, serial second polymerase chain reactions (PCRs) were performed to generate longer 5′-UTRs of RbcS1A::GFP with primers P2 and P3 using previous PCR products as template (for the nucleotide sequence of primers, see Supplementary Table S1). The 21 nt-long 5′-UTRs of Arabidopsis genes and their triple nucleotide substitution mutants were fused to GFP by PCR. For this, the 5′ primers (P4–P9) consisted of two regions: the 5′ region with the 21 nt-long 5′-UTRs or their mutant sequences, and the 20 nt-long 3′ region complementary to the 5′-end of the GFP-coding region. The 3′ primer was designed to contain 20 nts complementary to the 3′-end of the GFP-coding region. The PCR products were inserted into a pUC vector at the downstream region of the cauliflower mosaic virus (CaMV) 35S promoter. To generate pCAMBIA1300-UTR::GFP, the UTR::GFP fragment was obtained by digesting with XbaI/EcoRI and ligated to the binary vector pCAMBIA1300 digested with XbaI/EcoR1. For in vitro translation in Arabidopsis cell free extracts, the UTR::GFP fragments obtained by digesting with BamH1/XhoI were ligated to pCS2+ digested with BamHI/XhoI. The yeast phosphoribosylanthranilate isomerase (TRP1) coding region was obtained by PCR with primers P10 and P11 using pAS2-1 (Clontech) as template. The coding region of neomycin phosphotransferase II (NPTII) was prepared by PCR using primers P12 and P13 and the 5′-UTR::NPTII fragment was ligated to a pUC vector at the downstream region of the CaMV 35S promoter. All the constructs as well as the PCR products were confirmed by nucleotide sequencing.
Transient expression and immunoblot analysis
Plasmids were introduced into protoplasts by polyethylene glycol-mediated transformation (26,27). Transformed protoplasts were harvested 24 h after transformation and re-suspended in denaturation buffer (2.5% sodium dodecyl sulphate (SDS), 2% β-mercaptoethanol) and 5x sample buffer (250 mM Tris–Cl, pH 6.8., 0.5 dithiothreitol (DTT), 10% SDS, 0.05% bromophenol blue, 50% glycerol). After boiling for 7 min, the lysates were centrifuged at 10000g for 5 min to remove cellular debris. The expression level of proteins was analysed by immunoblotting using anti-GFP (Clontech), anti-GUS (Molecular Probes), anti-hemagglutinin (HA) (Roche Diagnostics), anti-hygromycin phosphotransferase (HPT) (Bio-application) or anti-NPTII (Bio-application) antibodies. The GUS construct co-transformed into protoplasts together with the UTR::GFP constructs was used as a control for transformation efficiency in Arabidopsis protoplasts. The HPT level was used as a control for the transient expression of the UTR::GFP constructs in Nicotiana benthamiana. Protein blots were developed with an ECL kit (Amersham Pharmacia Biotech), and images were obtained using the LAS3000 image capture system (FUJIFILM).
qRT-PCR analysis of transcript levels
Total RNA was extracted from Arabidopsis leaf tissue using the Qiagen RNeasy Plant Mini Kit and digested with TURBO DNase (Ambion). Total RNA was reverse-transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the SYBR Green Kit (Applied Biosystems) to detect the transcript level of GFP. Actin-2 (ACT2) was used as an internal control for qRT-PCR. Primers used for qRT-PCR were as follows: GFP_q5 and GFP_q3 for GFP, ACT2_q5 and ACT2_q3 for ACT2 (At3g18780), GUS_q5 and GUS_q3 for GUS (Supplementary Table S1).
Transient expression in leaves of Nicotiana benthamiana by Agrobacterium-mediated transformation
Agrobacterium tumefaciens strain LBA4404 harbouring pCAMBIA1300-UTR::GFP was grown overnight at 28°C until OD 1.0 at 600 nm in 10 ml LB broth supplemented with 50 μg/ml kanamycin. After pelleting by centrifugation (4000×g for 5 min at room temperature), Agrobacterium culture was re-suspended in 5 ml of infiltration buffer (10 mM MgCl2, 10 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.6, 100 μM acetosyringone) and mixed in a 1:1 ratio with a bacterial suspension carrying the coat protein of turnip crinkle virus (TCV-CP) viral silencing suppressor in the binary vector (28). The final mixture of Agrobacterium culture was infiltrated into the bottom side of fully expanded leaves of 4 week-old Nicotiana benthamiana plants. After infiltration, plants were placed in a green house at 20–22°C under a 16-h/8-h light/dark cycle and 70% relative humidity. At 4 days after infiltration, leaves were harvested and ground in liquid nitrogen. Tissue powder was re-suspended in protein extracting buffer (100 mM NaCl, 10 mM ethylenediaminetetraacetic acid, 200 mM Tris, 0.1% SDS, 200 mM sucrose, 14 mM β-mercaptoethanol, 0.05% Tween 20) and boiled for 7 min for immunoblot analysis.
In vitro transcription and translation using Arabidopsis cell extract and wheat germ extracts
The pCS2+ plasmid containing 5′-UTR::GFPs was linearized with ApaLI. Linearized DNA was used in the SP6 mMESSAGE mMACHINE® Kits (Ambion) to synthesize capped RNAs. Fifty femtomoles of RNA were used for in vitro translation using wheat germ extracts (Promega) and Arabidopsis cell extract (ACE) (29).
RESULTS
The immediate 5′-UTRs of Arabidopsis genes show a wide range of variation in translational efficiency
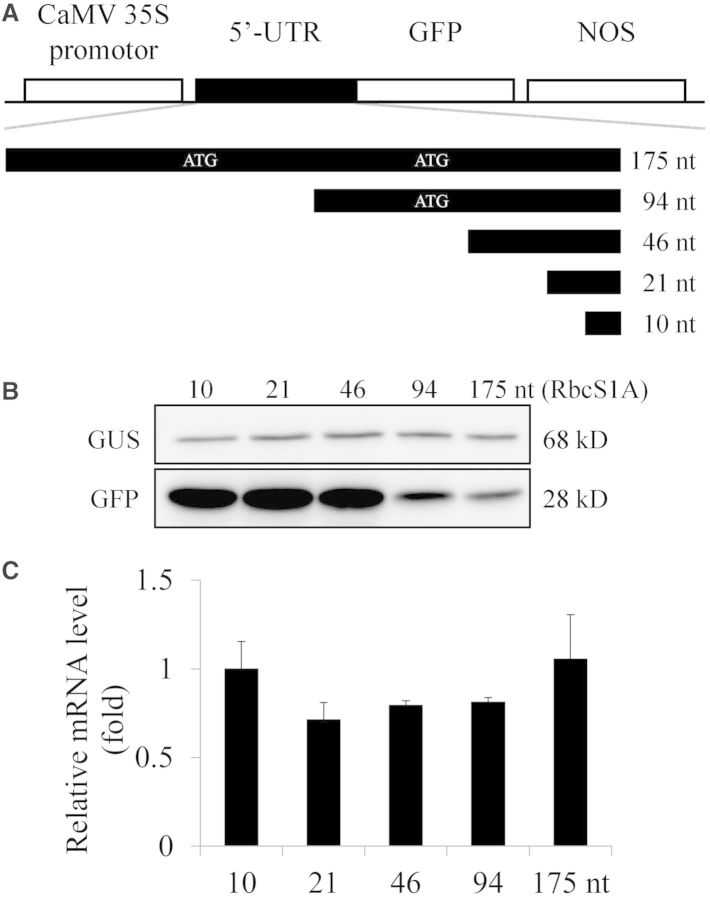
To gain an insight into the effect of the sequence context of the immediate upstream region from the AUG initiation codon on translational efficiency, we initially examined the effect of the 5′-UTR length on translational efficiency using the 5′-UTR of ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit 1A (RbcS1A, At1g67090) as a model sequence. The 5′-UTR of a gene varies greatly in length (30,31). In Arabidopsis, the average length of 5′-UTRs was 131 bases (32). The 5′-UTR of RbcS1A is 175 nucleotides. We generated serial deletion constructs with 10, 21, 46 and 94 nts from the AUG start codon. Each of these deletion constructs was placed between the CaMV 35S promoter and the AUG start codon of GFP (Figure 1A). These constructs were co-transformed into protoplasts together with a GUS (β-glucuronidase) construct used to normalize transformation efficiency, and the expression levels of GFP and GUS were determined by western blot analysis using anti-GFP and anti-GUS antibodies, respectively. The 10, 21 and 46 nt-long 5′-UTRs of RbcSA1 gave similar protein levels, indicating that the 10–46 nt region of the RbcS1A 5′-UTR from the −1 position shows similar translational efficiency. In contrast, the translational efficiency of full- or half-length RbcSA1 5′-UTR constructs was significantly lower than that of 10, 21 or 46 nt constructs (Figure 1B), indicating that the upstream half of RbcSA1 5′-UTR (−175 to −94) contains a sequence element that inhibits translation. The transcript levels of these constructs were nearly equal when examined by qRT-PCR (Figure 1C), confirming that the difference in the protein level reflects the translational efficiency of these constructs. Since the upstream AUG in the 5′-UTR often suppresses the translation (33), one possible explanation of the lower translational efficiency observed with the full- and half-length 5′-UTRs of RbcS1A is that the multiple upstream AUGs located in the 5′-UTR may negatively affect the translation of GFP.
Figure 1.
The effect of the RbcS1A 5′-UTR length on the translational efficiency. (A) Scheme of serial deletion constructs of the RbcS1A 5′-UTR. Black blocks in the serial deletion mutants represent RbcS1A 5′-UTRs. The length of 5′-UTRs is indicated at the end of each block (175 to 10 nt). The upstream ATG is shown in white. The deletion mutants of the RbcS1A 5′-UTR were placed in front of the GFP AUG codon. NOS, nos-terminator; nt, nucleotide. (B) The translational efficiency of various RbcS1A 5′-UTR::GFP constructs. The full-length or deletion mutants of the RbcS1A 5′-UTR fused to GFP were co-transformed into protoplasts together with a reference construct GUS, and GFP and GUS levels were determined by western blot analysis using anti-GFP and anti-GUS antibodies, respectively. The GUS levels were used to normalize the transformation efficiency. The number indicates the length of the RbcS1A 5′-UTR. (C) qRT-PCR analysis of GFP transcript levels. Total RNA from the transformed protoplasts was subjected to qRT-PCR for GFP and GUS transcripts. The GFP transcript levels were normalized using the co-expressed GUS transcript levels. ACT2 was used as an internal control for qRT-PCR. Error bar, standard deviation (n = 3).
Based on the results of RbcS1A 5′-UTR deletion constructs, we decided to use the 21 nt region from the position −1 of the 5′-UTR to examine the effect of the 5′-UTR sequence context on the translational efficiency. To include diverse 5′-UTR sequences in the analysis, the entire Arabidopsis genes were parsed against the nucleotide sequence of the RbcS1A 5′-UTR from positions −1 to −21 as a reference sequence. The sequence diversity was expressed with a similarity score by comparing the nucleotide sequence of a given 5′-UTR to that of RbcS1A. The 5′-UTRs that contained less than 21 nts were excluded from the analysis. Of the Arabidopsis genes, 75% of them (17 202 out of 22 998) contained more than 21 nts in the 5′-UTR. We selected 25 5′-UTRs (Table 1) with the similarity score ranging from 10 to 57%. These 5′-UTRs were placed at the upstream region of the GFP coding region (Figure 2A). Therefore, these constructs were identical to each other except the 21-nucleotide region immediately upstream from the AUG start codon. To determine the translational efficiency of individual 5′-UTRs, these constructs were co-transformed into protoplasts together with a reference construct, GUS, which was used to normalize the transformation efficiency, and the GFP and GUS levels were determined by western blot analysis using anti-GFP and anti-GUS antibodies, respectively (Figure 2B). The translational efficiency of these GFP constructs that had been normalized against the level of GUS ranged from 1 to 208% that of RbcS1A::GFP, indicating a wide range of variation in the translational efficiency among the 5′-UTRs (Figure 2C). To confirm that the difference in the translational efficiency of these constructs is not caused by the difference in the transcript levels, total RNA from the transformed protoplasts was used for qRT-PCR using specific primers for the GFP coding region. Although the 5′-UTRs of AT1G58420 and AT5G40850 induced dramatic differences in the GFP level, these constructs showed no significant differences in the transcript level, confirming that the difference in the GFP level stems from the difference in the translational efficiency (Figure 2D). These results strongly suggest that the nucleotide sequence immediately upstream of the AUG start codon greatly affects translational efficiency.
Table 1.
Nucleotide sequence of 5′-UTRs from various Arabidopsis genes used in this study
| Locus | GFP (%) | Sequence (−21 to −1) | Similarity (%) | Function | Length |
|---|---|---|---|---|---|
| AT1G67090 | 100 | CACAAAGAGTAAAGAAGAACA | 100 | Ribulose bisphosphate carboxylase small chain 1A | 175 |
| AT1G35720 | 192 | AACACTAAAAGTAGAAGAAAA | 57 | Annexin | 107 |
| AT5G45900 | 104 | CTCAGAAAGATAAGATCAGCC | 57 | Ubiquitin-like modifier-activating enzyme atg7 | 82 |
| AT5G61250 | 85 | AACCAATCGAAAGAAACCAAA | 52 | Heparanase-like protein 2 | 81 |
| AT5G46430 | 30 | CTCTAATCACCAGGAGTAAAA | 48 | 60S ribosomal protein L32-2 | 45 |
| AT5G47110 | 1 | GAGAGAGATCTTAACAAAAAA | 48 | Chlorophyll A-B binding family protein | 79 |
| AT1G03110 | 90 | TGTGTAACAACAACAACAACA | 43 | Transducin/WD-40 repeat-containing protein | 113 |
| AT3G12380 | 74 | CCGCAGTAGGAAGAGAAAGCC | 43 | Actin-related protein 5 | 522 |
| AT5G45910 | 43 | AAAAAAAAAAGAAATCATAAA | 43 | GDSL esterase/lipase | 37 |
| AT1G58420 | 208 | ATTATTACATCAAAACAAAAA | 38 | Uncharacterized conserved protein UCP031279 | 28 |
| AT5G44520 | 131 | CGTTCTTCCCACACAAAAAAA | 38 | NagB/RpiA/CoA transferase-like superfamily protein | 86 |
| AT1G07260 | 65 | GAGAGAAGAAAGAAGAAGACG | 38 | UDP-glycosyltransferase | 141 |
| AT3G55500 | 34 | CAATTAAAAATACTTACCAAA | 38 | Expansin-A16 | 80 |
| AT3G46230 | 2 | GCAAACAGAGTAAGCGAAACG | 38 | 17.4 kDa class I heat shock protein | 108 |
| AT2G36170 | 88 | GCGAAGAAGACGAACGCAAAG | 33 | Ubiquitin-60S ribosomal protein L40-1 | 59 |
| AT1G10660 | 33 | TTAGGACTGTATTGACTGGCC | 33 | Putative uncharacterized protein | 257 |
| AT4G14340 | 25 | ATCATCGGAATTCGGAAAAAG | 33 | Casein kinase 1-like protein 11 | 330 |
| AT1G49310 | 13 | AAAACAAAAGTTAAAGCAGAC | 33 | Putative uncharacterized protein | 42 |
| AT4G14360 | 55 | TTTATCTCAAATAAGAAGGCA | 29 | Probable methyltransferase PMT3 | 382 |
| AT1G28520 | 30 | GGTGGGGAGGTGAGATTTCTT | 29 | Transcription factor VOZ1 | 157 |
| AT1G20160 | 2 | TGATTAGGAAACTACAAAGCC | 29 | Subtilisin-like serine endopeptidase-like protein | 37 |
| AT5G37370 | 103 | CATTTTTCAATTTCATAAAAC | 24 | Pre-mRNA-splicing factor 38B | 148 |
| AT4G11320 | 1 | TTACTTTTAAGCCCAACAAAA | 24 | Probable cysteine proteinase | 82 |
| AT5G40850 | 10 | GGCGTGTGTGTGTGTTGTTGA | 19 | Urophorphyrin III methylase | 304 |
| AT1G06150 | 9 | GTGGTGAAGGGGAAGGTTTAG | 14 | Transcription factor EMB1444 | 503 |
| AT2G26080 | 19 | TTGTTTTTTTTTGGTTTGGTT | 10 | Glycine dehydrogenase | 42 |
Nucleotide sequences were obtained from 5′-UTRs (nucleotide positions −1 to −21) of 25 different genes. The translational efficiency of these 5′-UTRs was determined using GFP levels (GFP) (Figure 2B) and their relative expression levels were presented with the function (Function) of the encoded proteins and putative full length (Length) of their 5′-UTRs. The sequence similarity score is the percentage of how many nucleotides out of the 21 nt-long 5′-UTR are identical to that of the AT1G67090 (RbcS1A) 5′-UTR.
Figure 2.
The 21 nt region immediately upstream from the AUG start codon of various Arabidopsis genes shows a wide variation in translational efficiency. (A) Schematic presentation of fusion constructs. 5′-UTR, the 5′-untranslated region (165 nts) from the expression vector; 21 nt, 21 nt-long 5′-UTRs from various Arabidopsis genes; GFP-coding region; 3′-UTR, 3′-untranslated region from the expression vector. (B) Translation efficiency of the 5′-UTRs of various Arabidopsis genes. Reporter constructs were introduced into protoplasts together with a reference construct GUS and total protein extracts were subjected to western blot analysis using anti-GFP and anti-GUS antibodies. The 5′-UTR of Rbcs1A was used as a reference for the expression. (C) Quantification of translational efficiency. To quantify the translational efficiency, the signal intensity of immunoblots in (B) was quantified using the multi-gauge software equipped to the LAS3000 (FUJIFILM) and the GFP levels were normalized with the GUS level. (D) Quantification of transcriptional efficiency. To quantify the transcriptional efficiency, total RNA from the protoplasts transformed with two representative 5′-UTR constructs (asterisk in (C)) was used for qRT-PCR using specific primers for the GFP-coding region. The GUS transcript level was used as reference.
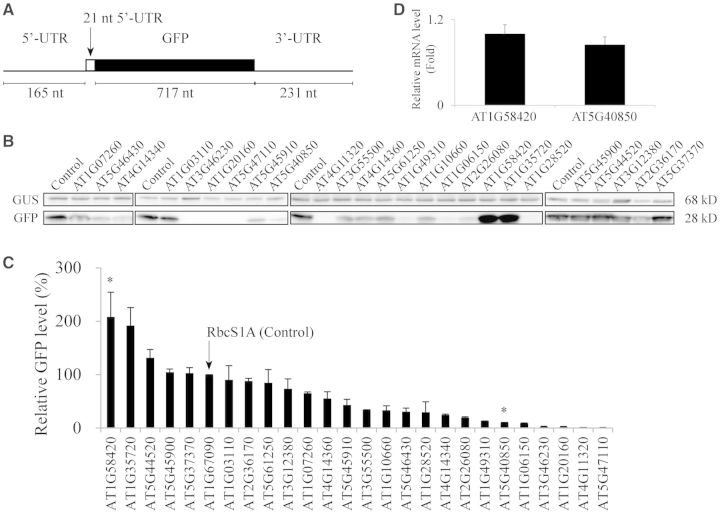
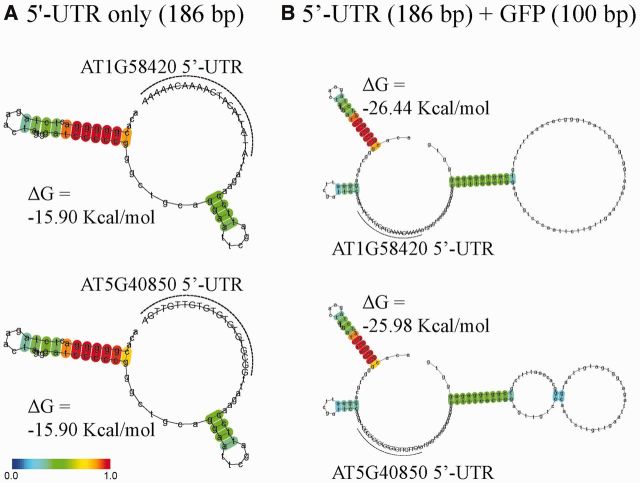
Next, we examined whether the secondary structure of the mRNA including the 5′-UTR and the coding region has any correlation with the translational efficiency. In previous studies, the secondary structure in the 5′-UTR as well as the 5′ side of the coding region affects the translational efficiency (34). We used the algorithm centroidfold (http://www.ncrna.org/centroidfold/) which highlights the best performance among similar methods (35,36). We analysed the secondary structure of the 5′-UTRs of GFP mRNA consisting of 165 nts of the 5′-UTR from the expression vector, 21 nts of the 5′-UTR from various Arabidopsis genes with or without the 100 nt GFP-coding region (Figure 3 and Supplementary Figure S1). Inclusion of the 100 nt GFP coding region did not cause any significant additional secondary structure (Figure 3B). Of the 25 5′-UTRs, 16 of them showed a nearly identical secondary structure. In this group of the 5′-UTRs, the major secondary structure was the 11 bp long hairpin structure (ΔG = −15.90 Kcal/mol) starting from the fourth nucleotide position from the cap (Figure 3A). Thus, this upstream hairpin structure was generated solely by the 5′-UTR from the vector sequence and the 21 nt 5′-UTRs from the Arabidopsis genes did not show any secondary structure. In contrast, in the other nine constructs, a proportion of the 21-nucleotide 5′-UTR region from the Arabidopsis genes was part of the hairpin structure. However, except for AT3G12380 5′-UTR, eight 5′-UTRs (AT1G10660, AT2G36170, AT4G11320, AT4G14340, AT5G44520, AT5G45900, AT5G47110 and AT1G20160) showed a rather low probability of forming a hairpin structure or had a small hairpin structure (Supplementary Figure S1). To test whether these additional hairpins have any inhibitory effect on the translational efficiency, we calculated the stabilizing energy resulting from the hairpin formation and found that ΔG ranged from −11.59 to −24.40 kcal/mol. Thus, the 5′-UTRs of AT2G36170, AT4G11320, AT4G14340, AT5G44520, AT5G45900, AT5G47110 and AT1G20160 showed a slightly more stable secondary structure than the first group of the 5′-UTRs with one hairpin structure originating from the vector sequence. However, the difference in ΔG was less than 5 kcal/mol. Moreover, the ΔG of the AT1G10660 5′-UTR was −11.59 kcal/mol. Thus, this 5′-UTR was even less stable than the first group of sixteen 5′-UTRs. In the case of the AT3G12380 5′-UTR (ΔG = −24.40 kcal/mol), this was more stable than the first group of the 5′-UTRs (ΔG = −15.90 kcal/mol). However, this difference in ΔG among the 5′-UTRs may not be significant. The hairpin structure with ΔG = −24.4 kcal/mol can be easily disrupted by ribosomes during translation and thus may not inhibit translation (37,38). Moreover, these 5′-UTRs did not show any correlation between the secondary structure formation and translation efficiency. The translational efficiency of the AT3G12380 5′-UTR that had the strongest secondary structure was 74% that of the RbcS1A 5′-UTR, whereas the translational efficiency of the AT1G10660 5′-UTR that had the weakest secondary structure was 33% that of the RbcS1A 5′-UTR. These results raised the possibility that the difference in the translational efficiency results from the nucleotide sequence difference in the 5′-UTRs. However, we cannot completely rule out the possibility that the hairpin structure may have an inhibitory effect on the translation.
Figure 3.
The secondary structural analysis of the AT1G58420 and AT5G40850 5′-UTRs as representatives of ‘good’ and ‘poor’ 5-UTRs, respectively. (A and B) The entire 5′-UTR consisting of the 165 nt-long 5′-UTR from the expression vector and the 21 nt region of the 5′-UTR from AT1G58420 or AT5G40850 (A), or together with 100 nt GFP-coding region (B) was analysed for the possible secondary structure using centroidfold (http://www.ncrna.org/centroidfold/). Heat colour gradation from blue to red represents the base-pairing probability from 0 to 1. The base-pairing probability is the probability that a pair of bases forms a base pair via hydrogen bonds in the secondary structure. The 21 nt 5′-UTRs from AT1G58420 and AT5G40850 are in capital letters and also indicated by broken lines.
Adenine residues in positions −1 to −5 from the AUG start codon are the most critical for translational efficiency
The 5′-UTRs that had a higher translational efficiency showed higher sequence similarities to the RbcS1A 5′-UTR. The majority of the 5′-UTRs have a purine base in the −3 position as predicted by the Kozak rule (18). However, as shown in the AT4G11320 5′-UTR, the presence of a purine base in the −3 position was not sufficient to give a high translational efficiency, raising the possibility that the plant sequence may not follow the Kozak rule. In addition, the 5′-UTRs that had high translational efficiency appeared to be rich in A nucleotide but devoid of C nucleotide. In particular, the AT1G35720 5′-UTR, one of the best 5′-UTR among the 25 5′-UTRs we analysed contained 14 As, 2 Cs, 3 Gs and 2 Ts, indicating a strong bias towards the A nucleotide (Table 1). In contrast, the AT4G40850 5′-UTR, one of the poorest 5′-UTRs, had a completely different nucleotide composition with 1 C, 10 Gs and 10 Ts, but devoid of the A nucleotide (Table 1), indicating that the A residue is crucial for higher translational efficiency. Interestingly, the omega leader sequence contains a region of poly(CAA), thereby having a higher content of the A residue (39).
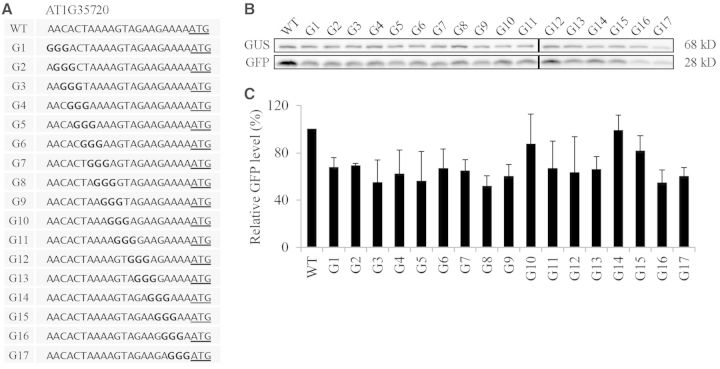
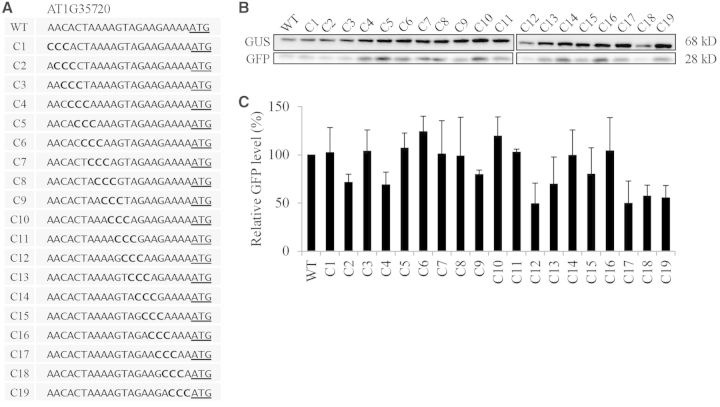
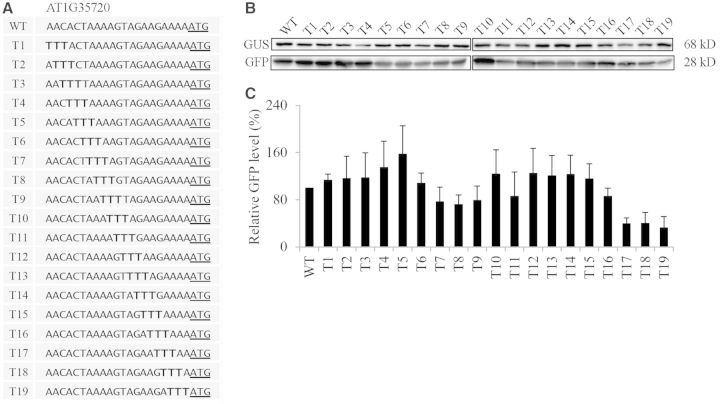
To examine the sequence context which underlies the high translational efficiency, we selected the AT1G35720 5′-UTR whose translational efficiency was 192% that of RbcS1A 5′-UTR. A previous study showed that substitution of three nucleotides in front of the AUG start codon (positions −3 to −1) affects the translation efficiency of a reporter gene (22). To elucidate the effect of individual nucleotides in the 21 nt 5′-UTR on translational efficiency, we generated serial nucleotide substitution mutants of the AT1G35720 5′-UTR by replacing three nucleotides simultaneously with triple A, C, G or T, starting from the 5′-end to the 3′-end of the 21 nt 5′-UTR (Figures 4–7). First, we examined the translational efficiency of triple A substitution mutants in protoplasts. The majority of them had a slightly higher translation efficiency compared to the wild-type 5′-UTR (Figure 4). In these mutants, when T or G nucleotides were substituted with A nucleotides the translational efficiency was increased, indicating that the A nucleotide in the 5′-UTR was favourable for the translational efficiency. Next, we examined the triple G substitution mutants for translational efficiency. Except for G14 that had a guanine substitution at the positions −4 to −6, all the triple G substitution mutants showed a 5–40% reduction in translational efficiency compared to the wild-type AT1G35720 5′-UTR. These results suggest that the G residue in the 21 nt 5′-UTR is not a favourable nucleotide for the translation (Figure 5). In the triple C substitution mutants, they were divided into two groups; one group of mutants including C1, C3, C5-C11 and C14-C16 did not show any significant change in translational efficiency and the other group of mutants containing C2, C4, C12, C13, C17-C19 had a 40–50% reduction in translational efficiency compared to the wild-type 5′-UTR. These results indicate that C residues in the 5′-UTR have a strong position-dependent effect on translational efficiency (Figure 6). In the case of triple T substitution mutants, they also showed a wide range of variation in translational efficiency from 32 to 158% depending on individual mutant constructs. For example, T16, T17 and T18 that had the triple T substitution at the 3′-end of the 5′-UTR showed an almost 70% decrease in the translational efficiency from the wild-type 5′-UTR. In contrast, mutants T7, T8 and T9 that had the triple T substitution in the middle of the 5′-UTR had only a minor reduction in translational efficiency. Moreover, a triple T substitution in the positions from −15 to −20 and from −5 to −10 of the 5′-UTR increased the translational efficiency to 158% of the wild-type 5′-UTR, indicating that the T nucleotide in the 5′-UTR affects the translation either favourably or unfavourably depending on the position in the 5′-UTR (Figure 7). These results strongly suggest that the sequence context immediately upstream of the AUG start codon is crucial in determining translational efficiency. Among four nucleotides, the A residue is the most favourable for translation, whereas the T residue is the least favourable.
Figure 4.
The A nucleotides introduced in the AT1G35720 5′-UTR are generally favourable for translational efficiency. (A) Sequences of triple A substitution mutants. Three nucleotides were sequentially substituted with three As in the 21 nt region of the AT1G35720 5′-UTR. (B) The effect of triple A substitutions on the translational efficiency. The mutant constructs of the AT1G35720 5′-UTR were co-transformed into protoplasts together with a GUS construct and protein extracts were subjected to western blot analysis using anti-GFP and anti-GUS antibodies. (C) Quantification of the translational efficiency. To quantify the translational efficiency, the signal intensity of immunoblots in (B) was quantified using the multi-gauge software equipped to the LAS3000 (FUJIFILM) and the GFP levels were normalized with the GUS levels. Error bar, standard deviation (n = 3).
Figure 5.
Triple G substitutions in the AT1G35720 5′-UTR largely cause a decrease in the translation efficiency. (A) Sequences of triple G substitution mutants. Three nucleotides were sequentially substituted with three Gs in the 21 nt region of the AT1G35720 5′-UTR. (B) The effect of triple G substitutions on the translational efficiency. The mutant constructs of the AT1G35720 5′-UTR were co-transformed into protoplasts together with a GUS construct and protein extracts were subjected to western blot analysis using anti-GFP and anti-GUS antibodies. (C) Quantification of the translational efficiency. To quantify the translational efficiency, the signal intensity of the immunoblots shown in (B) was quantified using the multi-gauge software equipped to the LAS3000 (FUJIFILM) and the GFP levels were normalized with the GUS levels. Error bar, standard deviation (n = 3).
Figure 6.
Triple C substitutions in the AT1G35720 5′-UTR cause suppression of the translational efficiency in a position-dependent manner. (A) Sequences of triple C substitution mutants. Three nucleotides were sequentially substituted with three Cs in the 21 nt region of the AT1G35720 5′-UTR. (B) The effect of triple C substitutions on the translational efficiency. The mutant constructs of the AT1G35720 5′-UTR were co-transformed into protoplasts together with a GUS construct and protein extracts were subjected to western blot analysis using anti-GFP and anti-GUS antibodies. (C) Quantification of the translational efficiency. To quantify the translational efficiency, the signal intensity of immunoblots in (B) was quantified using the multi-gauge software equipped to the LAS3000 and the GFP levels were normalized with the GUS levels. Error bar, standard deviation (n = 3).
Figure 7.
Triple T substitutions in the AT1G35720 5′-UTR cause a strong suppression or activation of the translational efficiency in a position-dependent manner. (A) Sequences of triple T substitution mutants. Three nucleotides were sequentially substituted with three Ts in the 21 nt region of the AT1G35720 5′-UTR. (B) The effect of triple T substitutions on the translational efficiency. The mutant constructs of the AT1G35720 5′-UTR were co-transformed into protoplasts together with a GUS construct and protein extracts were subjected to western blot analysis using anti-GFP and anti-GUS antibodies. (C) Quantification of the translational efficiency. To quantify the translational efficiency, the signal intensity of the immunoblots in (B) was quantified using the multi-gauge software equipped to the LAS3000 and the GFP levels were normalized with the GUS levels. Error bar, standard deviation (n = 3).
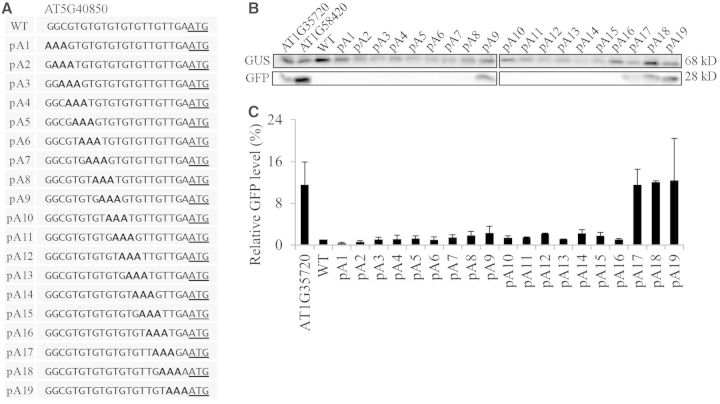
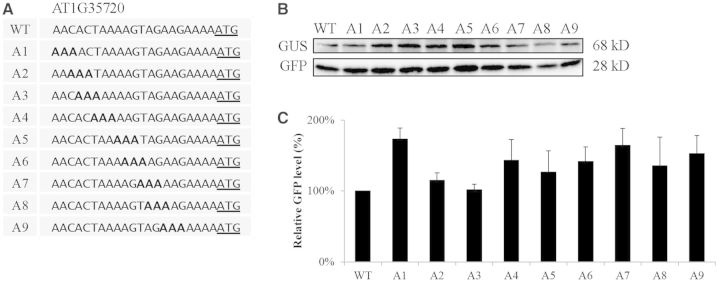
To further investigate how the sequence context influences the translational efficiency, we asked whether a ‘poor’ 5′-UTR can be converted into a ‘good’ 5′-UTR by changing its nucleotide sequence. To this end, we chose the AT4G40850 5′-UTR whose translational efficiency was only 10% of the RbcS1A 5′-UTR. Because the A residue is favourable for the translational efficiency, we generated serial triple A substitution mutants (Figure 8). These triple A mutants were fused to the GFP-coding region and the resulting constructs were co-transformed into protoplasts together with the reference construct GUS. The GFP and GUS levels were examined by western blot analysis using anti-GFP and anti-GUS antibodies, respectively, and the translational efficiency was determined by normalizing the GFP levels with the GUS levels. The triple A mutants of the AT4G40850 5′-UTR that had triple A substitutions in the region from positions −21 to −6 had only minor changes in the translational efficiency. Surprisingly however, when the nucleotide sequence GTTGA from the positions −5 to −1 was changed to AAAGA (pA17), GAAAA (pA18) or GTAAA (pA19), the GFP levels were greatly elevated to the levels which were similar to that of the ‘good’ 5′-UTR, AT1G35720 5′-UTR (Figure 8). These findings support the idea that the A nucleotides in the immediate upstream region (−5 to −1) from the AUG start codon are crucial for high translational efficiency. To further confirm this finding, we examined the secondary structure of the triple A substitution mutants using the algorithm centroidfold (35). These mutants did not show any difference in the secondary structure from the wild-type 5′-UTR (Supplementary Figure S2), indicating that the difference in the translational efficiency resulted from the sequence change in the mutant 5′-UTRs. Consistent with this finding, only the A nucleotide showed a slightly higher presentation in the 5′-UTR from positions −1 to −4 with the highest chance at the −3 position when 17202 5′-UTRs of Arabidopsis genes were analysed using weblogo (http://weblogo.berkeley.edu/) (Supplementary Figure S3) (40).
Figure 8.
Triple A substitutions in the region from positions −1 to −5 of the AT1G04850 5′-UTR cause a strong enhancement of translational efficiency. (A) Sequences of triple A substitutions in the 21 nt region of the AT1G04850 5′-UTR. Three nucleotides were sequentially substituted with three As and the resulting constructs were fused to GFP. (B) Translational efficiency of triple A substitution mutants. The wild-type AT1G04850 5′-UTR and its triple A substitution mutant constructs were introduced into protoplasts together with a GUS construct and expression levels of GFP and GUS were examined by western blot analysis using anti-GFP and anti-GUS antibodies, respectively. (C) Quantification of the GFP expression levels. The GFP levels were normalized using the GUS levels. Error bar, standard deviation (n = 3). The GFP level of the AT1G35720 5′-UTR construct was used as a control to compare elevated translational efficiency of pA17-pA19.
The effect of the sequence context in the immediate 5′-UTR on the translational efficiency is conserved in plants and is also independent of the coding region
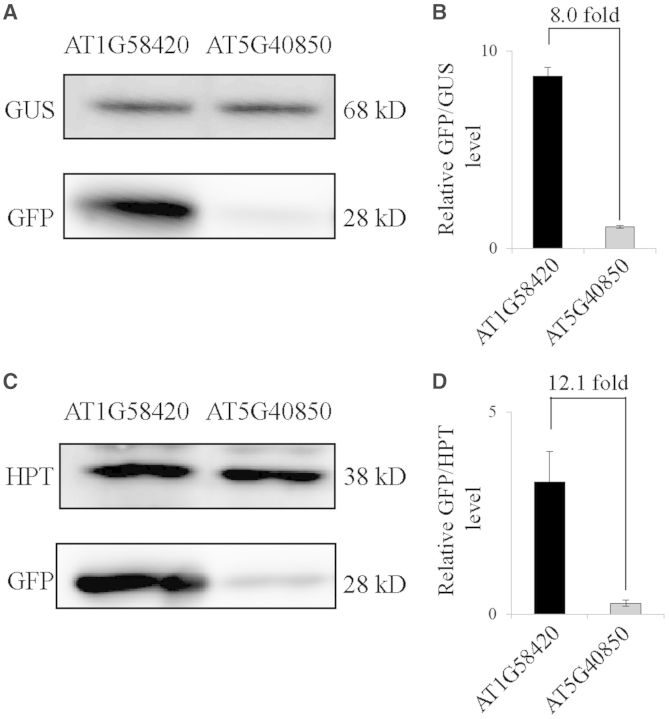
Next, to test whether the effect of the 5′-UTR sequence context on the translational efficiency observed in Arabidopsis protoplasts also applies to other plant species, we compared the translational efficiency of the AT1G58420 and AT4G40850 5′-UTRs in two different plants: Arabidopsis and tobacco. In Arabidopsis, the translational efficiency was determined in protoplasts, whereas the translational efficiency in tobacco leaf cells was determined by transient expression after Agrobacterium-mediated infiltration. The translational efficiency was determined by western blot analysis using anti-GFP antibody. To normalize the transformation efficiency, Arabidopsis and tobacco extracts were also subjected to western blot analysis using anti-GUS and anti-HPT antibodies, respectively. In Arabidopsis protoplasts, the translational efficiency of the AT1G58420 5′-UTR was 8-fold higher than that of the AT4G40850 5′-UTR (Figure 9A). In tobacco leaf tissues, the translational efficiency of the AT1G58420 5′-UTR was 12-fold higher than that of the AT4G40850 5′-UTR, indicating that the effect of sequence context on the translational efficiency is conserved between two different plant species (Figure 9C).
Figure 9.
The effect of the 5′-UTRs on the translational efficiency is conserved in different plant species. (A) GFP levels in Arabidopsis protoplasts. Reporter constructs of the AT1G58420 or AT1G04850 5′-UTRs were co-transformed into Arabidopsis protoplasts together with a GUS construct and the expression levels of GFP and GUS were determined by western blot analysis using anti-GFP and anti-GUS antibodies, respectively. (B) Quantification of translational efficiency. To quantify the translational efficiency, the signal intensity of the immunoblots in (A) was quantified using the multi-gauge software equipped to the LAS3000 and the GFP levels were normalized using the GUS levels. Error bar, standard deviation (n = 3). (C) GFP levels in tobacco leaf tissues. The GFP constructs with the 21 nt region of AT1G58420 or AT1G04850 5′-UTRs in a binary vector were transformed into Nicotiana benthamiana leaf tissues by Agrobacterium-mediated infiltration. The expression levels of GFP and HPT were determined by western blot analysis using anti-GFP and anti-HPT antibodies, respectively. HPT was used to normalize the transformation efficiency. (D) Quantification of translational efficiency. To quantify the translational efficiency, the signal intensity of the immunoblot in (C) was quantified using the multi-gauge software equipped to the LAS3000 and the GFP levels were normalized using the HPT levels. Error bar, standard deviation (n = 3).
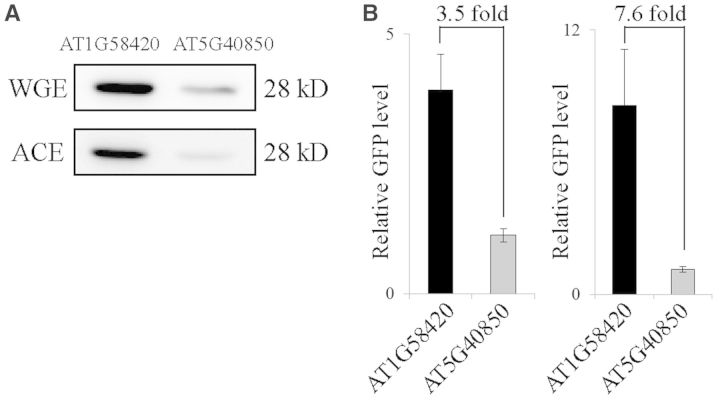
Next, we examined whether the effect of the sequence context on the translational efficiency is also operational in different translational systems. In the soybean cytosolic glutamine synthetase gene, the translational enhancing activity of the 5′-UTR is not maintained in in vitro translational systems (12). In addition, relative translational efficiency with a single mutation in only the −3 position of 5′-UTRs was also not consistent in two well-known in vitro systems: reticulocyte lysates and wheat germ extracts (41). We used ACEs (29) and wheat germ extracts for in vitro translation. Two constructs, AT1G58420 5′-UTR::GFP and AT4G40850 5′-UTR::GFP, were transcribed in vitro and their transcripts were used for translation in vitro. In ACEs and wheat germ extracts, the translational efficiency of the AT1G58420 5′-UTR was 7.5- and 3.5-fold higher than that of the AT4G40850 5′-UTR, respectively (Figure 10A), indicating that the effect of the sequence context in the 5′-UTR on translational efficiency was maintained in the in vitro systems and also in both dicot and monocot plants.
Figure 10.
The effect of the 5′-UTRs on the translational efficiency is independent of the translational system. (A) Western blot analysis of GFP levels. The AT1G58420 5′-UTR or AT1G04850 5′-UTR constructs were in vitro transcribed and 17 ng (50 fmol) of their transcripts were used for in vitro translation using ACE or wheat germ extracts. The expression levels of GFP were determined by western blot analysis using anti-GFP antibody. (B) Quantification of GFP levels. To quantify the translational efficiency, the signal intensity of the immunoblots in (A) was quantified using the multi-gauge software equipped to the LAS3000. Error bar, standard deviation (n = 3).
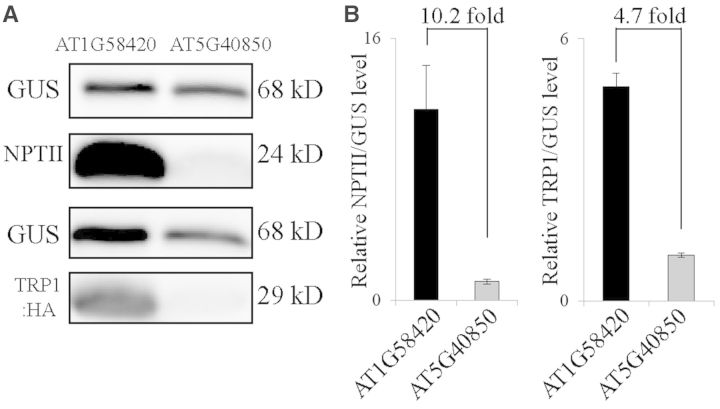
Another important issue in the effect of the 5′-UTR on the translational efficiency is whether the 5′-UTR has any dependency on the downstream coding region. To test this, we selected the AT1G58420 and AT4G40850 5′-UTRs as representatives of ‘good’ and ‘poor’ 5′-UTRs and fused them to two other coding regions, neomycin phosphotransferase II (NPTII) and yeast HA-tagged TRP1 (TRP1:HA). The resulting constructs were introduced into protoplasts and their expression levels were determined by western blot analysis using anti-NPTII and anti-HA antibodies. Again, both NPTII and TRP1:HA levels from the AT1G58420 5′-UTR were 10.2- and 4.7-fold higher than those from the AT4G40850 5′-UTR, respectively, confirming that the effect of the sequence context of the 5′-UTR on the translational efficiency is independent of the downstream coding region (Figure 11A).
Figure 11.
The effect of the 5′-UTRs on the translational efficiency is independent of the downstream coding region. (A) Western blot analysis of expressed proteins. The indicated 5′-UTR constructs were co-transformed into protoplasts together with a GUS construct and the expression of NPTII and TRP1:HA was determined by western blot analysis using anti-NTPII and anti-HA antibodies, respectively. In addition, GUS levels were detected with anti-GUS antibody. (B) Quantification of the translational efficiency. To quantify the translational efficiency, the signal intensity of the immunoblots in (A) was quantified using the multi-gauge software equipped to the LAS3000 and the NPTII and TRP1:HA levels were normalized using the GUS levels. Error bar, standard deviation (n = 3).
DISCUSSION
In this study, we systematically examined the effect of the nucleotide sequence context of the 5′-UTRs from Arabidopsis genes at the single nucleotide level and provide evidence that the nucleotides in positions −1 to −21 have a pronounced effect on the translational efficiency. This conclusion is based on the analysis of 25 greatly diverse 5′-UTRs of Arabidopsis genes on their ability to produce GFP proteins in protoplasts from leaf tissues of Arabidopsis. Of these 25 5′-UTRs, the difference in the translational efficiency between the strongest 5′-UTR and the weakest 5′-UTR was over 200-fold. The translational efficiency dictated by the sequence context of the 5′-UTR should be an intrinsic property of an mRNA. Therefore, our finding that Arabidopsis genes show great differences in translational efficiency raises many interesting questions as to how the protein levels of genes are determined in plant cells, why the 5′-UTR of certain mRNAs has the sequence context that is highly recalcitrant in translation, and whether transcriptional levels of genes have any correlation with their translational efficiency. Currently these questions remain unanswered.
Our finding that the nucleotide sequence of the 5′-UTR, in particular, the immediate upstream region from the nucleotide positions −1 to −5, is critical in translational efficiency is in a good agreement with the results from many previous studies that determined the effect of the sequence context of the 5′-UTR from nucleotide positions −1 to −4 and also at the nucleotide position +4 on the translational efficiency (20–23,39,41–43). However, in this study, we examined the effect of the sequence context in a much longer region (the 21 nt region from the positions −1 to −21) of the 25 5′-UTRs on the translational efficiency. As observed previously, the immediate upstream region from the nucleotide positions −1 to −4 was most critical in determining the translational efficiency. In addition, our study provided evidence that the region further upstream of the nucleotide position −4 can also significantly affect the translational efficiency depending on individual genes.
The 5′-UTRs can exert an effect on translational efficiency through multiple modes either positively or negatively by using specific sequence motifs, secondary structure or sequence context around the translational initiation site (5,44). Of these modes, the effect of the sequence context around the 5′-UTR has been extensively studied in animal cells (43). Indeed, the Kozak’s rule describes that A/G bases at the −3 position with surrounding pyrimidine residues and G at the +4 position are crucial for higher translational efficiency in animal genes. However, in plant genes, the consensus sequence, AAAA/CAAUGGC, of the translational initiation site which was obtained by analyzing 3643 plant genes does not conform to the Kozak sequence (19) although the consensus sequence also contains the conserved A and G residues at the −3 and +4 positions, respectively. However, it has not been assessed systematically how the consensus sequence is correlated with the level of translation in plant cells. In fact, a 28 nt-long synthetic 5′-UTR that serendipitously showed a high level of translational efficiency does not conform to the Kozak sequence, thus raising a question as to the importance of the Kozak sequence in the translational efficiency in plant systems (45). In fact, in plants, the A residue at positions −1 to −4 is most favourable in the translational efficiency (20,21,35,36,38–41). In this aspect, our results are consistent with these previous findings. Moreover, and further supporting with this idea, is that introduction of three As to a ‘poor’ 5′-UTR in the positions −1 to −5 greatly enhanced the translational efficiency.
Another important observation we made in this study is that the types of nucleotide and their position in the 21 nt-long 5′-UTR have different effects on the translational efficiency. Overall, the A residue was the most favourable nucleotide not only at positions −1 to −4 but also throughout the entire 21 nt region of the 5′-UTRs we examined. However, a previous study showed that the introduction of a stretch of A nucleotides in the 5′-UTR of the yeast alcohol oxidase gene greatly reduced the translational efficiency in a length-dependent manner (46), suggesting that introduction of poly(A) to the 5′-UTR is not always favourable for translation. In contrast, the T residue was the least favourable nucleotide in the 5′-UTRs. In particular, introduction of three Ts to a good 5′-UTR in positions −1 to −5 greatly suppressed the translational efficiency. However, in certain cases, the T residue located in the more 5′ side of the 21 nt-long 5′-UTR increased the translational efficiency, suggesting that the exact location of the T nucleotide is important for the translational efficiency. In the case of the C nucleotide, its effect on the translation was also position-dependent; introduction of three C residues in the positions −1 to −5 of the 5′-UTR greatly suppressed the translational efficiency, whereas the 3 C residues in the middle of the 21 nt region of the 5′-UTR did not affect or only slightly increased the translational efficiency. Introduction of G residues throughout the entire 21 nt region of the 5′-UTR generally suppressed the translational efficiency. These results strongly suggest that although the translational efficiency is in general most critically dependent on the immediate upstream region, for example the region from the positions −1 to −4, of the 5′-UTR from the AUG start codon, the translational efficiency of a particular 5′-UTR is determined by the sequence context of the entire nucleotide sequence of the 5′-UTR. Currently, however, it is not clearly understood at the molecular level how the different types of nucleotides affect differently the translational efficiency.
One of the most crucial events for the translation is the assembly of the initiation complex at the AUG start codon of an mRNA (5). Thus, one possible explanation for the differential effect of the nucleotide sequence context around the initiation site on the translation efficiency is that the assembly of the translation initiation complex is affected by the nucleotide sequence around the AUG start codon. One of most well-characterized sequence motifs in the 5′-UTR that affect the assembly of the initiation complex is the Shine-Dalgarno (SD) box in the 5′-UTR of prokaryotic mRNAs. The sequence in the SD box is involved in base-pairing with the 3′-end of 16S rRNA of ribosomes, thereby facilitates the formation of the initiation complex at the AUG start codon (47). However, the mechanism of initiation complex assembly in eukaryotic cells is different from that of prokaryotic cells. Multiple initiation factors are involved in assembly of the 40S ribosomal subunit in the AUG initiation codon. Of these initiation factors, eIF1A is involved in the selection of the initiation codon (48). Thus, the sequence context may play a role in the selection of the first AUG by eIF1A. In addition, other factors may also be involved in the 40S ribosomal subunit assembly. Interestingly, in the case of the omega leader sequence found in TMV, HSP101 is involved in the omega sequence-mediated translational enhancing (49). HSP101 directly binds to the poly(CAA) region of the omega leader sequence and mediates recruitment of two eukaryotic translational initiation factors, eIF4G and eIF3 to promote translation initiation (39). However, eIF4G binds the cap structure at the 5′-end of mRNA, thus playing a role in the initial recognition of mRNAs. Another important sequence element affecting the translational efficiency is the oligopyrimidine track in the 5′-end of the 5′-UTR which is present in many ribosomal protein genes. This oligopyrimidine track causes suppression of translation in both animal and plant cells (50). However, the oligopyrimidine track was located close to the 5′ cap structure in the 5′-UTR. Thus, the exact mechanism underlying the inhibitory effect of T or C residues in the oligopyrimidine track may not be the same as that of T or C residues in the positions −1 to −5 in this study. The sequence analysis of the 5′-UTRs of 15 971 Arabidopsis genes revealed that two sequence motifs, TAGGGTTT and AAAACCCT, are presented in many Arabidopsis genes, raising the possibility that these sequence elements may play a role in translational efficiency. Indeed, of the two sequence elements, the sequence motif TAGGGTTT has been shown to confer a higher translational efficiency (51). However, it is not known how these two sequence motifs confer their translational enhancing activity. One possibility is that an unknown factor may recognize these motifs. In contrast to these sequence motifs, the mechanism of how the secondary structure in the 5′-UTR affects the translational efficiency is well understood. It impedes the scanning process of the 40S subunit from the 5′ cap structure (52). Indeed, 5′-UTRs were, on average, less structured than the coding region in Arabidopsis and yeast transcripts (10,53). However, in contrast to this notion, Li et al. (53) recently reported results showing that the overall secondary structure of transcripts facilitates translation. This conclusion was based on the fact that the more structured transcripts were significantly more ribosome-associated than the less structured transcripts. These results raise the possibility that the secondary structure in the 5′-UTRs and coding region regulates the translation efficiency at multiple levels.
In summary, we have demonstrated here how in plants the sequence context around the initiation codon affects the translational efficiency. In particular, the presence of multiple A nucleotides at −1 to −5 from the AUG initiation codon is crucial for high translational efficiency. However, it remains elusive how this occurs at the molecular level. To elucidate the exact mechanism in detail, it will be imperative to identify the factor(s) involved in this process and to characterize them at the molecular and biochemical levels.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea [609004-05-4-SB240]. Funding for open access charge: Functional Genomics Center, Pohang University of Science and Technology, Republic of Korea.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Authors acknowledge the other lab members for the technical assistance and critical comments on the manuscript. We also thank Drs Katsunori Murota and Satoshi Naito (Hokkaido University, Japan) for their advice in preparation of Arabidopsis cell-free extracts.
REFERENCES
- 1.van der Velden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int. J. Biochem. Cell. Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- 2.Bashirullah A, Cooperstock RL, Lipshitz HD. Spatial and temporal control of RNA stability. Proc. Natl Acad. Sci. USA. 2001;98:7025–7028. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen RP. mRNA localization: message on the move. Nat. Rev. Mol. Cell. Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- 4.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 5.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bugaut A, Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lammich S, Kamp F, Wagner J, Nuscher B, Zilow S, Ludwig AK, Willem M, Haass C. Translational repression of the disintegrin and metalloprotease ADAM10 by a stable G-quadruplex secondary structure in its 5′-untranslated region. J. Biol. Chem. 2011;286:45063–45072. doi: 10.1074/jbc.M111.296921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agius P, Bennett KP, Zuker M. Comparing RNA secondary structures using a relaxed base-pair score. RNA. 2010;16:865–878. doi: 10.1261/rna.903510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schafer M, Nayernia K, Engel W, Schafer U. Translational control in spermatogenesis. Dev. Biol. 1995;172:344–352. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- 12.Ortega JL, Wilson OL, Sengupta-Gopalan C. The 5′ untranslated region of the soybean cytosolic glutamine synthetase beta(1) gene contains prokaryotic translation initiation signals and acts as a translational enhancer in plants. Mol. Genet. Genomics. 2012;287:881–893. doi: 10.1007/s00438-012-0724-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson SR. So you want to know if your message has an IRES? Wiley Interdiscip. Rev. RNA. 2012;3:697–705. doi: 10.1002/wrna.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng DW, Chandrasekharan MB, Hall TC. The 5′ UTR negatively regulates quantitative and spatial expression from the ABI3 promoter. Plant Mol. Biol. 2004;54:25–38. doi: 10.1023/B:PLAN.0000028767.06820.34. [DOI] [PubMed] [Google Scholar]
- 15.David-Assael O, Saul H, Saul V, Mizrachy-Dagri T, Berezin I, Brook E, Shaul O. Expression of AtMHX, an Arabidopsis vacuolar metal transporter, is repressed by the 5′ untranslated region of its gene. J. Exp. Bot. 2005;56:1039–1047. doi: 10.1093/jxb/eri097. [DOI] [PubMed] [Google Scholar]
- 16.Pichon X, Wilson LA, Stoneley M, Bastide A, King HA, Somers J, Willis AE. RNA binding protein/RNA element interactions and the control of translation. Curr. Protein Pept. Sci. 2012;13:294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi CP, Zhou H, Huang X, Chiang VL. Context sequences of translation initiation codon in plants. Plant Mol. Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- 20.Kawaguchi R, Bailey-Serres J. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 2005;33:955–965. doi: 10.1093/nar/gki240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuura H, Takenami S, Kubo Y, Ueda K, Ueda A, Yamaguchi M, Hirata K, Demura T, Kanaya S, Kato K. A computational and experimental approach reveals that the 5′-proximal region of the 5′-UTR has a Cis-regulatory signature responsible for heat stress-regulated mRNA translation in Arabidopsis. Plant Cell. Physiol. 2013;54:474–483. doi: 10.1093/pcp/pcs189. [DOI] [PubMed] [Google Scholar]
- 22.Sugio T, Matsuura H, Matsui T, Matsunaga M, Nosho T, Kanaya S, Shinmyo A, Kato K. Effect of the sequence context of the AUG initiation codon on the rate of translation in dicotyledonous and monocotyledonous plant cells. J. Biosci. Bioeng. 2010;109:170–173. doi: 10.1016/j.jbiosc.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Lukaszewicz M, Feuermann M, Jerouville B, Stas A, Boutry M. In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 2000;154:89–98. doi: 10.1016/s0168-9452(00)00195-3. [DOI] [PubMed] [Google Scholar]
- 24.Guerineau F, Lucy A, Mullineaux P. Effect of two consensus sequences preceding the translation initiator codon on gene expression in plant protoplasts. Plant Mol. Biol. 1992;18:815–818. doi: 10.1007/BF00020027. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008;36:861–871. doi: 10.1093/nar/gkm1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong GW, Hwang I. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell. 2001;13:1511–1526. doi: 10.1105/TPC.000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyunjong B, Lee DS, Hwang I. Dual targeting of xylanase to chloroplasts and peroxisomes as a means to increase protein accumulation in plant cells. J. Exp. Bot. 2006;57:161–169. doi: 10.1093/jxb/erj019. [DOI] [PubMed] [Google Scholar]
- 28.Qu F, Ren T, Morris TJ. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 2003;77:511–522. doi: 10.1128/JVI.77.1.511-522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murota K, Hagiwara-Komoda Y, Komoda K, Onouchi H, Ishikawa M, Naito S. Arabidopsis cell-free extract, ACE, a new in vitro translation system derived from Arabidopsis callus cultures. Plant Cell. Physiol. 2011;52:1443–1453. doi: 10.1093/pcp/pcr080. [DOI] [PubMed] [Google Scholar]
- 30.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- 32.Kim BH, Cai X, Vaughn JN, von Arnim AG. On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 2007;8:R60. doi: 10.1186/gb-2007-8-4-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang XQ, Rothnagel JA. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 2004;32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamada M, Kiryu H, Sato K, Mituyama T, Asai K. Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics. 2009;25:465–473. doi: 10.1093/bioinformatics/btn601. [DOI] [PubMed] [Google Scholar]
- 36.Puton T, Kozlowski LP, Rother KM, Bujnicki JM. CompaRNA: a server for continuous benchmarking of automated methods for RNA secondary structure prediction. Nucleic Acids Res. 2013;41:4307–4323. doi: 10.1093/nar/gkt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paek KY, Park SM, Hong KY, Jang SK. Cap-dependent translation without base-by-base scanning of an messenger ribonucleic acid. Nucleic Acids Res. 2012;40:7541–7551. doi: 10.1093/nar/gks471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 39.Gallie DR. The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 2002;30:3401–3411. doi: 10.1093/nar/gkf457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987;6:43–48. doi: 10.1002/j.1460-2075.1987.tb04716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elfakess R, Dikstein R. A translation initiation element specific to mRNAs with very short 5′UTR that also regulates transcription. PLoS One. 2008;3:e3094. doi: 10.1371/journal.pone.0003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi CP. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozak M. Some thoughts about translational regulation: forward and backward glances. J. Cell. Biochem. 2007;102:280–290. doi: 10.1002/jcb.21464. [DOI] [PubMed] [Google Scholar]
- 45.Kanoria S, Burma PK. A 28 nt long synthetic 5′UTR (synJ) as an enhancer of transgene expression in dicotyledonous plants. BMC Biotechnol. 2012;12:85. doi: 10.1186/1472-6750-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staley CA, Huang A, Nattestad M, Oshiro KT, Ray LE, Mulye T, Li ZH, Le T, Stephens JJ, Gomez SR, et al. Analysis of the 5′ untranslated region (5′UTR) of the alcohol oxidase 1 (AOX1) gene in recombinant protein expression in Pichia pastoris. Gene. 2012;496:118–127. doi: 10.1016/j.gene.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacob WF, Santer M, Dahlberg AE. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl Acad. Sci. USA. 1987;84:4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 49.Wells DR, Tanguay RL, Le H, Gallie DR. HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Genes Dev. 1998;12:3236–3251. doi: 10.1101/gad.12.20.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avni D, Shama S, Loreni F, Meyuhas O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol. Cell. Biol. 1994;14:3822–3833. doi: 10.1128/mcb.14.6.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu MJ, Wu SH, Chen HM, Wu SH. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol. Syst. Biol. 2012;8:566. doi: 10.1038/msb.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 53.Li F, Zheng Q, Vandivier LE, Willmann MR, Chen Y, Gregory BD. Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell. 2012;24:4346–4359. doi: 10.1105/tpc.112.104232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.