Abstract
The trigger loop (TL) in the RNA polymerase (RNAP) active center plays key roles in the reactions of nucleotide addition and RNA cleavage catalyzed by RNAP. The adjacent F loop (FL) was proposed to contribute to RNAP catalysis by modulating structural changes in the TL. Here, we investigate the interplay between these two elements during transcription by bacterial RNAP. Thermodynamic analysis of catalysis by RNAP variants with mutations in the TL and FL suggests that the TL is the key element required for temperature activation in RNAP catalysis, and that the FL promotes TL transitions during nucleotide addition. We reveal characteristic differences in the catalytic parameters between thermophilic Thermus aquaticus and mesophilic Deinococcus radiodurans RNAPs and identify the FL as an adaptable element responsible for the observed differеnces. Mutations in the FL also significantly affect the rate of intrinsic RNA cleavage in a TL-dependent manner. In contrast, much weaker effects of the FL and TL mutations on GreA-assisted RNA cleavage suggest that the FL-dependent TL transitions are not required for this reaction. Thus, functional interplay between the FL and TL is essential for various catalytic activities of RNAP and plays an adaptive role in catalysis by thermophilic and mesophilic enzymes.
INTRODUCTION
RNA synthesis by multisubunit RNA polymerases (RNAPs) is driven by coordinated conformational changes of several highly conserved structural elements of the active center, including the trigger loop (TL) and the bridge helix (BH) elements in the largest subunit of the enzyme (β' in bacteria; Figure 1) (1,4–8). During catalysis, the binding of the incoming NTP is accompanied by a transition of the TL from an open loop to a closed double α-helical conformation, in which the TL directly contacts the substrate in the insertion site of the active center and facilitates phosphodiester bond formation (Figure 1 and Supplementary Figure S1).
Figure 1.
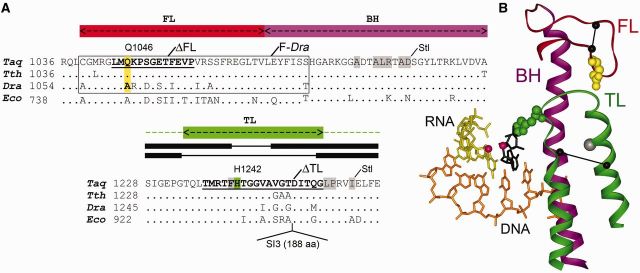
Structure of the key elements of the active center in thermophilic and mesophilic RNAPs. (A) Sequence alignments of the FL, BH and TL in the Taq, Dra, Thermus thermophilus (Tth) and E. coli (Eco) RNAPs. The region substituted in the F-Dra RNAP is boxed; the Q1046A substitution in the FL is yellow; H1242 in the TL is green; regions deleted in the ΔFL and ΔTL RNAPs are underlined. Amino acid residues contacted by Stl in Tth RNAP (1) are shown on a gray background. The α-helical and loop segments in the closed and open states of the TL are shown with thick and thin lines above the TL alignment. Position of the SI3 insertion in the TL in Eco RNAP is shown below the alignment. (B) Structure of the active center of Tth RNAP during nucleotide addition [2O5J, (1)]. The template DNA is orange, RNA is yellow, the FL is red, the BH is magenta, the TL is green and the incoming NTP is black. Catalytic Mg2+ ions are shown as red spheres. The TL residue H1242 that was shown to be directly involved in nucleotide addition and RNA cleavage (2,3) is shown in green; residue Q1046 in the FL is yellow. Positions of the FL and TL deletions are shown with lines connecting Cα atoms at the borders of the deletions. The position of the SI3 insertion in Eco RNAP is shown by a gray sphere.
Multisubunit RNAPs can also catalyze the removal of a short 3′-segment from the nascent RNA. This reaction is performed by the same active center and is preceded by backtracking of the transcription elongation complex (TEC), resulting in the extrusion of the RNA 3′-end through the secondary channel of RNAP (9). RNA cleavage is believed to play a role in reactivation of backtracked TECs and in transcription proofreading. In bacterial RNAP, the TL was shown to be directly involved in the intrinsic RNA cleavage (2). However, its conformation during this reaction must be different from that during nucleotide addition, since the positioning of the RNA 3′-end in the backtracked TECs in the secondary channel is incompatible with the folded TL (10,11). RNA cleavage is greatly stimulated by specialized Gre factors that bind within the secondary channel and functionally replace the TL in the cleavage reaction (12). Other factors that bind within the secondary channel, including Gfh1 (Gre factor homologue 1) and DksA, were proposed to affect the TL conformation, with possible implications for catalysis (13–15).
The catalytic cycle can be modulated by small molecules that bind at the RNAP active center. Antibiotic streptolydigin (Stl) binds at the middle of the BH and blocks the TL in a partially folded conformation, thereby freezing the active center with NTP in the pre-insertion site (Supplementary Figure S1B) (1). The RNAP II inhibitor α-amanitin binds the F loop (FL) and blocks the TL folding (5), and CBR compounds may act similarly, by binding to the FL and freezing an unproductive RNAP conformation (16). Recently, an RNAP inhibitor tagetitoxin was also proposed to affect TL folding (17–19). These observations, together with detrimental effects of some studied mutations in the TL on RNAP activity (2–4,7), demonstrate that changes in the structure and conformational mobility of the TL can critically affect RNAP catalysis and its regulation by various factors.
Previous studies revealed that RNAPs from thermophilic bacteria have higher temperature optima of activity than their mesophilic counterparts and are cold-sensitive (20–22). The cold-sensitivity of catalysis by thermophilic enzymes is generally proposed to result from their reduced conformational mobility at low temperatures, but identification of specific structural features of individual enzymes responsible for the cold sensitivity is often complicated (23–25). Using comparison of closely related thermophilic Thermus aquaticus (Taq) and mesophilic Deinococcus radiodurans (Dra) RNAPs, we identified the FL as the key RNAP element that largely determines the differences in the nucleotide addition between the Taq and Dra RNAPs. We proposed that the FL, which is located nearby the TL and BH, exerts its effects by altering structural transitions of the TL (Figure 1 and Supplementary Figure S1) (26). In this study, we analyze the interplay between the TL and FL during catalysis and demonstrate that the FL is an adaptable element of the tripartite TL-FL-BH structural unit that plays important roles in various RNAP activities through affecting the TL transitions.
MATERIALS AND METHODS
DNA and proteins
Wild-type core Taq RNAP with deletion of the β′ nonconserved domain (amino acids 159–453) was expressed from the plasmid pET28rpoABCd0ZTaq in the Escherichia coli BL21(DE3) strain as described previously (26,27). Mutations in the TL and FL in the Taq β′ subunit were obtained in the pET28rpoCd0ZTaq plasmid by site-directed mutagenesis and recloned into the expression plasmid. The wild-type and mutant Taq RNAPs were purified as described in (26,27). The Dra core RNAP was purified from Dra cells as described in (22). DNA and RNA oligonucleotides were purchased from Syntol (Moscow).
Analysis of nucleotide addition
The complexes of RNAP with the minimal nucleic acid scaffold were prepared as described previously (26). The 5′-labeled RNA, template and nontemplate DNA oligonucleotides were mixed at 10 µM final concentration in transcription buffer containing 40 mM Tris–HCl, pH7.9, 40 mM KCl and 10 mM MgCl2, heated to 65°C and slowly cooled down to 20°C. Core RNAP (100 nM) was mixed with the scaffold template (50 nM) and incubated for 10 min at 20°C; Stl was added to 100 µg/ml, when indicated. The samples were transferred to various temperatures and the transcription was started by addition of UTP (or CTP/dTTP in misincorporation experiments) at 1 mM. The reactions were quenched after different time intervals by addition of 0.5 M EDTA or urea/EDTA-containing stop buffer either by manual mixing or by using a RQF-3 mixer (KinTek, Austin, USA). The samples were separated by 23% PAGE and analyzed with PhosphorImager (GE Healthcare). Each experiment was independently repeated three to six times. The data were fit to a single exponential equation to calculate the kobs values of the reaction, by using GraFit software (Erithacus Software). The thermodynamic parameters of nucleotide addition were calculated in accordance to (23) using equations described in the Supplementary Text. In particular, the Ea values were calculated from the linear fits in Arrhenius plots of kobs values versus 1/T [equation (3)]. The kobs values were also used to directly calculate the ΔG# values using equation 4. The ΔH# and TΔS# were then calculated from the Ea and ΔG# values using equations 5 and 6.
RNA cleavage
RNA cleavage reactions were performed as described in (26). The TECs were assembled on a synthetic scaffold, shown in Figure 4A, containing 5′-labeled RNA in the absence of Mg2+ ions. RNA cleavage reactions were initiated by addition of MgCl2 to 10 mM and stopped after different time intervals by addition of the urea/EDTA containing stop buffer. For each RNAP, the data were fit to the single-exponential equation and normalized by the maximum cleavage efficiency.
Figure 4.

RNA cleavage by wild-type and mutant Taq RNAP variants. (A) Structure of the nucleic acid scaffold used in the cleavage experiments. The position of the RNA cleavage is indicated with an arrow. (B) Kinetics of RNA cleavage by various RNAPs at 20°C. Relative amounts of the 13-nt RNA cleavage product are shown. (C) Effects of GreA on RNA cleavage at 20°C. The rates of RNA cleavage by Taq RNAP mutants, measured either in the absence or in the presence of GreA, are shown relative to the rate of wild-type RNAP measured in the absence of GreA.
RESULTS
Effects of alterations in the FL and TL on the rate of nucleotide addition by Taq RNAP
To probe a hypothetical interplay between the FL and the TL during catalysis, we analyzed several Taq RNAP variants with mutations in the FL, the TL or both. In particular, we studied the following RNAPs: (i) ‘F-Dra’ with substitution of the FL and the BH with the sequence from Dra (residues 1039–1074, eight amino acid substitutions in the FL and a single substitution in the BH); (ii) Q1046A with substitution of a single residue in the FL, involved in direct interactions with the folded TL; (iii) ΔFL with a deletion of the central part of the FL (residues 1044–1056 replaced with Gly); (iv) ΔTL with a deletion of the flexible part of the TL (residues 1237–1255 replaced with three Ala); (v) ΔFL + ΔTL with deletions in both the FL and the TL (Figure 1A).
We measured the catalytic activity of the RNAP variants in the reaction of single nucleotide addition in TECs assembled on a minimal nucleic acid scaffold (Figure 2A). On this template, the TEC adopts a post-translocated conformation, thus allowing to specifically measure the catalytic rates of nucleotide addition, not accounting for possible differences in the translocation properties of different RNAP variants (26). We first compared the rates of UMP incorporation [kobs (s−1), the observed rate constants measured at 1 mM UTP] for all RNAP variants at 20°C. In accordance with the published data (26), Dra RNAP displayed a much higher rate of nucleotide addition than Taq RNAP (84 versus 0.26 s−1; 320-fold difference in the observed catalytic constants) (Figure 2B, Supplementary Table S1). This rate was comparable with the previously reported rates for mesophilic E. coli RNAP under similar conditions (30–80 s−1 at 23–25°C) (18,28). Substitution of the FL and a single residue in the BH in the F-Dra RNAP led to a significant (∼40-fold) increase in the rate of nucleotide addition, and a single Q1046A substitution resulted in a 3-fold increase. In contrast, the FL deletion in Taq RNAP strongly decreased (∼70-fold) the catalytic rate (Figure 2B). In agreement with the key role of the TL in catalysis, its deletion dramatically decreased the rate of UMP addition (∼15 000-fold; Figure 2B). Remarkably, combining the TL and the FL deletions not only did not exacerbate the catalytic defect, but increased the rate of catalysis relative to that of the ΔTL RNAP (Figure 2B). Thus, the deleterious effect of the FL deletion on catalysis is evident only in the presence of the TL.
Figure 2.
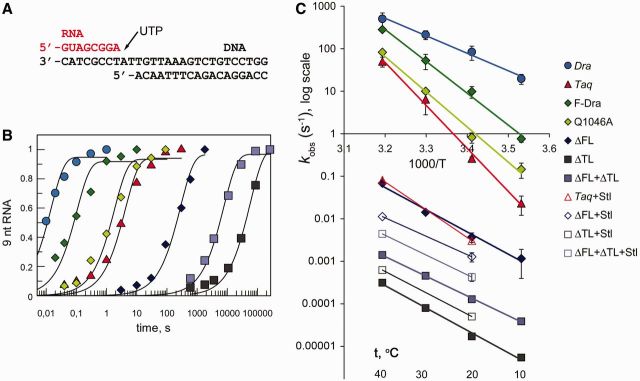
Nucleotide addition by the wild-type and mutant RNAP variants. (A) Schematics of the minimal nucleic acids scaffold used in the experiments. (B) Kinetics of UMP addition by various RNAPs at 20°C. Relative amounts of the 9-nt RNA extension product, normalized to maximal extension, are shown. The data for Dra, Taq and Q1046A RNAPs are from (26). (C) Arrhenius plots for the reaction of nucleotide addition. Corresponding reaction temperatures in °C are indicated at the bottom. Average values and standard deviations from three to five independent experiments are shown. Catalytic rates measured at 20 and 40°C in the presence of Stl are shown with open symbols (connected with lines for clarity).
Temperature dependence of activity of Taq RNAP and its mutant variants
To determine the temperature dependences of the nucleotide addition reaction for various RNAPs, we measured their catalytic rates at temperatures ranging from 10 to 40°C (Supplementary Table S1). Arrhenius plots [log(kobs) versus 1/T] of the resulting data for the wild-type Dra and Taq enzymes and for the mutant Taq RNAP variants are shown in Figure 2C. Within this temperature range, the plots for all RNAPs appeared as descending linear slopes, in accordance with the Arrhenius equation (see Supplementary Text). The differences in the catalytic rates between Taq and Dra RNAPs were larger at low temperatures (i.e. 850-fold at 10°C) than at high temperatures (10-fold at 40°C). It should be noted that we could not measure the catalytic rates at higher temperatures in our system because of thermal instability of the TECs assembled on the minimal nucleic acid scaffold. However, extrapolation of the plots to high temperatures suggests that the catalytic rate of Taq RNAP should reach the rate of Dra RNAP at ∼55–60°C, which is near the temperature optimum of Taq RNAP (22).
The steeper slope of the Arrhenius plot for Taq RNAP indicates a stronger temperature dependence of the reaction and corresponds to a much higher activation energy (Ea) for this RNAP (193.4 kJ/mol versus 78.8 kJ/mol for Dra RNAP, Table 1). The Taq RNAP variants with Dra-specific substitutions in the FL (F-Dra and Q1046A) revealed intermediate temperature dependences (Eas of 149.5 and 158.3 kJ/mol, respectively), and their Arrhenius plots were shifted toward the plot for Dra RNAP, corresponding to higher catalytic activities of these RNAPs, especially at low temperatures (Figure 2C). In contrast, the plots for the ΔFL, ΔTL and ΔFL + ΔTL RNAPs were all strongly shifted downward relative to the wild-type Taq RNAP, reflecting their highly reduced catalytic rates. At all temperatures, the ΔTL RNAP had the lowest catalytic rate, and combination of the FL and TL deletions in the ΔFL + ΔTL RNAP led to a several-fold increase in the rate of catalysis (Figure 2C). The activation energies for all three RNAPs (99.9, 100.8 and 88.7 kJ/mol, respectively) were much lower than the value for Taq RNAP, indicating that the catalysis by these RNAPs cannot be efficiently activated by temperature. Thus, the strong temperature-dependent activation of the Taq RNAP catalysis depends on both the TL and FL.
Table 1.
Activation parameters of the nucleotide addition reaction for wild-type and mutant RNAP variants at 20°C
| RNAP | kobs (s−1) | Ea (kJ/mol) | ΔG# (kJ/mol) | ΔΔG#,a (kJ/mol) | ΔH# (kJ/mol) | ΔΔH#,a (kJ/mol) | TΔS# (kJ/mol) | TΔΔS#,a (kJ/mol) |
|---|---|---|---|---|---|---|---|---|
| Dra | 83.9b | 78.8 | 60.9 | −14.1 | 76.4 | −114.6 | 15.5 | −100.5 |
| Taq | 0.26b | 193.4 | 75.0 | 0 | 191.0 | 0 | 116.0 | 0 |
| F-Dra | 9.9 | 149.5 | 66.2 | −8.9 | 147.1 | −43.9 | 80.9 | −35.1 |
| Q1046A | 0.82b | 158.3 | 72.2 | −2.8 | 155.9 | −35.1 | 83.6 | −32.3 |
| ΔFL | 0.0037 | 99.9 | 85.4 | 10.4 | 97.5 | −93.5 | 12.1 | −103.9 |
| ΔTL | 0.000017 | 100.8 | 98.5 | 23.5 | 98.4 | −92.6 | −0.12 | −116.1 |
| ΔTL + ΔFL | 0.000128 | 88.7 | 93.6 | 18.6 | 86.3 | −104.7 | −7.3 | −123.3 |
aDifferences in the thermodynamics parameters relative to wild-type Taq RNAP.
bData from (26).
Effects of the FL and TL mutations on the activation parameters of nucleotide addition by Taq RNAP
To understand the nature of the observed differences, we used the data on the kobs measurements to calculate the activation parameters (free energy, enthalpy and entropy of activation, connected by the equation ΔG# = ΔH# − TΔS#) of the nucleotide addition reaction for each RNAP at different temperatures. We also calculated the changes in these parameters (ΔΔG#, ΔΔH# and TΔΔS#) in comparison with wild-type Taq RNAP (see Supplementary Text and ‘Materials and Methods’ section for details). The resulting values are presented in Table 1 (for 20°C) and Supplementary Table S2 (for all temperatures). The plots of the ΔΔG#, ΔΔH# and TΔΔS# values versus temperature are shown in Supplementary Figure S2.
Consistent with variations in their catalytic rates, the ΔG# values were lowest for Dra, F-Dra and Q1046A RNAPs (ΔΔG# of −14.1, −8.9 and −2.8 kJ/mol relative to Taq RNAP at 20°C) and highest for the ΔFL, ΔTL and ΔFL + ΔTL RNAPs (ΔΔG# of 10.4, 23.5 and 18.6 kJ/mol, respectively), indicative of the highest energetic barriers to the activated states during catalysis in the latter three RNAPs (Table 1 and Supplementary Table S2). The favorable negative differences in the ΔG# values between Dra, F-Dra, Q1046A RNAPs and wild-type Taq RNAP decreased with temperature, due to the higher stimulatory effect of temperature on Taq RNAP (Supplementary Figure S2A). In contrast, the unfavorable positive differences in the ΔG# values between ΔFL, ΔTL, ΔFL + ΔTL RNAPs and Taq RNAP increased with temperature because these RNAPs were not activated by temperature to the same extent as wild-type Taq RNAP (Supplementary Figure S2A).
Taq RNAP was characterized by highly positive values of both ΔH# and TΔS# (Table 1 and Supplementary Table S2). Dra RNAP had a dramatically lower ΔH# value than Taq RNAP (ΔΔH# = −114.6 kJ/mol at 20°C). The observed decrease in the ΔH# value in the mesophilic Dra RNAP likely ensures its higher activity at low and moderate temperatures in comparison with the thermophilic Taq RNAP (see Supplementary Text and ‘Discussion’ section). The dramatic (favorable) loss in the activation enthalpy for Dra RNAP was partially compensated by a significant (unfavorable) decrease in the TΔS# value (TΔΔS# = −100.5 kJ/mol), a known enthalpy-entropy compensation phenomenon (see Supplementary Text). The resulting ΔΔG# values remained negative between 10 and 40°C, ensuring higher Dra RNAP activity in this temperature range (Supplementary Figure S2). However, the temperature-dependent gain in the entropic term was much higher for Taq RNAP than for Dra RNAP, reducing the difference in their catalytic rates with increasing temperature (Figure 2C, Supplementary Table S2 and Supplementary Figure S2B).
Similarly to Dra RNAP, F-Dra and Q1046A RNAPs were characterized by negative ΔΔH# values in comparison with wild-type Taq RNAP, although the differences were less dramatic (Table 1, Supplementary Figure S2 and Supplementary Table S2). Thus, the FL substitutions likely affect heat-dependent rearrangements of the active center during catalysis. As in the case of Dra RNAP, the decrease in ΔH# was partially compensated by a decrease in TΔS#. The resulting ΔΔG# values for these RNAPs relative to Taq RNAP remained negative between 10 and 40°C but became smaller on temperature increase.
Deletions of the FL and the TL resulted in dramatic unfavorable drops in the activation entropy in the mutant RNAPs in comparison with Taq RNAP (TΔΔS# = −103.9 and −116.1 kJ/mol at 20°C for ΔFL and ΔTL RNAPs, respectively; Table 1 and Supplementary Figure S2). This was partially compensated by a decrease in the activation enthalpy, which was similar for both enzymes (ΔΔH# = −93.5 and −92.6 kJ/mol for ΔFL and ΔTL RNAPs, respectively). The larger drop in the TΔS# value in the case of the ΔTL RNAP accounts for its much lower activity in comparison with the ΔFL RNAP (see Figure 2C). Deletion of the FL in the ΔTL background brought only minor changes of the activation parameters (Table 1). Thus, deletions of the TL and the FL likely result in the loss of major structural changes during catalysis that are associated with high values of the activation entropy and enthalpy in wild-type RNAP (see ‘Discussion’ section).
Roles of the FL and the TL in transcription inhibition by Stl
Antibiotic Stl was proposed to inhibit nucleotide addition through blocking TL folding and stabilizing the pre-insertion state of the active center (Supplementary Figure S1B) (1,29). Accordingly, Stl did not inhibit Taq RNAP with the TL deletion (29). We analyzed nucleotide addition by wild-type Taq RNAP and its variants with deletions in the FL and TL in the presence of Stl (100 µg/ml) at 20 and 40°C. As expected, Stl strongly inhibited UMP addition by wild-type Taq RNAP (Figure 2C and Supplementary Table S1), with significantly more efficient inhibition observed at 40°C (∼6250-fold versus ∼80-fold inhibition at 20°C), consistent with a view that Stl targets the TL-dependent catalysis, which is greatly activated by temperature in Taq RNAP. As we showed previously (26), ΔFL RNAP was only weakly inhibited by Stl (∼4.5- and 6-fold at 20 and 40°C, respectively; Supplementary Table S1). As a result, the nucleotide addition rates for the wild-type and ΔFL RNAPs in the presence of Stl became much more similar than in its absence (Figure 2C and Supplementary Table S1). In agreement with the published data (29), the ΔTL RNAP was not inhibited but instead was slightly activated by Stl (2.9- and 2.0-fold at 20 and 40°C). Finally, the ΔFL + ΔTL RNAP was activated by Stl to a similar extent (3.3- and 3.1-fold at 20 and 40°C; Figure 2C and Supplementary Table S1). Altogether, our results suggest that deletion of the FL significantly decreases the inhibition efficiency of Stl because it by itself disrupts the folding of the TL. Accordingly, in the absence of the TL, Stl does not inhibit NMP addition, independently of the FL presence.
Role of the FL in transcription fidelity
Previous studies revealed an important role of the TL in discrimination of nucleotide substrates (3,4,7). Mutations in the FL were also demonstrated to affect transcription fidelity (8,26). In this work, to highlight possible functional interaction between the FL and TL in nucleotide discrimination, we compared the fidelity of nucleotide incorporation by Taq RNAP variants with mutations in the FL, TL or both. The reactions were performed on the minimal nucleic acid scaffold with noncomplementary CTP and complementary dTTP. Wild-type Taq RNAP efficiently discriminated against these noncognate substrates (the rates of CMP and dTMP incorporations were ∼4500 and 14 000 times slower than the rate for correct UMP at 40°C) (Figure 3 and Supplementary Table S3). In accordance with published data (8), the F-Dra RNAP had a slightly improved fidelity (Figure 3). By contrast, the FL deletion resulted in ∼3-fold decrease in the efficiency of discrimination against CTP (now incorporated 1400 times slower than UMP) and significantly (∼70-fold) impaired discrimination against dTTP (incorporated 200 times slower than UMP; Figure 3 and Supplementary Table S3). Deletion of the TL had a qualitatively similar but stronger effect on transcription fidelity. In particular, the efficiency of discrimination against CTP and dTTP was decreased ∼20- and ∼1100-fold in comparison with wild-type RNAP, demonstrating a crucial role of TL in discrimination of ribo- and deoxyribonucleotides, in agreement with the published data (3). Deletion of the FL in the ΔTL RNAP did not further compromise fidelity (Figure 3 and Supplementary Table S3). Thus, the FL can affect the discrimination against noncognate substrates only in the presence of the TL.
Figure 3.
Fidelity of nucleotide addition by wild-type Taq RNAP and its variants with the FL and TL deletions. For each RNAP, the ratios of the incorporation rates for incorrect (CTP or dTTP) and correct (UTP) nucleotides are shown. All kobs values were measured at 40°C at 1 mM NTP concentrations.
The FL affects the rate of RNA cleavage through the TL
To reveal whether the FL can modulate TL-dependent RNA cleavage, we analyzed this reaction in TEC assembled on a synthetic nucleic acid scaffold with an unpaired RNA 3′-end, corresponding to the backtracked TEC that can form after nucleotide misincorporation (Figure 4A). Dra RNAP was found to have a significantly faster rate of RNA cleavage than Taq RNAP at 20°C (Figure 4B and Supplementary Table S4). At the same time, this difference was less pronounced than for the nucleotide addition reaction (8-fold versus 320-fold, see above). Both RNAPs had similar rates of RNA cleavage at 40°C (Supplementary Table S4), suggesting that the lower catalytic rate of Taq RNAP at 20°C is related to the temperature adaptation. Remarkably, the F-Dra RNAP displayed the same cleavage rate as Dra RNAP (Figure 4B), suggesting that the difference between Taq and Dra RNAPs is mainly explained by substitutions in the FL and, possibly, by a single substitution in the BH also present in the F-Dra RNAP. Deletion of the FL, on the contrary, resulted in a significant decrease in the RNA cleavage rate (∼7.5-fold at 20°C; Figure 4B and C and Supplementary Table S4), further supporting a role for the FL in the cleavage reaction.
Previously, ΔTL Taq RNAP was demonstrated to have greatly reduced RNA cleavage rates in TECs of a similar structure (3). We confirmed this result and found that the TL deletion led to a dramatic drop in the RNA cleavage activity (500-fold at 20°C; Figure 4B and C and Supplementary Table S4). The FL deletion did not further decrease the RNA cleavage rate in RNAP also lacking the TL (ΔFL + ΔTL, Figure 4B and C and Supplementary Table S4), further supporting our hypothesis that the FL acts through changes in the TL.
GreA stimulates RNA cleavage independently of the FL and TL
The RNA cleavage factor GreA was shown to substitute the TL in the RNAP active center during the cleavage reaction (12). We therefore expected that changes in the FL and the TL should not significantly affect GreA-dependent RNA cleavage by Taq RNAP. Indeed, we found that the rates of GreA-stimulated cleavage for RNAPs with mutations in the FL (F-Dra and ΔFL) were identical to, while the rate of the ΔTL RNAP was only six times slower than the rate of wild-type Taq RNAP (Figure 4C and Supplementary Table S4). Interestingly, the ΔFL + ΔTL RNAP displayed a larger decrease in the rate of GreA-dependent cleavage (∼30-fold in comparison with wild-type Taq RNAP, Figure 4C and Supplementary Table S4) suggesting that combination of these mutations may somehow interfere with GreA binding and/or function (see ‘Discussion’ section).
DISCUSSION
The main roles in catalysis by multisubunit RNAPs were shown to be played by a few structural elements of the active center, including catalytic magnesium ions (coordinated by conserved RNAP aspartates and the reaction substrates), the TL and the BH (2,3,6,7,30). Recently, we have identified an additional element involved in RNAP catalysis, the FL, which is conserved in bacterial and eukaryotic RNAPs and forms a common structural unit with the TL and BH (Figure 1) (8,26). In this study, we investigated the functional interplay between the TL and FL during RNAP catalysis and showed that the FL plays an essential role in TL-driven catalysis by promoting temperature-dependent TL transitions. Below, we briefly discuss the roles of the TL and FL in various RNAP activities.
Thermodynamics of nucleotide addition by bacterial RNAP and temperature adaptation of catalysis
Thermodynamic analysis of nucleotide addition by wild-type Dra and Taq RNAPs revealed characteristic differences in the catalytic parameters between the thermophilic and mesophilic enzymes. Taq RNAP is characterized by a much higher Ea value than Dra RNAP, reflecting its much stronger temperature dependence of catalysis, and by highly positive values of both activation enthalpy (ΔH#) and activation entropy (TΔS#), which correspond to larger differences in the enthalpy and entropy between the transition-state and the ground-state complexes (see Supplementary Text). Although straightforward structural interpretation of the ΔH# and TΔS# values may be complicated (see Supplementary Text), large values of the activation enthalpy and entropy measured for Taq RNAP indicate major heat-dependent but entropically favorable rearrangements of the TEC during catalysis. The available TEC structures of Tth RNAP, which is closely related to the Taq enzyme and has an essentially identical active center [Figure 1A; (31)], suggest that these changes likely include TL folding and accompanying changes in the downstream RNAP region, in particular, opening of the downstream β pincer (which connects to the folded TL through the β Fork2 region) and disordering of the β′ Jaw domain (Figure 5 and Supplementary Text) (1). Mutations in the TL and the FL may therefore influence catalysis by (i) directly affecting the TL folding and (ii) allosterically affecting TL-dependent TEC changes, in particular, in the downstream channel of RNAP. It should be noted that additional factors, such as dehydratation of the FL-TL interface during the TL folding, can also significantly contribute to the activation entropy and enthalpy of nucleotide addition.
Figure 5.
Conformational changes of RNAP coupled to the TL folding during nucleotide insertion. The structure of the TEC with NTP bound in the insertion site is shown [2O5J, (1)], as viewed roughly from the top of the BH. The color code corresponds to Figure 1. The downstream DNA duplex (dwDNA) is shown in light yellow. The β Fork2 is light blue, the downstream β pincer (residues 144–191 in Tth RNAP are shown) is black, the β segment (residues 300–331) connecting Fork2 and the pincer is gray. The jaw domain, disordered in the NTP insertion complex, is semitransparent. Position of the deletion in the FL (Δ1044–1056) is shown with a black line. Residue H1242 in the TL and residues E445 (interacts with the TL) and R432 (interacts with the FL) in Fork2 are shown as CPK models. Residue S1074 in the BH whose substitution was shown to affect catalysis in Taq RNAP (26) is also shown. The proposed direction of conformational changes that occur on NTP binding is indicated by gray arrows.
The strong temperature-dependent activation was lost in Taq RNAP lacking the TL, suggesting that it is the TL folding that is activated by temperature during nucleotide addition. The FL and TL deletions resulted in similar decreases in the activation energy, strongly supporting a model in which the FL is required for the TL-dependent temperature activation. The deletion of the TL led to a dramatic unfavorable drop in the activation entropy likely because it disrupted structural reorganization of TEC associated with the TL folding. The deletion of the FL had a similar, even though less dramatic, effect on the activation entropy. Thus, in the absence of the FL, the TL may still undergo structural changes promoting nucleotide addition, but with a much lower efficiency. From the energetics point of view, the TL and FL deletions might affect the ground state complex, the transition state complex or both. Elucidation of the exact effects of the mutations on the structures and energies of the ground state and the transition state complexes awaits in-depth structural and biochemical studies. However, the available data suggest that the FL stimulates the TL folding and associated downstream TEC rearrangements, likely through direct interactions with the TL in the transition state complex.
In comparison with Taq RNAP, Dra RNAP is characterized by much lower activation enthalpy, with a parallel decrease in the activation entropy. The decrease in the ΔH# value ensures the higher activity of Dra RNAP at lower temperatures, by decreasing the heat dependence of the reaction and lowering the heat energy required for the restructuring of the active center during catalysis. Similar trends were previously observed for many mesophilic and psychrophilic enzymes, whose increased activity at low temperatures was also explained by lowering of the ΔH# values of their respective reactions (see Supplementary Text) (23–25,32,33).
The catalytic differences between Dra and Taq RNAPs at least in part depend on the FL structure. Dra-specific substitutions in the FL in the F-Dra and Q1046A RNAPs resulted in a significant decrease in the activation enthalpy and stimulated RNAP activity at low temperatures. From the structural point of view, this can be likely explained by a more flexible FL structure in Dra RNAP that may promote the TL folding by facilitating structural changes required for the FL-TL contact formation in the transition state complex. In particular, substitution of a bulky Q1046 residue with alanine might ease the entry of the TL into the FL pocket. On the contrary, Dra-specific substitutions in the TL did not significantly change the catalytic rate of Taq RNAP (26), suggesting that the differences in the TL structure in Taq and Dra RNAPs do not affect the temperature dependence of catalysis.
In addition to the FL, other elements of the active center also likely contribute to the temperature adaptation of catalysis in thermophilic and mesophilic RNAPs. In particular, Dra-specific S1074T substitution in the BH (present in the F-Dra RNAP) was shown to increase the rate of catalysis by Taq RNAP at low temperatures (26). This substitution may affect the cooperative conformational changes in the TL-FL-BH unit and, probably, associated changes in the Fork2 and the downstream RNAP channel (Figure 5). Other Dra-specific features promoting low-temperature catalysis may include changes in the downstream DNA-RNAP interactions associated with the nucleotide addition (see Supplementary Text).
Roles of the FL and TL in RNA cleavage
Previously, we demonstrated that changes in the FL had only minor effect on RNA cleavage by Taq RNAP in TECs containing fully complementary RNA (26). In this work, we found that mutations in the FL more strongly affected cleavage of RNA containing a noncomplementary nucleotide at its 3′-end. Dra-specific substitutions in the FL and BH in the F-Dra RNAP fully accounted for the observed differences in the cleavage rates between Taq and Dra RNAPs. The inhibitory effect of the FL deletion on the cleavage depended on the TL, suggesting that the FL is important for specific TL transitions required for the cleavage reaction. The stronger effect of the FL mutations on the cleavage in TECs containing misincorporated nucleotides may therefore be connected to the proposed role of the TL in the coordination of the 3′-unpaired nucleotide in the RNA substrate (2). Three-dimensional structures of backtracked TECs of Saccharomyces cerevisiae RNAPII reveal that partially unfolded TL can directly contact the FL in the backtracked state (Supplementary Figure S3A) (10,11). Similar contacts between the TL and the FL in bacterial RNAP may influence the TL conformation during intrinsic RNA cleavage and may explain the importance of the FL for this reaction.
In contrast, the TL was shown to be dispensable for GreA-assisted RNA cleavage (12). Consistently, mutations in the TL [(12) and this study] and the FL did not significantly affect GreA-assisted cleavage in Taq RNAP. However, combination of the ΔTL and ΔFL mutations in the same RNAP significantly decreased the rate of GreA-dependent cleavage. In the structure of the Tth TEC in complex with Gfh1, the coiled-coil domain of Gfh1 homologous to GreA interacts with both the FL and the unfolded TL (Supplementary Figure S3B) (13). Simultaneous loss of these contacts in the ΔFL + ΔTL RNAP may impair GreA binding, explaining the importance of the TL and the FL for GreA-dependent cleavage.
In conclusion, we demonstrated that the FL promotes temperature-dependent TL transitions during nucleotide addition. Alterations in the FL structure can also affect nucleotide discrimination by RNAP, either increasing transcription fidelity or decreasing it; these effects are strictly TL-dependent, consistent with the reported role of the TL in nucleotide discrimination (3,4,7). Furthermore, mutations in the FL change the sensitivity of RNAP to antibiotic Stl that inhibits catalysis by restricting the TL mobility. The FL also modulates the TL-dependent RNA cleavage but the underlying mechanism remains to be elucidated. Thus, the FL is an adaptable element of RNAP that alters the catalytic properties of the enzyme without changing the conserved catalytic mechanism. Owing to its roles in various catalytic activities and its conservation in various bacterial lineages (8,26), the FL is also a promising target for development of novel antibiotics targeting bacterial RNAP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [34,35].
FUNDING
Russian Academy of Sciences Presidium Program in Molecular and Cellular Biology (to A.K.); Young Russian Scientists Grant Program of the President of Russian Federation [MK-7156.2012.4]; Russian Foundation for Basic Research [12-04-33187]; Federal Targeted Program ‘Scientific and scientific-pedagogical personnel of innovative Russia 2009-2013’ [8106 and 8475]; European Social Fund under the Global Grant measure [VP1-3.1-ŠMM-07-K-01-105 to S.K.]; National Institutes of Health [GM067153 to I.A.]. Funding for open access charge: Russian Foundation for Basic Research [12-04-33187].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 2.Yuzenkova Y, Zenkin N. Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis. Proc. Natl Acad. Sci. USA. 2010;107:10878–10883. doi: 10.1073/pnas.0914424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuzenkova Y, Bochkareva A, Tadigotla VR, Roghanian M, Zorov S, Severinov K, Zenkin N. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat. Struct. Mol. Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 6.Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu. Rev. Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat. Struct. Mol. Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miropolskaya N, Nikiforov V, Klimasauskas S, Artsimovitch I, Kulbachinskiy A. Modulation of RNA polymerase activity through trigger loop folding. Transcription. 2010;1:89–94. doi: 10.4161/trns.1.2.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 12.Roghanian M, Yuzenkova Y, Zenkin N. Controlled interplay between trigger loop and Gre factor in the RNA polymerase active centre. Nucleic Acids Res. 2011;39:4352–4359. doi: 10.1093/nar/gkq1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 14.Furman R, Sevostyanova A, Artsimovitch I. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res. 2012;40:3392–3402. doi: 10.1093/nar/gkr1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, Rutherford ST, Lee JH, Butcher SE, Gourse RL. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 2012;26:2634–2646. doi: 10.1101/gad.204693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artsimovitch I, Chu C, Lynch AS, Landick R. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science. 2003;302:650–654. doi: 10.1126/science.1087526. [DOI] [PubMed] [Google Scholar]
- 17.Artsimovitch I, Svetlov V, Nemetski SM, Epshtein V, Cardozo T, Nudler E. Tagetitoxin inhibits RNA polymerase through trapping of the trigger loop. J. Biol. Chem. 2011;286:40395–40400. doi: 10.1074/jbc.M111.300889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malinen AM, Turtola M, Parthiban M, Vainonen L, Johnson MS, Belogurov GA. Active site opening and closure control translocation of multisubunit RNA polymerase. Nucleic Acids Res. 2012;40:7442–7451. doi: 10.1093/nar/gks383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuzenkova Y, Roghanian M, Bochkareva A, Zenkin N. Tagetitoxin inhibits transcription by stabilizing pre-translocated state of the elongation complex. Nucleic Acids Res. 2013;41:9257–9265. doi: 10.1093/nar/gkt708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meier T, Schickor P, Wedel A, Cellai L, Heumann H. In vitro transcription close to the melting point of DNA: analysis of Thermotoga maritima RNA polymerase-promoter complexes at 75 degrees C using chemical probes. Nucleic Acids Res. 1995;23:988–994. doi: 10.1093/nar/23.6.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue Y, Hogan BP, Erie DA. Purification and initial characterization of RNA polymerase from Thermus thermophilus strain HB8. Biochemistry. 2000;39:14356–14362. doi: 10.1021/bi0012538. [DOI] [PubMed] [Google Scholar]
- 22.Kulbachinskiy A, Bass I, Bogdanova E, Goldfarb A, Nikiforov V. Cold sensitivity of thermophilic and mesophilic RNA polymerases. J. Bacteriol. 2004;186:7818–7820. doi: 10.1128/JB.186.22.7818-7820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lonhienne T, Gerday C, Feller G. Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim. Biophys. Acta. 2000;1543:1–10. doi: 10.1016/s0167-4838(00)00210-7. [DOI] [PubMed] [Google Scholar]
- 24.Fields PA. Review: Protein function at thermal extremes: balancing stability and flexibility. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2001;129:417–431. doi: 10.1016/s1095-6433(00)00359-7. [DOI] [PubMed] [Google Scholar]
- 25.Lam SY, Yeung RC, Yu TH, Sze KH, Wong KB. A rigidifying salt-bridge favors the activity of thermophilic enzyme at high temperatures at the expense of low-temperature activity. PLoS Biol. 2011;9:e1001027. doi: 10.1371/journal.pbio.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miropolskaya N, Artsimovitch I, Klimasauskas S, Nikiforov V, Kulbachinskiy A. Allosteric control of catalysis by the F loop of RNA polymerase. Proc. Natl Acad. Sci. USA. 2009;106:18942–18947. doi: 10.1073/pnas.0905402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuznedelov K, Lamour V, Patikoglou G, Chlenov M, Darst SA, Severinov K. Recombinant Thermus aquaticus RNA polymerase for structural studies. J. Mol. Biol. 2006;359:110–121. doi: 10.1016/j.jmb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Foster JE, Holmes SF, Erie DA. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell. 2001;106:243–252. doi: 10.1016/s0092-8674(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 29.Temiakov D, Zenkin N, Vassylyeva MN, Perederina A, Tahirov TH, Kashkina E, Savkina M, Zorov S, Nikiforov V, Igarashi N, et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 30.Sosunov V, Zorov S, Sosunova E, Nikolaev A, Zakeyeva I, Bass I, Goldfarb A, Nikiforov V, Severinov K, Mustaev A. The involvement of the aspartate triad of the active center in all catalytic activities of multisubunit RNA polymerase. Nucleic Acids Res. 2005;33:4202–4211. doi: 10.1093/nar/gki688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, Yasugi M, Arisaka F, Yamagishi A, Oshima T. Adaptation of a thermophilic enzyme, 3-isopropylmalate dehydrogenase, to low temperatures. Protein Eng. 2001;14:85–91. doi: 10.1093/protein/14.2.85. [DOI] [PubMed] [Google Scholar]
- 33.D'Amico S, Marx JC, Gerday C, Feller G. Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 2003;278:7891–7896. doi: 10.1074/jbc.M212508200. [DOI] [PubMed] [Google Scholar]
- 34.Collins T, Meuwis MA, Gerday C, Feller G. Activity, stability and flexibility in glycosidases adapted to extreme thermal environments. J. Mol. Biol. 2003;328:419–428. doi: 10.1016/s0022-2836(03)00287-0. [DOI] [PubMed] [Google Scholar]
- 35.Fouqueau T, Zeller ME, Cheung AC, Cramer P, Thomm M. The RNA polymerase trigger loop functions in all three phases of the transcription cycle. Nucleic Acids Res. 2013;41:7048–7059. doi: 10.1093/nar/gkt433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




