Abstract
Homologous recombination catalyzed by the RAD51 recombinase eliminates deleterious DNA lesions from the genome. In the presence of ATP, RAD51 forms a nucleoprotein filament on single-stranded DNA, termed the presynaptic filament, to initiate homologous recombination-mediated DNA double-strand break repair. The SWI5-SFR1 complex stabilizes the presynaptic filament and enhances its ability to mediate the homologous DNA pairing reaction. Here we characterize the RAD51 presynaptic filament stabilization function of the SWI5-SFR1 complex using optical tweezers. Biochemical experiments reveal that SWI5-SFR1 enhances ATP hydrolysis by single-stranded DNA-bound RAD51. Importantly, we show that SWI5-SFR1 acts by facilitating the release of ADP from the presynaptic filament. Our results thus provide mechanistic understanding of the function of SWI5-SFR1 in RAD51-mediated DNA recombination.
INTRODUCTION
Double-strand breaks (DSBs) are among the most deleterious chromosomal lesions that, if not eliminated promptly and in an error-free manner, will cause genome instability. Homologous recombination (HR) represents a major error-free pathway for DSB elimination (1,2). As such, HR is indispensable for the maintenance of genome integrity and cancer avoidance (3–5).
In DSB repair by HR, the ends of the DNA break are nucleolytically resected to yield 3′ single-stranded DNA (ssDNA) tails. The polymerization of RAD51 molecules on the ssDNA leads to the assembly of a right-handed helical protein filament known as the presynaptic filament. The ssDNA is held in an extended conformation in the presynaptic filament, being stretched by ∼50% relative to B form duplex DNA, with an axial rise of ∼5.0 Å per nucleotide (6–10). RAD51 has a DNA-stimulated ATPase activity, and although ATP binding is necessary for presynaptic filament assembly, ATP hydrolysis leads to the turnover of RAD51 protomers from DNA (7,11–15). Once assembled, the presynaptic filament engages duplex DNA and conducts a search for homology in the bound duplex. Invasion of the homologous DNA sequence leads to the formation of a DNA joint called the displacement loop, or D-loop. The D-loop structure is resolved by one of several pathways to complete the HR process (2,16).
As first revealed in genetic studies in the fission yeast Schizosaccharomyces pombe, swi5 and sfr1 mutants exhibit the same HR deficient phenotype and are clearly epistatic. These observations and companion biochemical analyses have provided strong evidence that Swi5 and Sfr1 proteins function in HR as a complex (17–19). The genes that encode mouse and human SWI5 and SFR1 proteins have been isolated and characterized recently (20,21). Swi5 or Sfr1 knockout in mouse embryonic stem cells or knockdown in human cell lines engenders sensitivity to DNA damaging agents such as etoposide and X-ray. Moreover, genetic and biochemical analyses have revealed that the mammalian SWI5 and SFR1 orthologs also function as a complex in RAD51-dependent HR and DNA repair (20,21).
We have recently devised procedures for the expression and purification of the mouse SWI5-SFR1 complex. Our biochemical and biophysical analyses have verified the heterodimeric nature of the complex and demonstrated a stimulatory effect that the complex exerts on the RAD51-mediated homologous DNA pairing reaction. Importantly, we have furnished evidence that enhancement of the RAD51 recombinase activity stems from a stabilizing effect of SWI5-SFR1 on the presynaptic filament (22). Here we present our single-molecule and biochemical analyses to delineate the action mechanism of SWI5-SFR1 in the homologous pairing reaction with the RAD51 presynaptic filament. Surprisingly, we have found that SWI5-SFR1 stimulates, rather than attenuates, ATP hydrolysis by the presynaptic filament. Importantly, we present evidence that SWI5-SFR1 promotes the release of ADP from the RAD51 presynaptic filament to help maintain the presynaptic filament in its active ATP-bound form. Our results thus unveil a novel action mechanism of a key, evolutionarily conserved RAD51 accessory factor.
MATERIALS AND METHODS
DNA substrates, construction of expression plasmids, protein expression and purification and biochemical assays were conducted as described in the Supplementary Data.
Single-molecule optical tweezers
Optical tweezers setup
Our optical tweezers instrument uses two lasers. A 1064-nm laser (CrytsaLaser) is used for beads trapping, whereas the 780-nm laser (CrytsaLaser) is for detection of bead centroid position. After modulating the laser intensities with half-wave plates and polarized beam splitters, the two-laser beams were aligned in the microscope (Nikon eclipse Ti) using the same path. An optical trap was then formed by strongly focusing the laser beams with a high N.A objective (Nikon Apo TIRF objective 100x, N.A. = 1.49). To enable a precise position control of samples, the reaction chamber was mounted to a 3D piezo-stage (Madcity Laboratory). A quadrant position detector (QPD, thorlab) was used to detect the bead position within the trap based on the forward scattering light of the detection laser.
Single-molecule optical tweezers experiments
A 1.26-µm streptavidin-coated polystyrene beads (Spherotech) were attached to one end of the dsDNA through a biotin-strepavidin linkage. The other DNA end was tethered to the coverslip surface through the digoxigenin and surface-immobilized anti-digoxigenin linkage. A force-extension assay was conducted to determine the contour lengths of the DNA tethers. First, potential tethers were identified by its Gaussian-shaped Brownian motion in the absence of any external force. Second, these tethers were moved to the proximity of the trap center, where trapping occurred automatically. The trapped microspheres were moved to a pre-set height of 1 μm. We first identified the tether anchoring point by a quick scan in x and y directions. A force-extension assay was carried out by moving the piezo stage in a 50-nm stepwise pattern in two orthogonal directions. During the stage movement, corresponding forces exerted on tethers were also recorded. The force-extension curve is fitted to a worm-like-chain model using a custom-written MATLAB program.
All force-extension assays were done at 23°C in the standard RAD51 reaction buffer [20 mM Tris–HCl (pH 7.5), 100 mM KCl, 2.5 mM MgCl2, glucose oxidase (165 U/ml), catalase (2170 U/ml), glucose (0.4% wt/wt), 10 mM dithiothreitol]. An oxygen scavenger system is used to remove potential oxygen damage to enzyme activity in the optical tweezers setup (23). Unless otherwise noted, all proteins were at 250 nM, and the nucleotide concentration was ATP (50 µM), AMP-PNP (50 µM) and ADP (2 mM). For the contour length histograms, lengths were measured every 2 min over a 30-min time frame and repeated at least three times for each condition. The contour length histograms were implemented with a bootstrapping analysis (R-software) that resamples the means of lengths 100 times based on 25 (dsDNA), 16 (RAD51-ATP), 15 (RAD51-ADP), 11 (RAD51-AMP-PNP) and 22 (RAD51-ATP-SWI5-SFR1) single-molecule measurements.
ATPase activity
To examine the effect of SWI5, SFR1, SWI5-SFR1 or RAD51AP1 on ATP hydrolysis by RAD51, DMC1, scRad51 or RecA, 1.6 µM of the recombinase were incubated in 12.5 µl of buffer A [35 mM Tris–HCl (pH 7.5), 1 mM DTT, 2.5 mM MgCl2, 35 mM KCl and 100 ng/µl BSA] containing the final concentration of 0.5 mM ATP and with or without 80mer Oligo 1 ssDNA or dsDNA (4.8 µM nucleotides and base pairs, respectively) at 37°C for 5 min, followed by the addition of the indicated amounts of SWI5, SFR1, SWI5-SFR1 or RAD51AP1 and a 5min incubation. After adding 0.3 µCi [γ-32P] ATP, 1.5 µl of aliquots of the reactions were removed at the indicated times and mixed with an equal volume of 500 mM EDTA to halt the reaction. The level of ATP hydrolysis was determined by thin layer chromatography in polyethyleneimine cellulose sheets (Fluka) with phosphorimaging analysis in a Personal FX phosphorimager using the Quantity One software (Bio-Rad), as described (24). The apparent velocity (Vapp.) was calculated before 10% of the radiolabeled ATP had been hydrolyzed.
Nitrocellulose filter binding assay
Measuring ADP–ATP exchange within the RAD51 presynaptic filament
Reaction mixtures were assembled in 25 µl of buffer A containing 3.2 µM RAD51, 9.6 µM nucleotides of ssDNA, if present, and either 3.2 µM [14C] ADP in the presence of ssDNA or 16 µM [14C] ADP in the absence of DNA. Reactions were incubated at 37°C for 30 min before the addition of the indicated amounts of SWI5, SFR1, SWI5-SFR1 or RAD51AP1 and 3.2 µM ATP, followed by a 5-min incubation. The reaction mixture was then passed through a nitrocellulose filter (Portran, Whatman), which had been presoaked with buffer [25 mM Tris–HCl (pH 7.5)], in a Minifold apparatus (Schleicher & Schuell) under vacuum. The level of ADP was determined by phosphorimaging analysis as described earlier in the text.
Measuring ADP release from the RAD51 filament
The experiments were set up with either RAD51-ssDNA or RAD51-dsDNA filament as described earlier in the text for the ADP–ATP exchange assay except for the omission of ATP.
Measuring ADP- and ATP-binding affinities of RAD51
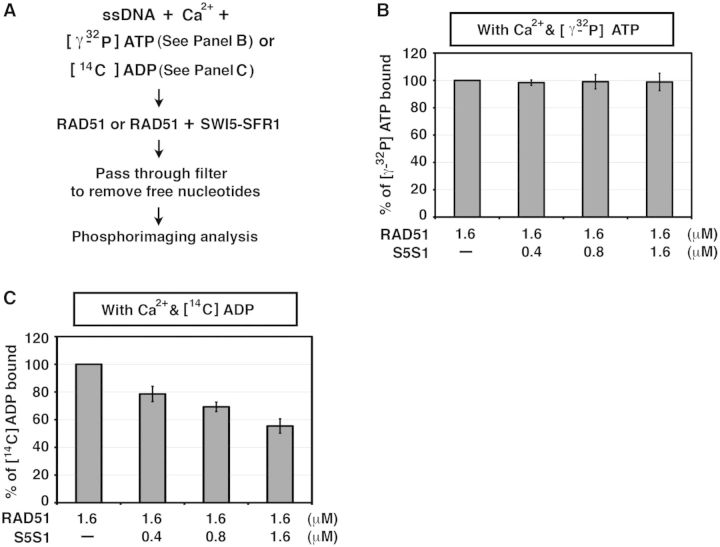
To monitor how SWI5-SFR1 affects the ADP- or ATP-binding affinity of RAD51, 1.6 µM RAD51 or pre-mixed RAD51 and SWI5-SFR1 were incubated in 25 µl of buffer B [35 mM Tris–HCl (pH 7.5), 1 mM DTT, 10 mM CaCl2, 35 mM KCl] containing 20 µM [14C] ADP or 20 µM ATP with 0.125 µCi [γ-32P] ATP and with 80mer Oligo 1 ssDNA or dsDNA (4.8 µM nucleotides and base pairs, respectively) at 37°C for 5 min. The reaction mixture was then filtered through nitrocellulose filters, and the level of ADP or ATP bound was determined by phosphorimaging analysis. We note that CaCl2 at 10 mM inhibits the ATPase activity of RAD51 efficiently but has little or no effect on the DNA strand exchange activity [(11); Supplementary Figure S5; and our unpublished data].
RESULTS
Single-molecule optical tweezers measurement provides evidence for stabilization of RAD51-DNA nucleoprotein filaments by SWI5-SFR1
Our previous studies provided evidence that SWI5-SFR1 helps stabilize the nucleoprotein filament of RAD51 (22). As DNA is stretched by as much as 50% within the nucleoprotein filament, we could follow the nucleoprotein filament stabilization function of SWI5-SFR1 by monitoring the contour length of individual RAD51-dsDNA nucleoprotein filaments in optical tweezers experiments. In these experiments, we fix dsDNA onto a coverslip slide through a digoxigenin/anti-digoxigenin linkage, and the distal biotin-labeled end of the DNA is attached to a streptavidin-labeled bead, which can be optically trapped by a 1064-nm laser with pico-Newton range force (Figure 1A). By moving the stage, we can alter the force exerted on the surface-bound DNA-bead complex and determine the DNA length change at different forces. This force-extension experiment returns the elasticity of the DNA molecules or the nucleoprotein filament based on the worm-like-chain model and provides the contour length of the nucleoprotein filament. As shown in Figure 1B, in the presence of RAD51 and ATP, RAD51 nucleoprotein filament showed the averaged contour length being 1.28-fold of DNA alone, indicating the dynamics of assembly and disassembly of the filament. In addition, we observed even longer averaged contour lengths (∼1.5-fold increase) of RAD51 nucleoprotein filament when SWI5-SFR1 complex was present, thus verifying the stabilization of the RAD51 filament by SWI5-SFR1 (Figure 1B). The contour length of the RAD51-SWI5-SFR1 filament of ∼1.5-fold that of naked B-form DNA is consistent with that of fully RAD51-coated DNA (6–8,10,13,15). We also measured the contour length of the RAD51 filament in the presence of the non-hydrolyzable nucleotide AMP-PNP to specifically prevent turnover of RAD51 protomers and confirmed the expected 1.5-fold extension of DNA contour length (Supplementary Figure S1). Consistent with a published study (13), we observed no significant extension of the contour length of the RAD51 filament with ADP as the nucleotide cofactor (Supplementary Figure S1). Altogether, the results from the single-molecule analysis suggested the dynamic nature of the RAD51-DNA filament assembled with ATP and the stabilizing effect of SWI5-SFR1 on the filament.
Figure 1.
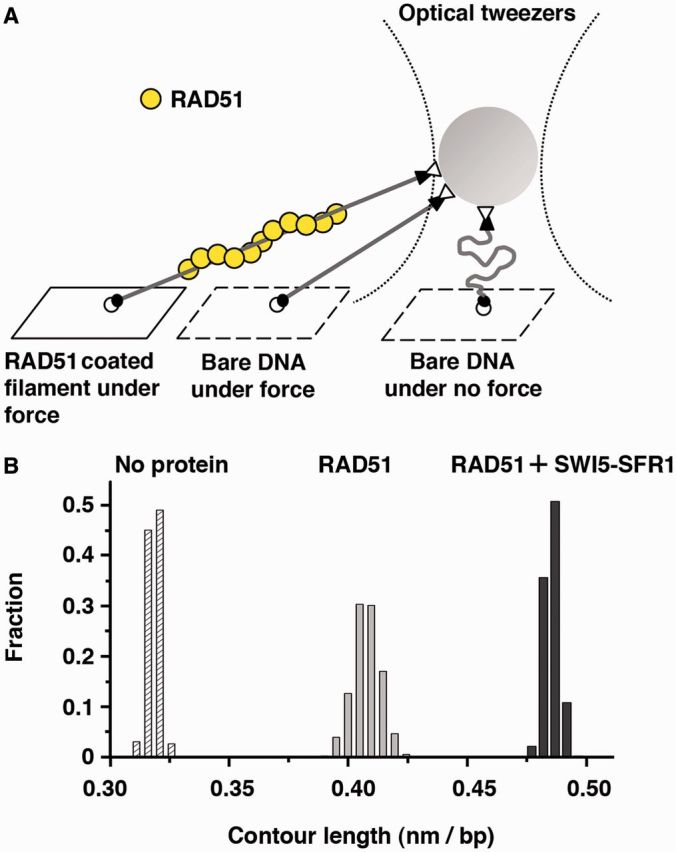
SWI5-SFR1 stabilizes RAD51 filament. (A) Schematic of the optical tweezers setup to monitor the presynaptic filament length. (B) Histograms of contour lengths of individual naked 6123-bp duplex DNA molecules alone, in the presence of RAD51 and ATP, or the mixture of RAD51, SWI5-SFR1 and ATP.
SWI5-SFR1 enhances ATP hydrolysis by the RAD51 presynaptic filament
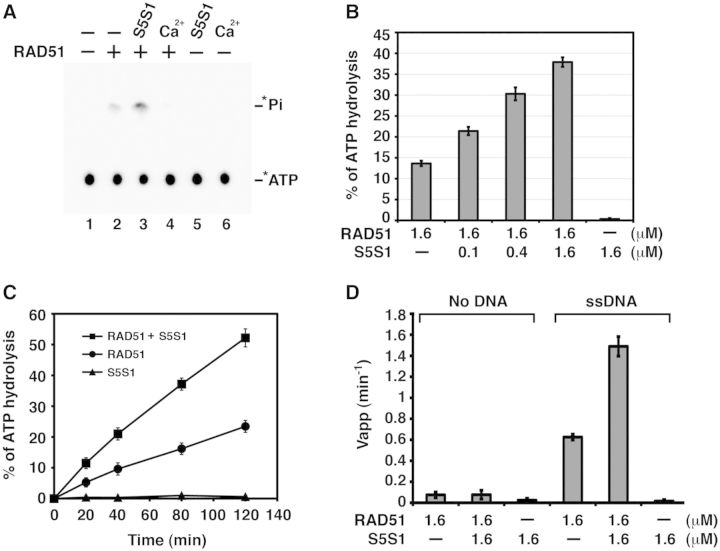
It has been well documented that the stability of RAD51 presynaptic filament is affected by ATP hydrolysis. Accordingly, the use of a non-hydrolyzable nucleotide (such as AMP-PNP) or addition of Ca2+ ions to retard ATP hydrolysis leads to stabilization of the RAD51 presynaptic filament (11,12). This raises the important question of whether SWI5-SFR1 stabilizes the RAD51 presynaptic filament by attenuating ATP hydrolysis by the filament. To examine this, presynaptic filaments of RAD51 were assembled on 80mer ssDNA and incubated with [γ-32P] ATP with or without SWI5-SFR1. Aliquots of the reaction mixtures were removed to determine the level of ATP hydrolysis by thin layer chromatography (24). Surprisingly, although the addition of Ca2+ led to a strong inhibition of ATP hydrolysis, SWI5-SFR1, in a concentration-dependent and time-dependent manner, significantly increased the amount of ATP hydrolyzed by RAD51 (Figure 2A, B and C). This stimulatory effect occurred only when DNA was present (Figure 2D). We note that the SWI5-SFR1 complex alone has no detectable ATPase activity (Figure 2C). To verify that the stimulatory effect of SWI5-SFR1 does not stem from the removal of secondary structure in the ssDNA, we examined ATP hydrolysis using poly-dT, which lacks secondary structure, to assemble the presynaptic filaments. In this case also, SWI5-SFR1 enhanced the DNA-stimulated ATPase activity of RAD51 to the same degree as when the 80mer ssDNA was used (Figure 2D and Supplementary Figure S2A).
Figure 2.
SWI5-SFR1 enhances RAD51 ATPase activity. (A) Thin layer chromatography to monitor the hydrolysis of [γ-32P] ATP by RAD51. The asterisk denotes the 32P label. Ca2+ inhibits ATP hydrolysis, whereas SWI5-SFR1 stimulates hydrolysis in a concentration-dependent (B) and time-dependent (C) manner. (D) The stimulatory effect of ATP hydrolysis by SWI5-SFR1 is dependent on ssDNA. (B–D) Error bars represent the standard deviation (±SD) calculated based on at least three independent experiments. Symbol: S5S1, SWI5-SFR1.
Enhancement of RAD51 ATPase activity requires a specific interaction of SWI5-SFR1 with RAD51
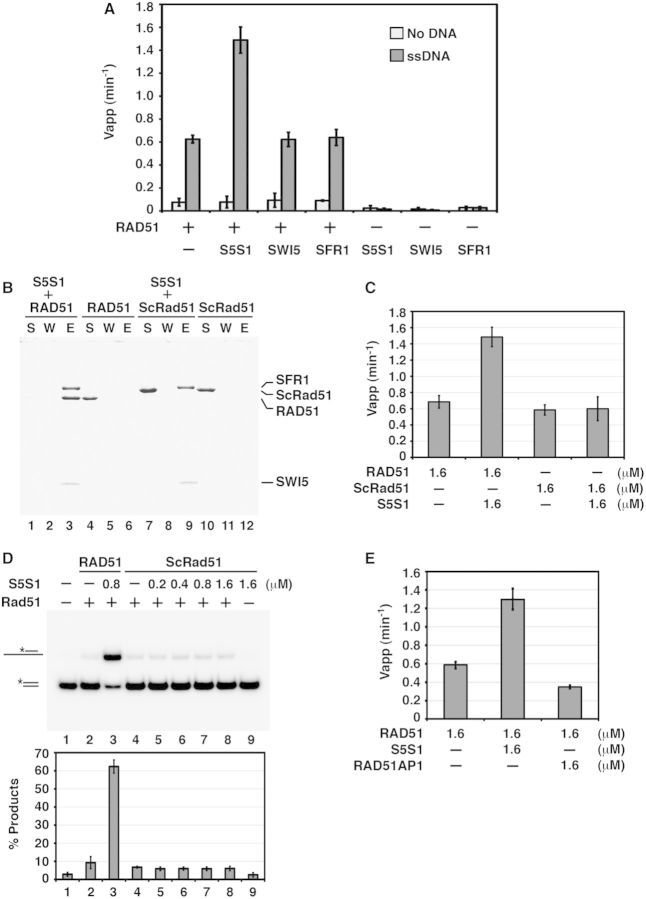
We demonstrated previously that while SWI5-SFR1 associates with RAD51 avidly, neither SWI5 nor SFR1 alone has such a capability (22). Importantly, neither SWI5 nor SFR1 exerts any effect on the DNA-stimulated ATPase activity of RAD51 (Figure 3A). SWI5-SFR1 does not interact with E. coli RecA (22) or Saccharomyces cerevisiae Rad51 (ScRad51; Figure 3B), and it has no effect on ATP hydrolysis by the presynaptic filament of either RecA (Supplementary Figure S2B) or ScRad51 (Figure 3C). As expected, there is no stimulation of ScRad51-mediated homologous DNA-pairing activity by mouse SWI5-SFR1 (Figure 3D). In summary, the results presented here provide strong evidence that SWI5-SFR1 enhances RAD51 ATP hydrolysis in a species-specific manner. Lastly, the results in Figure 3E demonstrate that mouse RAD51AP1, which physically interacts with RAD51 and enhances the RAD51 recombinase activity [(25,26); and our unpublished data], is devoid of the ability to enhance ATP hydrolysis by the RAD51 presynaptic filament (Figure 3E).
Figure 3.
Functional interactions between RAD51 and SWI5-SFR1 complex. (A) SWI5-SFR1 complex but not SWI5 or SFR1 enhances ATP hydrolysis by the RAD51 presynaptic filament. (B) No physical interaction was seen between mouse SWI5-SFR1 and S. cerevisiae Rad51 (ScRad51) by affinity pulldown. The supernatant (S), wash (W) and SDS eluate (E) from the pulldown reaction were analyzed by SDS–PAGE. (C) SWI5-SFR1 has no effect on ATP hydrolysis by the ScRad51 presynaptic filament. (D) SWI5-SFR1 has no effect on ScRad51-mediated homologous DNA pairing. The results were graphed. The asterisk denotes the 32P label in the DNA strand. (E) RAD51AP1 has no effect on ATP hydrolysis by the RAD51 presynaptic filament. (A, C, D and E) Error bars represent the standard deviation (±SD) calculated based on at least three independent experiments. Symbols: S5S1, SWI5-SFR1; ScRad51, S. cerevisiae Rad51.
SWI5-SFR1 facilitates ADP/ATP exchange in the RAD51 presynaptic filament
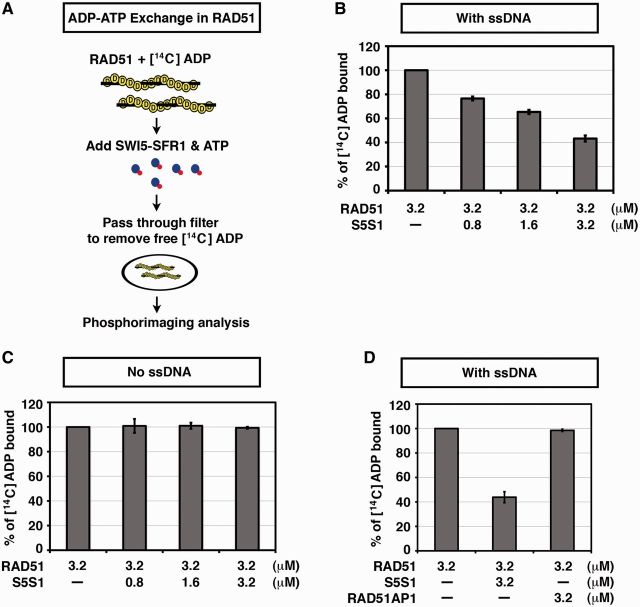
It has been shown that the RAD51-ATP-ssDNA filament is converted to the inactive RAD51-ADP-ssDNA form owing to ATP hydrolysis and the slow dissociation of ADP (11,13). We wanted to test whether SWI5-SFR1 functions to facilitate the exchange of ADP for ATP within the RAD51 presynaptic filament, as such a nucleotide exchange attribute could account for the enhancement of DNA-stimulated ATP hydrolysis by RAD51 as revealed earlier in the text (22,27). To do so, we used a nitrocellulose filter-binding assay to monitor ADP–ATP exchange within the RAD51 presynaptic filament with or without SWI5-SFR1 being present. Previously, by electron microscopy, an ability of RAD51 to form a compact filament on ssDNA was demonstrated (28). In our assay, ssDNA and radiolabeled [14C] ADP are pre-incubated with RAD51 to form the RAD51-ADP-ssDNA filament (28), and then a varying amount of the SWI5-SFR1 complex and cold ATP are added to the reaction for a varying time. After incubation, the reaction mixture is filtered through a nitrocellulose membrane trap to remove free [14C] ADP. Filters are air dried, and the amount of [14C] ADP is determined by phosphorimaging analysis (Figure 4A). As shown in Figure 4B, SWI5-SFR1 facilitates the [14C] ADP/ATP exchange within the RAD51 presynaptic filament in a dosage-dependent manner (Figure 4B). This stimulatory effect is unique between SWI5-SFR1 and the RAD51-ssDNA filament because (i) there was no enhancement of [14C] ADP/ATP exchange in RAD51 when ssDNA was absent (Figure 4C) and (ii) RAD51AP1 lacked the ability to stimulate [14C] ADP/ATP exchange (Figure 4D). We have not been able to examine whether SWI5-SFR1 would enhance ADP/ATP exchange in the RecA-ssDNA or yeast Rad51-ssDNA nucleoprotein filament because these filaments do not bind [14C] ADP well (unpublished data).
Figure 4.
SWI5-SFR1 but not RAD51AP1 mediates ADP–ATP exchange of RAD51 filament. (A) Schematic of the filter-binding assay to monitor ADP–ATP exchange within the RAD51-ssDNA presynaptic filament. (B) SWI5-SFR1 facilitates ADP–ATP exchange of RAD51 filament in a dosage-dependent manner. (C) There is no enhancement of nucleotide exchange by SWI5-SFR1 on the omission of ssDNA. (D) RAD51AP1 lacks the ability to facilitate ADP–ATP exchange within the RAD51 presynaptic filament. (B–D) Error bars represent the standard deviation (±SD) calculated based on at least three independent experiments. Symbol: S5S1, SWI5-SFR1.
To further delineate the action of SWI5-SFR1, we asked whether it possesses the ability to bind ADP or ATP. We note that SWI5-SFR1 lacks any nucleotide interaction motif in its primary sequence, and, as shown in Supplementary Figure S3, it does not bind either ATP or ADP (Supplementary Figure S3). The results presented in the next section further reveal that SWI5-SFR1 accelerates the release of ADP from the RAD51-ssDNA filament.
SWI5-SFR1 enhances ADP release from the RAD51 filament
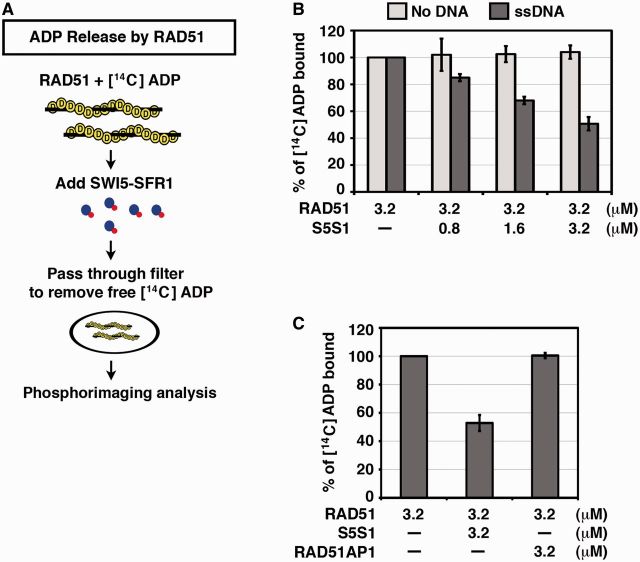
Within the RAD51 presynaptic filament, hydrolysis of ATP is followed by a relatively slow dissociation of ADP (11,13). In other words, ADP release is likely rate limiting in ADP/ATP exchange. For this reason, we used a filter-binding assay to ask whether SWI5-SFR1 facilitates ADP release from the RAD51 presynaptic filament (Figure 5A). We found that SWI5-SFR1, in a concentration-dependent manner, facilitates ADP release from the RAD51 filament (Figure 5B). Importantly, the effect of SWI5-SFR1 on ADP release from RAD51 is contingent on the presence of ssDNA (Figure 5B). In contrast, RAD51AP1 has no effect on the rate of ADP release from the RAD51 presynaptic filament (Figure 5C).
Figure 5.
SWI5-SFR1 expedites ADP release from the RAD51 presynaptic filament. (A) Schematic of the filter-binding assay to monitor ADP release from the RAD51 presynaptic filament. (B) SWI5-SFR1 facilitates ADP release from RAD51 presynaptic filament in a protein concentration- and ssDNA-dependent manner. (C) RAD51AP1 lacks the ability to facilitate ADP release from the presynaptic filament. (B and C) Error bars represent the standard deviation (±SD) calculated based on at least three independent experiments. Symbol: S5S1, SWI5-SFR1.
In addition to the previously mentioned approach (Figure 5A), we devised another assay to examine the enhancement of ADP release by SWI5-SFR1. In this assay (Supplementary Figure S4A), ssDNA and radiolabeled [α-32P] ATP are pre-incubated with RAD51 to form the RAD51-ATP-ssDNA filament. Following a 10-min incubation to convert >90% of ATP to ADP, as monitored by thin layer chromatography (Supplementary Figure S4B), a varying amount of SWI5-SFR1 is added to the reaction. After a 5-min incubation, the reaction mixture is filtered through a nitrocellulose membrane trap to remove free [α-32P] ADP. Filters are air dried, and the amount of [α-32P] ADP is determined by phosphorimaging analysis. This approach thus provides a RAD51-ADP-ssDNA substrate via the conversion of ATP to ADP by RAD51. Importantly, the results from this assay provided independent verification for the enhancement of ADP release from the RAD51 presynaptic filament by SWI5-SFR1 (Supplementary Figure S4C).
SWI5-SFR1 has no significant effect on ATP-binding affinity of RAD51 filament
We wished to determine whether SWI5-SFR1 alters the affinity of the RAD51 presynaptic filament for ATP. To do so, we sought to eliminate ATP hydrolysis as a complicating factor and then assess ATP binding by the presynaptic filament with or without SWI5-SFR1 present. As the DNA-stimulated ATPase of RAD51 is strongly attenuated by Ca2+ ions (11,12), we used the nitrocellulose filter-binding assay to monitor ATP binding in buffer supplemented with Ca2+ (Figure 6A). As shown in Supplementary Figure S5, little or no ATP hydrolysis occurred under the conditions used (Supplementary Figure S5). Importantly, we found that (i) SWI5-SFR1 has no significant effect on the affinity of the presynaptic filament for ATP (Figure 6B) and (ii) even with Ca2+ ions being present, SWI5-SFR1 still attenuates the ADP-binding ability of the presynaptic filament (Figure 6C).
Figure 6.
SWI5-SFR1 does not alter ATP-binding affinity of RAD51 presynaptic filament. (A) Schematic of the filter-binding assay to monitor the influence of SWI5-SFR1 on the nucleotide-binding affinity of RAD51 in the presence of Ca2+ ions. (B) SWI5-SFR1 has no significant effect on ATP binding by RAD51. (C) Even with Ca2+ present, attenuation of the ADP-binding affinity of RAD51 presynaptic filament by SWI5-SFR1 still occurred. (B and C) Error bars represent the standard deviation (±SD) calculated based on at least three independent experiments. Symbol: S5S1, SWI5-SFR1.
DISCUSSION
The function of SWI5-SFR1 in RAD51-mediated HR
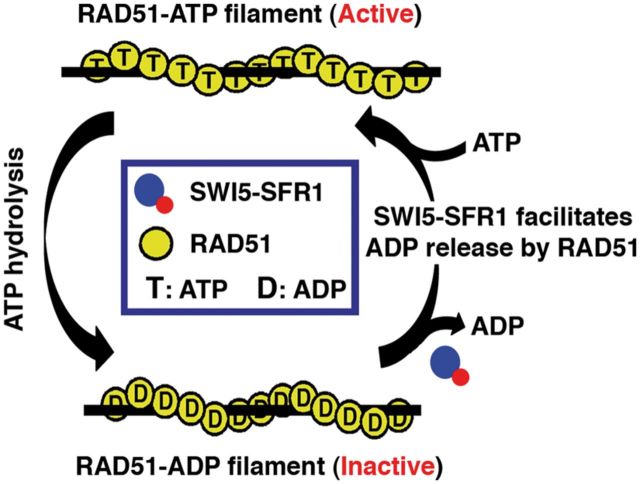
Our results help elucidate the role of SWI5-SFR1 in RAD51-mediated homologous DNA pairing. Specifically, we provide evidence for an ability of SWI5-SFR1 to facilitate ADP release from RAD51. We note that the stimulatory effect of SWI5-SFR1 on the RAD51 ATPase activity and ADP release from RAD51 is seen only when either ssDNA or dsDNA is present (Figures 2D and 5B; Supplementary Figure S6A and B), thus providing evidence that SWI5-SFR1 acts on the RAD51-DNA filament specifically. In contrast to the enhancement of ADP release, SWI5-SFR1 does not seem to affect the affinity of the RAD51 filament for ATP (Figure 6B and Supplementary Figure S6C). Based on these results, we suggest that the enhancement of ADP release from RAD51 presynaptic filament by SWI5-SFR1 helps maintain the catalytically active, ATP-bound form of the presynaptic filament during the HR reaction (Figure 7).
Figure 7.
Model depicting the mechanistic action of SWI5-SFR1 on RAD51 filament. Our results show that SWI5-SFR1 helps maintain the catalytically active ATP-bound state of the RAD51 presynaptic filament via enhancement of ADP release from the filament.
Implications for the yeast Swi5-Sfr1 orthologs
There are clear parallels between the mouse SWI5-SFR1 complex and its S. pombe ortholog. Specifically, both protein complexes stabilize the RAD51 presynaptic filament and stimulate RAD51-mediated homologous DNA pairing [(22,27,29); and this work], and like its mouse ortholog, the S. pombe Swi5-Sfr1 complex also elevates the ssDNA-stimulated ATPase activity of Rad51 [(27); and this work]. These results thus reveal an evolutionarily conserved function in the SWI5-SFR1 complex. Importantly, our study shows that SWI5-SFR1 helps maintain the RAD51 presynaptic filament in an active ATP-bound state by facilitating ADP release. It remains to be determined whether S. pombe Swi5-Sfr1 also acts in the same fashion as we have demonstrated for the mouse complex, i.e. in facilitating the release of ADP from the SpRad51 presynaptic filament. We note that recent biophysical, SAXS and X-ray crystallographic studies have shown that S. pombe Swi5-Sfr1 complex adopts an elongated dogleg-shaped structure. It has been suggested that the elongated structure allows Swi5-Sfr1 to fit into the helical groove of the presynaptic filament where it may act by locking the Rad51 protomers onto ssDNA (27,29–32). Future studies will determine whether mouse SWI5-SFR1 adopts the same elongated structure as its S. pombe counterpart and the validity of the mechanism that has been proposed.
Saccharomyces cerevisiae Mei5 and Sae3 proteins are the respective orthologs of Sfr1 and Swi5, and these proteins also form a stable complex (33,34). Even though Mei5-Sae3 appears to physically interact with Rad51 through Mei5, no enhancement of the Rad51 recombinase activity by this protein complex has been seen (34). Interestingly, the expression of the Mei5-Sae3 complex is restricted solely to meiosis (35,36), and published studies have provided evidence that Mei5-Sae3 functions specifically with the meiotic specific recombinase Dmc1 [(33,35–37); see next section].
Functional relationship of SWI5-SFR1, RAD51 and DMC1 in meiotic recombination
Genetic studies in S. pombe and S. cerevisiae have provided compelling evidence for a role of the Swi5-Sfr1 and Mei5-Sae3 complexes in meiotic recombination. Genetic ablation of these protein complexes leads to a sporulation defect and poor spore viability that stem from defective meiotic recombination (35,36,38,39). Importantly, S. pombe Swi5-Sfr1 physically interacts with Dmc1 and stimulates Dmc1-mediated DNA strand exchange, likely by facilitating the loading of Dmc1 onto ssDNA (27). We note that S. cerevisiae Mei5-Sae3 physically interacts with Dmc1 and functions as a mediator of this recombinase, specifically to overcome the inhibitory effect of RPA (Replication Protein A), the evolutionarily conserved single-strand DNA-binding protein, on the assembly of the Dmc1 presynaptic filament (33). However, mouse SWI5-SFR1 does not stimulate homologous DNA pairing mediated by DMC1, nor does it stabilize the DMC1 presynaptic filament or enhance ATP hydrolysis by the presynaptic filament (Supplementary Figure S7).
It has been known for some time that Rad51 regulates an attribute(s) of Dmc1 that is important for meiotic recombination (2,40–42). By the use of a separation-of-function mutant variant of Rad51 that can assemble into a helical filament on ssDNA but lacks homologous pairing activity, Cloud et al. (37) have recently provided evidence that (i) the recombinase activity of Rad51 is dispensable for meiotic recombination; (ii) Dmc1’s enzymatic activity is sufficient for interhomolog recombination; and most importantly, (iii) in addition to Mei5-Sae3, Rad51, but not its recombinase activity, is required for Dmc1 foci formation during meiosis (43). Cloud et al. (37) have also demonstrated that Mei5-Sae3 coordinates with Rad51 to stimulate Dmc1-mediated homologous DNA pairing. Given the previously mentioned findings on S. cerevisiae Mei5-Sae3, it will be important to examine whether and how the mammalian SWI5-SFR1 complex plays a role in meiotic HR by cooperating with RAD51 to facilitate DMC1-mediated homologous DNA pairing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [9, 22, 25 and 26].
FUNDING
Academia Sinica and Taiwan National Science Council [NSC 101-2311-B-002-019 and NSC 102-2628-B-002-044-MY3 to P.C.], and [NSC 100-2113-M-002-009-MY2 to H.W. L.]; and National Taiwan University [102R7848 and 102R7560-6 to P.C.]. Funding for open access charge: [NSC 102-2628-B-002-044-MY3].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 2.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 3.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 7.Cox MM. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- 8.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 9.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 10.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 11.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair (Amst) 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl Acad. Sci. USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–3302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson RB, Moses DN, Kwon Y, Chan P, Chi P, Klein H, Sung P, Greene EC. Structural transitions within human Rad51 nucleoprotein filaments. Proc. Natl Acad. Sci. USA. 2009;106:12688–12693. doi: 10.1073/pnas.0811465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair (Amst) 2010;9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl Acad. Sci. USA. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akamatsu Y, Tsutsui Y, Morishita T, Siddique MS, Kurokawa Y, Ikeguchi M, Yamao F, Arcangioli B, Iwasaki H. Fission yeast Swi5/Sfr1 and Rhp55/Rhp57 differentially regulate Rhp51-dependent recombination outcomes. EMBO J. 2007;26:1352–1362. doi: 10.1038/sj.emboj.7601582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haruta N, Akamatsu Y, Tsutsui Y, Kurokawa Y, Murayama Y, Arcangioli B, Iwasaki H. Fission yeast Swi5 protein, a novel DNA recombination mediator. DNA Repair (Amst) 2008;7:1–9. doi: 10.1016/j.dnarep.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Akamatsu Y, Jasin M. Role for the mammalian Swi5-Sfr1 complex in DNA strand break repair through homologous recombination. PLoS Genet. 2010;6:e1001160. doi: 10.1371/journal.pgen.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan J, Chen J. The role of the human SWI5-MEI5 complex in homologous recombination repair. J. Biol. Chem. 2011;286:9888–9893. doi: 10.1074/jbc.M110.207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai SP, Su GC, Lin SW, Chung CI, Xue X, Dunlop MH, Akamatsu Y, Jasin M, Sung P, Chi P. Rad51 presynaptic filament stabilization function of the mouse Swi5-Sfr1 heterodimeric complex. Nucleic Acids Res. 2012;40:6558–6569. doi: 10.1093/nar/gks305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuman KC, Chadd EH, Liou GF, Bergman K, Block SM. Characterization of photodamage to Escherichia coli in optical traps. Biophys. J. 1999;77:2856–2863. doi: 10.1016/S0006-3495(99)77117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 25.Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell. 2007;28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 26.Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell. 2007;28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat. Struct. Mol. Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- 28.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl Acad. Sci. USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurokawa Y, Murayama Y, Haruta-Takahashi N, Urabe I, Iwasaki H. Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biol. 2008;6:e88. doi: 10.1371/journal.pbio.0060088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kokabu Y, Murayama Y, Kuwabara N, Oroguchi T, Hashimoto H, Tsutsui Y, Nozaki N, Akashi S, Unzai S, Shimizu T, et al. The fission yeast Swi5-Sfr1 complex, an activator of Rad51 recombinase, forms an extremely elongated Dogleg-shaped structure. J. Biol. Chem. 2011;286:43569–43576. doi: 10.1074/jbc.M111.303339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuwabara N, Murayama Y, Hashimoto H, Kokabu Y, Ikeguchi M, Sato M, Mayanagi K, Tsutsui Y, Iwasaki H, Shimizu T. Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex. Structure. 2012;20:440–449. doi: 10.1016/j.str.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Saikusa K, Kuwabara N, Kokabu Y, Inoue Y, Sato M, Iwasaki H, Shimizu T, Ikeguchi M, Akashi S. Characterisation of an intrinsically disordered protein complex of Swi5-Sfr1 by ion mobility mass spectrometry and small-angle X-ray scattering. Analyst. 2013;138:1441–1449. doi: 10.1039/c2an35878f. [DOI] [PubMed] [Google Scholar]
- 33.Ferrari SR, Grubb J, Bishop DK. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J. Biol. Chem. 2009;284:11766–11770. doi: 10.1074/jbc.C900023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Say AF, Ledford LL, Sharma D, Singh AK, Leung WK, Sehorn HA, Tsubouchi H, Sung P, Sehorn MG. The budding yeast Mei5-Sae3 complex interacts with Rad51 and preferentially binds a DNA fork structure. DNA Repair (Amst) 2011;10:586–594. doi: 10.1016/j.dnarep.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayase A, Takagi M, Miyazaki T, Oshiumi H, Shinohara M, Shinohara A. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940. doi: 10.1016/j.cell.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 36.Tsubouchi H, Roeder GS. The budding yeast mei5 and sae3 proteins act together with dmc1 during meiotic recombination. Genetics. 2004;168:1219–1230. doi: 10.1534/genetics.103.025700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–1225. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellermeier C, Schmidt H, Smith GR. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics. 2004;168:1891–1898. doi: 10.1534/genetics.104.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okada T, Keeney S. Homologous recombination: needing to have my say. Curr. Biol. 2005;15:R200–R202. doi: 10.1016/j.cub.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Sheridan S, Bishop DK. Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 2006;20:1685–1691. doi: 10.1101/gad.1447606. [DOI] [PubMed] [Google Scholar]
- 42.Shinohara A, Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet. Genome Res. 2004;107:201–207. doi: 10.1159/000080598. [DOI] [PubMed] [Google Scholar]
- 43.Bishop DK. Rad51, the lead in mitotic recombinational DNA repair, plays a supporting role in budding yeast meiosis. Cell Cycle. 2012;11:4105–4106. doi: 10.4161/cc.22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.