Abstract
The c-myb proto-oncogene product (c-Myb) regulates both the proliferation and apoptosis of hematopoietic cells by inducing the transcription of a group of target genes. However, the biologically relevant molecular mechanisms that regulate c-Myb activity remain unclear. Here we report that c-Myb protein is phosphorylated and degraded by Wnt-1 signal via the pathway involving TAK1 (TGF-β-activated kinase), HIPK2 (homeodomain-interacting protein kinase 2), and NLK (Nemo-like kinase). Wnt-1 signal causes the nuclear entry of TAK1, which then activates HIPK2 and the mitogen-activated protein (MAP) kinase-like kinase NLK. NLK binds directly to c-Myb together with HIPK2, which results in the phosphorylation of c-Myb at multiple sites, followed by its ubiquitination and proteasome-dependent degradation. Furthermore, overexpression of NLK in M1 cells abrogates the ability of c-Myb to maintain the undifferentiated state of these cells. The down-regulation of Myb by Wnt-1 signal may play an important role in a variety of developmental steps.
Keywords: Myb, HIPK2, NLK, TAK1, phosphorylation, degradation
The c-myb proto-oncogene is the cellular progenitor of the v-myb oncogenes carried by the chicken retroviruses AMV (avian myeloblastosis virus) and E26 (Klempnauer et al. 1982). c-myb is highly expressed in immature hematopoietic cells, but this expression is turned off during terminal differentiation (Gonda and Metcalf 1984). Analysis of c-myb-deficient mice showed that c-myb is essential for the proliferation of immature hematopoietic cells and early T-cell development (Mucenski et al. 1991; Allen et al. 1999). The myb gene is well conserved in many species. For example, Drosophila melanogaster has one myb gene (dmyb) that is required in diverse cellular lineages throughout the course of development (Katzen et al. 1998; Okada et al. 2002). c-Myb is a transcriptional activator that recognizes a specific DNA sequence (Biedenkapp et al. 1988; Ness et al. 1989; Sakura et al. 1989; Weston and Bishop 1989). Several c-Myb target genes, including c-myc, are involved in cell cycle control, lineage commitment in differentiation, and blockage of apoptosis (Ness et al. 1989; Nakagoshi et al. 1992; Frampton et al. 1996; Taylor et al. 1996; Kowenz-Leutz et al. 1997; Schmidt et al. 2000). c-Myb has three functional domains that are responsible for DNA binding, transcriptional activation, and negative regulation, respectively (Sakura et al. 1989). The DNA-binding domain (DBD) in the N-terminal region of c-Myb consists of three imperfect tandem repeats of 51–52 amino acids, each containing a helix–turn–helix variation motif (Ogata et al. 1994). The transcriptional coactivator CBP binds to the transcriptional activation domain to mediate trans-activation (Dai et al. 1996). Deletion of the negative regulatory domain (NRD), which is located in the C-terminal portion of the molecule, increases both the trans-activation and transformation capacity of the molecule, which suggests that the NRD normally represses c-Myb activity (Sakura et al. 1989; Hu et al. 1991; Dubendorff et al. 1992; Kanei-Ishii et al. 1992).
The function of some nuclear oncogene products can be regulated by certain signaling pathways, including those using multiple kinases. For instance, Jun activity is enhanced by its phosphorylation by JNK/p38, whereas NF-κB is recruited into the nucleus when IκB is phosphorylated by IκB kinase and then degraded (for review, see Ghosh and Karin 2002; Weston and Davis 2002). In contrast, the specific signaling pathways that regulate c-Myb activity are at present unclear. It is possible that c-Myb activity may also be regulated by phosphorylation as casein kinase II phosphorylates c-Myb at a site upstream of the DBD and thereby inhibits its DNA binding (Lüscher et al. 1990). However, although the removal of this phosphorylation site was initially speculated to correlate with the oncogenic activation of c-Myb (Lüscher et al. 1990), it was shown later that it is actually the removal of the neighboring first DBD repeat, which stabilizes the Myb–DNA complex (Tanikawa et al. 1993), that is required for the oncogenic activation of c-Myb (Dini and Lipsick 1993). Also suggesting that c-Myb activity may be regulated by phosphorylation is that the 42-kD mitogen-activated protein kinase (p42MAPK) phosphorylates c-Myb in vitro at the Ser 528 residue in its NRD, and that the Ala mutant of this site has higher trans-activation activity than does wild-type c-Myb (Aziz et al. 1995). However, the biological significance of this phosphorylation event is at present unclear. Pim-1 kinase, the expression of which is induced by a number of cytokines, also enhances c-Myb activity in combination with the putative coactivator p100 (Leverson et al. 1998). However, it is unknown whether the specific cytokines that can induce Pim-1 kinase also enhance c-Myb. It is also not clear how Pim-1 kinase regulates the c-Myb activity.
The Wnt proteins belong to a large family of secreted signaling molecules that direct growth and cell fates in processes as diverse as embryonic segmentation, limb development, and CNS patterning (Wodarz and Nusse 1998; Peifer and Polakis 2000). When the Wnt molecules are secreted and bind to Frizzled (Fz) receptors and the LRP coreceptors (Wodarz and Nusse 1998; Peifer and Polakis 2000; Pandur and Kuhl 2001), the disheveled protein becomes phosphorylated and can then associate with axin and APC (adenomatous polyposis coli) to block glycogen synthetase kinase 3β (GSK3β) activity (Zeng et al. 1997; Behrens et al. 1998). In the absence of Wnt stimulation, GSK3β phosphorylates β-catenin, resulting in the ubiquitination of β-catenin and its destruction by the proteasome (Jiang and Struhl 1998; Winston et al. 1999). However, when the Wnt pathway is activated and GSK3β is inhibited, β-catenin accumulates and forms complexes with members of the TCF (T-cell factor)/LEF (lymphoid enhancer factor) transcription factor family (Behrens et al. 1996). These complexes then regulate the expression of various target genes (Behrens et al. 1996).
Recent evidence indicates that the Wnt/β-catenin signaling pathway is regulated by a mitogen-activated protein kinase (MAPK) pathway composed of the MAPKKK (MAP kinase kinase kinase)-type kinase transforming growth factor (TGF)-β-activated kinase (TAK1) and the MAP kinase (MAPK)-type kinase NLK (Nemo-like kinase). TAK1 is activated by various cytokines, including TGF-β family ligands and interleukin 1 (IL-1; Yamaguchi et al. 1995; Ninomiya-Tsuji et al. 1999). The TAB1 protein is a specific partner of TAK1 and promotes TAK1 autophosphorylation (Shibuya et al. 1996; Kishimoto et al. 2000). Recently, TAK1 and TAB1 homologs encoded by the mom-4 and tap-1 genes, respectively, have been identified in Caenorhabditis elegans (Meneghini et al. 1999). Genetic analysis has shown that MOM-4 and TAP-1 regulate Wnt signaling in C. elegans by activating the MAPK-like LIT-1, a C. elegans homolog of NLK (Meneghini et al. 1999; Rocheleau et al. 1999; Shin et al. 1999). This is consistent with the observation that coexpression of TAK1 and TAB1 in mammalian cells can activate NLK, and that the TAK1–NLK MAPK pathway regulates Wnt signaling by phosphorylating TCF in mammalian cells (Ishitani et al. 1999). However, the components of the signaling pathway between TAK1 and NLK have not yet been defined. In addition, it is not clear whether the TAK1–NLK pathway targets only the TCF/LEF transcription factors.
Here, we demonstrate that the c-Myb protein is phosphorylated and degraded via the pathway involving TAK1, HIPK2, and NLK upon Wnt-1 stimulation.
Results
c-Myb binds to two kinases, HIPK2 and NLK
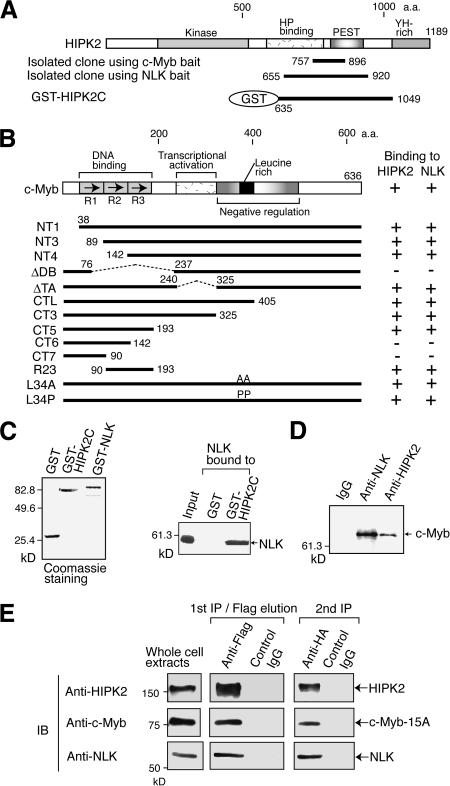
To identify the regulator that binds to c-Myb, we performed yeast two-hybrid screening of a mouse embryo library using the c-Myb mutant protein lacking its transcriptional activation domain (ΔTA) as the bait. This yielded 109 clones. Sequence analysis indicated that 30 of these encoded a fragment (amino acids 757–896) of homeodomain-interacting protein kinase 2 (HIPK2; Fig. 1A). HIPK2 was originally identified as Nkx-1.2 homeoprotein-interacting protein and contains a kinase domain (Kim et al. 1998). We also performed yeast two-hybrid screening to identify the NLK-binding protein by using the NLK protein lacking the N-terminal 123 amino acids. The three clones that were isolated also encoded a fragment (amino acids 655–920) of HIPK2 (Fig. 1A). These observations suggest that HIPK2 binds to c-Myb together with NLK. We then examined the in vitro binding of c-Myb to these two kinases. GST pull-down assays with GST–HIPK2 fusion protein (GST–HIPK2C) indicated that in vitro-translated c-Myb binds efficiently to HIPK2 (Supplementary Fig. 1; see also the summary of these results in Fig. 1B). GST pull-down assays with various c-Myb mutants also indicated that repeats 2 and 3 in the DBD of c-Myb were responsible for the binding to HIPK2. GST pull-down assays with GST–NLK also revealed that NLK binds to repeats 2 and 3 in the DBD of c-Myb (Supplementary Fig. 2; see also the summary of these results in Fig. 1B). In addition, we showed that in vitro-translated NLK also binds efficiently to the GST–HIPK2 fusion protein (Fig. 1C). That c-Myb, HIPK2, and NLK all interact directly with each other suggests that these two kinases bind together to c-Myb. To confirm that c-Myb and these two kinases associate in vivo, coimmunoprecipitation of endogenous proteins in Molt-4 cell lysates was performed (Fig. 1D). Anti-NLK and anti-HIPK2 antibodies coprecipitated c-Myb, whereas normal IgG did not. To further examine whether c-Myb, HIPK2, and NLK form a complex two-step coimmunoprecipitation was performed (Fig. 1E). In this experiment, the c-Myb 15A mutant, in which the Ser and Thr residues linked to Pro at 15 sites were mutated to Ala, was used. This mutant c-Myb is resistant to the HIPK2- and NLK-induced degradation. 293 cells were transfected with the expression plasmids for the c-Myb 15A mutant, Flag-HIPK2, and HA-NLK, and the prepared cell lyastes were first immunoprecipitated with anti-Flag antibody. This immunocomplex was eluted by anti-Flag peptide and confirmed to contain c-Myb and HA-NLK in addition to Flag-HIPK2. The Flag-peptide eluates were then immunoprecipitated with anti-HA or control IgG. The anti-HA immunocomplex contained c-Myb, but the IgG complex did not. Thus, c-Myb, HIPK2, and NLK can form a complex.
Figure 1.
HIPK2 and NLK bind to c-Myb. (A,B) HIPK2 and NLK bind to the DBD of c-Myb. (A) The domain structure of HIPK2 and the clones isolated by yeast two-hybrid screening using c-MybΔTA or NLK as bait. (B) The various c-Myb mutants used in the GST pull-down assays and the summary of the results of these assays (shown in Supplementary Figs. 1, 2). (C) NLK binds to HIPK2. (Left) The GST–HIPK2C and GST–NLK fusion proteins used in the binding assays were analyzed by SDS-PAGE followed by Coomassie blue staining. (Right) The binding of in vitro-translated NLK to GST–HIPK2C. In the input lanes, the amount of NLK was 10% of that used for the binding assay. (D) Endogenous c-Myb is coimmunoprecipitated with antibodies to NLK or HIPK2. Lysates from Molt-4 cells were precipitated with the antibody shown above, and the immunocomplexes were analyzed by Western blotting with the anti-c-Myb antibody. (E) Two-step coimmunoprecipitation of the complex containing HIPK2, c-Myb, and NLK. Lysates from 293 cells, which were transfected with plasmids to express Flag-HIPK2, c-Myb-15A, and HA-NLK, were first precipitated with anti-Flag antibody, eluted with Flag peptide, and eluates were analyzed by Western blotting with the antibodies indicated on the left. The eluates were then immunoprecipitated with anti-HA antibody or control IgG, followed by Western blotting.
HIPK2 functions between TAK1 and NLK
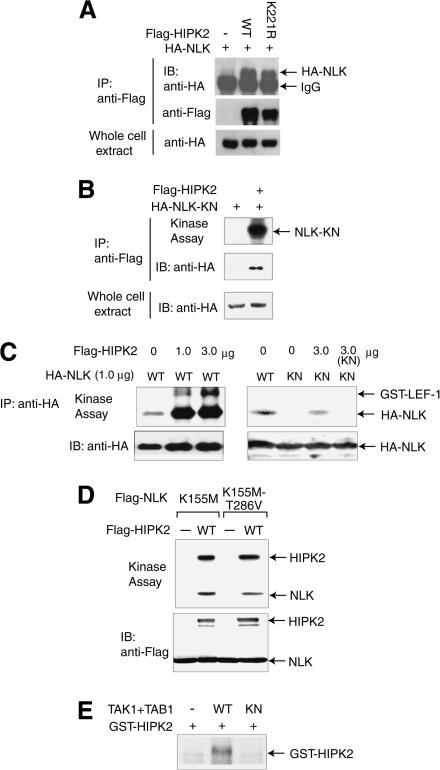
We previously demonstrated that the MAPKKK TAK1 activates the MAPK-like kinase NLK in a signaling pathway (Ishitani et al. 1999). However, TAK1 does not directly interact with NLK (Ishitani et al. 1999), suggesting that an unidentified molecule functions between TAK1 and NLK in the pathway. That HIPK2 interacts in vitro with NLK (Fig. 1C) suggests that HIPK2 may act upstream to activate NLK and thus may be the missing molecule in the TAK1–NLK signaling pathway. To explore this possibility, we investigated whether HIPK2 physically interacts with NLK within cultured cells (Fig. 2A). Flag-tagged HIPK2 was coexpressed together with HA-tagged NLK in 293 cells, and cell lysates were immunoprecipitated with anti-Flag antibody. Analysis of the immunoprecipitates by Western blotting with anti-HA antibody revealed that HIPK2 associates with NLK (Fig. 2A, top panel, middle lane). A kinase-negative form of HIPK2, HIPK2-K221R, also interacts with NLK (Fig. 2A, top panel, rightmost lane), indicating that HIPK2 binds to NLK in a manner that is independent of the kinase activity of HIPK2.
Figure 2.
HIPK2 functions between TAK1 and NLK in the signaling pathway. (A) NLK is coimmunoprecipitated with HIPK2. 293 cells were transfected with expression vectors encoding Flag-tagged wild-type (WT) or kinase-negative mutant (K221R) HIPK2 together with HA-NLK. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody, and the immunocomplexes were analyzed by Western blotting with anti-HA antibody (top) or anti-Flag antibody (middle). (Bottom) The amount of HA-NLK in the whole cell extracts is also shown. (B) HIPK2 phosphorylates NLK. 293 cells were transfected with expression vectors for Flag-HIPK2 together with HA-tagged kinase-negative NLK (HA-NLK-KN). (Top) Cell lysates were immunoprecipitated with anti-Flag antibody, and the immunocomplexes were subjected to the in vitro kinase assay. The amount of HA-NLK-KN in the anti-Flag immunoprecipitates (middle) and the whole-cell extracts (bottom) are shown. (C) HIPK2 enhances NLK kinase activity. 293 cells were transfected with expression vectors for wild-type or kinase-negative HA-NLK together with various amounts of Flag-HIPK2. (Top) Cell lysates were immunoprecipitated with anti-HA antibody and subjected to the in vitro kinase assay with bacterially expressed GST–LEF-1 as an exogenous substrate. (Bottom) The amount of immunoprecipitated HA-NLK is shown. (D) HIPK2 does not phosphorylate the site in the T-loop of NLK. 293 cells were transfected with expression vectors for kinase-negative (K155M) HA-NLK containing the normal or mutated (T286V) T-loop together with Flag-HIPK2. (Top) Cell lysates were immunoprecipitated with anti-HA antibody and subjected to the in vitro kinase assay. (Bottom) The amounts of immunoprecipitated HA-NLK and Flag-HIPK2 are shown. (E) TAK1 phosphorylates HIPK2. 293 cells were transfected with the expression vectors for TAB1 and HA-tagged wild-type (WT) or kinase-negative mutant (KN) TAK1 and immunoprecipitated with anti-HA antibody. The immunoprecipitates were subjected to an in vitro kinase assay using bacterially expressed GST–HIPK2 as a substrate.
We next tested whether HIPK2 phosphorylates NLK (Fig. 2B). HA-tagged kinase-negative NLK (HA-NLK-K155M) and Flag-HIPK2 were coexpressed in 293 cells. Cell lysates were immunoprecipitated with anti-Flag antibody, followed by immunoblotting and an in vitro kinase assay. NLK-K155M was detected in HIPK2 immunoprecipitates (Fig. 2B, middle panel, right lane). When the immunoprecipitates were incubated with [γ-32P]ATP, NLK-K155M became phosphorylated (Fig. 2B, top panel, right lane). Thus, HIPK2 appears to interact with and phosphorylate NLK. We then determined whether NLK can be activated by HIPK2 (Fig. 2C). To this end, wild-type or kinase-negative HA-NLK was coexpressed in 293 cells together with increasing amounts of Flag-HIPK2. HA-NLK protein was immunoprecipitated from the cell lysates with anti-HA antibody, and the ability of these precipitates to autophosphorylate was measured. HIPK2 weakly phosphorylated the kinase-negative NLK, whereas the degree of phosphorylation of wild-type NLK was much higher than that of kinase-negative NLK, indicating that NLK autophosphorylation was stimulated by HIPK2. The NLK immunoprecipitates were also assayed by using bacterially expressed GST–LEF-1 as a substrate to confirm that the enhanced autophosphorylation is a reflection of enhanced NLK activity. HIPK2 enhanced the NLK-mediated phosphorylation of GST–LEF-1, whereas the NLK–HIPK2 immunocomplex containing the kinase-negative NLK did not phosphorylate GST–LEF-1. Thus, HIPK2 is a direct activator of NLK. These observations suggest that HIPK2 participates in the TAK1–NLK signaling pathway as an intermediate kinase between TAK1 and NLK. When in vitro kinase assays were performed by using the HIPK2–NLK complex containing the NLK mutants, HIPK2 similarly phosphorylated the two kinase-negative mutants (K155M) containing normal or mutated phosphorylation site in the T-loop (T286V; Fig. 2D), suggesting that HIPK2 does not directly phosphorylate the site in the T-loop. Because the T-loop of NLK is required for the autophosphorylation (Brott et al. 1998), the phosphorylation of uncharacterized site(s) by HIPK2 may induces its autophosphorylation, which leads to the phosphorylation of the site in the T-loop.
We next tested whether TAK1 phosphorylates HIPK2 (Fig. 2E). We have previously shown that coexpression of TAK1 and the TAK1 binding protein TAB1 strongly activates TAK1 (Shibuya et al. 1996). Thus, active TAK1 was isolated from 293 cells expressing wild-type TAK1 and TAB1. The active TAK1 molecules were then incubated with [γ-32P]ATP and bacterially expressed GST–HIPK2. TAK1 phosphorylates HIPK2 in vitro (Fig. 2E, middle lane), and this phosphorylation is dependent on the kinase activity of TAK1 (Fig. 2E, rightmost lane). However, we failed to detect the activation of HIPK2 kinase activity by TAK1 (data not shown). Thus, it remains to be determined whether TAK1 is a direct activator of HIPK2.
The TAK1–HIPK2–NLK pathway inhibits c-Myb activity upon Wnt-1 stimulation
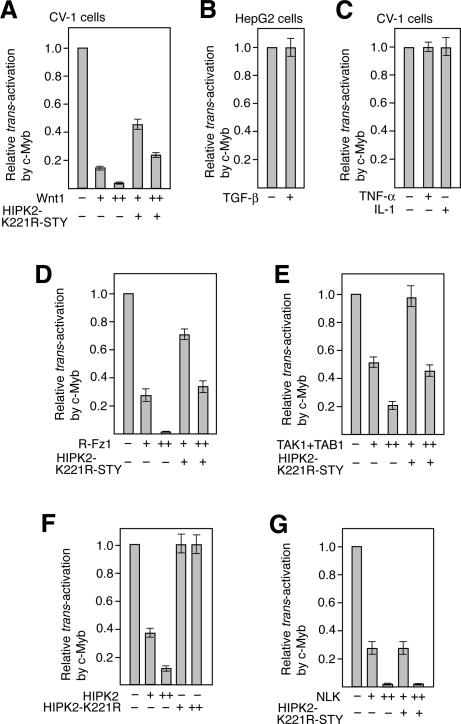
To examine whether the pathway involving TAK1, HIPK2, and NLK regulates c-Myb-induced transcriptional activation, we performed cotransfection assays using the CAT reporter containing the c-myc promoter (pc-mycCAT), which is one of the c-Myb targets (Fig. 3; Nakagoshi et al. 1992; Schmidt et al. 2000). TAK1 is activated by Wnt proteins, TGF-β, TNF-α, and IL-1 (Yamaguchi et al. 1995; Meneghini et al. 1999; Ninomiya-Tsuji et al. 1999; Rocheleau et al. 1999; Shin et al. 1999). Therefore, we first investigated the effect of these stimuli on c-Myb-dependent trans-activation in CV-1 cells. We also performed the same experiments with the TGF-β-responsive cell line HepG2. Coexpression of the pc-myc-CAT reporter with the c-Myb expression plasmid enhanced CAT expression ∼10.1- and 6.9-fold in CV-1 and HepG2 cells, respectively. When Wnt-1 was coexpressed, this c-Myb-dependent CAT expression was inhibited in a manner that was dependent on the dose of Wnt-1 (Fig. 3A). However, TGF-β, TNF-α, or IL-1 did not inhibit the c-Myb-dependent CAT expression from the c-myc promoter (Fig. 3B,C). The control experiments revealed that TGF-β treatment of HepG2 cells and TNF-α or IL-1 treatment of CV-1 cells induced the transcription from the promoters containing Smad3/4-binding and NF-κB-binding sites, respectively (data not shown), indicating that TGF-β, TNF-α, and IL-1 signalings are operative in these cells. When the dominant-negative form of HIPK2, K221R-STY, which has mutations in its kinase domain and putative activation loop, was expressed together with Wnt-1, the inhibitory effect of Wnt-1 on c-Myb-dependent CAT expression was partly suppressed (Fig. 3A). The reason why this mutant only partly suppressed the Wnt-1 effect is that this mutant protein is very unstable (data not shown). These observations suggest that the Wnt-1 signal negatively regulates c-Myb activity via HIPK2. We also wished to determine whether NLK participates in this signaling pathway by using the kinase-negative mutant of NLK as a dominant-negative form. However, this NLK mutant could not be used in this assay because it weakly inhibits c-Myb activity through its competition with the transcriptional coactivator CBP for binding to c-Myb (data not shown). However, we found that expression of rat Frizzled-1 (R-Fz1), which acts as the Wnt-1 receptor, TAK1/TAB1, HIPK2, and NLK also inhibited c-Myb-dependent CAT expression from the c-myc promoter (Fig. 3D–G). The inhibitory effect of R-Fz1, TAK1/TAB1, and HIPK2, but not NLK, was suppressed by coexpressing the kinase-negative HIPK2 mutant. These observations support the notion that HIPK2 acts downstream of Fz1 and TAK1/TAB1 but upstream of NLK in Wnt-1 signaling. Thus, c-Myb-dependent transcriptional activation is inhibited by the pathway involving TAK1, HIPK2, and NLK.
Figure 3.
The TAK1–HIPK2–NLK pathway negatively regulates the c-Myb-dependent activation of the c-myc promoter upon Wnt-1 stimulation. (A) Wnt-1 suppresses c-Myb activity via HIPK2. CV-1 cells were transfected with a mixture of the CAT reporter containing the c-myc promoter and either the c-Myb expression plasmid or the control plasmid with ([+] 0.5 μg; [++] 2.0 μg) or without (–) the Wnt-1 expression plasmid. In some cases, 8.0 μg of the plasmid that expresses the kinase-negative form of HIPK2 was added. CAT activity was measured, and the relative trans-activation capacity of c-Myb is indicated. Shown are the averages of three experiments. The degree of activation by c-Myb in CV-1 cells in the absence of Wnt-1 and/or HIPK2 overexpression was 10.1 ± 1.2-fold. (B,C) TGF-β, TNF-α, and IL-1 do not affect c-Myb-induced activation of the c-myc promoter. HepG2 (B) or CV-1 (C) cells were transfected with the c-myc-CAT reporter together with the c-Myb expression plasmid or the control plasmid. The transfected cells were treated with TGF-β, TNF-α, or IL-1, and CAT activity was measured. The degree of activation by c-Myb in HepG2 cells in the absence of cytokines was 6.9 ± 1.1-fold. (D–G) Wnt-1 signaling suppresses c-Myb activity via the TAK1–HIPK2–NLK pathway. Experiments similar to those described in A were performed except that plasmids expressing R-Fz1 (D), TAK1 and TAB1 (E), HIPK2 (F), or NLK (G) were used instead of the Wnt-1 expression plasmid; + and ++ indicate that 0.5 and 2.0 μg, respectively, of the R-Fz1, HIPK2, and NLK expression plasmids were used. Regarding the TAK1 and TAB1 plasmids, either 0.2 μg (+) or 0.5 μg (++) was used.
c-Myb protein degradation is induced by the pathway involving TAK1, HIPK2, and NLK
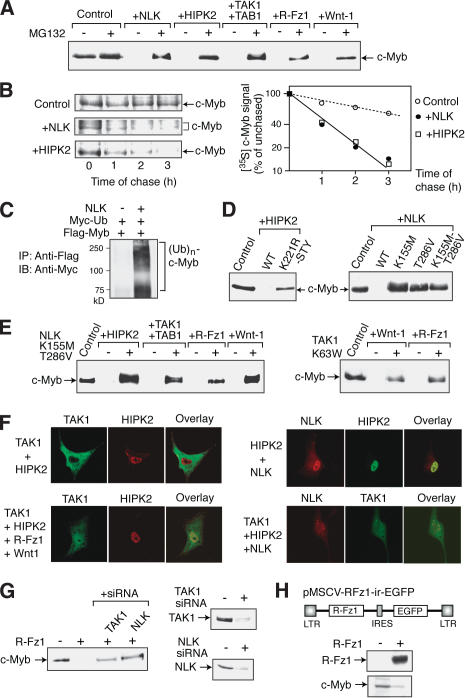
While investigating the mechanism by which NLK and HIPK2 inhibit c-Myb-dependent activation of the c-myc promoter, we found that coexpression of NLK or HIPK2 induces the degradation of the c-Myb protein. Transfection of CV-1 cells with the c-Myb expression plasmid together with the NLK or HIPK2 expression plasmid resulted in dramatically lower levels of c-Myb protein compared with that of control cells transfected with the c-Myb expression plasmid alone (Fig. 4A). The c-Myb protein levels recovered when the cotransfected cells were treated with the proteasome inhibitor MG132, which suggests that both HIPK2 and NLK induce c-Myb degradation. Similar observations were also made when c-Myb was coexpressed with TAK1/TAB1, R-Fz1, or Wnt-1 (Fig. 4A). The half-life of c-Myb in the presence or absence of NLK or HIPK2 was then assessed in transfected CV-1 cells by pulse-chase labeling (Fig. 4B). When NLK was coexpressed, several bands of c-Myb were observed. Overexpressed NLK may rapidly phosphorylate c-Myb, which may not be able to be so rapidly ubiquitinated and degraded. The presence of NLK or HIPK2 decreased the 3-h c-Myb half-life by ∼3.6-fold to 50 min.
Figure 4.
Induction of the degradation of c-Myb via the TAK1–HIPK2–NLK pathway upon Wnt-1 stimulation. (A) Coexpression of Wnt–NLK pathway components induces c-Myb degradation. CV-1 cells were transfected with the c-Myb expression plasmid and the internal control plasmid pact-β-gal together with a plasmid expressing the protein indicated above each lane. The transfected cells were treated with MG132 or control solvent, and total cell lysates were prepared and analyzed by Western blotting using anti-c-Myb antibody. (B) NLK and HIPK2 overexpression decrease the half-life of c-Myb. CV-1 cells were transfected with the Flag-c-Myb expression plasmid with or without the NLK or HIPK2 expression plasmid and pulse-chase labeled with [35S]methionine. At various times after the chase, c-Myb protein levels were examined by immunoprecipitation and plotted. (C) NLK enhances c-Myb ubiquitination. CV-1 cells were transfected with the Flag-c-Myb expression vector, pact-β-gal, and the Myc tag-linked ubiquitin expression plasmid with or without the plasmid that expresses NLK. Transfected cells were treated with MG132 before lysate preparation. Whole-cell lysates were immunoprecipitated with anti-Flag antibody, and the immunocomplexes were analyzed by Western blotting with anti-Myc antibody. (D) NLK and HIPK2 kinase activity is required for c-Myb degradation. CV-1 cells were transfected with the c-Myb expression plasmid and pact-β-gal together with the plasmid expressing the protein indicated above each lane. Whole-cell lysates were prepared and analyzed by Western blotting using anti-c-Myb antibody. (E) The kinase-negative mutant of NLK or TAK1 blocks Wnt-1-induced c-Myb degradation. CV-1 cells were transfected with the c-Myb expression plasmid and pact-β-gal together with the plasmid expressing the protein indicated above each lane and either the kinase-negative NLK (left) or TAK1 (right) expression plasmid (+) or the control empty vector (–). c-Myb protein levels were then analyzed as described above. (F) Nuclear entry of TAK1 upon Wnt-1 stimulation. CV-1 cells were transfected with the plasmids expressing the proteins shown on the left. The cells were then immunostained with the antibodies against the protein shown above, visualized by FITC- and rhodamine-conjugated secondary antibodies, and analyzed by confocal microscopy. In the panel indicated as overlay, the signals for both proteins are superimposed. (G, right) siRNAs directed against either TAK1 or NLK inhibit the R-Fz1-induced degradation of c-Myb protein. CV-1 cells were transfected with TAK1 or NLK siRNA duplex, and Western blotting was performed. (Left) CV-1 cells were transfected with the plasmid to express c-Myb and R-Fz1 together with TAK1 or NLK siRNA, and the c-Myb protein levels were assayed. (H) Down-regulation of c-Myb proteins by R-Fz1 in M1 cells. The retrovirus vector that express R-Fz1 together with EGFP is shown on the top. (Upper panel) CV-1 cells were transfected with the R-Fz1 virus DNA, and the expression of R-Fz1 was confirmed by Western blotting. (Lower panel) M1 cell clones infected with either of R-Fz1 virus or control empty virus were isolated and the expression of c-Myb proteins in typical clones was analyzed by Western blotting.
NLK could induce proteasome-dependent c-Myb degradation by enhancing the ubiquitination of c-Myb. To test this, CV-1 cells were transfected with a Flag-tag-linked c-Myb expression plasmid together with the Myc-tag-linked ubiquitin expression vector, and cell lysates were prepared after MG132 treatment (Fig. 4C). Flag-c-Myb was then immunoprecipitated by anti-Flag antibody and analyzed by Western blotting using anti-Myc antibody. Coexpression of c-Myb with NLK significantly enhanced c-Myb ubiquitination, which supports the notion that NLK induces proteasome-dependent c-Myb degradation.
To confirm that the kinase activities of HIPK2 and NLK are required for c-Myb degradation, we investigated the effect on c-Myb protein levels of various mutants of these two kinases (Fig. 4D). The HIPK2 mutant (K221R-STY) that has mutations in its kinase domain and putative activation loop failed to induce c-Myb degradation. In addition, the NLK mutants that have the mutation in either the kinase domain (K155M) or the activation loop (T286V), or both sites (K155M-T286V), also failed to induce c-Myb degradation.
To confirm that the Wnt-1–TAK1–HIPK2 signaling pathway induces c-Myb degradation via NLK, we examined the effect of the dominant-negative form of NLK (K155M-T286V; Fig. 4E, left). The decrease in the c-Myb protein levels due to HIPK2, TAK1/TAB1, R-Fz1, or Wnt-1 expression was abolished when the dominant-negative form of NLK was coexpressed. However, neither TGF-β nor IL-1 induced c-Myb degradation, although TGF-β and IL-1 are known to activate TAK1 (data not shown). Furthermore, the kinase-negative mutant of TAK1 also blocked the c-Myb degradation by R-Fz-1 or Wnt-1 (Fig. 4E, right), indicating that TAK1 is involved in the Wnt-1-induced c-Myb degradation. Thus, Wnt-1 induces c-Myb degradation via the pathway involving TAK1, HIPK2, and NLK.
When the subcellular localization of components of Wnt-1–NLK signaling was examined, TAK1 was in the cytosol in the absence of Wnt-1, R-Fz-1, or TAB1 (Fig. 4F, upper left). However, TAK1 was colocalized with HIPK2 in the nuclear dot-like structure in the presence of Wnt-1 signals (Fig. 4F, lower left), suggesting that Wnt-1 signal induces the nuclear entry of TAK1. Although NLK was almost uniformly distributed in the nucleoplasm when it was expressed alone (data not shown), NLK was observed in the nuclear dot-like structure with HIPK2 or TAK1 when they were coexpressed together (Fig. 4F, right). Thus, Wnt-1 induces the nuclear entry of TAK1, which then forms a complex with HIPK2 and NLK.
To further examine whether TAK1 and NLK is required for the Wnt-1-induced degradation of c-Myb protein, we used small inhibitory RNA (siRNA) for both kinases. Transfection of CV-1 cells with the siRNAs dramatically decreased the levels of endogenous TAK1 and NLK proteins (Fig. 2G, right). Cotransfection of CV-1 cells with these siRNAs apparently inhibited the R-Fz1-induced degradation of c-Myb proteins (Fig. 2G, left). We could not find the effective siRNA sequences for the monkey HIPK2 mRNA.
To further examine whether endogenous c-Myb proteins in hematopoietic cells are down-regulated by Wnt-1 signal, the myeloblastic cell line M1 was used. We generated M1 cell clones that constitutively express R-Fz1 using the retrovirus vector, and measured the levels of endogenous c-Myb proteins (Fig. 2H). The levels of c-Myb proteins in the R-Fz1-expressing M1 cells were significantly lower than that of control cells.
NLK directly phosphorylates c-Myb at multiple sites and induces its degradation
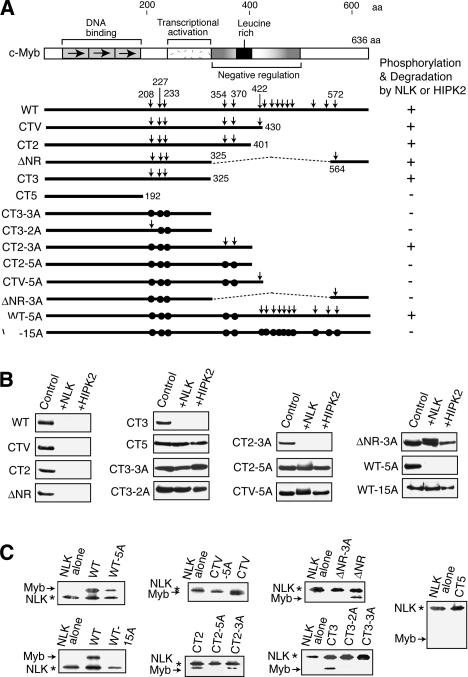
To further analyze the mechanism by which NLK provokes c-Myb degradation, we examined whether NLK directly phosphorylates c-Myb. To this end, we used various c-Myb mutants (Fig. 5A). Thus, CV-1 cells were transfected with a plasmid that expresses a particular form of c-Myb together with the NLK or HIPK2 expression plasmid, and c-Myb protein levels were analyzed by Western blotting (Fig. 5B). At the same time, to see how the phosphorylation of c-Myb correlated with the cellular levels of this protein, the in vitro phosphorylation of the various forms of c-Myb by NLK was investigated (Fig. 5C). Thus, 293 cells were transfected with either the Flag-c-Myb or the Flag-NLK expression plasmid, after which NLK or c-Myb was immunoprecipitated with anti-Flag antibody and then used in an in vitro kinase reaction with [γ-32P]-ATP.
Figure 5.
NLK phosphorylates c-Myb at multiple sites and thereby induces its degradation. (A) Domain structure of c-Myb and the various c-Myb mutants used. The Ser and Thr residues linked to Pro are shown by vertical arrows. The residues that were mutated to Ala are indicated by closed circles. The results of the c-Myb degradation (B) and phosphorylation (C) assays are summarized on the right. (B) Effect of c-Myb mutations on NLK-induced c-Myb degradation. CV-1 cells were transfected with the plasmid that expresses a particular form of c-Myb and pact-β-gal together with the plasmid that expresses NLK or HIPK2. Whole-cell lysates were prepared and analyzed by Western blotting using anti-c-Myb antibody. (C) Effect of c-Myb mutations on NLK-induced in vitro phosphorylation of c-Myb. Wild-type NLK or various forms of c-Myb were expressed in transfected 293T cells and immunoprecipitated. The amounts of each protein were analyzed by Western blotting, and normalized amounts of c-Myb and NLK were mixed and incubated with [γ-32P]ATP. The reactions were analyzed by SDS-PAGE followed by autoradiography. The NLK band is due to its autophosphorylation.
Regarding the three C-truncated mutants of c-Myb (CT2, CT3, and CT5), NLK and HIPK2 degraded CT2 and CT3 but not CT5. Consistent with this, NLK phosphorylated both CT2 and CT3 but not CT5. Thus, the region between amino acids 192 and 325 contains the NLK phosphorylation sites. NLK is a member of the MAP kinase super family that is known to phosphorylate the Ser or Thr residues next to Pro. The region between 192 and 325 contains three such sites, namely, Thr 208, Ser 227, and Ser 233. As expected, the CT3-3A mutant, in which all of these three residues were mutated to Ala, was neither degraded by NLK and HIPK2 nor phosphorylated by NLK. Interestingly, however, the CT2-3A mutant, which bears a 76-amino acid region downstream of the sequences contained in CT3-3A, was still phosphorylated by NLK and degraded by HIPK2 and NLK. There are two more candidate NLK phosphorylation sites in this 76-amino acid region. The CT2-5A mutant, in which all five Ser and Thr residues linked to Pro in the region downstream from amino acid 192 in CT2 molecule were mutated to Ala, was neither phosphorylated nor degraded by NLK. Thus, the two sites in the 76-amino acid region are also sites that can be phosphorylated. Furthermore, additional sites are also phosphorylated by NLK because the full-length c-Myb mutant (WT-5A), in which the five Ser and Thr residues between amino acids 192 and 401 are mutated to Ala, was still degraded by NLK and HIPK2 and phosphorylated by NLK. However, NLK did not phosphorylate and degrade the WT-15A mutant, in which all the Ser and Thr residues linked to Pro in the region downstream from amino acid 192 were mutated. Thus, NLK directly phosphorylates c-Myb at multiple sites and induces its degradation.
NLK decreases the ability of c-Myb to maintain M1 cells in an undifferentiated state
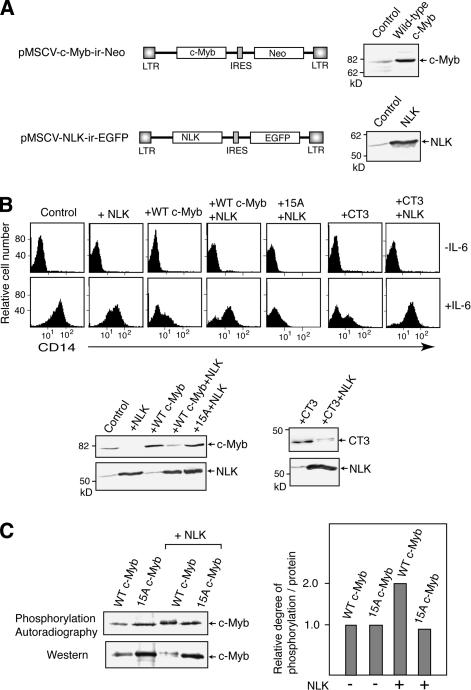
We next asked whether NLK plays a biological role as a negative regulator of c-Myb. To address this, we investigated the effect of overexpressing NLK on the capacity of c-Myb to maintain the undifferentiated state of the myeloblastic cell line M1 (Fig. 6). Upon IL-6 stimulation, M1 cells normally undergo terminal differentiation into a mature macrophage phenotype. However, constitutive expression of c-Myb in M1 cells will block this IL-6-induced differentiation (Selvakumaran et al. 1992). We generated M1 cell clones that constitutively express various forms of c-Myb together with or without NLK as well as control clones containing the empty vector (Fig. 6A). IL-6 treatment caused the control M1 cell clones to differentiate into macrophages, and the cell surface differentiation marker CD14 was expressed (Fig. 6B, upper panel, control). In contrast, the clones constitutively expressing wild-type or CT3 mutant of c-Myb did not efficiently express CD14 when IL-6 was added. However, when NLK was overexpressed in these latter cells, differentiation was induced in response to IL-6, resulting in an increased rate of CD14 positivity (Fig. 6B). However, NLK could not induce differentiation of the M1 cells expressing the 15A mutant, in which all the Ser and Thr residues linked to Pro were mutated (Fig. 6B). Thus, NLK blocks the wild-type c-Myb activity but not the activity of 15A mutant. Furthermore, the cells expressing wild-type c-Myb or CT3 started to differentiate, but this was not observed in the case of cells expressing the 15A mutant, suggesting that the 15A mutant acts in a dominant-negative manner to block differentiation. Western blotting analysis indicated that the levels of wild-type or CT3 c-Myb, but not 15A mutant, in M1 cell clones overexpressing NLK were decreased (Fig. 6B, lower panel). These results indicate that NLK induces M1 cell differentiation by promoting c-Myb protein degradation.
Figure 6.
NLK blocks the ability of c-Myb to maintain the undifferentiated state of M1 cells. (A) Retrovirus expression vectors used. The retrovirus vectors that express wild-type c-Myb together with the neo-resistance gene are shown on the top. The retrovirus vector that expresses NLK together with EGFP is indicated below. M1 cell clones infected with either of these viruses were isolated, and the expression of c-Myb or NLK proteins in typical clones was analyzed by Western blotting. (B) NLK induces the differentiation of M1 cells that overexpress c-Myb by promoting c-Myb degradation. (Top) M1 cell clones that constitutively express wild-type c-Myb, the CT3 mutant or the 15A mutant with or without NLK were treated with IL-6 or the control solvent and CD14 expression was examined by flow cytometry. (Bottom) The levels of c-Myb and NLK proteins in each cell clone were also analyzed by Western blotting. (C) In vivo phosphorylation of wild-type and 15A mutant of c-Myb. CV-1 cells were transfected with wild-type or 15A mutant of c-Myb expression vector and the internal control plasmid pact-β-gal together with the NLK or control vector. (Top left) Cells were labeled with [32P]orthophosphate, and c-Myb proteins were immunoprecipitated, analyzed by SDS-PAGE, and detected by autoradiography. The amounts of lysates used were normalized based on the transfection efficiency. (Bottom left) Cell lysates were also used for Western blotting to measure the c-Myb protein levels. (Right) Relative degree of phosphorylation per c-Myb protein is shown by a bar graph.
To confirm that the 15A mutant is not really phosphorylated by NLK in vivo, wild-type c-Myb or 15A mutant was coexpressed with or without NLK in CV-1 cells, and labeled with [32P]orthophosphate (Fig. 6C). Coexpression of NLK enhanced the degree of phosphorylation of wild-type c-Myb but not that of 15A mutant. Thus, NLK appears to block the c-Myb activity to maintain the undifferentiated state of M1 cells via directly phosphorylating the c-Myb protein.
Discussion
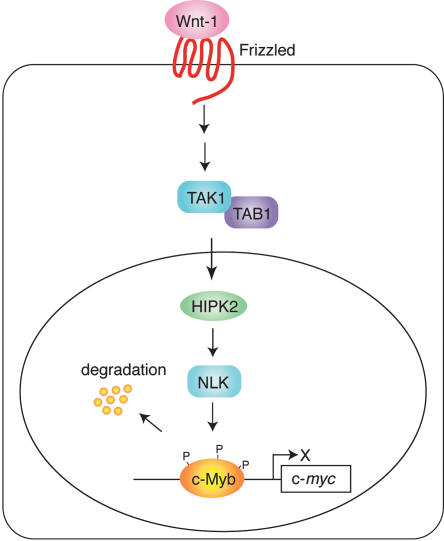
The present study has identified a signaling pathway that regulates c-Myb-dependent transcription. We showed that Wnt signaling travels via the pathway involving TAK1, HIPK2, and NLK to induce c-Myb protein degradation. This in turn inhibits c-Myb-dependent activation of c-myc promoter and may induce G1 arrest (Fig. 7). Overexpression of NLK in M1 cells abrogates the ability of c-Myb to maintain the undifferentiated state of these cells. These observations suggest that at least one way Wnt signaling controls the differentiation or apoptosis of many cell types, including hematopoietic cells by down-regulating c-Myb protein levels. Supporting this notion is the observation that Wnt signaling modulates the diversification of hematopoietic cells (Brandon et al. 2000). Recently Wnt signaling was shown to be critical for self-renewal of hematopoietic stem cells (Reya et al. 2003) or to suppress the cellular proliferation in hematopoietic tissue (Liang et al. 2003). Therefore, the Wnt-induced c-Myb degradation demonstrated in this study could play some role in the proliferation control of hematopoietic cells by Wnt signal.
Figure 7.
Degradation of c-Myb protein by Wnt-1 signal via the pathway involving TAK1, HIPK2, and NLK leads to G1 arrest.
We demonstrated that NLK directly phosphorylates c-Myb at multiple sites and that this leads to the ubiquitination and proteasome-dependent degradation of c-Myb. It is likely that this phosphorylation of c-Myb causes the recruitment of certain E3 ligase. We found that when NLK was overexpressed, c-Myb was phosphorylated at >10 sites. We also found that A-Myb and B-Myb, other vertebrate members of the Myb protein family, are also directly phosphorylated by NLK (data not shown). However, most of the NLK phosphorylation sites in the c-Myb molecule are not conserved in A-Myb and B-Myb (Mizuguchi et al. 1990), or the c-Myb of other species, including humans, chickens, and Drosophila. This suggests that the position of the NLK phosphorylations sites in the Myb molecule is not critical. It is likely that many sites in c-Myb can be phosphorylated by overexpressed NLK, and that phosphorylation at any one site may be sufficient to induce the degradation of c-Myb. However, we cannot exclude the possibility that some sites are preferentially phosphorylated by NLK in vivo, especially when only low levels of NLK are present. Notably, the NRD in the C-terminal half of c-Myb contains nine putative NLK phosphorylation sites. It was previously reported that oncogenic activation of c-Myb by C-terminal truncation is associated with the decreased proteolysis of c-Myb by the ubiquitin-26S proteasome pathway (Bies and Wolff 199). However, further analysis is required to determine whether the removal of the NRD makes Myb resistant to the degradation induced by the Wnt–NLK signaling pathway.
Both HIPK2 and NLK directly bind to the DBD of c-Myb. However, gel mobility shift assays revealed that this binding does not inhibit the binding of c-Myb to DNA because Myb–HIPK2 complexes bound to the DNA probe were observed (data not shown). This suggests that HIPK2 and NLK interact with the surface of the c-Myb DBD. Previously, HIPK2 was shown to interact with the corepressor Groucho and histone deacetylase (HDAC; Choi et al. 1999). At present, it is unknown whether HIPK2 can interact simultaneously with NLK and the Groucho–HDAC complex. Because not only NLK but also HIPK2 directly bind to c-Myb, we cannot exclude the possibility that HIPK2 recruits the Groucho–HDAC complex to c-Myb and inhibits its trans-activating capacity in addition to inducing its phosphorylation and degradation through NLK.
Recently, HIPK2 was reported to directly phosphorylate p53 at Ser 46 in UV-irradiated cells, which leads to the transcriptional activation of the p53AIP1 gene (D'Orazi et al. 2002; Hofmann et al. 2002). In addition, corepressor CtBP was also shown to be directly phosphorylated by HIPK2 in response to UV irradiation, leading to the proteasome-dependent degradation of CtBP (Zhang et al. 2003). In contrast to these reports, HIPK2 did not directly phosphorylate the c-Myb protein, although NLK efficiently phosphorylated c-Myb. Both HIPK2 and NLK phosphorylate the Ser or Thr residues next to Pro. Further study will be necessary to determine whether how HIPK2 selects the substrate proteins for phosphorylation, and whether NLK and TAK1, which are known to be activated by UV irradiation, are also involved in the UV-induced phosphorylation of p53 or CtBP.
In contrast to the Wnt-1–GSK3β–β-catenin pathway, the so-called noncanonical Wnt pathway mediated by the Wnt-5a subclass triggers intracellular Ca2+ release that activates Ca2+-sensitive enzymes (Kuhl et al. 2000). Recently, we (S.K. Kishida, J. Ninomiya-Tsuji, and K. Matsumoto, unpubl.) demonstrated that the Wnt/Ca2+ pathway activates NLK through TAK1 in 293 cells (Ishitani et al. 2003). We have also observed that c-Myb protein degradation is induced by overexpressing rat Fz2 receptor, a component of the Wnt/Ca2+ pathway (data not shown). It is likely that the particular Wnt family protein member that activates the TAK1–NLK pathway will depend on the cellular context. Further study will be required to understand the precise molecular mechanisms by which the various Wnt proteins regulate the activity of the Myb protein family members.
Materials and methods
Yeast two-hybrid screening and GST pull-down assays
Yeast two-hybrid screening was performed by using a mouse embryonic cDNA library as described (Nomura et al. 1999). The mouse c-Myb protein lacking the transcriptional activation domain (amino acids 240–324) or an NLK fragment (amino acids 1245–515) was used as a bait. GST pull-down assays were performed as described (Nomura et al. 1999). To increase the solubility of GST fusion proteins expressed in bacteria, the thioredoxin coexpression system (Yasukawa et al. 1995) was used. The binding buffer used consisted of 20 mM HEPES (pH 7.5), 1 mM DTT, 0.1% NP-40, and 150 mM (for the interaction between Myb and HIPK2) or 100 mM (for the interaction between HIPK2 and NLK) KCl. To determine the interaction between Myb and NLK, buffer B (20 mM HEPES at pH 7.5, 5 mM DTT, 0.01% NP-40, 0.01% BSA, and 100 mM KCl) was used.
Coimmunoprecipitation
Molt-4 and 293 cells were maintained in RPMI medium and high-glucose DMEM, respectively. Both media were supplemented with 10% fetal bovine serum and antibiotics. To coimmunoprecipitate the endogenous Myb, HIP2, and NLK proteins (Fig. 1D), Molt-4 cell lysates were prepared by pipetting in NETN buffer (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 10% glycerol, protease inhibitor cocktail). Rabbit antibodies against NLK (Ishitani et al. 1999) or HIPK2, which were raised against the GST–HIPK2 fusion protein containing amino acids 860–1141 of HIPK2, or normal rabbit IgG were used for immunoprecipitation. The immunocomplexes were Western blotted with the anti-Myb monoclonal antibody 1-1 and the Enhanced Chemiluminescence (ECL) Western Blotting System (Amersham). To investigate the in vivo interaction between HIPK2 and NLK (Fig. 2), 293 cells (1 × 106 cells per 100-mm dish) were transfected by the CaPO4 method with a total of 10 μg DNA consisting of various expression vectors. The transfectants were then incubated for 24–36 h and lysed in 0.3 mL 0.5% Triton X-100 lysis buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 12.5 mM β-glycerophosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 2 mM DTT, 1 mM sodium orthovanadate, 1 mM PMSF, and 20 mM aprotinin. The lysates were immunoprecipitated with 1 μg of the anti-Flag (M2, Sigma) or anti-HA (HA.11, BABCO) monoclonal antibodies and 20 μL of protein G-Sepharose (Pharmacia). The immunecomplexes were washed three times with wash buffer containing 20 mM HEPES (pH 7.4), 500 mM NaCl, and 10 mM MgCl2; rinsed once with rinse buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, and 10 mM MgCl2; and suspended in 30 μL rinse buffer. For immunoblotting, the immunoprecipitates or whole-cell lysates were resolved on SDS-PAGE and transferred to Hybond-P membranes (Amersham). The membranes were immunoblotted with various antibodies, and the bound antibodies were visualized by horseradish peroxidase-conjugated antibodies against rabbit or mouse IgG using the ECL Western Blotting System. For two-step coimmunoprecipitation, 293 cells (7 × 105 cells/10-cm dish) were transfected with mixture of the plasmids to express Flag-HIPK2 (2 μg), HA-NLK (1 μg), and c-Myb-15A (3 μg) using lipofectAMINE plus. Two-step coimmunoprecipitation was performed as described (Harada et al. 2003).
Kinase assay
To detect the HIPK2 and NLK kinase activity using GST–LEF-1 or GST–HIPK2 (Fig. 2), the immunoprecipitates were incubated with 5 μCi of [γ-32P]ATP (3,000 Ci/mmole) and 1 μg of GST–LEF-1 or GST–HIPK2 in 10 μL kinase buffer containing 10 mM HEPES (pH 7.4), 1 mM DTT, 5 mM MgCl2 for 2 min at 25°C. Samples were separated by SDS-PAGE and visualized by autoradiography. For phosphorylation of c-Myb by NLK (Fig. 5), 293T cells were transfected with the Flag-linked c-Myb expression plasmid (pact-Flag-c-Myb) or the Flag-NLK expression plasmid. The lysates were prepared from the transfected cells using NET buffer (20 mM Tris-HCl at pH 8.0, 1 mM EDTA, 0.5% NP-40, protease inhibitor cocktail) containing 150 mM NaCl and immunoprecipitated with anti-Flag antibody. The immunecomplexes were sequentially washed with washing buffer (20 mM Tris-HCl at pH 8.0, 1 mM EDTA, 1 M NaCl, 2% NP-40, and 10% glycerol) and then NLK buffer (10 mM HEPES at pH 7.4, 1 mM DTT, and 5 mM MgCl2). An aliquot of the c-Myb immunocomplexes was used for Western blotting. Flag-NLK proteins were eluted from the immunocomplexes with Flag peptides. To phosphorylate c-Myb proteins, the c-Myb immunocomplexes were mixed with the eluted Flag-NLK proteins and [γ-32P]-ATP and incubated for 2 h at 25°C. The amount of c-Myb proteins used in the kinase reaction was adjusted based on the Western blotting data. Phosphorylated proteins were analyzed by SDS-PAGE followed by autoradiography.
CAT reporter assays
By using the CaPO4 method, CV-1 cells (4 × 105 cells/100-mm dish) were transfected with a mixture of the pc-myc-CAT reporter containing the human c-myc promoter (4 μg; Nakagoshi et al. 1992); the c-Myb expression plasmid pact-c-myb (2 μg); the plasmid that expresses Wnt-1, R-Fz1, TAK1/TAB1, HIPK2; or the dominant-negative form of HIPK2, and the internal control plasmid pact-β-gal (0.3 μg). Forty hours after transfection, CAT assays were performed. In some cases, transfected cells were treated with TNF-α (100 ng/mL) for 24 h or IL-1β (12.5 ng/mL) for 24 h before lysate preparation. To examine the effect of TGF-β, HepG2 cells, which are TGF-β responsive, were transfected with a mixture of the pc-myc-CAT reporter (4 μg), the c-Myb expression plasmid pact-c-myb (2 μg), and the internal control plasmid pact-β-gal (0.3 μg), and the cells were treated with TGF-β1 (3 ng/mL) for 20 h before lysate preparation. The CAT activity was then measured. In the experiments assessing the effect of TGF-β, TNF-α, and IL-1, the total plasmid amounts used were adjusted to 10 μg by adding the empty vector. In other experiments, however, the total plasmid amount was adjusted to 16.3 μg.
Western blotting and ubiquitination assay
CV-1 cells were transfected with a mixture of the c-Myb expression plasmid (6 μg), the plasmid (2 μg) that expresses various components of the Wnt–TAK1–HIPK2–NLK pathway, and the internal control plasmid pact-β-gal (0.3 μg). Total plasmid amounts were adjusted to 8.3 μg by adding the empty plasmid. In some cases, cells were treated with MG132 (50 μM) for 7 h before lysate preparation. Forty hours after transfection, cells were lysed in SDS sample buffer with mild sonication and subjected to SDS-PAGE followed by Western blotting with mouse anti-Myb antibody and ECL detection reagents (Amersham). The anti-c-Myb monoclonal antibody (for Fig. 4) and the rabbit polyclonal antibody (for Fig. 5), which was raised against the GST–CT5 fusion protein, were used for Western blotting. Aliquots of the cells were used to determine the transfection efficiency by measuring β-galactosidase activity and the amounts of lysates used for Western blotting were normalized based on the β-galactosidase activity. To detect ubiquitinated c-Myb, CV-1 cells were transfected with the plasmids that express Flag-c-Myb (3 μg), Myc-ubiquitin (2 μg), and the internal control pact-β-gal (0.3 μg) together with the NLK expression plasmid or the control vector (2 μg). Theses cells were also treated with MG132 (50 μM) for 7 h before lysate preparation. Thirty hours after transfection, the cells were then scraped into 100 μL lysis buffer (10 mM Tris-HCl at pH 8.0, 500 mM NaCl, 2% SDS), heated for 10 min at 95°C, diluted with buffer lacking SDS to reduce the SDS concentration to 0.2%, and then sonicated mildly on ice. After preadsorption with protein G-Sepharose, anti-Flag M2 monoclonal antibody was used to precipitate the immunocomplexes, which were then used for Western blotting with anti-Myc antibody (MBL).
Pulse-chase experiments
CV-1 cells were transfected with the Flag-c-Myb (8 μg) expression vector together with the NLK expression plasmid or control DNA (2.5 μg), and pact-β-gal (0.5 μg). Twenty-four hours after transfection, cells were labeled with [35S]methionine and [35S]cysteine for 3 h, after which the radioactivity was chased at 1-h intervals. At various times, lysates were prepared by using the NETN buffer, and immunoprecipitated by anti-Flag antibody (M2, Sigma). The immunocomplexes were analyzed by SDS-PAGE followed by autoradiography.
Subcellular localization
CV-1 cells were transfected with the plasmids to express HA-TAK1, Flag-HIPK2, HIPK2, Flag-NLK, R-Fz1, and Wnt-1 in following combinations: HA-TAK1 (1 μg) and Flag-HIPK2 (1 μg; Fig. 4F, upper left); HA-TAK1 (1 μg), Flag-HIPK2 (1 μg), R-Fz1 (0.5 μg), and Wnt-1 (0.5 μg; Fig. 4F, lower left); Flag-NLK (1 μg) and HIPK2 (1 μg; Fig. 4F, upper right); and Flag-NLK (1 μg), HIPK2 (1 μg), and HA-TAK1 (1 μg; Fig. 4F, lower right). Immunostaining with anti-HA, anti-Flag and anti-HIPK2 antibodies and analysis by confocal microscopy were performed as described (Nomura et al. 1999).
siRNA
siRNA oligonucleotides for TAK1 was described previously (Takaesu et al. 2003). siRNA with two thymidine residues (dTdT) at the 3′ end of the sequence was designed for NLK: sense, 5′-GAAUAUCCGCUAAGGAUGC-3′. This sequence is conserved between human and mouse NLK mRNAs. CV-1 cells (4 × 105 cells per 100-mm dish) were transfected with 9 μg oligonucleotides using the CaPO4 method and, 44 h later, the levels of endogenous TAK1 and NLK proteins were assayed by Western blotting. To investigate the effect of siRNA on the R-Fz1-induced degradation of c-Myb protein, CV-1 cells were transfected with the c-Myb expression plasmid (6 μg), the R-Fz1 expression plasmid (1.5 μg), the internal control plasmid pact-β-gal (0.5 μg), and siRNA (9 μg).
Analysis of M1 cell clones
The murine myeloid leukemia cell line M1 was grown in DMEM medium containing 10% fetal bovine serum. The retroviral expression plasmids for R-Fz1, c-Myb, and NLK were constructed by using the murine stem cell virus (MSCV)-based retroviral vector and viruses were prepared as described (Shinagawa et al. 2001). To generate M1 cell clones that express R-Fz1 or c-Myb together with EGFP or the neomycin-resistance gene, M1 cells were infected with viruses and then selected by EGFP expression or growth in the presence of G418 (400 μg/mL). Several independent clones were isolated and characterized regarding the level of expression of R-Fz1 or Myb. The M1 cell clones containing the Myb expression vector or the control vector were then used to isolate several independent clones expressing NLK by selecting the EGFP-positive cells. M1 cell clones were induced to differentiate with IL-6 (50 ng/mL, Wako) after being seeding at a concentration of 1 × 105 cells/mL for 72 h. Cells were stained with a phycoerythrin (PE)-conjugated anti mouse CD14 monoclonal antibody (Pharmingen) on ice for 30 min after blocking with 3% FBS. The cells were then analyzed on a FACSCaliblur flow cytometer (Becton Dickinson).
In vivo phosphorylation of c-Myb
CV-1 cells were transfected with the Flag-c-Myb expression vector together with the internal control plasmid pact-β-gal, and labeled with 32P-orthophosphate for 3 h. Lysates were prepared from the transfected cells using RIPA buffer, and the c-Myb proteins were immunoprecipitated with anti-Flag antibody and analyzed by SDS-PAGE, followed by autoradiography or Western blotting using anti-Flag antibody (M2, Sigma). The amounts of lysates used were normalized based on the transfection efficiency.
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research and the Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Science and Technology of Japan, and by a grant from the Human Frontier Science Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1170604.
Corresponding author.
References
- Allen R.D., Bender, T.P., and Siu, G. 1999. c-Myb is essential for early T-cell development. Genes & Dev. 13: 1073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz N., Miglarese, M.R., Hendrickson, R.C., Shabanowitz, J., Sturgill, T.W., Hunt, D.F., and Bender, T.P. 1995. Modulation of c-Myb-induced transcription activation by a phosphorylation site near the negative regulatory domain. Proc. Natl. Acad. Sci. 92: 6429-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J., von Kries, J.P, Kuhl, M., Bruhn, L., Wedlich, D., Grosschedl, R., and Birchmeier, W. 1996. Functional interaction of β-catenin with the transcription factor LEF-1. Nature 382: 638-642. [DOI] [PubMed] [Google Scholar]
- Behrens J., Jerchow, B.A., Wurtele, M., Grimm, J., Asbrand, C., Wirtz, R., Kuhl, M., Wedlich, D., and Birchmeier, W. 1998. Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3β. Science 280: 596-599. [DOI] [PubMed] [Google Scholar]
- Biedenkapp H., Borgmeyer, U., Sippel, A.E., and Klempnauer, K.-H. 1988. Viral myb oncogene encodes a sequence-specific DNA binding activity. Nature 335: 835-837. [DOI] [PubMed] [Google Scholar]
- Bies J. and Wolff, L. 1997. Oncogenic activation of c-Myb by carboxyl-terminal truncation leads to decreased proteolysis by the ubiquitin–26S proteasome pathway. Oncogene 14: 203-212. [DOI] [PubMed] [Google Scholar]
- Brandon C., Eisenberg, L.M., and Eisenberg, C.A. 2000. WNT signaling modulates the diversification of hematopoietic cells. Blood 96: 4132-4141. [PubMed] [Google Scholar]
- Brott B.K., Pinsky, B.A., and Erikson, R.L. 1998. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc. Natl. Acad. Sci. 95: 963-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.Y., Kim, Y.H., Kwon, H.J., and Kim, Y. 1999. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem. 274: 33194-33197. [DOI] [PubMed] [Google Scholar]
- Dai P., Akimaru, H., Tanaka, Y., Hou, D.X., Yasukawa, T., Kanei-Ishii, C., Takahashi, T., and Ishii, S. 1996. CBP as a transcriptional coactivator of c-Myb. Genes & Dev. 10: 528-540. [DOI] [PubMed] [Google Scholar]
- Dini P.W. and Lipsick, J.S. 1993. Oncogenic truncation of the first repeat of c-Myb decreases DNA binding in vitro and in vivo. Mol. Cell. Biol. 13: 7334-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orazi G, Cecchinelli, B., Bruno, T., Manni, I., Higashimoto, Y., Saito, S., Gostissa, M., Coen, S., Marchetti, A., Del Sal, G., et al. 2002. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 4: 11-19. [DOI] [PubMed] [Google Scholar]
- Dubendorff J.W., Whittaker, L.J., Eltman, J.T., and Lipsick, J.S. 1992. Carboxyl-terminal elements of c-Myb negatively regulate transcriptional activation in cis and in trans. Genes & Dev. 6: 2524-2535. [DOI] [PubMed] [Google Scholar]
- Frampton J., Ramqvist, T., and Graf, T. 1996. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes & Dev. 10: 2720-2731. [DOI] [PubMed] [Google Scholar]
- Ghosh S. and Karin, M. 2002. Missing pieces in the NF-κB puzzle. Cell 109(Suppl): 81-96. [DOI] [PubMed] [Google Scholar]
- Gonda T.J. and Metcalf, D. 1984. Expression of myb, myc, and fos proto-oncogenes during the differentiation of a murine myeloid leukaemia. Nature 310: 249-251. [DOI] [PubMed] [Google Scholar]
- Harada J., Kokura, K., Kanei-Ishii, C., Nomura, T., Khan, M.M., Kim, Y., and Ishii, S. 2003. Requirement of the co-repressor homeodomain-interacting protein kinase 2 for ski-mediated inhibition of bone morphogenetic protein-induced transcriptional activation. J. Biol. Chem. 278: 38998-39005. [DOI] [PubMed] [Google Scholar]
- Hofmann T.G., Moller, A., Sirma, H., Zentgraf, H., Taya, Y., Droge, W., Will, H., and Schmitz, M.L. 2002. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4: 1-10. [DOI] [PubMed] [Google Scholar]
- Hu L.-Y., Ramsay, R.G., Kanei-Ishii, C., Ishii, S., and Gonda, T.J. 1991. Transformation by carboxyl-deleted Myb reflects increased transactivating capacity and disruption of a negative regulatory domain. Oncogene 6: 1549-1553. [PubMed] [Google Scholar]
- Ishitani T., Ninomiya-Tsuji, J., Nagai, S., Nishita, M., Meneghini, M., Barker, N., Waterman, M., Bowerman, B., Clevers, H., Shibuya, H., et al. 1999. The TAK1–NLK–MAPKrelated pathway antagonizes signalling between β-catenin and transcription factor TCF. Nature 399: 798-802. [DOI] [PubMed] [Google Scholar]
- Ishitani T., Kishida, S., Hyodo-Miura, J., Ueno, N., Yasuda, J., Waterman, M., Shibuya, H., Moon, R.T., Ninomiya-Tsuji, J., and Matsumoto, K. 2003. The TAK1–NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca2+ pathway to antagonize Wnt/β-catenin signaling. Mol. Cell. Biol. 23: 131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. and Struhl, G. 1998. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391: 493-496. [DOI] [PubMed] [Google Scholar]
- Kanei-Ishii C., MacMillan, E.M., Nomura, T., Sarai, A., Ramsay, R.G., Aimoto, S., Ishii, S., and Gonda, T.J. 1992. Transactivation and transformation by Myb are negatively regulated by a leucine-zipper structure. Proc. Natl. Acad. Sci. 89: 3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen A.L., Jackson, J., Harmon, B.P., Fung, S.M., Ramsay, G., and Bishop, J.M. 1998. Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes & Dev. 12: 831-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.H., Choi, C.Y., Lee, S.J., Conti, M.A., and Kim, Y. 1998. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J. Biol. Chem. 273: 25875-25879. [DOI] [PubMed] [Google Scholar]
- Kishimoto K., Matsumoto, K., and Ninomiya-Tsuji, J. 2000. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J. Biol. Chem. 275: 7359-7364. [DOI] [PubMed] [Google Scholar]
- Klempnauer K.-H., Gonda, T.J., and Bishop, J.M. 1982. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31: 453-463. [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E., Herr, P., Niss, K., and Leutz, A. 1997. The homeobox gene GBX2, a target of the myb oncogene, mediates autocrine growth and monocyte differentiation. Cell 91: 185-195. [DOI] [PubMed] [Google Scholar]
- Kuhl M., Sheldahl, L.C., Park, M., Miller, J.R., and Moon, R.T. 2000. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 16: 279-283. [DOI] [PubMed] [Google Scholar]
- Leverson J.D., Koskinen, P.J., Orrico, F.C., Rainio, E.M., Jalkanen, K.J., Dash, A.B., Eisenman, R.N., and Ness, S.A. 1998. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol. Cell 2: 417-425. [DOI] [PubMed] [Google Scholar]
- Liang H., Chen, Q., Coles, A.H., Anderson, S.J., Pihan, G., Bradley, A., Gerstein, R., Jurecic, R., and Jones, S.N. 2003. Wnt5a inhibits B cell proliferation and functions as a tumor suppressor in hematopoietic tissue. Cancer Cell 4: 349-360. [DOI] [PubMed] [Google Scholar]
- Lüscher B., Christenson, E., Litchfield, D.W., Krebs, E.G., and Eisenman, R.N. 1990. Myb DNA binding inhibited by phosphorylation at a site deleted during oncogenic activation. Nature 344: 517-522. [DOI] [PubMed] [Google Scholar]
- Meneghini M.D., Ishitani, T., Carter, J.C., Hisamoto, N., Ninomiya-Tsuji, J., Thorpe, C.J., Hamill, D.R., Matsumoto, K., and Bowerman, B. 1999. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature 399: 793-797. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Nakagoshi, H., Nagase, T., Nomura, N., Date, T., Ueno, Y., and Ishii, S. 1990. DNA binding activity and transcriptional activator function of the human B-myb protein compared with c-MYB. J. Biol. Chem. 265: 9280-9284. [PubMed] [Google Scholar]
- Mucenski M.L., McLain, K., Kier, A.B., Swerdlow, S.H., Schereiner, C.M., Miller, T.A., Pietryga, D.W., Scott, W.J., and Potter, S.S. 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65: 677-689. [DOI] [PubMed] [Google Scholar]
- Nakagoshi H., Kanei-Ishii, C., Sawazaki, T., Mizuguchi, G., and Ishii, S. 1992. Transcriptional activation of the c-myc gene by the c-myb and B-myb gene products. Oncogene 7: 1233-1240. [PubMed] [Google Scholar]
- Ness S.A., Marknell, A., and Graf, T. 1989. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 59: 1115-1125. [DOI] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z., and Matsumoto, K. 1999. The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398: 252-256. [DOI] [PubMed] [Google Scholar]
- Nomura T. Khan, M.M., Kaul, S.C., Dong, H.D., Wadhwa, R., Colmenares, C., Kohno, I., and Ishii, S. 1999. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes & Dev. 13: 412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K., Morikawa, S., Nakamura, H., Sekikawa, A., Inoue, T., Kanai, H., Sarai, A., Ishii, S., and Nishimura, Y. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79: 639-648. [DOI] [PubMed] [Google Scholar]
- Okada M., Akimaru, H., Hou, D.-X., Takahashi, T., and Ishii, S. 2002. Myb controls G2/M progression by inducing cyclin B expression in the Drosophila eye imaginal disc. EMBO J. 21: 675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur P. and Kuhl, M. 2001. An arrow for wingless to take-off. Bioessays 23: 207-210. [DOI] [PubMed] [Google Scholar]
- Peifer M. and Polakis, P. 2000. Wnt signaling in oncogenesis and embryogenesis: A look outside the nucleus. Science 287: 1606-1609. [DOI] [PubMed] [Google Scholar]
- Reya T., Duncan, A.W., Ailles, L., Domen, J., Scherer, D.C., Willert, K., Hintz, L., Nusse, R., and Weissman, I.L. 2003. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423: 409-414. [DOI] [PubMed] [Google Scholar]
- Rocheleau C.E., Yasuda, J., Shin, T.H., Lin, R., Sawa, H., Okano, H., Priess, J.R., Davis, R.J., and Mello, C.C. 1999. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell 97: 717-726. [DOI] [PubMed] [Google Scholar]
- Sakura H., Kanei-Ishii, C., Nagase, T., Nakagoshi, H., Gonda, T.J., and Ishii, S. 1989. Delineation of the three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc. Natl. Acad. Sci. 86: 5758-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Nazarov, V., Stevens, L., Watson, R., and Wolff, L. 2000. Regulation of the resident chromosomal copy of c-myc by c-Myb is involved in myeloid leukemogenesis. Mol. Cell. Biol. 20: 1970-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumaran M., Liebermann, D.A., and Hoffman-Liebermann, B. 1992. Deregulated c-myb disrupts interleukin-6- or leukemia inhibitory factor-induced myeloid differentiation prior to c-myc: Role in leukemogenesis. Mol. Cell. Biol. 12: 2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya H., Yamaguchi, K., Shirakabe, K., Tonegawa, A., Gotoh, Y., Ueno, N., Irie, K., Nishida, E., and Matsumoto, K. 1996. TAB1: An activator of the TAK1 MAPKKK in TGF-β signal transduction. Science 272: 1179-1182. [DOI] [PubMed] [Google Scholar]
- Shin T.H., Yasuda, J., Rocheleau, C.E., Lin, R., Soto, M., Bei, Y., Davis, R.J., and Mello, C.C. 1999. MOM-4, a MAP kinase kinase kinase-related protein, activates WRM-1/LIT-1 kinase to transduce anterior/posterior polarity signals in C. elegans. Mol. Cell 4: 275-280. [DOI] [PubMed] [Google Scholar]
- Shinagawa T., Nomura, T., Colmenares, C., Ohira, M., Nakagawara, A., and Shunsuke Ishii, S. 2001. Increased susceptibility to tumorigenesis of ski-deficient heterozygous mice. Oncogene 20: 8100-8108. [DOI] [PubMed] [Google Scholar]
- Takaesu G., Surabhi, R.M., Park, K.J., Ninomiya-Tsuji, J., Matsumoto, K., and Gaynor, R.B. 2003. TAK1 is critical for IκB kinase-mediated activation of the NF-κB pathway. J. Mol. Biol. 26: 105-115. [DOI] [PubMed] [Google Scholar]
- Tanikawa J., Yasukawa, T., Enari, M., Ogata, K., Nishimura, Y., Ishii, S. and Sarai, A. 1993. Recognition of specific DNA sequences by the c-myb proto-oncogene product: role of three repeat units in the DNA-binding domain. Proc. Natl. Acad. Sci. 90: 9320-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D., Badiani, P., and Weston, K. 1996. A dominant interfering Myb mutant causes apoptosis in T cells. Genes & Dev. 10 2732-2744. [DOI] [PubMed] [Google Scholar]
- Weston C.R. and Davis, R.J. 2002. The JNK signal transduction pathway. Curr. Opin. Genet. Dev. 12: 14-21. [DOI] [PubMed] [Google Scholar]
- Weston K. and Bishop, J.M. 1989. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell 58: 85-93. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Strack, P., Beer-Romero, P., Chu, C.Y., Elledge, S.J., and Harper, J.W. 1999. The SCFβ–TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes & Dev. 13: 270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A. and Nusse, R. 1998. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14: 59-88. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Shirakabe, K., Shibuya, H., Irie, K., Oishi, I., Ueno, N., Taniguchi, T., Nishida, E., and Matsumoto, K. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270: 2008-2011. [DOI] [PubMed] [Google Scholar]
- Yasukawa T., Kanei-Ishii, C., Maekawa, T., Fujimoto, J., Yamamoto, T., and Ishii, S. 1995. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J. Biol. Chem. 270: 25328-25331. [DOI] [PubMed] [Google Scholar]
- Zeng L., Fagotto, F., Zhang, T., Hsu, W., Vasicek, T.J., Perry III, W.L., Lee, J.J., Tilghman, S.M., Gumbiner, B.M., and Costantini, F. 1997. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 90: 181-192. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Yoshimatsu, Y., Hildebrand, J., Frisch, S.M., and Goodman, R.H. 2003. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 115: 177-186. [DOI] [PubMed] [Google Scholar]