Abstract
A major challenge for cancer genetics is to determine which low frequency somatic mutations are drivers of tumorigenesis. Here we interrogate the genomes of 7,651 diverse human cancers to identify novel drivers and find inactivating mutations in the homeodomain transcription factor CUX1 (cut-like homeobox 1) in ~1-5% of tumors. Meta-analysis of CUX1 mutational status in 2,519 cases of myeloid malignancies reveals disruptive mutations associated with poor survival, highlighting the clinical significance of CUX1 loss. In parallel, we validate CUX1 as a bona fide tumor suppressor using mouse transposon-mediated insertional mutagenesis and Drosophila cancer models. We demonstrate that CUX1 deficiency activates phosphoinositide 3-kinase (PI3K) signaling through direct transcriptional downregulation of the PI3K inhibitor PIK3IP1 (phosphoinositide-3-kinase interacting protein 1), leading to increased tumor growth, while exposing susceptibility to PI3K-AKT inhibition. Thus, our complementary approaches identify CUX1 as a new pan-driver of tumorigenesis and uncover a potential strategy for treating CUX1-mutant tumors.
It is acknowledged that most sequencing studies are only sufficiently sensitive to identify commonly mutated cancer genes, such that cancer drivers mutated at low frequency escape detection. To address this issue we used an extensive collection of 7,651 genome sequences (352 whole genomes, 7,299 exomes) derived from 28 tumor types to identify novel loss-of-function cancer driver genes (Supplementary Table 1). Our strategy, which involved searching for genes showing a significant enrichment for nonsense mutations (see Methods), identified 54 genes (q < 0.01). These included known drivers of tumorigenesis such as CDKN2A, TP53 and PTEN, and genes not previously implicated as recurrently-mutated tumor suppressors (e.g. ARHGAP35, LARP4B, AMOT and MGA), providing a hitherto unseen picture of the cancer gene landscape (Fig. 1a). Here we focussed on CUX1, which showed a 3.4-fold increase in the ratio of observed/expected nonsense mutations (q = 0.0006). Across all samples, nonsense and frameshift deletion/insertion lesions accounted for 21% of mutations in CUX1, with 51% of mutations being missense, around half of which were classed as disruptive by SIFT1 and PolyPhen-22 (Fig. 1b, c and Supplementary Table 2). Overall nonsense and frameshift mutations in CUX1 were detected in 1-5% of tumors, spanning many types, with the highest frequency occurring in endometrial cancer (Fig. 1c).
Figure 1. Identification and significance of CUX1 mutations in human cancer.
(a) Histogram showing genes with increased observed/expected nonsense mutations (Benjamini–Hochberg’s false discovery rate adjusted p-value, q < 0.01). Genes are ranked according to the observed/expected ratio based on a mutation-selection model as described in Methods and ref. 24. CUX1 is highlighted in red.
(b) Predicted consequences of CUX1 mutations identified from the analysis of 7,651 human cancer genomes are depicted across the CUX1p200 1505 amino acid (aa) isoform (NCBI, NP_853530). CUX1 protein domains are highlighted: ID, inhibitory domain; CC, coiled-coil; CUT, CUT DNA-binding; HD, homeodomain. Mutation type is indicated.
(c) Frequency of CUX1 mutations according to tumor type. Mutation type is indicated. Total number of genomes analyzed in each tumor type is shown above each column. CLL, chronic lymphocytic leukemia; adeno, adenocarcinoma; GBM, glioblastoma multiforme.
(d) Schematic of CUX1p200, CUX1p110 and CUX1p75 protein isoforms. C, CUT domain, blue; HD, homeodomain, red.
(e) Distribution and type of CUX1 mutations in myeloid malignancies. MDS-associated mutations9, solid symbols; AML-associated mutations, outlined symbols. Mutation type as in (b).
(f) Estimated survival curves for 598 patients with MDS or MDS/MPN overlap according to presence of CUX1-truncating mutations or −7/del(7q), generated using the Nelson-Aalen-Breslow estimator of hazard after correction for age and WHO subtype of disease.
(g) Estimated survival curves for 1477 patients with AML according to presence of CUX1-truncating mutations or −7/del(7q), generated as in (f).
The CUX1 transcription factor (also known as CUTL1 or CDP) contains four DNA-binding motifs – three CUT repeats and a C-terminal homeodomain3- and has numerous cellular functions4,5. Previous studies have identified multiple CUX1 isoforms with differing transcriptional activation properties; whereas full-length CUX1p200 acts predominantly as a transcriptional repressor, proteolytic cleavage of the N-terminus generates a truncated CUX1p110 isoform with transcriptional activation properties6. Additionally, a shorter pro-tumorigenic CUX1p75 isoform found in breast tumors and certain normal tissues e.g. thymus, can be generated by transcriptional initiation from within intron 20 (ref. 7) (Fig. 1d).
Intriguingly, most studies to date have attributed an oncogenic role for CUX1 in human cancer (Supplementary Note). Conversely, a recent gene expression study found a ~50% reduction in CUX1 levels in 11 cases of acute myeloid leukemia (AML) characterized by complete or partial monoallelic loss of chromosome 7q, where CUX1 resides, leading to the proposal that CUX1 is a haploinsufficient myeloid tumor suppressor8. To evaluate the role of CUX1 in myeloid cancers more comprehensively, we performed a meta-analysis of our sequencing studies that targeted CUX1 coding exons (ref. 9 and unpublished data; A.R.G., E.P., H.D., J.N., P.J.C. et al.). We found no mutations in exomes from 151 patients with myeloproliferative neoplasms (Supplementary Fig. 1a). In a targeted gene screen of 111 genes in 738 patients with myelodysplasia (MDS), a clonal pre-leukemic hematopoietic condition, and related myelodysplastic/myeloproliferative neoplasms (MDS/MPN) such as chronic myelomonocytic leukemia (CMML)9, we identified 22 CUX1 mutations including heterozygous inactivating (nonsense, frameshift and essential splice site) variants in 15 samples (2%) (Fig. 1e and Supplementary Table 3). An identical screen applied to 1,630 AML patients (Supplementary Fig. 1a) identified 10 CUX1 mutations (nine inactivating, one missense) (Fig. 1e and Supplementary Table 3). CUX1 mutations did not co-occur with −7/del(7q) abnormalities, consistent with the notion that CUX1 is a haploinsufficient tumor suppressor (Supplementary Table 3). However, one CMML patient harbored a homozygous nonsense CUX1 mutation, suggesting that CUX1 can also function rarely as a classical tumor suppressor, consistent with a case with myeloproliferative disease10. CUX1 mutations were found across most MDS subtypes, including refractory anemia – thought to represent an early stage in MDS evolution – suggesting that CUX1 mutations can arise around disease initiation (Supplementary Fig. 1b). CUX1 mutations were relatively frequent in CMML, accounting for 7/70 (10%) cases (Supplementary Fig. 1c). Given that the presence of −7/del(7q) lesions in myeloid malignancies is associated with poor prognosis11 we compared the overall survival of CUX1-truncating to −7/del(7q) cases in myeloid disease cohorts. In MDS and MDS/MPN, both CUX1 inactivation and −7/del(7q) were associated with poorer overall survival, even after correction for age and WHO category (p = 0.03 and p = 0.0004 respectively) (Fig. 1f). With AML, there was a tendency towards poorer survival in CUX1-truncating cases (p = 0.1), whereas −7/del(7q) was associated with a significantly worse overall outcome (p < 0.0001), after correction for age and randomized treatment (Fig. 1g). A larger cohort of AML cases will be required to assess further the impact of CUX1 mutations, but nonetheless our data suggest that CUX1 loss is an important independent prognostic indicator and determinant of unfavourable outcome in −7/del(7q) myeloid malignancies.
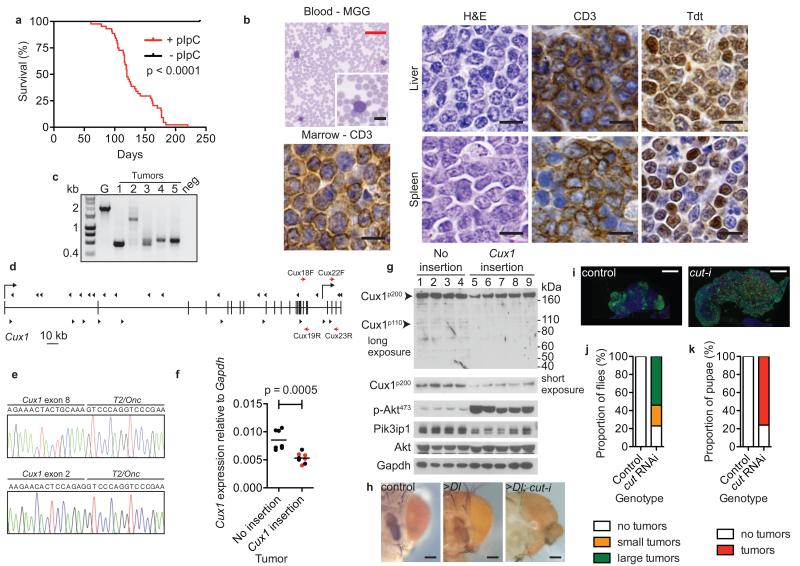
Although these genomic data strongly suggested that CUX1 functions as a tumor suppressor, we used cross-species oncogenomics approaches in mice and Drosophila to provide corroborative evidence. As we found CUX1 mutations in myeloid neoplasms, we speculated that CUX1 might be a conserved hematopoietic tumor suppressor, a notion supported by the myeloid hyperplasia phenotype of Cux1-deficient mice12. Previously, we have used transposon-mediated mutagenesis in mice as an unbiased forward-genetic approach to uncover novel cancer genes in a tissue-specific manner13. A screen performed in hematopoietic tissues using mice carrying Sleeping Beauty T2/Onc transposons14,15, a conditional transposase (CAGGS-SB11; SB11) allele13, and a polyinosinic–polycytidylic acid (pIpC)-inducible pan-hematopoietic Cre driver (Mx1-Cre)16 (Supplementary Fig. 2) led to T-cell lymphoblastic lymphoma (T-ALL) in >80% triple transgenic mice with a median survival of 120 days following pIpC injection (Fig. 2a). Tumors from these mice were CD3+;Tdt+ and showed clonal T-cell rearrangements confirming a diagnosis of T-ALL (Fig. 2b, c). Transposon insertion site analysis from 44 tumors revealed 90 genes associated with common insertion sites (CIS) (p < 0.05) (Supplementary Table 4). Strikingly, 20 (45%) tumors contained sense and/or anti-sense insertions in Cux1, including six tumors with two insertions and one tumor harboring three insertions (Fig. 2d). Insertions were also present in 14 key human T-ALL genes, highlighting the human relevance and sensitivity of our screen (Supplementary Fig. 3 and Supplementary Table 5). Although the bi-directional pattern of transposon insertions within Cux1 was typical of gene inactivation17, shorter, potentially oncogenic Cux1 isoforms could theoretically be produced where the transposon lands in a ‘forward’ orientation due to transcriptional activation by the MSCV promoter. Thus, we first confirmed the presence of truncating Cux1-T2/Onc fusion transcripts in independent tumors by RT-PCR (Fig. 2e and Supplementary Fig. 4), and then used qRT-PCR to demonstrate a ~50% reduction in Cux1 levels in tumors with Cux1 insertions compared with tumors without such insertions (Fig. 2f and Supplementary Fig. 5a). Immunoblotting confirmed a reduction in all Cux1 protein isoforms (Fig. 2g). There was no increase in the expression of Cux1I20, encoding oncogenic Cux1p75, in tumors with Cux1 insertions (Supplementary Fig. 5b). Additionally, Cux1 insertions were present in 59-79% of cells in three tumors analyzed, suggesting that Cux1 insertions arose in a dominant tumor clone (Supplementary Fig. 6). Residual Cux1 expression was detectable, even in tumors with two Cux1 insertions (Fig. 2f and Supplementary Fig. 5a, red points, Fig. 2g, lanes 6, 7), suggesting mono-allelic or bi-clonal insertion events. Altogether, these data indicate that Cux1 insertions led to predominantly heterozygous gene inactivation akin to the nature of most human CUX1 mutations. Lastly, to explore the degree of evolutionary conservation of CUX1 in tumorigenesis we used a Drosophila cancer model. Marked cell proliferation was observed after depletion of CUX1 (cut in Drosophila) either in the proliferating eye disc in conjunction with Delta (Dl) Notch-ligand expression (Fig. 2h and ref. 18) or alone in the lymph gland and larvae (Fig. 2i-k and ref. 8), indicating intrinsic cut tumor suppressive activity. Although the exact contribution of CUX1 in human T-ALL requires further investigation (Supplementary Note), our collective data support the hypothesis that CUX1 is an evolutionarily conserved tumor suppressor.
Figure 2. Identification of CUX1 as a tumor suppressor gene in mice and Drosophila.
(a) Kaplan-Meier survival curve of T2/Onc; SB11; Mx1-Cre transgenic mice injected with pIpC (red line, n = 44) or sham controls (black line, n = 7). Increased mortality in pIpC-injected mice (median survival 120 days; p < 0.0001, log-rank test).
(b) T-ALL phenotype of mouse transposon tumors. Blood smear stained with May-Grünwald-Giemsa (MGG). Liver and spleen sections stained with haematoxylin/eosin (H&E) showing blast cells. Immunohistochemistry performed on liver and spleen tissues to detect CD3 and Tdt (Terminal deoxynucleotidyl transferase) antigens. Bone marrow immunostaining showing infiltration with CD3+ T cells. Scale bars; black = 10 μm, red 50 μm.
(c) TCRβ gene rearrangements in transposon tumors by PCR analysis. ~2-kb germline (G) product obtained using wild-type mouse tail DNA. Smaller bands in mouse tumors 1-5 indicative of TCRβ gene rearrangement; neg, no DNA.
(d) Distribution of transposon insertions (arrowheads) across Cux1 locus. Vertical bars, exons. Direction of Cux1 transcription is left-to-right. Transcriptional start sites for Cux1p200 and Cux1p75 are shown by right-angled arrows from exon 1 and intron 20 respectively. Direction of arrowheads indicates orientation of transposon insertion with respect to MSCV promoter. Red arrows show location of primer pairs used for qRT-PCR: Cux18F-Cux19R, nucleotides 2884-2998; Cux22F-Cux23R, nucleotides 3484-3671 with respect to Cux1 transcript ENSMUST00000176172.
(e) Detection of splicing from Cux1 exons 8 and 2 to T2/Onc transposon by RT-PCR from two tumors, predicted to truncate the Cux1 transcript.
(f) Reduction in Cux1 levels in mouse transposon thymic tumors with Cux1 insertions by qRT-PCR using Cux18F-Cux19R primers that detect Cux1p200 and Cux1p110 transcripts. Data points are mean of triplicate assessments. Bar, mean of each group; p = 0.0005, t-test. Red, tumors with two Cux1 insertions.
(g) Immunoblot showing levels of indicated proteins in transposon tumors with and without Cux1 insertions.
(h) Knockdown of cut enhances overgrowth phenotype caused by Delta overexpression. Control showing normal adult eyes (ey-Gal4 UAS-GFP, UAS-GFPRNAi), slightly enlarged eyes caused by Delta overexpression (>Dl) (ey-Gal4 UAS-Dl, UAS-GFPRNAi) and dramatically altered eyes caused by Delta overexpression combined with cut-knockdown (>Dl; cut-i) (ey-Gal4 UAS-Dl, UAS-cutRNAi). Scale bar = 100 μm.
(i) Hyperplasia caused by ablation of cut in blood cell lineage. Differentiating blood cells (marked by pxn-Gal4 UAS-GFP) are restricted to the cortex in control lymph glands but are greatly expanded in the presence of cut RNAi (pxn-Gal4 UAS-GFP UAS-cutRNAi) causing overgrowth of the whole gland. Red, differentiating crystal cells; blue, nuclei labelled with DAPI. Scale bar = 50 μm.
(j, k) Percentage of adult females (j) and pupae (k) exhibiting visible melanotic tumors in control (pxn-Gal4 UAS-GFP UAS-white RNAi) (n = 70 and 31 respectively) and cut knockdown (pxn-Gal4 UAS-GFP UAS-cutRNAi) (n = 74 and 46 respectively) genotypes.
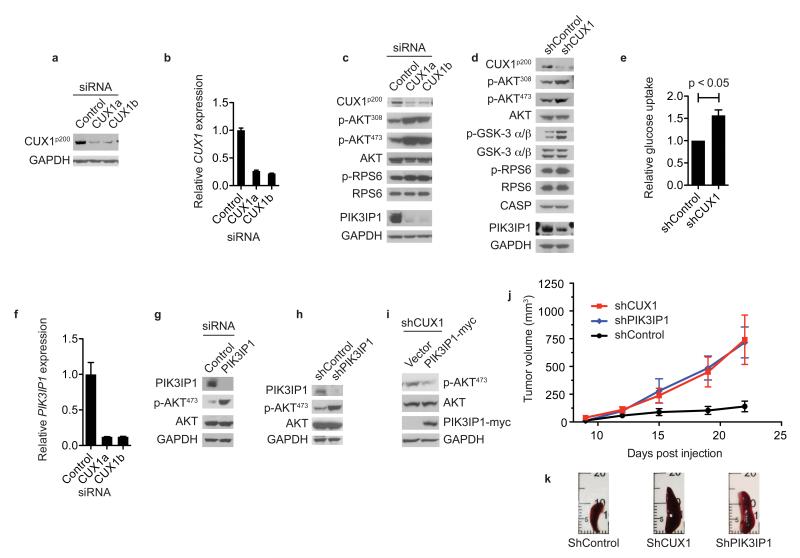
To explore the mechanism of tumor suppression by CUX1 we depleted CUX1 in human LOUCY T-ALL cells by RNA interference (Fig. 3a, b) and performed transcriptome profiling to identify 99 downregulated genes and 62 upregulated genes in CUX1-knockdown cells (fold change of >1.5, p < 0.005) (Supplementary Fig. 7 and Supplementary Table 6). Regulation of several targets was validated by qRT-PCR (Supplementary Fig. 7). Gene ontology analysis of dysregulated genes showed perturbation of a range of biological processes including cell cycle control and notably, regulatory components of the PI3K-signaling axis (Supplementary Fig. 7 and Supplementary Table 7).
Figure 3. CUX1 deficiency activates PI3K signaling associated with downregulation of the PI3K inhibitor, PIK3IP1.
(a, b) Confirmation of siRNA-mediated CUX1 knockdown in LOUCY cells by immunoblotting (a) and qRT-PCR (b) using 2 siRNAs (CUX1a, CUX1b) compared to control siRNA. GAPDH, loading control in (a). Data for (b), mean + s.d. from 3 independent experiments assessed in triplicate.
(c, d) PI3K signaling activation following CUX1 knockdown in LOUCY (c) and KE37 (d) cells assessed by immunoblotting using antibodies shown. CASP is a product of alternative splicing of the CUX1 locus from exon 14 onwards.
(e) Glucose uptake in KE37 cell lines transduced with indicated shRNA vectors (data normalized to control shRNA, showing mean + s.e.m. of 3 independent experiments performed in triplicate; p < 0.05, t-test).
(f) Reduced PIK3IP1 expression in CUX1-knockdown LOUCY cells by qRT-PCR (mean + s.d. of 2 independent experiments assessed in triplicate).
(g, h) Depletion of PIK3IP1 activates PI3K signaling. Lysates from PIK3IP1 siRNA-knockdown LOUCY cells (g) and PIK3IP1 shRNA-knockdown KE37 cells (h) were used for immunoblotting with indicated antibodies.
(i) PIK3IP1 suppresses AKT activation. Lysates from shCUX1 KE37 cells transduced with indicated vectors were used for immunoblotting with indicated antibodies.
(j) Enhanced tumorigenesis following CUX1 and PIK3IP1 depletion in vivo. NOD-SCID mice (n = 5 per group) injected subcutaneously with KE37 cells transduced with indicated vectors were monitored for tumor growth over 22 days. Data show mean ± s.e.m.
(k) Images of spleens from sacrificed mice from (j) at 22 days.
Given the importance of PI3K signaling in cancer, we assessed the effects of CUX1 loss on the PI3K pathway and found increased phosphorylation of AKT and RPS6 in CUX1-knockdown LOUCY cells (Fig. 3c). Stable shRNA-mediated knockdown of CUX1 in KE37 T-ALL cells also led to increased phosphorylation of AKT, its substrate GSKα/β (Fig. 3d), and to increased glucose uptake (Fig. 3e). Of note the most highly downregulated gene in our transcriptome analysis was PIK3IP1, whose protein product binds the p110 catalytic subunit of PI3K heterodimers, thereby antagonizing PI3K catalytic activity19,20. Consistent with these microarray data, and qRT-PCR (Fig. 3f), we found reduced PIK3IP1 protein levels in CUX1-depleted LOUCY (Fig. 3c) and KE37 cells (Fig. 3d). To determine whether downregulation of PIK3IP1 was responsible for activation of PI3K-AKT signaling in this context, we confirmed that PIK3IP1 knockdown in T-ALL cells increased phospho-AKT levels (Fig. 3g, h), and showed that enforced PIK3IP1 expression in CUX1-shRNA knockdown cells attenuated AKT activation (Fig. 3i). Subcutaneous injection of NOD-SCID immunodeficient mice with KE37 cells transduced with shRNA vectors that target CUX1 or PIK3IP1 resulted in the formation of larger tumors with systemic spread of tumor cells, indicating that CUX1 and PIK3IP1 are tumor suppressors in vivo (Fig. 3j, k and Supplementary Fig. 8).
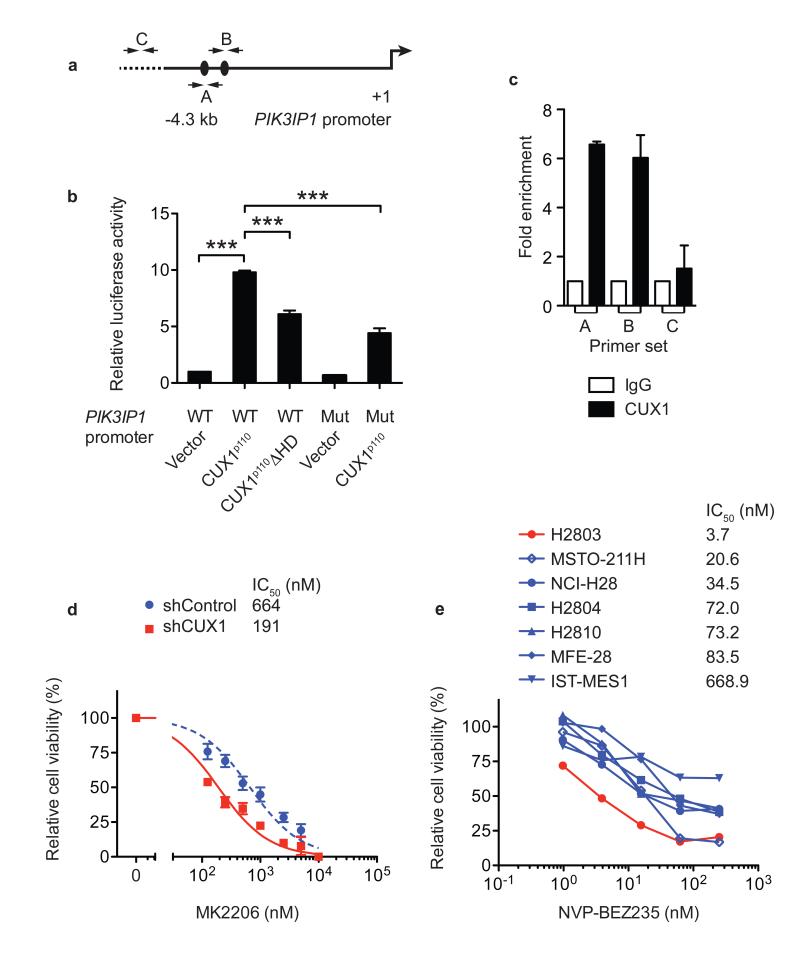
Previous studies have shown that CUX1 has a binding preference for ATC(G/A)AT consensus DNA sequences on target gene promoters21-23. To investigate whether CUX1 regulates PIK3IP1 expression directly, we inspected the PIK3IP1 genomic locus and found two putative CUX1-binding sites ~3 kb upstream of the PIK3IP1 transcriptional start site (Fig. 4a). By using a ~4.3-kb PIK3IP1 genomic fragment encompassing the two putative CUX1-binding sites in luciferase reporter assays, we found that exogenous CUX1p110 could transactivate PIK3IP1-luciferase expression around ~10-fold (Fig. 4b). Mutation of both putative CUX1-binding sites within the reporter construct, or expression of a homeodomain-deletion CUX1p110 mutant, led to a significant reduction of luciferase activity (Fig. 4b); the residual luciferase activity observed using these mutants was possibly attributable to additional, unidentified binding sites in the reporter. Subsequently, ChIP assays in LOUCY cells demonstrated direct binding of CUX1 to the PIK3IP1 promoter in vivo (Fig. 4c). Collectively, these data show that CUX1 modulates PI3K signaling through direct transcriptional regulation of PIK3IP1 expression (Supplementary Fig. 9). As most human CUX1 mutations appear to be heterozygous, we next investigated whether haploinsufficient CUX1 levels were sufficient to affect both PIK3IP1 expression and PI3K signaling. Consistently, mouse transposon tumors expressing ~50% Cux1 levels showed decreased expression of Pik3ip1 associated with higher phospho-Akt levels (Fig. 2g), and human SUP-T1 T-ALL cells, which harbor a heterozygous CUX1-truncating mutation, also displayed similar reciprocal alterations (Supplementary Fig. 10).
Figure 4. CUX1 directly regulates PIK3IP1 expression and CUX1 deficiency increases sensitivity to PI3K-AKT-mTOR inhibition.
(a) 4.3-kb genomic fragment upstream of PIK3IP1 transcriptional start site (+1) used for promoter luciferase assays. Black ovals, putative CUX1-binding sites (ATCAAT). A, B, C; primer-binding sites for qPCR in (c).
(b) Promoter luciferase assays in 293T cells using indicated PIK3IP1 promoter-luciferase constructs and CUX1 expression vectors. WT, wild-type; Mut, mutant; ΔHD, homeodomain-deletion mutant. Data were normalized to WT PIK3IP1 promoter + vector and represent mean + s.e.m. of 3 independent experiments assessed in triplicate. *** p < 0.001, t-test.
(c) ChIP-qPCR assays in LOUCY cells using indicated antibodies for regions shown in (a) (mean + s.e.m. of 3 independent experiments assessed in duplicate).
(d, e) Cell viability dose-response curves of KE37 shRNA vector-transduced cells (d) and human mesothelioma cell lines (e) treated with MK2206 or NVP-BEZ235. Data in (d) mean ± s.e.m. of triplicate experiments; (e) mean of duplicate experiments. IC50 for each cell line is shown.
Given that PI3K signaling is hyperactivated in CUX1-deficient cells we investigated the therapeutic merits of PI3K-AKT pathway inhibition. To minimise other genetic factors confounding response assessments, we evaluated the viability of KE37 cell lines, which were co-isogenic apart from carrying control or CUX1-shRNA knockdown vectors, exposed to a pan-AKT inhibitor (MK2206), or a dual PI3K/mTOR inhibitor (NVP-BEZ235). We also compared the responses to both compounds of H2803 mesothelioma cells, which harbor a focal homozygous deletion of CUX1 (Supplementary Fig. 11), with those of a panel of wild-type CUX1 mesothelioma cell lines. Remarkably, both CUX1-deficient cell lines showed increased sensitivity to drug treatment compared with controls, suggesting increased dependency on PI3K-AKT-mTOR signaling (Fig. 4d, e and Supplementary Fig. 12). Consistently, SUP-T1 cells were also more sensitive to MK2206 treatment compared with cells expressing higher CUX1 levels (Supplementary Fig. 10). Thus our data support an important role for CUX1 deficiency in PI3K signaling, but as CUX1 functions in multiple biological processes, other CUX1-mediated tumor suppressive mechanisms may also operate. Nonetheless, the susceptibility of CUX1-mutant cells to PI3K-AKT-mTOR pathway inhibition provides a new potential therapeutic avenue. Finally, while we identified CUX1 as a novel pan-driver of cancer in this study, our approach highlighted additional tumor suppressor candidates that warrant investigation in the future (Supplementary Note).
ONLINE METHODS
Collection of human cancer datasets
We curated freely available somatic mutations from three sources: the data portal of The Cancer Genome Atlas (TCGA); the data portal of the International Cancer Genome Consortium (ICGC), and from other published studies25-61. For all cancers used as part of our analysis, normal DNA had also been sequenced to establish the somatic origin of variants. Further, extensive filtering was performed to remove any residual germline mutations and technology-specific sequencing artifacts prior to analyzing the data. This involved filtering for germline mutations from dbSNP62, the 1000 genomes project63, and NHLBI GO Exome Sequencing Project64. Optimized minor allele frequency cut-offs were applied in order to avoid removing known driver mutations and variants were only excluded if they exactly matched a germline variant call. Technology-specific sequencing artifacts were filtered using a panel of matched BAM files (between 120-500 BAMs per technology) representing the sequence of unmatched germline controls. Any candidate somatic mutation present in at least three well-mapping reads in at least two normal BAM files was discarded. Curated somatic mutation sets were lifted between genomes using UCSC’s liftover tool. Ambiguously mapped calls were discarded.
Statistical analysis and human cancer datasets
To identify genes with a rate of missense, nonsense and splice site mutations higher than expected by chance, we used an adaptation of a previously described method24. Briefly, the rate of mutations is modelled as a Poisson process, with a rate given by a product of the mutation rate and the impact of selection. In particular, we use 12 parameters to describe the different rates of the 12 possible single nucleotide substitutions, 2 parameters to account for the CpG context-dependent effect on C>T transitions in each strand, and 3 selection parameters to measure the observed-over-expected ratio of missense (wMIS), nonsense (wNON) and essential splice site (wSPL) mutations. For example, the expected number of A>C missense mutations is modelled as λmissense,AtoC = (t)*(AtoC)*(wMIS)*(Lmissense,AtoC). “AtoC” refers to the relative rate of A>C transversions. “t” refers to the local mutation rate or the local density of mutations, which helps to control for gene-to-gene variations in coverage or in the background mutation rate. The inclusion of “t” makes one of the substitution rates unnecessary and so arbitrarily we fix GtoT = 1. “wMIS” represents the rate of missense mutation relative to synonymous mutations, and can be interpreted as the observed-over-expected ratio of missense mutations. “Lmissense,AtoC ” represents the number of sites that can suffer a missense A>C mutation in the gene sequence (thus accounting for the length and the sequence of the gene). The likelihood of observing a given number of missense A>C mutations in a particular gene (nmissense,AtoC), given the expected rate (λmissense,AtoC), is calculated as:
The likelihood of the entire model is the product of the individual likelihoods of each of the 14 mutation types:
This allows us to quantify separately the strength of selection at missense, nonsense and essential splice site mutations, avoiding the confounding effects of gene length, sequence composition and different rates of each substitution type. To obtain accurate estimates of the relative rates of each substitution type, the 14 rate parameters were estimated from the entire collection of mutations. These rates were then assumed constant and maximum-likelihood estimates for “t”, “wMIS”, “wNON” and “wSPLICE” were obtained for each gene. Likelihood Ratio Tests were then used to test deviations from neutrality (wMIS = 1, wNON = 1 or wSPL = 1). P-values were adjusted for multiple testing using Benjamini-Hochberg False Discovery Rate. In Fig. 1 only genes with wNON > 1 and q-value < 0.01 are shown, representing a set of candidate tumor suppressor genes.
Mouse strains and common insertion site (CIS) analysis
T2/Onc, CAGGS-SB11 and Mx1-Cre mouse strains were kindly provided by L. Collier, A. Uren and P. Liu respectively. The mice were on a predominantly C57BL/6J background. Mice were housed according to Home Office regulations (United Kingdom) and all procedures were performed with Home Office approval. At ~6 weeks of age, mice were randomly assigned to study groups and injected intraperitoneally with six doses of 250 μg pIpC (GE Healthcare) over two weeks to induce Cre recombinase expression. Controls were injected with PBS. Allocation to study groups was blinded. Genomic DNA was isolated from thymic tumors, bone marrow and spleens from sacrificed mice using a Puregene Core Kit A (Qiagen). To detect transposon mobilization, PCR was performed on genomic DNA using T2/onc-Excision-F and T2/onc-Excision-R primers. Oligonucleotides used in this study are listed in Supplementary Table 9. Splinkerette PCR and CIS analysis were performed as described previously13. Chromosome 1, harboring the transposon concatamer, was excluded from CIS analysis to account for local hopping.
RT-PCR detection of Cux1-T2/Onc and Pten-T2/Onc fusion mRNA in mouse transposon tumors
RNA was prepared from frozen suspensions of transposon tumors using an RNeasy Mini kit (Qiagen) and reverse transcribed into cDNA using Superscript III (Invitrogen) primed with random hexamers. PCR was performed using primer pairs mCuxEX2F/UnivRevT2onc1, mCux1ex8/UnivRevT2onc1 or mCuxEX19F/carpSA1 to detect Cux1-T2/Onc fusion transcripts. Primers mPtenex2 and CarpSA1 were used to detect Pten-T2/Onc fusion mRNA. PCR products were cloned using a TOPO TA cloning kit (Invitrogen) and individual clones were sequenced.
Mouse pathology
Air-dried blood smears were stained with MGG (Sigma). Mouse tissues were fixed in formalin and embedded in paraffin. Blocks were cut into 5 μm sections and stained with haematoxylin/eosin. For immunohistochemistry, antigen retrieval was performed by microwaving sections in 10 mM citric acid (pH 6.0). Endogenous peroxidases were quenched with 3% H2O2/PBS for 30 min. Sections were incubated with primary antibodies diluted in TBST for 1 h at room temperature. Primary antibodies included anti-CD3 (1:100; Dako, A0452), anti-Tdt (1:50; Dako, A3524), anti-PTEN (1:200; Cell Signaling, 9559) and anti-CUX1 (1:100; Santa Cruz, sc-6327). Following TBST washes, signal detection was accomplished using an Elite Vectastain ABC kit and DAB (Vector Laboratories). Sections were counterstained with haematoxylin before dehydration and mounting. Slide images were captured using a Nanozoomer digital slide scanner (Hamamatsu).
TCRβ gene rearrangement
PCR was performed on genomic tumor DNA using primers DB2.1 and JB2.7.
Drosophila cancer model
The mild enlarged eye phenotype produced by overexpression of the Notch ligand Delta (eyeless-Gal4 UAS-Dl) provides a sensitized background to identify tumor suppressors in Drosophila65. The effects of reducing cut expression were assayed by examining phenotypes in adult progeny (reared at 30°C from 48 h after egg lay) from eyeless-Gal4 UAS-Dl and UAS-cutRNAi (ref. 66) fly crosses. To investigate consequences of cut knock-down in Drosophila blood cells, lymph glands were dissected from pxnGal4/+; UAS-CD8GFP/UAS-cutRNAi larvae or control pxnGal4/+; UAS-CD8GFP/UAS-whiteRNAi (progeny were cultured at 30°C from 24-48 h onwards) and processed for immunofluorescence as described previously67. Primary antibodies were mouse anti-Lozenge (1:20; DSHB/Hybridoma Bank), goat anti-GFP (8:300; Abcam, ab6673). Secondary antibodies were FITC anti-goat (1:200) and Cy3 anti-mouse (1:100) from Jackson ImmunoResearch. Samples were mounted using Vectashield mounting medium containing DAPI and imaged using a Nikon D-ECLIPSE C1. Pupae and adult flies from the same genotypes were also scored for the presence of melanotic tumors (detectable as pigmented masses present beneath the cuticle of living specimens).
Gene expression profiling
RNA was extracted from siRNA-treated cells from 3 independent knockdown experiments using an RNeasy Mini Kit (Qiagen) and processed for transcriptome profiling using a HumanHT-12 v4 Expression BeadChip Kit (Illumina). Raw probe data were transformed into log-scale expression values using variance stabilization transformation68 and quantile normalized. Differentially expressed genes were identified by fitting an individual linear model for each gene across different sample groups69. GO analysis was performed using DAVID70.
Quantitative PCR
RNA was extracted from cell lines or mouse tumors using an RNeasy Mini kit (Qiagen). Contaminating DNA was digested using TURBO DNase (Ambion). Total RNA was reversed transcribed using RNA to cDNA EcoDry Premix (Clontech). Quantitative reverse transcriptase PCR (qRT-PCR) was carried out using SYBR-Green PCR Master Mix (Applied Biosystems) on a 7900HT Real-Time PCR machine (Applied Biosystems). Gene expression was normalized to GAPDH expression. A ΔΔCt loss of copy number approach was used to determine the burden of Cux1 transposon insertions within a given tumor sample. Quantitative PCR was performed on wild-type and tumor genomic DNA samples using primers flanking individual transposon insertion sites. The Sh2b2 gene, amplified with mSh2b2EX2QFand mSh2b2EX2QR primers, was used as a reference. The insertion specific primers used are shown in Supplementary Table 9.
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was performed to detect Cux1I20 mRNA species in mouse tumors using primers as described7. Gapdh expression was used as a control.
Immunoblotting
Cells were washed in PBS before resuspension in Cellytic (Sigma) supplemented with protease/phosphatase inhibitor cocktail (Cell Signaling). Cells were lysed using a TissueLyser LT (Qiagen). Samples were centrifuged at 13,000 rpm for 10 min to pellet insoluble debris. Protein concentrations were determined by Bradford assay. Equal amounts of protein were resolved on 4-12% Bis-Tris SDS-PAGE gels (Novex) and transferred to nitrocellulose membranes (GE Healthcare). Immunoblotting was performed according to standard techniques using primary antibodies including anti-CUX1 (ab73885, Abcam; sc-13024, Santa Cruz; ABE218, Millipore), anti-PIK3IP1 (16826-1-AP, Proteintech), anti-CASP (11733-1-AP, Proteintech), anti-phospho-AKTT308 (9275, Cell Signaling), anti-phospho-AKTS473 (9271, Cell Signaling), anti-AKT (9272, Cell Signaling), anti-phospho-GSK-α/β (9331, Cell Signaling), anti-GSK-α/β (5676, Cell Signaling), anti-phospho-RPS6 (2211, Cell Signaling), anti-RPS6 (2217, Cell Signaling), anti-GAPDH (2118, Cell Signaling) and anti-myc (2276, Cell Signaling). HRP-linked secondary antibodies were from Cell Signaling and Abcam. Chemiluminescent detection was performed using ECL Prime reagent (GE Healthcare).
Plasmids, shRNAs and siRNA
Oligonucleotides for shRNA knockdowns were cloned into PLKO.pig. For siRNA experiments, LOUCY cells were resuspended in Opti-MEM (Gibco) and electroporated in 4 mm cuvettes (300 V, 1000 μF; Genepulser XCell, Bio-Rad) using Stealth siRNAs (Ambion) against CUX1, PIK3IP1 or negative control duplexes. Cells were rested for 15 min following electroporation before dilution into normal growth media. Cells were processed for downstream analysis 48-72 h later. For PIK3IP1 expression, the PIK3IP1 coding sequence was amplified and myc-tagged by PCR using PIKRIf and PIKmycBglR using IMAGE clone 4180134 as template. The PCR product was digested with EcoRI and BglII before cloning into MSCV-neo (Clontech) digested with the same enzymes. All vectors were verified by sequencing.
Cell culture
LOUCY and KE37 human T-ALL cells originated from DSMZ and were grown at 37°C and 5% CO2 in RPMI supplemented with 20% or 10% fetal bovine serum (FBS) respectively, glutamine and antibiotics. 293T cells, which originated from ATCC, and Phoenix cells (from Clontech) were grown in DMEM with 10% FBS, glutamine and antibiotics. The identity of cell lines was confirmed prior to use, and all cell lines were mycoplasma tested before entering the laboratory. Cell lines injected into mice received a complete MAP/pathogen test.
Virus production and infection
For lentivirus production, HEK293T cells were seeded onto 15 cm plates and incubated overnight before calcium phosphate-mediated transfection with pLKO.pig shRNA vectors, pMDLg/prre, pRSVRev and pMD2-VSV-G. After 16 h, 5 mM sodium butyrate (Sigma) was added to maximize virus production. For retrovirus production, 5 × 106 Phoenix-Ampho cells were incubated overnight on 10 cm plates and transfected with MSCV expression vectors using calcium phosphate precipitation. Viral supernatants were harvested 48 h post transfection and passed through 0.45 μm filters before infection of target cells or storage at −80°C. Target cells were infected by incubation for 4-8 h with viral supernatant in the presence of 4 μg/ml polybrene. Cells transduced with PLKO.pig and MSCV vectors were selected with 6 μg/ml puromycin and 1 mg/ml G418 respectively.
Glucose uptake assay
Cells were washed in PBS and resuspended in glucose-free RPMI media (Gibco). 2.5 × 104 cells in 100 μl were dispensed in triplicate into 96 well plates. After 30 min, 100 μl glucose-free RPMI supplemented with 20% FBS and 2-NBD glucose (2-, deoxy-2-[(7-,nitro-2,1,3-benzoxadiazol-4-yl)amino]-D-glucose) was added to the cells. Glucose uptake was determined by fluorescence readings after 15 min according to Glucose Uptake Kit (Cayman) protocol.
Drug treatment assays
1 × 104 KE37 cells transduced with shRNA vectors were dispensed into 96 well plates in triplicate. Decreasing doses of MK2206 or NVP-BEZ235 were added to achieve final dose spectrum from 10 μM to 0 μM. Cells were incubated for 48 h before assessment of cell viability using CellTiter-Blue reagent (Promega). Dose-response curves were generated using Prism 5 software (GraphPad). For NVP-BEZ235 and MK2206 dose response curves on mesothelioma cell lines, cell viability was assessed 72 h after drug treatment using a previously described protocol71.
Tumor xenograft assay
3 × 106 cells in 100 μl of PBS were injected subcutaneously into the left flank of NOD-SCID mice (Jackson Laboratory). Mice were monitored daily for tumor development and the daily size of each tumor was measured using calipers. Tumor volume = 1/2 (length × width2). Mice were sacrificed after 3 weeks and necropsies were performed. Mice were ~6-weeks-old at the time of injection and were randomly assigned to each study group. Allocation to study groups was blinded.
PIK3IP1 promoter luciferase assay
To create PIK3IP1 promoter luciferase vectors, the genomic region upstream of the PIK3IP1 transcriptional start site was amplified by PCR using PIKpromSacF and PIKpromXhoR. The PCR product was digested with SacI and XhoI before cloning into pGL4.10[luc2] (Promega) digested with the same enzymes, creating pGL4.10-PIK3IP1promWT. The 2 putative CUX1-binding sites were mutated in pGL4.10-PIK3IP1promWT by 2 rounds of site-directed mutagenesis using PIKmut1F and PIKmut1R followed by PIKmut2F and PIKmut2R, leading to pGL4.10-PIK3IP1promMUT. 293T cells were seeded into 24 well plates the evening before transfection. Cells were transfected with empty vector (pXJ42), CUX1p110 (pXJ myc-CUX1p110-HA) or CUX1p110ΔHD (pXJ myc-CUX1p110ΔHD-HA) expression vectors along with pGL4.10-PIK3IP1promWT or pGL4.10-PIK3IP1promMUT and control Renilla luciferase expression vector (pGL4.75[hRluc/CMV]; Promega) using lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. Luciferase activity was assessed 24 h later using Dual-Luciferase Reporter Assay System (Promega). Transfections were performed in triplicate on three separate occasions. Luciferase signals were normalized to Renilla and empty vector signals. The CUX1 homeodomain truncated mutant (CUX1p110ΔHD) was generated by PCR using CUXmyc110f and CUXdHDHAr on pXJ myc-CUX1p110-HA. The PCR product was cloned into pXJ42. Vectors were confirmed by sequencing.
Chromatin immunoprecipitation (ChIP)
ChIP was performed on LOUCY cells using a Magna ChIP A/G Chromatin Immunoprecipitation Kit (Millipore) with minor modifications. 5 × 107 formaldehyde-fixed cells were treated with 0.125 M glycine before resuspension in 1 ml lysis buffer (5 mM Tris-HCl pH 8.0, 85 mM KCl, 1% NP40 and protease inhibitors [Roche]). After 10 min on ice, the nuclei were pelleted and resuspended in 0.5 ml nuclear lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.3% SDS and protease inhibitors [Roche]) before 20 cycles of 30 s on/off sonication using a Bioruptor Plus device (Diagenode). Insoluble material was removed by centrifugation. An aliquot of chromatin was used to determine satisfactory sonication and DNA concentration after cross-link reversal. 100 μg chromatin was diluted in dilution buffer (16.7 mM Tris-HCl pH 8.0, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100 and protease inhibitors [Roche]) before ChIP according to the kit protocol. Antibodies used for ChIP included anti-CUX1 (sc-6327X, Santa Cruz) and normal goat IgG (sc-2028, Santa Cruz). ChIP DNA samples were analyzed by qPCR using SYBR-Green PCR Master Mix (Applied Biosystems) on a 7900HT Real-Time PCR machine (Applied Biosystems). PIK3IP1 locus primers at sites A were PIKChIP1f/PIKChIP1r; B, PIKChIP2f/PIKChIP2r and C, PIKChIPcf/PIKChIPcr.
Statistical analyses
Student’s two-tailed unpaired t-tests were performed to assess statistical significance in pairwise comparisons. A log-rank (Mantel-Cox) test was performed on mouse transposon survival curves to determine significance. To evaluate the prognostic impact of CUX1-truncating mutations in the MDS and MDS/MPN cohorts, we used Cox proportional hazards models. We included correction for age and WHO subtype of disease. Overall survival was the end-point of interest. Survival analysis was restricted to cases with available outcome data. Likelihood ratio tests were used to assess the impact of CUX1 truncations and −7/del(7q) lesions on survival. To generate survival curves plotted in Fig. 1f, we used the Nelson-Aalen-Breslow estimator of hazard, with CUX1 and −7/del(7q) treated as strata, after correction for confounding variables as above.
Patient and cell line DNA sequencing
We performed a meta-analysis of sequencing studies that covered all CUX1 coding exons in myeloid malignancies. The three cohorts studied included: 1) exomes from a cohort of 151 patients with myeloproliferative neoplasms; 2) targeted sequencing of 111 genes in 738 patients with MDS and MDS/MPN overlap conditions9; 3) targeted sequencing of 111 genes in 1,630 patients with AML enrolled in the German AML Study Group trials 07-04 [ClinicalTrials.gov Identifier: NCT00151242] and HD98A72. Bioinformatic analysis of the sequencing data for the exomes was performed as previously published9. Human T-ALL cell lines (BE13, CCRF-CEM, CML-T1, KARPAS45, KE37, LOUCY, SUP-T1 and PF382) were sequenced by standard capillary techniques. Primer sequences are available on request.
Supplementary Material
ACKNOWLEDGEMENTS
The PLKO.pig vector was a kind gift from I. Lemischka. Plasmids pXJ42 and pXJ myc-CUX1p110-HA were kindly provided by A. Nepveu. We thank D. Pask, J. Carter and T. Hamilton for mouse husbandry. C.C.W and D.J.A are supported by Cancer Research UK and the Wellcome Trust. S.J.B. and S.L. are supported by the Medical Research Council. Work in the A.R.G. lab is supported by Leukemia and Lymphoma Research, Cancer Research UK, the Kay Kendall Leukaemia Fund, the NIHR Cambridge Biomedical Research Centre, the Cambridge Experimental Cancer Medicine Centre, and the Leukemia & Lymphoma Society of America. I.M. is supported by an EMBO Long-Term Fellowship. J.N. is supported by a Kay Kendall Leukaemia Fund Clinical Fellowship. U.M. is supported by a Cancer Research UK Clinician Scientist Fellowship.
Footnotes
Accession codes: Microarray data have been deposited in the ArrayExpress database under E-MTAB-1833 accession number.
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
REFERENCES
- 1.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 2.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neufeld EJ, Skalnik DG, Lievens PM, Orkin SH. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat. Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 4.Hulea L, Nepveu A. CUX1 transcription factors: from biochemical activities and cell-based assays to mouse models and human diseases. Gene. 2012;497:18–26. doi: 10.1016/j.gene.2012.01.039. [DOI] [PubMed] [Google Scholar]
- 5.Vadnais C, et al. CUX1 transcription factor is required for optimal ATM/ATR-mediated responses to DNA damage. Nucleic Acids Res. 2012;40:4483–4495. doi: 10.1093/nar/gks041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moon NS, et al. S phase-specific proteolytic cleavage is required to activate stable DNA binding by the CDP/Cut homeodomain protein. Mol. Cell. Biol. 2001;21:6332–6345. doi: 10.1128/MCB.21.18.6332-6345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goulet B, et al. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res. 2002;62:6625–6633. [PubMed] [Google Scholar]
- 8.McNerney ME, et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood. 2013;121:975–983. doi: 10.1182/blood-2012-04-426965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013 doi: 10.1182/blood-2013-08-518886. doi:10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klampfl T, et al. Genome integrity of myeloproliferative neoplasms in chronic phase and during disease progression. Blood. 2011;118:167–176. doi: 10.1182/blood-2011-01-331678. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg PL, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinclair AM, et al. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. 2001;98:3658–3667. doi: 10.1182/blood.v98.13.3658. [DOI] [PubMed] [Google Scholar]
- 13.March HN, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat. Genet. 2011;43:1202–1209. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 15.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 17.Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat Rev Cancer. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- 18.Zhai Z, et al. Antagonistic regulation of apoptosis and differentiation by the Cut transcription factor represents a tumor-suppressing mechanism in Drosophila. PLoS Genet. 2012;8:e1002582. doi: 10.1371/journal.pgen.1002582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, et al. PIK3IP1, a Negative Regulator of PI3K, Suppresses the Development of Hepatocellular Carcinoma. Cancer Res. 2008;68:5591–5598. doi: 10.1158/0008-5472.CAN-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z, et al. PI3K is negatively regulated by PIK3IP1, a novel p110 interacting protein. Biochemical and Biophysical Research Communications. 2007;358:66–72. doi: 10.1016/j.bbrc.2007.04.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrés V, Chiara MD, Mahdavi V. A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 1994;8:245–257. doi: 10.1101/gad.8.2.245. [DOI] [PubMed] [Google Scholar]
- 22.B Aufiero, E. J. N. S. H. O. Sequence-specific DNA binding of individual cut repeats of the human CCAAT displacement/cut homeodomain protein. Proc. Natl. Acad. Sci. U.S.A. 1994;91:7757. doi: 10.1073/pnas.91.16.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harada R, Bérubé G, Tamplin OJ, Denis-Larose C, Nepveu A. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol. Cell. Biol. 1995;15:129–140. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenman C, Wooster R, Futreal PA, Stratton MR, Easton DF. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics. 2006;173:2187–2198. doi: 10.1534/genetics.105.044677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 32.Govindan R, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiao Y, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones DTW, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Gallo M, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 2012;44:1310–1315. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, et al. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res. 2012;22:2315–2327. doi: 10.1101/gr.140988.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu P, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis. 2012;33:1270–1276. doi: 10.1093/carcin/bgs148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peifer M, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudin CM, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sausen M, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat. Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seo J-S, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah SP, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 56.Wei X, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu J, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc. Natl. Acad. Sci. U.S.A. 2011;108:21188–21193. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zang ZJ, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat. Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 60.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferres-Marco D, et al. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- 66.Grueber WB, Jan LY, Jan YN. Different Levels of the Homeodomain Protein Cut Regulate Distinct Dendrite Branching Patterns of Drosophila Multidendritic Neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- 67.Terriente-Felix A, et al. Notch cooperates with Lozenge/Runx to lock haemocytes into a differentiation programme. Development. 2013;140:926–937. doi: 10.1242/dev.086785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2007;36:e11–e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2007. pp. 397–420. [Google Scholar]
- 70.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 71.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlenk RF, et al. Prospective Evaluation of Allogeneic Hematopoietic Stem-Cell Transplantation From Matched Related and Matched Unrelated Donors in Younger Adults With High-Risk Acute Myeloid Leukemia: German-Austrian Trial AMLHD98A. J. Clin. Oncol. 2010;28:4642–4648. doi: 10.1200/JCO.2010.28.6856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.