Abstract
Background
Sinonasal undifferentiated carcinoma remains a poorly characterized malignancy at both the clinical and molecular level, and consequently the optimal treatment strategy remains undefined.
Methods
We utilized a mass spectroscopy-based approach (Sequenom™) to evaluate 95 hallmark single nucleotide variations within 12 oncogenes or tumor suppressor genes (AKT, BRAF, CDK4, Beta-catenin, EGFR, FBXW7, JAK2, c-KIT, KRAS, PDGFR, PI3K, VEGF) in 13 histologically confirmed SNUC cases.
Results
None of the samples demonstrated activating mutations in any of the 95 SNVs.
Conclusions
Select clinically relevant activating genomic mutations were not identified the 13 patient samples. However, polymorphisms were noted within the promoter region of VEGF. These may merit future study as predictive biomarkers for treatment response or overall survival. Additionally, future studies focusing on larger tumor sets and utilizing whole genome or exome sequencing may help define genetic aberrations in SNUC that can be clinically targeted with available or emerging biological agents.
Keywords: SNUC, Sinonasal Undifferentiated Carcinoma, Paranasal sinus tumors, VEGF, Sequenom
Introduction
Sinonasal undifferentiated carcinoma (SNUC) remains a poorly characterized malignancy at both the clinical and molecular level. It was first described as a unique clinical entity arising in the nasal cavity and paranasal sinuses by Frierson et al in 1986(1). Although of uncertain histogenesis, SNUC features unique clinicopathologic characteristics permitting its segregation from other types of epithelial and non-epithelial neoplasms within the sinonasal tract. It commonly presents with the rapid onset of epistaxis, nasal obstruction, proptosis, vision changes, and pain. It is defined by both an aggressive biologic phenotype with locally advanced disease on presentation(2–6) and characteristic histologic features(6–9).
In large part due to the rarity of the disease, the optimal treatment strategy for SNUC remains undefined. While nearly all studies show a survival benefit from multimodality therapy(2, 3, 10, 11), the timing of surgery, radiation, and chemotherapy along with the choice of chemotherapeutic agents remains unresolved and inconsistent across large tertiary referral centers(12). Additionally, despite aggressive multimodality therapy, the two-year survival rates in reported series to date range from 25% to 67% (2, 3, 10, 12).
Previous reports from studies of more common malignancies such as lung, breast, and melanoma have documented key mutations within oncogenes or tumor suppressor genes that increase susceptibility to molecularly targeted therapeutics(13). In this study we sought to determine if SNUC tumors harbored previously identified hotspot mutations within 12 oncogenes or tumor suppressor genes (AKT, BRAF, CDK4, Beta-catenin, EGFR, FBXW7, JAK2, c-KIT, KRAS, PDGFRA, PI3K) or single nucleotide polymorphisms (SNPs) in VEGF that might guide the selection of available molecularly targeted therapeutics.
Methods
Histology
Paraffin-embedded clinical specimens were obtained from the Head & Neck Surgery Tissue Resource and Pathology Core at The University of Texas MD Anderson Cancer Center under an Institutional Review Board approved protocol with the explicit informed consent of the research subjects. The diagnosis of SNUC was confirmed with H&E staining and a panel of immunohistochemical markers as previously described(8). 13 biologically distinct samples were derived from 12 patients treated at MDACC between 1996 and 2010. Patients 2, 3, 5, 6 and 7 had received initial induction chemotherapy prior to tissue acquisition. Patients 1, 4, 9, 10, 11, 12 had received radiation along with chemotherapy prior to tissue acquisition. Patient 8 had received no treatment prior to tissue acquisition.
Transmission Electron Microscopy
Transmission electron microscopy was performed by the High Resolution Electron Microscopy Facility at the MD Anderson Cancer Center. A sample was taken from central portion of a solid tumor from the sinonasal cavity specimen immediately after surgical extirpation and was preserved in a solution containing 3% glutaraldehyde, 2% formaldehyde, and 0.1 M cacodylate (pH 7.3). Ultrathin sections were cut with an LKB Ultracut microtome (Leica, Deerfield, IL, USA), stained with uranyl acetate and lead citrate in an LKB Ultrostainer, and examined with a JEM 1010 transmission electron microscope (JEOL, Peabody, MA) at an accelerating voltage of 80 kV. Digital images were obtained using the AMT imaging system (Advanced Microscopy Techniques Corp., Danvers, MA). EM analysis was supported by grant CA 16672 for the MDACC electron microscopy core facility.
DNA isolation
Ten- to twenty-micrometre sections were prepared and microscopic evaluation by a experienced pathologist confirmed tumor content in the section used. Genomic DNA was then isolated using the QIAmp DNA Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s guidelines by the Biospecimen Extraction Core Facility at MDACC, also supported by CA 16672.
Single Nucleotide Polymorphism Profiling
A mass spectroscopy-based approach evaluating SNVs and SNPs was used to detect 95 hallmark mutations within 12 oncogenes or tumor suppressor (TS) genes (Table 2.) as previously described(14). In brief, polymerase chain reaction (PCR) and extension primers were designed using Sequenom Inc. (San Diego, CA, USA) Assay Design. PCR-amplified DNA was cleaned using EXO-SAP (Sequenom), primer extended by IPLEX chemistry, desalted using Clean Resin (Sequenom), and spotted onto Spectrochip matrix chips using a nanodispenser (Samsung). Chips were run in duplicate on a SequenomMassArray MALDI-TOF MassArray system. SequenomTyper Software and visual inspection were used to interpret mass spectra. Reactions where more than 15% of the resultant mass ran in the mutant site in both reactions were scored as positive.
Table 2.
Single nucleotide variations (SNVs) in 95 hallmark mutations within 12 oncogenes or tumor suppressor genes (AKT, BRAF, CDK4, Beta-catenin, EGFR, FBXW7, JAK2, c-KIT, KRAS, PDGFRA, PI3K, VEGF) probed in this experiment.
| BRAF |
| BRAF_D594_1781A |
| BRAF_E586K_1756GA_Slp |
| BRAF_G464_1391G |
| BRAF_G466_1397G |
| BRAF_G466R_1396_GC |
| BRAF_G469_1407A |
| BRAF_K601E_AG |
| BRAF_K601N_A |
| BRAF_L597R_1790TG |
| BRAF_V600_1798G_1 |
| BRAF_V600_1799T_1 |
| BRAF_V600_1799T_2 |
| BRAF_V600_1800G |
| KRAS |
| KRAS_A146_436 |
| KRAS_G12_34G |
| KRAS_G12_35G |
| KRAS_G12_36T |
| KRAS_G13_37G |
| KRAS_G13_38G |
| KRAS_G13_39 |
| KRAS_Q61_181C |
| KRAS_Q61_182A |
| KRAS_Q61_183A |
| Beta-catenin |
| CTNNB1_D32_94G |
| CTNNB1_D32_95A |
| CTNNB1_G34_101G |
| CTNNB1_S33_97T |
| CTNNB1_S37_109T |
| CTNNB1_S37_110C |
| CTNNB1_S45_133T |
| CTNNB1_S45_134C |
| CTNNB1_T41_121A |
| EGFR |
| EGFR_G719_G2155 |
| EGFR_L858R_TG |
| EGFR_T790M_C2369T_Spl |
| VEGF |
| VEGF_5_1154_GA_ref |
| VEGF_5_1498_CT |
| VEGF_5_2573_CA |
| VEGF_5_634_GC_ref |
| cKIT |
| KIT_D816H_GC |
| KIT_D816V_AT |
| KIT_K642E_AG |
| KIT_L576P_TC |
| KIT_N556D_AG |
| KIT_R634W_CT |
| KIT_V559_T |
| KIT_V560D_TA_SPLICE |
| KIT_V825A_TC |
| KIT_Y553N_TA |
| PDGFR A |
| PDGFRA_D842_A2525T |
| PDGFRA_D842_G2524 |
| PDGFRA_E996K_G2986 |
| PDGFRA_N659K_C1977A |
| PDGFRA_N659Y_A1975T |
| PDGFRA_V561D_T1682A |
| PDGFRA_V824L_G2470C |
| FBXW7 |
| FBWX7_R465C_C1393T |
| FBWX7_R465H_G1394A |
| FBWX7_R479_G1436 |
| FBWX7_R505_G1514 |
| FBWX7_R505C_C1513T |
| FBWX7_S582L_C1745T |
| FBXW7_H460R_A1379G |
| PI3K |
| PIK3CA_A1046V |
| PIK3CA_C420R |
| PIK3CA_E110K |
| PIK3CA_E418K |
| PIK3CA_E453K |
| PIK3CA_E542_1624G |
| PIK3CA_E545_1633G |
| PIK3CA_E545_1634A |
| PIK3CA_E545_1635G |
| PIK3CA_F909L |
| PIK3CA_G1049R |
| PIK3CA_H1047 |
| PIK3CA_H1047_1 |
| PIK3CA_H1047Y |
| PIK3CA_H701P |
| PIK3CA_K111N |
| PIK3CA_M1043V |
| PIK3CA_N345K |
| PIK3CA_P539R |
| PIK3CA_Q060K |
| PIK3CA_Q546_1636C |
| PIK3CA_Q546_1637A |
| PIK3CA_R088Q |
| PIK3CA_S405F |
| PIK3CA_T1025_3073A |
| PIK3CA_Y1021_3061T |
| PIK3CA_Y1021C_3062 |
| AKT |
| AKT1_E17K_G49A |
| AKT2_E17K_G49A |
| AKT3_E17K_G49K |
| JAK2 |
| JAK2_V617F_G1849T |
| cDK4 |
| CDK4_R24C_C70T |
Results
Histology
Microscopic diagnosis of the SNUC tumors specimens was confirmed on the basis of:
Light microscopic evaluation in which the hematoxylin and eosin stained SNUC specimens demonstrated characteristic hypercellular proliferation with a trabecular growth pattern, medium to large pleomorphic and hyperchromatic nuclei, inconspicuous to prominent nucleoli, varying amount of eosinophilic cytoplasm, and a high nuclear-to-cytoplasmic ratio.
Immunohistochemistry: positive for pancytokeratin, and negative for synaptophysin, lymphoid markers, S100, and HMB45.
Transmission Electron Microscopy
Electron microscopy demonstrates undifferentiated polygonal cells with sparse intracellular membrane structures; and numerous polyribosomes, mitochondria (figure 1; insert A.), and abundant lipid-filled vacuoles. Occasional tonofilaments and microtubules are visible (figure 1; insert B.), along with membrane-bound, dense-core, neurosecretory granules (figure 1; insert C.). These findings appear similar to other limited reports on the ultrastructural features of SNUC(1).
Figure 1.
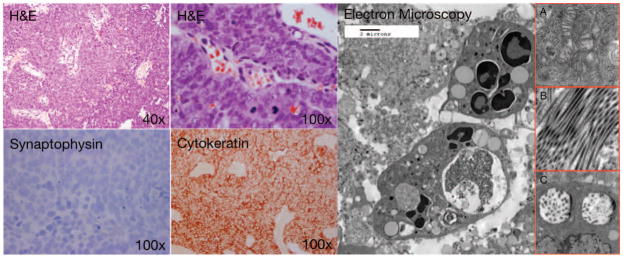
Histological examination of SNUC after H&E staining reveals characteristic hypercellular proliferation with a trabecular growth pattern, medium to large pleomorphic and hyperchromatic nuclei, inconspicuous to prominent nucleoli, varying amount of eosinophilic cytoplasm, high nuclear-to-cytoplasmic ratio. Immunohistochemistry is negative for synaptophysin, positive for pancytokeratin, negative for lymphoid markers (not shown), S100 (not shown), HMB45 (not shown). Electron microscopy demonstrates undifferentiated polygonal cells with sparse intracellular membrane structures; and numerous polyribosomes, mitochondria (figure 1, insert A.), and abundant lipid-filled vacuoles. Occasional tonofilaments and microtubules (figure 1, insert B.), are visible, along with membrane-bound, dense-core, neurosecretory granules (figure 1, insert C.),
Patient Demographics
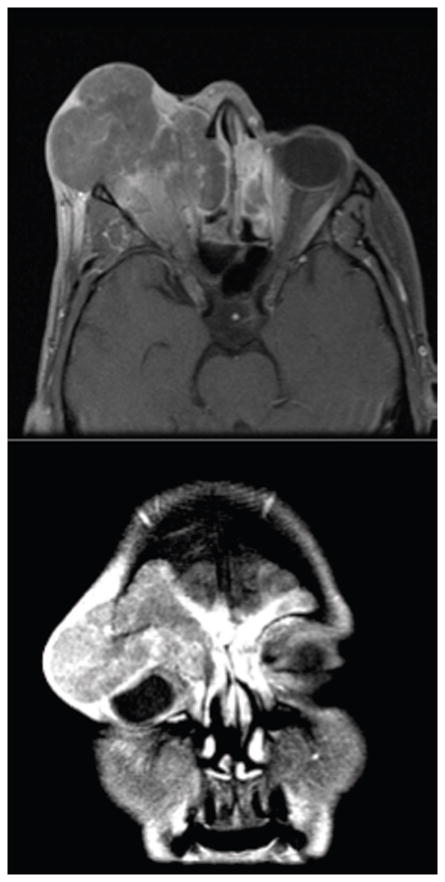
The demographics (Table 1.) show a relatively even breakdown between men (7) and women (5). The mean age at presentation was 55 years with a large range (30–83). The initial therapy was quite varied and demonstrates the divergent approaches to treatment seen at many outside institutions prior to their presentation at MDACC. The vast majority of disease was advanced on presentation. 11 of 12 patients presented with disease involving the orbit, orbital apex, dura, brain, and/or skin. Axial and coronal computed tomography images from a representative patient illustrate the extent of surrounding structural involvement (Figure 2.).
Table 1.
The demographics show a relatively even breakdown between men (7) and women (5). The mean age at presentation was 55 years with a large range (30–83). The initial therapy was quite varied and demonstrates the divergent approaches to treatment seen at many outside institutions prior to their presentation at MDACC. The vast majority of disease was advanced on presentation (T4). 11 of 12 patients presented with disease involving the orbit, orbital apex, dura, brain, and/or skin.
| Patient Characteristics | # (%) |
|---|---|
| Gender | |
| Male | 7 (58) |
| Female | 5 (42) |
| Age | |
| Mean age in years | 55.6 |
| Range in years | 30–83 |
| Initial Therapy | |
| Radiation | 2 (16) |
| Surgery | 2 (16) |
| Chemotherapy | 6 (50) |
| Unknown | 2 (16) |
| T and N classification
| |||||
|---|---|---|---|---|---|
| N0 | N1 | N2 | N3 | Total | |
| T3 | 1 | 0 | 0 | 0 | 1 |
| T4 | 10 | 0 | 1 | 0 | 11 |
| Total | 11 | 0 | 1 | 0 | 12 |
Figure 2.
The vast majority of disease was advanced on presentation. 11 of 12 patients presented with advanced disease involving the orbit, orbital apex, dura, brain, and/or skin. Axial and coronal computed tomography images from a representative patient illustrate the extent of surrounding structural involvement.
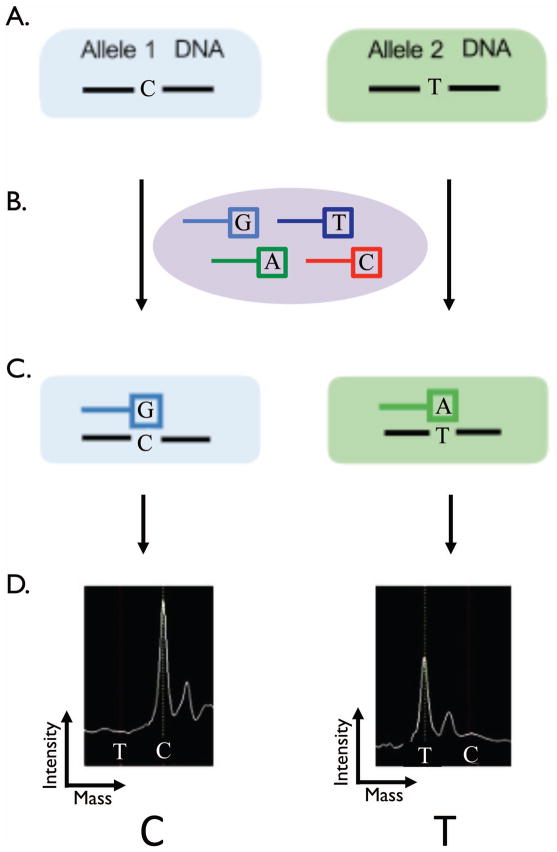
Assay Design
DNA was harvested from ten- to twenty-micrometre sections of paraffin embedded tumor. Figure 3 illustrates the principles of the assay. The Sequenom™ assay was preformed as previously described(14). In brief, the assay interrogates predefined single nucleotide base changes (Figure 3A.). The assays begins with and initial locus-specific PCR reaction, followed by single base extension using mass-modified dideoxynucleotide terminators of an oligonucleotide primer (ddATP*, ddGTP*, ddCTP*, ddTTP*) which anneals immediately upstream of the polymorphic site of interest (Figure 3B.). Allele-specific products with distinct masses are shown in (Figure 3C). Using MALDI-TOF mass spectrometry (Matrix-assisted laser desorption/ionization – time of flight) the distinct mass of the extended primer identifies the base change allele (Figure 3D.).
Figure 3.
The Sequenom™ assay interrogates predefined gene single nucleotide polymorphisms (A.). The assays begins with and initial locus-specific PCR reaction, followed by single base extension using mass-modified dideoxynucleotide terminators of an oligonucleotide primer (ddATP*, ddGTP*, ddCTP*, ddTTP*) which anneals immediately upstream of the polymorphic site of interest (B.). Allele-specific products with distinct masses (C.). Using MALDI-TOF mass spectrometry (Matrix-assisted laser desorption/ionization – time of flight) the distinct mass of the extended primer identifies the SNP allele (D.).
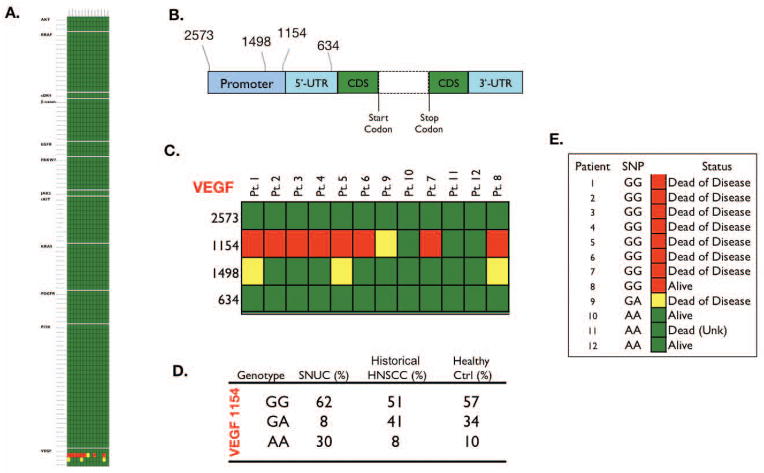
Genotyping
Table 2 lists the 95 SNVs probed in this experiment. None of the 13 samples demonstrated known activating mutations in any of the 95 SNVs, although nucleotide polymorphisms were noted at positions 1154 and 1498 in the promoter region of VEGF (Figure 4C). VEGF 1154 genotype frequencies of SNUC patients contrasted with a group of previously described(15) 100 HNSCC and healthy controls (Figure 4D.) showed a higher percentage of the GG and AA genotypes when compared with both a group of 100 H&NSCC, and a group of 100 normal controls. VEGF 1154 SNP genotypes and survival of 12 SNUC patients (Figure 4E.). Of the 3 patients alive at last contact, 2 possessed the historically protective AA genotype.
Figure 4.
None of the 13 samples demonstrated known activating mutations in any of the 95 SNVs (Figure 4A.). Structure of VEGF gene and position of SNPs relative to translation start site, dashed lines indicate the region consisting of coding sequence (CDS) and seven introns, UTR: untranslated region (Figure 4B.). Polymorphisms were noted in genotype 1154 and 1498 of VEGF (Figure 4C.). VEGF 1154 genotype frequencies of SNUC patients contrasted with a group of 100 HNSCC and healthy controls (Figure 4D.). VEGF 1154 SNP genotypes and survival of 12 SNUC patients (Figure 4E.). Of the 3 patients alive at last contact, 2 possessed the historically protective AA genotype.
Discussion
Many malignancies contain key mutations within oncogenes or tumor suppressor genes that increase susceptibility to targeted molecular therapeutics (13). KIT mutations in gastrointestinal stromal tumors (GISTs) (16), Her2/neu mutations in breast cancer (17), EGFR mutations in NSCLCs(18) and BRAF mutations in melanoma (19) mark some of the most convincing clinical examples. Conversely, some mutations portend targeted therapy failure. Inactivating PTEN mutations in glioblastomas predict resistance to erlotinib(20) and colorectal cancers harboring KRAS mutations are unresponsive to treatment with EGFR inhibitors(21).
Although little is known about the molecular pathogenesis of SNUC, an isolated previous study in 2009 explored the expression of 3 common oncogenic transmembrane tyrosine kinase receptors in SNUC. At the protein level, 8 of 11 samples over expressed c-KIT, 3 of 11 over expressed EGFR, and 0 of 11 overexpressed her2/neu(22). However, the overexpression at the protein level was not be demonstrated to result from activating mutations or gene amplification. The lack of activating mutations in c-KIT was confirmed in our study, as 0 of 10 c-KIT SNVs demonstrated activating mutations in any of the 13 SNUC patient samples. As a technique, Sequenom™ excels at detecting known hot-spot single-nucleotide substitutions. However in GIST, c-KIT is frequently activated by short deletions. Similar deletions in SNUC would not be detected with the methodology used in this experiment.
Interestingly, this phenomenon of c-KIT protein overexpression without activating mutations in the c-kit gene is found in many other tumor types. Aberrant epigenetic regulation and altered post-translational control are postulated to play a role, although the exact mechanism of overexpression in the absence of mutation is not well understood. However, in contrast to mutations, levels of KIT protein expression do not correlate with clinical responsiveness to small molecule KIT inhibitors in GIST or melanoma.
We did find polymorphisms in the VEGF gene at both 1154 & 1498 locations. VEGF signaling induces the proliferation, differentiation, and migration of vascular endothelial cells; increases the permeability of the capillaries; and enhances the survival of endothelial cells by preventing their apoptosis. In multiple studies, overexpression of VEGF in HNSCC was associated with metastasis, recurrence, and poor prognosis(15, 23)
Several single nucleotide polymorphisms have been described in the VEGF gene, some of which have been shown to be associated with differential expression of VEGF in vitro (24). Interestingly position 1154 is located in the VEGF promoter(24). The 1154GG genotype is associated with higher VEGF production (25)while the 1154AA genotype has been shown to be associated with a decreased risk for prostate cancer and less advanced melanoma(26–28). Interestingly, of the 3 patients alive at last contact, 2 possessed the historically protective AA genotype. The low total number of patients involved in this study prohibit adequately powering this relationship, and limit firm conclusions. However, the VEGF 1154 genotype may merit further analysis as a predictive biomarker for response to treatment and overall survival.
The relationship of the 1498 polymorphism with clinical outcome is less clear. There has been little literature demonstrating a relationship between treatment response or overall survival with polymorphisms in the 1498 VEGF SNP in head and neck malignancies. In prostate carcinoma the 1498 SNP was not related with risk or disease prognosis (i.e., with tumor grade, stage, Gleason scores)(29). In lung adenocarcinoma the 1498 SNP had no association with overall survival(30). However, in breast cancer the 1498 CC allele was associated with shorter overall and disease-free survival (age adjusted hazard ratio of 1.5 for both)(31). Although polymorphisms in 1498 didn’t appear to correlate with survival, the small number of patients in this rare disease prevent firm conclusions based on the data.
In this subset of 12 patients treated at MDACC with adequate archival tissue, the overall survival at last follow up was 25% (3/12). This is significantly lower than most contemporary reports as well as a larger series from our own institution(32), and likely relates to those patients for whom adequate tissue for molecular analysis was available. As the current treatment paradigm includes initial induction chemotherapy, followed by concomitant chemoradiotherapy, minimal residual tissue was typically available from patients who responded favorably to this regimen. Therefore, there was a bias to acquisition of tumors from those refractory to these non-surgical approaches that subsequently underwent attempted surgical salvage.
Recent investigation into a subset of EBV negative, undifferentiated carcinomas in the upper aerodigestive tract at the University of Virgina demonstrated that of 25 cases originally classified as SNUC, 2 (8%) showed IHC and FISH staining consistent with rearrangement of NUT and BRD4(33). Tumors harboring this genetic rearrangement have recently been grouped together as a clinical entity termed “aggressive midline carcinoma”(34). These tumors appear to arise in midline structures, especially the upper aerodigestive tract and thymus. Early reports define these tumors as histologically undifferentiated or with minimal focal squamous differentiation. Survival is typically less than 1 year(35, 36).
One of the limitations of the technology utilized in this work is its restricted ability to probe for previously defined single nucleotide variations. Large-scale genomic events such as the NUT gene translocation on chromosome 15q14 were not evaluated. It will be interesting to investigate the status of the NUT translocation in future studies on expanded clinical samples with next generation molecular arrays.
Select clinically relevant activating genomic mutations were not identified in any of 13 SNUC patient samples. However, nucleotide polymorphisms were noted within the promoter region of VEGF. These may merit future study as predictive biomarkers for treatment response or overall survival. Future studies focusing on larger tumor sets and utilizing whole genome or exome sequencing may help to define additional genetic aberrations in SNUC that can be clinically targeted with available or emerging biological agents. Additionally, the recent establishment of two novel, highly tumorigenic SNUC cell lines(37) should greatly facilitate efforts at molecularly targeted approaches for this deadly disease.
Acknowledgments
This work was supported by the Pittsburgh Foundation Study of Sinonasal Malignancies, The RNR Cross Foundation, The Charlie And Daneen Steifel Chair in Cancer Research, and a National Cancer Institute-Cancer Center Core Grant (NCI CA16672).
Footnotes
We report no financial relationships that would impart a conflict of interest. This manuscript has been read and approved by all authors, and the requirements for authorship as stated in the Cancer editorial policies have been met. This manuscript is not under consideration elsewhere.
References
- 1.Frierson HF, Jr, Mills SE, Fechner RE, Taxy JB, Levine PA. Sinonasal undifferentiated carcinoma. An aggressive neoplasm derived from schneiderian epithelium and distinct from olfactory neuroblastoma. Am J Surg Pathol. 1986;10(11):771–9. [PubMed] [Google Scholar]
- 2.Lin EM, Sparano A, Spalding A, et al. Sinonasal undifferentiated carcinoma: a 13-year experience at a single institution. Skull Base. 2010;20(2):61–7. doi: 10.1055/s-0029-1236165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musy PY, Reibel JF, Levine PA. Sinonasal undifferentiated carcinoma: the search for a better outcome. Laryngoscope. 2002;112(8 Pt 1):1450–5. doi: 10.1097/00005537-200208000-00023. [DOI] [PubMed] [Google Scholar]
- 4.Righi PD, Francis F, Aron BS, Weitzner S, Wilson KM, Gluckman J. Sinonasal undifferentiated carcinoma: a 10-year experience. Am J Otolaryngol. 1996;17(3):167–71. doi: 10.1016/s0196-0709(96)90055-1. [DOI] [PubMed] [Google Scholar]
- 5.Jeng YM, Sung MT, Fang CL, et al. Sinonasal undifferentiated carcinoma and nasopharyngeal-type undifferentiated carcinoma: two clinically, biologically, and histopathologically distinct entities. Am J Surg Pathol. 2002;26(3):371–6. doi: 10.1097/00000478-200203000-00012. [DOI] [PubMed] [Google Scholar]
- 6.Levine PA, Frierson HF, Jr, Stewart FM, Mills SE, Fechner RE, Cantrell RW. Sinonasal undifferentiated carcinoma: a distinctive and highly aggressive neoplasm. Laryngoscope. 1987;97(8 Pt 1):905–8. [PubMed] [Google Scholar]
- 7.Gaffey MJ, Frierson HF, Weiss LM, Barber CM, Baber GB, Stoler MH. Human papillomavirus and Epstein-Barr virus in sinonasal Schneiderian papillomas. An in situ hybridization and polymerase chain reaction study. Am J Clin Pathol. 1996;106(4):475–82. doi: 10.1093/ajcp/106.4.475. [DOI] [PubMed] [Google Scholar]
- 8.Cohen ZR, Marmor E, Fuller GN, DeMonte F. Misdiagnosis of olfactory neuroblastoma. Neurosurg Focus. 2002;12(5):e3. doi: 10.3171/foc.2002.12.5.4. [DOI] [PubMed] [Google Scholar]
- 9.Barnes L, Eveson JW, Reichart P, Sidransky D. Pathology and Genetics of Head and Neck Tumours. Lyon, France: IARC Press; 2005. World Health Organization Classification of Tumours; p. 430. [Google Scholar]
- 10.Rischin D, Porceddu S, Peters L, Martin J, Corry J, Weih L. Promising results with chemoradiation in patients with sinonasal undifferentiated carcinoma. Head Neck. 2004;26(5):435–41. doi: 10.1002/hed.10396. [DOI] [PubMed] [Google Scholar]
- 11.Tanzler ED, Morris CG, Orlando CA, Werning JW, Mendenhall WM. Management of sinonasal undifferentiated carcinoma. Head Neck. 2008;30(5):595–9. doi: 10.1002/hed.20748. [DOI] [PubMed] [Google Scholar]
- 12.Reiersen DA, Pahilan ME, Devaiah AK. Meta-analysis of Treatment Outcomes for Sinonasal Undifferentiated Carcinoma. Otolaryngol Head Neck Surg. 2012 doi: 10.1177/0194599812440932. [DOI] [PubMed] [Google Scholar]
- 13.MacConaill LE, Campbell CD, Kehoe SM, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4(11):e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39(3):347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz MT, Biselli PM, Maniglia JV, Pavarino-Bertelli EC, Goloni-Bertollo EM. Genetic variability of vascular endothelial growth factor and prognosis of head and neck cancer in a Brazilian population. Braz J Med Biol Res. 2010;43(2):127–33. doi: 10.1590/s0100-879x2009007500036. [DOI] [PubMed] [Google Scholar]
- 16.Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–9. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 17.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 18.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 19.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loupakis F, Pollina L, Stasi I, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol. 2009;27(16):2622–9. doi: 10.1200/JCO.2008.20.2796. [DOI] [PubMed] [Google Scholar]
- 21.Khambata-Ford S, Garrett CR, Meropol NJ, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25(22):3230–7. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 22.Chernock RD, Perry A, Pfeifer JD, Holden JA, Lewis JS., Jr Receptor tyrosine kinases in sinonasal undifferentiated carcinomas--evaluation for EGFR, c-KIT, and HER2/neu expression. Head Neck. 2009;31(7):919–27. doi: 10.1002/hed.21061. [DOI] [PubMed] [Google Scholar]
- 23.Unal ZN, Unal M, Bagdatoglu OT, Polat G, Atik U. Genetic polymorphism of VEGF-1154 (A/G) in laryngeal squamous cell carcinoma. Arch Med Res. 2008;39(2):209–11. doi: 10.1016/j.arcmed.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12(8):1232–5. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 25.Awata T, Inoue K, Kurihara S, et al. A common polymorphism in the 5′-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes. 2002;51(5):1635–9. doi: 10.2337/diabetes.51.5.1635. [DOI] [PubMed] [Google Scholar]
- 26.Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, Sivridis E. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004;46(3):293–8. doi: 10.1016/j.lungcan.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 27.Lee SJ, Lee SY, Jeon HS, et al. Vascular endothelial growth factor gene polymorphisms and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(3):571–5. doi: 10.1158/1055-9965.EPI-04-0472. [DOI] [PubMed] [Google Scholar]
- 28.McCarron SL, Edwards S, Evans PR, et al. Influence of cytokine gene polymorphisms on the development of prostate cancer. Cancer Res. 2002;62(12):3369–72. [PubMed] [Google Scholar]
- 29.Langsenlehner T, Langsenlehner U, Renner W, et al. Single nucleotide polymorphisms and haplotypes in the gene for vascular endothelial growth factor and risk of prostate cancer. Eur J Cancer. 2008;44(11):1572–6. doi: 10.1016/j.ejca.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Heist RS, Zhai R, Liu G, et al. VEGF polymorphisms and survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2008;26(6):856–62. doi: 10.1200/JCO.2007.13.5947. [DOI] [PubMed] [Google Scholar]
- 31.Lu H, Shu XO, Cui Y, et al. Association of genetic polymorphisms in the VEGF gene with breast cancer survival. Cancer Res. 2005;65(12):5015–9. doi: 10.1158/0008-5472.CAN-04-2786. [DOI] [PubMed] [Google Scholar]
- 32.Rosenthal DI, Barker JL, Jr, El-Naggar AK, et al. Sinonasal malignancies with neuroendocrine differentiation: patterns of failure according to histologic phenotype. Cancer. 2004;101(11):2567–73. doi: 10.1002/cncr.20693. [DOI] [PubMed] [Google Scholar]
- 33.Stelow EB, Bellizzi AM, Taneja K, et al. NUT rearrangement in undifferentiated carcinomas of the upper aerodigestive tract. Am J Surg Pathol. 2008;32(6):828–34. doi: 10.1097/PAS.0b013e31815a3900. [DOI] [PubMed] [Google Scholar]
- 34.French CA, Miyoshi I, Aster JC, et al. BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19) Am J Pathol. 2001;159(6):1987–92. doi: 10.1016/S0002-9440(10)63049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engleson J, Soller M, Panagopoulos I, Dahlen A, Dictor M, Jerkeman M. Midline carcinoma with t(15;19) and BRD4-NUT fusion oncogene in a 30-year-old female with response to docetaxel and radiotherapy. BMC Cancer. 2006;6:69. doi: 10.1186/1471-2407-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.French CA, Kutok JL, Faquin WC, et al. Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol. 2004;22(20):4135–9. doi: 10.1200/JCO.2004.02.107. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi Y, Kupferman ME, Bell D, et al. Establishment and characterization of novel cell lines from sinonasal undifferentiated carcinoma. Clin Cancer Res. 2012;18(22):6178–87. doi: 10.1158/1078-0432.CCR-12-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]